Abstract
Cyclin dependent kinase (CDK) inhibitors, such as flavopiridol, demonstrate significant single-agent activity in chronic lymphocytic leukemia (CLL), but the mechanism of action in these nonproliferating cells is unclear. Here we demonstrate that CLL cells undergo autophagy after treatment with therapeutic agents, including fludarabine, CAL-101, and flavopiridol as well as the endoplasmic reticulum (ER) stress-inducing agent thapsigargin. The addition of chloroquine or siRNA against autophagy components enhanced the cytotoxic effects of flavopiridol and thapsigargin, but not the other agents. Similar to thapsigargin, flavopiridol robustly induces a distinct pattern of ER stress in CLL cells that contributes to cell death through IRE1-mediated activation of ASK1 and possibly downstream caspases. Both autophagy and ER stress were documented in tumor cells from CLL patients receiving flavopiridol. Thus, CLL cells undergo autophagy after multiple stimuli, including therapeutic agents, but only with ER stress mediators and CDK inhibitors is autophagy a mechanism of resistance to cell death. These findings collectively demonstrate, for the first time, a novel mechanism of action (ER stress) and drug resistance (autophagy) for CDK inhibitors, such as flavopiridol in CLL, and provide avenues for new therapeutic combination approaches in this disease.
Introduction
Chronic lymphocytic leukemia (CLL) is a progressive B-cell malignancy that demonstrates significant heterogeneity with respect to biology as well as progression-free and overall survival.1 Because of the lack of survival advantage with early treatment, therapy for CLL is delayed until symptoms develop.2 The lack of curative and effective therapy for all genetic subsets of CLL has fueled investigation of new therapeutic approaches for this disease. A notable advancement in this effort has been the introduction of cyclin-dependent kinase (CDK) inhibitors. Flavopiridol was the first broad inhibitor of CDK enzymes3,4 that entered clinical development for CLL. Preclinical studies by several groups demonstrated that flavopiridol mediates potent apoptosis in CLL cells that occurs independent of del(17p13.1) or loss of p53 function.5,6 Further studies in CLL and other leukemias suggested that flavopiridol mediates its effects through activation of p38MAPK or JNK1, or through inhibition of CDK9 and RNA transcription with subsequent depletion of short half-life antiapoptotic proteins.4,7,8 These preclinical studies prompted clinical investigation of flavopiridol in relapsed and refractory CLL. Significant activity was observed in up to 50% of patients with refractory CLL, with a dose-limiting side effect of hyperacute tumor lysis syndrome.9,10 This effect is not observed with any other CLL therapy to date, suggesting that flavopiridol uses a novel mechanism of action compared with currently available treatments. Although down-regulation of short half-life pro-survival proteins, such as Mcl-1, represents a viable hypothesis,11 detailed characterization of early events in leukemia cells obtained serially from CLL patients receiving flavopiridol demonstrated no correlation of Mcl-1 changes with treatment response,12 most likely because of the complexity of pathways targeted by flavopiridol in addition to Mcl-1. This prompted our group to hypothesize that this agent uses an alternative cytotoxic mechanism.
In studying the mechanism of action of flavopiridol, our group demonstrated by electron microscopy that in vitro treatment of CLL patient cells with flavopiridol promoted the appearance of double-membrane structures suggestive of autophagy.13 Autophagy is an intracellular process that plays a role in normal cell homeostasis. In this process, misfolded proteins, damaged/aged organelles, or other intracellular components are sequestered inside a double-membrane vesicle called an “autophagosome,” which then fuses with lysosomes to allow degradation of its contents.14,15 In both normal and transformed cells, autophagy occurs after multiple different stimuli, including starvation, endoplasmic reticulum (ER) stress, reactive oxygen species stress, or pharmacologic inhibition of mammalian target of rapamycin (mTOR) or class I phosphatidylinositol 3-kinase (class I PI3K).16,17 After activation of this pathway, autophagosome formation is initiated by class III PI3K, which forms a complex with Beclin-1 (ATG6) and Barkor (Beclin 1-associated autophagy related key regulator, homolog of ATG14).16,18 ATG4 cleaves a carboxi-termini arginine off of LC3 I (16-kDa isoform), thus exposing a glycine residue where phosphatidylethanolamine binds to produce LC3 II (14-kDa isoform); LC3-II is activated by ATG7 and transferred to ATG3, which conjugates it to phosphatidylethanolamine. Subsequently, LC3-II is recruited to the forming autophagosome membrane with the help of the ATG12-ATG5 complex, contributing to the expansion of the autophagosome. Finally, autophagosome-lysosome fusion results in degradation of the enclosed cellular proteins.19 In the majority of settings autophagy is protective,20 but with excessive autophagy, an autophagocytic type II death via necrosis has been reported.21,22 However, the decision point in this process is unclear, and no investigation has examined in one tumor type the effect of multiple autophagy-inducing stimuli on cell death. Here we demonstrate that autophagy induced by ER stress, but not other mechanisms, protects CLL cells from cell death and that the CDK inhibitor flavopiridol robustly induces both ER stress and a protective autophagic response. These observations prompted detailed study of ER stress as a novel mechanism of CDK inhibitor-mediated cell death. These studies demonstrate, for the first time, that flavopiridol induces robust ER stress in vitro and in patients and that subsequent cell death is dependent on IRE1-induced ASK1 activation and downstream caspase 4. Collectively, these findings point to a new mechanism of action of flavopiridol and possibly other CDK inhibitors as well as strategies to overcome resistance to these treatments.
Methods
Patients, cell separation, culture conditions, and reagents
For both in vivo and in vitro studies, written, informed consent was obtained in accordance with the Declaration of Helsinki to procure cells from patients with previously diagnosed CLL as defined by the modified National Cancer Institute criteria.23 Approval was obtained from The Ohio State University (OSU) institutional review board. CD19+ cells from CLL patients, and normal volunteers were selected and maintained in culture as previously described by our group.24 Flavopiridol was obtained from the National Cancer Institute. Rapamycin was purchased from EMD-Calbiochem. Chloroquine, thapsigargin, F-ara-A (9-β-D-arabinofuranosyl-2-fluoroadenine 5′-phosphate), chlorambucil, and tunicamycin were obtained from Sigma-Aldrich, rituximab from Genentech, and CAL-101 from Calistoga Pharmaceuticals.
Confocal immunofluorescence microscopy of fixed cells
See supplemental Methods (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Quantification of immunofluorescence data.
LC3 intensity and number of dots per cell were both assessed in the fluorescence images. We used Metamorph Version 7.0 for LC3 intensity; and for cell counts and measurement of correlation index, the program used is based on MatLab Version 7.9.0.529 (R2009b) software, and it was developed by members of Department of Biomedical Informatics, OSU. For more details, see supplemental Methods.
Immunoblot analysis and coimmunoprecipitation
Proteins extracted from whole-cell lysates (50 μg/lane) were separated on polyacrylamide gels and transferred on nitrocellulose membrane. Antibodies used for immunoblots included LC3, IRE1α, TRAF2, PI3K III, ATG4C, ATG5-12, UVRAG, Beclin-1, GRP78 (Cell Signaling), GAPDH, Nrf2, ASK1, ULK1 (Santa Cruz Biotechnology), and Bif-1, ATG14-Barkor (Sigma-Aldrich). Species-appropriate secondary antibodies were obtained from BioRad. After antibody incubations, proteins were detected using a chemiluminescent substrate (BioRad). For coimmunoprecipitations, TrueBlot beads (eBioscience) and One-hour IP Western kit (GenScript) were used according to manufacturer protocols. Quantification was done using FluorChem Q (Alpha Innotech).
