Abstract
The chemotaxis of Vibrio cholerae, the causative agent of cholera, has been implicated in pathogenicity. The bacterium has more than 40 genes for methyl-accepting chemotaxis protein (MCP)-like proteins (MLPs). In this study, we found that glycine and at least 18 l-amino acids, including serine, arginine, asparagine, and proline, serve as attractants to the classical biotype strain O395N1. Based on the sequence comparison with Vibrio parahaemolyticus, we speculated that at least 17 MLPs of V. cholerae may mediate chemotactic responses. Among them, Mlp24 (previously named McpX) is required for the production of cholera toxin upon mouse infection. mlp24 deletion strains of both classical and El Tor biotypes showed defects in taxis toward several amino acids, which were complemented by the expression of Mlp24. These amino acids enhanced methylation of Mlp24. Serine, arginine, asparagine, and proline were shown to bind directly to the periplasmic fragment of Mlp24. The structural information of its closest homolog, Mlp37, predicts that Mlp24 has two potential ligand-binding pockets per subunit, the membrane distal of which was suggested, by mutational analyses, to be involved in sensing of amino acids. These results suggest that Mlp24 is a chemoreceptor for multiple amino acids, including serine, arginine, and asparagine, which were previously shown to stimulate the expression of several virulence factors, implying that taxis toward a set of amino acids plays critical roles in pathogenicity of V. cholerae.
INTRODUCTION
Toxigenic Vibrio cholerae, the causative agent of cholera, is a Gram-negative, highly motile bacterium with a single polar flagellum. The bacterium inhabits nutrient-poor aquatic environments such as rivers and estuaries or occasionally intrudes into the nutrient-rich lumen of the human gastrointestinal tract (38). Chemotaxis of V. cholerae toward favorable locations is proposed to play critical roles in its survival (6) and pathogenicity (2, 5, 8, 13, 14, 15, 16, 17, 20, 21, 30, 31). However, the external signals and cellular factors that mediate chemotaxis of V. cholerae are only poorly understood.
The genome sequence of V. cholerae (El Tor biotype [26]) predicts that the bacterium has three sets of chemotaxis signaling proteins (Che proteins) and 45 proteins with significant homology to chemoreceptors, also known as methyl-accepting chemotaxis proteins (MCPs). (A member of the latter family within V. cholerae is hereafter referred to as an MCP-like protein [MLP].) The functions of Che proteins and MCPs are best understood in Escherichia coli (4, 24), providing the framework for understanding those of homologs in other bacteria. Environmental signals (e.g., serine and aspartate) are sensed by four MCPs (25). An MCP forms a ternary complex with the adaptor CheW and the histidine kinase CheA. CheA is activated when it is coupled to an unliganded MCP. CheA autophosphorylates and then donates its phosphoryl group to the response regulator CheY. Binding of phospho-CheY to the flagellar motor induces its clockwise (CW) rotation. Attractant binding to an MCP inactivates CheA, which decreases the cellular level of phospho-CheY, leading to counterclockwise (CCW) rotation of the flagellar motor. Persistent stimulation attenuates the initial response, an essential process of which is called adaptation and involves reversible methylation of the relevant MCP (catalyzed by the methyltransferase CheR and the methylesterase CheB). Increased methylation of the MCP reactivates CheA to counteract the inhibitory effect of the attractant.
In the genome of V. cholerae (El Tor biotype), each set of che genes is linked together to form a cluster: cluster I contains cheY1 (VC1395), cheA1 (VC1397), cheY2 (VC1398), cheR1 (VC1399), cheB1 (VC1401), and putative cheW (VC1402); cluster II contains cheW1 (VC2059), cheB2 (VC2062), cheA2 (VC2063), cheZ (VC2064), and cheY3 (VC2065); and cluster III contains cheB3 (VCA1089), cheD (VCA1090), cheR3 (VCA1091), cheW2 (VCA1093), cheW3 (VCA1094), cheA3 (VCA1095), and cheY4 (VCA1096). The cheR2 (VC2201) gene is in the fla gene cluster located just next to the che cluster II. Clusters I and II as well as the fla gene cluster are located on chromosome I, whereas cluster III is on chromosome II. Since the products of the genes in a particular cluster are known or hypothesized to constitute a coherent signaling system, we hereafter refer to the three Che systems according to their gene clusters (note that CheR2 is assigned to system II). Out of the three Che systems, only system II is involved in flagellum-mediated chemotaxis (19, 27). The other two systems are likely to be involved in other cellular functions. However, their roles have been elusive, since neither deletion nor overexpression of genes of clusters I and III produces any detectable phenotype. In addition, chromosome I contains, outside any che or fla gene cluster, a pair of genes encoding a non-CheA histidine kinase (VC1315) and a putative CheY protein (VC1316), which are not believed to be involved in chemotaxis (27).
In this study, the 45 mlp genes are named mlp1 through mlp45 on the basis of genome order in the large and small chromosomes. Among them, mlp13 (VC1394), mlp14 (VC1403), mlp15 (VC1405), and mlp16 (VC1406) are located in che cluster I, and mlp44 (VCA1088) and mlp45 (VCA1092) are located in cluster III. The others are distributed throughout the genome, with roughly half being located on either of the two chromosomes. For most MLPs, it has not been established which is coupled to which CheA homolog.
Previous studies revealed that three MLPs (Mlp7 [previously named TcpI], Mlp8 [previously named AcfB], and Mlp30 [previously named HlyB]) are involved in pathogenicity (2, 13, 14, 23, 29) and that several che genes of cluster II (cheZ, cheA2, and cheY3) as well as a gene (VC2161) encoding a putative chemoreceptor (previously named mcpX, which is renamed mlp24 in this paper) are required for the expression of the genes encoding cholera toxin (CT) and its positive regulator ToxT upon mouse infection (31). Several mlp genes (mlp2 [VC0216], mlp29 [VCA0176], and mlp42 [VCA1056]) are expressed upon human infection (21). However, how these MLPs and Che proteins contribute to pathogenicity remains to be elucidated.
This study aimed to identify the chemoreceptors involved in taxis toward amino acids. The results from our in silico, in vivo, and in vitro analyses of Mlp24, which has been implicated in pathogenicity, strongly argue that it serves as a chemoreceptor for various amino acids, including serine, arginine, and asparagine, which are known to stimulate the expression of several virulence factors under the control of the ToxR regulon (35).
MATERIALS AND METHODS
Strains and plasmids.
