Abstract
Silk fibroin has been successfully used as a biomaterial for tissue regeneration. In order to prepare silk fibroin biomaterials for human implantation a series of processing steps are required to purify the protein. Degumming to remove inflammatory sericin is a crucial step related to biocompatibility and variability in the material. Detailed characterization of silk fibroin degumming is reported. The degumming conditions significantly affected cell viability on the silk fibroin material and the ability to form three-dimensional porous scaffolds from the silk fibroin, but did not affect macrophage activation or β-sheet content in the materials formed. Methods are also provided to determine the content of residual sericin in silk fibroin solutions and to assess changes in silk fibroin molecular weight. Amino acid composition analysis was used to detect sericin residuals in silk solutions with a detection limit between 1.0% and 10% wt/wt, while fluorescence spectroscopy was used to reproducibly distinguish between silk samples with different molecular weights. Both methods are simple and require minimal sample volume, providing useful quality control tools for silk fibroin preparation processes.
Keywords: silk fibroin, sericin, biomaterial, biocompatibility, regenerative medicine
Introduction
Important considerations in engineering biomaterials for regenerative medicine applications include biocompatibility and reproducibility of the material preparation process. For tissue-engineered biomaterial devices to be used in a clinical setting, surgical implant standards must be met as established by the International Standards Organization (ISO) and the American Society for Testing and Materials (ASTM). In terms of biocompatibility, biomaterials must undergo a series of tests outlined by the ISO 10993, to provide evidence of biomaterial safety upon implantation. In terms of reproducibility, the process for manufacturing the biomaterial should adhere to ISO 9001:2008, which delineates a method for implementing quality management systems. For both biocompatibility and reproducibility, it is therefore important to have a well characterized process for preparing the biomaterial for use in downstream biomedical technologies.
Silk fibroin, a protein polymer, is being used to develop a variety of biomedical devices and regeneration technologies. Silk fibroin is harvested from Bombyx mori silkworm cocoons and assembled into a spectrum of material formats using a variety of fabrication techniques1. The silk fibroin is present within B. mori silkworm cocoons as a double-stranded fiber, which is coated with glue-like proteins called sericins. The raw silk fiber mass is composed of about 20–30% sericin and 70–80% silk fibroin with trace amounts of waxes and carbohydrates2. To prepare pure silk fibroin solution for silk-based biomaterials, silkworm cocoons undergo a degumming procedure to separate the silk fibroin fiber from the sericin glue, a critical step, since (1) residual sericin causes inflammatory responses3 and (2) non-degummed fibers are resistant to solubilization4.
Silk fibroin is composed of a heavy protein chain (~390 kDa molecular weight) and a light protein chain (~26 kDa molecular weight) which are connected by a disulfide linkage5,6. The heavy chain consists of a block co-polymer arrangement of primarily hydrophobic amino acids and is the source of the robust mechanical properties7. The light chain consists of about 47% hydrophobic amino acid residues5 and is crucial for proper cellular secretion of the heavy chain8. The heavy chain amino acid sequence primarily consists of Gly–X repeats where X is typically Ala, Ser, or Tyr. The G-X motif exists in repeating hexamer units of GAGAGS and GAGAGY6. The silk fibroin molecule contains 12 domains of these repeating hexamer units that are separated by 11 short linker sequences, which are nonrepetitive and contain amino acid residues such as Asn, Asp, Glu, Lys, Phe, Pro, Thr, and Trp9,10. Silk fibroin can exist as three structural morphologies termed silk I, II, and III where silk I is a water soluble form and silk II is an insoluble form consisting of extended β-sheets. The silk III structure is helical and is observed at the air-water interface11. In the silk II form, the 12 repetitive domains form anti-parallel β-sheets stabilized by hydrogen bonding6.
Sericins are a family of proteins with molecular weights ranging from 20 kDa to 400 kDa depending on gene coding and post-translational modifications12,13. Three different sericins have been isolated with molecular weights of 150, 250, and 400 kDa13. The primary amino acid sequence of most sericins contains a 38-amino acid repeat14 composed of mostly Ser, Gly, Asn, and Asp13,15 and a random coil secondary structure16.
A variety of silk fiber degumming methods have been reported, including the use of boric acidsodium borate buffer17, succinic acid17, urea4, and enzymes such as endopeptidase, papain, and pepsin derived from bacteria, plants and mold respectively18. It is also common to use 0.02 M sodium carbonate solution at 100°C for 30–60 minutes to accomplish degumming19. All of the degumming methods employ denaturants (i.e. heat and pH) to help remove the sericin from the fibers.
A few previous studies have examined the effect of degumming on the silk fibroin protein. One study compared the effect of different degumming protocols on silk fiber tensile properties17. The degumming reagents investigated were boric acid-sodium borate buffer, sodium carbonate, urea and succinic acid. The authors found that the degummed fibers had decreased elasticity and a lowered yield point compared to the non-degummed fibers. They also found that the sodium carbonate method resulted in the largest decrease in elasticity and yield point. The authors concluded that in the process of degumming silk fibers, intermolecular hydrogen and/or van der Waals bonds were disrupted. The sodium carbonate method also altered the molecular weight of the silk fibroin17. To further elucidate the effect of sodium carbonate degumming on silk fibroin, silkworm cocoons were degummed for a variety of times, followed by gel electrophoresis on the solubilized silk fibroin protein4. The silk fibroin heavy chain extracted directly from silk worm glands appeared as a band at 390 kDa; however, when extruded silk fibers were degummed with sodium carbonate the heavy chain band was less visible and a smear of protein appeared below 390 kDa. As the degumming time increased the smear appeared further down the gel, indicating that the silk fibroin protein was being degraded into lower molecular weight fragments4.
The effect of degumming conditions on silk fibroin protein has also been shown not only to impact mechanical properties, but also to affect cell function. For example, intact silk fibroin proteins taken directly from the silkworm gland increased human skin fibroblast proliferation when added to the culture media by 150% compared to the tissue culture polystyrene (TCP) control after two days of cell culture20. In contrast, silk fibroin that had been degummed for one hour decreased cell proliferation compared to the TCP control. It was hypothesized that decreased fibroblast proliferation on the degraded silk fibroin protein was due to the release of small peptide fragments that somehow negatively impacted cell viability20.
The goal of the present study was to further characterize the effect of degumming on silk fibroin protein properties, including issues of biocompatibility and reproducibility related to materials formation. We built on the findings from prior studies cited above with a focus on the functional outcomes of the changes in silk fibroin in terms of biocompatibility and formation of useful biomaterial products from the silk. Of particular interest were endothelial cell viability, macrophage activation, and silk fibroin three-dimensional (3D) porous scaffold formation related to the process variables. For product development purposes it is not only important to understand the effect of biomaterial processing parameters on polymer features, but also to establish appropriate analytical protocols to efficiently detect the effects of these parameters on the end product. Therefore, an additional goal of the present work was to establish methods for assessing the changes in the silk protein as a function of degumming time for quality control purposes.
Methods
Silk Fiber Degumming and Solubilization
Five grams of B. mori silk worm cocoons were immersed into 1 L of boiling 0.02 M Na2CO3 solution for 5, 30, or 60 minutes. These samples will be hereafter referred to as 5 mb, 30 mb, and 60 mb, where ‘mb’ stands for ‘minutes-boiled’. The degummed fibers were collected and rinsed with distilled water three times then air dried. The fibers were solubilized in 9.3 M LiBr (20% w/v) at 60°C for four hours. A volume of 15 mL of this solution was then dialyzed against 1 L of distilled water (water was changed after 1, 3, 6, 24, 36, and 48 hours) with a regenerated cellulose membrane (3,500 MWCO, Slide-A-Lyzer, Pierce, Rockford, IL). LiBr was judged to have been fully removed from the silk solution when the conductivity of the dialysis water dropped below 5 µS·cm−1. The solubilized silk fibroin protein solution was centrifuged to remove insoluble particulates and stored at 4°C. Protein concentration was determined by drying a known volume of the silk solution under a hood for 12 hours and assessing the mass of the remaining solids19.
Gel electrophoresis
Silk fibroin protein electrophoretic mobility was visualized with sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE). For each sample, 5 mg of silk protein was loaded into a 3–8% Tris Acetate Novex gel (NuPAGE, Invitrogen, Carlsbad, CA) under reducing conditions. The gel was then stained with a Colloidal Blue staining kit (Invitrogen). Protein distribution was quantified by taking densiometric measurements along the length of the gel (ImageJ, NIH, Bethesda, MD). To determine whether degumming time specifically decreased the molecular weight of the silk fibroin protein, 15 mL of 6% w/v silk fibroin solution was redialyzed against 1 L of 8 M urea for three days to fully denature the protein. Samples were rerun with SDS-PAGE and stained with a Colloidal Blue staining kit (Invitrogen).