Caspase activity assays
The presence of active caspase enzymes was determined by the 7-amino-4-trifluoromethylcoumarin assay as previously described.25 For more details, see supplemental Methods.
Cell viability
Percent of live cells was determined by staining with annexin V–FITC and propidium iodide (PI), followed by flow cytometry using a Beckman-Coulter Cytomics FC500 cytometer. For more details, see supplemental Methods.
siRNA experiments
CLL cells were suspended in buffer V (Amaxa kit; Lonza), and nucleofection was performed using program U016 (5 μg siRNA and 1 × 107 cells per cuvette). Scrambled siRNA and siRNAs to ATG5, ATG7, and ASK1 were purchased from Sigma-Aldrich. Cells were immediately suspended in warm, complete media supplemented with 10% human serum, and RNA was extracted after 36 hours. For CDK1 and CDK5, siRNAs were purchased from Applied Biosystems. Transfections were done using 5nM siRNA per cuvette, and RNA was harvested at 48 hours.
ChIP assays
EZ Magna ChIP kit from Millipore was used according to the protocol suggested by the manufacturer. In brief, cells were treated with 37% formaldehyde to ensure crosslinking of proteins to DNA. Cells were lysed and sonicated to shear the chromatin to fragments of approximately 500 bp. Sheared chromatin was incubated with ATF6 antibody (LifeSpan Biosciences), followed by precipitation, washing, and reversing of the crosslinks. Purified DNA was then analyzed by real-time quantitative PCR using primers to the GRP78 promoter region.26 Primers were designed using Primer 3 software and purchased from IDT.
PCR and real-time quantitative PCR
RNA was extracted using TRIzol reagent (Invitrogen) and purified using QIAGEN RNeasy columns (QIAGEN). cDNA was prepared with SuperScript First-Strand Synthesis System (Invitrogen). Real-time PCR was performed using primers purchased from Applied Biosystems. Detection was performed using an ABI Prism 7700 sequence detection system (Applied Biosystems). Average relative expression (treatment compared with vehicle) was normalized to the internal control gene CD52. Primers to detect spliced XBP127 were designed using Primer Version 3 software and purchased from IDT. Samples were analyzed on 10% acrylamide gels.
Statistical analyses
All analyses were performed using SAS/STAT Version 9.2 (SAS Institute). See supplemental Methods for more details.
Results
CLL cells express autophagy-related proteins and are susceptible to autophagy induced by starvation or pharmacologic agents
Given the absence of reports on autophagy in CLL, we first examined whether CLL cells express the critical components of this pathway. As shown in supplemental Figure 1, autophagy pathway proteins, including ATG family members, are clearly expressed in primary CLL cells at levels comparable with normal B cells. ATG4C, UVRAG, and ULK1 were increased in CLL samples.
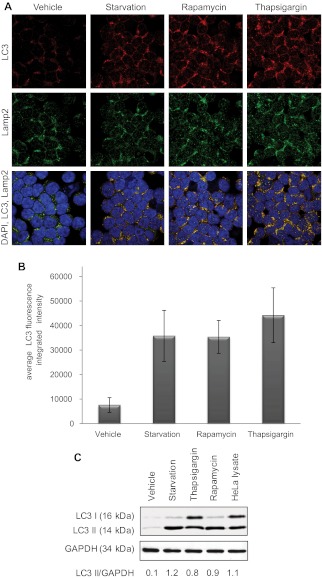
We next determined whether CLL cells undergo autophagy with well-characterized initiators of this process. As shown by confocal fluorescence microscopy in Figure 1A and numerically quantified in Figure 1B, CLL patient samples (n = 6) undergoing 4 hours of starvation or 4 hours of incubation with thapsigargin or rapamycin show significant (P < .0001 treatment vs vehicle-treated for all conditions) increase in LC3 aggregation, indicating increased autophagosome formation. The anti-LC3 antibody used here to detect autophagosomes is the same as the one used next for the immunoblot experiment; it detects uncleaved LC3B, LC3-I, and LC3-II. Formation of autophagosomes was also confirmed by immunoblot,28 showing increased LC3-II (Figure 1C). Autophagosome accumulation can be caused either by induction of their formation or by inhibition of fusion with lysosomes. In Figure 1A, autophagosome-lysosome fusion is demonstrated by colocalization of LC3 and the lysosome marker Lamp2, thereby suggesting full execution of autophagy flux. Collectively, these studies suggest that autophagy occurs in CLL cells after classic stimuli.
Figure 1.
Autophagy in CLL cells. (A) Confocal fluorescence microscopy for CLL cells untreated or after 4-hour starvation in HBSS or 4-hour treatment with rapamycin (5μM) or thapsigargin (1μM). LC3 (red) shows autophagosomes. Lamp2 (green) shows lysosomes. 4,6-diamidino-2-phenylindole (blue) shows nuclei. Images were collected with 60× objective and 4× optical zoom. (B) Quantification of LC3 immunofluorescence intensity. Integrated intensity was averaged in 5 microscopic fields relative to number of cells per field and then averaged in 6 CLL patients' cells. Differences from vehicle-treated were significant (P < .0001). (C) Immunoblot for LC3B I and II in CLL cells (representative of 4 experiments). GAPDH was used as loading control.
Inhibition of ER stress-induced autophagy, but not mTOR inhibitor-, CAL-101-, or fludarabine-induced autophagy, enhances CLL cell death
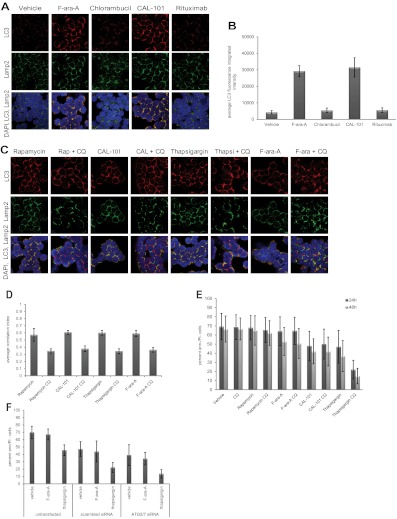
In normal and cancer cells, autophagy has been demonstrated to promote either survival or cell death, depending on context.29 We next sought to determine whether autophagy protects CLL cells from death, using autophagy stimuli examined in Figure 1 and also agents used to treat this disease. As shown in Figure 2A and quantified in Figure 2B, we observed by confocal immunofluorescence microscopy a significant (P < .0001; n = 6) induction of autophagy with either the metabolically active form of fludarabine F-ara-A or the PI3K-δ isoform inhibitor CAL-101,25 as evidenced by autophagosome accumulation and subsequent fusion with lysosomes. In contrast, neither chlorambucil nor rituximab, 2 other agents used in CLL therapy, produced this effect. This suggests that autophagy is not a uniform occurrence with all therapeutic agents used in CLL therapy.
Figure 2.