The classical biotype strain O395N1 is wild type for chemotaxis (Che+), and its derivative VcheA2 lacks the cheA2 gene (19). The El Tor biotype strain AC-V66 is Che+, and its derivative AC-V1400 lacks the mlp24 gene (31). The E. coli strain HCB436 (42) lacks CheB and CheR as well as the chemoreceptors. The E. coli strain SM10λpir (35) was used for mobilization of suicide plasmids into V. cholerae. Plasmid pTWV228 (TaKaRa Bio) is a pBR322-based vector carrying the bla gene. Plasmid pKY704 (43) is a suicide vector carrying the sacB gene. Plasmid pGEX-6P-2 (GE Healthcare) is an expression vector for producing a glutathione S-transferase (GST)-fused protein.
Plasmid constructions.
Plasmid pAH901 was constructed by cloning the FLAG sequence between the SphI and HindIII sites of pTWV228. For cloning of the mlp24 gene, the coding region was amplified by PCR from genomic DNA of V. cholerae strain O395N1 to introduce restriction sites at the 5′ and 3′ ends. The resulting DNA fragment was digested with the relevant restriction enzymes and cloned into pAH901, yielding pMlp24, which encodes Mlp24 fused with a FLAG tag at its carboxy terminus. Site-directed mutagenesis of the mlp24 gene was carried out with a QuikChange II site-directed mutagenesis kit (Agilent Technologies/Stratagene, CA), using pMlp24 as a template. The DNA fragment encoding the entire periplasmic domain (residues 29 to 274) of Mlp24 was amplified and subcloned into the expression vector pGEX-6P-2 so that the GST-encoding sequence was fused in frame to the 3′ end of the Mlp24-encoding sequence to yield pGEX-Mlp24p. Nucleotide sequences of the cloned and mutated genes were confirmed by the dideoxy-chain termination method, using the BigDye Terminator v3.1 cycle sequencing kit (Applied Biosystems).
Strain construction.
The mlp24 deletion allele was constructed by using two-step PCR, and the sequence that lacks mlp24 was cloned into pKY704. SM10λpir cells carrying the plasmid were conjugated with O395N1 cells to introduce the deletion allele into the genome of O395N1 by allelic exchange on a selection plate (LB agar with 10% sucrose and without NaCl) to yield Vmlp24 (Δmlp24). The double mutant lacking the mlp24 and mlp37 genes was constructed similarly and named Vmlp201 (Δmlp24 Δmlp37). The introduced allele was confirmed by PCR analyses.
Methylation assay.
Receptor methylation was examined by immunoblotting. An overnight culture of cells in TG medium (1% tryptone, 0.5% NaCl, 0.5% glycerol) was diluted 1:30 into fresh TG medium. After cells were cultured at 30°C for 6 h, harvested, and washed with TM buffer (50 mM Tris-HCl [pH 7.4], 5 mM glucose, 5 mM MgCl2), an amino acid (only l-amino acids and the nonracemic amino acid glycine were used in this study) was added to cells resuspended in the motility medium (TMN buffer; 50 mM Tris-HCl [pH 7.4], 5 mM glucose, 100 mM NaCl, 5 mM MgCl2), and the suspension was incubated at 30°C for 30 min. Cells were collected by centrifugation and suspended in SDS loading buffer (67 mM Tris-HCl [pH 6.8], 8% glycerol, 1% SDS, 0.003% bromophenol blue) supplemented with 7.7% 2-mercaptoethanol. Samples were boiled for 3 min and subjected to SDS-PAGE followed by immunoblotting with anti-FLAG M2 antibody (Sigma).
Capillary assay.
Chemotactic ability was examined by a capillary assay (1). An overnight culture of V. cholerae cells grown in TG medium at 30°C was diluted 1:30 into fresh TG medium, shaken at 30°C for 6 h, harvested, and washed with TM buffer (pH 7.4). Cells were resuspended in TMN buffer (pH 7.4) (to an optical density at 600 nm [OD600] of 0.1). After preincubation of cells at 30°C for 1 h, a capillary containing an amino acid was inserted into the cell suspension and the suspension incubated for another 1 h. The bacteria in the capillary were counted by plating serial dilutions on LB agar. For the competition, cells were preincubated in the presence of an attractant, and then a capillary containing a second attractant as well as the first attractant was inserted.
Preparation of the periplasmic fragment of Mlp24.
Hereafter, the periplasmic fragment of Mlp24 is referred to as Mlp24p. Purification of GST-fused Mlp24p (encoded in plasmid pGEX-Mlp24p) was carried out essentially as described previously (40). In brief, the protein was overproduced in receptorless HCB436 cells. After sonication, the cell lysate was subjected to low- and high-speed centrifugation to remove cell debris and insoluble fractions, respectively. The protein was purified with glutathione-affinity column chromatography and treated with a site-specific protease to remove the GST part. The purified protein was dialyzed against the cleavage buffer and used for the binding assay after dilution.
Isothermal titration calorimetry (ITC).
Titrations of Mlp24p with a ligand amino acid were carried out on a VP-ITC microcalorimeter (MicroCal Inc., Northampton, MA). During titration, the sample was held at 25°C in a stirred (307 rpm) reaction cell (1.4 ml). An injection series (1.5 μl for the first injection and 3 μl each for later injections) was carried out using a 250-μl syringe filled with amino acid solution (30 injections with 240-s intervals). Data points were averaged and stored at 2-s intervals. The data were analyzed using an Origin-ITC software package (MicroCal Inc.).
Statistical analysis.
To evaluate statistical significance, triplicate data were analyzed by statistical hypothesis testing (Kolmogorov-Smirnov test, F-test, and t test) using application program “R” (37a).
RESULTS
Characterization of amino acid taxis of classical biotype V. cholerae strain O395N1.
Vibrio cholerae strain P (O1 serogroup, classical biotype, Ogawa serotype) is known to show positive chemotaxis toward all of the 20 proteinogenic amino acids (16). We have been using a different strain, strain O395N1 (O1 serogroup, classical biotype, Ogawa serotype), to examine roles of the multiple chemotaxis-related genes (19, 27). We therefore tested which amino acids serve as attractants for strain O395N1 by using a capillary assay. First, we optimized the conditions for a capillary assay by examining serine taxis of O395N1 (Che+). The Che+ strain was attracted by serine with the peak concentration of 10 mM (Fig. 1A). This value is comparable to that of E. coli (34). In contrast, the cheA2 deletion (ΔcheA2) derivative VcheA2 (19), which lacks the only cheA homolog involved in chemotaxis, was not significantly attracted by serine at any concentration tested (Fig. 1A). By using the same setup, we examined whether the Che+ strain is attracted by the other 19 amino acids at 10 mM (Table 1). These amino acids elicited attractant responses to some extent, with some being strong (e.g., glycine, alanine, arginine, cysteine, and asparagine) and others being weak (e.g., leucine, tryptophan, and isoleucine). For tyrosine, no significant response was detected.