Gel Permeation (Size Exclusion) Chromatography
Silk fibroin solution was diluted to 0.1% w/v in PBS and loaded onto an Agilent Bio-SEC3 size exclusion column (4.6 × 300 mm, 300 Å pore size) (Agilent Technologies, Santa Clara, CA). PBS was used as the mobile phase with a 0.5 mL/min flow rate maintained at 25°C. Protein released from the column was measured with UV-Vis at 210 nm and plotted as a function of time. The smaller the protein molecular weight, the longer the protein was retained in the column.
FTIR Analysis
β-sheet crystal content was measured according to previously published work21. Briefly, 1% w/v silk fibroin solutions were cast on Teflon-coated polystyrene dishes and air dried for 12 hours. To induce β-sheet crystallization, the films were soaked in methanol for four days. Prior to FTIR analysis, films were air dried for 12 hours and placed in a vacuum for 12 hours to remove residual water from the samples. FTIR measurements were performed with a Bruker Equinox 55/S FTIR spectrometer (Bruker Optics, Billerica MA). Each measurement was taken with 132 scans ranging from 400 to 4000 cm−1 at 4 cm−1 resolution. The spectra were fourier self-deconvoluted (FSD) with Opus 5.5 spectroscopy software (Bruker Optics) using the method reported in Hu et al. (2006). The FSD FTIR spectra were fitted with Gaussian profiles in the amide I region between 1595–1705 cm−1. The integral of the Gaussian profiles (termed ‘bands’) was used to determine the β-sheet fraction of the amide I region. Bands between 1617–1637 cm−1 and the band between 1697–1703 cm−1 were attributed to β-sheet crystals. The ratio of these bands to the total amide I bands is reported as the β-sheet fraction21.
Thermogravimetric Analysis
Thermogravimetric analysis (TGA) (TA Instruments Q500) was used to measure weight changes of silk films assembled from 1% w/v silk fibroin solutions. TGA curves were obtained under nitrogen atmosphere with a gas flow of 50 mL/min. Two methods were used in this study to understand the thermal degradation properties of these materials. The analysis was first performed by heating the sample from 25°C to 400°C at a rate of 5°C/min. Silk film weight loss was recorded as a function of temperature. Second, the instrument was directly heated to 250°C, and isothermally held for three hours while the weight loss for each sample was recorded as a function of time.
Scaffold Assembly and Analysis
Silk fibroin solutions were assembled into aqueous-derived scaffolds according to previously published methods22. Briefly, 4 g of NaCl particles with diameters of 500–600 µm were sifted into polyethylene cylindrical molds (Ø=1.8 cm, height=2.0 cm) containing 2 mL of 6% w/v silk fibroin solution. The molds were covered and left at room temperature for 48 hours. After 48 hours, the cylindrical molds were immersed in distilled water for two days to dissolve away the NaCl, leaving behind a porous silk scaffold. To prepare scaffolds for scanning electron microscopy (SEM) and mercury intrusion porosimetry (MIP), scaffolds were subjected to a series of graded ethanol washes and dried with a critical point dryer (CPD) (Tousimis Autosamdri815, Rockville, MD) to ensure preservation of scaffold architecture. For SEM, samples were sputter coated with platinum/palladium and imaged with a high resolution field emission scanning electron microscope (Supra55VP, Zeiss, Oberkochen, Germany). For MIP, samples were loaded into glass sample cells with 0.5 cc stem volumes and subsequently analyzed using a PoreMaster 33 porosimeter (Quantachrome Instruments, Boynton Beach, FL). The sample cells were evacuated to a pressure of ~50 millitorr, after which mercury was introduced and pressurized to 50 psi pneumatically. Each sample cell was then transferred to an oil-filled hydraulic chamber for further intrusion over a pressure range of 20 to 33,000 psi. PoreMaster for Windows 5.10 (Quantachrome) was used for all data collection and analysis, including the determination of pore size, pore size distribution, and total intruded volume. To determine porosity, the density of the silk scaffold was assumed to be 1.3 g/cm3 based on previously reported literature23.
Endothelial Cell Viability Assay
Samples were prepared by casting 1% w/v silk fibroin films in the bottom of 24-well, polystyrene tissue culture plates (BD Bioscience, San Diego, CA). β-sheet crystallization was induced with methanol, making the films insoluble, after which they were sterilized with 30 minutes of UV light exposure. Human dermal microvascular endothelial cells (hMVECs) (P5–P8, Lonza, Basel, Switzerland) in Microvascular Endothelial Cell Growth Medium-2 (Lonza) and 5% fetal bovine serum (Lonza) were initially seeded at a density of 10,000 cells/cm2 in 1 mL of media and incubated at 5% CO2 and 37°C. Media was exchanged every 2–3 days. At day seven, the cells were analyzed for cell viability using the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay (Invitrogen). Cells were incubated with a 0.5 mg/mL MTT dye in serum-free media for two hours. The converted MTT dye was solubilized with DMSO and the absorbance was measured at 540 nm (normalized to 670 nm). hMVEC viability was determined as the average of four replicates and normalized to the TCP control.
Macrophage Activation
Silk samples were prepared by casting 1% w/v silk fibroin films in the bottom of 96-well, polystyrene tissue culture plates (BD Bioscience) and treated with methanol and sterilized with 30 minutes of UV light exposure. TCP and collagen samples were used as negative controls for macrophage activation. Collagen films were prepared by mixing a 3.44 mg/mL collagen solution (BD Bioscience) with PBS (1:10 collagen solution:PBS volume ratio) and 1 M NaOH (1:40 collagen solution:NaOH solution volume ratio). 100µL of this solution was dispensed in a 96-well tissue plate and allowed to dry overnight in a sterile hood. A human monocytic cell line, THP-124 (American Type Culture Collection, Manassas, VA) in RPMI1640 medium supplemented with 10% fetal bovine serum (Invitrogen) and antibiotics (50 U/L penicillin G (sodium salt), 50 mg/mL streptomycin) (Invitrogen) was initially seeded at a density of 1x106 cells/well in 200 µL of media and incubated at 5% CO2 and 37°C. On the same day of seeding, the THP-1 cells were differentiated into human macrophages with RPMI 1640 media supplemented with 5 ng/ml phorbo112-myristate 13-acetate (EMD Chemicals, Gibbstown, NJ). Two days after differentiation, the culture media was changed with RPMI 1640 media or media supplemented with lipopolysaccharide (LPS) (referred to ‘−LPS’ and ‘+LPS’, respectively). For +LPS media, a commercial LPS (Escherichia coli O26, Difco, Lawrence, KS) was added to the culture media at a final concentration of 100 ng/ml. The +LPS samples were included as positive controls for macrophage activation and TNF-α release. One day after LPS stimulation, macrophage tumor necrosis factor-α (TNF-α) release was determined with an ELISA kit (R&D Systems, Minneapolis, MN) with a detection limit of 5 pg/mL. TNF-α reaction was measured at 450 nm (normalized to 570 nm) and converted to pg/mL with a standard curve covering concentrations of 62.5–2,000 pg/mL. TNF-α release was normalized to sample volume and cell count as determined by the alamarBlue assay (Invitrogen). All cell culture measurements were carried out three times with four replicates each.
Urea Degumming for Sericin Isolation
Sericin was isolated from the silk fiber using a method adapted from Takasu et al. (2002). B. mori silkworm cocoons (0.5 g) were immersed in 20 mL of 8 M urea solution at 80°C for five minutes. The solution was cooled and the silk fibers were removed by centrifugation. Three times the volume of ethanol was added to the supernatant, which caused the sericin to precipitate. The precipitate was collected by centrifugation and air dried. The sericin pellet was dissolved in 1 mL of 9.3 M LiBr and dialyzed against distilled water (water was changed after 1, 3, 6, 24, 36, and 48 hours) with a regenerated cellulose membrane (3,500 molecular weight cut-off, Pierce). LiBr was judged to have been fully removed from the silk solution when the conductivity of the dialysis water dropped below 5 µS/cm. Protein concentration was determined by drying a known volume of the sericin solution under a hood for 12 hours and assessing the mass of the remaining solids. The isolated sericin was mixed at varying concentrations with 60 mb silk fibroin solution, lyophilized, and analyzed for amino acid content. The 60 mb silk fibroin solution was used because it is considered to be completely devoid of sericin, which was confirmed with in vivo studies that it does not cause chronic inflammation25.
