Summary
Secreted phospholipase B1 (CnPlb1) is essential for dissemination of Cryptococcus neoformans to the central nervous system (CNS) yet essential components of its secretion machinery remain to be elucidated. Using gene deletion analysis we demonstrate that CnPlb1 secretion is dependent on the CnSEC14 product, CnSec14-1p. CnSec14-1p is a homologue of the phosphatidylinositol transfer protein (PITP) ScSec14p, which is essential for secretion and viability in Saccharomyces cerevisiae. In contrast to CnPlb1, neither laccase 1 (Lac1)-induced melanization within the cell wall nor capsule induction were negatively impacted in CnSEC14-1 deletion mutants (CnΔsec14-1 and CnΔsec14-1CnΔsfh5). Similar to the CnPLB1 deletion mutant (CnΔplb1), CnΔsec14-1 was hypo-virulent in mice and did not disseminate to the CNS by day 14 post infection. Furthermore, macrophage expulsion of live CnΔsec14-1 and CnΔplb1 (vomocytosis) was reduced. Individual deletion of CnSEC14-2, a closely-related CnSEC14-1 homologue, and CnSFH5, a distantly-related SEC fourteen-like homologue, did not abrogate CnPlb1 secretion or virulence. However, reconstitution of CnΔsec14-1 with CnSEC14-1 or CnSEC14-2 restored both phenotypes, consistent with functional genetic redundancy. We conclude that CnPlb1 secretion is SEC14-dependent and that C. neoformans preferentially exports virulence determinants to the cell periphery via distinct pathways. We also demonstrate that CnPlb1 secretion is essential for vomocytosis.
Introduction
Cryptococcus neoformans is a facultative intracellular basidiomycete responsible for an estimated one million cases of meningitis and 675,000 deaths per year in HIV-infected patients alone (Park et al., 2009). Although predominantly an environmental saprophyte, C. neoformans causes systemic infections in humans and may have evolved as a pathogen capable of surviving macrophage defence mechanisms, via engaging in phagocytic interactions with soil amoeba (Alvarez & Casadevall, 2006). After inhalation, C. neoformans establishes a lung infection and can disseminate to other organs, especially the CNS, where it causes severe life-threatening meningoencephalitis.
Three major virulence mechanisms of C. neoformans are laccase 1 (Lac1)-mediated production of the stress-protecting pigment, melanin (Zhu et al., 2001), elaboration of the polysaccharide capsule, which prevents yeast desiccation and has immunomodulatory properties (Zaragoza et al., 2009) and secretion of phospholipase B1 (CnPlb1) (Cox et al., 2001, Noverr et al., 2003, Santangelo et al., 2004). Secretion of CnPlb1 into host tissue has been inferred by detection of CnPlb1 breakdown products in cryptococcal lung lesions using magnetic resonance spectroscopy (Wright et al., 2002) and detection of specific antibody in serum from infected patients (Santangelo et al., 2005).
CnPlb1 contains phospholipase B (PLB) and lysophospholipase (LPL) activities that hydrolyse one (LPL) or both (PLB) ester linkages on membrane phospholipids, respectively (Chen et al., 2000), releasing fatty acids as a potential energy source (Wright et al., 2007). CnPlb1 also contains lysophospholipase transacylase (LPTA) activity which is essential for the incorporation of macrophage-derived arachidonic acid into cryptococcal lipids during cryptococcus-phagocyte interactions (Wright et al., 2007). This arachidonic acid serves as a reservoir for eicosanoid production by C. neoformans which suppresses macrophage activity, thus diminishing the host immune response (Noverr et al., 2003). Targeted disruption of the CnPlb1 encoding gene (CnPLB1), which abolishes secretion of all three phospholipase activities, reduces virulence in animal models and prevents dissemination of cryptococci to the CNS (Cox et al., 2001, Noverr et al., 2003). C. neoformams also survives and replicates within the phagolysosome of lung macrophages (Feldmesser et al., 2000), the acidic pH of which is optimal for CnPlb1 activity (Chen et al., 2000). C. neoformans escape from macrophages occurs via a lytic process or via a non-lytic extrusion mechanism that maintains the viability of both C. neoformans and the macrophage (Ma et al., 2006, Alvarez & Casadevall, 2006).
The externalization and/or secretion of Lac1, capsular glucuronoxylomannan (GXM) and CnPlb1 is dependent on fungal secretion pathways. Recent studies in C. neoformans defined preferential pathways for export of Lac1 and GXM, which utilise SEC6-dependent exosomes of the lysosomal trafficking pathway and SAV1(SEC4)-dependent vesicles, respectively (Panepinto et al., 2009, Yoneda & Doering, 2006). Although the glycosylphosphatidylinositol (GPI) anchor regulates the cellular export and secretion of CnPlb1 (Djordjevic et al., 2005, Siafakas et al., 2007) and N-linked glycosylation is an essential requirement (Turner et al., 2006), components of CnPlb1-specific secretion machinery are unknown. Despite these studies in C. neoformans, much of our knowledge about secretion in fungi is derived from the non-pathogenic model ascomycete, S. cerevisiae (Harsay & Bretscher, 1995, Muniz & Riezman, 2000, Vorisek, 2000). One key component is the phosphatidylinositol transfer protein (PITP), ScSec14p, which regulates trans-Golgi export pathways and is essential for viability (Bankaitis et al., 1989, Curwin et al., 2009). ScSec14p links protein traffic and Golgi lipid homeostasis via its lipid sensory function (Bankaitis et al., 2010). However the role of SEC14 homologues in secretory processes in pathogenic fungi remains to be elucidated.
We have identified three SEC14 homologues in C. neoformans: CnSEC14-1 and CnSEC14-2 which are highly homologous to ScSEC14, and a SEC14-like homologue (CnSFH5) which is most homologous to ScSFH-5. Using gene deletion analysis we investigated the role of SEC14 homologues in secretion of CnPlb1, melanization and capsule induction and how this impacted on virulence using a murine model of cryptococcosis. We also compared the rate of non-lytic expulsion of the CnSEC14-1 deletion mutant from macrophage phagolysosomes (vomocytosis), with that of CnΔplb1. Our results reveal that secretion of CnPlb1, but not melanization and capsule induction, is CnSEC14-1-dependent, that reduced CnPlb1 secretion in CnSEC14-1 deficient mutants coincides with attenuated virulence and dissemination in an animal model, and that CnPlb1 secretion is an essential requirement for vomocytosis.
Results
Identification of SEC14 homologues in C. neoformans
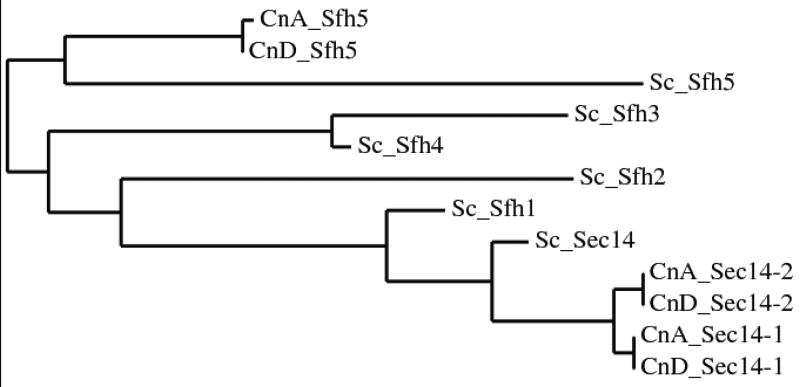
The putative cryptococcal Sec14p homologues were identified by BLAST searching the C. neoformans Serotype D database with S. cerevisiae Sec14p and its homologues. Two putative CnSec14 phosphatidylinositol transfer proteins (PITPs), CNG04130 (designated CnSec14-1p) and CNA00270 (designated CnSec14-2p), sharing a 59% and 71% similarity with ScSec14p, respectively, were identified. In addition to ScSec14p, S. cerevisiae has five Sec fourteen like homologues (Sfh) which share a 42-79% similarity with ScSec14p. A third putative PITP (CNE04320) sharing a 36% similarity with ScSec14p (E-value 8.0E-05) was also identified in serotype D. This protein was most similar (47%) to ScSfh5p and was named CnSfh5p (Figure 1). All three cryptococcal proteins contain the cellular retinaldehyde-binding protein (CRALBP) and TRIO guanine exchange factor structural domain (CRAL/TRIO) typical of PITPs. Due to the greater degree of similarity between CnSec14-1p and CnSec14-2p (86%) compared to ScSec14p and ScSfh1p (79%), the cryptococcal genes were designated CnSEC14-1 and CnSEC14-2 in preference to using the SFH nomenclature. The serotype D amino acid sequences were used in a BLAST search of the serotype A database and sequences CNAG_03153 (chromosome 8), CNAG_00036 (chromosome 1) and CNAG_02104 (chromosome 6) were retrieved and designated CnSec14-1p, CnSec14-2p and CnSfh5p based on homology. A phylogenetic tree depicting the Sec14p similarities among C. neoformans serotypes and S. cerevisiae is shown in Figure 1.
Figure 1. Phylogenetic tree summarizing Sec14p similarities.
The tree was created using a Web-based service at http://www.phylogeny.fr/version2_cgi/advanced.cgi (ClustalW alignment, default curation, phylogeny and tree rendering). The similarities of Cryptococcus neoformans serotype A and D sequences are compared to those of Saccharomyces cerevisiae.