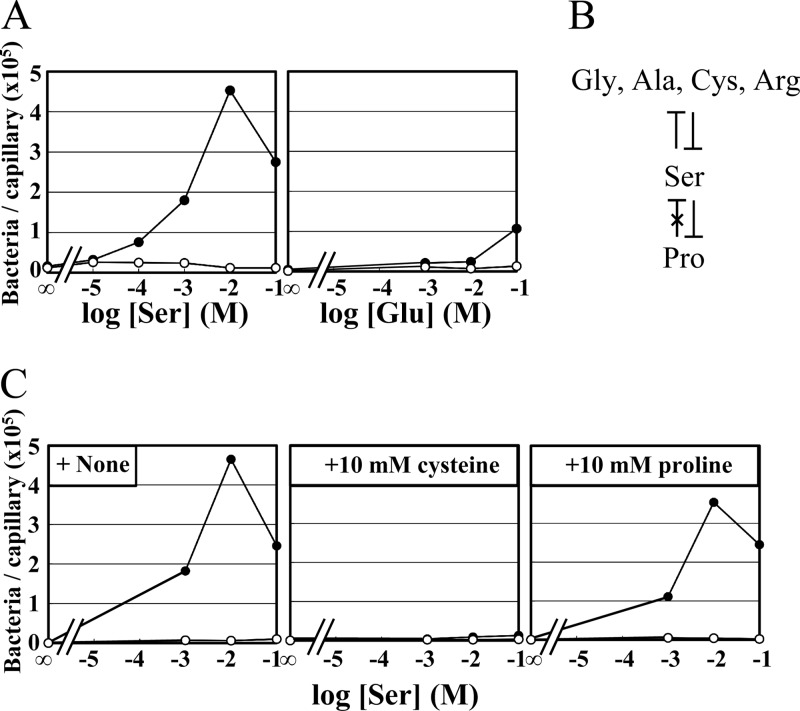
Fig 1.
Chemotactic responses of the classical V. cholerae strain to amino acids. (A) Capillary assay of responses of strain O395N1 to serine (attractant) and glutamate (nonattractant). Closed circles, O395N1 (Che+); open circles, VcheA2 (ΔcheA2). (B) Summary of the results of competition assays. Preincubation with proline failed to inhibit serine taxis, while that with serine inhibited proline taxis. (C) Competition between different attractants in chemotaxis of V. cholerae. O395N1 (closed circles) and VcheA2 (open circles) were preincubated with TMN buffer (+ None), 10 mM cysteine, and 10 mM proline and were examined for responses to serine.
Table 1.
Chemotactic responses of classical V. cholerae strains: O395N1 carrying pAH901 (Che+), Vmlp24 carrying pAH901 (Δmlp24), and Vmlp24 carrying pMlp24 (Δmlp24/pMlp24)
| Amino acid | No. of cells in the capillary with 10 mM amino acid (×105)a |
||
|---|---|---|---|
| Che+b | Δmlp24c | Δmlp24/ pMlp24d | |
| Serine | 855 ± 174* | 479 ± 77* | 934 ± 68** |
| Glycine | 586 ± 74** | 239 ± 52** | 633 ± 113** |
| Alanine | 541 ± 42** | 221 ± 60** | 700 ± 16** |
| Cysteine | 522 ± 97* | 149 ± 22** | 605 ± 18** |
| Arginine | 504 ± 45** | 203 ± 41** | 184 ± 45 |
| Asparagine | 427 ± 60** | 168 ± 29** | 638 ± 30** |
| Histidine | 370 ± 96* | 65 ± 19** | 333 ± 48** |
| Threonine | 344 ± 24** | 53 ± 6** | 459 ± 51** |
| Lysine | 328 ± 63* | 131 ± 11** | 394 ± 60** |
| Glutamine | 323 ± 40** | 103 ± 75* | 350 ± 89** |
| Proline | 322 ± 5** | 91 ± 17** | 557 ± 32** |
| Methionine | 288 ± 84* | 154 ± 48 | 413 ± 47** |
| Valine | 253 ± 17** | 184 ± 67 | 405 ± 71* |
| Phenylalanine | 154 ± 47* | 41 ± 5 | 289 ± 11** |
| Glutamic acid | 137 ± 37** | 96 ± 33 | 104 ± 59 |
| Aspartic acid | 120 ± 35* | 214 ± 36 | 135 ± 50 |
| Leucine | 111 ± 23* | 112 ± 12 | 347 ± 142 |
| Tryptophan | 103 ± 19* | 74 ± 10 | 60 ± 28 |
| Isoleucine | 101 ± 22* | 81 ± 21 | 295 ± 94* |
| Tyrosine | 68 ± 22 | 38 ± 13 | 213 ± 18** |
| None | 28 ± 2 | 22 ± 14 | 16 ± 4 |
Means ± standard deviations of triplicate data are shown.
Values significantly higher than that of “none” at a P value of <0.05 (*) or <0.01 (**) in the t test are marked.
For each amino acid, values were tested in comparison with those of “Che+” and are marked if they are significantly lower at a P value of <0.05 (*) or <0.01 (**) in the t test.
For each amino acid, values were tested in comparison with those of “Δmlp24” and are marked if they are significantly higher at a P value of <0.05 (*) or <0.01 (**) in the t test.
Competition between different attractants.
To gain insights into receptors responsible for amino acid taxis, we examined whether an attractant response to one amino acid is inhibited by preincubation with another amino acid. Such inhibition would likely be due to titration of a shared binding site and/or to methylation (desensitization) of a chemoreceptor that detects both amino acids. V. cholerae cells were suspended in motility medium containing one attractant. After 1 h of incubation at 30°C (allowing cells to adapt to the first attractant), a capillary containing a mixture of another attractant and the original one was inserted into the suspension, which was then incubated for another hour. For simplicity, we examined competition between serine and the other amino acids. Typical results are shown in Fig. 1C. Taxis to cysteine was inhibited by serine and vice versa. In contrast, proline failed to inhibit taxis to serine, whereas serine inhibited taxis to proline. Similar experiments were carried out using alanine, arginine, and glycine. As summarized in Fig. 1B, serine inhibited taxis to alanine, arginine, and glycine and vice versa. The simplest interpretation of these results may be that the V. cholerae strain has at least two classes of amino acid chemoreceptors. However, we cannot rule out the possibility that the noncompeting attractants bind to different sites of the same receptor.
Mlp24 is a candidate for the receptor involved in taxis to amino acids.