Amino Acid Composition Analysis
Amino acid analysis was performed at the W.M. Keck Foundation Biotechnology Resource Laboratory at Yale University (New Haven, CT). Degummed silk fibers were hydrolyzed in vacuo for 16 hours at 115°C in 6 N HCl. Samples were then quantified using a Hitachi L-8900PH amino acid analyzer (ion-exchange separation, post-column derivatization with ninhydrin) (Hitachi, Schaumburg IL). Data were analyzed with Perkin Elmer/Nelson data acquisition software (Artisan Scientific, Champaign, IL). In this study cysteine, methionine, and tryptophan were eliminated from the analysis because cysteine residues co-elute with proline and methionine residues co-elute with aspartic acid, while tryptophan is poorly hydrolyzed by HCl. It is important to note that during the hydrolysis step, glutamine and asparagine are converted to glutamic acid and aspartic acid, respectively. Therefore in the analysis glutamine and glutamic acid are collectively abbreviated as ‘glx’ or ‘B’ and asparagines and aspartic acid are collectively abbreviated to ‘asx’ or ‘Z’.
Fluorescence of Silk Fibroin Solutions
Silk fibroin solutions were diluted to 0.1% w/v and a 1 mL volume was loaded into a fluorescence spectrophotometer (F4500, Hitachi). Samples were maintained at 25°C and excited at 280 nm while an emission spectrum scan was collected from 290–400 nm with a 700 V photomultiplier tube. The scan speed was 1,200 nm/min with an excitation and emission slit of 5 nm. Fluorescent emission spectra were collected with a 1 nm resolution. The emission spectra were peak normalized and the emission intensity at 330 nm was compared between the different solutions.
Statistical Analyses
Unless otherwise noted, a two-sample t-test was performed when appropriate, using a significance of p values <0.05. Amino acid compositions were compared using a paired t-test with a significant p value of <0.01.
Results
Effect of degumming on material properties
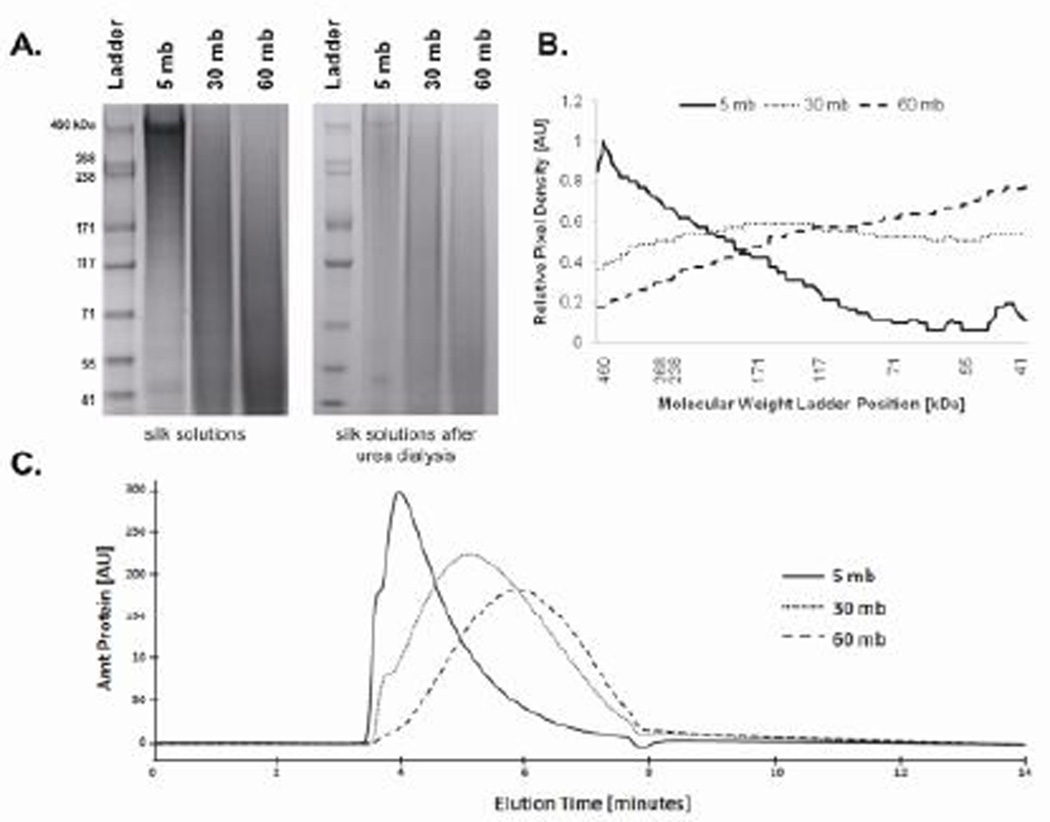
Differences in degumming duration in 0.02 M Na2CO3 resulted in changes in electrophoretic mobility. SDS-PAGE showed that each protein sample existed as a smear and that as degumming time increased, the smear moved down the gel (Figure 1A). Densiometric analysis showed that the bulk of the 5 mb sample was concentrated at the high molecular weight markers, while the smear of the 60 mb sample was found near the lower molecular weight markers. The 30 mb sample had a broad densiometric distribution with a peak around 100 kDa (Figure 1B). Since electrophoretic mobility is a function of both protein size and folding, samples were then dialyzed against 8 M urea for three days to fully denature the protein. This resulted in no change to the protein distributions on the gels (Figure 1A). GPC was also performed to confirm the SDS-PAGE results (Figure 1C). The peak of the 5 mb sample eluted at 4.0 minutes while the 30 and 60 mb samples were at 5.1 and 5.9 minutes, respectively, indicating that as degumming time increased, molecular weight decreased.
Figure 1.
Molecular weight analysis of silk fibroin solutions. (A) SDS-PAGE showed that the 5 mb sample had a visible band around the highest molecular weight marker, while the 30 mb and 60 mb samples were visible as smears lower down the gel (mb = minutes boiled). (B) Densiometric analysis of protein distribution along the gel showed that the 30 mb sample had a peak around 150 kDa, while the 60 mb peak was around 50 kDa. (C) Gel permeation chromatography showed that the 5 mb sample was first to elute from the column, followed by the 30 mb and then the 60 mb sample. The peak of the 5 mb sample eluted at 4.0 minutes, the 30 mb sample at 5.1 minutes, and the 60 mb sample at 5.9 minutes.
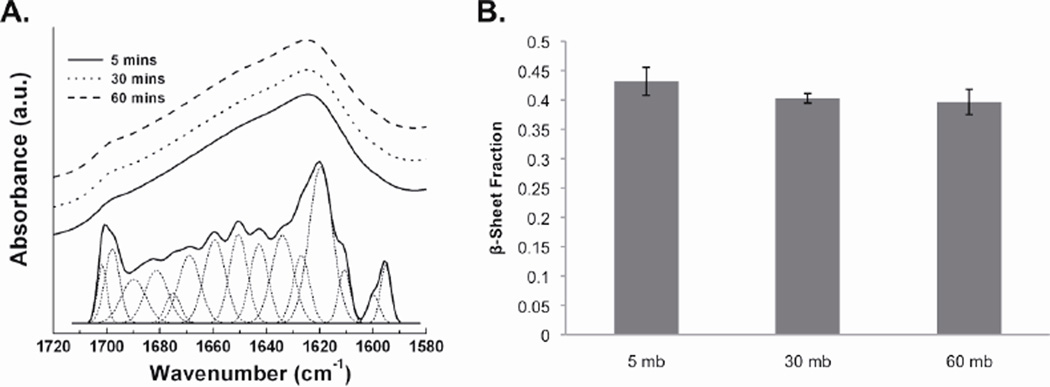
β-sheet crystal content of silk fibroin films formed from the degummed silk solutions was determined. FTIR measurements and FSD analysis showed no difference in β-sheet content between the 5, 30, and 60 mb silk films (p>0.01) (Figure 2). The average β-sheet fraction for all samples was between 0.40 and 0.45, which is typical for silk films treated with methanol to induce crystalline β-sheet21. These data indicate that differences in molecular weight did not impact β-sheet crystal assembly caused by up to 60 minutes of boiling in 0.02 M Na2CO3.
Figure 2.
Effect of degumming on FTIR spectra and β-sheet content of silk films. (A) The FTIR spectra for the amide I region are depicted along with an example FSD and the fitted Gaussian profiles. There were no discernable differences between the three FTIR spectra. (B) FTIR measurements and FSD analysis showed no difference (p>0.01, n=3) between the β-sheet content of the films formed from 5, 30, and 60 mb silk solutions. Error bars represent the standard deviation of the mean.
