Cortical neurons have complex, highly branched dendritic trees that are essential for their function. However, the signaling pathways and mechanisms that regulate dendritic branching in vivo are poorly understood. In this study by Birchmeier and colleagues, the authors demonstrate that inactivation of the RhoGTPase Cdc42 is necessary for the branching of dendritic processes. The authors find that Cdc42 inactivation is regulated by NOMA-GAP, a novel Cdc42-specific GTPase-activating protein whose expression is switched on only in post-mitotic neurons during cortical development. Thus, this study delineates novel mechanisms regulating dendritic branching and neuronal complexity in the cerebral cortex.
Keywords: Cdc42, NOMA-GAP, cofilin, dendritic branching, dendritic complexity, neocortical development, neuronal differentiation
Abstract
Neocortical neurons have highly branched dendritic trees that are essential for their function. Indeed, defects in dendritic arborization are associated with human neurodevelopmental disorders. The molecular mechanisms regulating dendritic arbor complexity, however, are still poorly understood. Here, we uncover the molecular basis for the regulation of dendritic branching during cortical development. We show that during development, dendritic branching requires post-mitotic suppression of the RhoGTPase Cdc42. By generating genetically modified mice, we demonstrate that this is catalyzed in vivo by the novel Cdc42-GAP NOMA-GAP. Loss of NOMA-GAP leads to decreased neocortical volume, associated specifically with profound oversimplification of cortical dendritic arborization and hyperactivation of Cdc42. Remarkably, dendritic complexity and cortical thickness can be partially restored by genetic reduction of post-mitotic Cdc42 levels. Furthermore, we identify the actin regulator cofilin as a key regulator of dendritic complexity in vivo. Cofilin activation during late cortical development depends on NOMA-GAP expression and subsequent inhibition of Cdc42. Strikingly, in utero expression of active cofilin is sufficient to restore postnatal dendritic complexity in NOMA-GAP-deficient animals. Our findings define a novel cell-intrinsic mechanism to regulate dendritic branching and thus neuronal complexity in the cerebral cortex.
The neocortex is involved in processing sensory information and maintaining cognitive function. The dendritic arbor is the site of information input and processing of the neuron. The complexity and scope of the dendritic arbor are subtype-specific and determine the richness of neuronal interactions. During cortical development, the six cortical layers are set up in an inside-out fashion by the radial migration of neurons that are sequentially generated in the ventricular zone (VZ) and subventricular zone (SVZ) (for review, see Molyneaux et al. 2007). Differentiation of these neurons occurs through distinct space- and time-separable steps. After birth, in the VZ/SVZ, neurons undergo a multipolar stage characterized by the formation and short extension of multiple short processes. This is followed by polarization of the newly born neurons, axon determination, and migration out of the SVZ along radial glia processes. Axon extension starts during this migratory phase. Unknown mechanisms prevent dendritic branching during these early phases so that dendritic arborization is only initiated after the neuron has reached its correct position in the cortical plate. Finally, in postnatal stages, following dendritic arborization, synaptic contacts between neurons are established, and dendritic spines are formed along the branched dendritic tree.
Considerable attention has recently been paid to how progenitor function and neuronal fate specification are regulated during cortical development (for review, see Molyneaux et al. 2007). Several primarily in vitro studies have also started to address the mechanisms that regulate the initial determination and outgrowth of neurites as well as the subsequent axonal extension. The mechanisms that regulate dendritic branching in vivo, however, are not understood. Evidence from Drosophila screens suggests that specific transcription factors exist that specify dendritic branching independently of axon formation (Parrish et al. 2006). The targets of these transcription factors and their downstream signaling are not known. It is clear, however, that not only are axon extension and dendritic branching initiated at different stages of neuronal differentiation, but these structures also have distinct organization, subcellular composition, and regulatory mechanisms.
In humans, dendritic arborization starts prenatally, after neurons reach their destined position in the neocortex, and continues during early childhood (for review, see Jan and Jan 2010). Alterations in dendritic tree morphology have been reported in human neurological diseases, in particular those associated with mental retardation. Studies using Golgi staining of post-mortem material from children with mental retardation-associated syndromes report defects in dendritic branching, indicating a need to uncover the molecular mechanisms that regulate this process (Marin-Padilla 1972; Huttenlocher 1974; Takashima et al. 1989; Bauman et al. 1995; Armstrong et al. 1998; Schule et al. 2008; Judas et al. 2009).
Cdc42 is a RhoGTPase whose activity is required during early phases of neuronal differentiation to promote progenitor cell polarity and the number and initial lengthening of neurites sprouting from newly born neurons (Threadgill et al. 1997; Irie and Yamaguchi 2002; Scott et al. 2003; Schwamborn and Puschel 2004; Cappello et al. 2006). It has also been suggested to have positive roles during axonal pathfinding and in the formation of spines (Irie and Yamaguchi 2002; Scott et al. 2003). Its role, if any, during dendritic branching is not known. Like other RhoGTPases, Cdc42 activity is tightly regulated by multiple guanine nucleotide exchange factors (GEFs), guanine nucleotide dissociation inhibitors (GDIs), and GTPase-activating proteins (GAPs) that promote hydrolysis of GTP and inactivation of the protein. These regulators usually show activity against multiple RhoGTPase family members. However, restricted tissue expression and context-specific recruitment of GAPs in particular could enable fine regulation of particular Rho family members.
Recently, we described a novel neuronal GAP, NOMA-GAP, that shows specific activity toward the Rho family member Cdc42 (Rosario et al. 2007). By generating transgenic animals and in utero electroporations, we now show that inhibition of Cdc42 signaling is critical to allow dendritic branching of upper-layer neurons during cortical development. This is achieved through post-mitotic expression of NOMA-GAP. Our data further show that suppression of Cdc42 signaling by NOMA-GAP in vivo enables activation of the actin-binding protein cofilin during late cortical development. Finally, using in utero electroporation, we demonstrate that activation of cofilin by NOMA-GAP is a key step for induction of dendritic branching. Our data thus provide a novel molecular signaling cascade responsible for the intrinsic regulation of dendritic branching during cortical development.
Results
NOMA-GAP regulates neocortical thickness
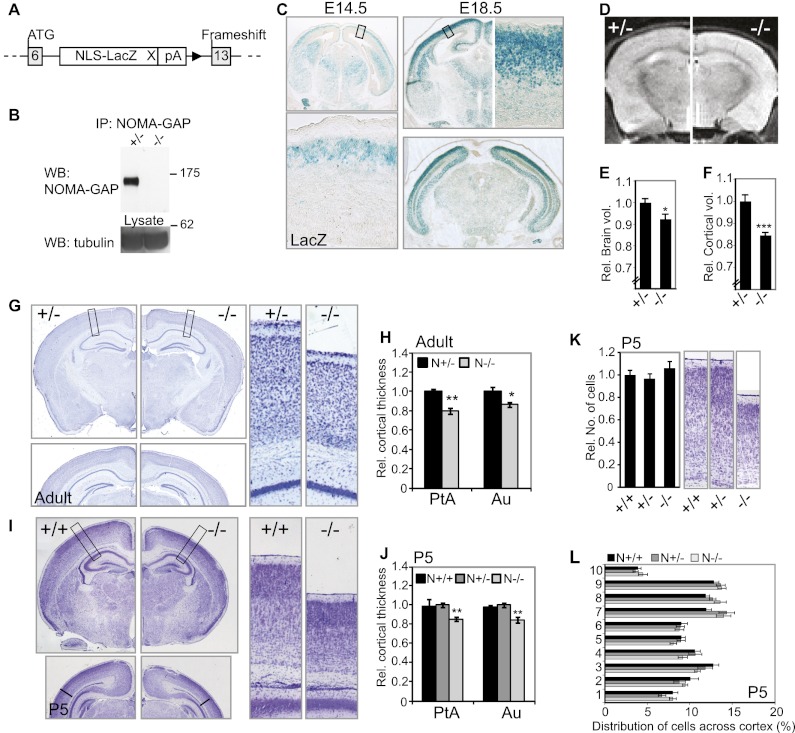
Previously, we identified a neuronal Cdc42-specific GAP, NOMA-GAP, that is strongly expressed in the adult murine neocortex (Rosario et al. 2007). To analyze the role of NOMA-GAP during cortical development, we generated NOMA-GAP-deficient mice by replacing exons 7–12 of Arhgap33, the gene encoding NOMA-GAP, with a nuclear-localized β-galactosidase (β-gal) gene, followed by insertion of stop codons and resulting in a frameshift (Fig. 1A; Supplemental Fig. 1A). One isoform of NOMA-GAP has been predicted to exist in mice, but two alternatively spliced isoforms, which use alternative start codons in exons 0 and 6, respectively, have been predicted in primates. Exons 7–12 code for the SH3 domain and the N-terminal half of the RhoGAP domain and are present in both isoforms. Replacement of exons 7–12 with the LacZ cassette resulted in the loss of NOMA-GAP expression, as shown by immunoprecipitation from whole-brain lysates using an antibody directed against the C terminus of NOMA-GAP (Fig. 1B). Analysis of cortical lysates from NOMA-GAP-deficient (referred to as N−/−) animals by Western blotting and immunoprecipitation using an antibody directed against the N terminus (exon 2) also failed to detect truncated forms of NOMA-GAP, suggesting that these mice are null (Supplemental Fig. 1E–G). However, we cannot rule out possible expression of a 31-amino-acid peptide coded by exon 6. Specific expression of the reporter gene under the control of the NOMA-GAP promoter was confirmed by LacZ assays in adult mouse brains and embryonic day 13.5 (E13.5) embryos (Supplemental Fig. 1B,C). In accordance with our previous in situ hybridization results (Rosario et al. 2007), NOMA-GAP promoter activity is restricted to the nervous system and, in the adult mouse brain, is present in all layers of the cerebral neocortex as well as in the olfactory bulb, the hippocampus, the striatum, the Purkinje cell layer of the cerebellum, and distinct cell populations throughout the brain (Supplemental Fig. 1B). We used β-gal activity to further characterize NOMA-GAP expression during cortical development (Fig. 1C). Expression of β-gal is first seen around E14.5 in the cortical plate and increases within the neocortex throughout late embryogenesis. No expression is seen in the VZ or SVZ. Expression of β-gal does not appear to be regionalized, although dorsal cortical areas show stronger reporter activity at E18.5 than lateral areas (Fig. 1C).
Figure 1.
