Abstract
Genetic evidence suggests that plant peroxisomes are the site of fatty acid β-oxidation and conversion of the endogenous auxin indole-3-butyric acid (IBA) to the active hormone indole-3-acetic acid. Arabidopsis mutants that are IBA resistant and sucrose dependent during early development are likely to have defects in β-oxidation of both IBA and fatty acids. Several of these mutants have lesions in peroxisomal protein genes. Here, we describe the Arabidopsis pex6 mutant, which is resistant to the inhibitory effects of IBA on root elongation and the stimulatory effects of IBA on lateral root formation. pex6 also is sucrose dependent during early seedling development and smaller and more pale green than WT throughout development. PEX6 encodes an apparent ATPase similar to yeast and human proteins required for peroxisomal biogenesis, and a human PEX6 cDNA can rescue the Arabidopsis pex6 mutant. The pex6 mutant has reduced levels of the peroxisomal matrix protein receptor PEX5, and pex6 defects can be partially rescued by PEX5 overexpression. These results suggest that PEX6 may facilitate PEX5 recycling and thereby promote peroxisomal matrix protein import.
In eukaryotes, peroxisomes serve to compartmentalize several metabolic processes, including fatty acid β-oxidation and H2O2 inactivation by catalase (1). In plants, leaf peroxisomes act with chloroplasts and mitochondria in photorespiration (1) and catabolism of branched-chain amino acids (2), whereas specialized peroxisomes called glyoxysomes contain glyoxylate cycle enzymes (1, 3, 4). Plant peroxisomes also are implicated in developmental events, including photomorphogenesis (5) and lateral root formation (6), and jasmonic acid synthesis required for wounding responses (7).
Identification of genes altered in peroxisome-defective yeast mutants and humans with peroxisomal biogenesis disorders has implicated >20 peroxin (PEX) proteins in peroxisomal biogenesis and function (8-10). Phenotypic characterization of the mutants and biochemical studies of the proteins have linked many PEXs to particular aspects of peroxisome biogenesis, maintenance, and matrix protein import.
Two peroxisomal targeting signals (PTSs) direct matrix-bound proteins from the cytoplasm into peroxisomes: PTS1 consists of Ser-Lys-Leu (SKL), or related variants, at the extreme C terminus of the protein, whereas PTS2 is a nonapeptide found near the N terminus (1, 9, 11). Proteins destined for the peroxisomal matrix are bound by the PEX5 or PEX7 receptors in the cytoplasm and escorted into peroxisomes (1, 9, 11). Recent evidence suggests that the receptor-matrix protein complex dissociates in the matrix after import, releasing the matrix proteins, and the receptors are recycled to the cytoplasm for another round of import (11, 12). Certain PEXs are clearly implicated in docking of the receptor-matrix protein complex at the membrane and translocation into the matrix (1, 8, 9), whereas the roles of others are less well defined. For instance, PEX1 and PEX6 are interacting ATPases required for matrix protein import (13-15); the exact role of these proteins in peroxisomal function, however, remains unclear.
Much of what is known about plant peroxisome biogenesis is based on similarities to yeast or human systems. Sequence comparisons indicate that many, but not all, PEX proteins have plausible homologs in Arabidopsis (8, 16), and several Arabidopsis PEX proteins have been described (5, 17-24). The identification of Arabidopsis peroxisome-defective mutants confirms the importance of plant peroxisomal functions and provides an unbiased method to identify important peroxisomal proteins (25, 26). Genetic evidence in Arabidopsis suggests that conversion of the endogenous auxin indole-3-butyric acid (IBA) to free indole-3-acetic acid is an important peroxisomal function that occurs in a mechanism analogous to fatty acid β-oxidation (24, 25). We have identified a group of Arabidopsis IBA-response mutants that are resistant to the inhibitory effects of IBA on root elongation (6, 24, 27). Many of these mutants also have peroxisome-defective phenotypes, including slowed long-chain fatty acid catabolism during germination and sucrose-dependent seedling development, especially in the absence of photosynthesis (6, 24, 27).
In this work, we demonstrate that a defect in the Arabidopsis PEX6 gene disrupts IBA responses and has a strong peroxisome-defective phenotype. PEX6 encodes an apparent ATPase similar to yeast and human proteins necessary for peroxisomal function. Our analysis of the pex6 mutant indicates that PEX6 may facilitate PEX5 recycling.
Materials and Methods
Phenotypic Analysis. pex6 was identified in an IBA-response screen of ethyl methanesulfonate-mutagenized Columbia (Col-0) seeds; the mutant was originally designated B11 (24). Seeds were surface-sterilized (28) and plated on plant nutrient medium (PN) (29) solidified with 0.6% agar and containing sucrose and hormones as indicated. All phenotypic assays were conducted at least twice with similar results on backcrossed mutant lines.
Genetic Analysis and Mutant Complementation. pex6 was outcrossed to Wassilewskija for mapping. F2 seedlings were grown on 15 μM IBA, DNA was isolated from resistant individuals (30), and the defect was mapped by using published markers (31). A candidate gene (PEX6, At1g03000) was PCR-amplified from mutant DNA, and overlapping fragments covering the gene from 145 bp upstream of the putative translation start site to 193 bp down-stream of the stop codon were sequenced.