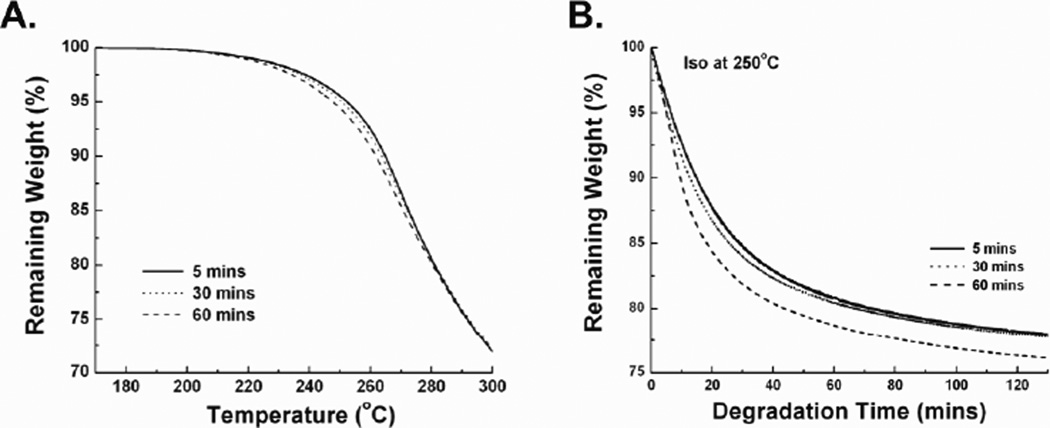
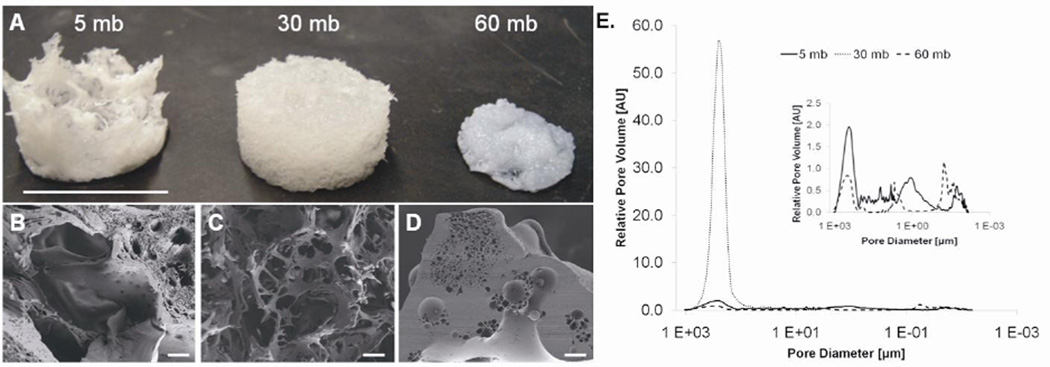
Despite similar β-sheet content, the samples had different material properties and morphologies as determined by thermal degradation (Figure 3) and scaffold assembly (Figure 4). The TGA results showed that under a ramped heating regime from 25°C to 400°C, the three silk fibroin samples exhibited different rates of degradation with the greatest difference around 250°C (Figure 3A). To further elucidate the difference between the degradation profiles, samples were isothermally degraded at 250°C, which showed the initial degradation step of the protein backbone was faster for the 60 mb case and slowest for the 5 mb sample. However, after this event the curves run more or less parallel, indicating that once the initial step is complete, the degradation rate is more or less unchanged versus degumming time (Figure 3B). After 15 minutes of an isothermal hold at 250°C the 5 mb, 30 mb, and 60 mb samples lost 9.97%, 11.2%, and 13.8% of their original weight, respectively. Scaffold assembly from the three silk fibroin solutions resulted in distinct differences in macroscopic shape, pore size, and porosity. The 30 mb sample formed a homogenous cylindrical sponge, while the 5 mb sample did not form an intact sponge. The 60 mb sample had a similar bulk morphology compared to the 30 mb sample but was much smaller in size (Figure 4A). SEM images qualitatively showed distinct differences in pore size and interconnectivity between the three samples (Figure 4B-D). Both the 5 mb and 60 mb samples had a large range of pore sizes but the 60 mb sample displayed less pore interconnectivity. The 30 mb sponge had homogenous pores sizes that were highly interconnected. MIP found that both the 5 mb and 60 mb samples had a tri-modal distribution of pore sizes, where pores were typically between 200–500 µm, 1–5 µm, and 0.02–0.06 µm (Figure 4E inset). Consistent with the SEM images shown, the 5 mb samples were observed to have more pore volume in the 200–500 µm pore size range, while the pore volume of the 60 mb samples was roughly equally distributed between the three aforementioned modes. In contrast, the 30 mb sample was observed to have a narrow distribution of pores in the range between 100–350 µm. In addition there was a difference in porosity between samples. Consistent with physical observations of the samples, the 60 mb sample was observed to have the lowest porosity (46%), followed by the 5 mb sample (64%) and finally the 30 mb sample (91%).
Figure 3.
TGA thermograms of silk films with variable degumming duration. (A) Weight loss during 5°C/min ramped heating from 170°C to 300°C. (B) Weight loss during isothermal heating at 250°C for 130 minutes. Both heating regimes showed that increased degumming duration decreased thermal stability.
Figure 4.
The effect of degumming on 3D porous silk scaffolds. (A) Degumming duration had an observable effect on porous silk scaffold assembly. Both the size and shape of the scaffolds was affected by how long the silk fibroin was degummed (scale bar = 1.5 cm). (B-D) Characteristic SEM images showed that the 5 mb, 30 mb, and 60 mb sponges had different pore sizes and interconnectivity (scale bars = 100 µm). (E) MIP analysis shows that the 5 mb and 60 mb samples had a tri-modal distribution of pore sizes (inset) while the 30 mb sample had a monomodal pore size distribution and substantially higher porosity.
Effect of degumming on biocompatibility
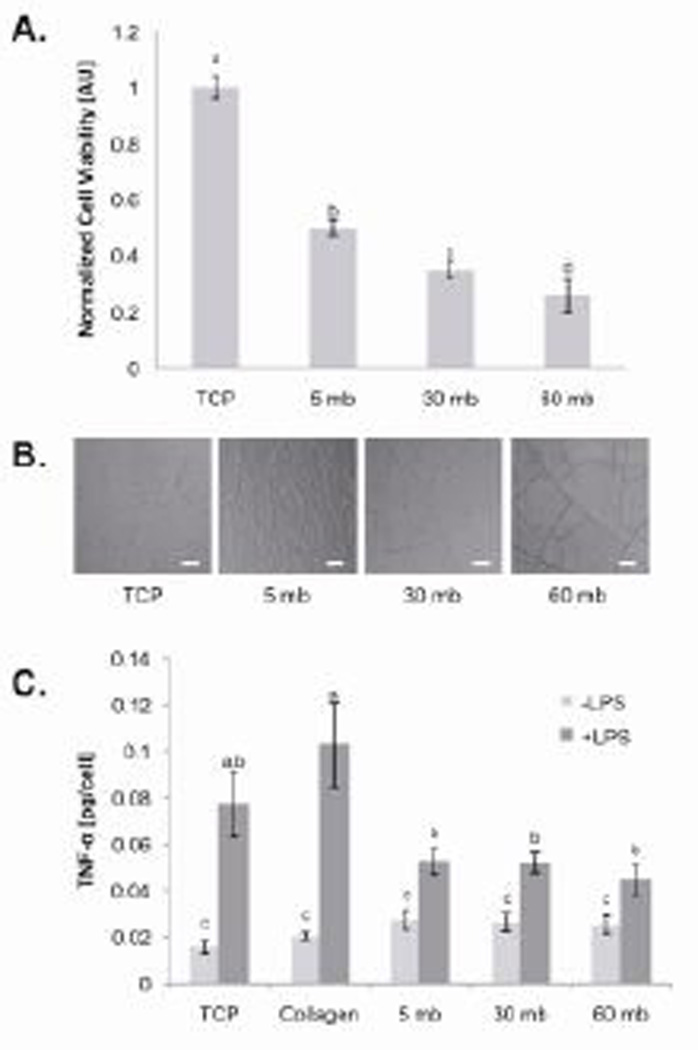
The MTT assay showed that as the duration of degumming increased, endothelial cell viability decreased (Figure 5A). Between the TCP control and the 5 mb sample, cell viability decreased 50%, whereas cell viability on the 60 mb samples decreased 75% compared to the TCP control. There was also a difference in cell viability between the 5 mb and 30 mb samples (p< 0.01) and between the 30 mb and 60 mb samples (p<0.05). Between the 5 mb and 30 mb samples, cell viability decreased from 50% to 35% (relative to the control) and between the 30 mb and 60 mb sample cell viability decreased from 35% to 25% (relative to the control).
Figure 5.
Cellular responses to differently degummed silk fibroin films. (A) As degumming time increased, hMVEC viability decreased with a difference between cell viability on the 5, 30, and 60 mb films (*p< 0.05; ^p< 0.01, n=4). Error bars represent the standard deviation of the mean. (B) Degumming time showed no significant effect on TNF-α release from -LPS stimulated human macrophages (*p>0.01). Furthermore, there was a slight increase in TNF-α released from -LPS stimulated human macrophages on the silk fibroin films compared to the negative controls (TCP and collagen). TNF-α released from +LPS was similar between the silk samples (^p>0.01), which was significantly less compared to the negative controls. Error bars represent the standard error of the mean.