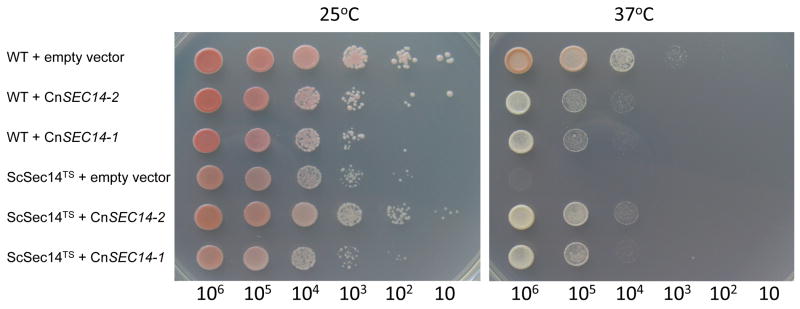
To determine whether the CnSEC14 homologues identified have a similar function to their S. cerevisiae orthologues, we exploited a SEC14 temperature sensitive-mutant of S. cerevisiae, (ScSEC14TS) which is permissive to growth at 25-30°C but cannot survive at 37°C. WT S. cerevisiae and ScSEC14TS were transformed with pCXJ28 (empty vector) or empty vector containing CnSEC14-1 or CnSEC14-2 cDNA. The results in Figure 2 show that all strains grew at 25°C, with CnSEC14-2-transformed cells growing slightly better than CnSEC14-1-transformed cells in both WT and ScSEC14TS. Overexpression of CnSEC14-1 in WT reduced growth slightly consistent with mild toxicity. As expected, ScSEC14TS transformed with empty vector did not grow at 37°C. However, growth at 37°C was restored in ScSEC14TS by overexpression of either CnSEC14-2 or CnSEC14-2, confirming that both CnSEC14-1 and CnSEC14-2 have a similar function and are orthologues of ScSEC14.
Figure 2. Overexpression of CnSEC14-1 and CnSEC14-2 in an ScSEC14TS mutant restores growth at the restrictive temperature (37 °C).
WT S. cerevisiae (M2915-6A) and ScSEC14TS were transformed with pCXJ28 (empty vector) or with pCXJ28 containing CnSEC14-1 or CnSEC14-2 cDNA. CnSEC14 expression was induced by incorporating galactose as the sole carbon source. The growth of serial 10 fold dilutions of each strain was compared at 25°C and 37°C.
CnSEC14 gene deletion in C. neoformans
CnSEC14-1, CnSEC14-2 and CnSFH5 were deleted in C. neoformans var. grubii strain H99 as described in Experimental Procedures and Supporting Information (Figure S1, Table S1), creating CnΔsec14-1, CnΔsec14-2 and CnΔsfh5, respectively. CnSFH5 was also deleted in combination with either CnSEC14-1 or CnSEC14-2, creating CnΔsec14-1/CnΔsfh5 and CnΔsec14-2/CnΔsfh5. However, in contrast to CnΔsec14-1/CnΔsfh5 and CnΔsec14-2/CnΔsfh5 disruption of CnSEC14-2 in CnΔsec14-1 to produce the CnΔsec14-1/CnΔsec14-2 double deletion mutant, did not produce any transformants despite three attempts with the use of controls. This is presumably because the absence of both CnSEC14-1 and CnSEC14-2 is lethal. CnΔsec14-1 was also reconstituted with either CnSEC14-1 or its closely related CnSEC14-2 homologue, creating CnΔsec14-1/CnSEC14-1 and CnΔsec14-1/CnSEC14-2, respectively. It is important to note that in CnΔsec14-1/CnSEC14-2, CnSEC14-2 is ectopically over expressed in addition to being expressed from its endogenous genetic locus. In contrast, CnSEC14-1 is only ectopically overexpressed in CnΔsec14-1/CnSEC14-1. Gene deletion at only the correct locus and genetic reintegration of the full-length gene into the deletion mutants, were confirmed by PCR and Southern hybridization (Supporting Information Figure S2, Table S3).
CnSEC14 gene deletion and phospholipase B1 secretion in C. neoformans
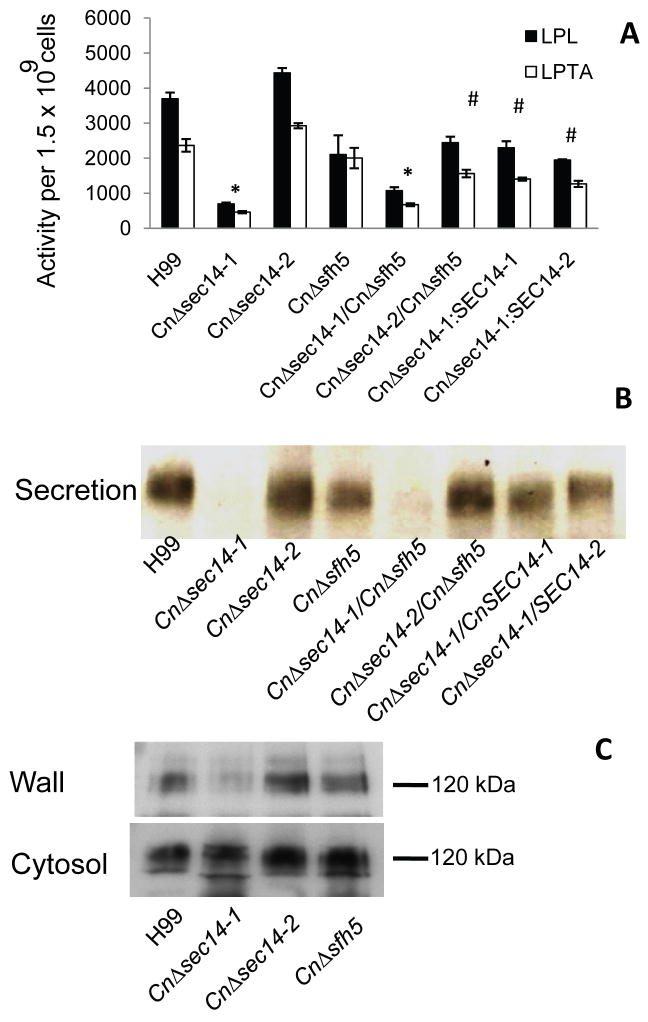
To determine whether CnSEC14-1 or its homologues, play a role in CnPlb1 secretion, CnPlb1 activity (measured as LPL and LPTA) and protein levels were determined in YNB culture supernatants using a radiometric enzyme assay and anti-Plb1 western blotting, respectively (Figure 3). CnΔsec14-1 and CnΔsec14-1/CnΔsfh5 secreted a reduced amount of enzyme protein (Figure 3B) and LPL and LPTA activities were ∼5-fold and 3.5-fold lower than in the H99 WT, respectively (Figure 3A). In contrast, secretion of active CnPlb1 by CnΔsec14-2 was not reduced. Genetic reconstitution of CnΔsec14-1 with CnSEC14-1 or CnSEC14-2 partially restored the activity of secreted CnPlb1 back to H99 WT levels.
Figure 3. CnPlb1 export to the cell wall, and secretion, in C. neoformans, is CnSEC14-1 dependent.
The supernatants (secretions) from 24 h C. neoformans YNB broth cultures were assessed for CnPlb1 activity (LPL and LPTA) by radiometric enzyme assay (A) and CnPlb1 protein by anti-Plb1 western blotting (B). In (A), total LPL/LPTA activity is expressed as Units (nmoles/per min) of substrate hydrolysed/reformed per 1.5 × 109 cells. * indicates statistically significant differences (p<0.05 unpaired T test) in secreted LPL and LPTA activity associated with CnΔsec14-1, relative to WT and the other strains. # indicates statistically significant differences (p<0.05 unpaired T test) in secreted LPL and LPTA activity associated with CnΔsec14-2/CnΔsfh5 and the CnΔsec14-1 reconstituted strains, relative to WT and CnΔsec14-2. In (B) secretions prepared from 2.5 × 108 cells were analysed by SDS PAGE and anti-Plb1 western blotting. Total protein secreted by equal cell numbers from each strain are: WT(344ug); CnΔsec14-1(150ug); CnΔsec14-1/CnΔsfh5(160ug); CnΔsec14-2(403ug); CnΔsfh5(314ug); CnΔsec14-2/CnΔsfh5 (308ug); CnΔsec14-1/CnSEC14-1(308 ug), CnΔsec14-1/CnSEC14-2(189 ug). (C) For CnΔsec14-1, CnΔsec14-2 and CnΔsfh5, CnPlb1 in cell wall and cytosolic fractions prepared from the secreting cells in (A) and (B) was detected by anti-Plb1 western blotting as in (B). The MW of Plb1 125 kDa) was in agreement with the expected MW of glycosylated protein.
In addition to being secreted, some CnPlb1 remains associated with cell wall β-glucans by the GPI anchor remnant (Djordjevic et al., 2005, Siafakas et al., 2007). CnPlb1 levels in β 1-3-glucanase treated cell walls were therefore assessed by western blotting (Figure 3C) to determine whether the reduction in CnPlb1 secretion coincides with the reduced presence of Plb1 in the cell wall. Relative to the H99 WT, CnΔsec14-2 and CnΔsfh5, cell wall-associated CnPlb1 was in fact reduced in CnΔsec14-1 CnPlb1 was present in the CnΔsec14-1 cytosol confirming that CnPlb1 is being synthesized. It has been demonstrated that yeast strains with disrupted genes that encode GPI anchored proteins have cell wall defects as exhibited by their sensitivity to SDS and calcofluor white (Plaine et al., 2008). Thus we examined the sensitivity of CnΔsec14-1 to growth in the presence of each inhibitor. Similar to the PLB1 deletion mutant, CnΔplb1 (Siafakas et al., 2007) CnΔsec14-1 and CnΔsec14-1/CnΔsfh5 were sensitive to SDS and exhibited clumping (Supporting Information Figure S3 A and B). Impaired growth in SDS correlates with enhanced accessibility of the detergent to the membrane as a result of a compromised cell wall barrier. Furthermore, all three mutants were sensitive to calcofluor white (Figure S3 C). The cell wall defect is putatively due to the absence of GPI anchored CnPlb1 in the cell wall. All phenotypes were restored in CnSEC14-1 by genetic reconstitution with CnSEC14-1, or restored/partially restored by genetic reconstitution with CnSEC14-2 (Figure S3). Despite the cell wall defects in CnΔplb1, CnΔsec14-1 and CnΔsec14-1/CnΔsfh5, all strains exhibited comparable growth to H99 WT at 37°C (Cox et al., 2001, Noverr et al., 2003) (Figure S3).
