Abstract
Borrelia burgdorferi is the tick-borne bacterium that causes the multistage inflammatory disease Lyme disease. B. burgdorferi has a reduced genome and lacks the enzymes required for de novo synthesis of purines for synthesis of RNA and DNA. Therefore, this obligate pathogen is dependent upon the tick vector and mammalian host environments for salvage of purine bases for nucleic acid biosynthesis. This pathway is vital for B. burgdorferi survival throughout its infectious cycle, as key enzymes in the purine salvage pathway are essential for the ability of the spirochete to infect mice and critical for spirochete replication in the tick. The transport of preformed purines into the spirochete is the first step in the purine salvage pathway and may represent a novel therapeutic target and/or means to deliver antispirochete molecules to the pathogen. However, the transport systems critical for purine salvage by B. burgdorferi have yet to be identified. Herein, we demonstrate that the genes bbb22 and bbb23, present on B. burgdorferi's essential plasmid circular plasmid 26 (cp26), encode key purine transport proteins. BBB22 and/or BBB23 is essential for hypoxanthine transport and contributes to the transport of adenine and guanine. Furthermore, B. burgdorferi lacking bbb22-23 was noninfectious in mice up to a dose of 1 × 107 spirochetes. Together, our data establish that bbb22-23 encode purine permeases critical for B. burgdorferi mammalian infectivity, suggesting that this transport system may serve as a novel antimicrobial target for the treatment of Lyme disease.
INTRODUCTION
Borrelia burgdorferi, the pathogenic spirochete that causes Lyme disease, is an obligate parasite that cycles in nature between the tick vector, Ixodes sp., and mammalian host, typically a small rodent. Transmission of B. burgdorferi to humans by tick bite, although not part of the zoonotic infectious cycle of the spirochete, results in Lyme disease. Unlike many other bacterial pathogens, B. burgdorferi is not known to harbor classic virulence factors, such as toxins or secretion systems to deliver bacterial effector molecules (8, 13). Clinical manifestations associated with Lyme disease are believed to result from the immune response to the spirochetal infection (39). Therefore, genes encoding physiological functions that allow for growth within the infected host are important virulence determinants (18, 19, 27), as survival is a critical component of B. burgdorferi pathogenesis.
B. burgdorferi lacks the enzymes required for de novo synthesis of purines and therefore relies absolutely upon salvage of purines from its environment for nucleic acid biosynthesis (3, 19, 22, 26). B. burgdorferi has evolved a novel purine salvage pathway that, in addition to scavenge of purine bases, involves the salvage of deoxynucleotides from the host environment. Purine metabolism in B. burgdorferi also utilizes interconversion of purine bases to deoxynucleosides by the deoxyribosyltransferase BB0426 and the catalysis of IMP to GMP and dIMP to dGMP by GuaA and GuaB, respectively (22). This pathway is vital for B. burgdorferi survival throughout its infectious cycle, as key enzymes in this purine salvage pathway are essential for the ability of the spirochete to infect mice and critical for spirochete replication in the tick (18, 19). The transport of preformed purines into the spirochete represents the first step in the purine salvage pathway; however, the transport systems critical for purine salvage by B. burgdorferi have yet to be identified.
Circular plasmid 26 (cp26) is the only plasmid that is present in all B. burgdorferi isolates examined to date and is not lost during in vitro propagation. This plasmid carries genes essential for survival of the spirochete during both in vitro and in vivo growth (7, 17). Cp26 harbors the essential gene resT, which encodes the telomere resolvase required for replication of linear plasmids and the linear chromosome (7), and the ospC gene, which is critical for mammalian infection (15, 34, 35, 38). Furthermore, cp26 carries the genes encoding IMP dehydrogenase (GuaB) and GMP synthase (GuaA), two key enzymes in the B. burgdorferi purine salvage pathway, described above (19, 22, 24). In addition to genes involved in peptide transport (oppAIV) (4), chitobiose transport (chbA, chbB, and chbC) (36), and glucose transport (bbb29) (7), cp26 harbors two adjacent genes, bbb22 and bbb23, both of which are hypothesized to encode putative purine permeases (7).
Herein we establish that the BBB22 and BBB23 proteins function to transport hypoxanthine, adenine, and guanine. Moreover, our results demonstrate that this transport system is absolutely required for survival of B. burgdorferi during mammalian infection.
MATERIALS AND METHODS
Bacterial clones and growth conditions.
All low-passage-number B. burgdorferi clones were derived from infectious clone A3-68ΔBBE02, which lacks cp9, lp56, and the gene bbe02 on lp25 (28). B. burgdorferi was grown in liquid Barbour-Stoenner-Kelly (BSK) II medium supplemented with gelatin and 6% rabbit serum (2) and plated in solid BSK medium as previously described (29, 30). All spirochete cultures were grown at 35°C and supplemented with 2.5% CO2. Kanamycin was used at 200 μg/ml, streptomycin at 50 μg/ml, and gentamicin at 40 μg/ml, when appropriate. All cloning steps were carried out using Escherichia coli DH5α. E. coli cultures were grown in LB broth or on LB agar plates containing 50 μg/ml kanamycin, 300 μg/ml spectinomycin, or 10 μg/ml gentamicin.
Deletion of bbb22-23.
A 3.8-kb DNA fragment encompassing the BBB22 and BBB23 open reading frames (ORFs) along with 500-bp flanking regions from both sides was amplified from B. burgdorferi B31 clone A3 genomic DNA using the Expand Long PCR system (Roche) and primers 1 and 2 (Table 1) and was cloned into the TOPO XL vector (Invitrogen), creating plasmid TOPO XL bbb22-23+500. The 2.8-kb DNA sequence containing the BBB22 and BBB23 ORFs was removed by inverse PCR using the Expand Long PCR system (Roche) and primers 7 and 8, generating linear pΔbbb22-23 with SalI ends. A spectinomycin and streptomycin resistance cassette, flaBp-aadA (17) with XhoI ends, was amplified using Taq polymerase and primers 3 and 4. The flaBp-aadA product was treated with XhoI and ligated with SalI- and DpnI-digested linear pΔbbb22-23, yielding the allelic-exchange plasmid pΔbbb22-23-flaBp-aadA. B. burgdorferi A3-68ΔBBE02 was transformed with 20 μg of pΔbbb22-23-flaBp-aadA purified from E. coli as previously described (12, 14, 30). Streptomycin-resistant colonies were confirmed to be true transformants by PCR using primer pairs 1 and 2 and 5 and 6. Positive Δbbb22-23-flaBp-aadA clones were screened with a panel of primers for the presence of all of the B. burgdorferi plasmids of the parent A3-68ΔBBE02 clone (28), and a single clone was selected for further experiments.
Table 1.