Inflammatory potential was assessed in vitro with THP-1 human macrophages based on TNF-α release. All three silk fibroin samples under the –LPS condition elicited similar TNF-α expression (p> 0.01) (Figure 5B). Compared to the negative controls (TCP and collagen) the silk samples elicited slightly higher TNF-α expression (p< 0.01). Under the +LPS condition, the three silk samples had similar macrophage activation, which was significantly less compared to the negative controls. Thus, it appears that the degumming duration did not significantly affect the inflammatory potential of the silk samples.
Effect of degumming on sample reproducibility
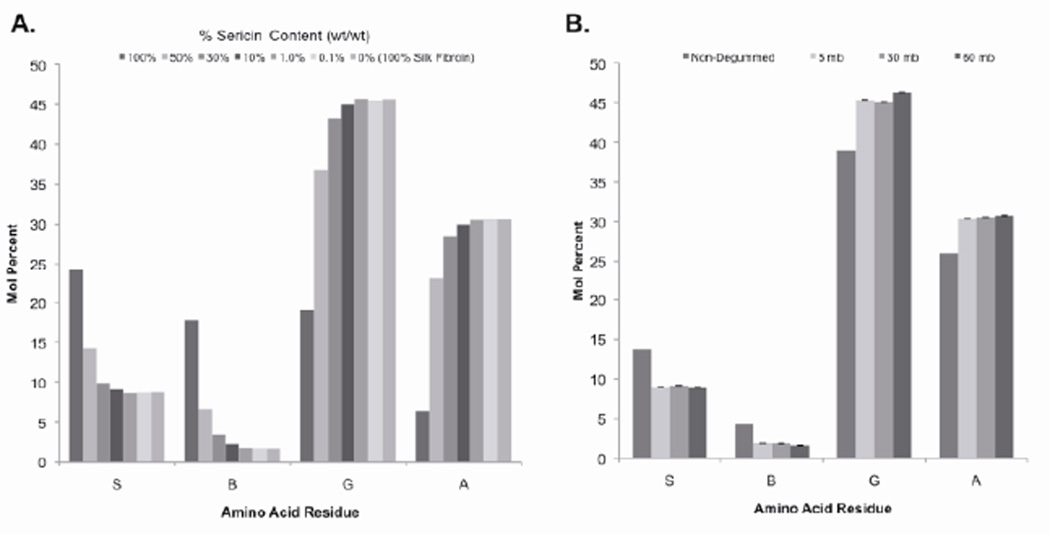
The amino acid composition of degummed silk fibroin was determined in an effort to measure residual sericin content. First, the amino acid compositions of silk fibroin samples containing different amounts of sericin were analyzed based on fibroin solutions doped with known amounts of sericin solution. For each amino acid residue (except phenylalanine, which was consistent between samples), changes in concentration were observed as the sericin content increased (Figure 6). For example, as the sericin content increased in the silk fibroin solution, threonine and serine concentrations increased from 0.9 to 6.9 mol% and 8.8 to 24.2 mol%, respectively, while the glycine and alanine concentrations decreased from 45.7 to 19.0 mol% and 30.6 to 6.4 mol%, respectively. The serine concentration decreased the most with increasing sericin content and the glycine concentration increased the most as sericin content increased. The ratio of the molar concentrations of serine and glycine residues was calculated in order to establish a metric for comparing amino acid composition between silk fibroin samples. In a 100% sericin sample the molar ratio of serine to glycine was 1.27, while in a 100% silk fibroin sample the molar ratio was 0.19 (Figure 6). The molar ratio of serine to glycine for the 1.0%, 0.1%, and 0% w/w sericin content samples all had the same value of 0.19. The 10% w/w sample had a value of 0.20, the 30% w/w sample had a value of 0.29, and the 50% w/w sample had a value of 0.39. The mole ratio of serine to glycine in the 5, 30, and 60 mb samples was 0.19 +/− 0.005 (Figure 7).
Figure 6.
Amino acid composition of silk fibroin solutions with different amounts of sericin. The ratio of mole percent of serine and glycine residues was used to compare the different samples, with pure silk fibroin samples having a ratio of 0.19 while pure sericin samples had a ratio of 1.27. The detection limit of this method is between 1.0% and 10% w/w of sericin content.
Figure 7.
Effect of degumming on silk solution amino acid composition. There was no difference in amino acid composition between the 5, 30, and 60 mb samples (p>0.01, n=3). Standard error bars have been omitted for easier viewing.
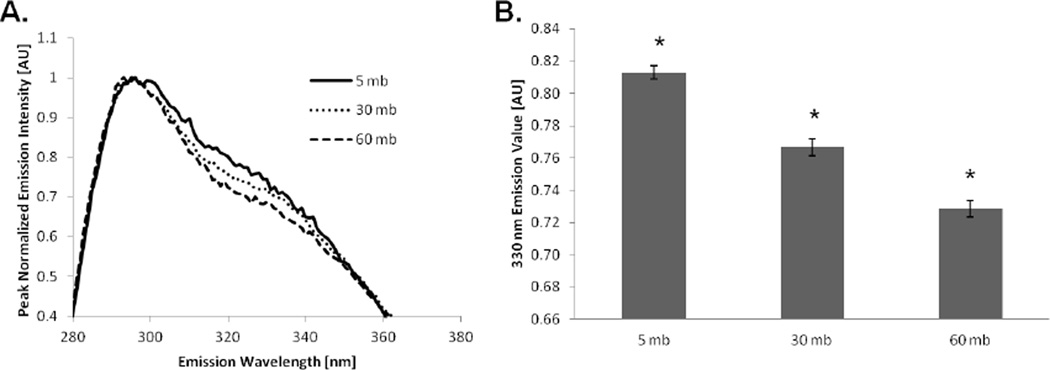
Silk fibroin solutions degummed for different times and analyzed via fluorescence emission spectroscopy at 280 nm yielded distinct emission spectra with a peak at 307 ± 1 nm. To compare the emission spectra between samples, each spectrum was normalized to the peak value at 307 nm, which revealed that with increased degumming duration the emission value at 330 nm decreased relative to the peak value at 307 nm (p<0.01) (Figure 8). The emission value at 330 nm between the three samples were as follows; the 5 mb samples had an emission value of 0.81 ± 0.01, the 30 mb sample 0.76 ± 0.01, and the 60 mb sample had the lowest value of 0.72 ± 0.01.
Figure 8.
(A) Fluorescence emission spectra of silk solutions from silk fibers degummed for different durations. Each spectrum has a peak at 307 nm and a shoulder around 330 nm. (B) The emission value at 330 nm for each sample was found to be different (*p<0.01, n=8). Since silk solutions degummed for 5, 30, and 60 minutes have similar amino acid sequences, the difference in emission spectra was not due to residual sericin but due to the difference in molecular weight distribution. The error bars represent the standard error of the mean.
Discussion
Effect of silk fibroin degradation on β-sheet content, thermal degradation, and porous scaffold assembly
The duration of degumming affected electrophoretic mobility of the fibroin as visualized by SDS-PAGE. This result was expected because a similar trend was observed in previous publications,4,20. The observed difference in protein mobility was previously reported to be due to differences in protein degradation; however, electrophoretic mobility is a function of both protein size and protein folding. To isolate the cause of the change in protein mobility, silk fibroin solutions were fully denatured with 8 M urea and the same electrophoretic mobility was observed with SDS-PAGE, thereby substantiating the claim that longer degumming times progressively degrade the protein. GPC results confirmed this conclusion.