A PEX6 genomic clone was constructed by subcloning a 6.6-kb XmnI fragment (1.8-kb 5′ UTR and 0.4-kb 3′ UTR) from bacterial artificial chromosome F10O3 into SmaI-cut pBluescript II KS+. A BamHI/SalI fragment containing PEX6 was subcloned into the plant transformation vector pBIN19 (32) cut with the same enzymes to give pBIN-PEX6. The 35S-HsPEX6 clone was constructed by subcloning a SalI/NotI fragment containing a full-length human PEX6 cDNA (I.M.A.G.E. Consortium, Lawrence Livermore National Laboratory, Berkeley, CA, clone ID 5140908) into the plant transformation vector 35SpBARN (33) cut with XhoI and NotI. pBIN-PEX6, 35S-HsPEX6, and 35S-PEX5 (24) were electroporated into Agrobacterium tumefaciens strain GV3101 (34) and transformed into pex6 plants (35). Transformants were selected for the ability to develop on PN in the dark or PN containing 15 mM sucrose plus 12 μg/ml kanamycin or 7.5 μg/ml glufosinate ammonium (Basta), and homozygous progeny were used for phenotypic assays.
GFP Analysis. We obtained a cytoplasmically targeted GFP construct optimized for plant use (36, 37). To make a peroxisome-targeted line, we performed oligonucleotide-directed mutagenesis (38) with the oligonucleotide 5′-GGATGAACTATACAAAAGCAAGCTTTAAGAGCTCGAATTTCCC-3′, inserting a C-terminal peroxisomal signal “SKL” (underlined) immediately preceding the termination codon (bold). The GFP-SKL gene was subcloned into the pBICaMV vector (39) behind the constitutive 35S promoter to give 35S-GFP-SKL.
The GFP construct was transformed into WT Col-0 plants. Homozygous transformants were crossed to the pex6 and pxa1 (6) mutants and homozygous pex6 (pBIN-PEX6) rescue plants. To examine GFP expression, seeds were plated on PN with 15 mM sucrose and incubated under white light overnight, then for 4 days in the dark. Whole plants were mounted on slides, and GFP localization in seedling root hairs was examined by using a Zeiss Axioplan 2 fluorescence microscope equipped with a narrow-band GFP filter set (41020, Chroma Technology, Rockingham, VT).
PEX5 Protein Analysis. Seeds were surface-sterilized and placed under white light for 2 days in water. Protein samples were prepared by grinding seedlings on ice with a motorized pestle, adding an equal volume of 2× NuPAGE LDS-sample buffer with reducing agent (Invitrogen), and heating to 80°C for 10 min. Proteins were separated by gel electrophoresis using NuPAGE 10% Bis-Tris gels and transferred at 24 V for 1 h to Hybond ECL nitrocellulose membrane (Amersham Pharmacia) by using NuPAGE transfer buffer according to the manufacturer's instructions.
A rabbit antibody (α-PEX5) was generated against the C-terminal region of PEX5 (A713-L728, ACESRNLDLLQKEFPL) and affinity-purified by Bethyl Laboratories (Montgomery, TX). For immunoblotting, membranes were incubated in blocking buffer (8% milk in TBS-T) (38) for 2 h, then with α-PEX5 (0.05 mg/ml in blocking buffer) overnight at 4°C, followed by horseradish peroxidase (HRP)-linked goat α-rabbit IgG (Santa Cruz Biotechnology) diluted 1:500 in blocking buffer for 1 h. Control blots used α-HSC70 (SPA-795; StressGen Biotechnologies, Victoria, Canada) at a 1:1,500 dilution. HRP was visualized by using LumiGLO reagent (Cell Signaling Technology, Beverly, MA).
Results
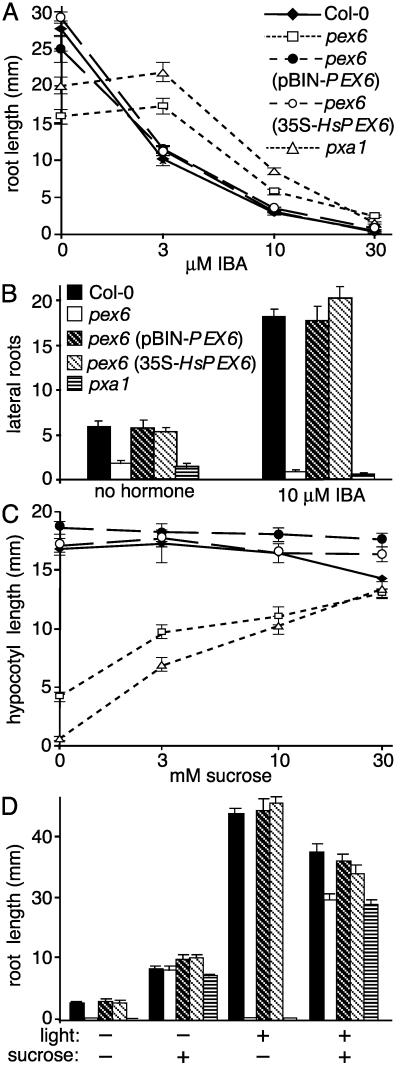
The pex6 Mutant Displays Peroxisome-Defective Phenotypes. pex6 was identified in a screen for IBA-response mutants (24). The mutant is resistant to root elongation inhibition by IBA (Fig. 1A), but responds normally to indole-3-acetic acid (IAA) (24). Moreover, pex6 fails to respond to the stimulatory effects of IBA on lateral root initiation (Fig. 1B), but makes lateral roots in response to IAA (24). These phenotypes indicate that pex6 is not generally defective in lateral root initiation or auxin responsiveness, but is specifically defective in IBA responses.