CnSEC14 is not required for melanin or capsule production in C. neoformans
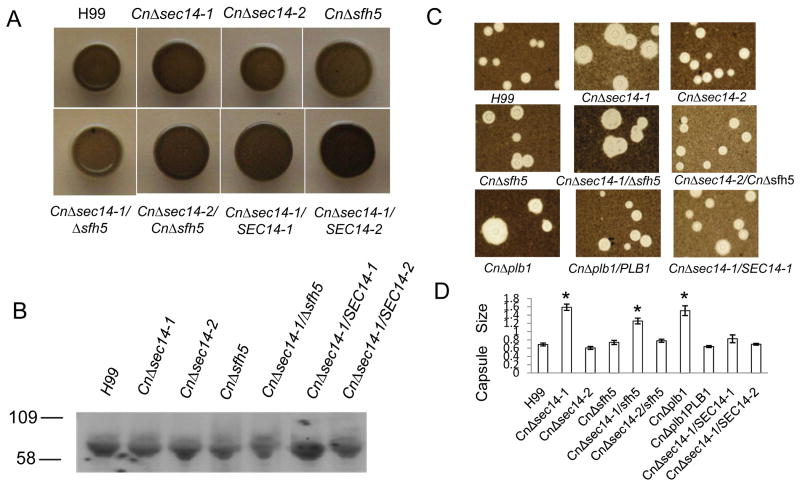
Lac1-induced melanin production is dependent on Lac1 transport to the cell periphery via the lysosomal secretory pathway (Panepinto et al., 2009). In the cell wall, Lac1 synthesizes melanin from externally-derived phenolic compounds such as L-DOPA, which is present in mammalian brain. To determine whether CnSEC14-1 or its homologues affect Lac1 transport to the cell wall, melanin production was measured following growth on L-DOPA agar. The extent of melanin pigmentation was similar in H99 WT and all CnSEC14 mutants (Figure 4A). Additionally, similar amounts of Lac1 were detected in cell walls by western blotting (Figure 4B).
Figure 4. CnSEC14-1 is not required for Lac1-induced melanin production or capsule formation in C. neoformans.
(A) H99 WT and mutant strains were grown overnight in YPD broth and 106 cells were spotted onto DOPA agar and incubated at 30°C for 3 days to allow melanin pigment production (B) Cell wall fractions prepared from equal numbers of cells grown as described in Figure 2 were subjected to western blotting using anti-Lac1 antibodies. Lac1 bands (∼75 kDa) were detected by ECL following exposure to hyper film (C) Following induction of capsule, cells were stained with India ink and visualized by light microscopy (1000× mag). In (D), the mean capsule/cell body size ratio (n=13-34) and standard error are shown. *indicates that strains in which CnPlb1 secretion was absent (CnΔplb1) or attenuated (CnΔsec14-1, CnΔsec14-1/sfh5) had larger capsule/cell body ratios than other mutants and H99 [P<0.05, One way ANOVA]. Due to space restrictions CnΔsec14-1/SEC14-2 is not shown in C but, as indicated in D, has a similar ratio to H99.
Capsule formation is dependent on vesicular transport of GXM to the cell periphery, which is blocked in SAV1(SEC4)-deficient mutants (Yoneda & Doering, 2006). To determine whether capsule formation is also dependent on an intact CnSEC14-1 dependent pathway, capsule size was measured following growth under capsule inducing conditions (Figure C and D). Capsule was not reduced in any of the mutants. Rather, strains in which CnPlb1 secretion was absent (CnΔplb1)(Cox et al., 2001) or attenuated (CnΔsec14-1, CnΔsec14-1/sfh5) (Figure 3), had larger capsule/cell body ratios than all the other strains.
Effect of CnSEC14/SFH deletion on C. neoformans virulence
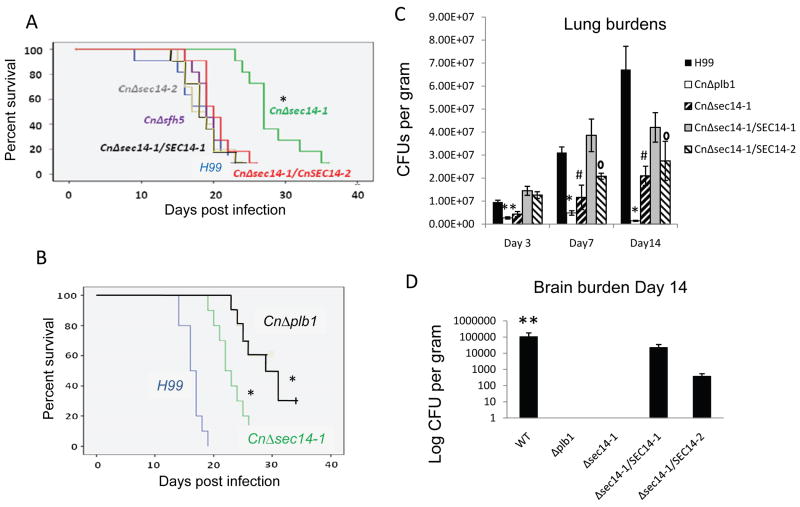
The virulence of the CnSEC14/SFH5 deletion mutants was tested in an inhalation model of cryptococcosis. Only mice infected with CnΔsec14-1 [and CnΔsec14-1/CnΔsfh5 (not shown)] survived longer (median survival 27 and 24 days) than H99 WT-infected mice (median survival 19 days), (Figure 5A). Prolonged survival correlated with reduced CnPlb1 secretion (Figure 3). The median survival of mice infected with CnΔsec14-2, CnΔsfh5 and CnΔsec14-2/CnΔsfh5 was similar to that of H99 WT (P>0.05). Reconstitution of CnΔsec14-1 with CnSEC14-1 or CnSEC14-2 restored virulence. The effect on virulence of CnΔsec14-1, in which CnPlb1 secretion is blocked, was also compared to that of CnΔplb1, which neither synthesizes nor secretes CnPlb1 (Cox et al., 2001, Siafakas et al., 2007) (Figure 5B). Although CnΔplb1 was previously found to be hypo-virulent in animal models, these studies were performed in different mouse strains and/or using different inoculation routes/doses (Cox et al., 2001, Noverr et al., 2003, Santangelo et al., 2004). Mice infected with CnΔplb1 survived a median of 7 days longer than mice infected with CnΔsec14-1. We note in Fig 5B, that the median survival times for mice infected with H99 WT and CnΔsec14-1 (Day 16 and 22, respectively) were shorter than in Fig. 5A (Day 19 and 27, respectively) and that the difference in the median survival time between mice infected with H99 WT and CnΔsec14-1 was 6 and 8 days, respectively. The differences may reflect batch to batch variation in mice susceptibility to C. neoformans infection. The CnΔplb1/CnPLB1 reconstituted strain has previously been shown to exhibit virulence similar to that of H99 WT and thus was not tested here (Cox et al., 2001, Noverr et al., 2003, Santangelo et al., 2004).
Figure 5. CnSEC14-1 is required for virulence.
Survival study (A and B) Mice were inoculated at T=0 and euthanized after losing 20% of their starting weight. Mice asymptomatic at 34 days post infection were also euthanized. An estimation of survival differences (log-rank test) using the Kaplan-Meier method was performed where a P-value <0.05 was considered statistically significant. *indicates that the different survival time for that group, relative to all other infection groups, is statistically significant. Organ burden (C and D) Groups of 9 mice (n=3 for each group) were inoculated with the indicated strains, as above. Three mice from each group were euthanazed 4, 7, and 14 days post inoculation. Lungs (C) and brains (D) were homogenised and CFUs per gram of tissue determined. No dissemination to the brain was observed on day 3 and 7 for any strain (not shown). By 14 days post-infection, only H99, CnΔsec14-1/CnSEC14-1 and CnΔsec14-1/CnSEC14-2 had disseminated. * indicates that organ burdens are statistically different to H99 and the reconstituted strain (where relevant), # indicates that organ burdens are statistically different to H99, the reconstituted strain and CnΔplb1, ° indicates statistically significant difference to WT and ** indicates a statistically significant difference to both deletion mutants but not to CnΔsec14-1/CnSEC14-1 or CnΔsec14-1/CnSEC14-2 (statistical significance is indicated by a P value <0.05 using an unpaired T-test and loge values).