Primers and probes used in this study
| Primer or probe | Designation | Sequence (5′–3′) |
|---|---|---|
| 1 | BBB22 − 500 | CAGTTAAAGAGCTTACAAGCCC |
| 2 | BBB23 + 500 | AGGATACCTTGCAATATGATCC |
| 3 | aadA-3′ Xho1 | CCGCTCGAGTTATTTGCCGACCTACCTTGG |
| 4 | flaB- 5′ Xho1 | CCGCTCGAGCTGTCGCCTCTTGTGGCTTC |
| 5 | BBB22-3′ HindIII | CCCAAGCTTGACTCTTTTTTATGATTTATAATTTAGG |
| 6 | BBB23-5′ BamHI | CGGGATCCTATTTTCAAACTTTACCTGACAGCG |
| 7 | BBB22F (17918) Sall 3′ | ACGCGTCGACCCTAAATTATAAATCATAAAAAAGAGTC |
| 8 | BBB23R (20824) Sall 5′ | ACGCGTCGACAATAATTATTAGGCTTTTAATTCTTTTTAAAGGCC |
| 9 | BBB22-Fwd TaqMan | AAGAAATTATCGGTGGTATTACCACTTT |
| 10 | BBB22-Rev TaqMan | CAATTGGCATACCTGTGCTAGATAATAT |
| 11 | BBB23-Fwd TaqMan | GCACTAGTTACCGCAACCTGTCT |
| 12 | BBB23-Rev TaqMan | CTCATTCCAGAAGCCAATGCT |
| 13 | BBB22 Probe | AGCATGGCATACATAATAGCTGTTAATCCGGC |
| 14 | BBB23 Probe | CTAATGGGACTTTATACCAATACGCCTT |
Complementation of the Δbbb22-23 mutant.
A DNA fragment encompassing the bbb22-23 genes and upstream putative promoter sequence was amplified from B31 A3 genomic DNA using Phusion enzyme (Thermo Scientific) and primers 5 and 6 (Table 1), which introduced BamHI and HindIII sites at the 5′ and 3′ ends, respectively. The BamHI- and HindIII-digested PCR product was ligated into BamHI- and HindIII-cut B. burgdorferi shuttle vector pBSV2G (12). The plasmid structure and sequence were confirmed by restriction digest and DNA sequence analysis. The Δbbb22-23 mutant was transformed with 20 μg of pBSV2G bbb22-23 or pBSV2G isolated from E. coli and positive transformants selected as previously described (18). The clones that retained the B. burgdorferi plasmid content of the parent clone were selected for use in further experiments.
In vitro growth analysis.
Wild-type (A3-68ΔBBE02), Δbbb22-23/vector, and Δbbb22-23/bbb22-23+ spirochetes were inoculated in triplicate at a density of 105 spirochetes/ml in 5 ml of BSK II medium. Spirochete densities were determined every 24 h under dark-field microscopy using a Petroff-Hausser chamber over the course of 96 h.
RNA isolation from in vitro-grown spirochetes.
Wild-type (B31 A3) spirochetes were grown in triplicate in 5 ml of BSK II medium (pH 7.5) at 35°C to a density of 2 × 108 spirochetes/ml. A total of 1 × 107 spirochetes were harvested from each culture and resuspended in 25 μl of RNAlater (Life Technologies) and stored at −80°C until processing. Total RNA was isolated using TRIzol reagent (Life Technologies) according to the manufacturer's instructions. RNA was resuspended in 100 μl of diethyl pyrocarbonate (DEPC)-treated distilled water (dH2O). RNA was treated with TURBO DNase (Life Technologies) to remove any contaminating genomic DNA. A total of 1 μl of Riboguard (40 U/μl) RNase inhibitor (Epicentre) was added to all samples and RNA stored at −80°C.
RNA isolation from infected mouse tissue.
B. burgdorferi-infected mouse hearts (see “Mouse infection experiments” below) were manually macerated on ice using sterile scalpels and transferred to a 2-ml tube containing lysing Matrix D (MP Biomedicals). A total of 1 ml of RNApro solution (FastRNA Pro Green kit; MP Biomedicals) was added to each sample on ice. Tissues were homogenized using a PowerGen high-throughput homogenizer (Fisher Scientific) following six cycles of beating for 45 s and 2 min on ice. Samples were centrifuged at 15,300 × g for 5 min at 4°C. The upper aqueous phase was transferred to new tubes and incubated for 5 min at room temperature. A total of 500 μl of 1-bromo-3-chloropropane (Sigma) and 45 μl of 5 M sodium acetate were added to each sample, and samples were incubated for an additional 5 min at room temperature. Samples were centrifuged at 15,300 × g for 5 min at 4°C. The upper aqueous phase was transferred to new tubes and RNA precipitated with the addition of 500 μl of absolute ethanol and 1 μl of GlycoBlue (Life Technologies). RNA was pelleted by centrifugation at 15,300 × g for 10 min at 4°C. RNA was then washed with 70% ethanol in DEPC-treated H2O and resuspended in 100 μl of DEPC-treated H2O. Finally, RNA was treated with TURBO DNase (Life Technologies) to remove any contaminating genomic DNA. A total of 1 μl of Riboguard (40 U/μl) RNase inhibitor (Epicentre) was added to all samples and RNA stored at −80°C.
Gene expression analysis.
cDNA was synthesized from all RNA samples using the qScript Flex cDNA synthesis kit (Quanta BioSciences) with random hexamer primers according to the manufacturer's instructions. Parallel cDNA reactions were carried out in the absence of reverse transcriptase. Real-time quantitative PCRs (qPCRs) were prepared using 5 μl of cDNA and Perfecta qPCR Fast Mix (Quanta BioSciences). Using an Applied Biosystems 7500 instrument, samples were assayed for the flaB (19), bbb22, and bbb23 transcripts using primer pair 9 and 10 (Table 1), primer pair 11 and 12, and probes 13 and 14, respectively. The flaB transcript was used as the endogenous reference to which the transcripts of the target genes were normalized. The bbb22 and bbb23 primers were confirmed to be specific for their respective gene targets. The amounts of the flaB, bbb22, and bbb23 transcripts were determined using a standard curve for each gene target that was generated using genomic DNA isolated from 106,105, 104, 103, 102, and 101 spirochetes. Samples were analyzed in triplicate with at least two biological replicates, and gene expression was reported as the number of gene transcripts per flaB mRNA copy. The amplification of samples lacking reverse transcriptase was similar to that of the no-template control. Data sets were compared using the nonparametric Kruskal-Wallis test with Dunn's multiple-comparison test using GraphPad Prism 5.0 for Windows (GraphPad Software).
Radioactive-transport assays.
