Abstract
Factors and mechanisms determining the differences in virulence and host specificity between the zoonotic agents Chlamydia psittaci and Chlamydia abortus are still largely unknown. In the present study, two strains were compared for their invasiveness, virulence, and capability of eliciting an immune response in chicken embryos. On breeding day 10, embryonated chicken eggs were inoculated with 5 × 104 inclusion-forming units. As shown by immunohistochemistry and quantitative real-time PCR, C. psittaci displayed a significantly better capability of disseminating in the chorioallantoic membrane (CAM) and internal organs than C. abortus. The higher infectious potential of C. psittaci in birds was underlined by significantly higher mRNA expression rates of essential chlamydial genes, such as incA, groEL (in CAM, liver, and spleen), cpaf, and ftsW (in CAM). Although the immune responses to both pathogens were similar, C. psittaci elicited higher macrophage numbers and a stronger expression of a subset of immune-related proteins. The data imply that invasiveness of Chlamydia spp. and propagation in the host are not solely dependent on the level of host immune response but, even to a greater extent, on the expression of bacterial factors related to virulence. The fact that C. psittaci has coped far better than C. abortus with the avian embryo's response by upregulating essential genes may be a key to understanding the mechanisms underlying host adaptation and etiopathology.
INTRODUCTION
The animal pathogens Chlamydia psittaci and Chlamydia abortus both possess a significant and well-documented zoonotic potential (3, 42, 53). At the same time, there are marked differences in host specificity, virulence, tissue tropism, and clinical manifestations of these obligate intracellular bacteria (25). In the case of C. psittaci, psittacine birds, poultry, and wild birds are the preferred hosts (23), but cattle, sheep, swine, horses, dogs, and cats can also be infected (33). Typical symptoms of psittacosis in birds include respiratory distress, diarrhea, conjunctivitis, and rhinitis, with lesions mostly confined to spleen, liver, and air sac, while the immune response is distinguished by lymphocyte depletion and macrophage influx. Chronic infection leads to characteristic diffuse granulomatous or pyogranulomatous inflammation of serosal surfaces. In contrast, C. abortus is primarily adapted to sheep and goats, where it can induce abortion, fetal death, and genital disorders (41). The agent also infects cattle, swine, and horses and rarely infects birds, but clinical manifestations are less frequent than in small ruminants. A high number of mononuclear cells expressing tumor necrosis factor alpha (TNF-α) in infected placental arterioles and arteries is deemed to be crucial for the infection resulting in abortion (25). Nevertheless, the reasons for the unequal host ranges and etiopathologies of these two closely related chlamydial species have yet to be identified.
For their intracellular survival, chlamydial pathogens have developed sophisticated mechanisms to evade lytic damage along the intracellular pathway in the host cell (18). Thus, chlamydiae express a broad variety of potential virulence factors, some of which are assumed to be involved in processes linked to the regulation of host adaptation. Of special importance may be proteins that (i) confer bacterial viability and proliferation activity (e.g., FtsW) (13), (ii) block NF-κB nuclear translocation, phagosome-lysosome fusion (e.g., IncA) (17), and/or apoptosis (e.g., CPAF) in host cells (6), or (iii) potentially affect the production of important host cell immune mediators (43).
In addition to the bacterial virulence properties, the quality and intensity of the host-mediated immune response could be accountable for the observed variations in host specificity of individual Chlamydia species and strains. Studies using gene knockout mice showed that the absence of certain cytokines can increase (gamma interferon [IFN-γ] or interleukin-12 [IL-12]) or decrease (IL-10) the bacterial load and clinical severity in Chlamydia infection (43). The activity of immune-related GTPase was also suggested to be responsible for different host specificities of Chlamydia trachomatis, Chlamydia pneumoniae, and C. psittaci toward mice and humans (12, 29, 49).
