Abstract
Mycobacterium avium subsp. paratuberculosis is the causative agent of Johne's disease in cattle and may have implications for human health. Establishment of chronic infection by M. avium subsp. paratuberculosis depends on its subversion of host immune responses. This includes blocking the ability of infected macrophages to be activated by gamma interferon (IFN-γ) for clearance of this intracellular pathogen. To define the mechanism by which M. avium subsp. paratuberculosis subverts this critical host cell function, patterns of signal transduction to IFN-γ stimulation of uninfected and M. avium subsp. paratuberculosis-infected bovine monocytes were determined through bovine-specific peptide arrays for kinome analysis. Pathway analysis of the kinome data indicated activation of the JAK-STAT pathway, a hallmark of IFN-γ signaling, in uninfected monocytes. In contrast, IFN-γ stimulation of M. avium subsp. paratuberculosis-infected monocytes failed to induce patterns of peptide phosphorylation consistent with JAK-STAT activation. The inability of IFN-γ to induce differential phosphorylation of peptides corresponding to early JAK-STAT intermediates in infected monocytes indicates that M. avium subsp. paratuberculosis blocks responsiveness at, or near, the IFN-γ receptor. Consistent with this hypothesis, increased expression of negative regulators of the IFN-γ receptors SOCS1 and SOCS3 as well as decreased expression of IFN-γ receptor chains 1 and 2 is observed in M. avium subsp. paratuberculosis-infected monocytes. These patterns of expression are functionally consistent with the kinome data and offer a mechanistic explanation for this critical M. avium subsp. paratuberculosis behavior. Understanding this mechanism may contribute to the rational design of more effective vaccines and/or therapeutics for Johne's disease.
INTRODUCTION
Mycobacterium avium subsp. paratuberculosis is the causative agent of Johne's disease, a chronic inflammatory disorder of the gastrointestinal tract of ruminants (29, 55). Johne's disease is of considerable economic importance to the dairy industry as it is responsible for the highest average production losses among five production-limiting diseases (11, 56). There is additional growing concern that M. avium subsp. paratuberculosis may be a causative or contributing factor to Crohn's disease in humans. While this link has yet to be conclusively determined (coincidence does not indicate causation), there is considerable circumstantial evidence implicating M. avium subsp. paratuberculosis in Crohn's disease (24, 42, 50). M. avium subsp. paratuberculosis has also been implicated as a trigger for type 1 diabetes (46) and ulcerative colitis (45). The potential zoonotic threat and realized economic impact of Johne's disease have energized efforts for development of effective disease management strategies. The limited success of these efforts to date indicates that greater understanding of the biology and virulence mechanisms of M. avium subsp. paratuberculosis is required to adopt a more strategic approach to the design of vaccines or other therapeutics. Specifically, an understanding of the mechanisms by which M. avium subsp. paratuberculosis subverts host immune responses could form the basis for development of a successful vaccine and/or other therapeutics.
When colonizing the bovine host, M. avium subsp. paratuberculosis establishes persistent infections within host macrophages in the small intestine. This requires M. avium subsp. paratuberculosis to subvert the normal functions of the macrophage which would result in destruction of the internalized bacteria (57, 59). In doing so, M. avium subsp. paratuberculosis is able to convert the cells which would normally be responsible for destruction of an invading pathogen into a protected haven from the host immune response. Subversion of macrophage function appears to involve a number of mechanisms. For example, M. avium subsp. paratuberculosis has been well characterized for its ability to block maturation of the phagolysosomes (26). The bacterium also appears to interfere with other host processes which are equally essential for effective clearance of intracellular pathogens, including blocking the ability of the infected host to utilize gamma interferon (IFN-γ) for induction of protective innate immune responses. It has been observed that cattle in the excretory, subclinical stage of Johne's disease have increased IFN-γ at the site of infection (53) as well as higher IFN-γ production in culture supernatants after stimulation of peripheral blood mononuclear cells (PBMC) with M. avium subsp. paratuberculosis antigens (14). M. avium subsp. paratuberculosis appears to block the ability of the infected cells to respond to IFN-γ; macrophages pretreated with IFN-γ effectively clear mycobacteria (9, 25) while the same treatment, given postinfection, is unable to achieve efficient destruction of the bacterium (17, 48). Collectively, these results indicate an inability of the infected animals to respond to, rather than produce, IFN-γ. The mechanism(s) by which M. avium subsp. paratuberculosis blocks host cell IFN-γ responsiveness has yet to be determined.
Gamma interferon plays a central role in the immune defense against a variety of intracellular pathogens including mycobacteria (18, 19). Mice deficient in IFN-γ show increased susceptibility to intracellular pathogens (13, 15), and humans with mutations of the IFN-γ receptor are susceptible to infection with low-virulence mycobacterial strains and suffer severe and recurrent episodes of tuberculosis (20, 34). IFN-γ is released from T cells and natural killer cells to activate targets cells through a high-affinity receptor composed of two chains: IFN-γ receptor 1 (IFNGR1) and IFN-γ receptor 2 (IFNGR2). Signal transduction by IFN-γ is classically associated with a specific Janus family kinase-signal transducer and activator of transcription (JAK-STAT) signaling cascade (5, 16). Ligand binding by the IFN-γ receptor causes phosphorylation of Jak1 and Jak2 with subsequent phosphorylation of IFNGR1 (6, 32). Phosphorylation of IFNGR1 results in recruitment and phosphorylation of STAT1, which translocates to the nucleus to activate transcription of IFN-γ-inducible genes (28). IFN-γ acts primarily through regulation of gene expression to induce macrophages to kill intracellular pathogens.
To avoid destruction by the ensuing host defense response, a number of viral and bacterial pathogens have evolved strategies to block the IFN-γ responsiveness of infected cells. Different pathogens disrupt this host response at a variety of levels, which range from actions targeted to specific gene products to general inhibition of IFN-γ signaling. Furthermore, these studies indicate that JAK-STAT signaling can be inhibited at a number of levels, including at the receptor, intermediate signal molecules, or final effectors. At the receptor, a number of pathogens decrease expression of IFNGR1, IFNGR2, or both; Trypanosoma cruzi (35) and Leishmania donovani (47) decrease expression of IFNGR1, adenovirus decreases expression of IFNGR2, and Mycobacterium avium decreases expression of both IFNGR1 and -R2 (31). IFN-γ responsiveness can also be dampened by reducing the quantity or activation status of JAK-STAT pathway intermediates; human cytomegalovirus targets JAK kinases for degradation (40), mumps virus reduces levels of STAT1 (27), varicella-zoster virus reduces levels of Jak2 and STAT1 (1), and L. donovani activates protein tyrosine phosphatase SHP-1 for dephosphorylation and inactivation of Jak2 (7). Microbial pathogens also target the JAK-STAT transcriptional effectors; adenovirus inhibits IFN-γ-induced gene expression through direct interaction with cellular transcription factors (21, 38). Collectively, targeted disruption of the IFN-γ response by a variety of viral and bacterial pathogens emphasizes the importance of this system in the host defense against intracellular pathogens. The diverse mechanisms employed suggest that different points of intervention are more appropriate, or easily achieved, by different pathogens.
In this report we investigate patterns of host signal transduction in uninfected and M. avium subsp. paratuberculosis-infected bovine monocytes in response to IFN-γ stimulation. Analysis is performed through a novel bovine-specific peptide array for kinome analysis. Pathway analysis of the kinome data indicates activation of the JAK-STAT pathway, a hallmark of IFN-γ signaling, in uninfected monocytes. In contrast, IFN-γ stimulation of M. avium subsp. paratuberculosis-infected monocytes fails to induce patterns of peptide phosphorylation consistent with JAK-STAT activation. The inability of IFN-γ to induce differential phosphorylation of peptides corresponding to early JAK-STAT intermediates in infected monocytes indicates that M. avium subsp. paratuberculosis blocks responsiveness at, or near, the IFN-γ receptor. Consistent with this hypothesis, we report increased expression of negative regulators of the IFN-γ receptor SOCS1 and, to a lesser extent, SOCS3, as well as decreased expression of IFNGR1 and IFNGR2 in M. avium subsp. paratuberculosis-infected monocytes. These responses are anticipated to contribute to the inability of M. avium subsp. paratuberculosis-infected cells to be activated by IFN-γ. This, in turn, contributes to the ability of M. avium subsp. paratuberculosis to convert cells which would normally be responsible for mediating bacterial clearance into protected havens for bacterial proliferation.