B. burgdorferi clones were grown in 40 ml of BSK II medium to a density of 8 × 107 spirochetes/ml, washed twice with HN buffer (50 mM HEPES, 50 mM NaCl [pH 7.6]), and resuspended in a final volume of 2.5 ml of HN buffer. Aliquots of 500 μl each (∼5 × 108 spirochetes) of spirochetes were combined with 20 μl of 300 mM glucose (6 mM final concentration) and an additional 480 μl of HN buffer. Spirochetes were preincubated at 37°C for 30 min. A 250 nM concentration of [3H]hypoxanthine monohydrochloride (specific activity, 20 Ci/mmol) (PerkinElmer) or 33 nM [2,8-3H]adenine (specific activity, 30 Ci/mmol) (PerkinElmer) was added to the reaction mixture, and 100-μl aliquots of the reaction mixture (5 × 107 spirochetes) were removed at various time points following the addition of radioactivity, filtered onto cellulose acetate filter membranes using a 10-place filtration manifold (Hoefer, Inc.), and immediately washed with 10 ml of HN buffer. Filter-bound spirochetes were collected in scintillation vials containing 3 ml of ScintiVerse BD cocktail scintillation fluid (Fisher Scientific) and radioactivity counted in a Microbeta2 with an efficiency of 52% (PerkinElmer). The rates of uptake of hypoxanthine and adenine (picomoles/107 spirochetes/minute) were calculated by first converting counts per minute (cpm) to picomoles assuming 1 cpm = disintegrations per minute (dpm) × an efficiency of 0.52 and 1 Ci = 2.2 × 1012 dpm. The rates corresponding to the time points for the linear portion of each uptake curve were averaged to determine the average rate of uptake.
Spirochetes for [3H]hypoxanthine monohydrochloride uptake assays in the presence of cold competitors were grown and prepared as described above. Stock solutions (1 mM) of each cold competitor—hypoxanthine, adenine, guanine, xanthine, cytosine, thymine, uracil, ascorbic acid, and 5′-deoxy-5′-(methylthio)adenosine (MTA)—were prepared in 8 mM NaOH. Reactions were initiated with the addition of 50 μl of competitor (50 μM final concentration, 200-fold excess) or 50 μl of 8 mM NaOH alone and 250 nM [3H]hypoxanthine monohydrochloride. A 100-μl aliquot (5 × 107 spirochetes) of each reaction mixture was removed 10 min following the addition of radioactivity, filtered onto cellulose acetate filter membranes using a 10-place filtration manifold (Hoefer, Inc.), and immediately washed with 10 ml of HN buffer. Filter-bound spirochetes were collected in scintillation vials containing 3 ml of ScintiVerse BD cocktail scintillation fluid (Fisher Scientific) and radioactivity counted in a Microbeta2 (PerkinElmer). A3-68ΔBBE02 and A3-68ΔBBE02 heated to 95°C for 10 min served as the wild-type control and the heat-killed negative control, respectively, for all transport assays. Each transport assay was repeated in triplicate.
Mouse infection experiments.
The University of Central Florida (UCF) is accredited by the International Association for Assessment and Accreditation of Laboratory Animal Care. Protocols for all animal experiments were prepared according to the guidelines of the National Institutes of Health and approved by UCF's Institutional Animal Care and Use Committee. For gene expression studies, four mice (C3H/HeN, 6- to 8-week-old females; Harlan) were needle inoculated, 80% intraperitoneally and 20% subcutaneously, with B. burgdorferi clone B31 A3 at a dose of 1 × 105 spirochetes. For infectivity studies, groups of 6 mice each (C3H/HeN, 6- to 8-week-old females; Harlan) were needle inoculated, 80% intraperitoneally and 20% subcutaneously, with clone Δbbb22-23/vector or Δbbb22-23/bbb22-23+ at a dose of 1 × 104 or 1 × 107 spirochetes. The number of spirochetes inoculated into mice was determined using a Petroff-Hausser counting chamber and verified by CFU counts in solid BSK medium. Ten colonies per inoculum were verified by PCR for the presence of the virulence plasmids lp25, lp28-1, and lp36 in at least 90% of the individuals in the population. Further, the total plasmid content of each inoculum was confirmed to be as expected (18). Mouse infection was assessed 3 weeks postinoculation by serology and reisolation of spirochetes from ear, bladder, and joint tissues, as previously described (18). Heart tissues were harvested from B31 A3-inoculated mice for RNA isolation and gene expression studies.
RESULTS
BBB22 and BBB23 open reading frames on cp26.
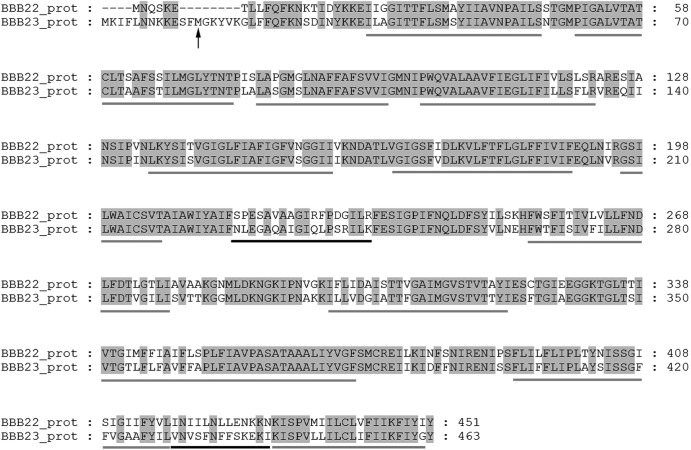
The BBB22 and BBB23 open reading frames (ORFs) are in the same orientation and separated by 109 bp (8, 13). The bbb22 and bbb23 genes (1,356 bp and 1,392 bp, respectively) and their annotated proteins (451 amino acids and 463 amino acids, respectively) are 79.8% and 78.3% identical at the nucleotide and amino acid levels, respectively, suggesting that these genes may be paralogs and their encoded proteins may have overlapping functions in B. burgdorferi. Both BBB22 and BBB23 are annotated as guanine and xanthine permeases and have a conserved domain, COG2252 (xanthine, uracil, and vitamin C permease), that is a distant subfamily of the NCBI CD superfamily cI00967 NAT (nucleobase and ascorbate transporter) family (23). BBB22 and BBB23, similar to other COG2252 domain proteins, are predicted to both localize to the bacterial inner membrane and contain 12 transmembrane domains (9, 20, 21, 40, 42) (Fig. 1). Current annotation of the BBB23 ORF suggests that translation of the BBB23 protein initiates at the alternative start codon TTG and produces a 463-amino-acid protein that is 12 amino acids longer than the predicted 451-amino-acid size for the BBB22 protein (8, 13) (Fig. 1). Upon further analysis of the BBB23 ORF sequence, we identified an ATG start codon 36 nucleotides within the current annotation of the BBB23 ORF, which is preceded by a reasonable Shine-Dalgarno sequence 5 bp upstream. Our prediction of this ATG residue as the putative true start codon for the BBB23 protein resulted in the additional prediction that the BBB23 ORF produces a 451-amino-acid protein, as is predicted for BBB22.
Fig 1.
The BBB22 and BBB23 proteins demonstrate a high level of identity. Shown is an amino acid sequence alignment of the BBB22 (NP_047008.1) and BBB23 (NP_047009.2) proteins, according to current annotation. Identical amino acids are shaded. Amino acid sequences were aligned using the Clustal W algorithm in the MegAlign program from the DNASTAR Lasergene suite. The putative sequences of the 12 transmembrane domains were predicted using the TMHMM server, version 2.0 (http://www.cbs.dtu.dk/services/TMHMM/), and are indicated with gray underlines. Divergent sequences in putative periplasmic loops 4 and 6 are indicated with black underlines. An alternative prediction for the start codon of BBB23 is indicated with a black arrow.