A more detailed understanding of the specific actions launched by chlamydial pathogens and the immune response elicited in the host will contribute to better control of these infections in humans and animals. Unfortunately, the unavailability of genetically manipulated Chlamydia strains enormously hampers the study of molecular mechanisms. Moreover, the zoonotic potential of both C. psittaci and C. abortus infections makes in vivo studies very demanding in terms of safety precautions, so investigators have typically resorted to cell culture models (reviewed in reference 45) or, to a lesser extent, experiments in mice using Chlamydia muridarum (28) and C. trachomatis challenge strains (52). In this context, infection of the chorioallantoic membrane (CAM) of developing chicken embryos is a promising alternative. The chicken embryo model has been used in bacteriology and virology for a long time and represents an inexpensive, easily manageable, and comparatively safe in vivo system. We considered it a valuable tool to investigate the innate immune response and inflammatory reaction to chlamydial infection that should also be suitable for comparison of the virulence properties of different strains and the study of host-pathogen interactions. The CAM as a convenient inoculation site for Chlamydia spp. was first described more than 70 years ago (8). Indeed, it resembles a natural infection across epithelial layers, and more recent studies showed that the CAM was capable of eliciting an inflammatory response similar to that of mammalian epithelial cells (50).
The present study was undertaken to identify molecular processes on the pathogen's side in conjunction with the host's response that could explain differences in host adaptation between chlamydial species. The chicken embryo model provided the possibility to compare the virulence properties of C. psittaci and C. abortus, i.e., their capability to disseminate and colonize, in the face of the chicken embryo's immune defense.
MATERIALS AND METHODS
Chicken eggs.
Fertilized White Leghorn chicken eggs purchased from Geflügel GmbH Bornitz (Bornitz, Germany) were incubated at 37.8°C and a relative humidity of 60%, with rotation. The first day of incubation was defined as embryonic developmental day 1 (ED 1). Between ED 10 and ED 18, the vitality of the chicken embryos was determined by candling once daily.
Bacterial strains.
C. psittaci strain DC15, isolated from an aborted calf fetus in Germany in 2002 (21), and C. abortus type strain S26/3, isolated from a case of ovine abortion (27), were used in this study. Buffalo green monkey (BGM) cells were infected and cultured at 37°C and 5% CO2 in Eagle's minimal essential medium (Cambrex Biosciences, Verviers, Belgium) supplemented with 5% fetal bovine serum (Sigma-Aldrich, Munich, Germany) and 2 mM glutamine (Sigma-Aldrich). After 48 h, chlamydiae were harvested using a cell scraper, aliquoted, and stored at −80°C. After 6 weeks of storage, the chlamydial titer was determined by reinfection of BGM cell culture and by counting of inclusion-forming units (IFU) according to standard methodology (21).
Inoculation of embryonated eggs.
Inoculation was conducted using the false air sac method (22) at ED 10. For infection, the air sac of the egg was moved from the side to the top, and the bacterial suspension (5 × 104 IFU/egg) or BGM cell preparation (mock infection) was transferred onto the chorioallantoic membrane (CAM). The total number of eggs used was 120 for C. psittaci experiments, 104 for C. abortus experiments, and 96 for mock infections.
To determine the survival rates of chicken embryos after Chlamydia infection, eggs were inoculated with different doses of the chlamydial strains (from 5 × 106 IFU to 5 × 103 IFU; 20 eggs per infection dose). The vitality of the chicken embryos was controlled by candling once daily up to 8 days postinfection (dpi), and the survival data were presented as Kaplan-Meier curves (using Graph Pad Prism version 5.0).
Reference experiments (cell culture control).
BGM cells in Trac bottles with glass coverslips (Bibby Sterilin Ltd., Staffordshire, United Kingdom) containing 1 ml medium were inoculated with 5 × 103 IFU C. psittaci DC15 and C. abortus S26/3 by centrifugation at 3,400 × g and 37°C for 1 h, followed by a 1-h incubation at 37°C and 5% CO2. The supernatant was removed, and 1 ml of medium as described above was added. Incubation was maintained for 12, 24, 36, 48, 60, and 72 hpi, followed by harvesting using sonication (Branson 450D Sonifier; Branson, Danbury, CT). Each batch (bottle) of a total of six series was done in triplicate, i.e., two for RNA and one for DNA extraction. DNA and RNA extracts of each sample were subjected to real-time PCR (for determination of genome copies) and quantitative reverse transcription-PCR (for determination of mRNA expression rates).