Loss of NOMA-GAP leads to cortical thinning. (A) Schema of the targeted NOMA-GAP gene generated. Exons 7–12 of the murine gene encoding NOMA-GAP were replaced with a nuclear-localized β-gal gene. For details, refer to Supplemental Figure 1A. (B) Deletion of exons 7–12 results in loss of NOMA-GAP expression. NOMA-GAP was immunoprecipitated from heterozygote (+/−) and mutant (−/−) whole-brain extracts derived from P0 littermate animals and was detected using an antibody against the C terminus of NOMA-GAP. (C) NOMA-GAP is expressed in the cortical plate but is absent from the progenitor zone during cortical development. Analysis of LacZ reporter activity in coronal sections of E14.5 and E18.5 N+/− embryos. (D) MRI of brains of living NOMA-GAP-deficient adult animals shows cortical thinning but no alteration in overall brain structure. (E) Quantification of the average total brain volume per animal derived from analysis of 500-μm-spaced MRI scans. n = 5 N−/− and 4 N+/−; P = 0.03. (F) Quantification of total cortical volume of adult mouse brains. P = 0.0008. (G,H) Adult N−/− mice have a thinner cortex. The thickness of different cortical regions was determined from 40-μm Nissl-stained coronal sections of adult mouse brains. Representative images of the Nissl-stained sections used are shown in G. Magnifications are shown in the right panels. The relative thickness of the parietal (PtA) and auditory (Au) cortex is shown in H. Student's t-tests were used to compare N+/− and N−/− animals. PPtA = 0.002; PAu = 0.02; n = 9. Additional measured regions are shown in Supplemental Figure 1I. (I,J) Newborn NOMA-GAP-deficient mice have thinner cortices. The relative thickness of different cortical regions was determined for P5 mice using matched Nissl-stained 10-μm coronal sections of littermate animals. n = 4 N−/−, 5 N+/−, and 2 N+/+; PPtA = 0.0026, PAu = 0.004. Representative images of the sections used for quantification are shown in I. The relative thickness of the parietal (PtA) and auditory (Au) cortex is shown in J. (K,L) Loss of NOMA-GAP does not affect the number or distribution of cells in the somatosensory cortex of newborn mice. (K) The relative number of cortical cells in the barrel field region of the primary somatosensory cortex of P5 animals was determined. n = 3 N+/+, 3 N+/−, and 4 N−/− animals; PANOVA = 0.5. Representative images of the sections used for counting are shown in the right panels. (L) Distribution of cells across the cortex at P5. The cortex was divided into 10 equal-sized bins, and the proportion of cells in each bin was determined. Layer II/III corresponds to bins 8–9. PANOVA = 0.4 ± 0.25 across measured bins.
NOMA-GAP-deficient mice are born at the expected Mendelian ratio (24.7% mutant mice from heterozygotic matings; n = 142) and are indistinguishable from their littermates at birth in terms of appearance and total body weight (Supplemental Fig. 1D). Magnetic resonance imaging (MRI) scans of living adult mice show that overall brain structure is conserved (Fig. 1D). Volumetric measurements, however, indicate a small (7.7%) reduction in total brain volume in NOMA-GAP-deficient mice (Fig. 1E). Total sagittal brain length is unaltered (data not shown). Analysis of the brain volume profile shows that reduction in brain volume is predominantly found in the forebrain region (Supplemental Fig. 1H). We therefore measured the cortical volume of these animals from MRI. Figure 1F shows that loss of NOMA-GAP expression results in a strong reduction (15.6%) in total cortical volume in adult mice. As cortical thinning was visible from the MRI analysis (Fig. 1D), we measured the thickness of various cortical regions in matched Nissl-stained paraffin sections of adult mouse brains (Fig. 1G,H; Supplemental Fig. 1I). Loss of NOMA-GAP leads to a reduction in cortical thickness in dorsal and dorsolateral cortical areas; namely, in the secondary motor (11%), somatosensory (7%–9%), parietal (11%–20%), retrosplenial granular (8%–14%), and visual and auditory (12%–14%) cortex.
Region-dependent cortical thinning (in the range of 2%–7%, with the parietal cortex usually being most affected) has been observed in multiple human neurological disorders, including childhood neurodevelopmental disorders such as autism and attention-deficient disorder, where it correlates with disease severity (Shaw et al. 2006; Wallace et al. 2010). Reduction in cortical thickness may be a consequence of developmental defects or degenerative shrinkage during adult life. We therefore measured neocortical thickness in 5-d-old mice (Fig. 1I,J). NOMA-GAP-deficient newborn mice show strongly reduced cortical thickness in the parietal (16%–18%), somtosensory (22%), and auditory (13%) regions, suggesting that cortical thinning is due to developmental processes (Fig. 1J; data not shown).
NOMA-GAP is not required for early stages of neuronal development
We analyzed whether reduced cortical volume in N−/− mice was due to a reduction in cell number. We found that although NOMA-GAP-deficient newborn mice have a visibly thinner neocortex, the cell number and distribution across the cortex is not altered (Fig. 1K,L). In addition, we did not detect changes in cell death in N−/− animals (Supplemental Fig. 2). BrdU pulse labeling experiments and staining for radial glia also failed to reveal any alterations in the radial migration of pyramidal neurons or in radial glia structure upon loss of NOMA-GAP (Supplemental Fig. 3A–C). Furthermore, loss of NOMA-GAP did not affect the specification or location of the different cortical layer neuronal subtypes, although compression of the layers was observed postnatally (Supplemental Fig. 3E,F). This suggests that early events in cortical development, including neuronal birth, fate determination, and migration, are not regulated by NOMA-GAP.
Axon extension is initiated during radial migration. Previously, using the PC12 pheochromocytoma cell line, we showed that NOMA-GAP promotes the lengthening of the longest neurite that is destined to become the axon (Rosario et al. 2007). We therefore used several methods, including injection of the lipophilic tracer DiI and staining for axonal projections, to map the major cortical connections (cortico–cortico, cortico–thalamic, and cortico–spinal tract) in NOMA-GAP-deficient mice (Supplemental Fig. 4). We could detect no defects in axon pathfinding nor any changes in cortico–cortico or cortico–thalamic projections and only a mild delay in projections along the cortico–spinal tract in NOMA-GAP-deficient animals. These data suggest that either NOMA-GAP does not play a significant role in cortical axonal outgrowth and pathfinding or, in vivo, other molecules can compensate for NOMA-GAP function in these events.
NOMA-GAP is required for dendritic branching in vivo
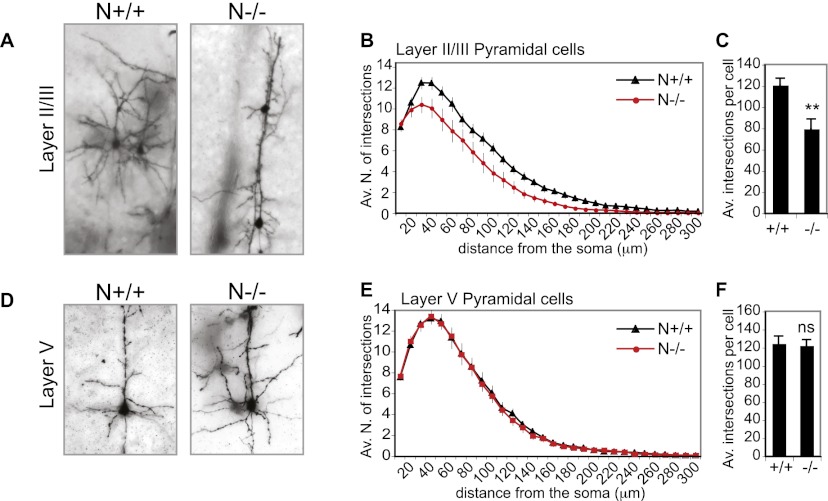
Cortical thinning may arise from defective formation of the dendritic tree. To assess whether NOMA-GAP regulates dendritic arborization in vivo, we analyzed Golgi-stained slices of adult mouse brains. Golgi staining showed that NOMA-GAP-deficient upper-layer neurons possess very simplified dendritic trees (Fig. 2A; Supplemental Movies 1,2). Sholl analysis (Sholl 1953) was performed on layer II/III or layer V neurons. This analysis quantifies the complexity of the dendritic arbor by determining the number of dendrites that cross (intersect) each of 30 10-μm-spaced concentric circles centered on the soma. Sholl analysis of layer II/III pyramidal neurons revealed a strong reduction in the dendritic complexity of N−/− neurons, as seen by the downward shift of the intersection curve and the reduction in the average total number of intersections per cell (Fig. 2B,C). No difference, however, was observed in the number of primary dendrites emerging from the soma (number of dendrites at 10 μm from the somal center) (Fig. 2B). Analysis of upper-layer somatosensory neurons randomly labeled by injection of Lucifer yellow dye confirmed these observations (data not shown). We also analyzed the dendritic complexity of deep layer V neurons to a distance of up to 300 μm from the soma, which excludes apical branching. Unlike upper-layer neurons, we did not observe any changes in the complexity of layer V neurons upon loss of NOMA-GAP (Fig. 2E,F).
Figure 2.
NOMA-GAP regulates dendritic complexity in vivo. (A–C) Loss of NOMA-GAP severely reduces the dendritic complexity of upper-layer cortical neurons. (A) Golgi staining of P25–P26 animals shows decreased dendritic arborization in N−/− upper-layer neurons. Representative average projections of Z-stack images of Golgi-stained upper-layer somatosensory cortex neurons are shown. The Z-stack images of these and other examples are available in the Supplemental Material (Supplemental Movies 1, 2). (B) Sholl analysis of Golgi-stained wild-type and NOMA-GAP-deficient layer II/III pyramidal neurons shows a reduction in dendritic complexity upon loss of NOMA-GAP. The average number of total intersections per cell are shown in C. n = 64 N+/+ and 36 N−/− cells from five and four independent animals, respectively; P = 0.004. (D–F) Sholl analysis of layer V neurons shows no significant difference in the complexity of NOMA-GAP mutant and wild-type neurons to a distance of 300 μm. Golgi-stained layer V neurons are shown in D. The Sholl profiles and average total intersections per cell for layer V neurons are shown in E and F, respectively. n = 37 N+/+ and 33 N−/− cells from four and three independent animals, respectively; P = 0.8.
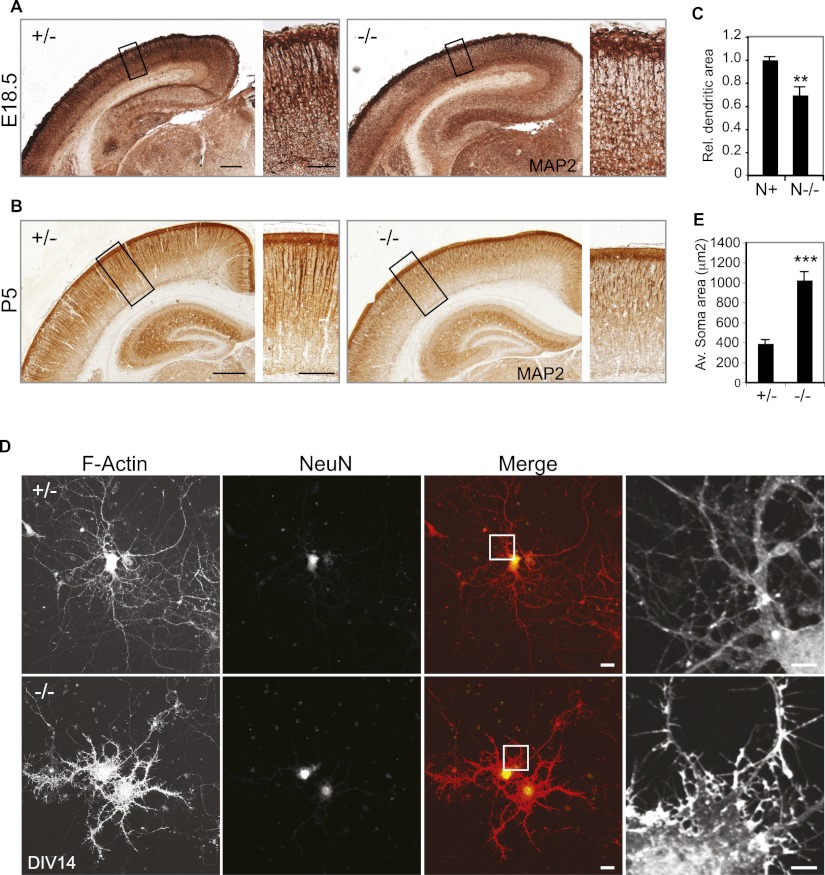
Dendritic extension and branching starts during late embryogenesis, after axogenesis and termination of migration, and continues in the postnatal period. We assessed the extent of dendritic development in the brains of late embryonic and newborn mice (E18.5 and postnatal day 5 [P5]) by staining for the dendritic protein MAP2 (Fig. 3A,B). At both developmental stages, we observed weaker cortical MAP2 staining in N−/− animals, particularly in the upper layers (Fig. 3A,B). Quantification of the relative area occupied by dendrites in the upper cortical layers of newborn animals indicates that loss of NOMA-GAP results in a strong reduction in dendritic processes (31%) (Fig. 3C).