The genome sequence of the El Tor biotype strain suggests that V. cholerae has 45 MLPs which contain the highly conserved domain (HCD) (26). However, no ligand for any MLP has been identified to date except that a periplasmic binding protein (with its ligand-bound form) has been suggested to serve as a ligand for Mlp8 (37), nor have any MLPs been shown to function as chemoreceptors in flagellum-mediated chemotaxis. We therefore performed in silico analysis to identify such MLPs and their putative ligands. We found that 17 MLPs of V. cholerae have their closely related homologs in Vibrio parahaemolyticus (data not shown), which has only one set of Che proteins with high similarities to those of Che system II of V. cholerae. Therefore, we suspect that these MLPs mediate flagellum-mediated chemotactic responses. Moreover, the deduced periplasmic domains of six MLPs (Mlp6 [VC0514], Mlp8 [VC0840 and AcfB], Mlp10 [VC1289], Mlp24 [VC2161], Mlp37 [VCA0923], and Mlp43 [VCA1069]) were found to contain a Cache domain, which has been implicated in binding to various small molecules among various sensors (3). A well-conserved sequence (P-X-X-D-Y-D-P-R-X-R-P-W-Y-X-D-A-[K/V]) was found just before the Cache domain (named a pre-Cache motif) (Fig. 2). The Cache domain is conserved among the extracellular or periplasmic portions of bacterial chemotaxis receptors from various Gram-positive and -negative species, spirochetes, Thermotoga, and Archaea (3). The pre-Cache motif was also found to be conserved among MCPs of various bacteria. In particular, PctA, PctB, and PctC of Pseudomonas aeruginosa (39) and McpB of Bacillus subtilis (22, 36), which are involved in amino acid taxis, have both the Cache domain and the pre-Cache motif. The Cache domain and the pre-Cache motif, though they are well conserved among MLPs, do not serve as separate domains but, rather, constitute two tandem PAS-like domains (44) as judged from the three-dimensional structure of the periplasmic domain of Mlp37 (VCA0923) of V. cholerae (Protein Data Bank [PDB] identification [ID], 3C8C). Each subunit of the homodimeric structure consists of two tandem PAS-like domains, providing two potential ligand-binding pockets (referred to as pockets I and II), representing a novel family of sensor domains of bacterial receptors (18, 44). Sequence similarities among MLPs of this family predict that they also share structural and functional homologies. Therefore, we suspect that this family of proteins in V. cholerae may contain one or more MLPs involved in chemotactic responses toward amino acids. We then focused on one of the candidates, Mlp24 (previously named McpX), which is required upon infection for full expression of the ctxAB and toxT genes, which encode CT and the transcriptional regulator, respectively (31). The mlp24 gene (VC2161) encodes a polypeptide with a calculated molecular mass of 67.7 kDa. Based on the sequence alignment of Mlp24 with Tsr of E. coli, Mlp24 contains two methylation helices as well as the HCD sequence (32). The amino sequences of Mlp24 from the El Tor (AC-V66) and the classical biotype (O395N1) were identical.
Fig 2.
Alignment of the periplasmic domains of MCPs from various bacterial species. Amino acid sequences of MLPs were aligned by using the ClustalW program, version 1.4. The Cache domain (3), consisting of the N- and C-terminal parts, and the pre-Cache motif identified in this study are indicated by separate lines above the sequences. White letters on a black background indicate residues identical to those of Mlp24. The names of the proteins are shown with the abbreviations of the species: Agrc, Agrobacterium tumefaciens; Bs, Bacillus subtilis; Cv, Chromobacterium violaceum; Vp, Vibrio parahaemolyticus; Vv, Vibrio vulnificus; Pa, Pseudononas aeruginosa; Pbpra, Photobacterium profundum; Smc, Sinorhizobium; Yp, Yersinia pestis.
Effects of an mlp24 deletion on amino acid taxis of classical biotype V. cholerae.
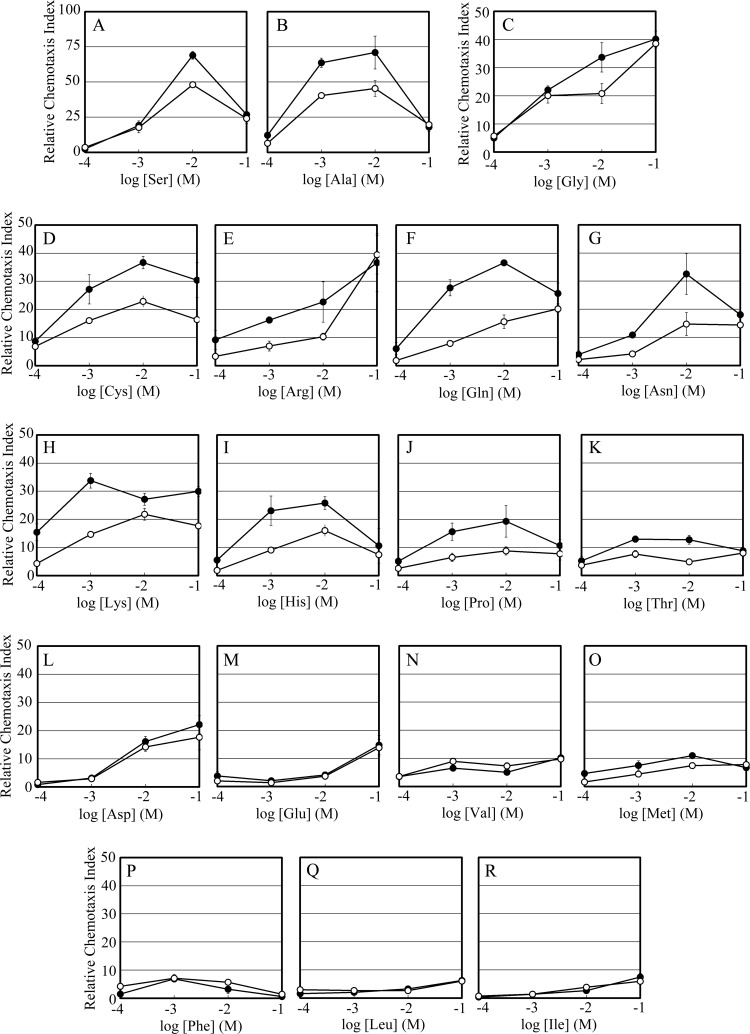
To test whether Mlp24 mediates taxis toward amino acids, we disrupted the mlp24 gene of the classical strain O395N1 and examined the resulting strain, Vmlp24, for its chemotactic abilities (Table 1; Fig. 3). The Δmlp24 mutant showed significantly weaker responses than its parent to 11 amino acids at 10 mM: serine, glycine, alanine, cysteine, arginine, asparagine, histidine, threonine, lysine, glutamine, and proline (Table 1). Further assays with a wider range of amino acid concentrations confirmed that Mlp24 is involved in attractant responses to these amino acids (Fig. 3A to K). Responses to the other amino acids, including the poorly soluble amino acids tyrosine and tryptophan, were not significantly affected by the mlp24 deletion at any concentration tested (data not shown). Note that attractant responses at higher concentrations of aspartate and glutamate are not supposed to be physiologically relevant, considering that asparagine and glutamine are reasonably strong attractants. Moreover, these responses, if any, were not affected by the Δmlp24 mutation.