Quantification of chlamydial organisms.
To quantify chlamydial genome copies in the course of infection, samples from CAM, liver, spleen, bursa of Fabricius, stomach, intestine, heart, and brain were collected from six animals per group at 1, 2, 3, 4, 6, and 8 dpi and stored at −20°C. Total DNA was extracted from whole organs (but no more than 100 mg tissue) using the High Pure PCR template preparation kit (Roche Diagnostics, Mannheim, Germany), eluted in 200 μl Tris buffer (10 mM, pH 7.2), and stored at −20°C. Quantitative real-time PCR targeting the 23S rRNA gene of Chlamydiaceae was conducted as described by Ehricht et al. (16), with an internal amplification control included (34). The chlamydial load of a sample was determined by the MxPro4 software of the thermocycler (Mx3000P; Agilent, Böblingen, Germany). Each run on a microtiter plate included a copy standard, i.e., a series of duplicate 10-fold dilutions of Chlamydia spp. DNA containing 0.27 × 105 to 2.7 × 105 target copies per μl, from which the instrument calculated a calibration curve. The final load was calculated in genome copies per 1 mg tissue.
Recovery of C. psittaci and C. abortus.
To check the viability of the Chlamydia strains in the course of infection, CAM and liver samples were collected at 4 and 8 dpi (four animals per day and strain), homogenized, and cultured on BGM cells. After 44 h of cultivation, cells were fixed with methanol and stained with an antilipopolysaccharide (anti-LPS) Chlamydiaceae-specific fluorescein isothiocyanate-labeled antibody (IMAGEN Chlamydia kit; Oxoid, Wesel, Germany). Finally, the sections were mounted with glycerol-1,4-diazabicyclo-(2,2,2)-octane (DAPCO; Sigma). Inclusion-containing cells were visualized using a TCS-SP2 confocal microscope (Leica, Bensheim, Germany). Fluorescence images sequentially registered in channels at 488 nm (Chlamydia displayed in green) and 543 nm (BGM cells displayed in red) were merged.
Immunohistochemistry.
To examine Chlamydia dissemination and immune cell influx into different organs of the chicken embryo, frozen sections of the organs (CAM, liver, spleen, bursa of Fabricius, intestine, and heart) from four animals per time point (1, 2, 3, 4, 6, and 8 dpi) and group were prepared and stained as described previously (4).
Cryostat sections of 7-μm thickness were fixed with acetone and incubated with the appropriate unlabeled primary monoclonal antibodies against γδ T cells (TCR1), αβ T cells (TCR2) (both from Southern Biotechnology Associates, Eching, Germany), or monocytes/macrophages (CVI 68.1; Institute for Animal Science and Health, Lelystad, The Netherlands).
Bacteria were stained using monoclonal anti-Chlamydia antibody BDI 168 (Santa Cruz Biotechnology, Heidelberg, Germany). For visualization, sections were incubated with a goat anti-mouse immunoglobulin labeled with horseradish peroxidase (Dako, Hamburg, Germany). The enzyme-linked antibody was detected by its reaction with 3,3′-diaminobenzidine (DAB; Dako). Sections were counterstained with hematoxylin (Carl Roth, Karlsruhe, Germany) and mounted with Canada balsam (Riedel de Haen AG, Seelze-Hannover, Germany). As a negative control, the primary antibodies were replaced with phosphate-buffered saline (PBS).
Image analysis.
The quantification of immune cells in the organs of each embryo was conducted by means of the image analysis system CELL (Olympus, Münster, Germany).
Numbers of immunohistochemically stained αβ T cells, γδ T cells, and macrophages were determined in organs of the chicken embryo. Immune cells were counted interactively by drawing five regions of interest in CAM, liver, bursa of Fabricius, intestine, and heart. In spleen, the whole organ was evaluated. In C. abortus-infected animals, only CAM, liver, and spleen were analyzed. The numbers of immune cells were calculated per 1 mm2. Macrophage and T cell numbers were calculated as fold increases in infected organs compared to their mock-infected counterparts.