Specificity and role of autophagy in CLL cells. (A) Confocal fluorescence microscopy of CLL cells incubated 4 hours with or without F-ara-A (5μM), chlorambucil (20μM), CAL-101 (1μM), or rituximab (10 μg/mL). LC3 (red) shows autophagosomes. Lamp2 (green) shows lysosomes. 4,6-diamidino-2-phenylindole (blue) shows nuclei. Images were collected with 60× objective and 4× optical zoom, using Olympus Fluoview 1000 Laser Scanning Confocal microscope. (B) Quantification of LC3 fluorescence in CLL cells from (A; n = 6). LC3 fluorescence increases for F-ara-A and CAL-101 treatments were significant (P < .0001). (C) Confocal fluorescence microscopy of CLL cells treated 4 hours with agent alone versus agent + chloroquine (CQ, 0.5μM). (D) Quantification of correlation index for LC3 and Lamp2 in CLL cells (n = 5) treated with rapamycin (5μM), CAL-101 (1μM), thapsigargin (1μM), F-ara-A (5μM), with or without CQ (0.5μM). Correlations of LC3 and Lamp2 were significant (rapamycin, P = .0004; thapsigargin, P = .0012; CAL-101, P = .0002; F-ara-A, P = .0001). (E) Viability at 24 hours and 48 hours, shown as percent of annexin (ann) negative and PI-negative cells (n = 8) by flow cytometry. Cytotoxicity of thapsigargin was significantly enhanced by CQ addition (P = .0008). (F) Viability at 24 hours in CLL cells (n = 5) untransfected or transfected with scrambled siRNA or ATG5/7 siRNA and treated with F-ara-A (5μM) or thapsigargin (1μM). Thapsigargin cytotoxicity significantly increased in ATG5/7 siRNA samples (P = .0047).
To investigate the impact of autophagy in either promoting or protecting CLL cells from death, we used chloroquine, an agent that prevents fusion of the autophagosome with the lysosome.30 In these experiments, chloroquine did indeed inhibit autophagosome-lysosome fusion at 0.1-0.5μM, concentrations attainable in patients receiving this for malaria treatment31 (supplemental Figure 2). In Figure 2C and D, we demonstrate by confocal microscopy and quantification of images that, in CLL patient cells (n = 6), the addition of chloroquine to rapamycin (P = .0004), thapsigargin (P = .0012), CAL-101 (P = .0002), or fludarabine (P = .0001) prevents fusion of autophagosomes with lysosomes at 4 hours compared with each agent alone.
Interestingly, chloroquine neither enhanced nor diminished cell death promoted by rapamycin, CAL-101, or fludarabine, nor did it affect cell viability by itself. In contrast, cell death caused by the ER stress-inducing agent thapsigargin was significantly enhanced by the addition of chloroquine (P = .0008; Figure 2E). To confirm that the enhanced killing in the presence of chloroquine was not the result of an off-target effect, siRNA directed at ATG5/ATG7 was used. As shown in supplemental Figure 3, ATG5/ATG7 expression in CLL cells was significantly reduced (P < .0001) with ATG5/ATG7 siRNA. In these cells, cytotoxicity of thapsigargin was significantly (P = .0047) enhanced compared with cells transfected with the scrambled siRNA control (Figure 2F). Together, these studies indicate that autophagy induced with traditional stimuli and common CLL therapeutics does not serve as a mechanism of resistance in CLL cells. In contrast, ER stress-induced autophagy does antagonize death induced by thapsigargin and potentially other ER stress-inducing drugs.
Flavopiridol promotes autophagy in vitro and in vivo in CLL cells
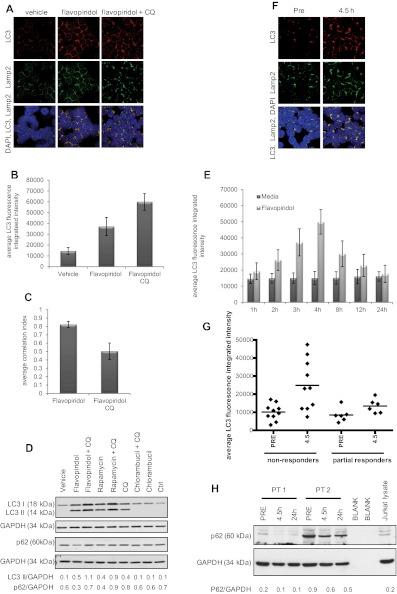
Prior studies by our group demonstrated the presence of double-membrane structures in flavopiridol-treated CLL cells, suggestive of autophagy.13 To confirm this, CLL cells (n = 6) were incubated 4 hours with or without flavopiridol and examined by immunofluorescence microscopy. Significant (P = .0001) formation of autophagosomes was observed in flavopiridol-treated cells compared with the vehicle control (Figure 3A-B). Concurrently, unrestricted autophagy flux was demonstrated by colocalization of LC3 and Lamp2 (quantified in Figure 3C). Induction of autophagy in flavopiridol-treated cells was also shown by immunoblot for LC3 and signaling adaptor protein p62/SQSTM1 (Figure 3D). p62/SQSTM1 is a target of the autophagic process32; thus, a decrease of this protein suggests induction of the autophagy flux. Consistent with this, p62/SQSTM1 levels are decreased in the presence of flavopiridol. However, it has been shown that levels of p62/SQSTM1 are not regulated exclusively by autophagy; therefore, p62/SQSTM1 cannot be taken as an absolute marker of autophagy flux.33 Autophagy flux was examined as previously described34 also in a time course with flavopiridol (Figure 3E), which revealed a significant (P = .0008) accumulation of autophagosomes (increased LC3 fluorescence intensity) up to 4 hours of treatment, followed by a decrease in LC3 intensity after 8 hours of treatment, indicative of autophagosome degradation by the lysosomal enzymes.
Figure 3.
Autophagy in CLL cells treated with the CDK inhibitor flavopiridol. (A) Confocal fluorescence microscopy for CLL cells untreated or treated 4 hours with flavopiridol (2μM) or flavopiridol + CQ (0.5μM). Samples were visualized as in Figures 1 and 2. (B) Quantification of LC3 immunofluorescence in CLL cells from panel A. LC3 fluorescence was increased by flavopiridol and further by the addition of CQ (P < .0001 for the increase with flavopiridol vs vehicle, the increase with flavopiridol + CQ vs vehicle, and the increase of flavopiridol + CQ vs flavopiridol). (C) Quantification of LC3-Lamp2 colocalization by average correlation index in 6 CLL samples treated with flavopiridol or flavopiridol + CQ as in panel A. The decreased colocalization with CQ was significant (P < .0001). (D) Immunoblot for LC3B I and II and p62, representative of 3 experiments. (E) Quantification of LC3 immunofluorescence in a time course with flavopiridol. After incubations, CLL cells (n = 3) were collected by cytospin at the specified time points. Samples collected after the 4-hour time point were washed and resuspended in media without flavopiridol for the remainder of the incubation. The increase in LC3 fluorescence flavopiridol compared with vehicle was significant (P = .001) at 3, 4, and 8 hours and decreased to levels comparable with vehicle alone at 12 and 24 hours. (F) Confocal fluorescence microscopy of CLL cells obtained from patients treated with flavopiridol. Samples were collected before treatment and at the end of flavopiridol infusion (4.5 hours) and visualized as in previous figures. (G) Average of LC3 fluorescence in CLL cells (n = 16) collected from patients treated with flavopiridol, showing LC3 intensity before treatment (PRE) and at the end of flavopiridol infusion (4.5 hours). Overall, LC3 intensity was 1.44 times higher in nonresponders compared with responders (95% CI, 1.07-1.95; P = .0183). Although the increase in intensity 4.5 hours versus before treatment was slightly higher for nonresponders compared with responders (fold change of 2.51 vs 1.82), it was not significantly higher (P = .3445). (H) Immunoblot for p62 in samples from CLL patients treated with flavopiridol. Samples were collected before treatment, at the end of flavopiridol infusion (4.5 hours), and at 24 hours from beginning of infusion. Data shown are representative of samples from 8 patients.