Fig 3.
Chemotactic responses of the classical strain of V. cholerae and its Δmlp24 mutant toward amino acids indicated in each panel. Closed circles, O395N1; open circles, Δmlp24. Relative chemotaxis index is defined as a ratio of the number of bacteria in the capillary of interest to that in the control capillary filled with buffer in each set of experiments. Standard errors of triplicate data for each concentration are shown.
For complementation, we cloned the mlp24 gene from the classical strain. The coding region of mlp24 was amplified and cloned into the expression vector pAH901, so that the FLAG tag sequence was fused to the C terminus of Mlp24. O395N1-24 cells were transformed with the resulting plasmid, pMlp24. The expression of the Mlp24-FLAG protein complemented the defects in amino acid taxis of the Δmlp24 mutant except for arginine (Table 1). Methylation of Mlp24-FLAG was also examined to further confirm the attractant sensing. Methylation of an MCP increases its mobility upon SDS-PAGE, which can be detected by immunoblotting (7, 9, 10, 12). Mlp24 became more methylated in the presence of arginine, proline, and serine, whereas glutamate had no effect (Fig. 4A). Based on all these results, we conclude that Mlp24 mediates taxis toward 11 amino acids: serine, glycine, alanine, cysteine, arginine, asparagine, histidine, threonine, lysine, glutamine, and proline. Because the introduction of pMlp24 significantly enhanced taxis of the Δmlp24 strain toward methionine, valine, phenylalanine, isoleucine, and tyrosine, Mlp24 may be able to mediate taxis toward these amino acids, though it does not contribute to the tactic ability of the wild type toward them under the conditions tested. It should also be noted that there may be an additional MLP(s) involved in serine taxis because the Δmlp24 mutant retained a weak but significant chemotactic response to serine and several other amino acids. This is consistent with the results of competition assays in which threonine and proline did not inhibit serine taxis, whereas serine inhibited taxis to threonine and proline (Fig. 1C). In fact, we found that Mlp37, the closest homolog of Mlp24 among all V. cholerae MLPs, also mediates taxis to a similar but distinct set of amino acids (data not shown). The existence of multiple chemoreceptors involved in amino acid taxis can also account for the swarm assay of the Δmlp24 strain in tryptone semisolid agar, in which the Δmlp24 mutant showed a wild-type level of swarming (data not shown).
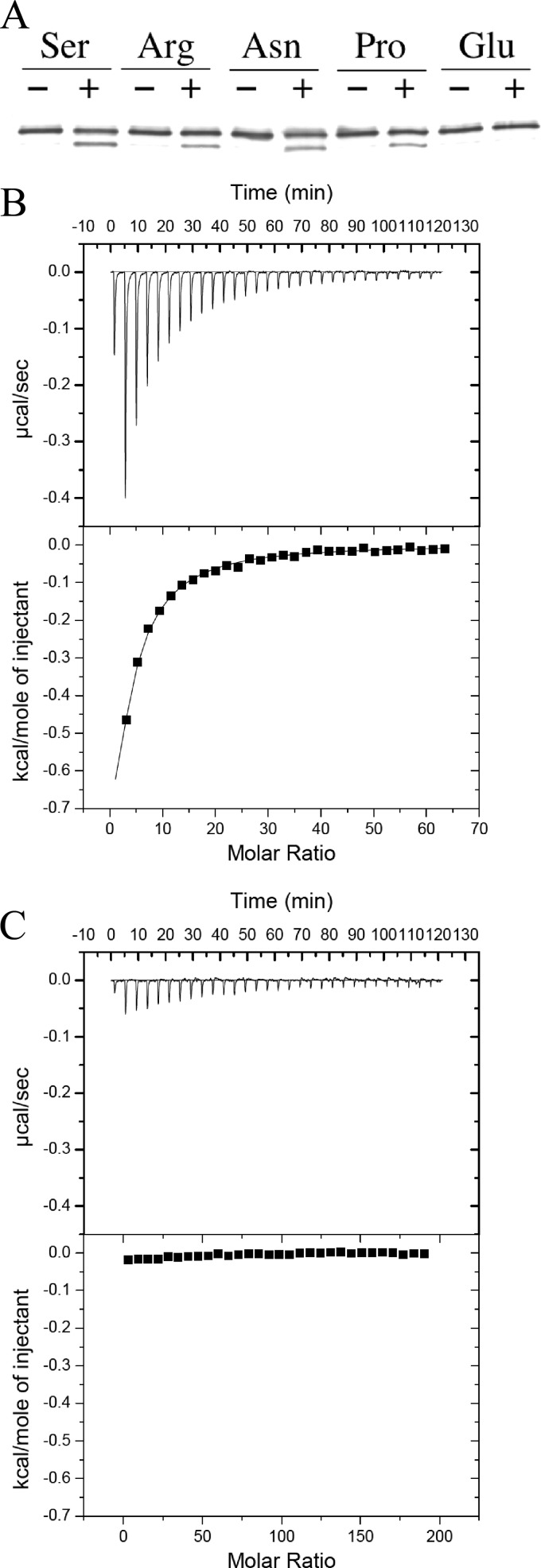
Fig 4.
Methylation and ligand-binding assays of Mlp24. (A) Methylation patterns of Mlp24 in the presence and absence of various amino acids. O395N1 cells (classical) expressing Mlp24-FLAG were incubated with (+) or without (−) serine (Ser), arginine (Arg), asparagine (Asn), proline (Pro), and glutamate (Glu), and Mlp24-FLAG was detected by immunoblotting with anti-FLAG antibody. (B and C) Binding of serine to the periplasmic fragment of Mlp24 (Mlp24p). ITC measurements of 10 μM Mlp24p were carried out with 10 mM serine (attractant [B]) and 30 mM glutamic acid (nonattractant [C]). Enthalpy changes per mole were plotted as a function of the molar ratio of serine to Mlp24p. The binding parameters calculated from the data are described in the text.
Characterization of an mlp24 deletion mutant from an El Tor strain.
