The maize transcription factor KNOTTED 1 (KN1) is an important factor in controlling the balance of organ initiation and meristem renewal during plant development. In this first ChIP-seq study of a maize transcription factor, Hake and colleagues identify the direct targets of KN1 and, together with in vitro data, determine the rules for KN1 binding, providing insights into KN1's orchestration of gene hierarchical regulatory networks.
Keywords: KNOX transcription factors, chromatin immunoprecipitation, gene regulation, maize
Abstract
KNOTTED1 (KN1)-like homeobox (KNOX) transcription factors function in plant meristems, self-renewing structures consisting of stem cells and their immediate daughters. We defined the KN1 cistrome in maize inflorescences and found that KN1 binds to several thousand loci, including 643 genes that are modulated in one or multiple tissues. These KN1 direct targets are strongly enriched for transcription factors (including other homeobox genes) and genes participating in hormonal pathways, most significantly auxin, demonstrating that KN1 plays a key role in orchestrating the upper levels of a hierarchical gene regulatory network that impacts plant meristem identity and function.
Plant organs initiate through an iterative process at the shoot apical meristem (SAM), a pluripotent stem cell population that generates the above-ground body of vascular plants. As leaves initiate from the flanks of the SAM, a proper balance between organ initiation and meristem renewal is critical for normal development. Transcription factors (TFs) belonging to the class I KNOTTED1 (KN1)-like homeobox (KNOX) family play important roles in this balance. KNOX proteins are members of the three-amino-acid loop extension (TALE) superfamily of homeodomain TFs, which function during development in all eukaryotic lineages (Hay and Tsiantis 2010). KNOX proteins favor cytokinin (CK) accumulation through the positive regulation of CK synthesis genes (Jasinski et al. 2005; Yanai et al. 2005). High gibberellin (GA) and auxin signaling are required for cell differentiation and elongation in organs. Excessive GA negatively affects SAM maintenance (Jasinski et al. 2005), and KNOX TFs are expected to maintain low quantities of active GA through direct repression of GA 20-oxidase (synthesis) and up-regulation of GA 2-oxidase (catabolism) (Sakamoto et al. 2001; Chen et al. 2004; Bolduc and Hake 2009).
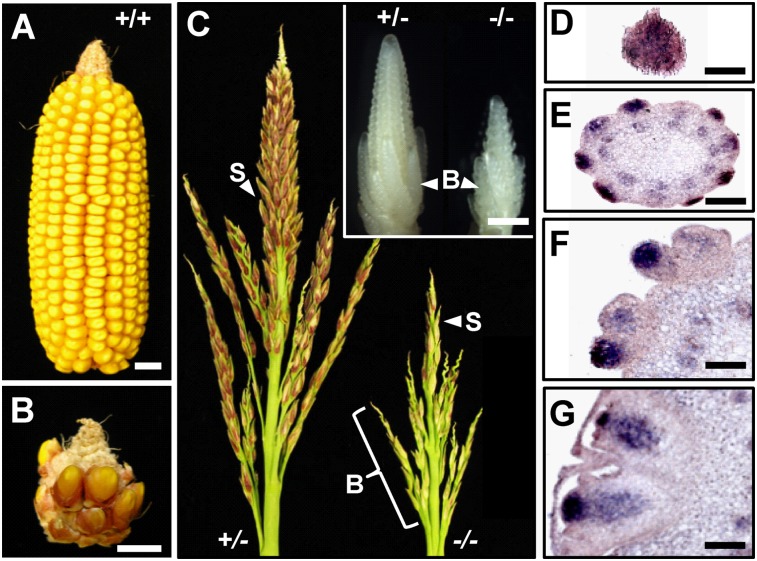
Maize kn1 was the first plant TALE gene to be identified as a result of its gain-of-function phenotype (Vollbrecht et al. 1991). kn1 is expressed in all shoot meristems but is specifically down-regulated in cells destined to become primordia (Jackson et al. 1994). kn1 loss-of-function plants occasionally lack shoots (Vollbrecht et al. 2000) but are mostly impaired in reproductive development (Fig. 1A–C; Kerstetter et al. 1997). When ectopically expressed in developing leaves, KN1 induces cell proliferation, resulting in the formation of knots or flaps, depending on the localization of kn1 misexpression (Smith et al. 1992; Ramirez et al. 2009).
Figure 1.
kn1 loss-of-function phenotypes. (A,B) Female inflorescence (ear) from wild-type (+/+) (A) and the null kn1-e1 (−/−) (B) allele, which produces a small cob with poor kernel yield. (C) Male inflorescence (tassel). kn1-e1 tassels have fewer long branches (B) and fewer spikelets (S). (Inset) Immature tassels at the stage used for ChIP and RNA-seq. Immature ears are similar but lack long branches. (D–G) kn1 in situ hybridization in transverse sections of a wild-type ear, going from the tip (D) toward the base (G). A specific signal corresponds to the dark-blue staining. Bars: A–C, 1 cm; C, inset, 1 mm; D–G, 0.1 mm.