Fig. 1.
pex6 peroxisome-defective phenotypes. (A) Root elongation on IBA. Seeds were plated on PN plus 15 mM sucrose and the indicated IBA concentrations and grown under yellow filters for 8 days. (B) Lateral root initiation on 15 mM sucrose. Five-day-old seedlings were transferred from hormone-free medium to medium containing no hormone or IBA. Lateral roots were counted after 5 additional days. (C and D) Sucrose dependence during seedling development. (C) Hypocotyl elongation on increasing sucrose concentrations in the dark. Seeds were plated on the indicated medium and incubated under white light overnight, followed by 5 days in the dark. Symbols are as in A.(D) Root elongation on 0 or 30 mM sucrose. Seeds were plated and grown in the dark as in C or in yellow light for 13 days when root lengths were measured. Symbols are as in B. Error bars indicate SEM (n ≥ 9). The peroxisome-defective pxa1 mutant (6) is shown for comparison.
Like a subset of IBA-response mutants (6, 24, 27), pex6 displays peroxisome-defective phenotypes. In oilseed plants like Arabidopsis, long-chain fatty acids stored in seeds are β-oxidized during germination to provide energy; mutants defective in peroxisomal β-oxidation do not develop normally without exogenous sucrose (40). Whereas WT plants develop similarly with and without sucrose, pex6 has dramatic defects in hypocotyl and root elongation in the absence of sucrose, but partially recovers when sucrose is supplied (Fig. 1 C and D). A more severe β-oxidation mutant, pxa1 (6), is shown for comparison. Both the IBA resistance and sucrose dependence of pex6 are recessive (data not shown).
pex6 also has obvious seedling and adult phenotypes. Even with supplemental sucrose, the mutant seedling has a shorter root (Fig. 1D) and hypocotyl (data not shown) than WT. pex6 plants grown in soil have smaller rosettes, fewer rosette leaves, and shorter primary inflorescence stems than WT (Fig. 2 and Table 1). In addition, the mutant has shorter siliques (Fig. 2F) containing fewer seeds than WT, resulting in decreased fecundity. The mutant is pale green in color (Fig. 2), similar to mutants defective in photorespiration, a peroxisomal process requiring PTS1 import (41). Similarly reduced pigmentation is seen in the ped2/pex14 mutant (18) but not in pxa1, which is defective in a peroxisomal transporter but not matrix protein import (6).
Fig. 2.
pex6 growth defects. (A) Twenty-nine-day-old Col-0, pex6 (two plants), pex6 (35S-PEX5) lines D4 and Z31, and pex6 (pBIN-PEX6) plants (left to right). (B) Twenty-one-day-old Col-0 (left), pex6 (center), and pex6 (35S-HsPEX6) (right) plants. (C) Forty-five-day-old Col-0 (left) and pex6 (right) rosettes. (D) Forty-one-day-old Col-0, pex6, pex6 (pBIN-PEX6), pex6 (35S-HsPEX6), and Col-0 (35S-PEX5) plants (left to right). (E) Forty-one-day-old Col-0, pex6, and pex6 (35S-PEX5) lines D4 and Z31 (left to right). (F) Close-up of the two longest WT (left) and pex6 (right) siliques from mature plants.
Table 1. pex6 growth defects.
| Rosette leaves, n
|
Rosette diameter, cm
|
Inflorescence height, cm
|
||||
|---|---|---|---|---|---|---|
| Days after sowing | Col-0 | pex6 | Col-0 | pex6 | Col-0 | pex6 |
| 17 | 9.2 ± 0.2 | 8.3 ± 0.2 | 1.9 ± 0.1 | 1.0 ± 0.1 | 0 | 0 |
| 25 | 16.3 ± 0.9 | 11.2 ± 0.4 | 3.8 ± 0.2 | 1.4 ± 0.1 | 0.1 ± 0.1 | 0.2 ± 0.1 |
| 32 | 34.3 ± 1.6 | 17.5 ± 1.3 | 5.7 ± 0.3 | 2.2 ± 0.3 | 5.0 ± 0.7 | 2.5 ± 0.7 |
| 41 | 41.8 ± 1.6 | 21.7 ± 1.6 | 6.4 ± 0.2 | 2.2 ± 0.4 | 24.4 ± 1.2 | 10.3 ± 0.6 |
WT Col-0 and pex6 were grown on hormone-free medium with 15 mM sucrose for 10 days, then transferred to soil. Numbers represent the means ± standard errors (n ≥ 9).
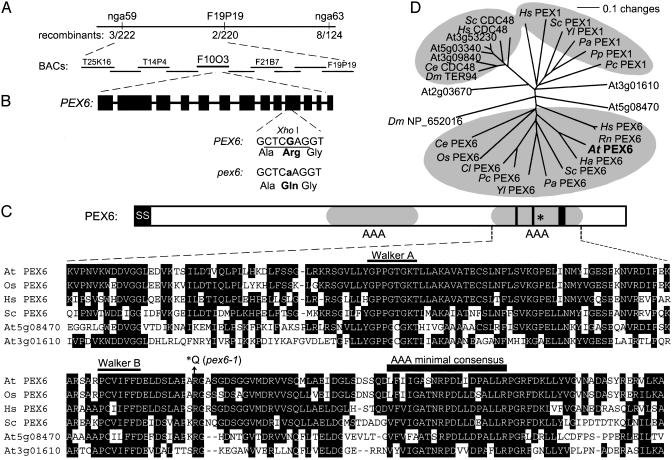
Positional Cloning of the Gene Defective in pex6. Using recombination mapping, we localized the gene defective in pex6 to the top of chromosome 1 (Fig. 3A). Within this region, we identified a candidate gene (At1g03000) that encodes a protein resembling human (42) and sunflower (19) PEX6 proteins, which are ATPases previously implicated in peroxisomal function. Sequencing this gene from mutant genomic DNA revealed a G-to-A mutation that replaces a conserved Arg with a Gln residue (Fig. 3B).