The bbb22-23 genes are essential for B. burgdorferi mouse infectivity.
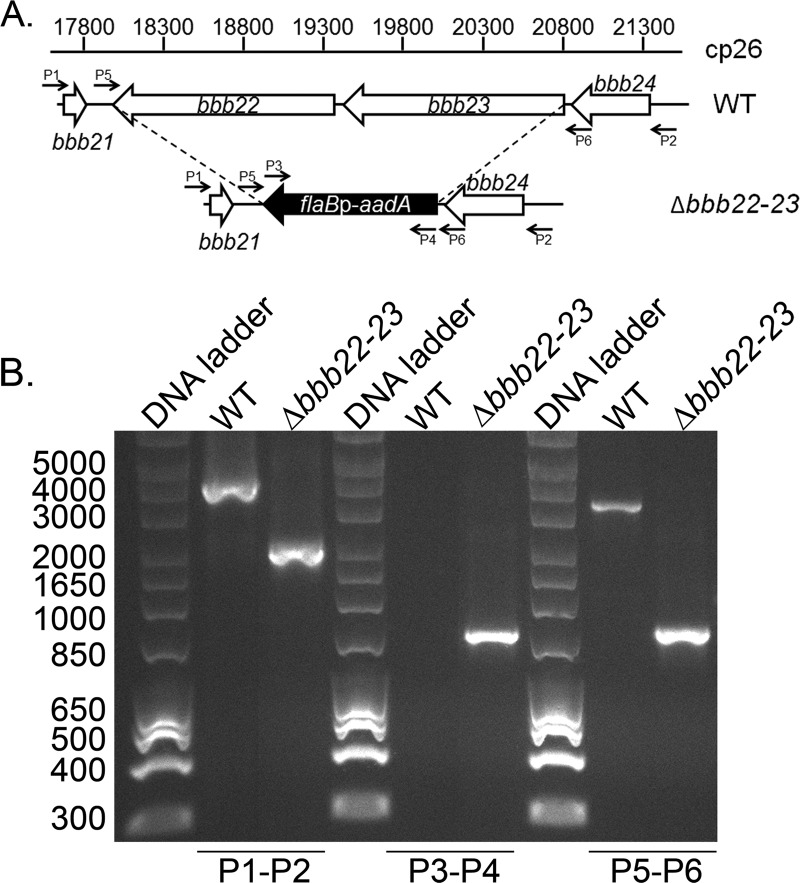
Given the high degree of amino acid identity between the BBB22 and BBB23 proteins, it is reasonable to predict that these two proteins could have overlapping functions and that in the absence of one, the other may be able to functionally compensate. Moreover, we have found that both genes are expressed during mammalian infection (Fig. 2), suggesting that both proteins may be important for survival in the mouse. Therefore, in order to examine the combined functional role of the BBB22 and BBB23 proteins during mammalian infection, the sequence encompassing both open reading frames was deleted from low-passage, infectious B. burgdorferi and replaced with the flaBp-aadA spectinomycin and streptomycin resistance cassette (17) by allelic exchange (Fig. 3). Successful recovery of Δbbb22-23 spirochetes demonstrates that the function of the encoded proteins is not essential for B. burgdorferi survival in vitro. The Δbbb22-23 spirochetes were genetically complemented in trans on the shuttle vector pBSV2G (12) with wild-type copies of the bbb22 and bbb23 genes and102 bp of the 5′ upstream sequence encompassing the presumed promoter of bbb23. In vitro growth analysis demonstrated no difference in growth between Δbbb22-23, Δbbb22-23/bbb22-23+, and wild-type spirochetes over the course of 96 h (data not shown), suggesting that the functions of the gene products are not required for spirochete replication in nutrient-rich BSK II in vitro growth medium.
Fig 2.
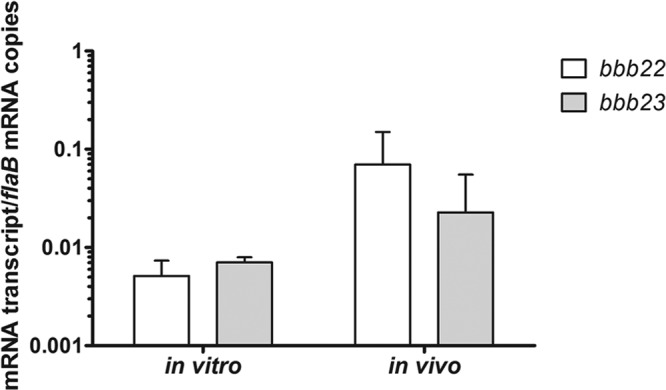
The bbb22 and bbb23 genes are expressed during murine infection. Spirochete RNA was isolated from log-phase (3 × 107 spirochetes/ml) B. burgdorferi clone B31 A3 grown in BSK II medium, pH 7.5, at 35°C (in vitro). Total RNA was isolated from spirochete-infected heart tissue harvested 2 weeks after intradermal inoculation of C3H/HeN mice with 1 × 105 B. burgdorferi clone B31 A3 spirochetes (in vivo). Expression of genes flaB, bbb22, and bbb23 was quantified by reverse transcriptase qPCR. The data are expressed as the number of bbb22 or bbb23 gene transcripts per number of flaB mRNA copies. The in vitro data represent the averages of two biological replicates. The in vivo data represent the averages of four biological replicates. Error bars represent the standard deviations from the means. All data sets were compared pairwise using the Kruskal-Wallis nonparametric rank test followed by Dunn's multiple-comparison test (GraphPad Prism 5.0) and were found to have no statistical difference.
Fig 3.
Generation of the Δbbb22-23 mutant in B. burgdorferi. (A) Schematic representation of the wild type (WT) and Δbbb22-23 loci on cp26. The sequence of the BBB22-23 open reading frame was replaced with a flaBp-aadA antibiotic resistance cassette. Locations of primers for analysis of the mutant clones are indicated with small arrows and labels P1 to P6. Primer sequences are listed in Table 1. The boundaries of the deletion construct are indicated by broken lines. (B) PCR analysis of mutant clones. Genomic DNA isolated from WT and Δbbb22-23 spirochetes served as the template DNA for PCR analyses. DNA templates are indicated across the top of the gel image. The oligonucleotide pairs used to amplify specific DNA sequences are indicated below the gel image and correspond to target sequences as shown in panel A.
The contribution of the bbb22-23 genes to B. burgdorferi mammalian infectivity was assessed by inoculation of mice with mutant spirochetes lacking bbb22-23 or the isogenic bbb22-23+ complemented clone at a dose of 1 × 104 spirochetes. None of the mice inoculated with the Δbbb22-23 mutant spirochetes became infected, as assessed by serology and reisolation of spirochetes from tissues 3 weeks postinoculation (Table 2). Conversely, six out of six mice inoculated with the bbb22-23+ complemented clone were seropositive, and spirochetes were reisolated from tissues (Table 2). To further explore the role of these genes in B. burgdorferi murine infection, mice were inoculated with 1 × 107 mutant or complemented spirochetes. Strikingly, no mice inoculated with spirochetes lacking bbb22-23 were infected, even at this high dose, whereas all mice inoculated with the complemented spirochetes became infected (Table 2). Together, these data demonstrate that bbb22 and/or bbb23 is absolutely required for B. burgdorferi mouse infectivity by needle inoculation.