MATERIALS AND METHODS
Isolation of bovine blood monocytes.
Blood was collected from three cattle (9-month-old Charolais-cross steers, designated animals 89, 136, and 148) by venipuncture using tubes containing EDTA as an anticoagulant. Blood was transferred to 50-ml polypropylene tubes and centrifuged at 1,400 × g for 20 min at 20°C. White blood cells were isolated from the buffy coat and mixed with Ca2+- and Mg2+-free phosphate-buffered saline (PBSA) to a final volume of 35 ml. The cell suspension was layered onto 15 ml of 54% isotonic Percoll (Amersham Biosciences/GE Health Care) and centrifuged at 2,000 × g for 20 min at 20°C. Peripheral blood mononuclear cells (PBMC) from the Percoll-PBSA interface were collected and washed three times with cold PBSA. Monocytes were purified from isolated PBMC by magnetic cell sorting (MACS) purification using CD14+ microbeads (Miltenyi Biotec Inc., Auburn, CA). Monocytes (>95% pure) were plated at 5 × 106 cells/well in six-well plates in RPMI 1640 medium (Gibco) supplemented with 10% fetal bovine serum (Gibco). Isolated monocytes were rested overnight prior to stimulation.
Infection of bovine monocytes with M. avium subsp. paratuberculosis.
M. avium subsp. paratuberculosis K10 culture was incubated at 37°C on Middlebrook 7H10 agar (Difco Labs, Detroit, MI) with enrichment medium composed of oleic acid, albumin, dextrose, and catalase (OADC; Difco Labs, Detroit, MI) and mycobactin J (Allied Monitor Inc., Fayette, MO). After 3 to 4 weeks of growth, colonies were transferred to Middlebrook 7H9 broth (Difco Labs, Detroit, MI) containing 0.05% Tween 80 (Sigma Chemical Co., St. Louis, MO), OADC enrichment medium, and mycobactin J and incubated at 37°C for 5 days to achieve log-phase growth. The number of CFU was determined using the pelleted wet weight method. Briefly, a 50-ml centrifuge tube was weighed prior to the addition of 50 ml of a 5-day liquid M. avium subsp. paratuberculosis culture. The culture was centrifuged at 3,400 × g for 30 min. Supernatant was decanted, and the pellet was dried for 30 min. Tube weight was then recorded, and pellet weight was determined according to Hines et al. (30), whereby 1 mg of M. avium subsp. paratuberculosis pellet is equal to 107 CFU. The M. avium subsp. paratuberculosis pellet was then resuspended in the appropriate volume of cell culture medium to achieve a 5:1 multiplicity of infection (MOI). Appropriate bacterial loads were added to each well containing 5 million monocytes/well. Plates, both infected and control, were spun at 300 × g for 2 min. All plates were incubated for 3 h at 37°C. Medium was removed, and cells were washed three times with warm RPMI 1640 medium. Cells were rested overnight at 37°C prior to stimulation with 10 ng/ml recombinant bovine IFN-γ for 1 h. Cells were treated with 10 ng/ml IFN-γ and then harvested at the appropriate time for either kinome or quantitative reverse transcription-PCR (qRT-PCR) analysis. Kinome analysis was performed 1 h after infection while qRT-PCR was performed after both 1 h and 18 h of infection.
RNA extraction.
Total RNA extraction was performed as per the RNeasy Mini Kit protocol (Qiagen). Briefly, 1 ml of Buffer RLT supplemented with beta-mercaptoethanol was added to each well for 5 min. Cells were collected in a 2-ml tube, vortexed briefly, and stored at −80°C until further processing. Homogenization of samples was achieved by running samples through a QIAshredder (Qiagen). Molecular-grade ethanol was added to each sample before the sample was run through an RNeasy Mini Spin Column. An optional DNase treatment was performed on each sample by the addition of a DNase solution (Qiagen) to the column and allowing the solution to sit for 15 min. Three washes were performed, followed by elution in nuclease-free water. Each sample was quantified and checked for purity using a 2100 Bioanalyzer (Agilent Technologies, Inc.).
Preparation of a cDNA library.
RNA (200 ng) was converted to cDNA by adding 8 μl of 2× RT buffer and 2 μl of RT enzyme (Invitrogen) to a total volume of 10 μl. A master mix of buffer and enzyme was made to eliminate pipetting errors. Samples were placed in a thermocycler under the following conditions: 25°C for 5 min, 50°C for 60 min, and 70°C for 15 min. RNA template was removed by the addition of 1 μl of Escherichia coli RNase H for 20 min. cDNA was stored at −20°C.
qRT-PCR.
Each reaction mixture for qRT-PCR included 9 μl of iQ SYBR green Master Mix (Bio-Rad), 3 μl of primer mix (3.3 μM), 2 μl of nuclease-free water, and 1 μl of cDNA for a total of a 15-μl reaction volume. Thermocycler conditions were as follows: cycle 1, 55°C for 2 min; cycle 2, 95°C for 8.5 min; cycle 3, 95°C for 15 s, 55°C for 30 s, and 72°C for 30 s; cycle 4, 55°C for 10 s. The set point temperature was increased by 1°C after cycle 2. Results were analyzed using the 2−ΔΔCT method (where CT is threshold cycle) described in Applied Biosystems User Bulletin 2 (4).
Monocyte TNF-α release in response to IFN-γ stimulation.
Purified monocytes (uninfected and M. avium subsp. paratuberculosis infected) were prepared as described earlier. Recombinant bovine IFN-γ (Ciba-Geigi) was added at a final concentration of 10 ng/ml. Plates were returned to an incubator overnight. Supernatant was collected from each well, diluted (1:2), and used for enzyme-linked immunosorbent assays (ELISAs) for bovine tumor necrosis factor alpha (TNF-α) as per Ellis et al. (23).
Cytospins.
Cells were harvested using a trypsin-ethylenediamine tetraacetic acid (Versene) solution. The cells were prepped for cytospins by centrifugation at 325 × g for 5 min. Cells were resuspended in 200 μl of PBSA–0.1% EDTA. Cytospins were performed by adding 100 μl of cell suspension to the apparatus and spinning the samples at 1,000 rpm for 3 min onto a glass slide. Slides were allowed to dry overnight in a fume hood. Cells were heat fixed to slides by briefly passing the samples through a flame. Slides were placed over boiling water and stained with carbol fuchsin for 5 min and rinsed, and acid destaining solution was briefly added to each slide before slides were rinsed with water. Slides were counterstained using methylene blue (Sigma) for 1 min and rinsed with water. Slides were allowed to dry overnight in a fume hood. The next day, each cytospot was fixed using Entellen New Rapid Mounting Medium (EMScience) with a coverslip. Cells were observed on a light microscope under oil immersion (magnification, ×100).
Peptide arrays.
Design, construction, and application of the peptide arrays were based upon a previously reported protocol with modifications (33). Notably, the kinome experiments for all the animals were performed simultaneously in a single run, minimizing the possibility of technical variances in the analysis. Briefly, approximately 10 × 106 cells were collected, pelleted, and lysed by addition of 100 μl of lysis buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl,1 mM EDTA, 1 mM EGTA, 1% Triton, 2.5 mM sodium pyrophosphate, 1 mM Na3VO4,1 mM NaF,1 μg/ml leupeptin, 1 g/ml aprotinin, 1 mM phenylmethylsulfonyl fluoride [PMSF]) (all products from Sigma-Aldrich unless indicated). Cells were incubated on ice for 10 min and spun in a microcentrifuge for 10 min at 4°C. A 70-μl aliquot of this supernatant was mixed with 10 μl of activation mix (50% glycerol, 500 μM ATP [New England BioLabs, Pickering, ON, Canada], 60 mM MgCl2, 0.05% [vol/vol] Brij 35, 0.25 mg/ml bovine serum albumin [BSA]) and incubated on the array for 2 h at 37°C. Arrays were then washed with phosphate-buffered saline (PBS)–1% Triton.
Slides were submerged in phospho-specific fluorescent ProQ Diamond Phosphoprotein Stain (Invitrogen) with agitation for 1 h. Arrays were then washed three times in destaining solution containing 20% acetonitrile (EMD Biosciences, VWR Distributor, Mississauga, ON, Canada) and 50 mM sodium acetate (Sigma) at pH 4.0 for 10 min. A final wash was done with distilled deionized H2O. Arrays were air dried for 20 min and then centrifuged at 300 × g for 2 min to remove any remaining moisture from the array. Arrays were read using a GenePix Professional 4200A microarray scanner (MDS Analytical Technologies, Toronto, ON, Canada) at 532 to 560 nm with a 580-nm filter to detect dye fluorescence. Images were collected using GenePix, version 6.0, software (MDS), and the spot intensity signal was collected as the mean of pixel intensity using local feature background intensity calculation with the default scanner saturation level.