Despite a deep appreciation for the importance of KNOX TFs in plant development, we know very little about the genes under KNOX regulation. We identified genes differentially expressed between normal and kn1 mutants (RNA sequencing [RNA-seq]) and compared this data set with KN1 genome-wide occupancy (chromatin immunoprecipitation [ChIP] coupled with deep sequencing [ChIP-seq]). Modulated and bound genes are enriched in TFs and hormone metabolism, most notably auxin, thus highlighting a pivotal role for KN1 in the regulatory network controlling maize development.
Results and Discussion
KN1 binds to thousands of loci in the maize genome
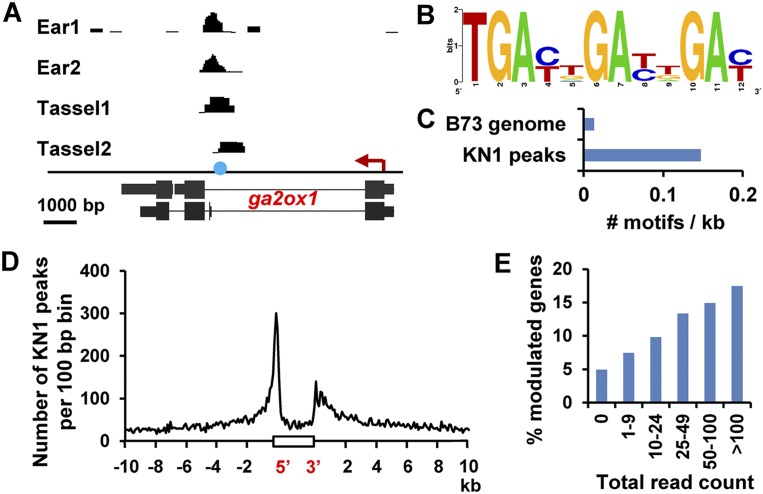
To elucidate the molecular link between KNOX activities and meristem maintenance, we sought to define a KNOX cistrome. We previously showed direct regulation of KN1 binding to the GA catabolism gene ga2ox1 using ChIP with antibodies recognizing the native KN1 protein (Bolduc and Hake 2009). Using this reliable assay, ChIP-seq was performed on two biological replicates of immature ears expressing kn1 in a developmental series of meristem types (Fig. 1D–G), in addition to a small-scale experiment on two immature tassel samples (Supplemental Table 1). Several thousand genomic regions were significantly enriched in each of the two ear samples (Supplemental Fig. 1; Supplemental Table 2). The known target ga2ox1 was enriched in all four samples (Fig. 2A), providing a positive control for the ChIP-seq and data analysis.
Figure 2.
ChIP-seq results. (A) ChIP-seq identified KN1 binding in ga2ox1. The blue circle indicates the position of the previously mapped KN1-binding motif, and the red arrow indicates gene orientation. (B,C) ga2ox1-like binding motif (B) is significantly over-represented (P-value < 1 × 10−100) in KN1-bound loci compared with the B73 genome (C). (D) Distribution of KN1 peaks around transcribed genes. A representative gene is shown as a white box on the X-axis. (E) Relationship between the total read count and the proportion of modulated genes in all tissues.
To validate the newly identified KN1-bound regions, such as those identified in the KNOX gene liguleless3 (lg3) (Supplemental Fig. 2A), we performed quantitative PCR (qPCR) on ChIP DNA (ChIP-qPCR) obtained from wild-type and kn1-null tissues (Supplemental Figs. 2–5). Enrichment was detected in the regions covered by peaks but not upstream and downstream, indicating that the ChIP-seq accurately identified the spatial distribution of KN1 binding in vivo. Furthermore, genomic regions that were enriched for KN1 binding in ears were enriched in meristems from wild-type tassels and SAM but not from kn1-null meristems. Thus, the bound regions identified in ears are KN1-specific and representative of KN1 binding in other tissues as well. To minimize false positive identification, we concentrated our analysis on the 6511 peaks that showed overlap between the two ears samples (referred to as “high-confidence” KN1-bound loci) (Supplemental Material). Among these, 879 loci were also identified in at least one tassel ChIP-seq experiment (Supplemental Fig. 1). However, the number of KN1 targets shared between ears and tassels is likely much larger, since all 28 ChIP-qPCR-validated loci identified exclusively as peaks in ears also showed enrichment in tassels (Supplemental Fig. 4; Supplemental Table 3).
KN1 binds in vitro to cis-regulatory elements present in the high-confidence KN1-bound loci
KNOX proteins bind sequences containing a TGAC core in vitro, but specific cis-regulatory elements recognized in vivo by KNOX proteins have been identified in only four direct targets (Sakamoto et al. 2001; Chen et al. 2004; Bolduc and Hake 2009; Spinelli et al. 2011). Starting from the KN1-bound regions, we computationally identified several motifs consisting of two “NGAC” sequences separated by one or two residues, reminiscent of the KN1-binding site found in the ga2ox1 gene (two TGAC motifs separated by four residues). However, in vitro KN1 binding was not detected for any of the three motifs tested, and binding was severely impaired for probes derived from the ga2ox1-binding site with a gap <3 base pairs (bp) (Supplemental Fig. 6; Supplemental Table 4), indicating that the spacing between the two TGAC motifs is important for efficient KN1 binding in vitro.