Table 2.
bbb22 and/or bbb23 is required for B. burgdorferi mouse infectivity
| Clone | Spirochete dose per mouse | No. of infected mice/totala |
|---|---|---|
| Δbbb22-23/pBSV2G | 1 × 104 | 0/6 |
| 1 × 107 | 0/6 | |
| Δbbb22-23/pBSV2G bbb22-23 | 1 × 104 | 6/6 |
| 1 × 107 | 6/6 |
Number of infected mice per number of mice analyzed, determined 3 weeks postinoculation by positive serology against B. burgdorferi proteins and reisolation of spirochetes from ear, bladder, and joint tissues.
BBB22 and/or BBB23 are hypoxanthine, adenine, and guanine transporters.
Transport and incorporation of hypoxanthine and adenine by wild-type B. burgdorferi have been demonstrated previously (3, 22, 26). Contrary to the annotated function of COG2252 domain proteins, none of the biochemically characterized members of this family has been shown to transport xanthine, uracil, or vitamin C. Instead, they have all been found to function as purine permeases capable of transporting adenine, hypoxanthine, and/or guanine (5, 6, 9, 33), supporting the hypothesis that BBB22 and BBB23 are responsible for transport of purines in B. burgdorferi. BBB22-23+ and BBB22-23− spirochetes were examined for transport of hypoxanthine and adenine using a radioactive filter-binding assay (37). Transport of [3H]hypoxanthine was found to be dependent on the BBB22 and/or BBB23 protein, as the Δbbb22-23 mutant clone, similar to heat-killed wild-type spirochetes, was incapable of hypoxanthine uptake (Fig. 4A). Conversely, wild-type and complemented spirochetes demonstrated increased accumulation of [3H]hypoxanthine over a period of approximately 15 min (Fig. 4A). Unlike transport of [3H]hypoxanthine, transport of [3H]adenine was not completely BBB22 and/or BBB23 dependent (Fig. 4B). The Δbbb22-23 mutant clone demonstrated transport of [3H]adenine. The average rate of [3H]adenine uptake for the Δbbb22-23 mutant spirochetes (0.015 ± 0.006 pmol/107 spirochetes/min) was reduced 68-fold relative to that of wild-type and complemented spirochetes (0.99 ± 0.02 pmol/107 spirochetes/min). A period of approximately 15 min was required for the mutant spirochetes to accumulate a maximum level of [3H]adenine equivalent to that of spirochetes harboring functional BBB22-23 proteins. Wild-type and complemented spirochetes achieved maximum [3H]adenine uptake within the first 1 min of the assay (Fig. 4B). The average rate of [3H]adenine uptake by wild-type and complemented spirochetes was 29-fold higher than the average rate of [3H]hypoxanthine uptake by the same two clones (0.035 ± 0.005 pmol/107 spirochetes/min). [3H]adenine transport by wild-type spirochetes was greater than 80% inhibited in the presence of a 200-fold excess of unlabeled adenine (data not shown). Together, these data demonstrate that BBB22 and/or BBB23 is responsible for salvage of hypoxanthine; these proteins contribute to uptake of adenine by B. burgdorferi, but the spirochetes appear to harbor additional transport systems important for this function.
Fig 4.
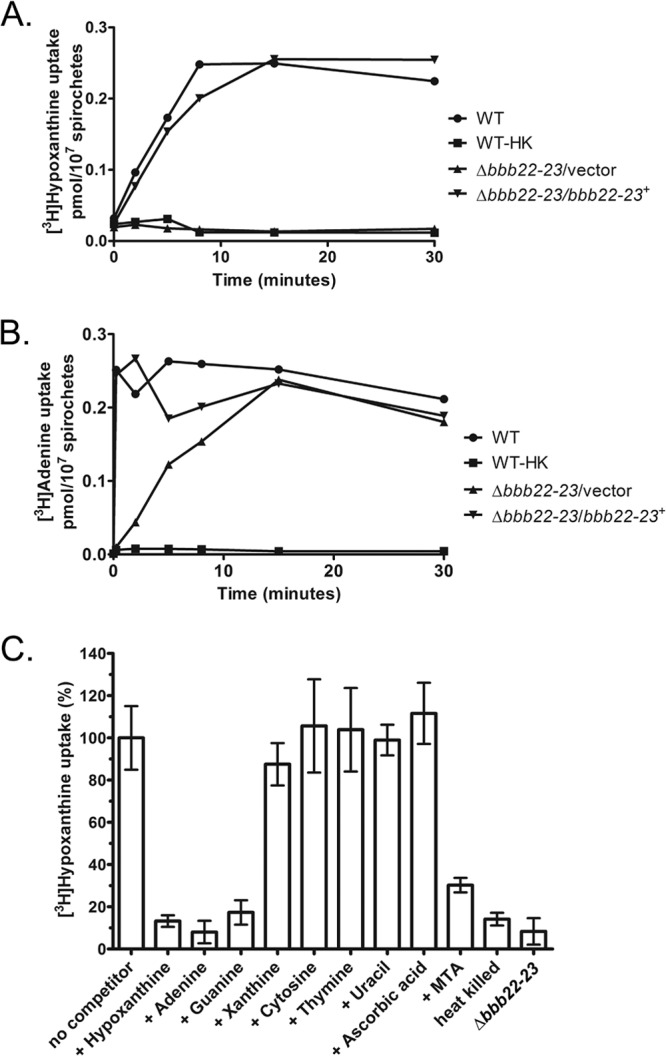
BBB22 and/or BBB23 are hypoxanthine, adenine, and guanine permeases. [3H]hypoxanthine uptake (A) and [3H]adenine uptake (B) by wild-type (WT), heat-killed WT (WT-HK), Δbbb22-23 mutant (Δbbb22-23/vector), and Δbbb22-23 complemented (Δbbb22-23/bbb22-23+) spirochetes were measured over a 30-min time course. Uptake of radiolabeled purine is expressed in picomoles/107 spirochetes/minute. The final concentrations of labeled hypoxanthine and adenine in the reactions were 250 nM and 33 nM, respectively. The specific activities of labeled hypoxanthine and adenine were 20 Ci/mmol and 30 Ci/mmol, respectively. Data shown are representative results of one of three replicate experiments. (C) [3H]hypoxanthine (250 nM, 20 Ci/mmol) uptake by wild-type spirochetes was measured in the absence (no competitor) or presence of a 200-fold excess (50 μM) of unlabeled competitor as indicated. Heat-killed wild-type (heat killed) and Δbbb22-23 mutant (Δbbb22-23) spirochetes in the absence of competitor served as negative controls. Radioactivity uptake was measured 10 min after the addition of [3H]hypoxanthine with or without unlabeled competitor. Radioactivity uptake by wild-type spirochetes in the absence of competitor was taken as 100%. All other data are represented as the percent uptake relative to that by wild-type spirochetes in the absence of competitor. Error bars represent the standard deviations of triplicate experiments. Statistical analyses were performed using the unpaired t test (GraphPad Prism 5.0).