Transmission electron microscopy.
After C. psittaci infection, the CAM was collected from two representative embryos at 1, 2, 3, 4, 5, 6, 7, and 8 dpi and washed in 0.1 M sodium cacodylate buffer containing 1% sucrose (pH 7.2). Ultrathin sections were prepared from CAM samples fixed with glutaraldehyde (2.5% in sodium cacodylate buffer) and osmium tetroxide (1% in sodium cacodylate buffer). The samples were dehydrated in rising ethanol concentrations and infiltrated with araldite resin (Bodo Möller Chemie, Offenbach, Germany). The curing of the resin was performed for 48 h at 60°C. The embedded samples were ultrathin sectioned with a LKB 8800A Ultratome III (LKB Produkter AB, Bromma, Sweden), picked onto Formvar-coated grids (Planto, Wetzlar, Germany), and contrasted with lead citrate for viewing in an EM 902A electron microscope (Zeiss, Oberkochen, Germany) using a 1 k FastScan charge-coupled-device (CCD) camera (TVIPS, Munich, Germany).
Quantitative real-time reverse transcription (RT)-PCR.
Samples (six per group) collected from CAM at 1, 2, 3, 4, 6, and 8 dpi and from liver and spleen at 3, 4, 6, and 8 dpi were submerged for 24 h in RNAlater (Qiagen, Hilden, Germany) and kept frozen at −20°C. Total RNA was extracted using the RNeasy minikit (Qiagen). Because of the small size of the spleen and generally low RNA amounts recovered from liver, total RNAs from 3 dpi and 4 dpi were pooled to yield six samples (3 and 4 dpi). Residual DNA was digested using the RNase-free DNase set (Qiagen). Extracted RNA was eluted in 50 μl RNase-free water (Qiagen) and stored at −80°C. Quantity and quality of mRNA were checked by spectral analysis (BioPhotometer, Eppendorf, Hamburg, Germany). Samples had to fulfill the purity criteria of an A260/A280 of 1.8 to 2.2 and an A260/A230 of >2 to be included in the study.
Avian-specific primers for IL-12β (p40), IL-18, lipopolysaccharide-induced tumor necrosis factor alpha factor (LITAF), inducible nitric oxide synthase (iNOS), MIP-1β (5), IFN-γ, IL-8, lymphotactin (10), Toll-like receptor 4 (TLR4), and IL-4 (36), as well as primers for the chlamydial genes 16S rRNA, groEL, incA, cpaf, and ftsW (21) of C. psittaci and C. abortus, were developed in previous studies by our group. All primers, including newly developed ones for IL-1β, IL-6, IL-10, IL-15, and IL-17α, as well as for chlamydial genes ompA and rpoN [sigma factor54], are given in Supplement S1 in the supplemental material. All Chlamydia-specific primers were verified by BLAST analysis against the chicken genome to rule out cross-reactivity with host genes. Each primer pair was tested by serial dilution of template RNA to ensure optimized amplification and comparable efficiencies of the RT-PCR.
To determine mRNA expression rates, the QuantiFAST SYBR green real-time one-step RT-PCR kit (Qiagen) was used as described previously (5). The Mx3000P real-time PCR thermocycler (Agilent) was used with the following temperature/time profile: 50°C/10 min and 95°C/5 min, 45 cycles of 95°C/10 s, and the optimized annealing temperature (see Supplement S1 in the supplemental material) for 30 s. To check the amplified products for specificity, subsequent melting curve analysis was conducted as follows: 95°C/60 s, 55°C/30 s, and 95°C/30 s. The threshold method was used for relative quantification of mRNA levels (35). Normalization of target genes was performed using glycerinaldehyde-3-phosphate (GAPDH) as an endogenous standard for chicken genes and chlamydial 16S rRNA for Chlamydia genes. Results were expressed as fold changes (2−ΔΔCT method) (35) from the amount of transcripts detected after mock infection or in the chlamydial inocula prior to transfer onto the CAM.