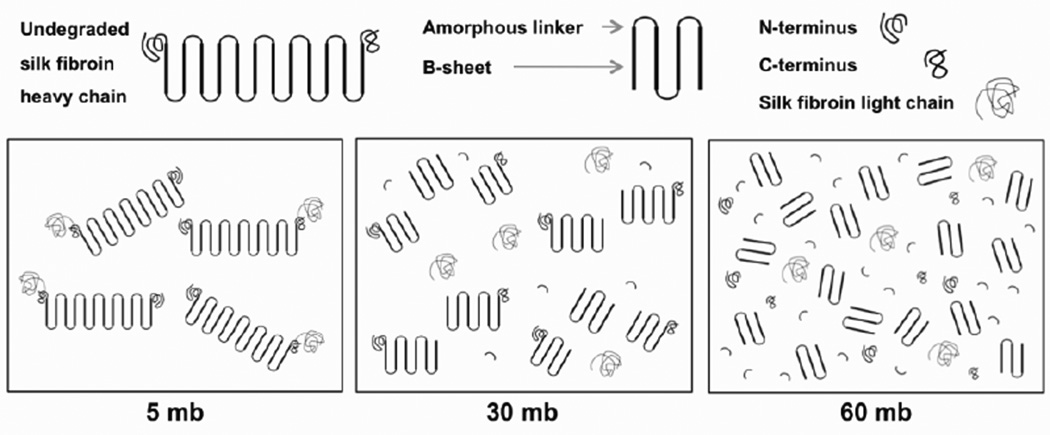
A change in β-sheet content between the 5, 30, and 60 mb samples would indicate that degumming affected the capacity for silk fibroin protein to self-assemble and transition from the silk I to silk II conformation. Since polymer crystallization can affect macroscopic structural properties, it was important to elucidate the relationship between degumming and silk fibroin β- sheet content. β-sheet content between the silk fibroin samples was expected to change because of differences in molecular weight. Small protein fragments can assemble more easily than larger ones, favoring an increase in β-sheet content with longer degumming times. Alternatively, if degumming caused degradation within the β-sheet region of the protein, we might instead expect β-sheet content to decrease with longer degumming. However, although longer degumming durations increasingly degraded the silk fibroin protein there was no effect on β-sheet content. This result suggests that the silk fibroin amino acid Gly–X repeating hexamer domains, which are the domains involved in the β-sheet secondary structure, are not affected during the degumming process. The non-repetitive, amorphous regions and N- and C-terminal domains are therefore the likely location of silk fibroin degradation during degumming (Figure 9). As illustrated in our model, the β-sheet content is the same between silk samples but due to degradation of the amorphous linkers and N- and C- terminal domains, the β-sheets are less confined in terms of relative organization in the 30 mb and 60 mb samples. For future work, it would be useful to understand the kinetics of β-sheet crystallization as a function of the degumming process, related to the molecular weight and fragment sequence. In a previous model for the kinetics of silk fibroin β-sheet crystallization, it was shown that the process was controlled by geometric patterns of phase-separated crystallizable and uncrystallizable regions of the silk fibroin protein chains26. Deviations from the model as a result of degumming duration may further elucidate silk fibroin fragment interactions and folding into the β-sheet crystal under different degraded states.
Despite having similar β-sheet content, the 5, 30, and 60 mb samples showed different thermal degradation profiles and scaffold assembly. In a previous study, silk films that contained higher β-sheet content were more resistant to thermal degradation27. In this study we find that thermal degradation properties are also dependant on molecular weight distribution. This is inconsistent with thermal degradation via random chain scission, which would be expected to occur in the same fashion regardless of molecular weight. All other things being equal, this implies that thermal degradation of the silk proteins occurs primarily via end-group initiated depolymerization. Nevertheless, further study is needed to confirm that no other confounding factors exist.
The differences in scaffold assembly may be due to differences in how the silk fibroin fragments fold and interact with other fragments during the 48 hours of incubation with NaCl crystals. The fact that the 60 mb sponge was much lower in porosity is an indication that smaller protein fragments are unable to assemble into robust macroscopic structures under the scaffold forming conditions that were used. The observed difference in 3D porous scaffold assembly, pore morphology, pore size, and porosity suggests that degumming needs to be highly controlled in order to provide reproducible materials. Previous work in our lab showed that tissue formation within silk fibroin scaffolds is highly dependent on scaffold structural properties such as mechanical stiffness, surface roughness, and porosity28. Logically, this means that slight changes in degumming may significantly affect tissue in-growth upon implantation. Another issue to consider is the effect of degumming on the long-term performance and degradation of silk fibroin in vivo. Wang et al. compared aqueous-derived and hexafluoroisopropanol (HFIP)-derived silk sponges and found that after implantation the aqueous-derived scaffold degraded sooner than the HFIP-derived sponges. With respect to the results presented in this study, differences in structural integrity after implantation may arise from differences in molecular weight. For example, polymer materials with lower molecular weights are more susceptible to enzymatic degradation and hydrolysis29. With these scaffold design criteria in mind, degumming could be used to tailor morphologic characteristics and the degradation profile of silk fibroin biomaterials to match the specific needs for the tissue being regenerated.
Effect of silk fibroin degradation on cell viability and macrophage activation
Based on previously published results hMVEC viability was expected to decrease with increased duration of degumming20. Characterizing hMVEC viability was of particular interest because this cell type is a key mediator in inflammatory responses30. A previous study reported that silk fibroin degummed with 0.05% Na2CO3 for 5, 10, and 30 minutes promoted human skin fibroblast cell viability compared to TCP controls. However, when the degumming time was increased to 60 minutes, cell viability was significantly hindered20. In the present study hMVEC viability followed a similar trend in that increased degumming decreased cell viability, however none of the silk fibroin samples promoted cell viability compared to the TCP control. This may be due to the fact that this study used a different cell type for their viability studies. Tsubouchi et al. (2003) reported that the decrease in cell viability on silk films degummed for longer durations was due to the release of small protein fragments. Based on our model of protein degradation we believe that differences in cell viability may be more in part due to differences in how proteins adsorb to the silk film surface, thereby affecting cell adhesion and proliferation. The way in which proteins adsorb to biomaterial surfaces has been shown previously to affect subsequent cellular responses31,32. Furthermore, protein adsorption has also been shown to be mediated by biomaterial surface charge33,34,35,36. For example, bovine serum albumin (BSA) has shown a much higher affinity for binding to hydrophobic surfaces but a tendency to denature following adsorption; on hydrophilic surfaces, however, its structure is preserved35. In our case, the short amorphous linker and N- and C- terminal regions contain a higher percentage of hydrophilic amino acid residues compared to the hydrophobic β-sheet crystal region. Upon the disruption of these regions as a result of longer degumming, the silk film surface charges are redistributed and may thereby change how proteins in the cell culture media adsorb to the silk film. Elucidating the mechanisms by which proteins adsorb and cells adhere to silk film surfaces is an important topic for further investigation.
Based on results published previously, the 5 mb sample was expected to promote more TNF-α release in conjunction with the +LPS condition compared to the 30 mb and 60 mb samples, due to the presence of residual sericin37. In this previous study, in which a murine macrophage cell line was used, sericin-coated silk fibers elicited a macrophage response when simultaneously treated with +LPS. However, the results we present here, in which a human macrophage cell line was used, indicate that silk fibroin degummed for five minutes elicits similar macrophage activation under the –LPS and +LPS conditions as the 30 and 60 mb samples. Since residual sericin on silk fibers is has been correlated to inflammation38,39,40, our result suggests that 5, 30, and 60 mb silk fibroin have similar low sericin content. The correlation of these results with the extent of sericin removal was therefore further investigated.
Detecting residual sericin and silk fibroin degradation
The residual sericin content in silk fibroin solutions was an important factor in assessing biocompatibility of silk fibroin-based technologies, based on the data above as well as prior literature. While sericin on its own has been shown to be biocompatible and antiinflammatory 41,42,43,44, the biocompatibility of silk fibroin-sericin combinations have only been partly characterized37,45. In vitro skin fibroblasts cultured on silk-sericin films showed decreased cell attachment and growth rates versus both 100% sericin and 100% silk films, which had similarly higher cell attachment and growth rates45. Furthermore, the presence of sericin on silk sutures caused in vivo Type I allergic responses38,39,40. These examples support the need to develop a method for efficiently detecting residual sericin in silk fibroin solutions. We report the use of the molar ratio of serine to glycine as determined by amino acid composition analysis to detect residual sericin content in silk fibroin solutions. Silk fibroin solutions that contained 1.0%, 0.1%, and 0% w/w residual sericin had a molar ratio of 0.19,while a silk fibroin solution that contained 10% w/w residual sericin had a molar ratio of 0.20, thus the lowest amount of sericin that can be detected with this method is between 1.0% and 10% w/w. The mole ratio of serine to glycine in the 5, 30, and 60 mb samples was 0.19 +/− 0.005, indicating all samples contained 10% w/w or less of sericin. The use of amino acid composition analysis to measure sericin content in silk fibroin solutions with a detection limit between 1.0% and 10% w/w sericin content was therefore developed. Previous studies have reported silk fibroin and sericin amino acid compositions13,15,46,47 but none of these studies used composition analysis to measure residual sericin content. Other reported methods for detecting residual sericin involve dyes that specifically target the sericin and not the fibroin, however, these dyes show poor differentiation capabilities48. Additionally, dyeing methods can only offer qualitative information at a single point along the silk fiber. In contrast we present a tool for quantitatively analyzing the amino acid composition of a silk fibroin solution containing an unknown content of sericin, offering a predictive method to assess biocompatibility of silk fibroin. However, more studies to correlate the in vitro data reported here with in vivo outcomes will be needed.