Fig. 3.
PEX6 positional cloning. (A) Recombination mapping localized the pex6 defect on the top of chromosome 1. A homolog of yeast and mammalian PEX6 genes is present on the F10O3 bacterial artificial chromosome (BAC). (B) Structure of the Arabidopsis PEX6 gene. Rectangles indicate exons, and thin lines indicate introns. pex6-1 has a G-to-A mutation at position 3725 (where position 1 is the A of the initiator ATG), causing an Arg-to-Gln change and destroying a XhoI site. (C) Protein structure and partial alignment of PEX6 showing the AAA modules (gray ovals) and the AAA consensus motif (thick bar) and canonical Walker A and B boxes (thin lines) in the second module. smart prediction programs (57, 58) suggest that Arabidopsis PEX6 has a signal sequence (SS) from amino acids 1-32 (black box). The alignment shows the second AAA module of PEX6 proteins from Arabidopsis (At, amino acids 693-882), rice (Os, amino acids 571-760; 44% identical overall, 89% in this region), human (Hs, amino acids 698-885; 30%/74% identical), and Saccharomyces cerevisiae (Sc, amino acids 725-914; 25%/66% identical), with Arabidopsis PEX1/At5g08470 (amino acids 837-1023; 19%/50% identical) and At3g01610 (amino acids 404-589; 24%/46% identical). Sequences were aligned with the megalign program (DNASTAR, Madison, WI) by using the clustalw method. (D) Phylogenetic tree of PEX6 relatives. The tree reconstructs the evolutionary relationship between PEX6 family members from C with additional proteins from Colletotrichum lagenarium (Cl), Pennicillium chrysogenum (Pc), sunflower (Ha; ref. 19), Caenorhabditis elegans (Ce), Pichia angusta (Pa), P. pastoris (Pp), Y. lipolytica (Yl), rat (Rn), and fruit fly (Dm). The unrooted phylogram was generated with paup 4.0b5 (59). The bootstrap method was performed for 100 replicates with a distance optimality criterion, and all characters were weighted equally.
To verify that the nucleotide change in the pex6 mutant causes the IBA-response mutant phenotype, we transformed a genomic fragment containing the WT PEX6 gene including its promoter into mutant plants and assessed complementation. This construct (pBIN-PEX6) alleviates the pex6 growth defects (Figs. 1 C and D and 2) and restores IBA sensitivity to root elongation (Fig. 1A) and lateral root initiation (Fig. 1B), confirming that we have identified the gene responsible for the pex6 phenotypes.
Human PEX6 Functionally Complements the Arabidopsis pex6 Mutant. Fig. 3C shows a partial alignment of the Arabidopsis PEX6 with homologs from other organisms. To determine whether the human and plant proteins function similarly, we transformed pex6 mutant plants with a human PEX6 cDNA driven by the cauliflower mosaic virus 35S promoter. This construct rescued the pex6 phenotypes similarly to the genomic Arabidopsis PEX6 construct, restoring WT sensitivity to IBA in root elongation (Fig. 1A) and lateral root initiation (Fig. 1B), imparting sucrose independence during seedling development (Fig. 1 C and D), and rescuing the size and pigmentation defects of mature pex6 plants (Fig. 2 B and D).
Visualizing pex6 Peroxisomes. To visualize peroxisomes in the pex6 mutant, we examined expression of a GFP containing a PTS (GFP-SKL) by using fluorescence microscopy. In WT, GFP-SKL was present in a distinct, punctate fluorescence pattern (Fig. 4A), consistent with previously reported peroxisomal localization patterns for similar constructs (5, 18).
Fig. 4.
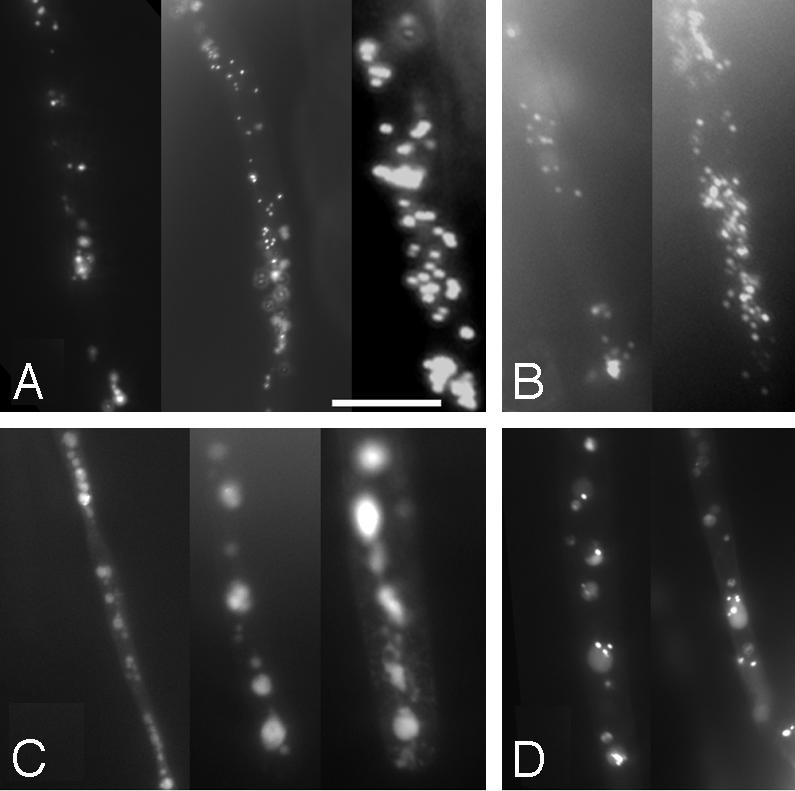
GFP-SKL localization in the pex6 mutant. Root hairs of 5-day-old dark-grown seedlings were examined by using fluorescence microscopy for expression of a peroxisomally targeted GFP (GFP-SKL) in WT (A), pxa1 (B), pex6 (C), and pex6 (pBIN-PEX6) rescue lines (D). All images are shown at the same magnification. (Scale bar: 25 μm.)