Figure 3.
NOMA-GAP regulates dendritic development. (A,B) Loss of NOMA-GAP reduces cortical MAP2 staining. MAP2 staining (brown) in 10-μm coronal sections of littermate E18.5 (A) and P5 (B) brains is shown. Magnifications of the upper cortical layers are shown on the right. Bars: A, 200 μm, 50 μm B, 500 μm, 200 μm. (C) Quantification of the relative dendritic area in the upper layer of P4–P5 animals. n = 5 N+ and 4 N−/− animals; P = 0.0018. (D,E) Loss of NOMA-GAP induces morphological changes and loss of dendritic complexity in mature primary cortical neurons at DIV14. Staining for F-actin (red) and the neuronal marker NeuN (green) is shown for cells derived from littermate N+/− and N−/− E16.5 embryos. Bar, 20 μm. Magnification of the F-actin staining is shown on the right. Bar, 5 μm. (E) The somal area is significantly increased in DIV14 N−/− neurons. n = 21 N+/− and 31 N−/− cells; P = 1.7 × 10−8.
We next prepared primary cultures of cortical neurons from N−/− and control littermate E16.5 embryos and analyzed their morphology upon maturity (14 d in vitro [DIV14]) (Fig. 3D). Surprisingly, staining for F-actin revealed that loss of NOMA-GAP induces strong alterations in cellular morphology. N−/− primary cortical neurons showed a marked reduction in dendritic arborization but also extensive formation of actin-rich lamellipodia and filopodia. Neuronal identity of these cells was confirmed by staining for the neuronal marker NeuN (Fig. 3D). Quantification of soma size revealed a 2.6-fold increase in soma area, resulting from the lamellipodial formation (Fig. 3E).
NOMA-GAP is a major regulator of the Cdc42/PAK pathway in the developing neocortex
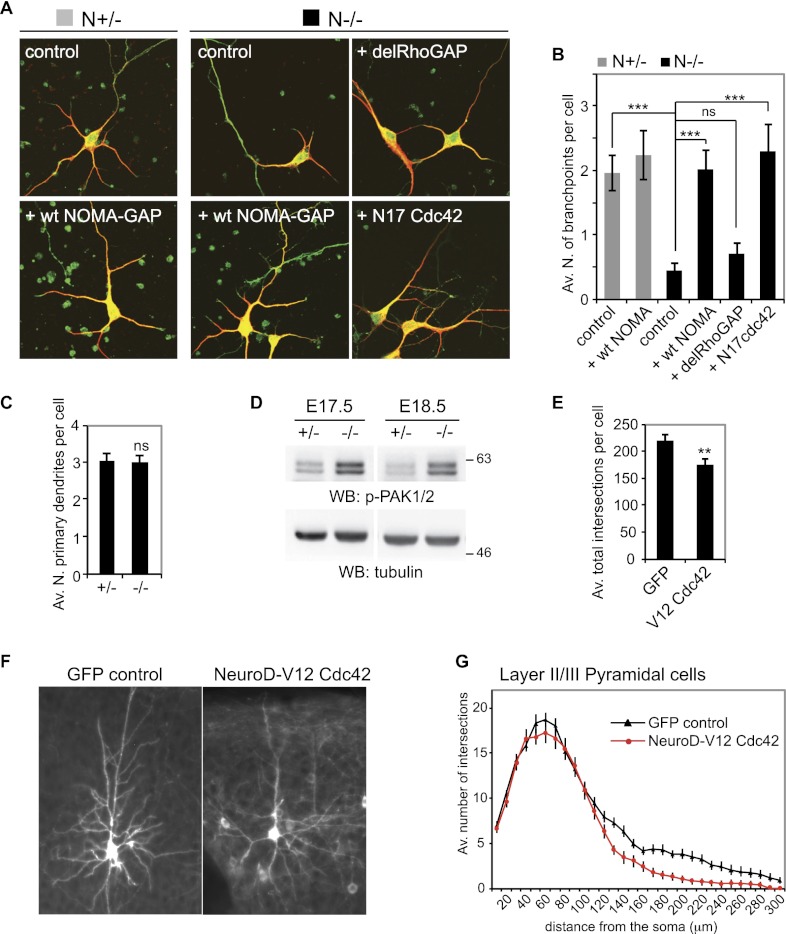
Next we analyzed the requirements for dendritic branching more closely by transfection of primary cortical cultures derived from NOMA-GAP-deficient and control littermate embryos. Cells were transfected with empty vector or the indicated constructs together with a GFP transfection marker, maintained for 5 d in culture, and stained for GFP and the dendritic protein MAP2. During this time, cortical neurons derived from heterozygotic and wild-type animals start dendritic branching but do not reach maturity or initiate synaptic contact (Fig. 4A; data not shown). N−/− neurons, on the other hand, show oversimplified dendritic trees with little/no branching (Fig. 4A, quantified in B). Somal flattening is not yet apparent at this stage. As was seen in vivo, the number of primary dendrites is not affected by loss of NOMA-GAP (Fig. 4C). Dendritic branching could, however, be rescued by re-expression of full-length NOMA-GAP in N−/− cells (Fig. 4A,B). Expression of NOMA-GAP in heterozygotic cells did not alter dendritic branching, suggesting haplosufficiency. Together with the in vivo analysis of N−/− neurons, these data show that cortical dendritic branching, but not initiation, is dependent on NOMA-GAP function.
Figure 4.
NOMA-GAP is a major negative regulator of Cdc42. (A,B) NOMA-GAP regulates dendritic branching through Cdc42. Cortical cells derived from N−/− or N+/− littermate embryos were transfected at DIV1 with control empty vector or the indicated expression constructs. Staining for MAP2 (red) and the GFP transfection marker (green) at DIV5 is shown in A. The average number of branchpoints per cell for a representative experiment is shown in B. n = 30, 30, 32, 32, 31, and 39 cells for the conditions from left to right; PANOVA = 5 × 10−8; PN+/− vs. N−/− = 4 × 10−6 ; PN−/− vs. N−/− wt = 1 × 10−6; PN−/− vs. N−/− del = 0.2; PN−/− vs. N−/− N17 = 2 × 10−5. (C) The number of primary dendrites per cell is not affected by loss of NOMA-GAP. Quantification was carried out using DIV5 cells derived from littermate animals. n = 29 N+/− and 31 N−/− cells; P = 0.99. (D) Loss of NOMA-GAP results in PAK1/2 kinase hyperactivation during late cortical development. Phosphorylated PAK proteins were detected by Western blotting of cortical lysates derived from littermate embryos at E17.5 and E18.5. (E–G) Hyperactivation of Cdc42 inhibits dendritic branching in vivo. V12 Cdc42 was expressed under the control of the NeuroD1 promoter in post-mitotic pyramidal cells by in utero electroporation of wild-type NMRI strain E15.5 embryos. Coelectroporation of a membrane-targeted GFP construct was used for visualization of dendritic processes. Analysis was carried out postnatally in cortical slices of P23 animals. Representative average Z-stack projection images of GFP fluorescence of electroporated upper-layer cortical neurons in cortical slices derived from P23 animals are shown in F. Sholl analysis was performed on these neurons. The average total number of intersections per cell are shown in E (P = 0.0048), and the Sholl profile is shown in G. Eighteen and 17 cells from three and two independent electroporations were analyzed for GFP control and NeuroD-V12 Cdc42-expressing neurons, respectively.
Rho family GTPases have been shown to regulate cell morphology through regulation of the actin cytoskeleton in various cell types (for review, see Newey et al. 2005). In particular, prolonged or constitutive activation of Cdc42 can induce the formation of filopodia and lamellipodia in different cell types (Nobes and Hall 1995; Price et al. 1998; Kurokawa et al. 2004). Previously, we showed that NOMA-GAP can function as a specific GAP for the Rho family member Cdc42 (Rosario et al. 2007). Since NOMA-GAP-deficient cortical neurons show extensive filopodia and lamellipodia formation upon long-term culture (Fig. 3D), we decided to address whether Cdc42-GAP function is necessary for NOMA-GAP-induced dendritic branching. We therefore generated a deletion mutant of NOMA-GAP lacking the RhoGAP domain (delRhoGAP) (Fig. 4A,B). Expression of the RhoGAP deletion mutant of NOMA-GAP failed to rescue dendritic branching in N−/− neurons, suggesting that GAP activity is essential for NOMA-GAP function.
Post-mitotic elevated Cdc42 activity inhibits dendritic branching in vivo
We hypothesized that impaired dendritic branching and thus complexity of N−/− neurons could result from loss of Cdc42-GAP activity and thus from aberrant activation of the Cdc42 signaling pathway in these cells. To test this, we used a dominant-negative mutant of Cdc42, N17 Cdc42, to inhibit Cdc42 signaling in N−/− primary neurons (Fig. 4A,B, last panel). Expression of N17 Cdc42 in N−/− neurons restored levels of dendritic branching, suggesting that the loss of dendritic complexity in N−/− neurons results from impaired regulation of Cdc42 signaling.
We then analyzed whether Cdc42 signaling is altered in the developing neocortex of NOMA-GAP-deficient mice by assessing the level of phosphorylation (and thus activation) of the Cdc42 effectors PAK serine/threonine kinases in embryonic cortical lysates (Fig. 4D). A robust hyperactivation of PAK1/2 was detected in the cortex of N−/− embryos, indicating that NOMA-GAP is a major regulator of the Cdc42/PAK pathway during cortical development.
Cdc42 activity is known to be required for early cortical development (Cappello et al. 2006), but the possible consequences of elevated Cdc42 signaling during cortical development have not been addressed. We asked whether hyperactivation of Cdc42 influences dendritic branching in vivo by expressing a constitutively active mutant of Cdc42 together with a membrane-targeted GFP marker in the developing cortex of wild-type mice by in utero electroporation at E15.5. V12 Cdc42 was expressed under the control of a post-mitotic neuronal promoter (NeuroD1) (de Anda et al. 2010). Brains were collected from postnatal animals (P23) and analyzed for GFP fluorescence. Sholl analysis of GFP-expressing layer II/III neurons at P23 revealed a significant reduction in dendritic branching and complexity (Fig. 4E–G), suggesting that elevated Cdc42 activity can suppress dendritic branching.