We also searched for the presence of a ga2ox1-like binding motif in the entire set of high-confidence bound loci (see the Supplemental Material). The motif was found 11 times more frequently in the KN1-bound loci compared with the maize genome (Fig. 2B,C), indicating that this cis-regulatory element is a common KN1-binding motif but not the only one, since it was identified in only 474 loci (7%). These results reinforce the idea that KNOX TFs have degenerate binding sites and acquire specificity through cooperation with binding partners, as found in animals (Moens and Selleri 2006). KNOX proteins recognize DNA as heterodimers with BEL1-LIKE HOMEOBOX (BLH) proteins (for review, see Hay and Tsiantis 2010), and the 17 BLH genes in maize likely provide ample opportunity for increased specificity. The different protein complexes could translate to different binding motifs, as found for Oct-1 and Ubx (Huang and Herr 1996; Slattery et al. 2011), and would explain our failed attempt at identifying de novo a unifying consensus binding site that relates to KN1's in vitro binding properties.
KN1 binds near genes and potential distal enhancers
To establish which genes are potential KN1 direct targets, each high-confidence bound locus was assigned to the closest gene, resulting in a total of 5118 genes, including 1026 that are associated with two or more loci. In 81% of the high-confidence KN1-bound loci (4274 genes), peaks are located within 10 kb of genes, mainly in the 5′ or 3′ regions, but also in introns and exons (Fig. 2D; Supplemental Fig. 7). However, among the more distant peaks, one was identified in a region that behaves as an enhancer for expression of teosinte branched1 (Studer et al. 2011), a key gene in maize domestication (Doebley et al. 1997). KN1 recognizes in vitro a motif present in this region that is conserved in maize landraces, modern inbreds, and teosinte (Supplemental Figs. 6C, 8). These data suggest that distantly located KN1-bound regions may represent enhancers important for gene regulation. The Drosophila TALE protein Prep1 was reported to activate pax6 via binding to an enhancer necessary for lens-specific expression (Rowan et al. 2010), suggesting that plant TALE proteins such as KN1 could similarly regulate their targets through enhancer binding.
Deciphering the genes that are bound and modulated
To test whether the bound genes are under KN1 regulation, we obtained genome-wide quantitative expression data (RNA-seq) for tissues where KN1 naturally accumulates at high levels (immature ears and tassels, and SAM), and compared kn1-e1 loss-of-function plants with their normal siblings. We also took advantage of semidominant Kn1 mutants that misexpress kn1 in leaf primordia (Smith et al. 1992). When analyzed by qRT–PCR, ga2ox1 and lg3 showed a gradient of accumulation in Kn1-N/+ and Kn1-N/Kn1-N leaf primordia proportional to kn1 overexpression (Supplemental Fig. 9). No obvious morphological differences were visible between wild-type and Kn1-N mutants at the stage sampled, indicating that differential gene expression should be primarily due to early KN1 activity, rather than secondary effects resulting from altered cell fate. We considered as differentially expressed genes showing an adjusted P-value of <0.01, regardless of their fold change, resulting in 1267 modulated genes in ears, 754 in tassels, and 14 in shoot apices (Table 1). We detected 1336 genes differentially expressed between wild-type and Kn1-N/Kn1-N leaves, with 23 of these also showing a difference between wild-type and the mild Kn1-N/+ leaves.
Table 1.