The hypovirulence of CnΔsec14-1 and CnΔplb1 relative to WT, correlated with reduced lung burdens at 3, 7 and 14 days post-infection and failure to disseminate to the brain within 14 days (Figure 5C and D). However by Day 14 the organ burdens for CnΔsec14-1 were significantly higher than those of CnΔplb1. Reconstitution of CnΔsec14-1 with CnSEC14-1 restored lung organ burdens back to H99 WT levels at Day 3, 7 and 14. However, reconstitution of CnΔsec14-1 with CnSEC14-2 only restored lung organ burdens back to H99 WT levels at Day 3. By Day 14 only H99 WT, CnΔsec14-1/SEC14-1 and CnΔsec14-1/SEC14-2 had disseminated to the CNS (Figure 5D). No dissemination was observed on day 3 and 7 for any strain (not shown). The brains of two animals were harvested and cultured upon succumbing to illness (day 19 for CnΔSec14-1 and day 25 for CnΔplb1). In agreement with previous findings, CnΔplb1 was absent from brain tissue. However, cryptococci (1.18 × 105 CFUs per gram) were cultured from brains of animals infected with CnΔsec14-1 (not shown), indicating that dissemination did eventually occur.
CnSec14 and CnPlb1 are required for efficient vomocytosis
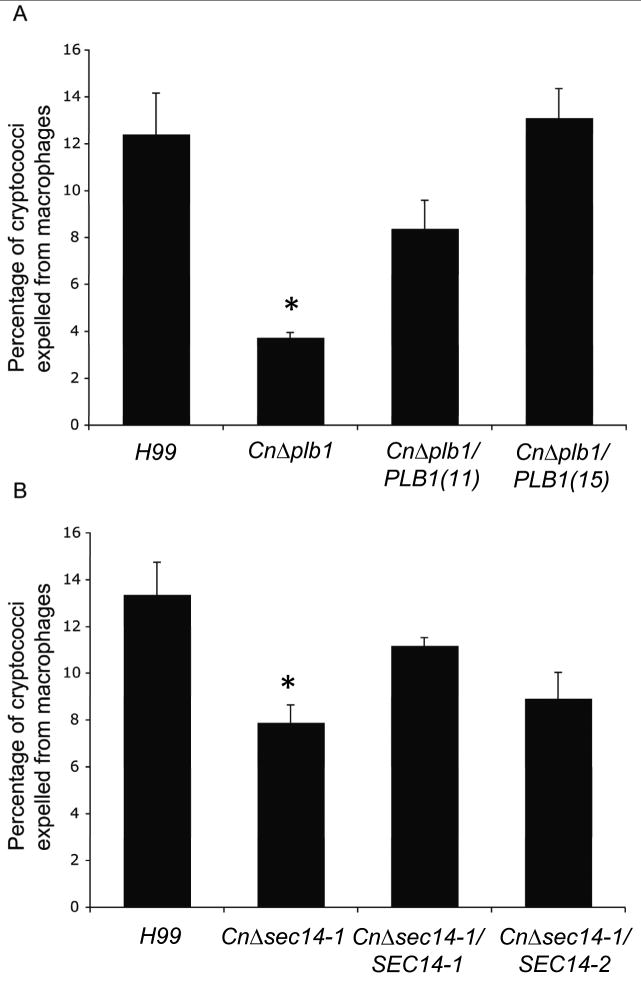
C. neoformans can escape macrophage killing via a phagolysosomal extrusion process known as vomocytosis. As CnPlb1 is essential for creating a macrophage-derived reservoir of arachidonic acid for eicosanoid production which can alter macrophage function (Wright et al., 2007, Noverr et al., 2003) we hypothesized that vomocytosis would be altered in CnPlb1-deficient strains of cryptococci. Vomocytosis was reduced by ∼60% in CnΔplb1 and fully restored in the reconstituted strain (Figure 6A). In CnΔsec14-1, vomocytosis was reduced by ∼40%; it was partially restored by reconstitution with CnSEC14-1 (Figure 6B). However, reconstitution of CnΔsec14-1 with CnSEC14-2 showed little difference with CnΔsec14-1 (P=0.09) and, although lower than that of H99 and CnΔsec14-1/CnSEC14-1, this difference was not statistically significant (P=0.34).
Figure 6. Absent or attenuated CnPlb1 secretion coincides with reduced expulsion of C. neoformans from macrophages.
The percentage of cryptococci expelled was calculated from counts of expulsion events over 18 h time lapse movies. With both CnΔplb1 (A) and CnΔsec14-1 (B) the incidence of expulsion was significantly reduced* when compared to the parental H99 strain (P=6×10-5 and P= 4.6×10-3 respectively). Reconstitution of CnΔplb1 with CnPLB1 [CnΔplb1/PLB1(11), CnΔplb1/PLB1(15)] and of CnΔsec14-1 with CnSEC14-1, rescued the expulsion phenotype [H99 vs CnΔplb1/PLB1(11) (P= 0.181); H99 vs CnΔplb1/PLB1(15) (P=0.806); CnΔplb1 vs CnΔplb1/PLB1(11) (P=0.00851); CnΔplb1 vs CnΔplb1/PLB1(15) (P=0.00002), H99 vs CnΔsec14-1/CnSEC14-1 (P=0.563); CnΔsec14-1 vs CnΔsec14-1/CnSEC14-1 (P=0.167). Each strain was tested at least three times independently. Error bars are SD.
Investigating potential compensatory mechanisms for the absence of CnSEC14
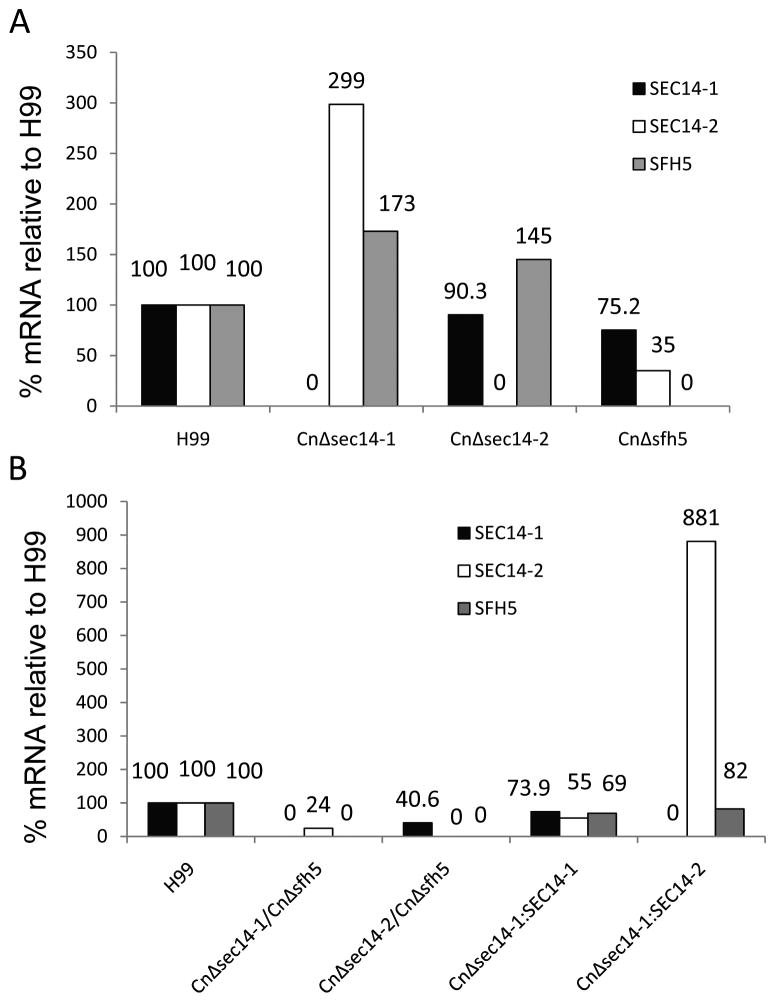
In contrast to S. cerevisiae (Bankaitis et al., 1989) and C. albicans (Monteoliva et al., 1996), deletion of CnSEC14 homologues is not lethal. Thus we investigated whether expression of other SEC14 homologues was up regulated in the absence of CnSEC14-1 as part of a potential compensation mechanism. qRT-PCR was used to quantify the expression of CnSEC14-1, CnSEC14-2 and CnSFH5 in all mutant strains (Figure 7). As expected, the respective mRNAs corresponding to the gene deleted, were absent from CnΔsec14-1, CnΔsec14-2 and CnΔsfh5 (Fig.7A) and CnΔsec14-1/CnΔsfh5 (Fig. 7B). In CnΔsec14-1, levels of CnSEC14-2 and CnSFH5 mRNA were increased 3-fold and 1.7-fold, respectively. In CnΔsec14-2, CnΔsfh5 and CnΔsec14-2/CnΔsfh5, levels of CnSEC14-1 mRNA were reduced by 10%, 25% and 60% respectively. While the CnSEC14-1 mRNA level in the CnΔsec14-2/CnΔsfh5 mutant was only 40% of the H99 WT, this was sufficient to achieve a full virulence phenotype. In fact, when CnSEC14-1 mRNA was inhibited by RNAi in CnΔsec14-2, a CnSEC14-1 mRNA level as low as 18% of the H99 WT (Supporting Information, Figure S4) was sufficient to achieve a H99 WT phenotype (not shown). Reconstitution of CnΔsec14-1 with CnSEC14-1 restored CnSEC14-1 mRNA to 75% of the H99 WT level, and was sufficient to restore all virulence phenotypes. Reconstitution of CnΔsec14-1 with CnSEC14-2, which also restored virulence, led to a 9-fold increase in expression of CnSEC14-2 mRNA (Figure 7B). This is most likely due to the presence of more than one copy of CnSEC14-2 in CnΔsec14-1/SEC14-2 (the native gene and the ectopically-introduced copy).
Figure 7. Comparison of CnSEC14/SFH mRNA in H99 and the various deletion mutants by qRTPCR.