GFP-SKL in the pex6 mutant retained punctate fluorescence, but there appeared to be fewer fluorescing regions in the mutant, many of which were larger than WT (Fig. 4C).Thisresultissimilartothat seen in human fibroblast lines defective in PEX6, which contain only ≈20% as many peroxisomes that are two to four times larger than WT (43, 44). In contrast, GFP-SKL fluorescence was indistinguishable from WT in pex6 (pBIN-PEX6) lines (Fig. 4D), indicating that the genomic construct restores WT fluorescence patterns in the mutant. We analyzed the pxa1 mutant for comparison; this mutant has a similar β-oxidation phenotype as pex6 (Fig. 1) and is likely to be defective in peroxisomal substrate import (6). pxa1 has WT GFP-SKL localization (Fig. 4B), suggesting that matrix protein import is unaffected in pxa1 and indicating that the abnormal GFP-SKL fluorescence seen in pex6 is not general to all peroxisome-defective mutants.
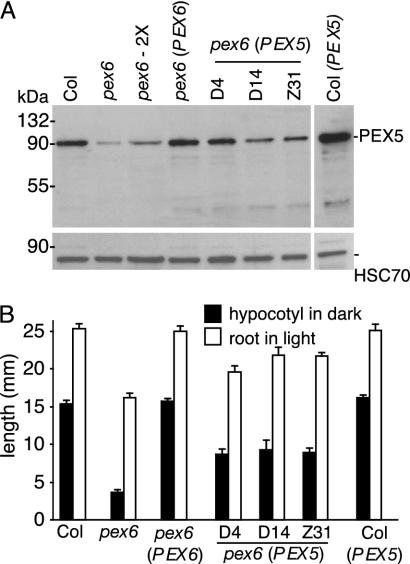
Reduced Levels of the Matrix Protein Receptor PEX5 in the pex6 Mutant. The precise role of PEX6 in peroxisomal processes has not been determined in any system. Immunofluorescence studies indicate that human fibroblast and Pichia pastoris pex6 mutants have reduced PEX5 levels (42, 45, 46). One hypothesis is that PEX6 functions in recycling PEX5 to the cytoplasm after peroxisomal matrix protein import. This hypothesis predicts that in a pex6 mutant PEX5 might perform one round of import and then be trapped in the peroxisome, where it is sequestered or degraded. Therefore, in the pex6 mutant, PEX5 would have limited function, resulting in slowed matrix protein import and peroxisome-defective phenotypes. To determine whether the Arabidopsis pex6 mutant has reduced PEX5 levels, we generated a peptide antibody that recognizes PEX5 and compared PEX5 levels in 2-day-old WT, pex6, and pex6 lines transformed with either the pBIN-PEX6 genomic construct or a 35S-PEX5 overexpression construct. We found that the Arabidopsis pex6 mutant has reduced PEX5 protein levels (≈10% of WT; Fig. 5A). Transforming the mutant with either the genomic PEX6 construct or the PEX5 overexpression construct increased PEX5 levels (Fig. 5A).
Fig. 5.
PEX5 expression in the pex6 mutant. (A) PEX5 protein levels are decreased in pex6. Western blotting usingα-PEX5 (Upper) to examine PEX5 levels or α-HSC70 (Lower) as a loading control was performed on protein from 2-day-old WT (Col), pex6, pex6 (pBIN-PEX6), three homozygous pex6 lines expressing 35S-PEX5 (D4, D14, and Z31), and Col-0 (35S-PEX5) seedlings. The pex6-2X lane was loaded with 2 vol of protein. Positions of molecular mass markers (kDa) are indicated on the left. (B) Overexpression of PEX5 can partially rescue the sucrose dependence and root growth defects of pex6. Lines from A were examined for hypocotyl elongation in the dark in the absence of sucrose (filled bars) or root elongation in the light on 15 mM sucrose (open bars) as described in Fig. 1. Error bars indicate SEM (n ≥ 13).
To determine whether restoring PEX5 levels in pex6 could restore peroxisome functions, we examined the phenotypes of pex6 plants expressing 35S-PEX5. This construct partially rescued pex6 growth defects, including hypocotyl and root length (Fig. 5B) and the adult fertility, size, and pigmentation phenotypes (Fig. 2 A and E), suggesting that PEX5 becomes limiting in the pex6 mutant. Interestingly, PEX5 overexpression did not restore normal sensitivity to IBA (data not shown), consistent with previous observations that IBA responses are more stringent measures of peroxisomal function than sucrose dependence (6).
Discussion
We have identified an Arabidopsis pex6 mutant in a screen for IBA-response mutants. pex6 requires sucrose during early development (Fig. 1 C and D), reflecting slowed rates of peroxisomal fatty acid β-oxidation (24). In addition, pex6 is resistant to the inhibitory effects of IBA on root elongation and the stimulatory effects of IBA on lateral root initiation (Fig. 1 A and B), presumably reflecting defects in peroxisomal IBA β-oxidation. Interestingly, whereas the sucrose dependence of pex6 may be weaker than the peroxisome-defective mutant pxa1 (Fig. 1C), pex6 has more obvious developmental delays and pigmentation and fertility defects (Fig. 2 and Table 1) than pxa1 (6). These phenotypes suggest globally defective peroxisomal function in the mutant and reveal roles for PEX6 throughout plant development.