Proportion of modulated genes that are bound by KN1
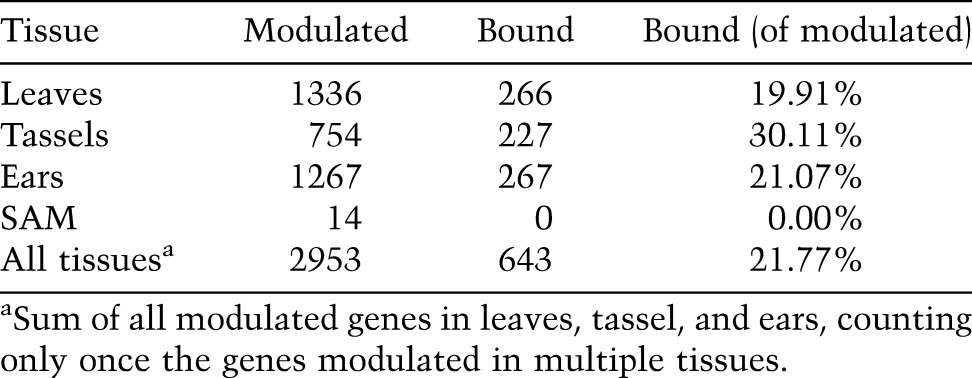
We then asked whether the genes identified as bound by KN1 in ears were modulated in the kn1-e1 or Kn1-N backgrounds, taking advantage of the quantitative aspect of ChIP-seq. For each gene, we compiled the number of reads at peaks up to 10 kb upstream and downstream. The total number of reads from all four biological replicates (total read count [TRC]) was used as a quantitative measure of KN1 occupancy (see the Supplemental Material). Among the genes that have no peaks within 10 kb, only 4.9% were differentially expressed in at least one tissue (Fig. 2E). The proportion of differentially expressed genes increases with the TRC and reaches 17.5% when considering genes with a TRC of >100. Thus, there is a positive correlation between KN1 occupancy and the proportion of modulated genes. These results are in agreement with the continuous network hypothesis (Biggin 2011), which states that TFs bind over a quantitative series of DNA occupancy levels that span functional, quasifunctional, and nonfunctional DNA-binding events. Genes more highly occupied are likely to be more biologically relevant targets. We thus restricted our analysis to modulated genes with a TRC of ≥25 (see the Supplemental Material). Using these criteria, 20%–30% of the genes initially identified as differentially expressed in leaves, tassels, or ears are considered bound by KN1 (Table 1). Of note, the absence of modulation for many of the KN1-bound genes may be due to nonfunctional DNA-binding events, but the redundancy of KNOX TFs is likely an important factor. Other KNOX proteins in kn1-e1 meristems may partially compensate for the absence of KN1. In addition, kn1 is misexpressed only in a few cells in Kn1-N leaves, making the detection of small differences difficult.
KN1 regulates other TFs
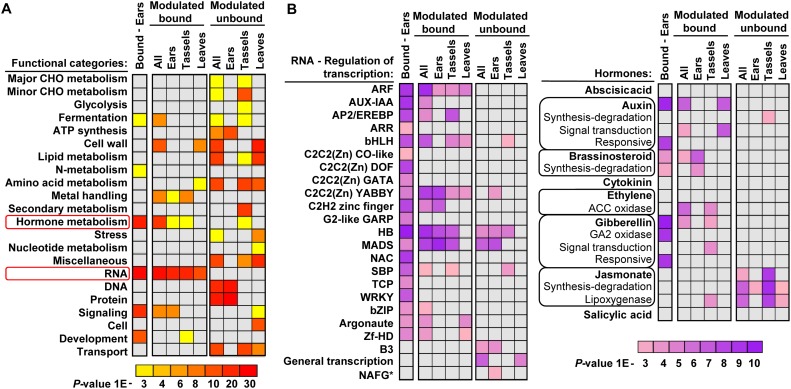
We assigned each gene to functional categories (Thimm et al. 2004) and compared the ones enriched among the bound and modulated genes (direct targets) with the categories enriched among the indirect targets, represented by the modulated genes that did not match our stringent criteria for KN1 binding. The “RNA” category presents the most significant enrichment in all three tissues in the KN1-bound and modulated genes but is not enriched among the indirect targets (Fig. 3A). The enriched subcategories comprise several families of TFs, with the homeobox (HB), MADS, auxin response factor (ARF), YABBY, and basic helix–loop–helix (bHLH) families showing the most significant enrichment (Fig. 3B).
Figure 3.
Functional categories of KN1 targets. Distribution of categories is shown for genes located within 10 kb of high-confidence KN1-bound loci (Ears—Bound) and modulated genes (bound vs. unbound). (A) Enrichments for all main categories. (B) Enrichments for subcategories in the “RNA—regulation of transcription” (left) and “hormone metabolism” (right) categories. (*) Nucleosome assembly factor group.
The HB family is particularly preponderant among the bound TFs and includes 10 knox genes but also several blh, hd-zipIII, etc. (Supplemental Table 5). Among the knox genes, KN1 positively regulates lg3, lg4a, and knox8 but negatively regulates gn1 (Supplemental Fig. 10). lg3 and lg4a are induced in Kn1-N leaves, suggesting that the dominant Kn1 phenotypes result from the cooperative work of all three KNOX proteins. Interestingly, the knox8 rice ortholog OSH43 is also a direct (positive) target of the KN1 ortholog OSH1 (Tsuda et al. 2011). Furthermore, KN1 binds within its own gene, in an intron where transposon insertions lead to dominant Kn1 mutants (Greene et al. 1994). The importance of autofeedback regulation mediated by cis-regulatory elements in this intron has been demonstrated in rice for OSH1 (Tsuda et al. 2011).