(A) H99 WT and the single deletion mutants and (B) H99 and the double deletion mutants and reconstituted strains. For each cDNA, a CnSEC14/SFH and actin (ACT1) PCR were performed in separate reactions and the amount of each product was determined from a standard curve. The CnSEC14/SFH:ACT1 ratio for each strain was expressed as a percentage relative to the CnSEC14/SFH:ACT1 ratio in H99WT which was set at 100%. The Y-axis designates the % of mRNA in each mutant, relative to WT. All PCR reactions were performed at least in duplicate. Some strains (CnΔsec14-1 and CnΔsec14-2) were analysed 3 times as they were used as controls for the different batches of assays performed and consistently showed no SEC14-1 and SEC14-2 mRNA, respectively.
Discussion
We have demonstrated that the cell wall association and secretion of the key cryptococcal virulence factor, CnPlb1, is CnSEC14-1 dependent. In contrast, neither the presence of Lac1 in the cell wall and its ability to synthesize melanin, nor the externalization of capsule, are dependent on SEC14-1. These results are consistent with the presence of distinct secretion pathways for virulence determinants in C. neoformans.
Our data demonstrates that two strains defective (CnΔsec14-1) or deficient (CnΔplb1) in CnPlb1 secretion have similar phenotypes in vitro (sensitivity to two cell wall perturbing agents, hyper encapsulation and reduced vomocytosis) and are hypovirulent in an mouse inhalation model of cryptococcosis. Attenuated virulence of both strains coincided with reduced lung burdens and failure to disseminate to the CNS by day 14 post infection. Our evidence suggests that the reduced presence of CnPlb1 in CnΔsec14-1 supernatants and the cell walls is due to reduced CnPlb1 traffic from the Golgi apparatus to the cell wall, rather than the lack of attachment of exported CnPlb1 to a defective cell wall, as the latter would have resulted in hyper secretion. Furthermore, similar to CnΔplb1 the reduced presence of GPI-anchored CnPlb1 in CnΔsec14-1 cell walls is most likely to be a factor contributing to the cell wall defect. Although CnΔsec14-1 and CnΔplb1 shared similar phenotypes, some phenotypes were more severe in CnΔplb1. In particular CnΔplb1 was less virulent than CnΔsec14-1, failed to disseminate to the CNS over the entire infection time course and was expelled from macrophages less efficiently than CnΔsec14-1. These differences may be attributed to the fact that secretion of CnPlb1, and potentially other proteins, is not blocked completely in CnΔsec14-1, potentially due to functional redundancy of SEC14 homologues. Alternatively, the possibility exists for CnPlb1 acting co-ordinately with CnSec14-1 to regulate Golgi phosphatidylcholine levels which are important for vesicle formation and secretion. Sec14p in S. cerevisiae has been reported to down regulate ScPlb1-mediated turnover of phosphatidylcholine (Schnabl et al., 2003). Thus the lack of CnPlb1 in CnΔplb1 could alter Golgi lipid levels and thereby influence the traffic of other proteins, contributing to the increased severity of the phenotype compared to CnΔsec14-1.
Sec4/Sav1 and Sec6-dependent intracellular vesicular trafficking pathways have been described for transport of capsular polysaccharide (GXM) and Lac1, respectively (Yoneda & Doering, 2006, Panepinto et al., 2009) and there is evidence that the Sec6-dependent intracellular vesicles are the precursors for extracellular exosomes containing Lac1 and urease (Panepinto et al., 2009). PI3 kinase (VPS34), which has been implicated in protein transport to the multivesicular body (the source of exosomes), is also essential for Lac1 activity (Hu et al., 2008). We therefore conclude that, as Lac1 traffic to the cell wall is not disrupted in CnΔsec14-1, CnSEC14-1 does not regulate protein traffic from the lysosome to the plasma membrane in C. neoformans. Clearly, cryptococcal virulence factors are transported to the cell periphery via more than one pathway.
The apparent exclusivity of the CnSEC14—dependent pathway for export of CnPlb1 may be a consequence of the protein nature of the cargo. In S. cerevisiae GPI anchored proteins are sequestered into ER-derived ergosterol and sphingolipid-enriched membrane vesicles (Muniz & Riezman, 2000, Sutterlin et al., 1997) which are the precursors of lipid rafts in the plasma membrane (Bagnat et al., 2000, Klemm et al., 2009) and we demonstrated the association of CnPlb1 with cryptococcal membranes possessing raft like properties (Siafakas et al., 2006). However, we also found the non-GPI anchored protein, superoxide dismutase (SOD), sequestered within rafts (Siafakas et al., 2006) arguing against raft-like vesicles being the primary transporters of GPI anchored proteins in C. neoformans. In S. cerevisiae there is support for the presence of three parallel pathways of protein exit from the ER; one specific for GPI anchored proteins and controlled by the p24 family member Emp24 and possibly by the inositol deacylase Bst1, the GPI anchor-remodelling phospholipase Per1 and the COPII components Sec12 and Sec16; and the other two for non-anchored and transmembrane proteins (Castillon et al., 2009). Whether CnSec14 homologues in C. neoformans preferentially regulate cellular export of all GPI-anchored proteins or all proteins not exported via the lysosomal pathway remains to be determined. Finally, our data that CnSEC14-1 regulates distinct protein export pathways in C. neoformans is supported by a recent finding in S. cerevisiae where ScSEC14 was found to regulate export of Bgl2, but not Hsp150, Cts1, Scw4, Scw10, Exg1, Cis3 and Ygp1 (Curwin et al., 2009). Interestingly, none of these proteins are predicted to be GPI anchored. Furthermore, ScSEC14 also regulates retrograde protein traffic from the endosome to the trans-Golgi (Curwin et al., 2009).
In contrast to S. cerevisiae (Bankaitis et al., 1989) and C. albicans (Monteoliva et al., 1996), deletion of CnSEC14 homologues is not lethal. The most likely explanation for a viable C. neoformans phenotype following deletion of CnSEC14-1 is that other SEC14 homologues, and in particular CnSEC14-2, compensate in part for the absence of CnSEC14-1. The CnSEC14-2 compensatory effect is further supported by our inability to create a viable CnΔsec14-1/CnΔsec14-2 mutant. We observed that both CnSEC14-1 and CnSFH5 have an effect on CnSEC14-2 mRNA levels. Notably CnSEC14-2 mRNA increased 3-fold in CnΔsec14-1, but did not coincide with virulence restoration, and increased 9-fold in CnΔsec14-1/CnSEC14-2 but did coincide with virulence restoration. The 9-fold increase is most likely due to the presence of two copies of CnSEC14-2 contributing to the final mRNA level. However, CnSEC14-2 levels dropped to 24% in CnΔsec14-1/CnΔsfh5, coinciding with this strain being hypovirulent, and 35% in the virulent CnΔsfh5 strain, consistent with the presence of CnSFH5 having a positive influence on CnSEC14-2 gene expression. The results also indicate that CnSEC14-2 mRNA levels as low as 24% are sufficient for C. neoformans viability in the absence of the two SEC14 homologues. Interestingly, attempts to suppress CnSEC14-1 mRNA in CnΔsec14-2 using RNAi, failed to further compromise the CnΔsec14-1 phenotype despite this methodology reducing the CnSEC14-1 mRNA level to as low as 18% of the H99 WT. This finding raises the question as to why CnSEC14-1 mRNA levels are so high in the first place, an observation that has also been made for SEC14 mRNA in S. cerevisiae (Bankaitis et al., 2010). We also observed that over expression of CnSEC14-1, but not CnSEC14-2, was growth inhibitory to S. cerevisiae.
CnPlb1 is essential for efficient replication of C. neoformans at its primary site of infection (the lungs), and for entry into the blood stream and dissemination to the brain (Cox et al., 2001, Santangelo et al., 2004). We now demonstrate that CnΔsec14-1 is hypovirulent in mice. This may be due in part to low levels of CnPlb1 secretion. CnPlb1 can facilitate cryptococcal entry into the lungs by hydrolysis of dipalmitoyl phosphatidylcholine (PC) in lung surfactant (Santangelo et al., 1999) and/or promote cryptococcal adherence to pulmonary epithelium (Ganendren et al., 2006). C. neoformams also survives and replicates within the acidic phagolysosome of alveolar and interstitial macrophages following phagocytosis (Feldmesser et al., 2000), an environment conducive to optimal CnPlb1 enzyme activity (Chen et al., 2000).
In experimental mice, inhaled cryptoccoci are rapidly ingested by alveolar and pulmonary macrophages (Santangelo et al., 2004) and a role for mononuclear phagocytes in conveying cryptococci through the blood stream and across the blood brain barrier has recently been demonstrated (Charlier et al., 2009). Macrophages expel phagolysosomes containing viable cryptococci via a non-lytic extrusion process called vomocytosis which does not trigger host cell death (Ma et al., 2007, Ma et al., 2006, Alvarez & Casadevall, 2006, Alvarez & Casadevall, 2007, Johnston & May, 2010). Vomocytosis from lung or mononuclear macrophages may serve as a mechanism for escape of C. neoformans into the blood stream allowing dissemination to the CNS. In this study, we found that vomocytosis was markedly impaired when CnPlb1 secretion was absent or attenuated in CnΔplb1 and CnΔsec14-1, respectively, an effect that may be related to the role of CnPlb1 in eicosanoid production and suppression of macrophage function (Wright et al., 2007, Noverr et al., 2003) or intracellular proliferation of C. neoformans at early time points post-phagocytosis (Cox et al., 2001). This reduced rate of vomocytosis coincided with reduced or absent rates of dissemination to the CNS. Other factors reported to be important in vomocytosis are depolymerization of the actin cytoskeleton (Jones, 2007, Alvarez & Casadevall, 2006, Ma et al., 2006, Johnston & May, 2010).