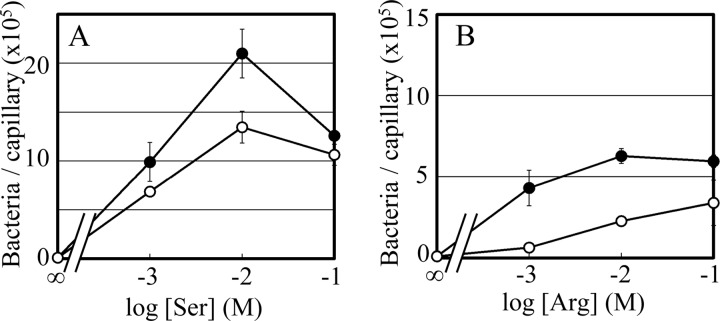
To test whether Mlp24 mediates amino acid taxis also in the El Tor biotype, we examined chemotactic responses of the Che+ strain AC-V66 and its mlp24 deletion derivative AC-V1400. As has been reported previously (31), the Δmlp24 mutant of the El Tor biotype swarms in semisolid agar to essentially the same extent as its parental strain (data not shown). Capillary assay, however, demonstrated significant defects of the Δmlp24 mutant strain: it showed significantly weaker responses than did its parent to serine (Fig. 5A), arginine (Fig. 5B), alanine, asparagine, glutamine, lysine, proline, and threonine (data not shown). The mlp24 gene cloned from the classical strain O395N1, which encodes an amino acid sequence identical to that of El Tor biotype, fully or partially complemented the defects in amino acid taxis (except for threonine) of the Δmlp24 strain (data not shown). Methylation patterns of Mlp24-FLAG in AC-V1400 cells were essentially the same as those in the classical biotype strain (data not shown). These results demonstrate that Mlp24 mediates attractant responses to various amino acids also in the El Tor biotype, although there might be minor differences between the two biotypes.
Fig 5.
Chemotactic responses of an El Tor biotype strain of V. cholerae and its Δmlp24 mutant toward serine (A) and arginine (B). Closed circles, AC-V66 and O395N1; open circles, its mlp24-deletion derivative AC-V1400 (Δmlp24). Standard errors of triplicate data for each concentration are shown.
Amino acid binding of the periplasmic fragment of Mlp24.
To test whether the set of amino acids that serves as attractants for V. cholerae binds directly to Mlp24, we employed isothermal titration calorimetry (ITC) with a periplasmic fragment of Mlp24 (Mlp24p). Construction and purification of Mlp24p were carried out as described in Materials and Methods. Mlp24p was then subjected to ITC analyses. The titration curve with serine (Fig. 4B) shows a saturation of heat release after 10 or more injections. This result suggests direct binding of serine to Mlp24p with a dissociation constant (Kd) of 71.9 μM, calculated by assuming that Mlp24p binds one serine molecule per monomer. ITC analyses also suggested that arginine, asparagine, and proline bind directly to Mlp24p with Kd values of 2.5 μM, 42.9 μM, and 74.6 μM, respectively (titration curves not shown). In contrast, the addition of glutamate to Mlp24p elicited no significant release or uptake of heat (Fig. 4C), a result which is consistent with the fact that this amino acid did not elicit a significant attractant response. It is striking that arginine had an affinity 1 order of magnitude higher than that of serine, a result which seems contradictory to the finding that the former elicited weaker attractant responses (Table 1). However, lower concentrations of arginine showed stronger attractant activity (Fig. 6C). Therefore, 10 mM arginine in the capillary may be too high a concentration in that bacteria swimming uphill of the concentration gradient may be concentrated primarily in the region just outside the mouth of the capillary, resulting in lower cell numbers in the capillary.
Fig 6.
Mutations introduced into the potential ligand-binding pockets of Mlp24. (A and B) Side chains of the residues constituting the potential binding pockets I (A) and II (B) of Mlp37 (PDB ID, 3C8C). The alanine molecule (shown with a space filling model) in pocket I (A) exists in the crystal. Residues conserved in Mlp24 are circled. Those corresponding to the Mlp24 mutations constructed in this study are marked with asterisks: *1, Y120A; *2, R125A; *3, Y192A; and *4, H205A. (C) Capillary assays of Vmlp201 (Δmlp24 Δmlp37) cells (derived from O395N1) carrying the vector (pAH901, open bars) or the plasmid encoding wild-type or mutant Mlp24 (wild type, hatched bars; Y120A, dotted bars; R125A, checked bars; Y192A, cross-hatched bars; H205A, closed bars). Capillaries were filled with TMN buffers containing each amino acid (1, 10, or 100 mM) shown at the bottom.
Mutational analyses of two potential ligand-binding pockets of Mlp24.
Each subunit of the homodimeric periplasmic domain of Mlp37 (PDB ID, 3C8C), the closest homolog of Mlp24, consists of two tandem PAS-like domains, providing two potential ligand-binding pockets: membrane-distal and -proximal ones are referred to as pockets I and II, respectively. Since Mlp37 and Mlp24 share 59% identity across their periplasmic domains, Mlp24 is expected to have a similar structure. We used this information to examine which pocket(s) of Mlp24 is responsible for amino acid sensing by introducing an alanine substitution for putative ligand-binding residues in pocket I (Y120A and R125A [Fig. 6A]) or pocket II (Y192A and H205A [Fig. 6B]). For capillary assays, we used the double mutant strain Vmlp201 (Δmlp24 Δmlp37) as a host to reduce residual amino acid taxis. All the mutant proteins were expressed essentially at the wild-type levels (data not shown). As shown in Fig. 6C, both pocket I mutants (Y120A and R125A) showed defects in mediating taxis toward serine, alanine, arginine, asparagine, and proline, suggesting the involvement of pocket I in amino acid recognition. In particular, the Y120A mutant mediated significant levels of taxis to arginine at higher concentrations (10 and 100 mM), suggesting that the affinity of the mutant to arginine is lower than that of wild-type Mlp24. Cells carrying these mutations showed better performances toward a high (normally inhibitory) concentration of arginine (100 mM) than cells carrying wild-type Mlp24, implying reduced affinities of these mutants for arginine. In contrast, the two pocket II mutations, Y192A and H205A, had much milder, if any, effects (Fig. 6C). These results suggest that at least pocket I is involved in sensing of the amino acid attractants.
DISCUSSION
In this study, we demonstrated that the V. cholerae classical biotype strain O395N1 is attracted by 19 of 20 proteinaceous amino acids and that Mlp24 mediates taxis toward at least 11 of them, i.e., serine, alanine, glycine, cysteine, arginine, glutamine, asparagine, lysine, histidine, proline, and threonine. No significant taxis toward tyrosine was detected. The apparent discrepancy with the reference (16) which reported that all 20 amino acid serve as attractants might be due to the differences in the experimental conditions, the strain, or both.