Forty-seven KN1-bound genes were comodulated in both ears and tassels (Supplemental Table 6), with the most significant enrichment for genes involved in regulation of transcription (P-value = 1.2 × 10−7). This list comprises 18 TFs, including members of the MADS-box family with similarity to SEPALLATA TFs known to participate in protein complexes with related MADS proteins (Thompson and Hake 2009). Considering the involvement of MADS TFs in floral development, these targets may be partly responsible for the strong floral defects in kn1 loss-of-function ears (Kerstetter et al. 1997). Another interesting case is the yabby genes belonging to the same clade as FIL (Juarez et al. 2004) that are down-regulated in ears and tassels and up-regulated in Kn1-N/Kn1-N leaves (Supplemental Fig. 10), indicative of a KN1-mediated positive regulation. YABBY TFs are important for leaf polarity and lamina growth (Sarojam et al. 2010), and misregulation of these proximally expressed genes (Juarez et al. 2004) may contribute to the presence of proximal structures in the distal part of the leaf in Kn1 gain-of-function alleles (Ramirez et al. 2009). Other genes involved in the specification of organ boundaries, such as NAC (NAM-ATAF1,2-CUC2) TFs, are targeted by SHOOTMERISTEMLESS in Arabidopsis, including the direct regulation of CUC1 (Spinelli et al. 2011). Similarly, KN1 binds to many maize NAC genes, although this class of TFs is not significantly enriched among the direct targets.
Other KN1-bound transcriptional regulators not classified as TFs were modulated in several tissues. Of particular interest is GRMZM2G079080, a gene nearly identical to ARGONAUTE10 (AGO10) (also known as PINHEAD) (Supplemental Fig. 10), which in Arabidopsis is critical for SAM development (Lynn et al. 1999). AGO proteins are the catalytic components of the RNA-induced silencing complex (RISC) responsible for small RNA-mediated gene silencing (Mallory and Vaucheret 2010). Three other KN1-bound AGO genes were identified as positively modulated in Kn1-N leaves or negatively modulated in kn1-e1 ears, indicating a general trend for a KN1-mediated positive regulation of this class of genes.
KN1 directly regulates genes in hormonal pathways
Our ChIP-seq results highlight the preferential binding of KN1 near genes belonging to the GA, brassinosteroid (BR), and auxin pathways, but modulated genes are mainly enriched for the auxin pathway (Fig. 3B; Supplemental Table 7). Although not enriched as a group, our results indicate that KN1 could promote a higher CK accumulation in Kn1-N leaves by positively regulating a gene similar to the rice CK synthesis gene LONELY GUY (Supplemental Fig. 10; Kurakawa et al. 2007). KN1 also targets genes involved in CK perception and signaling, including a gene nearly identical to the CK receptor WOODEN LEG that is down-regulated in kn1-e1 tassel primordia, suggesting that KN1 positively regulates CK perception. Thus, KN1 could positively influence CK synthesis and signaling by directly targeting only a few key players in the CK pathway.
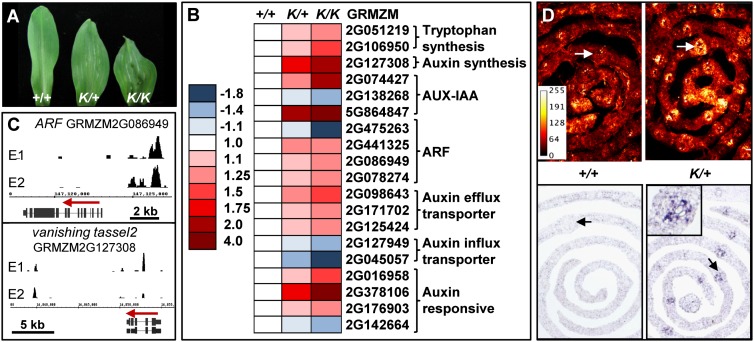
Among all of the bound hormonal genes, differential gene regulation occurs preferentially for the auxin-related genes, particularly in leaves (Figs. 3B, 4A,B). This enrichment is even more robust if we consider the TFs involved in auxin signaling: AUX-IAA and ARF. Indeed, KN1 binds to nearly half of the AUX-IAA and ARF genes annotated in the maize genome (Supplemental Table 7), with some of them showing differential expression in Kn1-N leaves. Beside these TFs, KN1 binds near genes coding for enzymes that synthesize auxin and maintain its homeostasis and auxin influx and efflux transporters, as well as AFB-like genes that are characterized as F-box auxin coreceptors in Arabidopsis (Chapman and Estelle 2009). Among these, modulation occurs in Kn1-N leaves for auxin efflux transporters of the PIN and ABCB families (up-regulated) and auxin influx carriers similar to AUX1 and LAX3 (down-regulated) (Fig. 4B). KN1-bound genes involved in the synthesis of auxin and its precursor, tryptophan, are also up-regulated in leaves, indicating that KN1 directly controls the auxin pathway at all levels. For example, the vanishing tassel2 (vt2) gene, identified through its mutant phenotype as a tryptophan aminotransferase (Phillips et al. 2011), is significantly up-regulated in Kn1-N leaves (Fig. 4B,C). Thus, KN1 directly modulates genes involved in the synthesis, transport, and signaling of auxin.
Figure 4.