Data analysis: data sets.
The data set contains the signal intensities associated with each of 300 peptides for the monocytes from three animals under four different treatments. The treatments included IFN-γ treatment alone, M. avium subsp. paratuberculosis infection alone, and M. avium subsp. paratuberculosis infection followed by IFN-γ treatment (M. avium subsp. paratuberculosis-IFN-γ) or no treatment. For each animal and each treatment, there are three intra-array replicates. All data processing and analysis were done as per Li et al. (37), with the following study specifics.
Animal-animal variability analysis.
For each of the 300 peptides, an F test was used to determine whether there are significant differences among the three animals under the same treatment conditions. Therefore, 300 F tests were carried out for a single treatment on the three animals, and 1,200 tests in total were carried out for all four treatments (i.e., 300 peptides times four treatments).
Treatment-treatment variability analysis.
Peptides identified by the F test as having consistent patterns of response to the various treatments across the three animals were subjected to a paired t test to compare their signal intensities under a treatment condition with those under control conditions. For each animal-independent peptide, the responses from all three animals were pooled to increase the statistical confidence. Three tests were performed for each peptide, specifically, IFN versus untreated monocytes, M. avium subsp. paratuberculosis-IFN versus untreated monocytes, and M. avium subsp. paratuberculosis infection versus untreated monocytes. Peptides with significant (P < 0.20) changes in phosphorylation were identified. This level of significance was chosen to retain as much data as possible and thus facilitate subsequent pathway analysis.
Cluster analysis.
The preprocessed M. avium subsp. paratuberculosis data were subjected to hierarchical clustering and principal component analysis (PCA) to cluster peptide response profiles across animal-treatment combinations. For each of the 300 peptides in a single treatment and animal, the average was taken over the three variance stabilization and normalization (VSN)-transformed replicates. For hierarchical clustering, each animal/treatment vector was considered a singleton (i.e., a cluster with a single element) at the initial stage of the clustering. The two most similar clusters were merged, and the distances between the newly merged clusters and the remaining clusters were updated, iteratively. The method, as described by Eisen et al. (22), used the following calculation: average linkage + (1 − Pearson correlation) (44). The method takes the average over the merged (i.e., the most correlated) kinome profiles and updates the distances between the merged clusters and other clusters by recalculating the correlations between them.
PCA was applied to the M. avium subsp. paratuberculosis data both before and after subtraction of biological controls. In either case, the first two principal components, namely, PC1 and PC2, which account for the largest variability within the sample data, were used to cluster the animal/treatment data points.
Pathway analysis of differentially phosphorylated peptides.
InnateDB is a publically available resource which, based on levels of either differential expression or phosphorylation, predicts biological pathways based on experiment fold change data sets (39). Pathways are assigned a probability value (P) based on the number of proteins present for a particular pathway as well as the degree to which they are differentially expressed or modified relative to a control condition. For our investigation, input data were limited to those peptides selected in the treatment-treatment variability analysis (above). Since InnateDB requires fold change (FC) values as input (with P values optional), the differences between the VSN-transformed intensities under control and treatment conditions are converted to fold change values by the following formula: 2d where d = averagetreatment − averagecontrol, where average is the average of the transformed signals for each peptide.
RESULTS
Infection of bovine monocytes with M. avium subsp. paratuberculosis.
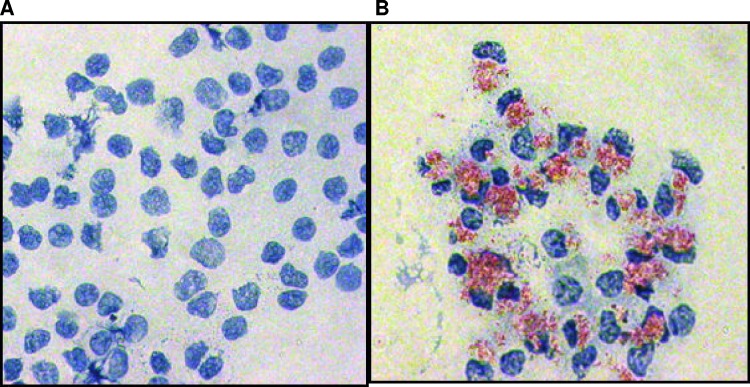
Following the in vitro infection of purified bovine monocytes, cytospins were performed to confirm and quantify the extent of M. avium subsp. paratuberculosis uptake (Fig. 1). Over three replicate experiments, there was an infection efficiency of 93% ± 4%. The in vitro infection of purified monocytes with M. avium subsp. paratuberculosis therefore provides a cell population of sufficient quantity and homogeneity, with respect to cell type and infection status, for kinome analysis. Having a homologous cell population ensures that any detected kinase activity was a result of changes in the cells of interest, which cannot be known in a mixed cell population. Isolating a single cell population can result in intracellular changes, including activation of the monocytes; resting the cells overnight returns the cells to baseline levels before treatment is given (58). In order to minimize the possibility of technical variances in the kinome, experiments for all animals were preformed simultaneously in a single run.
Fig 1.
Infection of bovine monocytes with M. avium subsp. paratuberculosis. Whole-blood-isolated bovine monocytes were infected with M. avium subsp. paratuberculosis at an MOI of 5:1. Cells were heat fixed to slides, M. avium subsp. paratuberculosis was stained with carbol fuchsin, and cells were counterstained with methylene blue. Thus, red indicates M. avium subsp. paratuberculosis bacteria, and blue indicates bovine monocytes. The figure shows stained cells of both uninfected (A) and infected (B) samples under oil immersion at a magnification of ×100.
Analysis of animal-animal variability.
In an outbred species, such as cattle, a degree of variability in biological responses is anticipated. To identify core, conserved biological processes, the kinome data from the three animals were analyzed to determine animal-dependent and animal-independent responses. Under the same treatment condition, any peptides with P values less than 0.01 were considered animal dependent. By this criterion only eight peptides appear to be animal dependent in all three treatments relative to the controls. Two hundred peptides elicited similar responses across all three treatments, regardless of the choice of animal. Ninety-two peptides were not conclusive in that P values for those peptides were not consistently greater than or less than 0.01 across all three treatments relative to the control. Examining peptides within each treatment revealed that 37, 52, and 43 peptides had significantly different reactions in the treatments of M. avium subsp. paratuberculosis-infected monocytes treated with IFN-γ, monocytes treated with IFN-γ, and monocytes infected with M. avium subsp. paratuberculosis, respectively. Interestingly, this indicates that there is more variation in how animals respond to IFN-γ stimulation than to M. avium subsp. paratuberculosis infection. This may be due to the number of phosphorylation events being affected by the treatment with IFN-γ compared to an infection with M. avium subsp. paratuberculosis. It is logical to conclude that an infection with M. avium subsp. paratuberculosis will induce more changes in a wider variety of kinases than a treatment with IFN-γ, which acts on a small subset of pathways. Thus, the remaining kinases not affected by either the treatment with IFN-γ or the infection will display more variability based on the biological context of the animal.
Analysis of treatment-treatment variability.
To identify peptides with significant changes in phosphorylation status relative to the control under the various treatment conditions, the 200 peptides identified as consistently regulated across the three animals were subjected to a paired t test. A listing of the differentially phosphorylated peptides as a result of M. avium subsp. paratuberculosis infection, treatment with M. avium subsp. paratuberculosis-IFN, and treatment with IFN alone relative to their corresponding controls is included in Table S1 in the supplemental material.
Cluster analysis of the various treatments/infections.
The kinome data sets were subjected to cluster analysis for comparison and visualization of patterns of response of the different animals to the different treatments. To this end, principal component analysis (PCA) to cluster responses was applied to the data sets with and without subtraction of the corresponding biological controls. The data were analyzed in this way to consider both the absolute kinome profile of each animal under each treatment condition (without subtraction of biological controls) and the dynamic response of each animal to each treatment (with subtraction of biological controls).