To determine whether flavopiridol-mediated autophagy is the result of inhibition of CDK versus an alternative kinase, we next examined CDK1 and CDK5, both targeted by flavopiridol and previously demonstrated to prevent autophagy initiation through phosphorylation of Vps34.35 CDK5 was found to be expressed in CLL patient cells, whereas CDK1 was below the level of detection both by real-time RT-PCR (CT values ∼ 32-34) and by immunoblot (supplemental Figure 4B). Cells transfected with siRNA to CDK5 (P = .0003) exhibited a decrease in CDK5 expression (supplemental Figure 4A-B) and an increase in autophagy (supplemental Figure 4C) compared with cells transfected with scrambled siRNA, indicating that flavopiridol-induced autophagy in CLL cells is mediated at least in part via CDK5 inhibition.
We next sought to determine whether induction of autophagy occurred in CLL patients treated with flavopiridol, using samples from patients enrolled on clinical trial OSU-0491 (NCI 7000; www.clinicaltrials.gov; #NCT00098371), a phase 2 multicenter study of flavopiridol administered as 30-minute loading dose followed by 4-hour continuous infusion in patients with previously treated CLL. In this trial, CLL cells were collected before and immediately after therapy (4.5 hours), day 1 (n = 16). By confocal fluorescence microscopy, both increased autophagosome formation (P = .0011) and fusion with lysosomes are observed (Figure 3F). Analysis of LC3 integrated intensity showed that overall, LC3 intensity was 1.44 times higher in nonresponders compared with responders (95% CI, 1.07-1.95; P = .0183). Although the increase in intensity in samples collected at 4.5 hours after beginning of flavopiridol infusion versus pretreatment was slightly higher for nonresponders compared with responders (fold change 2.51 vs 1.82), it was not significantly higher (P = .3445; Figure 3G). We also confirmed the increase in autophagy flux after flavopiridol therapy by p62/SQSTM1 immunoblot, which shows a decrease in protein expression in samples collected at 4.5 hours into the flavopiridol infusion (Figure 3H), supporting the observations in the CLL primary cells treated in vitro (Figure 3D). To confirm that the changes seen with LC3 and p62/SQSTM1 at protein level are indeed the result of differences in the rate of degradation and not because of transcription regulation, we tested LC3 and p62/SQSTM1 by RT-PCR (n = 5 for both in vitro and in vivo data), and we observed no significant changes relative to vehicle or pretreatment samples, as shown in supplemental Figure 8A through D. Collectively, these studies provide evidence that our in vitro observations are relevant to the clinical setting.
Inhibition of autophagy enhances flavopiridol cytotoxicity
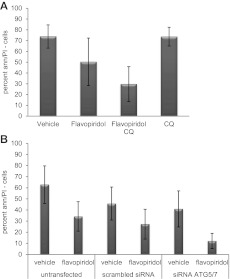
As shown Figure 4A, cotreatment with chloroquine significantly enhances the cytotoxicity of flavopiridol in CLL cells (n = 15; P = .001). Because chloroquine could affect other cell processes in addition to autophagy, we next tested whether knockdown of ATG5/ATG7 by siRNA enhanced flavopiridol cytotoxicity. After ATG5/ATG7 knockdown by transfection of cells with combination of siRNA to ATG5 and to ATG7 (supplemental Figure 3B), flavopiridol-mediated cell death was significantly increased (P = .004; Figure 4B) as previously observed with thapsigargin.
Figure 4.
Inhibition of flavopiridol-induced autophagy in CLL cells. (A) Viability of CLL cells (n = 15) treated with flavopiridol and CQ. Live cells are shown by percent of annexin-negative and PI-negative cells at 24 hours. Cells were incubated with flavopiridol for 4 hours; CQ was left on cells for 24 hours continuously. Viability decreased more with flavopiridol plus CQ versus flavopiridol alone (P = .001). (B) Viability in CLL samples (n = 10) untransfected or transfected with scrambled or combination ATG5/7 siRNA and then treated with flavopiridol (2μM) for 4 hours. Treatments began 24 hours after transfection. Percent of cells negative for annexin V and PI was measured 24 hours from flavopiridol treatment (48 hours after transfection). In flavopiridol-treated cells, viability was significantly lower in the presence of combination ATG5/7 siRNA versus the scrambled control (P = .004).
Flavopiridol induces ER stress in CLL cells in vitro and in vivo
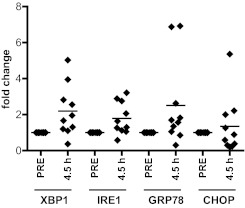
As flavopiridol-treated CLL cells show an autophagy induction pattern similar to ER stress agents, we hypothesized that flavopiridol induces ER stress, a mechanism of action not previously ascribed to CDK inhibitors. We observe a significant, time-dependent increase in expression of ER stress genes XBP1, IRE1, and GRP78 in CLL cells treated with flavopiridol as well as the ER stress-inducing agents thapsigargin and tunicamycin (supplemental Figure 5A-C). Importantly, significant increases in XBP1, IRE1, and GRP78 genes were also observed in samples obtained from CLL patients treated with flavopiridol; cells were collected before treatment (pre) and immediately at the end of flavopiridol infusion (4.5 hours, n = 10; P = .0008 for IRE1, P = .0016 for XBP1, P = .033 for GRP78) but not in CHOP (P = .292; Figure 5). To determine whether the ER stress induced by flavopiridol in CLL cells is the result of CDK5 inhibition, CLL cells were transfected with siRNA to CDK5. Compared with the scrambled control, CDK5 siRNA transfection resulted in increased expression of XBP1, GRP78, and IRE1 (P = .0007, .00036, and .0002, respectively), as was observed in cells incubated with flavopiridol (supplemental Figure 5D-F). For all our real-time RT-PCR experiments, we used CD52 as control housekeeping gene (GAPDH and TBP showed significant variability in the presence of flavopiridol).
Figure 5.
Flavopiridol induces ER stress in CLL cells. Real-time RT-PCR for XBP1, IRE1, GRP78, and CHOP in CLL cells (n = 10) collected from patients undergoing flavopiridol treatment. P values were calculated for comparisons between gene expression in pretreatment samples versus samples collected at the end of infusion (4.5 hours). P = .0008 for IRE1. P = .0016 for XBP1. P = .033 for GRP78. For all real-time RT-PCR experiments described here and in the following figures, we used CD52 as control housekeeping gene.
ER stress response in CLL cells is dysfunctional
Because our initial ER stress studies in CLL cells showed that one of the ER stress markers, CHOP, does not increase like the other markers examined, we decided to further characterize this pathway in CLL cells. To this end, we examined the activity of the unfolded protein response (UPR) initiators PERK, IRE1, and ATF6. PERK was minimally active after thapsigargin and flavopiridol treatment, as assessed by phosphorylation of its downstream target eIF2α. As a control, cell lines treated with flavopiridol or thapsigargin showed dramatic eIF2α phosphorylation (supplemental Figure 6A). Similarly, no evidence of IRE1 activity was noted after flavopiridol or thapsigargin treatment, as determined by a general deficiency in splicing of the IRE1 substrate XBP1 (8 of 10 CLL samples tested). Furthermore, neither thapsigargin nor tunicamycin induced XBP1 splicing in any of the samples analyzed (supplemental Figure 6B). In contrast, B cells from healthy donors and multiple cell lines (HeLa, 293T, RAW 264.7, 697) treated with flavopiridol or ER stress inducers showed XBP1 splicing. This finding suggests the UPR response machinery in CLL cells is at least partially dysfunctional.