In summary, we have defined a CnSEC14-1-dependent CnPlb1 secretion pathway in C. neoformans which is not utilised for export of two other major virulence determinants, Lac1 and capsular material. We have shown that the two CnPlb1 secretion defective strains, CnΔsec14-1 and CnΔplb1, in which CnPlb1 activity is attenuated either by deleting CnPLB1 or by blocking CnPlb1 secretion via deletion of CnSEC14-1, are hypovirulent in mice as a consequence of reduced or absent disseminationto the brain. We also present evidence for the first time, that cryptococcal expulsion from macrophages is CnPlb1-dependent. Reduced vomocytosis in the CnPlb1 deficient mutants may contribute to the failure of C. neoformans to disseminate to the CNS.
Experimental Procedures
Identification of SEC14 homologues in C. neoformans
The putative amino acid sequences of Sec14-1p and its homologues in C. neoformans var neoformans (Serotype D) were identified by performing a BLAST search of the C. neoformans Serotype D database (http://genome.slu.edu/blast.html) using the S. cerevisiae ScSec14, ScSfh1 and ScSfh5 sequences as a query. The serotype A homologues were then identified by performing a BLAST search of the C. neoformans var. grubii strain H99 protein databases (http://www.broadinstitute.org/annotation/genome/cryptococcus_neoformans/MultiHome.html) using the serotype D sequences as a query. GenBank accession numbers assigned to the three serotype A sequences are as follows: CnSEC14-1: HM567405; CnSEC14-2: HM567406; CnSFH5: HM567407.
Strains, Media and Plasmids
Wild type (WT) C. neoformans var. grubii strain H99 (serotype A, MATα, ATCC 208821), and the CnΔplb1 and CnΔplb1/PLB1 deletion mutants, also derived from H99 (Cox et al., 2001), were used in this study. The strains CnΔsec14-1, CnΔsec14-2, CnΔsfh5, CnΔsec14-1/CnΔsfh5, CnΔsec14-2/CnΔsfh5, CnΔsec14-1/CnSEC14-1, CnΔsec14-1/CnSEC14-2 were also created from H99. All strains were cultured in yeast extract-peptone-dextrose (YPD) medium with the addition of 2% Bacto agar when solid medium was required. H99 transformants containing genes disrupted with a neomycin resistance cassette (Neor) or nourseothricin resistance cassette (Natr) were initially grown on YPD agar plates containing 1 M sorbitol for 3 h and subsequently transferred onto YPD agar supplemented with 200 μg/mL geneticin (G418; Invitrogen, CA, USA) or 100 μg/mL nourseothricin (clonNAT; Werner BioAgents, Jena, Germany), respectively. Stable transformants were maintained in YPD medium. Sabouraud dextrose (SAB) agar, SAB Brain Heart Infusion Agar Plus Drugs [SABD-5 g neopeptone (Difco), 20 g glucose, 26 g Brain Heart Infusion Agar (Difco), 7.5 g agar, 0.025 g gentamicin and 0.25 g chloramphenicol/L] and synthetic Yeast Nitrogen Base (YNB) medium were used in some experiments as indicated. DOPA agar, consisting of glucose-free asparagine medium (1-g/L L-asparagine, 0.5-g/L MgSO4, 3-g/L KH2PO4, 1-mg/L thiamine) plus 1 mM L-DOPA, was used to demonstrate melanin pigment production. Capsules were induced on RPMI agar (RPMI, 2% glucose, 0.165M MOPS pH7, 1.5% bacto agar, 0.03% glutamine) by incubation at 37°C in a 5% CO2 atmosphere for 3 days. Plasmids pJAF and pCH233, were a gift from John Perfect. Saccharomyces cerevisiae wild type strain, M2915-6A (MATa, leu2, ade2, ura3), and the temperature sensitive SEC14 (TS) mutant purchased from ATCC® 208248™ [MATa sec14-3 ade2-1(ochre mutation) trp1-delta ura3-52 mal SUC2 CUP1 gal2], were grown in YPD or YNB without uracil following transformation with the pCXJ28 constructs.
CnSEC14 complementation in S. cerevisiae
CnSEC14 cDNA cloning
The cell pellet from an overnight YPD culture of H99 was centrifuged, frozen at -80°C, lyophilized and disrupted by vortexing with glass beads (2 mm). RNA was extracted using TRI reagent according to the manufacturer (Sigma-Aldrich, MO, USA) and 5 μg was treated with DNaseI (Invitrogen, CA, USA). cDNA was generated using the Superscript III First-Strand Synthesis kit (Invitrogen, CA, USA). CnSEC14-1 and CnSEC14-2 (CNAG_03153 and CNAG_00036.2, respectively, Broad Institute Cryptococcus neoformans var. grubii strain H99 Database www.broadinstitute.org/annotation/genome/cryptococcus_neoformans) were amplified from cDNA, using sense/antisense primers 5′-ATGGCCACCACAGACTTCCT-3′ / 5′-CTAAACACCTCCGGCAGCCT-3′ and 5′-ATGAGCGCCTCTGACCCTCT-3′/ 5′-TTAAACAGCCGTCGCGGTAT-3′, respectively. The PCR products were cloned into pCR®2.1-TOPO® (Invitrogen, CA, USA). DNA sequencing of the clones revealed at least five CnSEC14-2 splice variants with the expected size (0.8 kb). Using the flanking EcoRI sites in pCR®2.1-TOPO®, the CnSEC14-2 cDNA splice variant encoding a protein with the highest amino acid similarity to the predicted CnSec14-2 and ScSec14 sequences (96% and 57% respectively), and the CnSEC14-1 cDNA were subcloned into the S. cerevisiae galactose-inducible expression vector, pCXJ28, creating pCXJ28_CnSEC14-2 and pCXJ28_CnSEC14-1, respectively.
CnSEC14 complementation of an S. cerevisiae SEC14TS mutant
Wild type S. cerevisiae strain M2915-6A and the ScSEC14TS mutant were transformed with pCXJ28 (empty vector) or pCXJ28_CnSEC14-1/CnSEC14-2 using the lithium actate/polyethylene glycol method (Burke et al., 2000) and transformants were selected on uracil dropout medium containing 2% glucose. Complementation was deemed to be successful by the growth of the CnSEC14-transformed Scsec14TS mutant at the restrictive temperature (37°C) after 4 days on the selection plates supplemented with 2% galactose instead of glucose to induce expression of the heterologous gene.
Targeted SEC14 gene deletion in C. neoformans
Gene deletion strategy
Native CnSEC14-1, CnSEC14-2 and CnSFH5, were selectively deleted in C. neoformans strain H99, individually and in combination, by homologous recombination with exogenously-introduced hybrid gene constructs. The constructs contained an antibiotic resistance marker flanked by the 5′ and 3′ UTR of the gene to be deleted, and were created by overlap PCR (Davidson et al., 2002) as described previously (Chayakulkeeree et al., 2008). The hybrid gene construction strategy and the biololistic homologous recombination method are described, in detail, in Supporting Information (Figure S1), with the oligonucleotide primers listed in Table S1. Gene Reconstitution As only CnΔsec14-1 displayed attenuated pathogenicity relative to H99 WT, it was genetically-reconstituted with either CnSEC14-1 or CnSEC14-2 to satisfy the molecular Koch's postulates of virulence (Falkow, 1988) and to investigate whether CnSEC14-2 can complement CnSEC14-1 function as described in Supporting Information (Figure S1). Targeted gene deletion and gene reconstitution were confirmed by Southern blotting as described previously (Chayakulkeeree et al., 2008). Genomic DNA was prepared from H99 WT, CnΔsec14-1, CnΔsec14-2, CnΔsfh5, CnΔsec14-1/CnΔsfh5, CnΔsec14-2/CnΔsfh5, CnΔsec14-1/CnSEC14-1 and CnΔsec14-1/CnSEC14-2, digested and probed as described (Figure S2, Table S3 Supporting Information).
Quantitative real time-PCR (qRT-PCR)
RNA from C. neoformans strains (H99 and deletion mutants) was prepared, DNaseI treated and used as a template for oligo-dT primed cDNA synthesis as described above. cDNA (5 μl) diluted 1:25, was used as a template for qRT-PCR using SYBR Green Supermix (Invitrogen, CA, USA) and various primer sets; CnSEC14-1: (SEC141-RTF and SEC141-RTR); CnSEC14-2: (SEC142-RTF and SEC142-RTR); CnSFH5: (SFH5-RTF and SFH5-RTR) and ACT1: (ACT1-RTF and ACT1-RTR), each at a final concentration of 0.1 μM. Primer sequences for qRT-PCR are shown in Table S2, Supporting Information. Using the standard curve method, each specific mRNA was normalized to the housekeeping gene, ACT1, using the MxPro 3005p Real-time PCR System (Stratagene, CA, USA). PCR conditions were: 50°C for 2 min followed by 95°C denaturation for 2 min, and 45 cycles consisting of denaturation at 95°C for 15 sec and annealing/extension at 60° C for 45 sec. A melt curve was also performed from 60°C to 95°C with fluorescence monitored at 0.5°C intervals to establish presence of a single PCR product. Data were evaluated with the MxPro software (Stratagene, CA, USA).
Melanin production
C. neoformans strains (H99 and deletion mutants) were grown overnight at 30°C in YPD broth and the cells were pelleted by centrifugation, washed twice with sterile PBS and resuspended in PBS at 106 cells/5 ul. Aliquots (5 ul) were spotted onto DOPA agar (Missall et al., 2005) and plates were incubated at 30° C for 4 days.