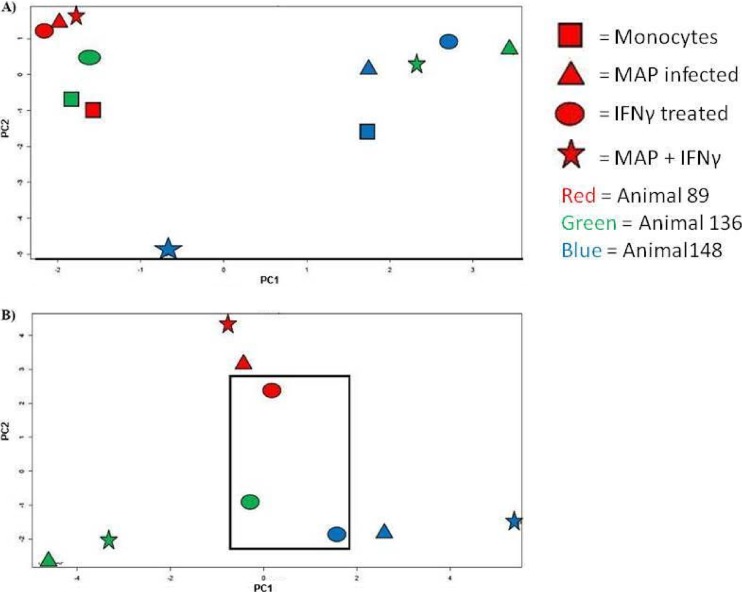
PCA clustering without subtracting the biological controls results in a seemingly random arrangement with respect to animals and treatment conditions (Fig. 2A). This is not unanticipated as within an outbred bovine population different baselines of cellular activity due to genetic, developmental, and/or environmental factors may impact baseline cellular kinase activity. These factors may also influence the dynamic responses of the animals to the stimuli, in particular, responses to a complex and multifaceted stimulus like bacterial infection.
Fig 2.
Principle component cluster analysis of kinome data. Kinome data sets were subjected to PCA cluster analysis. Individual treatment conditions are indicated. The rectangle indicates a conserved clustering of responses of uninfected monocytes to IFN-γ stimulation prior to subtraction of biological controls (A) and with subtraction of biological controls (B).
By subtracting the corresponding biological controls for each animal, it is possible to determine the dynamic response of the monocytes of each animal to each treatment condition. Investigating the kinome data in this manner reveals a strong pattern of clustering based first upon animals and subsequently upon treatment conditions (Fig. 2B). That consistent clustering on the basis of animal is discernible was surprising based on the previous results in data processing showing that 200 of the 300 peptides were found to be animal independent at a 0.01 level of significance. Animal dependence was verified by using hierarchical clustering, calculated as average linkage + (1 − Pearson correlation) (see Fig. S1 in the supplemental material). This animal dependence may reflect cumulative differences of the selected peptides, which together contributed to the notable variations among animals.
One possible reason for the strong clustering by animal was the stringent level of confidence used in the animal-animal variability analysis. Having a lower significance level (e.g., 5%) would mean that more peptides would be determined to have different expression levels across animals and would be eliminated from further analysis. This may result in less prominent clustering by animal but would result in fewer peptides being considered in the pathway analysis.
Within animals there is further clustering corresponding to conserved responses to the treatment conditions. For example, while the data sets for each of the three animals separate to different quadrants of the plot, there is conserved clustering of the responses of uninfected monocytes to IFN-γ stimulation to a distinct subcluster at the center of the plot. This indicates that in spite of animal-specific variances in the baseline kinomic profiles, there is a conserved and consistent response to IFN-γ across the biological replicates (Fig. 2B). Through these approaches, following subtraction of the biological controls, there is a strong tendency for the data sets to cluster first on the basis of animal, followed by secondary clustering based on the particular treatment condition. In particular, in two of the three animals, the responses of the M. avium subsp. paratuberculosis-infected monocytes to IFN-γ stimulation cluster more closely to the responses of M. avium subsp. paratuberculosis-infected monocytes rather than the IFN-γ-stimulated monocytes (see Fig. S1 in the supplemental material. This suggests that M. avium subsp. paratuberculosis blocks the ability of the cells to undergo the same changes in signal transduction observed in the uninfected cells and is consistent with the IFN-γ unresponsiveness (Fig. 2).
Pathways implicated by peptide array data.
The kinome data were subjected to pathway overrepresentation analysis to determine which cellular pathways/processes are activated under the different treatment conditions. To ensure that the identified pathways represent conserved and consistent biological responses, input data were limited to peptides with a consistent pattern of differential phosphorylation across the three biological replicates in at least one of the treatment sets (P > 0.01) as well as significant (P < 0.20) changes in phosphorylation level relative to the control treatment. These select data from the three animals were merged to generate a representative data set for each treatment condition and analyzed through InnateDB (39).
For uninfected monocytes stimulated with IFN-γ, a number of pathways were suggested to be activated with a high degree of confidence (P < 0.05). Most notably, this included five pathways directly associated with activation and regulation of JAK-STAT (Table 1). Specifically, there are 26 peptides on the array representing JAK-STAT intermediates. Following IFN-γ stimulation of uninfected monocytes, 16 of these show significant differential phosphorylation relative to the unstimulated monocytes: 15 with increased phosphorylation and 1 with decreased phosphorylation (Table 1). JAK-STAT is well defined for its role in mediating cellular responses to IFN-γ (5, 32). The identification of this pathway in monocytes following IFN-γ stimulation provides confidence in the ability of the arrays to detect and reflect biological responses. Specifically, there is a high degree of confidence (P < 0.002) for activation of the JAK-STAT pathway following IFN-γ treatment of the uninfected monocytes (Table 1). In contrast, for the same treatment of the M. avium subsp. paratuberculosis-infected monocytes, the confidence in activation of JAK-STAT is extremely low (P < 0.831), and only four of the peptides representing JAK-STAT signaling proteins show increased phosphorylation.
Table 1.
Pathway analysis of bovine monocytes and M. avium subsp. paratuberculosis-infected bovine monocytes in response to IFN-γ stimulationa
| Functional group | Pathway | Total no. of peptidesb | Peptide phosphorylation in: |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Uninfected monocytes |
M. avium subsp. paratuberculosis-infected monocytes |
|||||||||
| Upregulated |
Downregulated |
Upregulated |
Downregulated |
|||||||
| No. of peptides | P | No. of peptides | P | No. of peptides | P | No. of peptides | P | |||
| JAK-STAT signaling | JAK-STAT signaling pathway | 16 | 15 | 0.002 | 1 | 0.999 | 4 | 0.831 | 7 | 0.309 |
| JAK-STAT pathway/regulation | 12 | 11 | 0.006 | 1 | 0.998 | 1 | 0.840 | 2 | 0.532 | |
| Gene expression of SOCS | 6 | 6 | 0.025 | 0 | 1 | 1 | 0.915 | 3 | 0.340 | |
| Gene expression of SOCS1 | 6 | 6 | 0.025 | 0 | 1 | 1 | 0.915 | 3 | 0.340 | |
| Gene expression of SOCS3 | 6 | 6 | 0.025 | 0 | 1 | 1 | 0.915 | 3 | 0.340 | |
| Cytokine/chemokine signaling | IL-27-mediated signaling events | 6 | 6 | 0.025 | 0 | 1 | 1 | 0.915 | 3 | 0.340 |
| IL-12-mediated signaling events | 14 | 12 | 0.013 | 2 | 0.990 | 2 | 0.910 | 5 | 0.251 | |
| IL-22 soluble receptor signaling | 5 | 5 | 0.047 | 0 | 1 | 1 | 0.915 | 3 | 0.340 | |
| Androgen receptor | 11 | 10 | 0.081 | 1 | 0.999 | 4 | 0.247 | 2 | 0.919 | |
| IFN-β enhancer information processing | 6 | 6 | 0.025 | 0 | 1 | 0 | 1 | 0 | 1 | |
| TGF-β signalingc | TGF-β (canonical) | 8 | 7 | 0.139 | 1 | 0.990 | 3 | 0.236 | 1 | 0.952 |
| TGF-β signaling pathway | 5 | 5 | 0.045 | 0 | 1 | 2 | 0.425 | 1 | 0.895 | |
| TGF-β receptor | 15 | 11 | 0.273 | 4 | 0.957 | 5 | 0.061 | 0 | 1 | |
| NF-kB signaling | NF-κb activation | 10 | 9 | 0.012 | 2 | 0.994 | 2 | 0.425 | 1 | 0.895 |
| Other | Toll-like receptor signaling pathway | 25 | 18 | 0.044 | 3 | 0.999 | 5 | 0.373 | 4 | 0.802 |
Based on levels of differential expression or phosphorylation, InnateDB, a publically available pathway analysis tool (39), is able to predict pathways which are consistent with the experimental data. Pathways are assigned a probability value (P) based on the number of proteins present for a particular pathway. Output also includes the number of the uploaded proteins associated with a particular pathway as well as a subset of those which are differentially phosphorylated. For our investigation fold change cutoffs are set at a P value of <0.2 for confidence of difference between treatment and monocyte control groups. The number of peptides with increased or decreased phosphorylation with respect to the control condition is indicated.