Positional cloning of the gene defective in the mutant revealed a missense mutation in PEX6 (Fig. 3). Arabidopsis PEX6 is 30% identical to the previously characterized human PEX6, which can functionally complement the Arabidopsis pex6 mutant defects (Fig. 1). PEX6 contains an AAA module, found in ATPases associated with various cellular activities. AAA proteins comprise a distinct subset of ATPases that may act in protein folding or as protein-linked membrane protein clamps and have widely diverged roles including protein sorting, vesicular secretion, and cell division (47, 48).
Yeast have ≈200 apparent ATPases defined by the AAA module, a 200- to 250-aa motif containing an ATP-binding domain and an adjacent consensus region (Fig. 3C and refs. 47-49). AAA-domain proteins fall into 17 subfamilies; subfamily 2 proteins have been implicated in peroxisomal processes (49). Arabidopsis PEX6 and other subfamily 2 members contain two AAA modules; similar to other PEX6 proteins (49), only the second module has canonical Walker A and B ATP-binding site motifs. Fig. 3C shows an alignment of this second conserved region with Arabidopsis, rice, human, and yeast proteins. Within this interval, Arabidopsis PEX6 is 74% identical to human PEX6, compared to 30% over the whole protein. The pex6 mutation alters an Arg residue immediately after the Walker B box (Fig. 3C) that is present in all characterized PEX6 proteins and numerous other AAA proteins (49).
The PEX6-interacting protein PEX1 is a second AAA protein required for peroxisome biogenesis in yeast and mammals (13-15). A cDNA encoding a PEX1-like protein (At5g08470) has been cloned from Arabidopsis (21). At3g01610 encodes a second Arabidopsis protein similar to PEX1 in the AAA module but more similar to CDC48 overall. Like PEX6, these proteins have two AAA domains, but the sequences are more diverged outside of this region. Phylogenetic analysis reveals that Arabidopsis PEX6 is more closely related to PEX6 proteins from other organisms than to other Arabidopsis AAA proteins or to PEX1 proteins from other organisms (Fig. 3D).
In humans, PEX6 disruption is lethal. Human fibroblasts defective in PEX6 and yeast pex6 mutants synthesize peroxisomal membranes and import peroxisomal membrane proteins, but are defective in matrix protein import of both PTS1 and PTS2 proteins (42, 50). In addition, human fibroblasts and P. pastoris pex6 mutants have only ≈5-15% of WT PEX5 levels because of an increased PEX5 degradation rate (42, 45, 46), suggesting that PEX6 may act with PEX5 in peroxisomal matrix protein import. However, human PEX5 and PEX6 proteins do not physically associate in vitro (42).
Based on the defects in matrix protein import and the decreased PEX5 levels in pex6 mutants, PEX6 has been hypothesized to act in the final steps of peroxisomal matrix protein import (45). PEX6 is proposed to act in recycling PTS receptors from the peroxisome to the cytoplasm after each round of import (12, 42, 45, 46). This proposed role is similar to that of the CDC48 AAA protein, which is required for protein retrotranslocation out of the ER (51). CDC48, PEX1, and PEX6 are approximately equally diverged from each other (Fig. 3D) and are more closely related to one another than to other AAA proteins (49).
Because some AAA proteins act in membrane fusion, an alternative hypothesis is that PEX6 acts in membrane fusion events during peroxisomal formation and enlargement (52). Yarrowia lipolytica pex6 mutants have mislocalized peroxisomal membrane proteins and accumulate ER membranes and small peroxisomal vesicles (52). Although PEX6 antibodies can inhibit fusion events in vitro, analysis of yeast deletion mutants indicate that PEX6 is not essential for membrane fusion (53).
These opposing models suggest that PEX6 may act differently in different species. Our data support a role for Arabidopsis PEX6 in peroxisomal matrix protein import. Levels of the matrix protein receptor PEX5 are reduced in the Arabidopsis pex6 mutant and restoring PEX5 levels can partially rescue the pex6 mutant (Fig. 5), supporting a role for PEX6 in PEX5 recycling. If PEX5 can complete one round of import, but is not recycled back to the cytoplasm, matrix protein import will decline, leading to decreased fatty acid β-oxidation and sucrose dependence. Perhaps PEX5 overexpression increases matrix protein import, even if PEX5 is not efficiently recycled, partially restoring matrix protein import and increasing peroxisome function. The abnormal fluorescence pattern seen with GFP-SKL lines suggests that there may be fewer, larger peroxisomes in pex6 than in WT (Fig. 4). This result is consistent with previous analyses of peroxisome size and abundance in human fibroblasts defective in PEX6, where fewer, larger peroxisomes were observed (43, 44). In addition, this phenotype was seen in several other mutants with matrix protein import defects (43). An alternate explanation for the pex6 peroxisome defects is that peroxisome biogenesis is defective because of membrane fusion defects. However, if peroxisomes were not made or were enlarged because of a pex6 defect in membrane fusion, it seems unlikely that PEX5 overexpression would rescue pex6.