Inactivation of Cdc42 by NOMA-GAP is necessary for dendritic branching
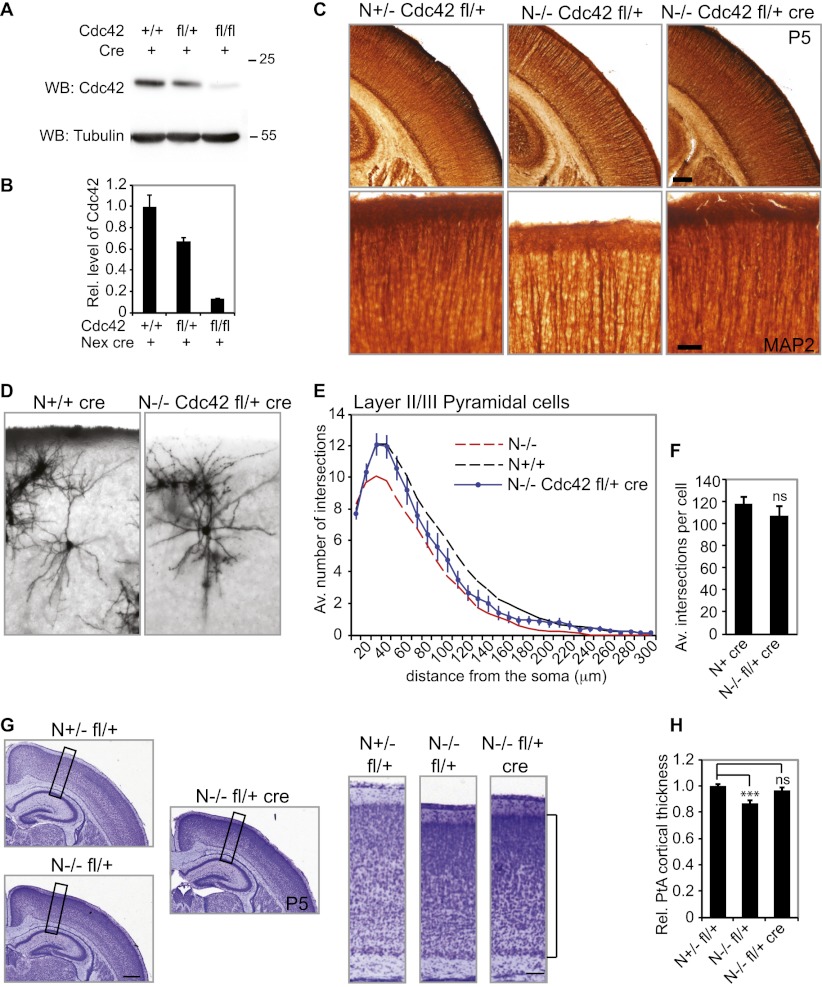
Our data suggest that aberrant activation of Cdc42 during embryonic development disturbs cytoskeletal dynamics and thus dendritic complexity in NOMA-GAP-deficient mice. To test this directly, we decreased Cdc42 expression in vivo by cre-mediated heterozygous deletion of one floxed Cdc42 allele (fl/+). Cdc42 is needed for the function of neuronal precursor cells (Cappello et al. 2006), which do not express NOMA-GAP. In order to delete Cdc42 only in post-mitotic neurons, we used the Nex1-cre recombinase mouse line. Cre recombinase is expressed specifically in post-mitotic pyramidal neurons of the developing neocortex in this mouse line and is absent from neural precursors, interneurons, oligodendrocytes, and astrocytes (Goebbels et al. 2006). Analysis of Cdc42 protein levels shows that expression of Nex1-driven cre recombinase reduces the levels of Cdc42 protein in the neocortex of E17 embryos (Fig. 5A,B). NOMA-GAP-deficient mice with only one copy of Cdc42 (N−/− fl/+ cre) were born at the expected Mendelian ratio and were indistinguishable from wild-type animals (data not shown). We analyzed dendritic development in these mice by MAP2 staining of P5 littermate animals (Fig. 5C). As previously seen, MAP2 staining was strongly reduced in the upper cortical layers of NOMA-GAP-deficient animals (N−/− Cdc42 fl/+). Apical dendrites originating in deeper layers appear unaltered. Post-mitotic heterozygous deletion of one Cdc42 allele in NOMA-GAP-deficient mice (N−/− Cdc42 fl/+ cre mice), however, restored the intensity of MAP2 staining in the upper cortical layers, suggesting that dendritic development was restored in these animals (Fig. 5C). To confirm this, the dendritic complexity of mature upper-layer neurons was analyzed using Golgi-stained sections of P25–P26 brains (Fig. 5D–F; Supplemental Movies 1–4). Expression of cre alone did not affect dendritic complexity (Supplemental Fig. 5). Post-mitotic deletion of one Cdc42 allele, however, partially restored dendritic branching and arbor complexity in NOMA-GAP-deficient mice, as seen through the upward shift of the Sholl profile of N−/− Cdc42 fl/+ cre neurons as well as the normalization of the average total number of intersections per cell (Fig. 5E,F, respectively). Post-mitotic excision of the Cdc42 allele did not affect the number of primary dendrites, suggesting that this event is determined very early in cortical development before Nex-induced cre expression (Fig. 5E, 10-μm distance). Furthermore, we determined the cortical plate thickness of the lateral parietal cortex of P4–P6 N−/− Cdc42 fl/+ cre animals relative to the littermate N+/− fl/+ control animals (Fig. 5G,H). Heterozygotic deletion of one Cdc42 allele significantly improved parietal cortical thickness in NOMA-GAP-deficient animals (Fig. 5H). These data are consistent with our observations using primary cell cultures and indicate that post-mitotic inhibition of Cdc42, mediated by NOMA-GAP, is a necessary requirement for dendritic branching and thus cortical development.
Figure 5.
NOMA-GAP regulates dendritic branching through inhibition of Cdc42 signaling. (A) Cdc42 expression can be decreased in vivo by expression of cre recombinase under the control of the Nex promoter. Cortical lysates derived from E17 embryos were Western-blotted for Cdc42 and tubulin. (B) Quantification of the reduction in Cdc42 protein expression in the cortex of E17–E18 embryos with floxed (fl) Cdc42 gene and Nex-regulated cre recombinase expression. n = 2, 3, and 1 animals; PANOVA = 0.01. (C) Post-mitotic Nex-cre-mediated excision of one Cdc42 allele rescues dendritic MAP2 staining in N−/− animals. Thirty-micrometer vibratome brain sections of littermate P5 animals were stained for MAP2 (brown). Magnifications of the upper cortical layers are shown in the bottom panels. (D–F) Dendritic complexity in N−/− mice can be rescued by post-mitotic excision of one Cdc42 allele. (D) Representative average projections of Z-stack images of Golgi-stained upper-layer pyramidal cortical neurons derived from P25–P26 animals are shown. Additional Z-stack images are available in the Supplemental Material (Supplemental Movies 3, 4). (E) Sholl profile of Golgi-stained P25–P26 N−/− Cdc42 fl/+ cre upper-layer pyramidal neurons indicates strong improvement in dendritic complexity following reduction of Cdc42 levels. n = 18 N+ cre and 26 N−/− fl/+ cre cells. Sholl profiles for N−/− and N+/+ have been described in Figure 2. The average total number of intersections per cell is shown in F. P = 0.4. (G,H) Post-mitotic excision of one Cdc42 allele improves the thickness of the cortical plate in newborn N−/− animals. (G) Nissl-stained sections of P5 littermate animals are shown. The boxed region is shown on the right. The cortical plate is marked by a bracket. Bars, 500 μm and 100 μm, respectively. (H) Relative thickness of the lateral parietal cortex of N−/− Cdc42 fl/+ cre P4–P6 animals. Measurements were made relative to littermate N+/− fl/+ animals. n = 11 N+/− fl/+, 10 N−/− fl/+, and 12 N+/− fl/+ cre animals; PANOVA = 4.9 × 10−4; PN+/− fl/+ vs. N−/− fl/+ = 5.8 × 10−5 ; PN+/− fl/+ vs. N−/− fl/+ cre = 0.27.
Cofilin promotes cortical dendritic complexity
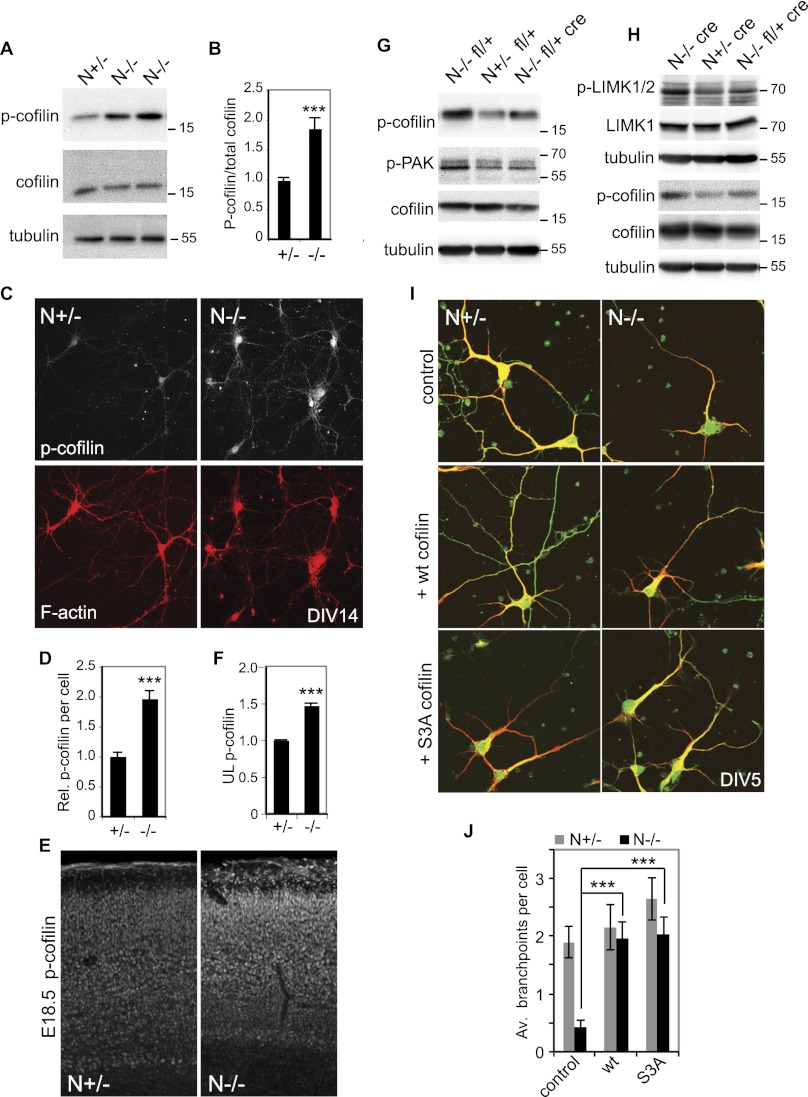
The molecular mechanism by which dendritic complexity is regulated in vivo was further analyzed. Changes in the activation of signaling proteins are often accompanied by changes in phosphorylation state. Phosphorylated proteins were immunoprecipitated from embryonic N−/− and wild-type littermate neocortices and identified by mass spectrophotometry (Supplemental Material). One protein that was more abundant in the phospho-immunoprecipitations of N−/− embryos was the actin polymerization regulator cofilin. We were unable to identify the site phosphorylated in cofilin by mass spectrophotometry. However, Ser 3 has been reported to be the major regulatory phosphorylation site in cofilin (Moriyama et al. 1996). We therefore analyzed whether cofilin Ser 3 phosphorylation was altered in the N−/− animals by Western blotting of E17 cortical lysates (Fig. 6A,B). Levels of specific Ser3 cofilin phosphorylation were found to be elevated 1.9-fold in lysates derived from the neocortex of N−/− animals (Fig. 6B). Cofilin phosphorylation was analyzed at a cellular level by staining mature (DIV14) cultured primary cortical cells derived from N−/− and N+/− littermate embryos (Fig. 6C,D). Cofilin has been shown to translocate between the cytoplasm and nucleus (Ohta et al. 1989). Phospho-Ser3 cofilin staining was observed in the nucleus of both N+/− and N−/− cells and also in the surrounding cytoplasm (Fig. 6C). Consistent with our observations using cortical lysates, quantification of somal phospho-cofilin levels revealed an almost twofold up-regulation of cofilin phosphorylation in N−/− cells (Fig. 6D). Finally, the distribution of cofilin phosphorylation in the developing neocortex was investigated (Fig. 6E). As previously reported for wild-type animals (Chai et al. 2009), control N+/− E18.5 embryos show only weak phospho-cofilin staining in the cortical plate and stronger staining in the marginal zone. NOMA-GAP-deficient embryos, however, showed strong up-regulation of cofilin phosphorylation in the cortical plate (Fig. 6E). Cofilin phosphorylation was especially marked in the upper layers of N−/− animals (1.5-fold increase) (Fig. 6F). Phosphorylation of cofilin at Ser 3 negatively regulates cofilin function (Moriyama et al. 1996). These data thus suggest that NOMA-GAP is a major stimulator of cofilin activity in the developing neocortex.