To further elucidate the function of the BBB22-23 transport system, we took advantage of the finding that BBB22 and/or BBB23 is the only transporter system for hypoxanthine uptake. The ability of these proteins to transport other substrates was determined by measuring [3H]hypoxanthine uptake in the presence of a 200-fold excess of unlabeled competitors, including purine derivatives, pyrimidines, and ascorbic acid. As expected, unlabeled adenine and hypoxanthine competed for [3H]hypoxanthine uptake. The percent [3H]hypoxanthine uptake in the presence of unlabeled adenine or hypoxanthine was not statistically different from that of heat-killed and Δbbb22-23 mutant spirochetes (adenine, P = 0.1 and P = 0.95, respectively; hypoxanthine, P = 0.69 and P = 0.27, respectively; unpaired t test) (Fig. 4C). Furthermore, these analyses established that guanine is an additional substrate for BBB22 and/or BBB23, as the percent [3H]hypoxanthine uptake in the presence of excess unlabeled guanine was similar to that of heat-killed and Δbbb22-23 mutant spirochetes (P = 0.38 and P = 0.11, respectively; unpaired t test) (Fig. 4C). The presence of xanthine, cytosine, thymine, uracil, or ascorbic acid had no inhibitory effect on [3H]hypoxanthine uptake, indicating that these metabolites are not transported by BBB22 or BBB23 (Fig. 4C). All in all, these data clearly establish BBB22 and/or BBB23 function as a hypoxanthine, adenine, and guanine permease.
5′-Methylthioadenosine (MTA) is a sulfur-containing nucleoside produced from S-adenosylmethionine (SAM) during polyamine synthesis in mammalian tissues (1) and polyamine, autoinducer-1, and biotin synthesis in bacteria (11). Bacterial 5′-methylthioadenosine/S-adenosylhomosysteine (MTA/SAH) nucleosidases (Pfs) catalyze the cleavage of MTA into 5′-methylthioribose (MTR) and adenine (25). Borrelia burgdorferi harbors three pfs homologs, BB0588 (bgp or pfs-2), BB0375 (pfs-1), and BBI06 (mtnN). Bgp and Mtn are believed to be present on the surface of the spirochete, whereas Pfs-1 is predicted to localize to the cytoplasm (11, 25). Both Bgp and Pfs-1 have demonstrated MTA nucleosidase activity (25), and together these proteins are predicted to contribute to adenine salvage from spirochete and host metabolism (11). To test the hypothesis that MTA may provide a source of adenine for salvage by B. burgdorferi, MTA was examined for its ability to compete for [3H]hypoxanthine transport by BBB22-23. In the presence of MTA, [3H]hypoxanthine transport was significantly reduced relative to transport in the absence of competitor (P = 0.0006; unpaired t test); however, MTA was not sufficient to reduce [3H]hypoxanthine transport levels equivalent to that of heat-killed or Δbbb22-23 mutant spirochetes (P = 0.0012 and P = 0.0030, respectively; unpaired t test) (Fig. 4C). These data support the hypothesis that exogenous MTA can serve as a source of adenine for salvage by B. burgdorferi.
DISCUSSION
During its infectious cycle, B. burgdorferi experiences environments with potentially distinct purine availability. Hypoxanthine is the most abundant purine in mammalian blood (16), and it is presumed to be available for salvage by B. burgdorferi during the blood meal of an infected tick and during the spirochete's transient presence in the mammalian bloodstream. During mammalian infection, B. burgdorferi resides in various tissues which have been shown to be rich in adenine (41). Guanine is present at low levels in mammalian blood and tissues (16, 41). It has been demonstrated previously that B. burgdorferi is able to transport and incorporate adenine, hypoxanthine, nucleosides, and deoxynucleosides into DNA and RNA (22, 26). However, little to no transport of guanine or xanthine has been previously observed for B. burgdorferi (26). We now establish that the homologous proteins BBB22 and/or BBB23, encoded on cp26, transport hypoxanthine, adenine, and guanine and are necessary for B. burgdorferi mammalian infection. The bbb22 and/or bbb23 gene product was found to be essential for hypoxanthine uptake. At least one of these transporters is also important for adenine uptake; however, our data revealed that an additional, as-yet-unknown, protein(s) also contributes to this function. Nonetheless, the function of the additional adenine transporter(s) was not sufficient to allow full or even attenuated survival of Δbbb22-23 mutant B. burgdorferi in the mouse, as spirochetes lacking only bbb22-23 were unable to infect mice up to a dose of 1 × 107 bacteria. These data suggest that transport of hypoxanthine, rather than adenine, may be critical during the initial stages of B. burgdorferi mammalian infectivity, consistent with the transient dissemination of spirochetes through the blood early in infection, while adenine salvage may be critical for persistent survival of B. burgdorferi in tissues. In addition to direct scavenge of adenine from host tissues, our data suggest that catabolism of MTA produced by the host may provide a source of exogenous adenine for purine salvage. B. burgdorferi harbors two MTA nucleosidase homologs, Bgp and BBI06, which have been shown and predicted, respectively, to localize to the spirochete's outer surface, suggesting that the biological role of these enzymes is to catabolize host-derived MTA (25). Spirochetes lacking bgp maintain wild-type virulence in mice (25), indicating that BBI06 is able to compensate for the loss of Bgp activity or that MTA catabolism contributes little to B. burgdorferi survival in the mammal. Hypoxanthine transport by BBB22 and/or BBB23 was competitively inhibited by guanine, demonstrating for the first time that B. burgdorferi is able to transport guanine, and this likely occurs via BBB22 and/or BBB23. Contrary to annotation of BBB22 and BBB23 as xanthine, uracil, and vitamin C permeases, none of these metabolites was able to compete for hypoxanthine transport. Furthermore, BBB22 and BBB23 function did not include pyrimidine transport, as neither cytosine nor thymine competed for hypoxanthine transport. These data are consistent with the function of the protein members of a new class of nucleobase transporters containing COG2252 defined by the Aspergillus nidulans hypoxanthine, adenine, and guanine permease, AzgA (9). Additional characterized members of this subfamily include the E. coli adenine permease, PurP (5, 6), and the two Bacillus subtilis guanine and hypoxanthine permeases, PbuG and PbuO (31–33).