During the degumming process, the temperature and the alkalinity of the sodium carbonate solution degrades the silk fibroin protein through heat and hydrolysis18. The results we present in this paper show that degumming has a significant effect on cell culture viability and downstream scaffold formation, due to silk fibroin protein degradation. Slight changes in degumming duration could potentially compromise the reproducibility of silk fibroin solutions between batches. To ensure that the silk solution meets a target molecular weight distribution, it is important to have a method to quickly and reproducibly measure silk fibroin degradation before using the material in any downstream biomedical technology. In this study we report the use of fluorescence spectroscopy to detect changes in silk fibroin degradation. In previous studies our lab used fluorescence spectroscopy to detect changes in the β-sheet content, molecular orientation, and hydration of silk fibroin constructs49,50. These studies found the emission spectra of silk fibroin solutions to be distinct from silk fibroin hydrogels, sponges, and fibers due to differences in crosslinking and intermolecular and intra-molecular interactions. In particular, as silk from solution assembles into higher order structures the fluorescence contribution from tyrosine residues decreases due to increasing proximity to tryptophan residues (and the resultant resonance energy transfer) as β-sheet crystals form. In the present study we adapted this method to assess protein degradation of the silk fibroin solutions. The observed spectra consisted of a main peak at approximately 307 nm and a prominent shoulder in the 330 nm region of the spectrum. These features are attributed primarily to tyrosine and tryptophan fluorescence, respectively49. We found that silk solutions degummed for different durations have distinct emission spectra, resulting from different relative levels of tryptophan to tyrosine fluorescence. Specifically, the less degraded samples had higher tryptophan to tyrosine fluorescence than the highly degraded samples. That is, as silk fibroin degradation increased, the relative tryptophan fluorescence of the solution decreased. The analysis time is on the order of minutes, and the analysis requires only 1 mL of a 0.1% w/v solution, and can distinguish between 5, 30, and 60 mb samples with a standard error of 0.01. It is likely that highly degraded silk fibroin solution exhibits lower tryptophan to tyrosine fluorescence because the probability for fluorescence resonance energy transfer between these two residues decreases in the smaller peptide chains. Given the successful use of fluorescence spectroscopy to detect changes in silk protein conformation it seems logical that fluorescence spectroscopy can also detect changes in silk protein degradation. This approach in essence provides a direct method for screening silk fibroin solutions for their molecular weight characteristics based on endogenous optical signals.
Conclusions
The effect of degumming on silk fibroin was characterized based on molecular weight, materials processing and in vitro biocompatibility. Increased degumming duration degraded silk fibroin causing the molecular weight to decrease. However, differences in degumming time, and the differences this imparts to protein molecular weight, do not affect in vitro macrophage activation. Although molecular weight was not shown to affect β-sheet crystal content, it did have a significant effect on thermal stability and three-dimensional scaffold assembly. These results therefore warranted the development of a method able to quickly monitor and assess the effect of degumming duration on silk fibroin molecular weight. We report the use of fluorescence spectroscopy to rapidly and reproducibly measure the extent of protein degradation within a silk fibroin solution. To ensure biocompatibility of silk fibroin, a method was developed to quantitatively measure residual sericin content in silk fibroin solutions based on amino acid content. Methods that quantitatively measure residual sericin content and silk fibroin degradation, two important tools for monitoring the quality and reproducibility of silk fibroin solutions, are presented in detail. These will be indispensible tools during silk fibroin biomedical product development.
Figure 9.
Silk fibroin degradation model. The data indicate that silk fibroin degradation occurs in the amorphous linkers and N- and C- terminus regions and not in the crystalline region, as would be expected. The model illustrates that β-sheet content stays constant while the degraded protein fragments in the 30 mb and 60 mb samples reassemble, which can account for the differences observed in thermal degradation, scaffold morphology, and cell viability (mb = minutes boiled).
Acknowledgements
We thank the NIH Tissue Engineering Resource Center (TERC) (NIH P41 EB002520) and the National Science Foundation Graduate Research Fellowship Program (NSF DGE 0806676) for support. The authors also thank Myron Crawford and Fernando Pineda for their assistance with the amino acid composition analysis, and Marc Keirstead and Tuna Yucel for assistance with gel permeation chromatography. This work was performed in part at the Center for Nanoscale Systems (CNS), a member of the National Nanotechnology Infrastructure Network (NNIN), which is supported by the National Science Foundation under NSF award no. ECS-0335765. CNS is part of the Faculty of Arts and Sciences at Harvard University.
References
- 1.Vepari C, Kaplan DL. Silk as a biomaterial. Prog Polym Sci. 2007;32:991–1007. doi: 10.1016/j.progpolymsci.2007.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Matthews JM. The textile fibres: their physical, microscopical and chemical properties. New York: J. Wiley & sons; 1913. pp. 63–96. [Google Scholar]
- 3.Altman GH, Diaz F, Jakuba C, Calabro T, Horan RL, Chen J, Lu H, Richmond J, Kaplan DL. Silk-based biomaterials. Biomaterials. 2003;24:401–416. doi: 10.1016/s0142-9612(02)00353-8. [DOI] [PubMed] [Google Scholar]
- 4.Yamada H, Nakao H, Takasu Y, Tsubouchi K. Preparation of undegraded native molecular fibroin solution from silkworm cocoons. Mater Sci Eng, C. 2001;14:41–46. [Google Scholar]
- 5.Yamaguchi K, Kikuchi Y, Takagi T, Kikuchi A, Oyama F, Shimura K, Mizuno S. Primary structure of the silk fibroin light chain determined by cDNA sequencing and peptide analysis. J Mol Biol. 1989;210:127–139. doi: 10.1016/0022-2836(89)90295-7. [DOI] [PubMed] [Google Scholar]
- 6.Zhou CZ, Confalonieri F, Jacquet M, Perasso R, Li ZG, Janin J. Silk fibroin: structural implications of a remarkable amino acid sequence. Proteins: Struct, Funct, Bioinf. 2001;44:119–122. doi: 10.1002/prot.1078. [DOI] [PubMed] [Google Scholar]
- 7.Horan RL, Antle K, Collette AL, Wang Y, Huang J, Moreau JE, Volloch V, Kaplan DL, Altman GH. In vitro degradation of silk fibroin. Biomaterials. 2005;26:3385–3393. doi: 10.1016/j.biomaterials.2004.09.020. [DOI] [PubMed] [Google Scholar]
- 8.Mori K, Tanaka K, Kikuchi Y, Waga M, Waga S, Mizuno S. Production of a chimeric fibroin light-chain polypeptide in a fibroin secretion-deficient naked pupa mutant of the silkworm Bombyx mori. J Mol Biol. 1995;251:217–228. doi: 10.1006/jmbi.1995.0429. [DOI] [PubMed] [Google Scholar]
- 9.Mita K, Ichimura S, James TC. Highly repetitive structure and its organization of the silk fibroin gene. J Mol Evol. 1994;38:583–592. doi: 10.1007/BF00175878. [DOI] [PubMed] [Google Scholar]
- 10.Ha SW, Gracz HS, Tonelli AE, Hudson SM. Structural study of irregular amino acid sequences in the heavy chain of Bombyx mori silk fibroin. Biomacromolecules. 2005;6:2563–2569. doi: 10.1021/bm050294m. [DOI] [PubMed] [Google Scholar]
- 11.Valluzzi R, Gido SP, Zhang W, Muller WS, Kaplan DL. Trigonal crystal structure of bombyx mori silk incorporating a threefold helical chain conformation found at the air-water interface. Macromolecules. 1996;29:8606–8614. [Google Scholar]
- 12.Sprague KU. Bombyx mori silk proteins: Characterization of large polypeptides. Biochemistry. 1975;14:925–931. doi: 10.1021/bi00676a008. [DOI] [PubMed] [Google Scholar]