Figure 6.
Cofilin is regulated by NOMA-GAP during cortical development. (A,B) Loss of NOMA-GAP leads to an increase in cortical cofilin phosphorylation at Ser 3. (A) Total and phospho-Ser 3 cofilin levels were detected by Western blot of E17.5 cortical lysates derived from littermate embryos. (B) Quantification of specific phospho-Ser 3 cofilin levels from Western blot analysis. Phospho-cofilin/total cofilin values were calculated relative to the average value for N+/− littermate controls. The change in specific cofilin phosphorylation was then compared across different litters. n = 14 +/− and 16 −/− animals; P = 1.4 × 10−4. (C,D) Elevation of phospho-Ser 3 cofilin levels in N−/− cortical cells. (C) Cortical cells derived from littermate E16.5 embryos were cultured for 14 d and stained in parallel for phospho-cofilin (white) and F-actin (red). (D) Quantification of the average relative levels of phospho-Ser cofilin in the soma of N+/− and N−/− DIV14 cortical cells. n = 35 +/− and 31 −/− cells. P = 6.6 × 10−9. (E,F) Phospho-Ser 3 cofilin levels are increased in the neocortex of N−/− mice. (E) Fifty-micrometer vibratome coronal brain sections of littermate E18.5 embryos were stained in parallel for phospho-Ser 3 cofilin. (F) Quantification of phospho-Ser 3 cofilin in the upper cortical layer of E18.5 animals. The relative mean staining intensity in the upper cortical layers of three pairs of littermate heterozygote and mutant embryos is shown. P = 7.7 × 10−5. (G,H) Cortical cofilin, PAK, and LIMK phosphorylation are regulated by Cdc42 downstream from NOMA-GAP. Representative Western blots of cortical lysates derived from littermate E17.5 embryos were blotted for phospho-Ser 3 cofilin (p-cofilin), phospho-PAK (p-PAK), and/or phospho-LIMK (p-LIMK1/2) and LIMK1 as indicated. Detection of phospho and total forms of cofilin and LIMK were carried out on duplicate Western blots. The tubulin loading controls shown were carried out on the membranes with anti-phospho antibodies. (I,J) Expression of cofilin promotes dendritic branching in N−/− cortical cells. (I) E16.5 primary cortical cells were transfected 1 d after plating with empty vector (control), wild-type cofilin, or S3A cofilin-expressing constructs together with a myr-GFP-expressing construct. Samples were stained at DIV5 for MAP2 (red) and GFP (green). (J) Quantification of the number of branchpoints per cell. n = 30, 32, 32, 38, 34, and 35 cells for the conditions from left to right. PANOVA = 4.2 × 10−5; PN−/− control vs. wt cofilin = 7.9 × 10−5; PN−/− control vs. SA = 7.7 × 10−6; PN+/− control vs. wild-type cofilin = 0.6.
To address whether cofilin inactivation in the neocortex of NOMA-GAP mice results from aberrant Cdc42 activation, we asked whether cortical cofilin activation could be restored by decreasing post-mitotic Cdc42 levels in vivo through post-mitotic cre-mediated excision of one Cdc42 allele (Fig. 6G,H). As previously seen, cofilin phosphorylation is elevated in the cortex of N−/− littermate animals either containing one floxed Cdc42 allele or expressing cre under the Nex promoter (Fig. 6G,H, respectively). In vivo cre-mediated heterozygotic excision of one Cdc42 allele, however, restored cofilin phosphorylation levels in N−/− animals (Fig. 6G,H). These data indicate that during cortical development, cofilin activation is positively regulated by NOMA-GAP through inhibition of Cdc42.
Cofilin phosphorylation at Ser 3 can be catalyzed by the kinases LIMK1/2 (Arber et al. 1998; Yang et al. 1998; Edwards et al. 1999). Furthermore, in epithelial and fibroblast cell lines, the Cdc42 effectors PAK kinases have been shown to be able to activate LIMK (Edwards et al. 1999; Dan et al. 2001). We therefore tested whether, during cortical development, cofilin activation downstream from NOMA-GAP was regulated through Cdc42/PAK signaling. The levels of PAK and LIMK phosphorylation, a measure of their activity, were analyzed by Western blotting of cortical lysates derived from N−/− Cdc42 fl/+ cre and control animals (Fig. 6G,H). Phosphorylation levels of both PAK and LIMK were increased in the neocortex of N−/− animals. Heterozygotic deletion of Cdc42 in N−/− mice restored the activity level of these kinases, suggesting that during cortical development, PAK and LIMK activities can be regulated by Cdc42 downstream from NOMA-GAP.
The role of cofilin during dendritic branching has not been previously addressed. To analyze whether loss of dendritic complexity in N−/− neurons was caused by the inappropriate inhibition of cofilin in these cells, we expressed wild-type or a phosphorylation-resistant mutant of cofilin (S3A cofilin) in primary cortical neuronal cultures. The number of dendritic branchpoints per cell was quantified at DIV5 (Fig. 6I,J). Expression of cofilin in heterozygotic cortical neurons had no significant effect on the level of dendritic branching. Expression of either wild-type or S3A cofilin in N−/− cortical neurons, however, restored the number of dendritic branchpoints in these cells (Fig. 6I,J).
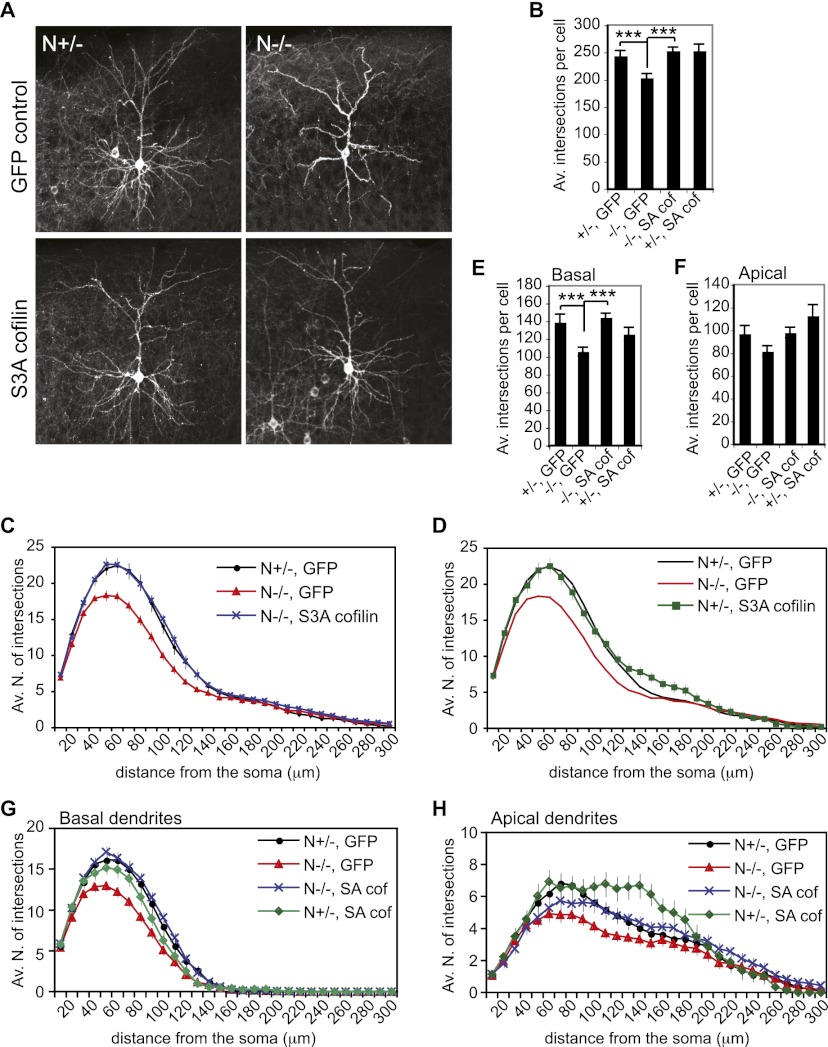
Finally, we investigated whether dendritic complexity is regulated by cofilin in vivo (Fig. 7; Supplemental Movies 5–8). We expressed S3A cofilin in the cortex of N−/− and littermate N+/− embryos by in utero electroporation. Coelectroporation of an expression construct for plasma membrane-targeted GFP was used as a control and to visualize expressing cells. Embryos were electroporated at E15.5 to label the late-born upper-layer neurons in the somatosensory cortex, and brains were collected postnatally from P23 animals. As seen with Lucifer yellow- and Golgi-stained samples (Fig. 2), loss of NOMA-GAP significantly reduces the branching of dendrites and results in a strong reduction in arbor complexity (Fig. 7A–C). Separate analysis of apical and basal dendrites indicates that the complexity of basal dendrites, which contributes 57% of total dendritic complexity, is particularly strongly affected by loss of NOMA-GAP (Fig. 7E–H). Expression of S3A cofilin in control N+/− neurons does not significantly affect overall dendritic complexity, although premature branching of the apical dendrite was observed in these cells (Fig. 7D,H). Remarkably, in vivo expression of S3A cofilin in N−/− neurons strongly promoted dendritic branching and was sufficient to restore overall dendritic complexity (Fig. 7A–C; Supplemental Movies 5–8). Rescue of dendritic complexity in N−/− cells was particularly strong in the basal dendritic compartment (Fig. 7E,G). These data show that cofilin activity is required in vivo for dendritic branching of upper-layer cortical neurons. Furthermore, the data indicate that NOMA-GAP is a critical regulator of cofilin activity and therefore neuronal arbor complexity during neocortical development.
Figure 7.