The diverse phenotypes of the Arabidopsis pex6 mutant suggest PEX6 importance in a variety of peroxisomal functions, including fatty acid and IBA β-oxidation in glyoxysomes and photorespiration in leaf peroxisomes. Interestingly, null mutations of Arabidopsis PEX2 or PEX10 genes confer embryonic lethality (5, 54, 55), suggesting that functioning peroxisomes are required for embryogenesis. Although a sequence-tagged insertion mutant collection (56) reports a putative PEX6 insertion (SALK_087302), we were unable to verify the presence of this insert (unpublished work); isolation of null pex6 alleles will require other approaches. Regardless of the phenotype of the pex6 null allele, the viable but peroxisome-defective pex6-1 missense allele described here will be a valuable tool to further elucidate the molecular mechanism of PEX6 function and peroxisomal processes in plants.
Acknowledgments
We thank Illeana Silva for assistance characterizing pex6 adult defects, the Arabidopsis Biological Resource Center (Ohio State University, Columbus) for the GFP and F10O3 clones, and Raquel Adham, Melanie Monroe-Augustus, Rebekah Rampey, and Andrew Woodward for critical comments on the manuscript. This work was supported by National Science Foundation Grants IBN-9982611 and 0315596 and Robert A. Welch Foundation Grant C-1309.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: IBA, indole-3-butyric acid; PEX, peroxin; PN, plant nutrient medium; PTS, peroxisomal targeting signal.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AY333116).
References
- 1.Olsen, L. J. (1998) Plant Mol. Biol. 38, 163-189. [PubMed] [Google Scholar]
- 2.Graham, I. A. & Eastmond, P. J. (2002) Prog. Lipid Res. 41, 156-181. [DOI] [PubMed] [Google Scholar]
- 3.Dey, P. M. & Harborne, J. B. (1997) Plant Biochemistry (Academic, San Diego).
- 4.Johnson, T. L. & Olsen, L. J. (2001) Plant Physiol. 127, 731-739. [PMC free article] [PubMed] [Google Scholar]
- 5.Hu, J., Aguirre, M., Peto, C., Alonso, J., Ecker, J. & Chory, J. (2002) Science 297, 405-409. [DOI] [PubMed] [Google Scholar]
- 6.Zolman, B. K., Silva, I. D. & Bartel, B. (2001) Plant Physiol. 127, 1266-1278. [PMC free article] [PubMed] [Google Scholar]
- 7.Weber, H. (2002) Trends Plant Sci. 7, 217-224. [DOI] [PubMed] [Google Scholar]
- 8.Charlton, W. & Lopez-Huertas, E. (2002) in Plant Peroxisomes, eds. Baker, A. & Graham, I. A. (Kluwer, Dordrecht, The Netherlands), pp. 385-426.
- 9.Subramani, S. (1998) Physiol. Rev. 78, 171-188. [DOI] [PubMed] [Google Scholar]
- 10.Tabak, H. F., Braakman, I. & Distel, B. (1999) Trends Cell Biol. 9, 447-453. [DOI] [PubMed] [Google Scholar]
- 11.Mullen, R. T. (2002) in Plant Peroxisomes, eds. Baker, A. & Graham, I. A. (Kluwer, Dordrecht, The Netherlands), pp. 339-383.
- 12.Dammai, V. & Subramani, S. (2001) Cell 105, 187-196. [DOI] [PubMed] [Google Scholar]
- 13.Faber, K. N., Heyman, J. A. & Subramani, S. (1998) Mol. Cell. Biol. 18, 936-943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Geisbrecht, B. V., Collins, C. S., Reuber, B. E. & Gould, S. J. (1998) Proc. Natl. Acad. Sci. USA 95, 8630-8635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kiel, J. A. K. W., Hilbrands, R. E., van der Klei, I. J., Rasmussen, S. W., Salomons, F. A., van der Heide, M., Faber, K. N., Cregg, J. M. & Veenhuis, M. (1999) Yeast 15, 1059-1078. [DOI] [PubMed] [Google Scholar]
- 16.Mullen, R. T., Flynn, C. R. & Trelease, R. N. (2001) Trends Plant Sci. 6, 256-261. [DOI] [PubMed] [Google Scholar]
- 17.Brickner, D. G., Brickner, J. H. & Olsen, L. J. (1998) Plant Physiol. 118, 330. [Google Scholar]
- 18.Hayashi, M., Nito, K., Toriyama-Kato, K., Kondo, M., Yamaya, T. & Nishimura, M. (2000) EMBO J. 19, 5701-5710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaplan, C. P., Thomas, J. E., Charlton, W. L. & Baker, A. (2001) Biochim. Biophys. Acta 1539, 173-180. [DOI] [PubMed] [Google Scholar]
- 20.Lin, Y., Sun, L., Nguyen, L. V., Rachubinski, R. A. & Goodman, H. M. (1999) Science 284, 328-330. [DOI] [PubMed] [Google Scholar]
- 21.Lopez-Huertas, E., Charlton, W. L., Johnson, B., Graham, I. A. & Baker, A. (2000) EMBO J. 19, 6770-6777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schumann, U., Gietl, C. & Schmid, M. (1999) Plant Physiol. 120, 339.10409053 [Google Scholar]
- 23.Schumann, U., Gietl, C. & Schmid, M. (1999) Plant Physiol. 119, 1147. [Google Scholar]
- 24.Zolman, B. K., Yoder, A. & Bartel, B. (2000) Genetics 156, 1323-1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bartel, B., LeClere, S., Magidin, M. & Zolman, B. K. (2001) J. Plant Growth Regul. 20, 198-216. [Google Scholar]
- 26.Hayashi, M. & Nishimura, M. (2002) in Plant Peroxisomes, eds. Baker, A. & Graham, I. A. (Kluwer, Dordrecht, The Netherlands), pp. 279-303.