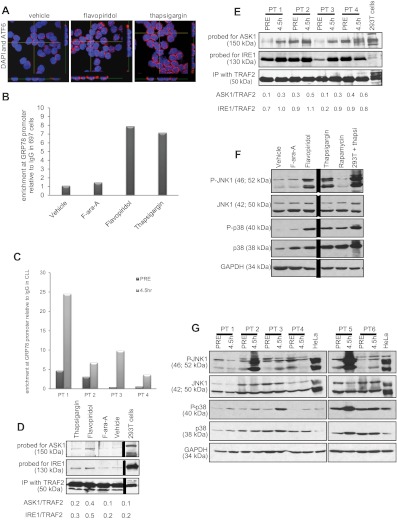
Although PERK activation and XBP1 splicing were not observed, the increases in XBP1, IRE1, and GRP78 expression in vitro and in vivo suggest that part of the UPR is activated with flavopiridol. GRP78 transcription is induced by ATF6, a transcription factor that translocates to the nucleus in the presence of ER stress.26 ATF6 nuclear translocation was therefore examined by confocal fluorescence microscopy. As shown in Figure 6A, increased nuclear translocation is observed in flavopiridol- or thapsigargin-treated CLL patient cells. This was complemented by enhanced binding of the GRP78 promoter region by ATF6, as determined by ChIP assay using anti-ATF6 antibody versus a nonspecific (IgG) control in 697 cells treated with flavopiridol or thapsigargin (Figure 6B). This increased promoter binding was not noted with F-ara-A. Importantly, results were similar in in vivo, in samples from CLL patients obtained pretreatment versus samples collected immediately after therapy (4.5 hours; Figure 6C). We also tested GRP78 protein expression by immunoblot, and we observed an increase in the presence of flavopiridol and thapsigargin (at 6 hours) but not in F-ara-A-treated CLL cells. We tested samples from 6 patients; supplemental Figure 6C shows one representative immunoblot of 3, each with samples from 2 CLL patients.
Figure 6.
ER stress in CLL cells. (A) Confocal fluorescence microscopy for ATF6. Z stacks were collected (0.4 μm per slice), and images were chosen from the middle of nuclei. Side views (across bottom and side of figures) are also shown to depict ATF6 nuclear localization in CLL cells treated with flavopiridol (2μM) or thapsigargin (1μM) for 4 hours. Results shown are representative of 4 experiments. (B) ChIP assay data showing enrichment of ATF6 at the GRP78 promoter region with flavopiridol and thapsigargin treatment in 697 cells. Values of enrichment at promoter are presented relative to the IgG negative control. (C) ChIP assay data showing enrichment of ATF6 at GRP78 promoter region in cells from CLL patients undergoing flavopiridol treatment, collected before or at the end of flavopiridol infusion (4.5 hours). (D-E) Coimmunoprecipitation assay for IRE1-ASK1-TRAF2 complex formation 697 cells (D) or samples from flavopiridol-treated CLL patients (E) were immunoprecipitated with TRAF2 and immunoblotted for ASK1 or IRE1. (F) Immunoblots for total and phosphorylated JNK1 and p38MAPK in CLL cells treated in vitro with flavopiridol (2μM), thapsigargin (1μM), F-ara-A (5μM), and rapamycin (5μM). Results shown are from one representative patient sample. (G) Immunoblots for total and phosphorylated JNK1 and p38MAPK in samples from 6 CLL patients treated with flavopiridol; samples were collected before treatment and at 4.5 hours into flavopiridol infusion.
In addition to ATF6 nuclear translocation and GRP78 transcriptional induction, another early event of UPR is the formation of a complex between IRE1, the adaptor molecule TRAF2, and the MAP kinase pathway member ASK1. We detected the formation of this complex by increased levels of ASK1 and IRE1 protein in TRAF2 coimmunoprecipitates in 697 cells treated with flavopiridol, and to a lesser extent with thapsigargin, as shown in Figure 6D. This same increase in IRE1 and ASK1 association with TRAF2 is noted in CLL patient cells in vivo after flavopiridol infusion (Figure 6E). Direct downstream effects of IRE1-TRAF2-ASK1 complex activity include phosphorylation of JNK1 and p38MAPK. Flavopiridol and thapsigargin treatment induced the phosphorylation of both JNK1 and p38MAPK in CLL cells, whereas F-ara-A did not (Figure 6F). Moreover, JNK1 and p38MAPK phosphorylation was observed in 5 of 10 CLL patients in samples obtained 4.5 hours after the beginning of flavopiridol infusion (Figure 6G). Although the cause of the JNK1 and p38MAPK phosphorylation variability among these clinical samples is unclear, it is possible that variations in sample processing could contribute to these results.
ER stress induced by flavopiridol as a mechanism of cell death
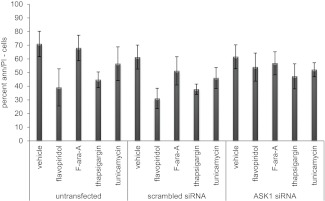
During ER stress-mediated cell death, ASK1 activates caspase 4.36,37 To determine whether the flavopiridol-induced IRE1-TRAF2-ASK1 complex contributes to cell death, we therefore investigated caspase 4 and its downstream target caspase 8. In CLL cells incubated with flavopiridol, thapsigargin, or tunicamycin, caspase 4 and caspase 8 activity increased 5- to 10-fold (n = 5; P = .0001) compared with cells incubated with vehicle or F-ara-A (supplemental Figure 7A-B). This increased caspase activity was successfully inhibited by the caspase 4 inhibitor Z-YVAD-FMK. Moreover, flavopiridol-mediated cell death was significantly (P = .012) reduced in the presence of Z-YVAD-FMK (supplemental Figure 7C). In samples from CLL patients treated with flavopiridol in the clinic, 13 of 20 showed at least a 2-fold increase in the activity of caspase 4 and caspase 8 at 4.5 hours after flavopiridol infusion (supplemental Figure 7E). To confirm the contribution of ASK1 activation to this process, cells were transfected with siRNA directed to ASK1 (efficacy shown in supplemental Figure 7D,F). These cells were significantly (P = .005) more resistant to flavopiridol treatment than cells transfected with scrambled siRNA (Figure 7). Moreover, we tested caspase 4 activity in siRNA ASK1 samples, and we observed the absence of activity in the presence of flavopiridol and thapsigargin, as opposed to scrambled and untransfected samples, adding more support to the idea of a possible connection between ASK1 and caspase 4 (supplemental Figure 7G). Collectively, these findings indicate that ER stress, via the IRE1-ASK1-JNK1-caspase 4 cascade, is a component of the mechanism of CLL cell death by flavopiridol.
Figure 7.
ER stress molecule ASK1 is necessary for flavopiridol-induced cell death. Viability of CLL cells (n = 5) untransfected or transfected with scrambled or ASK1 siRNA, then treated with flavopiridol (2μM), F-ara-A (5μM), thapsigargin (1μM), or tunicamycin (3 μg/mL) 24 hour after transfection. Flavopiridol cytotoxicity decreased in the presence of ASK1 siRNA (P = .0005).