Analysis of CnPlb1 and CnLac1 in cell walls by Western blotting
YNB broth-grown overnight cultures of C. neoformans (H99 and deletion mutants) prepared from equal cell numbers (1.5 × 109), were pelleted by centrifugation and the supernatants (representing the secreted CnPlb1 fraction) were dialysed and lyophilized. Cell wall fractions containing CnPlb1 and Laccase were prepared by disruption of the cell pellet using a MiniBeadbeater-8 cell disrupter and zirconia/silica beads, followed by differential centrifugation, as previously described (Siafakas et al., 2006). Briefly, cell lysates were centrifuged for 10 min (3,500 × g) and the supernatants were retained. Cell pellets were further disrupted by probe sonication, and recentrifuged as above. CnPlb1 and CnLac1 were released from the cell wall-enriched pellets by treatment with β-1,3-glucanase. The supernatants prepared from the cell lysates were combined and centrifuged [135,000 × g (45,000 rpm)] for 1 h at 4°C to obtain the cytosolic fraction (supernatant). CnPlb1 in the cell wall and cytosolic fractions were assessed by western blotting as previously described (Siafakas et al., 2006) using an anti-Plb1 peptide antibody (1 μg/ml). Laccase in the cell wall was detected using an anti-Lac1 antibody (2 μg/ml). Signals were detected by ECL.
Radiometric phospholipase activity assay
CnPlb1 specific activities (lysophospholipase [LPL] and lysophospholipase/transacylase [LPTA]) in lyophilized secretions prepared as described above, were assayed radiometrically as described previously (Chen et al., 2000). A unit of activity was defined as a micromole of the product, DPPC, formed in the LPTA assay or as a micromole of the substrate, LysoPC, degraded in the LPL assay at 37°C and pH 4.0.
Murine inhalation model of cryptococcosis
The procedures described using animals are approved and governed by the Sydney West Area Health Service Animal Ethics Committee, Department of Animal Care. Pathogenicity studies and organ burden analyses were conducted in 6- to 8-week-old BALB/c mice obtained from the Animal Resource Centre, Floreat Park, Western Australia. Animals were anaesthetized by inhalation of methoxyflurane. Survival study- Groups of 10 mice (H99 and deletion mutant strains) were inoculated intranasally with each C. neoformans strain (0.5 × 106 yeast cells in 50 μl saline) and observed daily for signs of ill-health. As CnΔsec14-1 and CnΔsec14-1/CnΔsfh5 exhibited clumping, one clump was counted as one cell. Mice which had loss 20% of their pre-infection weight, or which showed debilitating clinical signs prior to losing 20% of their pre-infection weight, were euthanized by CO2 inhalation followed by cervical dislocation. Otherwise they were sacrificed after 34 days. Organ burden- Groups of nine mice (H99 and deletion mutant strains) were inoculated intranasally with each C. neoformans strain (0.5 × 106 yeast in 50 μl saline). As CnΔsec14-1 exhibited clumping, one clump was counted as one cell. Three mice from each group were sacrificed 4, 7, and 14 days after inoculation (Day 0). Lungs and brains were harvested, weighed, homogenised and yeast colony forming units were quantified using SABD spread plates after 2-3 days incubation at 30°C.
Cryptococcus expulsion assay
J774A.1 macrophages were cultured in Dulbeccos's modified Eagles medium (DMEM) with 10% fetal calf serum (Gibco, Invitrogen, Paisley, UK), 50 U/ml penicillin and 50 μg/ml streptomycin at 37°C in a humidified atmosphere of 5% CO2. J774 (1×105) were plated in 24-well plates 18 h before phagocytosis of cryptococci. J774 cells were activated with PMA for 1 h before cryptococci (opsonised with mouse monoclonal anticapsular antibody, 18B7, provided by Arturo Casadevall) were added in DMEM for 2 h at 37°C. Time lapse images were captured on a Nikon TE2000 with Digital Sight DS-Qi1MC camera, 20× objective (Ph1 PLAN APO), using NIS elements AR software (Nikon, Richmond, UK). Images were captured every 90 sec for 18 h. The microscope stage was enclosed in a temperature controlled and humidified environmental chamber (Okolabs, NA, Italy) with 5% CO2 at 37°C. Expulsion of cryptococcal cells was scored as described previously (Ma et al., 2006).
Statistics
Student's t-test and one-way ANOVA were used to compare means between 2 groups and more than 2 groups of sample, respectively, where applicable, using SPSS version 16 statistical software. For the murine model of cryptococcosis, an estimation of differences in survival (log-rank test), using the Kaplan-Meier method, was performed and the survival curves plotted using the SPSS version 16 statistical software. In all cases, a P-value <0.05 was considered statistically significant. For the cryptococcal expulsion assay, statistical significance was calculated using Fisher's exact test.
Supplementary Material
Acknowledgments
This work was supported by the National Health and Medical Research Council of Australia (#632634), the University of Sydney Research Grant Scheme (2008, 2009) and the Westmead Hospital Charitable Trust (2008). RCM and SAJ are supported by the Medical Research Council (G0601171) and Wellcome Trust (WT088148MA). PRW is supported by an NIH grant (AI45995) and a Veteran's Administration Merit Review Award. MC is supported by the Endeavour International Postgraduate Research Scholarship (EIPRS), the International Postgraduate Award (IPA), the Postgraduate Scholarship in Fungal Pathogenesis and the Millennium Foundation Stipend Enhancement Grant. We thank the Broad Institute for providing C. neoformans Serotype A sequence information, Karen Bythe-Wilson for statistical analysis, Marina Barhoumah for technical assistance and Dr Ana Traven (Monash University, Melbourne, Australia) for helpful discussion.
References
- Alvarez M, Casadevall A. Phagosome extrusion and host-cell survival after Cryptococcus neoformans phagocytosis by macrophages. Curr Biol. 2006;16:2161–2165. doi: 10.1016/j.cub.2006.09.061. [DOI] [PubMed] [Google Scholar]
- Alvarez M, Casadevall A. Cell-to-cell spread and massive vacuole formation after Cryptococcus neoformans infection of murine macrophages. BMC Immunol. 2007;8:16. doi: 10.1186/1471-2172-8-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagnat M, Keranen S, Shevchenko A, Simons K. Lipid rafts function in biosynthetic delivery of proteins to the cell surface in yeast. Proc Natl Acad Sci U S A. 2000;97:3254–3259. doi: 10.1073/pnas.060034697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankaitis VA, Malehorn DE, Emr SD, Greene R. The Saccharomyces cerevisiae SEC14 gene encodes a cytosolic factor that is required for transport of secretory proteins from the yeast Golgi complex. J Cell Biol. 1989;108:1271–1281. doi: 10.1083/jcb.108.4.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankaitis VA, Mousley CJ, Schaaf G. The Sec14 superfamily and mechanisms for crosstalk between lipid metabolism and lipid signaling. Trends Biochem Sci. 2010;35:150–160. doi: 10.1016/j.tibs.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke D, Dawson D, Stearns T. Methods in yeast genetics: a Cold Spring Harbor Laboratory course manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, N.Y.: 2000. [Google Scholar]
- Castillon GA, Watanabe R, Taylor M, Schwabe TM, Riezman H. Concentration of GPI-anchored proteins upon ER exit in yeast. Traffic. 2009;10:186–200. doi: 10.1111/j.1600-0854.2008.00857.x. [DOI] [PubMed] [Google Scholar]
- Charlier C, Nielsen K, Daou S, Brigitte M, Chretien F, Dromer F. Evidence of a role for monocytes in dissemination and brain invasion by Cryptococcus neoformans. Infect Immun. 2009;77:120–127. doi: 10.1128/IAI.01065-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chayakulkeeree M, Sorrell TC, Siafakas AR, Wilson CF, Pantarat N, Gerik KJ, Boadle R, Djordjevic JT. Role and mechanism of phosphatidylinositol-specific phospholipase C in survival and virulence of Cryptococcus neoformans. Mol Microbiol. 2008;69:809–826. doi: 10.1111/j.1365-2958.2008.06310.x. [DOI] [PubMed] [Google Scholar]
- Chen SC, Wright LC, Golding JC, Sorrell TC. Purification and characterization of secretory phospholipase B, lysophospholipase and lysophospholipase/transacylase from a virulent strain of the pathogenic fungus Cryptococcus neoformans. Biochem J. 2000;347:431–439. doi: 10.1042/0264-6021:3470431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox GM, McDade HC, Chen SC, Tucker SC, Gottfredsson M, Wright LC, Sorrell TC, Leidich SD, Casadevall A, Ghannoum MA, Perfect JR. Extracellular phospholipase activity is a virulence factor for Cryptococcus neoformans. Mol Microbiol. 2001;39:166–175. doi: 10.1046/j.1365-2958.2001.02236.x. [DOI] [PubMed] [Google Scholar]