Cofilin promotes dendritic branching in vivo. In vivo expression of S3A cofilin rescues dendritic complexity in N−/− upper-layer cortical neurons. E15.5 embryos were electroporated in utero with a myr-GFP-expressing construct alone or together with a construct for S3A cofilin. GFP fluorescence was analyzed postnatally in sections of P23 brains. Representative average projection images of GFP fluorescence in electroporated cortical neurons are shown in A. Z-stack movies are available in the Supplemental Material (Supplemental Movies 5–8). Sholl analysis was performed on GFP-positive neurons. The average number of total intersections per cell is shown in B. PANOVA = 1.0 × 10−5 ; P+/− GFP vs. −/− GFP = 7.0 × 10−4; P−/− GFP vs. −/− SA = 3.5 × 10−6; P+/− GFP vs. +/− SA = 0.5. Sholl profiles for N−/− and N+/− animals electroporated with S3A cofilin are shown in C and D, respectively, together with GFP control electroporated animals. (E–H) The complexity of basal and apical dendrites was independently analyzed. The average total number of basal or apical intersections per cell is shown in E and F, respectively. (E) PANOVA=5.4 × 10−5; P+/− GFP vs. −/− GFP = 0.0009; P+/− GFP vs. −/− SA = 0.59; P−/− GFP vs. −/− SA = 4.7 × 10−7; P+/− GFP vs. +/− SA = 0.29. (F) PANOVA=9.0 × 10−3; P+/− GFP vs. −/− GFP = 0.06; P+/−GFP vs. −/− SA = 0.93; P−/− GFP vs. −/− SA = 0.02; P+/− GFP vs. +/− SA = 0.18. The Sholl profiles for basal and apical dendrites are shown in G and H, respectively. Four N−/− GFP, three N+/− GFP, three N−/− S3A cofilin, and two N+/− S3A cofilin animals were used for analysis. Ten cells were analyzed per animal.
Discussion
Dendritic arborization is a critical step in late neuronal differentiation and a direct prerequisite for complex neuronal communication. Dendrites are a characteristic of neurons and differ from other cellular processes such as axons in their subcellular composition, morphology, and function. Pyramidal neurons, the dominant cell type in the mammalian cerebral cortex, generate complex branched dendritic trees that form several thousand chemical synapses per cell. The formation of the branched dendritic arbor is one of the final steps in cortical development, occurring only after most surrounding neurons have been generated, have migrated to their final positions, and have extended axons. Little is known about how dendritic arborization is prevented during early developmental stages or how this temporal inhibition is lifted and branching is promoted once the neuron has reached the designated position in the neocortex.
We show for the first time that suppression of Cdc42 signaling is required to promote the branching of the dendritic tree in vivo and normal cortical thickness. During cortical development, this is mediated by post-mitotic expression of the recently identified Cdc42-GAP NOMA-GAP. NOMA-GAP expression thus acts as a molecular switch to initiate the generation of complex neuronal connectivity. Furthermore, we demonstrate that activation of the actin polymerization regulator cofilin is a key step in promoting dendritic branching and thus arbor complexity in vivo. Finally, we show that NOMA-GAP enables cofilin activation during late cortical development by suppression of Cdc42 activity.
Inactivation of Cdc42 by NOMA-GAP permits dendritic branching
We show that suppression of Cdc42 activity is an essential step for enabling dendritic branching and thus the complexity of cortical neurons in vivo. This unexpected result contrasts with the positive roles that have been attributed to Cdc42 during early cortical development (Threadgill et al. 1997; Cappello et al. 2006). Thus, while Cdc42 activity has been shown by various groups to be needed during early neuronal differentiation to promote the number and outgrowth of neurites in newly born neurons (Threadgill et al. 1997; Ruchhoeft et al. 1999; Rosario et al. 2007), we now show that Cdc42 inhibits later stages of neuronal differentiation; namely, the branching of the primary dendrites. Genetic inactivation of the Cdc42-GAP NOMA-GAP results in a reduction in cortical thickness of up to 22%, with no change in cell number or death. Cortical thinning is apparent at the end of the first postnatal week (Fig. 1) and is associated with profound impaired branching of the dendritic tree and hyperactivation of Cdc42 signaling (Figs. 2, 4D). We show that elevation of Cdc42 activity in post-mitotic neurons during cortical development inhibits dendritic branching in wild-type animals (Fig. 4F). Significantly, dendritic branching and cortical thickness could be improved in NOMA-GAP-deficient animals in vivo by post-mitotic reduction of Cdc42 expression (Figs. 4, 5H). These data demonstrate that NOMA-GAP is a major negative regulator of Cdc42 in vivo and, more importantly, that this regulation is essential to allow dendritic branching and normal cortical development.
Our data indicate that Cdc42 activity must be suppressed for cortical dendritic branching. The role of Cdc42 in mammalian dendritic branching has not been previously addressed independently from dendrite initiation. However, work using Drosophila Cdc42 mutants demonstrated that dendritic branching complexity is not altered by loss of drosCdc42 (Scott et al. 2003). Overexpression of dominant-active and dominant-negative mutants of Cdc42 have also been used to look at dendritic complexity in Xenopus neurons with contradictory results (Ruchhoeft et al. 1999; Li et al. 2000), which may result from differences in neuronal subtype analyzed or from the targeting of progenitor cells in these experiments. Furthermore, in concurrence with our observations using in utero electroporation of NeuroD1-driven V12 Cdc42, post-mitotic overexpression of a constitutively active mutant of drosCdc42 in Drosophila peripheral nervous system (PNS) neurons was reported to reduce branching and induce thickening and shortening of the primary dendritic branch (Gao et al. 1999), alterations that are reminiscent of the morphological changes we observed in long-term cultures of primary N−/− cortical neurons (Fig. 3D). Cdc42 thus has functions distinct from those of the related RhoGTPase Rac, which can positively regulate dendritic branching (Lee et al. 2003; Rosso et al. 2005). Indeed, these proteins elicit distinct morphological alterations in the large variety of cell types analyzed so far (for review, see Etienne-Manneville and Hall 2002).
Post-mitotic onset of NOMA-GAP expression signifies that the important early functions of Cdc42, such as the regulation of progenitor function and induction of neurite outgrowth, are not affected. Indeed, loss of NOMA-GAP has little effect on cell number, neuronal fate determination, or migration (Fig. 1; Supplemental Fig. 3). In addition, loss of NOMA-GAP has no effect on the number of primary neurites or dendrites in embryonic cultured cortical neurons or mature cortical neurons, respectively, in vivo, indicating that NOMA-GAP is not needed for these early differentiation steps in the cerebral cortex (Figs. 2, 4C). Moreover, while in vivo loss of NOMA-GAP results in a profound decrease in dendritic complexity, it has little effect on axon formation or connectivity (Supplemental Fig. 4). Indeed, as opposed to dendritic branching, Cdc42 activity is necessary for axon formation (for review, see Hall and Lalli 2010). These opposing roles of Cdc42 imply the requirement for a differential stage or location-specific regulation of this GTPase, such as is provided by NOMA-GAP during dendritic branching.
The dendritic complexity of upper-layer II/III neurons is strongly compromised by the genetic ablation of NOMA-GAP. Layer V neurons, however, did not appear to be affected to a distance of up to 300 μm from the soma. This difference may reflect molecular and/or environmental differences between these two classes of neurons. The complexity of the basal dendrites of layer V neurons, for example, has been shown to be regulated by calcium signaling (Redmond et al. 2002). Furthermore, the effects of different neurotrophins on dendritic growth are reported to be cortical layer-specific, with NT3 opposing brain-derived neurotrophic factor (BDNF) action in layer IV neurons and the opposite occurring in layer VI neurons (McAllister et al. 1997). We previously showed that NOMA-GAP can directly interact with the receptor for another neurotrophin, NGF, and become phosphorylated upon ligand binding (Rosario et al. 2007). It is possible that NOMA-GAP activity and/or downstream signaling are differentially regulated by extracellular signals, such as the neurotrophin family members, in a layer-specific fashion. The preferential up-regulation of phospho-Ser 3 cofilin in the upper cortical layers of N−/− animals also suggests the existence of layer-specific regulation of intracellular signaling (Fig. 6E). Such differences may be necessary for the establishment of distinct neuron subtype-specific dendritic branching patterns.
Dendritic complexity is likely to be regulated by NOMA-GAP in a cell-autonomous fashion, since compromised dendritic branching in E16.5 N−/− primary cortical cell cultures can be rescued by re-expression of NOMA-GAP (Fig. 4A). Rescue of dendritic branching can also be achieved in vivo by expression of active cofilin in individual upper-layer neurons of NOMA-GAP-deficient animals (Fig. 7), further arguing for cell-autonomous mechanisms of regulation of dendritic branching. Synaptic activity can regulate the extent of dendritic branching (Coleman and Riesen 1968). The presence of dendritic defects in the NOMA-GAP-deficient cortex and in primary cells at stages preceding synapse formation suggests that regulation of dendritic branching by NOMA-GAP is synapse-independent. However, we cannot exclude that other forms of neuronal activity contribute to NOMA-GAP-mediated dendritic complexity.
Post-mitotic heterozygous deletion of Cdc42 results in a partial rescue of dendritic complexity in N−/− mice (Fig. 5E). Rescue appears to be more complete proximal to the soma. This may reflect either the inability of this genetic manipulation to accurately mimic the wild-type spatial distribution of Cdc42 activation or a difference in how branching is regulated in the apical and basal compartments. Proximal complexity derives mostly from the branching of basal dendrites. Expression of S3A cofilin also results in a more complete rescue of basal as compared with apical complexity in N−/− animals (Fig. 7), suggesting that there may be molecular differences in how branching is regulated in these two dendritic compartments. Interestingly, apical and basal dendritic compartments appear to be differentially affected in different human neurodevelopmental conditions associated with mental retardation (for review, see Kaufmann and Moser 2000).
Cofilin promotes dendritic branching
Cofilin is a regulator of actin polymerization (for review, see Bernstein and Bamburg 2010), whose activity has been shown to regulate maintenance of the cortical progenitor pool (Bellenchi et al. 2007). Inactivation of cofilin, on the other hand, is necessary for various post-mitotic events; namely, for Reelin-mediated termination of neuronal radial migration, for repulsive axon guidance, and for the maturation of dendritic spines (Aizawa et al. 2001; Meng et al. 2004; Bellenchi et al. 2007; Chai et al. 2009; Shi et al. 2009).
We show for the first time that cofilin has a role in dendritic development, where it promotes branching. Indeed, exogenous expression of active cofilin is sufficient to rescue defective dendritic branching in N−/− animals and in cultured primary cortical neurons (Figs. 6, 7). Furthermore, we show that cofilin activity is positively regulated by NOMA-GAP during late cortical development. Loss of NOMA-GAP in vivo results in the inactivation of cofilin through Ser 3 phosphorylation in the cortex (Fig. 6).
The relationship between Cdc42 and cofilin appears to depend on the cellular context. While we show that cofilin is inactivated by Cdc42 during late cortical development, previous studies looking earlier at axon formation found that Cdc42 activates cofilin, possibly indirectly through regulation of Rho signaling (Chen et al. 2006; Garvalov et al. 2007). Overexpression studies in epithelial and fibroblast cell lines as well as in vitro assays, however, have indicated that Cdc42 can also mediate inhibition of cofilin through activation of LIMK by kinases such as PAK (Edwards et al. 1999; Sumi et al. 1999, 2001). We show that during late cortical development, Cdc42 predominantly acts to inhibit cofilin, most likely through activation of PAK and LIMK. Indeed cofilin, PAK and LIMK activities in the neocortex of N−/− embryos can be restored in vivo by genetic heterozygotic ablation of Cdc42 (Fig. 6G,H). Altogether, this suggests that Cdc42 and cofilin have flexible altering roles and relationships that enable the specific morphological alterations occurring at the different stages of neuronal differentiation.