Discussion
Since the initial reports that the CDK inhibitor flavopiridol was active as a therapeutic in CLL,5 significant effort has been put forth to understand why this agent potently induces cell death in growth-arrested, transformed lymphoid cells. As our earlier work suggested that flavopiridol caused changes consistent with autophagy in CLL patient cells,13 we performed a detailed examination of this pathway. Here we document that primary CLL cells express the critical components of the autophagy machinery and that autophagy can be robustly activated in these cells by commonly reported stimuli and also select CLL therapeutics, including fludarabine, CAL-101, and flavopiridol. Unexpectedly, the induction of autophagy offered little protection from cell death from most stimuli, including fludarabine and CAL-101. Flavopiridol and the ER stress-inducing agent thapsigargin were notable exceptions, where either pharmacologic or siRNA-mediated inhibition of autophagy enhanced cytotoxicity.
This observation prompted the hypothesis that an unappreciated mechanism of flavopiridol action was through ER stress. We demonstrate here that flavopiridol does indeed mediate robust ER stress in CLL cells but that this process is dysfunctional compared with normal B cells as characterized by the absence of PERK activation and XBP1 splicing. Nonetheless, flavopiridol-induced ER stress promotes cell death via IRE1-induced activation of ASK1 and caspase 4. Notably, activation of autophagy, ER stress, and downstream activation of caspase 4 was demonstrated in samples obtained from CLL patients receiving flavopiridol treatment. These collective findings document, for the first time, that CDK inhibitors promote ER stress and also identify autophagy as a mechanism of CDK inhibitor resistance that can be therapeutically targeted in resistant tumors.
Beyond identifying new mechanisms of action and resistance for CDK inhibitors, these studies have further relevance to the field of cancer. To our knowledge, this represents the first systematic investigation of autophagy in CLL or any other primary tumor cell with multiple inducers of this process. These results demonstrate that many stimuli, including common therapeutic agents, can promote autophagy in CLL tumor cells. However, for most of these stimuli, disruption of autophagy does not enhance cell death. Agents that induce ER stress proved an exception to this, indicating that, for ER stress mediated killing, the process of autophagy is a relevant resistance mechanism. The general applicability of this finding to other tumors is supported by several other studies in which autophagy is identified as a mechanism of drug resistance.38–40 Notably, each of the agents identified was shown to be an inducer of ER stress. These findings collectively provide justification for exploring combination strategies with CDK inhibitors, and potentially other ER stress inducers, with inhibitors of autophagy to enhance therapeutic benefit.
Multiple mechanisms of action have been suggested for flavopiridol and other CDK inhibitors.4,5,8 For example, flavopiridol has been shown to inhibit alternative targets, such as GSK3-β, in addition to multiple CDKs.4,41,42 However, no reports have associated this class of drugs with ER stress in CLL or other cancers. It is of interest that CDK1 and CDK5, both inhibited by flavopiridol, inhibit autophagy by phosphorylating Vps34, a necessary initiator of this process.35 In this work, we demonstrate that knockdown of CDK5 expression results in both induction of autophagy and increase in ER stress genes in CLL cells. ER stress is a natural cellular response to a variety of stimuli that leads to the UPR43–45 involving: (1) IRE1 activation that leads to XBP1 mRNA splicing; (2) ATF6 release from the ER membrane, followed by nuclear translocation and transcriptional up-regulation of ER response genes; and (3) PERK activation and phosphorylation of eIF2α, which leads to the inhibition of translation initiation.27 With continued ER stress, terminal UPR events occur that include release of ER-sequestered calcium, IRE1 association with TRAF2 and ASK1, and subsequent activation of multiple proapoptotic processes that ultimately lead to cell death.37,46,47 Limited studies have suggested that ER stress-inducing agents can promote apoptosis in CLL.48 Our data show that, in CLL, but not normal B cells or cell lines, the ER stress response is only partially functional as demonstrated by absence of XBP1 splicing and PERK activation. However, after treatment with flavopiridol or thapsigargin, CLL cells demonstrate ATF6 nuclear translocation and binding to GRP78 promoter as well as formation of the IRE1/TRAF2/ASK1 complex and activation of JNK1, p38, and caspase 4. The importance of the IRE1/TRAF2/ASK1 complex formation and downstream caspase 4 activation to flavopiridol cell death is demonstrated by both ASK1 siRNA knockdown and also caspase 4 inhibition, both of which antagonize flavopiridol-induced cell death. The clinical relevance of these in vitro findings was confirmed by examination of cells from patients receiving flavopiridol, in which these same biochemical events were noted.
Although our data are supportive of the mechanism of action put forward, there are some inconsistent results from prior published work and our findings. Specifically, although our data support ER stress induced by flavopiridol as one mechanism of action of this agent, the lack of correlation of response with down-regulation of Mcl-112 is problematic given that knockdown of this gene generally is associated with ER stress lethality.49 One potential explanation for lack of correlation of MCL-1 down-regulation is compensatory early rebound of this protein observed or alternative BCL-2 family protein member compensation. Downstream ER stress changes after flavopiridol treatment in vivo, including JNK and p38 phosphorylation, are not fully supportive, suggesting either inappropriate time assessment or alternative mechanisms of killing contributing to tumor elimination in a subset of patients treated with this agent. Further characterization of downstream ER stress pathway activation and its relationship to clinical response will be required to establish this as the driving mechanism of cell death induced by flavopiridol in CLL cells.
The complex network of signals induced by flavopiridol-mediated ER stress suggests novel approaches to enhance CDK inhibitor efficacy in CLL and potentially other cancers as well. Specifically, therapeutics, such as chloroquine, that block the autophagic process may convert a CDK inhibitor-resistant case to a responsive one. Notably, our studies with chloroquine were performed using concentrations that are physiologically attainable in patients receiving treatment for malaria. Other strategies to interfere with autophagy could include targeting ATG4 members or the Vps34/Beclin1/Barkor complex. Similarly, these results support the development of strategies to enhance activation of the late UPR response, including IRE1/TRAF2/ASK1 complex formation and activation of caspase 4. Given that thioredoxin directly binds to ASK1 and prevents its inclusion in this complex,50 pharmacologic agents depleting thioredoxin represent an attractive potential combination. Regardless, the clinical potential of CDK inhibitors in CLL and other malignancies cannot be ignored, and efforts focused on combination therapies targeting the autophagy and UPR networks will help to optimize their application in the treatment of cancer.
Supplementary Material
Acknowledgments
The authors thank the many patients for providing biologic samples for these studies and Dr. Brian Lannutti for providing CAL-101. All the immunofluorescence images were collected at the Confocal Microscopy Imaging Facility at The Ohio State University.
This work was supported by Specialized Center of Research from the Leukemia & Lymphoma Society SCORE Project 1, the National Cancer Institute (K12 CA133250, T32 CA09338, P50 CA140158, and P01 CA81534), and the D. Warren Brown Foundation. A.J.J. is a Paul Calabresi Scholar.
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: E.M. and D.M.L. designed, analyzed, and interpreted the data, drafted the manuscript, and provided critical revision; S.V.G., A.J.W.. S.E.M.H., L.L.S., and Y.-Y.Y. collected and analyzed the data; L.A., J.A.J., J.M.F., and K.A.B. provided clinical samples; X.Z. and A.L. analyzed the data; H.K. and M.G. designed, analyzed, and interpreted the data and drafted the manuscript; M.R.G., A.J.J., and J.C.B. conceived study design, analyzed and interpreted the data, and drafted the manuscript; and all authors gave final approval of the article.