- Curwin AJ, Fairn GD, McMaster CR. Phospholipid transfer protein Sec14 is required for trafficking from endosomes and regulates distinct trans-Golgi export pathways. J Biol Chem. 2009;284:7364–7375. doi: 10.1074/jbc.M808732200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson RC, Blankenship JR, Kraus PR, de Jesus Berrios M, Hull CM, D'Souza C, Wang P, Heitman J. A PCR-based strategy to generate integrative targeting alleles with large regions of homology. Microbiology. 2002;148:2607–2615. doi: 10.1099/00221287-148-8-2607. [DOI] [PubMed] [Google Scholar]
- Djordjevic JT, Del Poeta M, Sorrell TC, Turner KM, Wright LC. Secretion of cryptococcal phospholipase B1 (PLB1) is regulated by a glycosylphosphatidylinositol (GPI) anchor. Biochem J. 2005;389:803–812. doi: 10.1042/BJ20050063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falkow S. Molecular Koch's postulates applied to microbial pathogenicity. Rev Infect Dis. 1988;10(2):S274–276. doi: 10.1093/cid/10.supplement_2.s274. [DOI] [PubMed] [Google Scholar]
- Feldmesser M, Kress Y, Novikoff P, Casadevall A. Cryptococcus neoformans is a facultative intracellular pathogen in murine pulmonary infection. Infect Immun. 2000;68:4225–4237. doi: 10.1128/iai.68.7.4225-4237.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganendren R, Carter E, Sorrell T, Widmer F, Wright L. Phospholipase B activity enhances adhesion of Cryptococcus neoformans to a human lung epithelial cell line. Microbes Infect. 2006;8:1006–1015. doi: 10.1016/j.micinf.2005.10.018. [DOI] [PubMed] [Google Scholar]
- Harsay E, Bretscher A. Parallel secretory pathways to the cell surface in yeast. J Cell Biol. 1995;131:297–310. doi: 10.1083/jcb.131.2.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G, Hacham M, Waterman SR, Panepinto J, Shin S, Liu X, Gibbons J, Valyi-Nagy T, Obara K, Jaffe HA, Ohsumi Y, Williamson PR. PI3K signaling of autophagy is required for starvation tolerance and virulenceof Cryptococcus neoformans. J Clin Invest. 2008;118:1186–1197. doi: 10.1172/JCI32053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston SA, May RC. The human fungal pathogen Cryptococcus neoformans escapes macrophages by a phagosome emptying mechanism that is inhibited by Arp2/3 complex-mediated actin polymerisation. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1001041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones S. New way out for Cryptococcus. Nat Rev Microbiol. 2007;5 doi: 10.1038/nrmicro1583. [DOI] [Google Scholar]
- Klemm RW, Ejsing CS, Surma MA, Kaiser HJ, Gerl MJ, Sampaio JL, de Robillard Q, Ferguson C, Proszynski TJ, Shevchenko A, Simons K. Segregation of sphingolipids and sterols during formation of secretory vesicles at the trans-Golgi network. J Cell Biol. 2009;185:601–612. doi: 10.1083/jcb.200901145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H, Croudace JE, Lammas DA, May RC. Expulsion of live pathogenic yeast by macrophages. Curr Biol. 2006;16:2156–2160. doi: 10.1016/j.cub.2006.09.032. [DOI] [PubMed] [Google Scholar]
- Ma H, Croudace JE, Lammas DA, May RC. Direct cell-to-cell spread of a pathogenic yeast. BMC Immunol. 2007;8:15. doi: 10.1186/1471-2172-8-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Missall TA, Moran JM, Corbett JA, Lodge JK. Distinct stress responses of two functional laccases in Cryptococcus neoformans are revealed in the absence of the thiol-specific antioxidant Tsa1. Eukaryot Cell. 2005;4:202–208. doi: 10.1128/EC.4.1.202-208.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteoliva L, Sanchez M, Pla J, Gil C, Nombela C. Cloning of Candida albicans SEC14 gene homologue coding for a putative essential function. Yeast. 1996;12:1097–1105. doi: 10.1002/(SICI)1097-0061(19960915)12:11%3C1097::AID-YEA990%3E3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- Muniz M, Riezman H. Intracellular transport of GPI-anchored proteins. EMBO J. 2000;19:10–15. doi: 10.1093/emboj/19.1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noverr MC, Cox GM, Perfect JR, Huffnagle GB. Role of PLB1 in pulmonary inflammation and cryptococcal eicosanoid production. Infect Immun. 2003;71:1538–1547. doi: 10.1128/IAI.71.3.1538-1547.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panepinto J, Komperda K, Frases S, Park YD, Djordjevic JT, Casadevall A, Williamson PR. Sec6-dependent sorting of fungal extracellular exosomes and laccase of Cryptococcus neoformans. Mol Microbiol. 2009;71:1165–1176. doi: 10.1111/j.1365-2958.2008.06588.x. [DOI] [PubMed] [Google Scholar]
- Park BJ, Wannemuehler KA, Marston BJ, Govender N, Pappas PG, Chiller TM. Estimation of the current global burden of cryptococcal meningitis among persons living with HIV/AIDS. AIDS. 2009;23:525–530. doi: 10.1097/QAD.0b013e328322ffac. [DOI] [PubMed] [Google Scholar]
- Plaine A, Walker L, Da Costa G, Mora-Montes HM, McKinnon A, Gow NA, Gaillardin C, Munro CA, Richard ML. Functional analysis of Candida albicans GPI-anchored proteins roles in cell wall integrity and caspofungin sensitivity. Fungal Genet Biol. 2008;45:1404–1414. doi: 10.1016/j.fgb.2008.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santangelo R, Zoellner H, Sorrell T, Wilson C, Donald C, Djordjevic J, Shounan Y, Wright L. Role of extracellular phospholipases and mononuclear phagocytes in dissemination of cryptococcosis in a murine model. Infect Immun. 2004;72:2229–2239. doi: 10.1128/IAI.72.4.2229-2239.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santangelo RT, Chen SC, Sorrell TC, Wright LC. Detection of antibodies to phospholipase B in patients infected with Cryptococcus neoformans by enzyme-linked immunosorbent assay (ELISA) Med Mycol. 2005;43:335–341. doi: 10.1080/13693780412331282331. [DOI] [PubMed] [Google Scholar]
- Santangelo RT, Nouri-Sorkhabi MH, Sorrell TC, Cagney M, Chen SC, Kuchel PW, Wright LC. Biochemical and functional characterisation of secreted phospholipase activities from Cryptococcus neoformans in their naturally occurring state. J Med Microbiol. 1999;48:731–740. doi: 10.1099/00222615-48-8-731. [DOI] [PubMed] [Google Scholar]
- Schnabl M, Oskolkova OV, Holic R, Brezna B, Pichler H, Zagorsek M, Kohlwein SD, Paltauf F, Daum G, Griac P. Subcellular localization of yeast Sec14 homologues and their involvement in regulation of phospholipid turnover. Eur J Biochem. 2003;270:3133–3145. doi: 10.1046/j.1432-1033.2003.03688.x. [DOI] [PubMed] [Google Scholar]
- Siafakas AR, Sorrell TC, Wright LC, Wilson C, Larsen M, Boadle R, Williamson PR, Djordjevic JT. Cell wall-linked cryptococcal phospholipase B1 is a source of secreted enzyme and a determinant of cell wall integrity. J Biol Chem. 2007;282:37508–37514. doi: 10.1074/jbc.M707913200. [DOI] [PubMed] [Google Scholar]
- Siafakas AR, Wright LC, Sorrell TC, Djordjevic JT. Lipid rafts in Cryptococcus neoformans concentrate the virulence determinants phospholipase B1 and Cu/Zn superoxide dismutase. Eukaryot Cell. 2006;5:488–498. doi: 10.1128/EC.5.3.488-498.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutterlin C, Doering TL, Schimmoller F, Schroder S, Riezman H. Specific requirements for the ER to Golgi transport of GPI-anchored proteins in yeast. J Cell Sci. 1997;110(Pt 21):2703–2714. doi: 10.1242/jcs.110.21.2703. [DOI] [PubMed] [Google Scholar]
- Turner KM, Wright LC, Sorrell TC, Djordjevic JT. N-linked glycosylation sites affect secretion of cryptococcal phospholipase B1, irrespective of glycosylphosphatidylinositol anchoring. Biochim Biophys Acta. 2006;1760:1569–1579. doi: 10.1016/j.bbagen.2006.07.002. [DOI] [PubMed] [Google Scholar]
- Vorisek J. Functional morphology of the secretory pathway organelles in yeast. Microsc Res Tech. 2000;51:530–546. doi: 10.1002/1097-0029(20001215)51:6<530::AID-JEMT4>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- Wright L, Bubb W, Davidson J, Santangelo R, Krockenberger M, Himmelreich U, Sorrell T. Metabolites released by Cryptococcus neoformans var. neoformans and var. gattii differentially affect human neutrophil function. Microbes Infect. 2002;4:1427–1438. doi: 10.1016/s1286-4579(02)00024-2. [DOI] [PubMed] [Google Scholar]
- Wright LC, Santangelo RM, Ganendren R, Payne J, Djordjevic JT, Sorrell TC. Cryptococcal lipid metabolism: phospholipase B1 is implicated in transcellular metabolism of macrophage-derived lipids. Eukaryot Cell. 2007;6:37–47. doi: 10.1128/EC.00262-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneda A, Doering TL. A eukaryotic capsular polysaccharide is synthesized intracellularly and secreted via exocytosis. Mol Biol Cell. 2006;17:5131–5140. doi: 10.1091/mbc.E06-08-0701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaragoza O, Rodrigues ML, De Jesus M, Frases S, Dadachova E, Casadevall A. The capsule of the fungal pathogen Cryptococcus neoformans. Adv Appl Microbiol. 2009;68:133–216. doi: 10.1016/S0065-2164(09)01204-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Gibbons J, Garcia-Rivera J, Casadevall A, Williamson PR. Laccase of Cryptococcus neoformans is a cell wall-associated virulence factor. Infect Immun. 2001;69:5589–5596. doi: 10.1128/IAI.69.9.5589-5596.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.