In summary, our data provide a new insight into how the branching of primary dendrites can initially be prevented and subsequently promoted during cortical development. While activation of Cdc42 has been clearly shown to be required during early stages of cortical development, continued activation of Cdc42 in post-mitotic cells in the cortex interferes with dendritic arborization. Post-mitotic expression of the novel signaling molecule NOMA-GAP is a key event in this process, as it mediates the inhibition of Cdc42-initiated signaling. This initiates a cascade of signaling events that leads to the downstream activation of cofilin, a step that we show is key for the branching of primary dendrites and thus for arbor complexity in the mammalian neocortex. The molecular basis for the dendritic arbor defects commonly observed in many human neurodevelopmental disorders such as mental retardation and autism is mostly unknown. We believe that identification of this novel regulatory signaling cascade will aid understanding of the development of such conditions.
Materials and methods
Reagents
The following antibodies were used in this study: anti-cofilin, phospho-Ser 3 cofilin, phospho-Pak, LIMK1, and phospho-LIMK1/2 (Cell Signaling); anti-GFP and NOMA-GAP (Abcam); α-tubulin and MAP 2 (Sigma-Aldrich); anti-NeuN (Millipore); and anti-Cdc42 (BD Transduction Laboratories). In addition, the following cellular stains were used: phalloidin–tetramethylrhodamine B isothiocyanate, 4′,6-diamidino-2-phenylindole dihydrochloride, hematoxylin, and cresyl violet (Sigma-Aldrich).
V12 Cdc42 was subcloned into the multiple cloning site of NeuroD1 promoter-driven IRESmCherry pA vector (kindly provided by Goichi Miyoshi and Gordon Fishell, New York University Langone Medical Center). SA and wild-type cofilin expression constructs were kindly provided by Jun-Lin Guan (University of Michigan) and have been described previously (Yoo et al. 2009). The membrane-targeted GFP expression construct used is a myristoylated Venus-GFP driven from a β-actin promoter. All other plasmids have been previously described (Rosario et al. 2007).
Mice
All mouse experiments were carried out according to German law and were approved by Landesamt für Gesundheit und Soziales Berlin or Niedersächsisches Landesamt für Verbraucherschutz und Lebensmittelsicherheit. NOMA-GAP-null mutant mice were generated by replacing exons 7–12 of the NOMA-GAP gene Arhgap33 with a nuclear-localized LacZ reporter gene using homologous recombination in embryonic stem cells (Supplemental Fig. 1A). Floxed Cdc42 and Nex1-cre recombinase mouse lines were obtained from C. Brakebusch (University of Copenhagen) and K. Nave (Max Planck Institut für Experimentelle Medizin), respectively.
Primary neuron culture
Cortical neuronal cells were isolated from E16.5 mice and grown in neurobasal medium (Gibco) supplemented with 2% B27 (Invitrogen) and 200 mM glutamine (Gibco). For transfection, 1.5 × 105 cells were plated on poly-L-lysine-coated coverslips and transfected with Lipofectamine (Invitrogen) 1 d after plating, as indicated by the manufacturer.
MRI
MRI was performed using a 7 Tesla rodent scanner (Pharmascan 70/16, Bruker BioSpin). For imaging, a 1H-RF quadratur volume resonator with an inner diameter of 20 mm was used. A T2-weighted two-dimensional turbo spin echo sequence was carried out (imaging parameters TR/TE = 4059/36 msec, rare factor 8, four averages). Thirty-five axial slices with a slice thickness of 0.5 mm, a field of view of 2.60 × 2.60 cm, and a matrix of 256 × 256 were positioned over the whole brain. Data were acquired with the Bruker software Paravision 4.0. Images were blindly analyzed and quantified using ImageJ software. During the examinations, mice were placed on a heated circulating water blanket to ensure a constant body temperature of 37°C. Anesthesia was induced with 3% and maintained with 1.5%–2.0% isoflurane (Forene, Abbot) delivered in a O2/N2O mixture (30%/70%) via a facemask under constant ventilation monitoring (Small Animal Monitoring and Gating System, SA Instruments).
In utero electroporation
In utero electroporation were performed on E15.5 embryos as previously described (Saito 2006; Britanova et al. 2008). The brains of electroporated animals were collected at P23, fixed in 4% PFA/PBS, and sectioned at 100 μm using a vibratome.
LacZ assay
LacZ assay was carried out on 12-μm cryo sections post-fixed in 0.2% glutaraldehyde/PBS for 10 min or on whole embryos fixed for up to 2 h in 2%PFA/PBS. Samples were incubated in staining solution [10 mM K3Fe(CN)6, 10 mM K4Fe(CN)6, 1 mg/mL X-Gal] at 37°C until color developed.
Nissl staining
Nissl staining was performed in 1% cresyl violet acetate-buffered (pH 4.5) solution. The cortical thickness of matched sections was imaged, and the thickness of the cortical plate was measured perpendicular to the pial surface using the AxioVision 4.8 software on an Olympus BX60 microscope. Cell counting was carried out on 163-μm-wide regions of the somatosensory cortex of P5 mice, respectively, using ImageJ software. All measurements were carried out blind as to genotype. In all cases, measurements of right and left hemispheres were separately quantified to obtain an average value for each mouse.
Golgi impregnation
To analyze dendritic structure, P25–P26 female mice were decapitated in ether anesthesia, and the brains were quickly removed. Golgi impregnation of 200-μm sagittal sections was conducted using the FD Rapid GolgiStain kit (FD Neuro Technologies) according to the instructions. Briefly, the brain slices were immersed in the initial impregnation solution in darkness at room temperature. After 2 wk of incubation, the slices were transferred into solution C and incubated for 5 d at 4°C. Slices were then washed in ddH2O and transferred into a freshly prepared mixture of solutions D and E. After dehydration through graded ethanol, slices were cleared in xylene solution and mounted in Permount (Fisher Scientific).
Immunofluorescence and immunohistochemistry
All stainings were done in parallel on samples or cells derived from littermate animals. Primary neuron cultures and cryo sections were fixed in 4% PFA/PBS permeabilized in 0.1% TX100/PBS for 5–20 min, blocked in 10% fetal calf serum (FCS)/PBS for 30 min, and incubated with the indicated primary antibodies in PBS or 10% FCS/TNB buffer (0.1 M Tris HCl 150 mM NaCl at pH 7.5) overnight at 4°C, followed by the appropriate secondary antibody for 2 h. Vibratome sections were stained as free-floating sections. Sections for phospho-cofilin staining were permeabilized in a solution of 0.2% Triton X-100, 5% serum, and 1% bovine serum albumin in TNB buffer for 1 h, while sections for MAP2 staining were permeabilized in 0.4% Triton X-100/TNB for 20 min followed by blocking in 10% serum/TNB for 30 min. Sections were then incubated overnight at 4°C, with the respective primary antibodies diluted in 10% serum and 0.1% Triton X-100/TNB.
Detection of primary antibodies was carried out with appropriate peroxidase- or fluorochrome-coupled secondary antibodies from Jackson Laboratories. Detection of peroxidase-coupled secondary antibodies was carried out using the VectaStain ABC kit (Vector Laboratories).
Fluorescent images were captured on a confocal Zeiss LSM 5 exciter microscope, and staining intensity was quantified using the Zen 2009 software.
Sholl analysis
For analysis of Golgi-stained and fluorescent neurons, 3-μm-spaced Z-stack images of neurons in the specified layers (II/III or V) of dorsal cortical regions were taken with an Olympus IX81 microscope equipped with a F ViewII (sw) camera. Examples are shown as Supplemental Movies 1–8. Sholl analysis (Sholl 1953), was preformed blindly on the Z-stack images using 30 10-μm-spaced concentric circles, and intersections were counted using ImageJ software.
Western blotting
Freshly dissected cortices were lysed in Flag buffer (50 mM Tris at pH 7.5, 100 mM NaCl, 1 mM EDTA, 1% [v/v] Triton X-100) containing protease and phosphatase inhibitors (10 mg/mL leupeptin, 10 mg/mL aprotinin, 1 mM benzamidine, 5 mg/mL pepstatin, 0.5 mg/mL microcystin, 1 mM Na3VO4; Sigma-Aldrich) and centrifuged at 135000 rpm for 10 min. Protein concentrations were determined using the BCA protein assay kit (Thermo Scientific). Equal amounts of protein were separated by SDS-PAGE and transferred to Immobilon-P transfer membranes (Millipore). Proteins were detected by incubation with the indicated antibodies followed by incubation with the appropriate peroxidase-coupled secondary antibodies (Jackson Laboratory) and were visualized by chemiluminescence (ECL Western blotting detection reagents, GE Healthcare). Chemiluminescence was captured and quantified using the Image Lab software on a ChemiDoc XRS+ detector (Bio-Rad).
Statistics
Data are presented as mean ± standard error of the mean (SEM). Statistical significance was determined using ANOVA or two-tailed t-tests as indicated. Probability outcomes are summarized in graphs as follows: P < 0.05 (*), P < 0.01 (**), and P < 0.001 (***).
Acknowledgments
We thank D. Lajkó and M. Müller for valuable technical support. In addition, we thank G. Dittmar (Max Delbrück Center Mass Spectrometry Core Unit) for the mass spectrometric analysis. We also thank J. Schüler for advice on microscopy, and S. Müller for help with MRI. This study was supported by a Rahel Hirsch Fellowship (M.R.), the BIF Foundation (S.P.), and the DFG (RA 424/5 [R.J.] and SFB665, Heisenberg Program and Neurocure Cluster of Excellence [V.T.]).
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.191593.112.