- 13.Takasu Y, Yamada H, Tsubouchi K. Isolation of three main sericin components from the cocoon of the silkworm, bombyx mori. Biosci Biotechnol Biochem. 2002;66:2715–2718. doi: 10.1271/bbb.66.2715. [DOI] [PubMed] [Google Scholar]
- 14.Takasu Y, Yamada H, Saito H, Tsubouchi K. Characterization of bombyx mori sericins by the partial amino acid sequences. J Insect Biotechnol Sericol. 2005;74:103–109. [Google Scholar]
- 15.Dash R, Mukherjee S, Kundu S. Isolation, purification and characterization of silk protein sericin from cocoon peduncles of tropical tasar silkworm, antheraea mylitta. Int J Biol Macromol. 2006;38:255–258. doi: 10.1016/j.ijbiomac.2006.03.001. [DOI] [PubMed] [Google Scholar]
- 16.Teramoto H, Kakazu A, Asakura T. Native structure and degradation pattern of silk sericin studied by 13C NMR spectroscopy. Macromolecules. 2006;39:6–8. [Google Scholar]
- 17.Jiang P, Liu H, Wang C, Wu L, Huang J, Guo C. Tensile behavior and morphology of differently degummed silkworm (Bombyx mori) cocoon silk fibres. Mater Lett. 2006;60:919–925. [Google Scholar]
- 18.Freddi G, Mossotti R, Innocenti R. Degumming of silk fabric with several proteases. J Biotechnol. 2003;106:101–112. doi: 10.1016/j.jbiotec.2003.09.006. [DOI] [PubMed] [Google Scholar]
- 19.Sofia S, McCarthy MB, Gronowicz G, Kaplan DL. Functionalized silk-based biomaterials for bone formation. J Biomed Mater Res. 2001;54:139–148. doi: 10.1002/1097-4636(200101)54:1<139::aid-jbm17>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 20.Tsubouchi K, Nakao H, Igarashi Y, Takasu Y, Yamada H. Bombyx mori fibroin enhanced the proliferation of cultured human skin fibroblasts. J Insect Biotechnol Sericol. 2003;72:65–69. [Google Scholar]
- 21.Hu X, Kaplan D, Cebe P. Determining beta-sheet crystallinity in fibrous proteins by thermal analysis and infrared spectroscopy. Macromolecules. 2006;39:6161–6170. [Google Scholar]
- 22.Kim UJ, Park J, Joo Kim H, Wada M, Kaplan DL. Three-dimensional aqueous-derived biomaterial scaffolds from silk fibroin. Biomaterials. 2005;26:2775–2785. doi: 10.1016/j.biomaterials.2004.07.044. [DOI] [PubMed] [Google Scholar]
- 23.Minoura N, Tsukada M, Nagura M. Fine structure and oxygen permeability of silk fibroin membrane treated with methanol. Polymer. 1990;31:265–269. [Google Scholar]
- 24.Tsuchiya S, Yamabe M, Yamaguchi Y, Kobayashi Y, Konno T, Tada K. Establishment and characterization of a human acute monocytic leukemia cell line (THP-1) Int J Cancer. 1980;26:171–176. doi: 10.1002/ijc.2910260208. [DOI] [PubMed] [Google Scholar]
- 25.Meinel L, Hofmann S, Karageorgiou V, Kirker-Head C, McCool J, Gronowicz G, Zichner L, Langer R, Vunjak-Novakovic G, Kaplan DL. The inflammatory responses to silk films in vitro and in vivo. Biomaterials. 2005;26:147–155. doi: 10.1016/j.biomaterials.2004.02.047. [DOI] [PubMed] [Google Scholar]
- 26.Hu X, Lu Q, Kaplan DL, Cebe P. Microphase separation controlled β-sheet crystallization kinetics in fibrous proteins. Macromolecules. 2009;42:2079–2087. [Google Scholar]
- 27.Gil ES, Frankowski DJ, Bowman MK, Gozen AO, Hudson SM, Spontak RJ. Mixed protein blends composed of gelatin and bombyx mori silk fibroin: Effects of solvent-induced crystallization and composition. Biomacromolecules. 2006;7:728–735. doi: 10.1021/bm050622i. [DOI] [PubMed] [Google Scholar]
- 28.Wang Y, Rudym DD, Walsh A, Abrahamsen L, Kim HJ, Kim HS, Kirker-Head C, Kaplan DL. In vivo degradation of three-dimensional silk fibroin scaffolds. Biomaterials. 2008;29:3415–3428. doi: 10.1016/j.biomaterials.2008.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cao Y, Wang B. Biodegradation of silk biomaterials. Int J Mol Sci. 2009;10:1514. doi: 10.3390/ijms10041514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hakimi O, Gheysens T, Vollrath F, Grahn MF, Knight DP, Vadgama P. Modulation of cell growth on exposure to silkworm and spider silk fibers. J Biomed Mater Res, Part A. 2010;92:1366–1372. doi: 10.1002/jbm.a.32462. [DOI] [PubMed] [Google Scholar]
- 31.Anselme K. Osteoblast adhesion on biomaterials. Biomaterials. 2000;21:667–681. doi: 10.1016/s0142-9612(99)00242-2. [DOI] [PubMed] [Google Scholar]
- 32.Wilson CJ, Clegg RE, Leavesley DI, Pearcy MJ. Mediation of biomaterial-cell interactions by adsorbed proteins: a review. Tissue Eng. 2005;11:1–18. doi: 10.1089/ten.2005.11.1. [DOI] [PubMed] [Google Scholar]
- 33.Shelton R, Rasmussen A, Davies J. Protein adsorption at the interface between charged polymer substrata and migrating osteoblasts. Biomaterials. 1988;9:24–29. doi: 10.1016/0142-9612(88)90065-8. [DOI] [PubMed] [Google Scholar]
- 34.Altankov G, Groth T. Reorganization of substratum-bound fibronectin on hydrophilic and hydrophobic materials is related to biocompatibility. J Mater Sci Mater Med. 1994;5:732–737. [Google Scholar]
- 35.Roach P, Farrar D, Perry CC. Interpretation of protein adsorption: surface-induced conformational changes. J Am Chem Soc. 2005;127:8168–8173. doi: 10.1021/ja042898o. [DOI] [PubMed] [Google Scholar]
- 36.Michiardi A, Aparicio C, Ratner BD, Planell JA, Gil J. The influence of surface energy on competitive protein adsorption on oxidized NiTi surfaces. Biomaterials. 2007;28:586–594. doi: 10.1016/j.biomaterials.2006.09.040. [DOI] [PubMed] [Google Scholar]
- 37.Panilaitis B, Altman GH, Chen J, Jin HJ, Karageorgiou V, Kaplan DL. Macrophage responses to silk. Biomaterials. 2003;24:3079–3085. doi: 10.1016/s0142-9612(03)00158-3. [DOI] [PubMed] [Google Scholar]
- 38.Wen CM, Ye ST, Zhou LX, Yu Y. Silk-induced asthma in children: a report of 64 cases. Ann Allergy. 1990;65:375–378. [PubMed] [Google Scholar]
- 39.Hollander DH. Interstitial cystitis and silk allergy. Med Hypotheses. 1994;43:155–156. doi: 10.1016/0306-9877(94)90142-2. [DOI] [PubMed] [Google Scholar]
- 40.Zaoming W, Codina R, Fernandez-Caldas E, Lockey R. Partial characterization of the silk allergens in mulberry silk extract. J Invest Allerg Clin. 1996;6:237–241. [PubMed] [Google Scholar]
- 41.Terada S, Nishimura T, Sasaki M, Yamada H, Miki M. Sericin, a protein derived from silkworms, accelerates the proliferation of several mammalian cell lines including a hybridoma. Cytotechnology. 2002;40:3–12. doi: 10.1023/A:1023993400608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Terada S, Sasaki M, Yanagihara K, Yamada H. Preparation of silk protein sericin as mitogenic factor for better mammalian cell culture. J Biosci Bioeng. 2005;100:667–671. doi: 10.1263/jbb.100.667. [DOI] [PubMed] [Google Scholar]
- 43.Tsubouchi K, Igarashi Y, Takasu Y, Yamada H. Sericin enhances attachment of cultured human skin fibroblasts. Biosci Biotechnol Biochem. 2005;69:403–405. doi: 10.1271/bbb.69.403. [DOI] [PubMed] [Google Scholar]
- 44.Aramwit P, Sangcakul A. The effects of sericin cream on wound healing in rats. Biosci Biotechnol Biochem. 2007;71:2473–2477. doi: 10.1271/bbb.70243. [DOI] [PubMed] [Google Scholar]
- 45.Minoura N, Aiba SI, Gotoh Y, Tsukada M, Imai Y. Attachment and growth of cultured fibroblast cells on silk protein matrices. J Biomed Mater Res. 1995;29:1215–1221. doi: 10.1002/jbm.820291008. [DOI] [PubMed] [Google Scholar]
- 46.Takasu Y, Yamada H, Tamura T, Sezutsu H, Mita K, Tsubouchi K. Identification and characterization of a novel sericin gene expressed in the anterior middle silk gland of the silkworm bombyx mori. Insect Biochem Mol Biol. 2007;37:1234–1240. doi: 10.1016/j.ibmb.2007.07.009. [DOI] [PubMed] [Google Scholar]
- 47.Wu JH, Wang Z, Xu SY. Preparation and characterization of sericin powder extracted from silk industry wastewater. Food Chem. 2007;103:1255–1262. [Google Scholar]
- 48.Gulrajani M. Degumming of silk. Rev Prog Color Relat Top. 1992;22:79–89. [Google Scholar]
- 49.Georgakoudi I, Tsai I, Greiner C, Wong C, DeFelice J, Kaplan D. Intrinsic fluorescence changes associated with the conformational state of silk fibroin in biomaterial matrices. Biomacromolecules. 2004;5:718–726. doi: 10.1364/oe.15.001043. [DOI] [PubMed] [Google Scholar]
- 50.Rice WL, Firdous S, Gupta S, Hunter M, Foo CWP, Wang Y, Kim HJ, Kaplan DL, Georgakoudi I. Non-invasive characterization of structure and morphology of silk fibroin biomaterials using non-linear microscopy. Biomaterials. 2008;29:2015–2024. doi: 10.1016/j.biomaterials.2007.12.049. [DOI] [PMC free article] [PubMed] [Google Scholar]