It is likely that V. cholerae has multiple chemoreceptors for amino acids. Mlp24 and five other MLPs of V. cholerae, including Mlp37, and amino acid receptors of P. aeruginosa (39) and B. subtilis (22, 36) have the Cache domain (3) and the pre-Cache motif in their periplasmic domains, arguing that some of these five MLPs may be involved in amino acid taxis. The latter motif, in particular, appears to form a putative ligand-binding pocket of the first PAS-like domain in the Mlp37 structure (PDB ID, 3C8C). Indeed, our preliminary results suggest that Mlp37 mediates taxis toward amino acids, including serine, and that even the double mutant lacking both Mlp24 and Mlp37 shows a weak but significant taxis to serine and other amino acids. However, it should be noted that Mlp8 and Mlp10, which are homologs of Mlp24, do not mediate taxis toward any amino acid tested, at least under our experimental conditions, suggesting that the overall sequence and structural similarities of the periplasmic domain of an MLP to those of Mlp24 and Mlp37 per se do not necessarily guarantee its function as an amino acid chemoreceptor.
The ITC analyses with Mlp24p revealed that at least four amino acid attractants, i.e., serine, arginine, asparagine, and proline, bind directly to Mlp24, and therefore, periplasmic binding proteins are not required for sensing of these amino acids via Mlp24. The Kd values for these amino acids were similar to or 1 order of magnitude higher than the reported values of E. coli Tsr or Tar for serine or aspartate (11, 28, 33).
A series of experiments with Mlp24 point mutants, which inactivate either one of the two pockets of Mlp24, revealed the involvement of the first pocket in sensing of alanine, arginine, asparagine, proline, and serine. In contrast, the pocket II mutations had milder effects. These results are consistent with a recent study (18) on McpB of B. subtilis, suggesting that only the first PAS-like domain is responsible for asparagine binding.
One important question is why Mlp24 is required for the expression of the ctxAB operon in the infant mouse model of V. cholerae intestinal colonization (31). Certain amino acids (asparagine, arginine, glutamate, and serine) are known to stimulate the expression of ctxAB, tcpA, and ompU, all of which are under the control of the ToxR regulon (35). The results obtained in this study indicate that among these four amino acids, asparagine, arginine, and serine serve as attractants for Mlp24. Therefore, we hypothesize that Mlp24 is required to migrate toward amino acid-rich environments in the host intestine, where a sensor detects those amino acids to turn on the ctxAB operon through the virulence cascade consisting of ToxR, TcpP, and ToxT (35). One such candidate sensor is the VieS histidine kinase, which, together with the response regulator VieA, is required for amino acid stimulation of the ToxR regulon in minimal medium (41).
However, Mlp24 might have another function(s) in pathogenicity. The nonchemotactic point and deletion cheY3 mutants, which rotate their flagella exclusively counterclockwise, greatly outcompete the wild-type strain upon colonization in mouse intestine (8, 31). The wild-type strain and the cheY3 derivatives colonize in different segments of small intestine, where the wild type preferentially colonizes the lower half and the cheY3 mutant colonizes throughout (31). This increased colonization ability is believed to result from the motile but nonchemotactic phenotype of the cheY3 mutant (8). It is likely that cells lacking mlp24 behave differently than the cheY3 mutant in the intestine because they are competent for chemotaxis toward some attractants (or away from some repellents). In this scenario, fine-tuning of chemotaxis using Mlp24 may be required for colonization of specific sites where signals to induce the ToxR regulon are received. Thus, our finding that Mlp24 mediates taxis to several amino acids raises the possibility that this set of amino acids plays a critical role in pathogenicity of V. cholerae.
ACKNOWLEDGMENTS
We thank A. Tsuneshige of Hosei University for his generous help with ITC measurements and G. G. Hiremath of Hosei University, Y. Sowa of Hosei University, and T. Inaba of RIKEN (Wako City, Japan) for discussion and encouragement.
This work was partially supported by Grants-in-Aid for Scientific Research from the Japan Society for Promotion of Science and from the Ministry of Education, Culture, Sports, Science and Technology of Japan (MEXT) (to M.H. and I.K.) and the MEXT-supported Program for the Strategic Research Foundation at Private Universities, 2008–2012 (to I.K.), and by the Howard Hughes Medical Institute and the U.S. National Institutes of Health (AI45746) (to A.C.).
Footnotes
Published ahead of print 2 July 2012
REFERENCES
- 1. Adler J, Hazelbauer GL, Dahl MM. 1973. Chemotaxis toward sugars in Escherichia coli. J. Bacteriol. 115:824–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Alm RA, Manning PA. 1990. Characterization of the hlyB gene and its role in the production of the El Tor haemolysin of Vibrio cholerae O1. Mol. Microbiol. 4:413–425 [DOI] [PubMed] [Google Scholar]
- 3. Anantharaman V, Aravind L. 2000. Cache—a signaling domain common to animal Ca2+-channel subunits and a class of prokaryotic chemotaxis receptors. Trends Biochem. Sci. 25:535–537 [DOI] [PubMed] [Google Scholar]
- 4. Armitage JP. 1999. Bacterial tactic responses. Adv. Microb. Physiol. 41:229–289 [DOI] [PubMed] [Google Scholar]
- 5. Banerjee R, et al. 2002. Involvement of in vivo induced cheY-4 gene of Vibrio cholerae in motility, early adherence to intestinal epithelial cells and regulation of virulence factors. FEBS Lett. 532:221–226 [DOI] [PubMed] [Google Scholar]
- 6. Boin MA, Austin MJ, Hase CC. 2004. Chemotaxis in Vibrio cholerae. FEMS Microbiol. Lett. 239:1–8 [DOI] [PubMed] [Google Scholar]
- 7. Boyd A, Simon MI. 1980. Multiple electrophoretic forms of methyl-accepting chemotaxis proteins generated by stimulus-elicited methylation in Escherichia coli. J. Bacteriol. 143:809–815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Butler SM, Camilli A. 2004. Both chemotaxis and net motility greatly influence the infectivity of Vibrio cholerae. Proc. Natl. Acad. Sci. U. S. A. 101:5018–5023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chelsky D, Dahlquist FW. 1980. Structural studies of methyl-accepting chemotaxis proteins of Escherichia coli: evidence for multiple methylation sites. Proc. Natl. Acad. Sci. U. S. A. 77:2434–2438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. DeFranco AL, Koshland DE., Jr 1980. Multiple methylation in processing of sensory signals during bacterial chemotaxis. Proc. Natl. Acad. Sci. U. S. A. 77:2429–2433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dunten P, Koshland DE., Jr 1991. Tuning the responsiveness of a sensory receptor via covalent modification. J. Biol. Chem. 266:1491–1496 [PubMed] [Google Scholar]