Indicates the number of peptides differentially phosphorylated relating to the pathway.
Transforming growth factor β.
In addition to JAK-STAT, a number of other pathways relating to cytokine/chemokine signaling, as well as activation of the proinflammatory Toll-like receptor pathway, are observed. Many of these pathways, in particular those associated with cytokine/chemokine signaling, share signaling intermediates with IFN-γ-induced responses as well as overlap of their biological functions. Many of these responses would be anticipated as secondary events following IFN-γ stimulation. That activation of these secondary pathways is not observed in M. avium subsp. paratuberculosis-infected monocytes following IFN-γ stimulation would suggest that the pathogen blocks cellular responsiveness at, or near, the receptor rather than at intermediate or final effectors, which would provide an opportunity for activation of secondary signaling responses. Dampening of these responses may also help to facilitate intracellular survival of the pathogen.
Phosphorylation events within the JAK-STAT pathway.
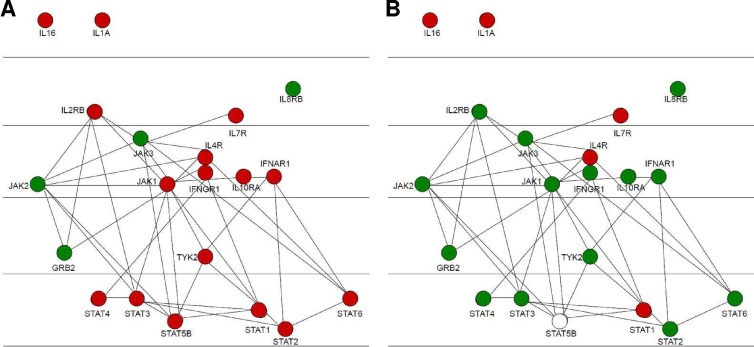
As JAK-STAT signaling events are represented quite comprehensively on the array, it is possible to investigate the specific level at which M. avium subsp. paratuberculosis blocks IFN-γ responsiveness. IFN-γ stimulation of the uninfected monocytes results in differential phosphorylation of numerous peptides corresponding to a variety of intermediates along the JAK-STAT pathway (Table 2 and Fig. 3). This includes phosphorylation events ranging from activation of IFNGR1 to differential phosphorylation of the final STAT effectors. The associated P values indicate the confidence of the fold change relative to the medium treatment. In contrast, following IFN-γ stimulation of the M. avium subsp. paratuberculosis-infected monocytes, there is an absence of signaling activity throughout the JAK-STAT pathway. This is observed as early as the IFN-γ receptor, suggesting that M. avium subsp. paratuberculosis blocks signaling events at a very early point (Table 2). A representation of the JAK-STAT pathway highlights activation of JAK-STAT by IFN-γ in uninfected monocytes (Fig. 3A) while M. avium subsp. paratuberculosis infection blocks IFN-γ responsiveness throughout the pathway beginning at the receptor (Fig. 3B).
Table 2.
Differential phosphorylation of peptides of the JAK-STAT pathway following IFN-γ stimulation of monocytes and M. avium subsp. paratuberculosis-infected monocytes
| Phosphoproteina | Relative phosphorylation inb: |
|||
|---|---|---|---|---|
| Uninfected monocytes |
M. avium subsp. paratuberculosis-infected monocytes |
|||
| Fold change | P value | Fold change | P value | |
| IFNAR1 | 1.14 | 0.04 | −1.17 | 0.23 |
| IFNGR1 | 1.09 | 0.26 | −1.07 | 0.27 |
| IL-10RA | 1.11 | 0.19 | −1.14 | 0.15 |
| IL-16 | 1.20 | 0.10 | 1.17 | 0.16 |
| IL-2RB | 1.10 | 0.24 | −1.09 | 0.27 |
| IL-4R | 1.16 | 0.14 | 1.08 | 0.31 |
| IL-6ST | 1.14 | 0.14 | 1.04 | 0.33 |
| IL-7R | 1.56 | 0.07 | 1.16 | 0.18 |
| JAK1 | 1.03 | 0.42 | −1.16 | 0.12 |
| JAK2 | −1.01 | 0.44 | −1.08 | 0.22 |
| JAK3 | −1.02 | 0.36 | −1.27 | 0.02 |
| STAT1 | 1.22 | 0.08 | 1.20 | 0.14 |
| STAT2 | 1.13 | 0.06 | −1.15 | 0.08 |
| STAT3 | 1.16 | 0.05 | −1.12 | 0.16 |
| STAT4 | 1.17 | 0.09 | 1.06 | 0.34 |
| STAT5B | 1.14 | 0.05 | 1.00 | 0.49 |
| STAT6 | 1.22 | 0.01 | −1.40 | 0.14 |
| Tyk2 | 1.04 | 0.16 | −1.03 | 0.43 |
The substrate protein with a peptide representing a phosphorylation site on the array. IL-10RA, IL-10 receptor A; IL-10RB, IL-10 receptor B; IL-6ST, IL-6 signal transducer.
Fold change is the relative change calculated by comparing the background-corrected and normalized signal values of these samples to the medium control. P values indicate how significant a difference there is between IFN-γ-treated cells and control cells.
Fig 3.
Signaling within the JAK-STAT pathway in bovine monocytes and M. avium subsp. paratuberculosis-infected bovine monocytes in response to IFN stimulation. Protein members of the JAK-STAT pathway are color coded with respect to fold change in differential phosphorylation. Red indicates increased phosphorylation, green indicates decreased phosphorylation, and white indicates no change. (A) Differential phosphorylation of JAK-STAT intermediates following IFN-γ in bovine monocytes. (B) Relative degrees of phosphorylation of M. avium subsp. paratuberculosis-infected versus uninfected bovine monocytes following IFN-γ stimulation. Diagrams were produced using the cytoscape visualization option of InnateDB.
Altered expression of SOCS and IFNGR in M. avium subsp. paratuberculosis-infected monocytes.
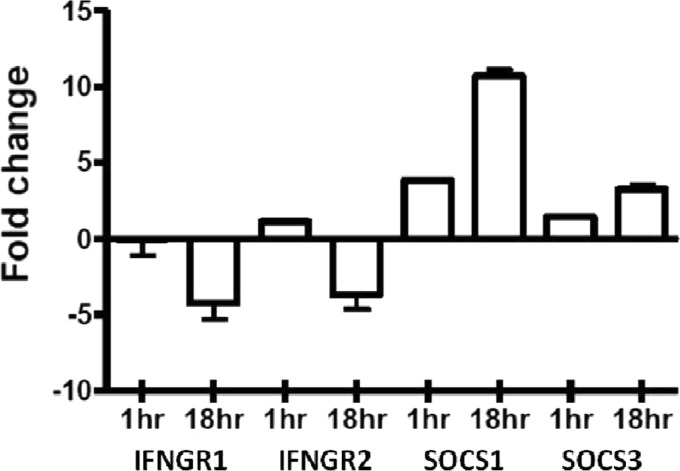
The absence of early JAK-STAT signal transduction activity in the infected monocytes in response to IFN-γ stimulation suggests that M. avium subsp. paratuberculosis influences host responsiveness at, or near, the IFN-γ receptor. This could result from decreased expression of the receptor or increased expression of an inhibitory factor which acts at the level of the receptor. Both mechanisms of JAK-STAT inhibition have been observed for other mycobacteria (31). In bovine monocytes M. avium subsp. paratuberculosis infection causes a decrease in expression (∼4-fold) for each of these chains (Fig. 4). Decreased expression of IFN-γ receptor chains has been reported for a number of pathogens which block cellular responsiveness to IFN-γ (31, 35, 47). Most notably, Mycobacterium avium, a closely related pathogen to M. avium subsp. paratuberculosis, decreases expression of both chains of the IFN-γ receptor, which is analogous to our observations.