KN1 targets the auxin pathway. (A) Kn1-N phenotype. First leaf from wild type (+/+), Kn1-N heterozygotes (K/+), and homozygotes (K/K) are shown. Kn1-N leaves are shorter and wider, and overall plant height is shorter in homozygotes. (B) Changes in gene expression for selected targets. All differences between homozygotes and wild type are statistically significant at a false discovery rate (FDR) of <0.01. (C) ChIP-seq identified KN1 binding in selected targets. Red arrows indicate gene orientation. Peak traces are shown for the two ear replicates (E1–E2). (D) Confocal images (maximum projection) of DR5rev∷mRFPer (top panels) or KN1 immunolocalization (bottom panels) in Kn1-N/+ plants and wild-type siblings (transverse sections). Increased fluorescence in the lateral veins (arrows) correlates with KN1 misexpression. A look-up table scale of fluorescence intensity in shown in the top left panel.
To understand how the observed differential gene expression affects auxin output, we compared DR5rev∷mRFPer accumulation (Gallavotti et al. 2008) between Kn1-N/+ and +/+ siblings. The DR5 reporter is more highly expressed in lateral veins of Kn1-N mutants at this stage, coincident with KN1 accumulation (Fig. 4D). In addition, RFP fluorescence was significantly higher in protein extracts from leaves of Kn1-N/+ than wild-type siblings (Supplemental Fig. 10C), indicating higher auxin signaling in the presence of KN1. Thus, by regulating many genes in the auxin pathway, KN1 has a positive impact on auxin signaling in leaves.
How does increased auxin signaling observed in leaves relate to KN1 activity in its normal context? KN1 accumulates in vasculature of unexpanded stems and immature inflorescences (Smith et al. 1992), and considering the requirement for auxin in vasculature development, it is likely that KN1 misexpression in leaf veins mimics its endogenous role in the vasculature. Furthermore, maize plants carrying mutations in the auxin pathway display small or nonexistent inflorescences (McSteen 2010; Phillips et al. 2011) that are very similar to the defect in lateral meristem initiation observed in kn1-null mutants (Kerstetter et al. 1997), suggesting that KN1 could directly contribute to the formation of lateral organs through specific spatiotemporal modulation of auxin synthesis, transport, and signaling. However, high auxin activity is observed at sites of organ initiation where KNOX genes are not expressed (Heisler et al. 2005; Gallavotti et al. 2008), thus questioning how KN1 would modulate auxin signaling in cells where it does not accumulate. It was recently shown that the apparent complexity of auxin response components in the Arabidopsis SAM can be simplified to two robust differential sensitivities to auxin, with the central zone having a lower sensitivity, and the peripheral zone having a higher one (Vernoux et al. 2011). KN1 accumulates in both central and peripheral zones of the SAM, and different spatiotemporal misexpression of KNOX genes in leaves leads to drastically different leaf phenotypes (Ramirez et al. 2009; Shani et al. 2009). Thus, KN1 activity within the SAM could also vary according to its spatiotemporal distribution, with a differential impact on auxin response components in the central and peripheral zones. KN1 is also present in a few cell layers at the base of initiating organ primordia (Jackson 2002), where it may contribute to establishing the proximal axis of the leaf. Regardless, it is important to remember that KNOX TFs also act through modulation of CK, GA, and potentially BR, all of which cross-talk with auxin (Jaillais and Chory 2010). A complex equilibrium between the different hormones is crucial for proper meristem maintenance and organ initiation, and our results support a model in which KN1 plays a major role in the establishment and maintenance of this equilibrium.
Considering the global changes that arise during the transition from a meristematic to a determinate fate, and the key role played by KN1 in that transition, the occupancy of thousands of loci, concomitant with the regulation of hundreds of genes, is not surprising. Our results show that KN1 stands at the top of an intricate regulatory network involving hormonal pathways and transcriptional regulators. This study provides a blueprint from which to improve desired traits, such as yield, in maize and related cereal crops, in addition to serving as a foundation for a systems approach to understanding meristem function in all plants.
Materials and methods
ChIP-seq and RNA-seq
All plant material was in the B73 background. ChIP was performed as described (Bolduc and Hake 2009), and libraries were sequenced on an Illumina Genome Analyzer II. Reads were aligned to the B73 maize reference genome version 2, and peak calling was performed with MACS using only the uniquely mapping reads. RNA-seq libraries were prepared from 1–2 μg of total RNA using the TruSeq kit (Illumina) and sequenced on an Illumina HiSeq 2000. Reads were aligned to the maize genome (B73 version 2, masked), and the number of reads in each gene model (filtered gene set 5b.60) was calculated. Differential gene expression was determined using the R bioconductor package DESeq. Analysis for ChIP-seq and RNA-seq is detailed in the Supplemental Material. The fully analyzed data set is available as a spreadsheet in Supplemental Table 11 and can be browsed at GRASSIUS (Yilmaz et al. 2009). Raw data are partially stored at Gene Expression Omnibus (GEO) (accession no. GSE38487 for RNA-seq, complete set; and GSE39161 for ChIP-seq, partial set) and at GRASSIUS (http://grassius.org/chipseqinfo.html?tf=KN1). ChIP-seq peaks from ears will also be available at MaizeGDB (http://gbrowse.maizegdb.org/gb2/gbrowse/maize_v2) (Sen at al. 2009). RNA-seq data can also be visualized at http://www.qteller.com.