- 12. Engström P, Hazelbauer GL. 1980. Multiple methylation of methyl-accepting chemotaxis proteins during adaptation of E. coli to chemical stimuli. Cell 20:165–171 [DOI] [PubMed] [Google Scholar]
- 13. Everiss KD, Hughes KJ, Kovach ME, Peterson KM. 1994. The Vibrio cholerae acfB colonization determinant encodes an inner membrane protein that is related to a family of signal-transducing proteins. Infect. Immun. 62:3289–3298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Everiss KD, Hughes KJ, Peterson KM. 1994. The accessory colonization factor and toxin-coregulated pilus gene clusters are physically linked on the Vibrio cholerae O395 chromosome. DNA Seq. 5:51–55 [DOI] [PubMed] [Google Scholar]
- 15. Freter R, Allweiss B, O'Brien PC, Halstead SA, Macsai MS. 1981. Role of chemotaxis in the association of motile bacteria with intestinal mucosa: in vitro studies. Infect. Immun. 34:241–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Freter R, O'Brien PC. 1981. Role of chemotaxis in the association of motile bacteria with intestinal mucosa: chemotactic responses of Vibrio cholerae and description of motile nonchemotactic mutants. Infect. Immun. 34:215–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Freter R, O'Brien PC, Macsai MS. 1981. Role of chemotaxis in the association of motile bacteria with intestinal mucosa: in vivo studies. Infect. Immun. 34:234–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Glekas GD, et al. 2010. A PAS domain binds asparagine in the chemotaxis receptor McpB in Bacillus subtilis. J. Biol. Chem. 285:1870–1878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gosink KK, Kobayashi R, Kawagishi I, Häse CC. 2002. Analyses of the roles of the three cheA homologs in chemotaxis of Vibrio cholerae. J. Bacteriol. 184:1767–1771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gupta S, Chowdhury R. 1997. Bile affects production of virulence factors and motility of Vibrio cholerae. Infect. Immun. 65:1131–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hang L, et al. 2003. Use of in vivo-induced antigen technology (IVIAT) to identify genes uniquely expressed during human infection with Vibrio cholerae. Proc. Natl. Acad. Sci. U. S. A. 100:8508–8513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hanlon DW, Ordal GW. 1994. Cloning and characterization of genes encoding methyl-accepting chemotaxis proteins in Bacillus subtilis. J. Biol. Chem. 269:14038–14046 [PubMed] [Google Scholar]
- 23. Harkey CW, Everiss KD, Peterson KM. 1994. The Vibrio cholerae toxin-coregulated-pilus gene tcpI encodes a homolog of methyl-accepting chemotaxis proteins. Infect. Immun. 62:2669–2678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hazelbauer GL, Falke JJ, Parkinson JS. 2008. Bacterial chemoreceptors: high-performance signaling in networked arrays. Trends Biochem. Sci. 33:9–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hedblom ML, Adler J. 1983. Chemotactic response of Escherichia coli to chemically synthesized amino acids. J. Bacteriol. 155:1463–1466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Heidelberg JF, et al. 2000. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature 406:477–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hyakutake A, et al. 2005. Only one of the five CheY homologs in Vibrio cholerae directly switches flagellar rotation. J. Bacteriol. 187:8403–8410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Iwama T, Homma M, Kawagishi I. 1997. Uncoupling of ligand-binding affinity of the bacterial serine chemoreceptor from methylation- and temperature-modulated signaling states. J. Biol. Chem. 272:13810–13815 [DOI] [PubMed] [Google Scholar]
- 29. Jeffery CJ, Koshland DE., Jr 1993. Vibrio cholerae hlyB is a member of the chemotaxis receptor gene family. Protein Sci. 2:1532–1535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Krukonis ES, DiRita VJ. 2003. From motility to virulence: sensing and responding to environmental signals in Vibrio cholerae. Curr. Opin. Microbiol. 6:186–190 [DOI] [PubMed] [Google Scholar]
- 31. Lee SH, Butler SM, Camilli A. 2001. Selection for in vivo regulators of bacterial virulence. Proc. Natl. Acad. Sci. U. S. A. 98:6889–6894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Le Moual H, Koshland DE., Jr 1996. Molecular evolution of the C-terminal cytoplasmic domain of a superfamily of bacterial receptors involved in taxis. J. Mol. Biol. 261:568–585 [DOI] [PubMed] [Google Scholar]
- 33. Lin LN, Li J, Brandts JF, Weis RM. 1994. The serine receptor of bacterial chemotaxis exhibits half-site saturation for serine binding. Biochemistry 33:6564–6570 [DOI] [PubMed] [Google Scholar]
- 34. Mesibov R, Adler J. 1972. Chemotaxis toward amino acids in Escherichia coli. J. Bacteriol. 112:315–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Miller VL, Mekalanos JJ. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 170:2575–2583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Müller J, Schiel S, Ordal GW, Saxild HH. 1997. Functional and genetic characterization of mcpC, which encodes a third methyl-accepting chemotaxis protein in Bacillus subtilis. Microbiology 143:3231–3240 [DOI] [PubMed] [Google Scholar]
- 37. Peterson KM. 2002. Expression of Vibrio cholerae virulence genes in response to environmental signals. Curr. Issues Intest. Microbiol. 3:29–38 [PubMed] [Google Scholar]
- 37a. R Development Core Team 2012. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: http://www.R-project.org/ [Google Scholar]
- 38. Reidl J, Klose KE. 2002. Vibrio cholerae and cholera: out of the water and into the host. FEMS Microbiol. Rev. 26:125–139 [DOI] [PubMed] [Google Scholar]
- 39. Taguchi K, Fukutomi H, Kuroda A, Kato J, Ohtake H. 1997. Genetic identification of chemotactic transducers for amino acids in Pseudomonas aeruginosa. Microbiology 143:3223–3229 [DOI] [PubMed] [Google Scholar]
- 40. Tajima H, et al. 2011. Ligand specificity determined by differentially arranged common ligand-binding residues in bacterial amino acid chemoreceptors Tsr and Tar. J. Biol. Chem. 286:42200–42210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tischler AD, Camilli A. 2005. Cyclic diguanylate regulates Vibrio cholerae virulence gene expression. Infect. Immun. 73:5873–5882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wolfe AJ, Berg HC. 1989. Migration of bacteria in semisolid agar. Proc. Natl. Acad. Sci. U. S. A. 86:6973–6977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Xu M, Yamamoto K, Honda T, Ming X. 1994. Construction and characterization of an isogenic mutant of Vibrio parahaemolyticus having a deletion in the thermostable direct hemolysin-related hemolysin gene. J. Bacteriol. 176:4757–4760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zhang Z, Hendrickson WA. 2010. Structural characterization of the predominant family of histidine kinase sensor domains. J. Mol. Biol. 400:335–353 [DOI] [PMC free article] [PubMed] [Google Scholar]