References
- Aizawa H, Wakatsuki S, Ishii A, Moriyama K, Sasaki Y, Ohashi K, Sekine-Aizawa Y, Sehara-Fujisawa A, Mizuno K, Goshima Y, et al. 2001. Phosphorylation of cofilin by LIM-kinase is necessary for semaphorin 3A-induced growth cone collapse. Nat Neurosci 4: 367–373 [DOI] [PubMed] [Google Scholar]
- Arber S, Barbayannis FA, Hanser H, Schneider C, Stanyon CA, Bernard O, Caroni P 1998. Regulation of actin dynamics through phosphorylation of cofilin by LIM-kinase. Nature 393: 805–809 [DOI] [PubMed] [Google Scholar]
- Armstrong DD, Dunn K, Antalffy B 1998. Decreased dendritic branching in frontal, motor and limbic cortex in Rett syndrome compared with trisomy 21. J Neuropathol Exp Neurol 57: 1013–1017 [DOI] [PubMed] [Google Scholar]
- Bauman ML, Kemper TL, Arin DM 1995. Pervasive neuroanatomic abnormalities of the brain in three cases of Rett's syndrome. Neurology 45: 1581–1586 [DOI] [PubMed] [Google Scholar]
- Bellenchi GC, Gurniak CB, Perlas E, Middei S, Ammassari-Teule M, Witke W 2007. N-cofilin is associated with neuronal migration disorders and cell cycle control in the cerebral cortex. Genes Dev 21: 2347–2357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BW, Bamburg JR 2010. ADF/cofilin: A functional node in cell biology. Trends Cell Biol 20: 187–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britanova O, de Juan Romero C, Cheung A, Kwan KY, Schwark M, Gyorgy A, Vogel T, Akopov S, Mitkovski M, Agoston D, et al. 2008. Satb2 is a postmitotic determinant for upper-layer neuron specification in the neocortex. Neuron 57: 378–392 [DOI] [PubMed] [Google Scholar]
- Cappello S, Attardo A, Wu X, Iwasato T, Itohara S, Wilsch-Brauninger M, Eilken HM, Rieger MA, Schroeder TT, Huttner WB, et al. 2006. The Rho-GTPase cdc42 regulates neural progenitor fate at the apical surface. Nat Neurosci 9: 1099–1107 [DOI] [PubMed] [Google Scholar]
- Chai X, Forster E, Zhao S, Bock HH, Frotscher M 2009. Reelin stabilizes the actin cytoskeleton of neuronal processes by inducing n-cofilin phosphorylation at serine3. J Neurosci 29: 288–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen TJ, Gehler S, Shaw AE, Bamburg JR, Letourneau PC 2006. Cdc42 participates in the regulation of ADF/cofilin and retinal growth cone filopodia by brain derived neurotrophic factor. J Neurobiol 66: 103–114 [DOI] [PubMed] [Google Scholar]
- Coleman PD, Riesen AH 1968. Environmental effects on cortical dendritic fields. I. Rearing in the dark. J Anat 102: 363–374 [PMC free article] [PubMed] [Google Scholar]
- Dan C, Kelly A, Bernard O, Minden A 2001. Cytoskeletal changes regulated by the PAK4 serine/threonine kinase are mediated by LIM kinase 1 and cofilin. J Biol Chem 276: 32115–32121 [DOI] [PubMed] [Google Scholar]
- de Anda FC, Meletis K, Ge X, Rei D, Tsai LH 2010. Centrosome motility is essential for initial axon formation in the neocortex. J Neurosci 30: 10391–10406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards DC, Sanders LC, Bokoch GM, Gill GN 1999. Activation of LIM-kinase by Pak1 couples Rac/Cdc42 GTPase signalling to actin cytoskeletal dynamics. Nat Cell Biol 1: 253–259 [DOI] [PubMed] [Google Scholar]
- Etienne-Manneville S, Hall A 2002. Rho GTPases in cell biology. Nature 420: 629–635 [DOI] [PubMed] [Google Scholar]
- Gao FB, Brenman JE, Jan LY, Jan YN 1999. Genes regulating dendritic outgrowth, branching, and routing in Drosophila. Genes Dev 13: 2549–2561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garvalov BK, Flynn KC, Neukirchen D, Meyn L, Teusch N, Wu X, Brakebusch C, Bamburg JR, Bradke F 2007. Cdc42 regulates cofilin during the establishment of neuronal polarity. J Neurosci 27: 13117–13129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goebbels S, Bormuth I, Bode U, Hermanson O, Schwab MH, Nave KA 2006. Genetic targeting of principal neurons in neocortex and hippocampus of NEX-Cre mice. Genesis 44: 611–621 [DOI] [PubMed] [Google Scholar]
- Hall A, Lalli G 2010. Rho and Ras GTPases in axon growth, guidance, and branching. Cold Spring Harb Perspect Biol 2: a001818 doi: 10.1101/cshperspect.a001818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttenlocher PR 1974. Dendritic development in neocortex of children with mental defect and infantile spasms. Neurology 24: 203–210 [DOI] [PubMed] [Google Scholar]
- Irie F, Yamaguchi Y 2002. EphB receptors regulate dendritic spine development via intersectin, Cdc42 and N-WASP. Nat Neurosci 5: 1117–1118 [DOI] [PubMed] [Google Scholar]
- Jan YN, Jan LY 2010. Branching out: Mechanisms of dendritic arborization. Nat Rev Neurosci 11: 316–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judas M, Sedmak G, Rados M, Sarnavka V, Fumic K, Willer T, Gross C, Hehr U, Strahl S, Cuk M, et al. 2009. POMT1-associated Walker-Warburg syndrome: A disorder of dendritic development of neocortical neurons. Neuropediatrics 40: 6–14 [DOI] [PubMed] [Google Scholar]
- Kaufmann WE, Moser HW 2000. Dendritic anomalies in disorders associated with mental retardation. Cereb Cortex 10: 981–991 [DOI] [PubMed] [Google Scholar]
- Kurokawa K, Itoh RE, Yoshizaki H, Nakamura YO, Matsuda M 2004. Coactivation of Rac1 and Cdc42 at lamellipodia and membrane ruffles induced by epidermal growth factor. Mol Biol Cell 15: 1003–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee A, Li W, Xu K, Bogert BA, Su K, Gao FB 2003. Control of dendritic development by the Drosophila fragile X-related gene involves the small GTPase Rac1. Development 130: 5543–5552 [DOI] [PubMed] [Google Scholar]
- Li Z, Van Aelst L, Cline HT 2000. Rho GTPases regulate distinct aspects of dendritic arbor growth in Xenopus central neurons in vivo. Nat Neurosci 3: 217–225 [DOI] [PubMed] [Google Scholar]
- Marin-Padilla M 1972. Structural abnormalities of the cerebral cortex in human chromosomal aberrations: A Golgi study. Brain Res 44: 625–629 [DOI] [PubMed] [Google Scholar]
- McAllister AK, Katz LC, Lo DC 1997. Opposing roles for endogenous BDNF and NT-3 in regulating cortical dendritic growth. Neuron 18: 767–778 [DOI] [PubMed] [Google Scholar]
- Meng Y, Takahashi H, Meng J, Zhang Y, Lu G, Asrar S, Nakamura T, Jia Z 2004. Regulation of ADF/cofilin phosphorylation and synaptic function by LIM-kinase. Neuropharmacology 47: 746–754 [DOI] [PubMed] [Google Scholar]
- Molyneaux BJ, Arlotta P, Menezes JR, Macklis JD 2007. Neuronal subtype specification in the cerebral cortex. Nat Rev Neurosci 8: 427–437 [DOI] [PubMed] [Google Scholar]
- Moriyama K, Iida K, Yahara I 1996. Phosphorylation of Ser-3 of cofilin regulates its essential function on actin. Genes Cells 1: 73–86 [DOI] [PubMed] [Google Scholar]
- Newey SE, Velamoor V, Govek EE, Van Aelst L 2005. Rho GTPases, dendritic structure, and mental retardation. J Neurobiol 64: 58–74 [DOI] [PubMed] [Google Scholar]
- Nobes CD, Hall A 1995. Rho, rac, and cdc42 GTPases regulate the assembly of multimolecular focal complexes associated with actin stress fibers, lamellipodia, and filopodia. Cell 81: 53–62 [DOI] [PubMed] [Google Scholar]
- Ohta Y, Nishida E, Sakai H, Miyamoto E 1989. Dephosphorylation of cofilin accompanies heat shock-induced nuclear accumulation of cofilin. J Biol Chem 264: 16143–16148 [PubMed] [Google Scholar]
- Parrish JZ, Kim MD, Jan LY, Jan YN 2006. Genome-wide analyses identify transcription factors required for proper morphogenesis of Drosophila sensory neuron dendrites. Genes Dev 20: 820–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price LS, Leng J, Schwartz MA, Bokoch GM 1998. Activation of Rac and Cdc42 by integrins mediates cell spreading. Mol Biol Cell 9: 1863–1871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redmond L, Kashani AH, Ghosh A 2002. Calcium regulation of dendritic growth via CaM kinase IV and CREB-mediated transcription. Neuron 34: 999–1010 [DOI] [PubMed] [Google Scholar]
- Rosario M, Franke R, Bednarski C, Birchmeier W 2007. The neurite outgrowth multiadaptor RhoGAP, NOMA-GAP, regulates neurite extension through SHP2 and Cdc42. J Cell Biol 178: 503–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosso SB, Sussman D, Wynshaw-Boris A, Salinas PC 2005. Wnt signaling through Dishevelled, Rac and JNK regulates dendritic development. Nat Neurosci 8: 34–42 [DOI] [PubMed] [Google Scholar]
- Ruchhoeft ML, Ohnuma S, McNeill L, Holt CE, Harris WA 1999. The neuronal architecture of Xenopus retinal ganglion cells is sculpted by rho-family GTPases in vivo. J Neurosci 19: 8454–8463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito T 2006. In vivo electroporation in the embryonic mouse central nervous system. Nat Protoc 1: 1552–1558 [DOI] [PubMed] [Google Scholar]
- Schule B, Armstrong DD, Vogel H, Oviedo A, Francke U 2008. Severe congenital encephalopathy caused by MECP2 null mutations in males: Central hypoxia and reduced neuronal dendritic structure. Clin Genet 74: 116–126 [DOI] [PubMed] [Google Scholar]
- Schwamborn JC, Puschel AW 2004. The sequential activity of the GTPases Rap1B and Cdc42 determines neuronal polarity. Nat Neurosci 7: 923–929 [DOI] [PubMed] [Google Scholar]
- Scott EK, Reuter JE, Luo L 2003. Small GTPase Cdc42 is required for multiple aspects of dendritic morphogenesis. J Neurosci 23: 3118–3123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw P, Lerch J, Greenstein D, Sharp W, Clasen L, Evans A, Giedd J, Castellanos FX, Rapoport J 2006. Longitudinal mapping of cortical thickness and clinical outcome in children and adolescents with attention-deficit/hyperactivity disorder. Arch Gen Psychiatry 63: 540–549 [DOI] [PubMed] [Google Scholar]
- Shi Y, Pontrello CG, DeFea KA, Reichardt LF, Ethell IM 2009. Focal adhesion kinase acts downstream of EphB receptors to maintain mature dendritic spines by regulating cofilin activity. J Neurosci 29: 8129–8142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sholl DA 1953. Dendritic organization in the neurons of the visual and motor cortices of the cat. J Anat 87: 387–406 [PMC free article] [PubMed] [Google Scholar]
- Sumi T, Matsumoto K, Takai Y, Nakamura T 1999. Cofilin phosphorylation and actin cytoskeletal dynamics regulated by rho- and Cdc42-activated LIM-kinase 2. J Cell Biol 147: 1519–1532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumi T, Matsumoto K, Shibuya A, Nakamura T 2001. Activation of LIM kinases by myotonic dystrophy kinase-related Cdc42-binding kinase α. J Biol Chem 276: 23092–23096 [DOI] [PubMed] [Google Scholar]
- Takashima S, Ieshima A, Nakamura H, Becker LE 1989. Dendrites, dementia and the Down syndrome. Brain Dev 11: 131–133 [DOI] [PubMed] [Google Scholar]
- Threadgill R, Bobb K, Ghosh A 1997. Regulation of dendritic growth and remodeling by Rho, Rac, and Cdc42. Neuron 19: 625–634 [DOI] [PubMed] [Google Scholar]
- Wallace GL, Dankner N, Kenworthy L, Giedd JN, Martin A 2010. Age-related temporal and parietal cortical thinning in autism spectrum disorders. Brain 133: 3745–3754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang N, Higuchi O, Ohashi K, Nagata K, Wada A, Kangawa K, Nishida E, Mizuno K 1998. Cofilin phosphorylation by LIM-kinase 1 and its role in Rac-mediated actin reorganization. Nature 393: 809–812 [DOI] [PubMed] [Google Scholar]
- Yoo Y, Ho HJ, Wang C, Guan JL 2009. Tyrosine phosphorylation of cofilin at Y68 by v-Src leads to its degradation through ubiquitin-proteasome pathway. Oncogene 29: 263–272 [DOI] [PMC free article] [PubMed] [Google Scholar]