Conflict-of-interest disclosure: E.M., D.M.L., A.J.J., M.R.G., and J.C.B. have filed a patent on the application of combined use of flavopiridol with autophagy antagonizing agents. The remaining authors declare no competing financial interests.
Correspondence: John C. Byrd, B302 Starling-Loving Hall, 320 West 10th Ave, Columbus, OH 43210, e-mail: john.byrd@osumc.edu; or Amy J. Johnson, The Ohio State University, CCC Bldg, Rm 455C, 410 W 12th Ave, Columbus, OH 43210; e-mail: amy.johnson@osumc.edu.
References
- 1.Rozman C, Montserrat E. Chronic lymphocytic leukemia. N Engl J Med. 1995;333(16):1052–1057. doi: 10.1056/NEJM199510193331606. [DOI] [PubMed] [Google Scholar]
- 2.Hallek M, Cheson BD, Catovsky D, et al. Guidelines for the diagnosis and treatment of chronic lymphocytic leukemia: a report from the International Workshop on Chronic Lymphocytic Leukemia updating the National Cancer Institute-Working Group 1996 guidelines. Blood. 2008;111(12):5446–5456. doi: 10.1182/blood-2007-06-093906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sedlacek H, Czech J, Naik R, et al. Flavopiridol (L86 8275; NSC 649890), a new kinase inhibitor for tumor therapy. Int J Oncol. 1996;9(6):1143–1168. doi: 10.3892/ijo.9.6.1143. [DOI] [PubMed] [Google Scholar]
- 4.Carlson BA, Dubay MM, Sausville EA, Brizuela L, Worland PJ. Flavopiridol induces G1 arrest with inhibition of cyclin-dependent kinase (CDK) 2 and CDK4 in human breast carcinoma cells. Cancer Res. 1996;56(13):2973–2978. [PubMed] [Google Scholar]
- 5.Byrd JC, Shinn C, Waselenko JK, et al. Flavopiridol induces apoptosis in chronic lymphocytic leukemia cells via activation of caspase-3 without evidence of bcl-2 modulation or dependence on functional p53. Blood. 1998;92(10):3804–3816. [PubMed] [Google Scholar]
- 6.Pepper C, Thomas A, Hoy T, et al. Leukemic and non-leukemic lymphocytes from patients with Li Fraumeni syndrome demonstrate loss of p53 function, Bcl-2 family dysregulation and intrinsic resistance to conventional chemotherapeutic drugs but not flavopiridol. Cell Cycle. 2003;2(1):53–58. [PubMed] [Google Scholar]
- 7.Dai Y, Rahmani M, Grant S. Proteasome inhibitors potentiate leukemic cell apoptosis induced by the cyclin-dependent kinase inhibitor flavopiridol through a SAPK/JNK- and NF-kappaB-dependent process. Oncogene. 2003;22(46):7108–7122. doi: 10.1038/sj.onc.1206863. [DOI] [PubMed] [Google Scholar]
- 8.Chen R, Keating MJ, Gandhi V, Plunkett W. Transcription inhibition by flavopiridol: mechanism of chronic lymphocytic leukemia cell death. Blood. 2005;106(7):2513–2519. doi: 10.1182/blood-2005-04-1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Byrd JC, Lin TS, Dalton JT, et al. Flavopiridol administered using a pharmacologically derived schedule is associated with marked clinical efficacy in refractory, genetically high-risk chronic lymphocytic leukemia. Blood. 2007;109(2):399–404. doi: 10.1182/blood-2006-05-020735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Christian BA, Grever MR, Byrd JC, Lin TS. Flavopiridol in chronic lymphocytic leukemia: a concise review. Clin Lymphoma Myeloma. 2009;9(Suppl 3):S179–S185. doi: 10.3816/CLM.2009.s.009. [DOI] [PubMed] [Google Scholar]
- 11.Ma Y, Cress WD, Haura EB. Flavopiridol-induced apoptosis is mediated through up-regulation of E2F1 and repression of Mcl-1. Mol Cancer Ther. 2003;2(1):73–81. [PubMed] [Google Scholar]
- 12.Woyach JA, Lozanski G, Ruppert AS, et al. Outcome of patients with relapsed or refractory chronic lymphocytic leukemia treated with flavopiridol: impact of genetic features. Leukemia. 2012;26(6):1442–1444. doi: 10.1038/leu.2011.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hussain SR, Lucas DM, Johnson AJ, et al. Flavopiridol causes early mitochondrial damage in chronic lymphocytic leukemia cells with impaired oxygen consumption and mobilization of intracellular calcium. Blood. 2008;111(6):3190–3199. doi: 10.1182/blood-2007-10-115733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stack JH, DeWald DB, Takegawa K, Emr SD. Vesicle-mediated protein transport: regulatory interactions between the Vps15 protein kinase and the Vps34 PtdIns 3-kinase essential for protein sorting to the vacuole in yeast. J Cell Biol. 1995;129(2):321–334. doi: 10.1083/jcb.129.2.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ericsson JL. Studies on induced cellular autophagy: I. Electron microscopy of cells with in vivo labelled lysosomes. Exp Cell Res. 1969;55(1):95–106. doi: 10.1016/0014-4827(69)90462-5. [DOI] [PubMed] [Google Scholar]
- 16.Tassa A, Roux MP, Attaix D, Bechet DM. Class III phosphoinositide 3-kinase–Beclin1 complex mediates the amino acid-dependent regulation of autophagy in C2C12 myotubes. Biochem J. 2003;376(3):577–586. doi: 10.1042/BJ20030826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Petiot A, Ogier-Denis E, Blommaart EF, Meijer AJ, Codogno P. Distinct classes of phosphatidylinositol 3′-kinases are involved in signaling pathways that control macroautophagy in HT-29 cells. J Biol Chem. 2000;275(2):992–998. doi: 10.1074/jbc.275.2.992. [DOI] [PubMed] [Google Scholar]
- 18.Sun Q, Fan W, Chen K, Ding X, Chen S, Zhong Q. Identification of Barkor as a mammalian autophagy-specific factor for Beclin 1 and class III phosphatidylinositol 3-kinase. Proc Natl Acad Sci U S A. 2008;105(49):19211–19216. doi: 10.1073/pnas.0810452105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cataldo AM, Hamilton DJ, Nixon RA. Lysosomal abnormalities in degenerating neurons link neuronal compromise to senile plaque development in Alzheimer disease. Brain Res. 1994;640(1):68–80. doi: 10.1016/0006-8993(94)91858-9. [DOI] [PubMed] [Google Scholar]
- 20.Klionsky DJ, Emr SD. Autophagy as a regulated pathway of cellular degradation. Science. 2000;290(5497):1717–1721. doi: 10.1126/science.290.5497.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Samara C, Syntichaki P, Tavernarakis N. Autophagy is required for necrotic cell death in Caenorhabditis elegans. Cell Death Differ. 2008;15(1):105–112. doi: 10.1038/sj.cdd.4402231. [DOI] [PubMed] [Google Scholar]
- 22.Mujumdar N, Saluja AK. Autophagy in pancreatic cancer: an emerging mechanism of cell death. Autophagy. 2010;6(7):997–998. doi: 10.4161/auto.6.7.13334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cheson BD, Bennett JM, Grever M, et al. National Cancer Institute-sponsored Working Group guidelines for chronic lymphocytic leukemia: revised guidelines for diagnosis and treatment. Blood. 1996;87(12):4990–4997. [PubMed] [Google Scholar]