Visualization of DR5rev∷mRFPer
Plants were hand-sectioned, mounted in water, and imaged with a Leica SP5 confocal microscope. Excitation was performed at 561 nm with a diode-pumped solid-state laser set at 10% power (pinhole set to 1 airy unit). For each sample, 27 Z sections were captured, keeping all settings identical between wild type and mutant. Images were processed with ImageJ.
Acknowledgments
We thank the Hake laboratory, J. Schnable, and A. Eveland for advice, discussion, and critical reading of the manuscript. The work was funded by NSF DBI-0604923 and ARS 5335-21000-013-00D to S.H., NSF DBI-0701405 and NSF IOS-1125620 to E.G., and NIH 5T32CA106196-05 to A.Y.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.193433.112.
References
- Biggin MD 2011. Animal transcription networks as highly connected, quantitative continua. Dev Cell 21: 611–626 [DOI] [PubMed] [Google Scholar]
- Bolduc N, Hake S 2009. The maize transcription factor KNOTTED1 directly regulates the gibberellin catabolism gene ga2ox1 Plant Cell. 21: 1647–1658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman EJ, Estelle M 2009. Mechanism of auxin-regulated gene expression in plants. Annu Rev Genet 43: 265–285 [DOI] [PubMed] [Google Scholar]
- Chen H, Banerjee AK, Hannapel DJ 2004. The tandem complex of BEL and KNOX partners is required for transcriptional repression of ga20ox1. Plant J 38: 276–284 [DOI] [PubMed] [Google Scholar]
- Doebley J, Stec A, Hubbard L 1997. The evolution of apical dominance in maize. Nature 386: 485–488 [DOI] [PubMed] [Google Scholar]
- Gallavotti A, Yang Y, Schmidt RJ, Jackson D 2008. The relationship between auxin transport and maize branching. Plant Physiol 147: 1913–1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene B, Walko R, Hake S 1994. Mutator insertions in an intron of the maize knotted1 gene result in dominant suppressible mutations. Genetics 138: 1275–1285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay A, Tsiantis M 2010. KNOX genes: Versatile regulators of plant development and diversity. Development 137: 3153–3165 [DOI] [PubMed] [Google Scholar]
- Heisler MG, Ohno C, Das P, Sieber P, Reddy GV, Long JA, Meyerowitz EM 2005. Patterns of auxin transport and gene expression during primordium development revealed by live imaging of the Arabidopsis inflorescence meristem. Curr Biol 15: 1899–1911 [DOI] [PubMed] [Google Scholar]
- Huang CC, Herr W 1996. Differential control of transcription by homologous homeodomain coregulators. Mol Cell Biol 16: 2967–2976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson D 2002. Double labeling of KNOTTED1 mRNA and protein reveals multiple potential sites of protein trafficking in the shoot apex. Plant Physiol 129: 1423–1429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson D, Veit B, Hake S 1994. Expression of maize KNOTTED1 related homeobox genes in the shoot apical meristem predicts patterns of morphogenesis in the vegetative shoot. Development 120: 405–413 [Google Scholar]
- Jaillais Y, Chory J 2010. Unraveling the paradoxes of plant hormone signaling integration. Nat Struct Mol Biol 17: 642–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasinski S, Piazza P, Craft J, Hay A, Woolley L, Rieu I, Phillips A, Hedden P, Tsiantis M 2005. KNOX action in Arabidopsis is mediated by coordinate regulation of cytokinin and gibberellin activities. Curr Biol 15: 1560–1565 [DOI] [PubMed] [Google Scholar]
- Juarez MT, Twigg RW, Timmermans MC 2004. Specification of adaxial cell fate during maize leaf development. Development 131: 4533–4544 [DOI] [PubMed] [Google Scholar]
- Kerstetter RA, Laudencia-Chingcuanco D, Smith LG, Hake S 1997. Loss-of-function mutations in the maize homeobox gene, knotted1, are defective in shoot meristem maintenance. Development 124: 3045–3054 [DOI] [PubMed] [Google Scholar]
- Kurakawa T, Ueda N, Maekawa M, Kobayashi K, Kojima M, Nagato Y, Sakakibara H, Kyozuka J 2007. Direct control of shoot meristem activity by a cytokinin-activating enzyme. Nature 445: 652–655 [DOI] [PubMed] [Google Scholar]
- Lynn K, Fernandez A, Aida M, Sedbrook J, Tasaka M, Masson P, Barton MK 1999. The PINHEAD/ZWILLE gene acts pleiotropically in Arabidopsis development and has overlapping functions with the ARGONAUTE1 gene. Development 126: 469–481 [DOI] [PubMed] [Google Scholar]
- Mallory A, Vaucheret H 2010. Form, function, and regulation of ARGONAUTE proteins. Plant Cell 22: 3879–3889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McSteen P 2010. Auxin and monocot development. Cold Spring Harb Perspect Biol 2: a001479 doi: 10.1101/cshperspect.a001479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moens CB, Selleri L 2006. Hox cofactors in vertebrate development. Dev Biol 291: 193–206 [DOI] [PubMed] [Google Scholar]