Despite the critical roles of BBB22 and/or BBB23 in purine uptake in vivo, spirochetes lacking bbb22-23 do not exhibit a growth defect in vitro. The rich, complex B. burgdorferi in vitro growth medium provides excess purines and is likely a source of nucleosides and deoxynucleosides. In the presence of excess nutrients, the unidentified, additional adenine transporter system may provide a mechanism for the spirochetes to salvage sufficient amounts of adenine for survival in the absence of BBB22 and BBB23. We have demonstrated previously that adenine is converted to hypoxanthine by the adenine deaminase activity of BBK17 (AdeC), which may relieve the requirement for hypoxanthine salvage in environments high in adenine (18). B. burgdorferi has been shown to import and incorporate into DNA and RNA exogenous nucleosides and deoxynucleosides (22). Direct salvage of nucleosides and deoxynucleosides by a transport system other than BBB22 and BBB23 may allow the spirochetes to bypass the requirement for hypoxanthine, adenine, and/or guanine transport to synthesize DNA and RNA. This mechanism has also been suggested to explain the absence of an in vitro growth defect for spirochetes lacking the in vivo essential, purine salvage genes guaA and guaB (19).
The high degree of sequence identity shared between bbb22 and bbb23 suggests that these two genes have evolved as the result of a duplication event. Moreover, this prediction raises the question of why B. burgdorferi has two purine permeases encoded by adjacent, paralogous genes on cp26. One possibility is that the two proteins have distinct substrate specificities. The BBB22 and BBB23 proteins are predicted to traverse the spirochete's inner membrane, and therefore, the amino acid sequences of the six periplasmic loops of each protein are likely important determinants of the substrates that are transported by these purine permeases. Two of the six putative periplasmic loops within BBB22 (amino acids 207 to 254 and 418 to 431) and BBB23 (amino acids 219 to 266 and 430 to 443) (Fig. 1) are predicted to be highly divergent between the two proteins, suggesting that these domains may contribute to differential substrate specificity between the two proteins, if any. Alternatively, BBB22 and BBB23 may have similar substrate specificities but each protein functions in response to distinct environmental cues. The sequences and synteny of the BBB22 and BBB23 homologs appear to be conserved among the Lyme disease Borrelia spp. Conversely, the relapsing fever spirochetes appear to only harbor distantly related BBB23-like sequences (data not shown). Furthermore, the Lyme disease spirochetes lack the key purine salvage enzymes ribonucleotide reductase, hypoxanthine-guanine phosphoribosyltransferase (Hpt), adenylosuccinate synthase (PurA), and adenylosuccinate lyase (PurB) (3, 22, 26), which are present in the relapsing-fever spirochetes (3, 22, 26), and the presence or absence of BBB22 and BBB23 homologs may reflect the distinct purine salvage requirements between the two groups of pathogens.
The bbb22 and bbb23 transcripts were detected in spirochetes grown in the in vivo mammalian host environment. Although not statistically significant, the data suggest that the levels of bbb22 and bbb23 expression in vivo, as determined by reverse transcription-quantitative real-time PCR, may be increased relative to the levels of expression of these genes under nutrient-rich in vitro growth conditions. Furthermore, these data imply that the expression of the encoded permeases may be upregulated in response to potentially nutrient-limited growth conditions in which purine salvage is likely critical for B. burgdorferi survival. Key purine salvage enzymes, IMP dehydrogenase (GuaB) and GMP synthase (GuaA) for the synthesis of GTP and dGTP, have been shown to be indispensable for B. burgdorferi survival throughout its infectious cycle (19), suggesting that the tick vector and mammalian host environments are limited for direct scavenge of GTP and dGTP for RNA and DNA biosynthesis. Moreover, despite evidence presented herein that guanine can be transported by BBB22 and/or BBB23, guanine and guanine nucleotide precursors are thought to be limiting for B. burgdorferi survival in vivo (19, 22). B. burgdorferi has evolved a unique purine salvage pathway in the absence of the “classic” purine salvage pathway enzymes (3, 22, 26), in order to generate dGTP from salvaged hypoxanthine or hypoxanthine derived from deaminated salvaged adenine (18, 19, 22). Based on these data, it is intriguing to speculate that expression of bbb22 and/or bbb23 may be controlled by the availability of guanine nucleotide precursors, leading to increased hypoxanthine and/or adenine transport when guanine nucleotide pools are low. In support of this notion, purine nucleotide metabolism in E. coli and B. subtilis has been shown to be controlled by the availability of purine nucleotides from the environment (5, 10, 33). The relative contributions of BBB22 and BBB23 individually to purine salvage and spirochete survival in vivo, as well as the regulatory mechanism(s) controlling bbb22 and bbb23 expression, are under investigation.
We have demonstrated that BBB22 and BBB23 function in the critical first step of the purine salvage pathway in B. burgdorferi and, in turn, are a gateway into the spirochete for the acquisition of essential nutrients. These transporters may also provide a novel means to deliver potential antispirochete molecules. Understanding the molecular determinants of substrate specificity and regulation of BBB22 and BBB23 may provide the framework for rational design of inhibitors or antimetabolites targeted against the B. burgdorferi purine salvage pathway, resulting in new treatments for Lyme disease.
ACKNOWLEDGMENTS
We thank T. Choudhury for technical assistance with RNA isolation from infected mouse tissues. Many thanks are extended to N. Parveen for insightful suggestions. We thank members of the M. Jewett and T. Jewett labs for helpful discussions. We thank T. Jewett for critical comments on the manuscript.
This work was supported by UCF startup funds and grant 5K22AI081739 from NIH, NIAID, to M.W.J.