Fig 4.
Altered expression of SOCS3 and IFNGR in response to M. avium subsp. paratuberculosis infection. RNA was extracted from bovine monocytes after either 1- or 18-h infection with M. avium subsp. paratuberculosis (MOI of 5:1). Relative expression of select genes was determined through qRT-PCR compared to time-matched uninfected monocytes.
The ability of infected cells to respond to IFN-γ could also reflect increased expression of the SOCS inhibitors. A number of viruses have been shown to abate IFN-γ responses through induced expression of these natural regulators of IFN-γ sensitivity including influenza A virus (43), herpes simplex virus 1 (60), hepatitis C virus (8), severe acute respiratory syndrome coronavirus (41), and respiratory syncytial virus (61). Following M. avium subsp. paratuberculosis infection, expression of SOCS1 and SOCS3 is upregulated in a time-dependent fashion. Unlike the expression of the IFN-γ receptor, M. avium subsp. paratuberculosis is able to impact expression of the SOCS proteins as early as 1 h after infection. This effect becomes even more pronounced at the 18-h time point, with SOCS1 expression increasing approximately 10-fold and SOCS3 expression increasing approximately 3-fold each relative to the uninfected cells.
IFN-γ-induced TNF-α release.
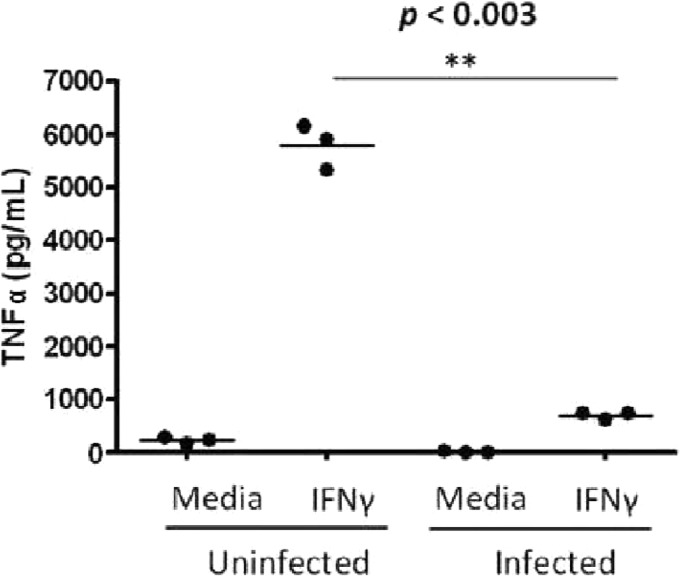
Collectively, the kinome data and the patterns of expression of the IFN-γ receptor and SOCS regulators predict that M. avium subsp. paratuberculosis infection of bovine monocytes will dampen the responsiveness of these cells to IFN-γ stimulation. Release of tumor necrosis factor alpha (TNF-α) is a well-established and easily quantified marker of macrophage activation by IFN-γ (12). For the uninfected monocytes, treatment with 10 ng/ml of bovine IFN-γ resulted in release of large quantities of TNF-α (Fig. 5) following overnight stimulation. In contrast, under identical stimulation conditions, there is minimal release of TNF-α from M. avium subsp. paratuberculosis-infected monocytes. While this assay took place overnight and while the peptide array experiment was conducted after 1 h of treatment, the ELISA is a measure of total release of TNF-α from the cells. Thus, any release of TNF-α from initial treatment to medium collection is measured. M. avium subsp. paratuberculosis infection inhibits any TNF-α release induced by IFN-γ. This phenotype is consistent with the responses anticipated by the kinome and gene expression data.
Fig 5.
IFN-γ-stimulated production of TNF-α in M. avium subsp. paratuberculosis-infected and noninfected bovine monocytes. The well-documented response of TNF-α release following IFN-γ stimulation is observed in uninfected cells. Upon infection the response is nearly completely eliminated.
DISCUSSION
Diseases caused by a relatively small number of Mycobacterium species are responsible for billions of dollars in economic losses in all major terrestrial and aquatic animal production systems as well as for a tremendous direct impact on human health. For example, tuberculosis resulting from infection by Mycobacterium tuberculosis is estimated to infect one-third of the world's population and is being reported with increasing prevalence in developed countries, including the emergence of drug resistant strains. Within the cattle industry, M. avium subsp. paratuberculosis shows ever-increasing rates of infection, and there is growing concern over food safety, with indications that M. avium subsp. paratuberculosis may represent a zoonotic threat. The lack of effective strategies to treat and/or prevent these diseases relates in large part to the ability of these pathogens to subvert host immune responses and establish chronic infections within host immune cells. While different species of mycobacteria utilize different specific strategies to achieve this objective, there is a common opinion that a better understanding of the pathogenic mechanism of these microbes is a first and essential step in the development of rational therapeutic strategies.
Host responses to infectious challenge are often regulated through phosphorylation. Moreover, the pathogenic mechanism of many mycobacteria involves production of a number of bacterially encoded, eukaryotic-like protein kinases, and phosphatases play essential roles in virulence and establishment of chronic infections (3). These findings highlight the importance of understanding the host-pathogen interaction of M. avium subsp. paratuberculosis from the perspective of dynamic patterns of phosphorylation. Additionally, the success of kinase inhibitors in treatment of other diseases has made the Mycobacterium kinases prime therapeutic targets (49). Until recently, however, the absence of bovine-specific research tools made it very difficult to conduct kinome analysis for Johne's disease. Our recent report on the development of species-specific peptide arrays for kinome analysis of nontraditional laboratory species (33) and analysis of such data (37) offer the opportunity to obtain more specific insights into host signaling responses to M. avium subsp. paratuberculosis infection in a relevant cell type.
Pathway analysis of the kinome data indicated activation of the JAK-STAT pathway, a hallmark of IFN-γ signaling, in uninfected but not M. avium subsp. paratuberculosis-infected monocytes. Specifically, the inability of IFN-γ to induce differential phosphorylation of peptides corresponding to early JAK-STAT intermediates in infected monocytes indicates that M. avium subsp. paratuberculosis rapidly, within 3 h, blocks responsiveness at, or near, the IFN-γ receptor. Consistent with this hypothesis, increased expression of negative regulators of the IFN-γ receptors SOCS1 and SOCS3, as well as decreased expression of IFN-γ receptor chains 1 and 2, is observed in M. avium subsp. paratuberculosis infection. These patterns of expression are functionally consistent with the kinome data and offer a mechanistic explanation for this critical M. avium subsp. paratuberculosis behavior. These mechanisms bear similarity to those reported for other pathogens in the targeted disruption of this critical host response. For example, the targeted downregulation of both chains of the IFN-γ receptor is also observed for the closely related Mycobacterium avium (31). Also in Mycobacterium tuberculosis, infection inhibition of IFN-γ signaling is independent of inhibition of STAT1 (54), as shown by the data in Table 2, STAT1 is not inhibited at all by M. avium subsp. paratuberculosis infection compared to monocytes treated with IFN-γ.
The independent assay used to confirm a loss of response to IFN-γ in infected monocytes was a measure of TNF-α release. The results indicated a significant release of TNF-α following IFN-γ treatment of monocytes but no release of TNF-α in similarly treated M. avium subsp. paratuberculosis-infected cells. This result agrees with previous studies of M. avium subsp. paratuberculosis infection and cytokine response. In Stabel (52), TNF production by clinically infected PBMC was reduced compared to production in subclinically infected cells following stimulation with both concanavalin A (ConA) and M. avium subsp. paratuberculosis sonicate. The author suggests, and we have provided supporting evidence in this study, that antigen-mediated cellular immune responses are abrogated in cows with a clinical infection of M. avium subsp. paratuberculosis. Conversely, Adams and Czuprynski (2) reported an increase in TNF-α production following stimulation with M. avium subsp. paratuberculosis and M. avium subsp. paratuberculosis cell wall components. There are several key differences in their study and ours which may explain the differences in results. Most importantly, they did not look at the response of M. avium subsp. paratuberculosis-infected cells but instead examined cells stimulated with either cell wall components or whole bacteria for relatively short time periods (1.5 h). Second, they used whole blood instead of a specific immune cell type, which makes it difficult to determine the source of the TNF-α. Finally, the TNF-α production was measured by mRNA levels, which may not correlate entirely with protein levels, especially in the context of a bacterial infection which may be inhibiting specific immune pathways. When Souza et al. (51) studied p38 mitogen-activated protein kinase (MAPK) phosphorylation and M. avium subsp. paratuberculosis infection in bovine monocytes, they found an increase in interleukin-10 (IL-10) and TNF-α mRNA expression following M. avium subsp. paratuberculosis infection. The induction of IL-10 was dependent on p38; when p38 was inhibited, a decrease in IL-10 expression and an increase in IL-12 expression were observed while there was no change in TNF-α expression. Since IL-10 is a known inhibitor of TNF-α and since IL-12 is a known inducer of TNF-α, the implication of these results is not clear. These results may reflect the measurement of mRNA expression levels instead of protein concentrations; it is possible that the increase in TNF-α expression was an initial response to infection which was then quickly suppressed by the activity of IL-10. It is also possible that IL-10 and IL-12 affected TNF-α protein concentration, which was not reflected by the mRNA levels.