- Phillips KA, Skirpan AL, Liu X, Christensen A, Slewinski TL, Hudson C, Barazesh S, Cohen JD, Malcomber S, McSteen P 2011. vanishing tassel2 encodes a grass-specific tryptophan aminotransferase required for vegetative and reproductive development in maize. Plant Cell 23: 550–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez J, Bolduc N, Lisch D, Hake S 2009. Distal expression of knotted1 in maize leaves leads to re-establishment of proximal/distal patterning and leaf dissection. Plant Physiol 151: 1878–1888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowan S, Siggers T, Lachke SA, Yue Y, Bulyk ML, Maas RL 2010. Precise temporal control of the eye regulatory gene Pax6 via enhancer-binding site affinity. Genes Dev 24: 980–985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto T, Kamiya N, Ueguehi-Tanaka M, Iwahori S, Matsuoka M 2001. KNOX homeodomain protein directly suppresses the expression of a gibberellin biosynthetic gene in the tobacco shoot apical meristem. Genes Dev 15: 581–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarojam R, Sappl PG, Goldshmidt A, Efroni I, Floyd SK, Eshed Y, Bowman JL 2010. Differentiating Arabidopsis shoots from leaves by combined YABBY activities. Plant Cell 22: 2113–2130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen TZ, Andorf CM, Schaeffer ML, Harper LC, Sparks ME, Duvick J, Brendel VP, Cannon E, Campbell DA, Lawrence CJ 2009. MaizeGDB becomes ‘sequence-centric.’ Database (Oxford) 2009: bap020 doi: 10.1093/database/bap020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shani E, Burko Y, Ben-Yaakov L, Berger Y, Amsellem Z, Goldshmidt A, Sharon E, Ori N 2009. Stage-specific regulation of Solanum lycopersicum leaf maturation by class 1 KNOTTED1-LIKE HOMEOBOX proteins. Plant Cell 21: 3078–3092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slattery M, Ma L, Negre N, White KP, Mann RS 2011. Genome-wide tissue-specific occupancy of the Hox protein Ultrabithorax and Hox cofactor Homothorax in Drosophila. PLoS ONE 6: e14686 doi: 10.1371/journal.pone.0014686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith LG, Greene B, Veit B, Hake S 1992. A dominant mutation in the maize homeobox gene, Knotted-1, causes its ectopic expression in leaf cells with altered fates. Development 116: 21–30 [DOI] [PubMed] [Google Scholar]
- Spinelli SV, Martin AP, Viola IL, Gonzalez DH, Palatnik JF 2011. A mechanistic link between STM and CUC1 during Arabidopsis development. Plant Physiol 156: 1894–1904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studer A, Zhao Q, Ross-Ibarra J, Doebley J 2011. Identification of a functional transposon insertion in the maize domestication gene tb1. Nat Genet 43: 1160–1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimm O, Blasing O, Gibon Y, Nagel A, Meyer S, Kruger P, Selbig J, Muller LA, Rhee SY, Stitt M 2004. MAPMAN: A user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J 37: 914–939 [DOI] [PubMed] [Google Scholar]
- Thompson BE, Hake S 2009. Translational biology: From Arabidopsis flowers to grass inflorescence architecture. Plant Physiol 149: 38–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda K, Ito Y, Sato Y, Kurata N 2011. Positive autoregulation of a KNOX gene is essential for shoot apical meristem maintenance in rice. Plant Cell 23: 4368–4381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernoux T, Brunoud G, Farcot E, Morin V, Van den Daele H, Legrand J, Oliva M, Das P, Larrieu A, Wells D, et al. 2011. The auxin signalling network translates dynamic input into robust patterning at the shoot apex. Mol Syst Biol 7: 508 doi: 10.1038/msb.2011.39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollbrecht E, Veit B, Sinha N, Hake S 1991. The developmental gene Knotted-1 is a member of a maize homeobox gene family. Nature 350: 241–243 [DOI] [PubMed] [Google Scholar]
- Vollbrecht E, Reiser L, Hake S 2000. Shoot meristem size is dependent on inbred background and presence of the maize homeobox gene, knotted1. Development 127: 3161–3172 [DOI] [PubMed] [Google Scholar]
- Yanai O, Shani E, Dolezal K, Tarkowski P, Sablowski R, Sandberg G, Samach A, Ori N 2005. Arabidopsis KNOXI proteins activate cytokinin biosynthesis. Curr Biol 15: 1566–1571 [DOI] [PubMed] [Google Scholar]
- Yilmaz A, Nishiyama Jr. MY, Fuentes BG, Souza GM, Janies D, Gray J, Grotewold E 2009. GRASSIUS: A platform for comparative regulatory genomics across the grasses. Plant Physiol 149: 171–180 [DOI] [PMC free article] [PubMed] [Google Scholar]