Footnotes
Published ahead of print 18 June 2012
REFERENCES
- 1. Avila MA, Garcia-Trevijano ER, Lu SC, Corrales FJ, Mato JM. 2004. Methylthioadenosine. Int. J. Biochem. Cell Biol. 36:2125–2130 [DOI] [PubMed] [Google Scholar]
- 2. Barbour AG. 1984. Isolation and cultivation of Lyme disease spirochetes. Yale J. Biol. Med. 57:521–525 [PMC free article] [PubMed] [Google Scholar]
- 3. Barbour AG, Putteet-Driver AD, Bunikis J. 2005. Horizontally acquired genes for purine salvage in Borrelia spp. causing relapsing fever. Infect. Immun. 73:6165–6168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bono JL, Tilly K, Stevenson B, Hogan D, Rosa P. 1998. Oligopeptide permease in Borrelia burgdorferi: putative peptide-binding components encoded by both chromosomal and plasmid loci. Microbiology 144(Pt 4):1033–1044 [DOI] [PubMed] [Google Scholar]
- 5. Burton K. 1994. Adenine transport in Escherichia coli. Proc. Biol. Sci. 255:153–157 [DOI] [PubMed] [Google Scholar]
- 6. Burton K. 1983. Transport of nucleic acid bases into Escherichia coli. J. Gen. Microbiol. 129:3505–3513 [DOI] [PubMed] [Google Scholar]
- 7. Byram R, Stewart PE, Rosa P. 2004. The essential nature of the ubiquitous 26-kilobase circular replicon of Borrelia burgdorferi. J. Bacteriol. 186:3561–3569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Casjens S, et al. 2000. A bacterial genome in flux: the twelve linear and nine circular extrachromosomal DNAs in an infectious isolate of the Lyme disease spirochete Borrelia burgdorferi. Mol. Microbiol. 35:490–516 [DOI] [PubMed] [Google Scholar]
- 9. Cecchetto G, Amillis S, Diallinas G, Scazzocchio C, Drevet C. 2004. The AzgA purine transporter of Aspergillus nidulans. Characterization of a protein belonging to a new phylogenetic cluster. J. Biol. Chem. 279:3132–3141 [DOI] [PubMed] [Google Scholar]
- 10. Cho BK, et al. 2011. The PurR regulon in Escherichia coli K-12 MG1655. Nucleic Acids Res. 39:6456–6464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cornell KA, Primus S, Martinez JA, Parveen N. 2009. Assessment of methylthioadenosine/S-adenosylhomocysteine nucleosidases of Borrelia burgdorferi as targets for novel antimicrobials using a novel high-throughput method. J. Antimicrob. Chemother. 63:1163–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Elias AF, Schmutzhard J, Stewart PE, Schwan TG, Rosa P. 2002. Population dynamics of a heterogeneous Borrelia burgdorferi B31 strain in an experimental mouse-tick infectious cycle. Wien. Klin. Wochenschr 114:557–561 [PubMed] [Google Scholar]
- 13. Fraser CM, et al. 1997. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature 390:580–586 [DOI] [PubMed] [Google Scholar]
- 14. Grimm D, et al. 2004. Experimental assessment of the roles of linear plasmids lp25 and lp28-1 of Borrelia burgdorferi throughout the infectious cycle. Infect. Immun. 72:5938–5946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Grimm D, et al. 2004. Outer-surface protein C of the Lyme disease spirochete: a protein induced in ticks for infection of mammals. Proc. Natl. Acad. Sci. U. S. A. 101:3142–3147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hartwick RA, Assenza SP, Brown PR. 1979. Identification and quantitation of nucleosides, bases and other UV-absorbing compounds in serum, using reversed-phase high-performance liquid chromatography. I. Chromatographic methodology. J. Chromatogr. 186:647–658 [DOI] [PubMed] [Google Scholar]
- 17. Jewett MW, et al. 2007. Genetic basis for retention of a critical virulence plasmid of Borrelia burgdorferi. Mol. Microbiol. 66:975–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jewett MW, et al. 2007. The critical role of the linear plasmid lp36 in the infectious cycle of Borrelia burgdorferi. Mol. Microbiol. 64:1358–1374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jewett MW, et al. 2009. GuaA and GuaB are essential for Borrelia burgdorferi survival in the tick-mouse infection cycle. J. Bacteriol. 191:6231–6241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Keseler IM, et al. 2011. EcoCyc: a comprehensive database of Escherichia coli biology. Nucleic Acids Res. 39(database issue):D583–D590 [Epub ahead of print.] doi:10.1093/nar/gkq1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Krogh A, Larsson B, von Heijne G, Sonnhammer EL. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305:567–580 [DOI] [PubMed] [Google Scholar]
- 22. Lawrence KA, Jewett MW, Rosa PA, Gherardini FC. 2009. Borrelia burgdorferi bb0426 encodes a 2′-deoxyribosyltransferase that plays a central role in purine salvage. Mol. Microbiol. 72:1517–1529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Marchler-Bauer A, et al. 2011. CDD: a conserved domain database for the functional annotation of proteins. Nucleic Acids Res. 39:D225–D229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Margolis N, Hogan D, Tilly K, Rosa PA. 1994. Plasmid location of Borrelia purine biosynthesis gene homologs. J. Bacteriol. 176:6427–6432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Parveen N, et al. 2006. Bgp, a secreted glycosaminoglycan-binding protein of Borrelia burgdorferi strain N40, displays nucleosidase activity and is not essential for infection of immunodeficient mice. Infect. Immun. 74:3016–3020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pettersson J, et al. 2007. Purine salvage pathways among Borrelia species. Infect. Immun. 75:3877–3884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Purser JE, et al. 2003. A plasmid-encoded nicotinamidase (PncA) is essential for infectivity of Borrelia burgdorferi in a mammalian host. Mol. Microbiol. 48:753–764 [DOI] [PubMed] [Google Scholar]
- 28. Rego RO, Bestor A, Rosa PA. 2011. Defining the plasmid-borne restriction-modification systems of the Lyme disease spirochete Borrelia burgdorferi. J. Bacteriol. 193:1161–1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rosa PA, Hogan D. 1992. Colony formation by Borrelia burgdorferi in solid medium: clonal analysis of osp locus variants, p 95–103 In Munderloh UG, Kurtti TJ. (ed), Proceeding of the First International Conference on Tick Borne Pathogens at the Host-Vector Interface University of Minnesota, St. Paul [Google Scholar]
- 30. Samuels DS. 1995. Electrotransformation of the spirochete Borrelia burgdorferi. Methods Mol. Biol. 47:253–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Saxild HH, Brunstedt K, Nielsen KI, Jarmer H, Nygaard P. 2001. Definition of the Bacillus subtilis PurR operator using genetic and bioinformatic tools and expansion of the PurR regulon with glyA, guaC, pbuG, xpt-pbuX, yqhZ-folD, and pbuO. J. Bacteriol. 183:6175–6183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Saxild HH, Nygaard P. 1988. Gene-enzyme relationships of the purine biosynthetic pathway in Bacillus subtilis. Mol. Gen. Genet. 211:160–167 [DOI] [PubMed] [Google Scholar]
- 33. Saxild HH, Nygaard P. 1987. Genetic and physiological characterization of Bacillus subtilis mutants resistant to purine analogs. J. Bacteriol. 169:2977–2983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Stewart PE, et al. 2006. Delineating the requirement for the Borrelia burgdorferi virulence factor OspC in the mammalian host. Infect. Immun. 74:3547–3553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tilly K, Bestor A, Jewett MW, Rosa P. 2007. Rapid clearance of Lyme disease spirochetes lacking OspC from skin. Infect. Immun. 75:1517–1519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tilly K, et al. 2001. Genetics and regulation of chitobiose utilization in Borrelia burgdorferi. J. Bacteriol. 183:5544–5553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tilly K, Grimm D, Bueschel DM, Krum JG, Rosa P. 2004. Infectious cycle analysis of a Borrelia burgdorferi mutant defective in transport of chitobiose, a tick cuticle component. Vector Borne Zoonotic Dis. 4:159–168 [DOI] [PubMed] [Google Scholar]
- 38. Tilly K, et al. 2006. Borrelia burgdorferi OspC protein required exclusively in a crucial early stage of mammalian infection. Infect. Immun. 74:3554–3564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tilly K, Rosa PA, Stewart PE. 2008. Biology of infection with Borrelia burgdorferi. Infect. Dis. Clin. North Am. 22:217–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. von Heijne G. 1992. Membrane protein structure prediction. Hydrophobicity analysis and the positive-inside rule. J. Mol. Biol. 225:487–494 [DOI] [PubMed] [Google Scholar]
- 41. Wishart DS, et al. 2007. HMDB: the Human Metabolome Database. Nucleic Acids Res. 35:D521–D526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yu NY, et al. 2010. PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics 26:1608–1615 [DOI] [PMC free article] [PubMed] [Google Scholar]