The ultimate result of studies such as this is to identify targets for therapeutic intervention. This strategy is exemplified by Kuijl et al. (36), who looked at the role of kinases in bacterial infection, including both salmonella and mycobacteria. Their strategy was one that we would advocate: looking at host kinases for insights into treatment of diseases. They uncovered the key signaling protein Akt involved in the phagosome-lysosome fusion. Inhibition of Akt resulted in reduced infection with both salmonella and mycobacteria. Interestingly our results show an increase in phosphorylation of Y326 of Akt1, a known site of Akt activation (see Tables S1 and S2 in the supplemental material) (10) following infection with M. avium subsp. paratuberculosis.
Collectively, this investigation offers specific insight into the pathogenic mechanisms of M. avium subsp. paratuberculosis which may be of value in the rational design of vaccines and/or therapeutics. This also offers further insight into the various and often redundant mechanisms used by viral and bacterial pathogens to block IFN-γ responsiveness to achieve chronic infections.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge the funding support provided by the Beef Cattle Research Council, Alberta Livestock Industry Development Fund, Saskatchewan Ministry of Agriculture and Natural Sciences, and the Engineering Research Council.
We declare that we have no competing interests.
Footnotes
Published ahead of print 11 June 2012
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1. Abendroth A, et al. 2000. Modulation of major histocompatibility class II protein expression by varicella-zoster virus. J. Virol. 74:1900–1907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Adams JL, Czuprynski CJ. 1995. Ex vivo induction of TNFa and IL-6 mRNA in bovine whole blood by Mycobacterium paratuberculosis and mycobacterial cell wall components. Microb. Pathog. 19:19–29 [DOI] [PubMed] [Google Scholar]
- 3. Alber T. 2009. Signaling mechanisms of the Mycobacterium tuberculosis receptor Ser/Thr protein kinases. Curr. Opin. Struct. Biol. 19:650–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Applied Biosystems 1997. User bulletin 2. ABI Prism 7700 sequence detection system: relative quantification of gene expression (P/N 4303859). Applied Biosystems, Foster City, CA [Google Scholar]
- 5. Bach EA, Aguet M, Schreiber RD. 1997. The IFN gamma receptor: a paradigm for cytokine receptor signaling. Annu. Rev. Immunol. 15:563–591 [DOI] [PubMed] [Google Scholar]
- 6. Bach EA, et al. 1996. Ligand-induced assembly and activation of the gamma interferon receptor in intact cells. Mol. Cell. Biol. 16:3214–3221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Blanchette J, Racette N, Faure R, Siminovitch KA, Olivier M. 1999. Leishmania-induced increases in activation of macrophage SHP-1 tyrosine phosphatase are associated with impaired IFN-gamma-triggered JAK2 activation. Eur. J. Immunol. 29:3737–3744 [DOI] [PubMed] [Google Scholar]
- 8. Bode JG, et al. 2003. IFN-alpha antagonistic activity of HCV core protein involves induction of suppressor of cytokine signaling-3. FASEB J. 17:488–490 [DOI] [PubMed] [Google Scholar]
- 9. Bonecini-Almeida MG, et al. 1998. Induction of in vitro human macrophage anti-Mycobacterium tuberculosis activity: requirement for IFN-gamma and primed lymphocytes. J. Immunol. 160:4490–4499 [PubMed] [Google Scholar]
- 10. Chen R, et al. 2001. Regulation of Akt/PKB activation by tyrosine phosphorylation. J. Biol. Chem. 276:31858–31862 [DOI] [PubMed] [Google Scholar]
- 11. Chi J, VanLeeuwen JA, Weersink A, Keefe GP. 2002. Direct production losses and treatment costs from bovine viral diarrhoea virus, bovine leukosis virus, Mycobacterium avium subspecies paratuberculosis, and Neospora caninum. Prev. Vet. Med. 55:137–153 [DOI] [PubMed] [Google Scholar]
- 12. Collart MA, Belin D, Vassalli JD, de Kossodo S, Vassalli P. 1986. Gamma interferon enhances macrophage transcription of the tumor necrosis factor/cachectin, interleukin 1, and urokinase genes, which are controlled by short-lived repressors. J. Exp. Med. 164:2113–2118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cooper AM, et al. 1993. Disseminated tuberculosis in interferon gamma gene disrupted mice. J. Exp. Med. 178:2243–2247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Coussens PM, Verman N, Coussens MA, Elftman MD, McNulty AM. 2004. Cytokine gene expression in peripheral blood mononuclear cells and tissues of cattle infected with Mycobacterium avium subsp. paratuberculosis: evidence for an inherent proinflammatory gene expression pattern. Infect. Immun. 72:1409–1422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dalton DK, et al. 1993. Multiple defects of immune cell function in mice with disrupted interferon-gamma genes. Science 259:1739–1742 [DOI] [PubMed] [Google Scholar]
- 16. Darnell JE. 1997. STATS and gene regulation. Science 277:1630–1635 [DOI] [PubMed] [Google Scholar]
- 17. Denis M, Gregg EO, Ghandirian E. 1990. Cytokine modulation of Mycobacterium tuberculosis growth in human macrophages. Int. J. Immunopharmacol. 12:721–727 [DOI] [PubMed] [Google Scholar]
- 18. Döffinger R, et al. 2000. Partial interferon-gamma receptor signaling chain deficiency in a patient with bacilli Calmette-Guerin Mycobacterium abscesses infection. J. Infect. Dis. 181:379–384 [DOI] [PubMed] [Google Scholar]
- 19. Dorman SE, Holland SM. 1998. Mutation in the signal-transducing chain of the interferon-gamma receptor and susceptibility to mycobacterial infection. J. Clin. Invest. 101:2364–2369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dupuis S, et al. 2000. Human interferon-gamma-mediated immunity is a genetically controlled continuous trait that determines the outcome of mycobacterial invasion. Immunol. Rev. 178:129–137 [DOI] [PubMed] [Google Scholar]
- 21. Eckner R, et al. 1994. Molecular cloning and functional analysis of the adenovirus E1A-associated 300-kD protein (p300) reveals a protein with properties of a transcriptional adaptor. Genes Dev. 8:869–884 [DOI] [PubMed] [Google Scholar]
- 22. Eisen MB, Spellman PT, Brown PO, Botstein D. 1998. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. U. S. A. 95:14863–14868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ellis JA, Godson D, Campos M, Sileghem M, Babiuk LA. 1993. Capture immunoassay for ruminant tumor necrosis factor-α: comparison with bioassay. Vet. Immunol. Immunopathol. 35:289–300 [DOI] [PubMed] [Google Scholar]
- 24. Eltholth MM, Marsh VR, Van Winden S, Guitian FJ. 2009. Contamination of food products with Mycobacterium avium paratuberculosis: a systematic review. J. Appl. Microbiol. 107:1061–1071 [DOI] [PubMed] [Google Scholar]
- 25. Flynn JL, et al. 1993. An essential role for interferon gamma in resistance to Mycobacterium tuberculosis infection. J. Exp. Med. 178:2249–2254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fratti RA, Backer JM, Gruenberg J, Corvera S, Deretic V. 2001. Role of phosphatidylinositol 3-kinase and Rab5 effectors in phagosomal biogenesis and mycobacterial phagosome maturation arrest. J. Cell Biol. 154:631–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fujii N, Yokosawa N, Shirakawa S. 1999. Suppression of interferon response gene expression in cells persistently infected with mumps virus, and restoration from its suppression by treatment with ribavirin. Virus Res. 65:175–185 [DOI] [PubMed] [Google Scholar]