- 24.Hertlein E, Wagner AJ, Jones J, et al. 17-DMAG targets the nuclear factor-kappaB family of proteins to induce apoptosis in chronic lymphocytic leukemia: clinical implications of HSP90 inhibition. Blood. 2010;116(1):45–53. doi: 10.1182/blood-2010-01-263756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Herman SE, Gordon AL, Wagner AJ, et al. Phosphatidylinositol 3-kinase-delta inhibitor CAL-101 shows promising preclinical activity in chronic lymphocytic leukemia by antagonizing intrinsic and extrinsic cellular survival signals. Blood. 2010;116(12):2078–2088. doi: 10.1182/blood-2010-02-271171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li M, Baumeister P, Roy B, et al. ATF6 as a transcription activator of the endoplasmic reticulum stress element: thapsigargin stress-induced changes and synergistic interactions with NF-Y and YY1. Mol Cell Biol. 2000;20(14):5096–5106. doi: 10.1128/mcb.20.14.5096-5106.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yoshida H, Matsui T, Yamamoto A, Okada T, Mori K. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell. 2001;107(7):881–891. doi: 10.1016/s0092-8674(01)00611-0. [DOI] [PubMed] [Google Scholar]
- 28.Mizushima N, Yoshimori T. How to interpret LC3 immunoblotting. Autophagy. 2007;3(6):542–545. doi: 10.4161/auto.4600. [DOI] [PubMed] [Google Scholar]
- 29.Dalby KN, Tekedereli I, Lopez-Berestein G, Ozpolat B. Targeting the prodeath and prosurvival functions of autophagy as novel therapeutic strategies in cancer. Autophagy. 2010;6(3):322–329. doi: 10.4161/auto.6.3.11625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hamano T, Gendron TF, Causevic E, et al. Autophagic-lysosomal perturbation enhances tau aggregation in transfectants with induced wild-type tau expression. Eur J Neurosci. 2008;27(5):1119–1130. doi: 10.1111/j.1460-9568.2008.06084.x. [DOI] [PubMed] [Google Scholar]
- 31.Projean D, Baune B, Farinotti R, et al. In vitro metabolism of chloroquine: identification of CYP2C8, CYP3A4, and CYP2D6 as the main isoforms catalyzing N-desethylchloroquine formation. Drug Metab Dispos. 2003;31(6):748–754. doi: 10.1124/dmd.31.6.748. [DOI] [PubMed] [Google Scholar]
- 32.Bjorkoy G, Lamark T, Brech A, et al. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol. 2005;171(4):603–614. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jain A, Lamark T, Sjottem E, et al. p62/SQSTM1 is a target gene for transcription factor NRF2 and creates a positive feedback loop by inducing antioxidant response element-driven gene transcription. J Biol Chem. 2010;285(29):22576–22591. doi: 10.1074/jbc.M110.118976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klionsky DJ, Abeliovich H, Agostinis P, et al. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy. 2008;4(2):151–175. doi: 10.4161/auto.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Furuya T, Kim M, Lipinski M, et al. Negative regulation of Vps34 by Cdk mediated phosphorylation. Mol Cell. 38(4):500–511. doi: 10.1016/j.molcel.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hitomi J, Katayama T, Eguchi Y, et al. Involvement of caspase-4 in endoplasmic reticulum stress-induced apoptosis and Abeta-induced cell death. J Cell Biol. 2004;165(3):347–356. doi: 10.1083/jcb.200310015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nishitoh H, Matsuzawa A, Tobiume K, et al. ASK1 is essential for endoplasmic reticulum stress-induced neuronal cell death triggered by expanded polyglutamine repeats. Genes Dev. 2002;16(11):1345–1355. doi: 10.1101/gad.992302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bellodi C, Lidonnici MR, Hamilton A, et al. Targeting autophagy potentiates tyrosine kinase inhibitor-induced cell death in Philadelphia chromosome-positive cells, including primary CML stem cells. J Clin Invest. 2009;119(5):1109–1123. doi: 10.1172/JCI35660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eisenberg-Lerner A, Kimchi A. The paradox of autophagy and its implication in cancer etiology and therapy. Apoptosis. 2009;14(4):376–391. doi: 10.1007/s10495-008-0307-5. [DOI] [PubMed] [Google Scholar]
- 40.Yorimitsu T, Nair U, Yang Z, Klionsky DJ. Endoplasmic reticulum stress triggers autophagy. J Biol Chem. 2006;281(40):30299–30304. doi: 10.1074/jbc.M607007200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.de Azevedo WF, Jr, Canduri F, da Silveira NJ. Structural basis for inhibition of cyclin-dependent kinase 9 by flavopiridol. Biochem Biophys Res Commun. 2002;293(1):566–571. doi: 10.1016/S0006-291X(02)00266-8. [DOI] [PubMed] [Google Scholar]
- 42.Leclerc S, Garnier M, Hoessel R, et al. Indirubins inhibit glycogen synthase kinase-3 beta and CDK5/p25, two protein kinases involved in abnormal tau phosphorylation in Alzheimer's disease: a property common to most cyclin-dependent kinase inhibitors? J Biol Chem. 2001;276(1):251–260. doi: 10.1074/jbc.M002466200. [DOI] [PubMed] [Google Scholar]
- 43.Xu C, Bailly-Maitre B, Reed JC. Endoplasmic reticulum stress: cell life and death decisions. J Clin Invest. 2005;115(10):2656–2664. doi: 10.1172/JCI26373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kadowaki H, Nishitoh H, Ichijo H. Survival and apoptosis signals in ER stress: the role of protein kinases. J Chem Neuroanat. 2004;28(1):93–100. doi: 10.1016/j.jchemneu.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 45.Marciniak SJ, Ron D. Endoplasmic reticulum stress signaling in disease. Physiol Rev. 2006;86(4):1133–1149. doi: 10.1152/physrev.00015.2006. [DOI] [PubMed] [Google Scholar]
- 46.Urano F, Wang X, Bertolotti A, et al. Coupling of stress in the ER to activation of JNK protein kinases by transmembrane protein kinase IRE1. Science. 2000;287(5453):664–666. doi: 10.1126/science.287.5453.664. [DOI] [PubMed] [Google Scholar]
- 47.Li G, Mongillo M, Chin KT, et al. Role of ERO1-alpha-mediated stimulation of inositol 1,4,5-triphosphate receptor activity in endoplasmic reticulum stress-induced apoptosis. J Cell Biol. 2009;186(6):783–792. doi: 10.1083/jcb.200904060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rosati E, Sabatini R, Rampino G, et al. Novel targets for endoplasmic reticulum stress-induced apoptosis in B-CLL. Blood. 2010;116(15):2713–2723. doi: 10.1182/blood-2010-03-275628. [DOI] [PubMed] [Google Scholar]
- 49.Rahmani M, Mayo M, Dash R, et al. Melanoma differentiation associated gene-7/interleukin-24 potently induces apoptosis in human myeloid leukemia cells through a process regulated by endoplasmic reticulum stress. Mol Pharmacol. 2010;78(6):1096–1104. doi: 10.1124/mol.110.068007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu Y, Min W. Thioredoxin promotes ASK1 ubiquitination and degradation to inhibit ASK1-mediated apoptosis in a redox activity-independent manner. Circ Res. 2002;90(12):1259–1266. doi: 10.1161/01.res.0000022160.64355.62. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.