- 27.Zolman, B. K., Monroe-Augustus, M., Thompson, B., Hawes, J. W., Krukenberg, K. A., Matsuda, S. P. T. & Bartel, B. (2001) J. Biol. Chem. 276, 31037-31046. [DOI] [PubMed] [Google Scholar]
- 28.Last, R. L. & Fink, G. R. (1988) Science 240, 305-310. [DOI] [PubMed] [Google Scholar]
- 29.Haughn, G. W. & Somerville, C. (1986) Mol. Gen. Genet. 204, 430-434. [Google Scholar]
- 30.Celenza, J. L., Grisafi, P. L. & Fink, G. R. (1995) Genes Dev. 9, 2131-2142. [DOI] [PubMed] [Google Scholar]
- 31.Bell, C. J. & Ecker, J. R. (1994) Genomics 19, 137-144. [DOI] [PubMed] [Google Scholar]
- 32.Bevan, M. (1984) Nucleic Acids Res. 12, 8711-8721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.LeClere, S. & Bartel, B. (2001) Plant Mol. Biol. 46, 695-703. [DOI] [PubMed] [Google Scholar]
- 34.Koncz, C. & Schell, J. (1986) Mol. Gen. Genet. 204, 383-396. [Google Scholar]
- 35.Clough, S. J. & Bent, A. F. (1998) Plant J. 16, 735-743. [DOI] [PubMed] [Google Scholar]
- 36.Davis, S. J. & Vierstra, R. D. (1998) Plant Mol. Biol. 36, 521-528. [DOI] [PubMed] [Google Scholar]
- 37.Haseloff, J. & Amos, B. (1995) Trends Genet. 11, 328-329. [DOI] [PubMed] [Google Scholar]
- 38.Ausubel, F., Brent, R., Kingston, R. E., Moore, D. E., Seidman, J. G., Smith, J. A. & Struhl, K. (1995) Short Protocols for Molecular Biology (Wiley, New York).
- 39.Hull, A. K., Vij, R. & Celenza, J. L. (2000) Proc. Natl. Acad. Sci. USA 97, 2379-2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hayashi, M., Toriyama, K., Kondo, M. & Nishimura, M. (1998) Plant Cell 10, 183-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reumann, S. (2002) in Plant Peroxisomes, eds. Baker, A. & Graham, I. A. (Kluwer, Dordrecht, The Netherlands), pp. 141-190.
- 42.Yahraus, T., Braverman, N., Dodt, G., Kalish, J. E., Morrell, J. C., Moser, H. W., Valle, D. & Gould, S. J. (1996) EMBO J. 15, 2914-2923. [PMC free article] [PubMed] [Google Scholar]
- 43.Chang, C. C., South, S. T., Warren, D. S., Jones, J., Moser, A. B. & Moser, H. W. (1999) J. Cell. Sci. 112, 1579-1590. [DOI] [PubMed] [Google Scholar]
- 44.Santos, M. J., Imanaka, T., Shio, H. & Lazarow, P. B. (1988) J. Biol. Chem. 263, 10502-10509. [PubMed] [Google Scholar]
- 45.Collins, C. S., Kalish, J. E., Morrell, J. C., McCaffery, J. M. & Gould, S. J. (2000) Mol. Cell. Biol. 20, 7516-7526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dodt, G. & Gould, S. J. (1996) J. Cell. Biol. 135, 1763-1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Confalonieri, F. & Duguet, M. (1995) BioEssays 17, 639-650. [DOI] [PubMed] [Google Scholar]
- 48.Patel, S. & Latterich, M. (1998) Trends Cell Biol. 8, 65-71. [PubMed] [Google Scholar]
- 49.Beyer, A. (1997) Protein Sci. 6, 2043-2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reuber, B. E., Germain-Lee, E., Collins, C. S., Morrell, J. C., Ameritunga, R., Moser, H. W., Valle, D. & Gould, S. J. (1997) Nat. Genet. 17, 445-448. [DOI] [PubMed] [Google Scholar]
- 51.Lord, J. M., Ceriotti, A. & Roberts, L. M. (2002) Curr. Biol. 12, R182-R184. [DOI] [PubMed] [Google Scholar]
- 52.Titorenko, V. I. & Rachubinski, R. A. (1998) Mol. Cell. Biol. 18, 2789-2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Titorenko, V. I. & Rachubinski, R. A. (2000) J. Cell. Biol. 150, 881-886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schumann, U., Wanner, G., Veenhuis, M., Schmid, M. & Gietl, C. (2003) Proc. Natl. Acad. Sci. USA 100, 9626-9631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sparkes, I. A., Brandizzi, F., Slocombe, S. P., El-Shami, M., Hawes, C. & Baker, A. (2003) Plant Physiol. 133, 1809-1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Alonso, J. M., Stepanova, A. N., Leisse, T. J., Kim, C. J., Chen, H., Shinn, P., Stevenson, D. K., Zimmerman, J., Barajas, P., Cheuk, R., et al. (2003) Science 301, 653-657. [DOI] [PubMed] [Google Scholar]
- 57.Schultz, J., Copley, R. R., Doerks, T., Ponting, C. P. & Bork, P. (2000) Nucleic Acids Res. 28, 231-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schultz, J., Milpetz, F., Bork, P. & Ponting, C. P. (1998) Proc. Natl. Acad. Sci. USA 95, 5857-5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Swofford, D. L. (2000) paup*: Phylogenetic Analysis Using Parsimony (and Other Methods) (Sinauer, Sunderland, MA).