- 28. Greenlund AC, et al. 1995. Stat recruitment by tyrosine-phosphorylated cytokine receptors: an ordered reversible affinity-driven process. Immunity 2:677–687 [DOI] [PubMed] [Google Scholar]
- 29. Harris NB, Barletta RG. 2001. Mycobacterium avium subsp. Paratuberculosis in veterinary medicine. Clin. Microbiol. Rev. 14:489–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hines ME, et al. 2007. Efficacy of spheroplastic and cell-wall competent vaccines for Mycobacterium avium subsp. Paratuberculosis in experimentally challenged baby goats. Vet. Microbiol. 120:261–283 [DOI] [PubMed] [Google Scholar]
- 31. Hussain S, Zwilling BS, Lafuse WP. 1999. Mycobacterium avium infection of mouse macrophages inhibits IFN-gamma Janus kinase-STAT signaling and gene induction by down-regulation of the IFN-gamma receptor. J. Immunol. 163:2041–2048 [PubMed] [Google Scholar]
- 32. Igarashi K, et al. 1994. Interferon-gamma induces tyrosine phosphorylation of interferon-gamma receptor and regulated association of protein tyrosine kinases, Jak1 and Jak2, with its receptor. J. Biol. Chem. 269:14333–14336 [PubMed] [Google Scholar]
- 33. Jalal S, et al. 2009. Genome to kinome: species-specific arrays for kinome analysis. Sci. Signal. 2:pl1 doi:10.1126/scisignal.254pl1 [DOI] [PubMed] [Google Scholar]
- 34. Jouanguy E, et al. 1999. A human IFNGR1 small deletion hotspot associated with dominant susceptibility to mycobacterial infection. Nat. Genet. 21:370–378 [DOI] [PubMed] [Google Scholar]
- 35. Kierszenbaum F, Mejia Lopez H, Tanner MK, Sztein MB. 1995. Trypanosoma cruzi-induced decrease in the level of interferon-gamma receptor expression by resting and activated human blood lymphocytes. Parasite Immunol. 17:207–214 [DOI] [PubMed] [Google Scholar]
- 36. Kuijl C, et al. 2007. Intracellular bacterial growth is controlled by a kinase network around PKB/AKT1. Nature 450:725–730 [DOI] [PubMed] [Google Scholar]
- 37. Li Y, et al. 2012. A systematic approach for analysis of peptide array kinome data. Sci. Signal. 5:pl2 doi:10.1126/scisignal.2002429 [DOI] [PubMed] [Google Scholar]
- 38. Look DC, et al. 1998. Direct suppression of Stat1 function during adenoviral infection. Immunity 9:871–880 [DOI] [PubMed] [Google Scholar]
- 39. Lynn DJ, et al. 2008. InnateDB: facilitating systems-level analysis of the mammalian innate immune response. Mol. Syst. Biol. 4:218 doi:10.1038/msb.2008.55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Miller DM, et al. 1998. Human cytomegalovirus inhibits major histocompatibility complex II expression by disruption of the Jak/Stat pathway. J. Exp. Med. 187:675–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Okabayashi T, et al. 2006. Cytokine regulation in SARS coronavirus infection compared to other respiratory virus infections. J. Med. Virol. 78:417–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Olsen I, et al. 2009. Isolation of Mycobacterium avium subspecies paratuberculosis reactive CD4 T cells from intestinal biopsies of Crohn's disease patients. PLoS One 4:e5641 doi:10.1371/journal.pone.0005641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pauli EK, et al. 2008. Influenza A virus inhibits type I IFN signaling via NF-κB dependent induction of SOCS-3 expression. PLoS Pathog. 4:e000196 doi:10.1371/journal.ppat.1000196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Pearson K. 1896. Mathematical contributions to the theory of evolution. III. Regression, heredity and panmixia. Philos. Trans. R. Soc. Lond. A 187:253–318 [Google Scholar]
- 45. Pierce ES. 2010. Ulcerative colitis and Crohn's disease: is Mycobacterium avium subspecies paratuberculosis the common villain? Gut Pathog. 2:21 doi:10.1186/1757-4749-2-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Rani PS, Sechi LA, Ahmed N. 2010. Mycobacterium avium subsp. paratuberculosis is a trigger of type-1 diabetes: destination Sardinia, or beyond. Gut Pathog. 2:1 doi:10.1186/1757-4749-2-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ray M, Gam AA, Boykins RA, Kenney RT. 2000. Inhibition of interferon-gamma signaling by Leishmania donovani. J. Infect. Dis. 181:1121–1128 [DOI] [PubMed] [Google Scholar]
- 48. Robertson AK, Andrew PW. 1991. Interferon gamma fails to activate human monocyte-derived macrophages to kill or inhibit the replication of a non-pathogenic mycobacterial species. Microb. Pathog. 11:283–288 [DOI] [PubMed] [Google Scholar]
- 49. Schreiber M, Res Matter IA. 2009. Protein kinases as antibacterial targets. Curr. Opin. Cell Biol. 2:325–330 [DOI] [PubMed] [Google Scholar]
- 50. Shin AR, et al. 2010. Identification of seroreactive proteins in the culture filtrate antigen of Mycobacterium avium ssp. paratuberculosis human isolates to sera from Crohn's disease patients. FEMS Immunol. Med. Microbiol. 58:128–137 [DOI] [PubMed] [Google Scholar]
- 51. Souza CD, Evanson OA, Weiss DJ. 2006. Mitogen activated protein kinase p38 pathway is an important component of the anti-inflammatory response in Mycobacterium avium subsp. paratuberculosis-infected bovine monocytes. Microb. Pathog. 41:59–66 [DOI] [PubMed] [Google Scholar]
- 52. Stabel JR. 2000. Cytokine secretion by peripheral blood mononuclear cells from cows infected with Mycobacterium paratuberculosis. Am. J. Vet. Res. 61:754–760 [DOI] [PubMed] [Google Scholar]
- 53. Sweeney RW, Jones DE, Habecker P, Scott P. 1998. Interferon-gamma and interleukin 4 gene expression in cows infected with Mycobacterium paratuberculosis. Am. J. Vet. Res. 59:842–847 [PubMed] [Google Scholar]
- 54. Ting L, Kim AC, Cattamanchi A, Ernst JD. 1999. Mycobacterium tuberculosis inhibits IFN-gamma transcriptional responses without inhibiting activation of STAT1. J. Immunol. 163:3898–3906 [PubMed] [Google Scholar]
- 55. Tiwari A, VanLeeuwen JA, McKenna LS, Keefe GP, Barkema HW. 2006. Johne's disease in Canada part I: clinical symptoms, pathophysiology, diagnosis, and prevalence in dairy herds. Can. Vet. J. 47:874–882 [PMC free article] [PubMed] [Google Scholar]
- 56. VanLeeuwen JA, Keefe GP, Tremblay R, Power C, Wichtel JJ. 2001. Seroprevalence of infection with Mycobacterium avium subspecies paratuberculosis, bovine leukemia virus, and bovine viral diarrhea virus in maritime Canada dairy cattle. Can. Vet. J. 42:193–198 [PMC free article] [PubMed] [Google Scholar]
- 57. Weiss DJ, Souza CD. 2008. Review paper: modulation of mononuclear phagocyte function by Mycobacterium avium subsp. paratuberculosis. Vet. Pathol. 45:829–841 [DOI] [PubMed] [Google Scholar]
- 58. Wilson HL, et al. 2005. Molecular analyses of disease pathogenesis: application of bovine microarrays. Vet. Immunol. Immunopathol. 105:277–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Woo SR, Heintz JA, Albrecht R, Barletta RG, Czuprynski CJ. 2007. Life and death in bovine monocytes: the fate of Mycobacterium avium subsp. paratuberculosis. Microb. Pathog. 43:106–113 [DOI] [PubMed] [Google Scholar]
- 60. Yokota S, Yokosawa N, Okabayashi T, Suzutani T, Fujii N. 2005. Induction of suppressor of cytokine signaling-3 by herpes simplex virus type 1 confers efficient viral replication. Virology 338:173–181 [DOI] [PubMed] [Google Scholar]
- 61. Zhao DC, Yan T, Li L, You S, Zhang C. 2007. Respiratory syncytial virus inhibits interferon-alpha-inducible signaling in macrophage-like U937 cells. J. Infect. 54:393–398 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.