Statistical analysis.
The normally distributed data of quantitative real-time RT-PCR were statistically evaluated using the Student t test. This test was also used for comparison of two independent samples to evaluate differences between the C. psittaci and C. abortus infection experiments, as well as between infected eggs and controls. Statistical analysis of data from immunohistochemistry and PCR quantification of chlamydial genome copies was conducted using the unpaired Mann-Whitney U test. P values of ≤0.05 were considered significant, and P values of ≤0.01 were regarded as highly significant.
RESULTS
Survival rate of infected embryos.
Daily observation of embryos revealed differences in mortality rates between C. psittaci and C. abortus infection experiments. Inoculation with 7 × 104 IFU of C. psittaci per egg and with larger amounts led to the death of all embryos until 8 dpi. A slight reduction of the dose to 5 × 104 IFU/egg allowed a survival rate of 70%, and 5 × 103 IFU/egg resulted in 95% survival at 8 dpi. In contrast, the infection with C. abortus generally caused a lower mortality rate, with no embryos dying up to 4 dpi at any of the doses applied. Inoculation with 1 × 106 IFU/egg resulted in 60% living embryos at 8 dpi. To ensure comparability, a dose of 5 × 104 IFU/egg was used in all further experiments of this study. More details are given in Fig. 1.
Fig 1.
Kaplan-Meier curves showing the survival of chicken embryos after infection with C. psittaci or C. abortus. On developmental day 10, embryos (n = 20 per group) were inoculated with different amounts of Chlamydia (IFU/egg) and their vitality was determined by daily candling.
Dissemination of chlamydiae in internal organs.
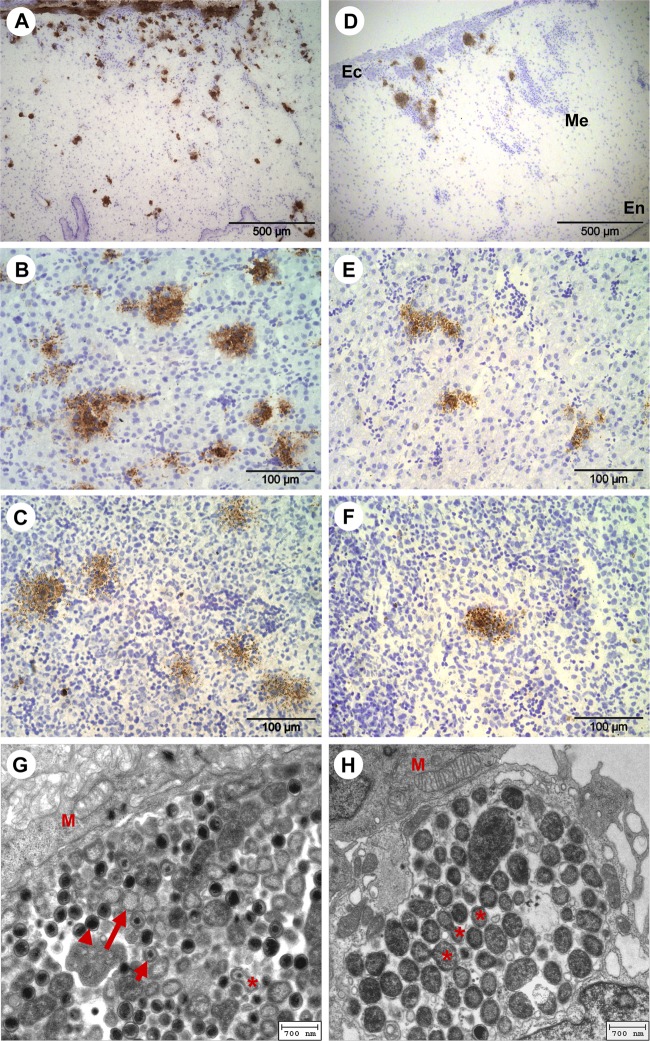
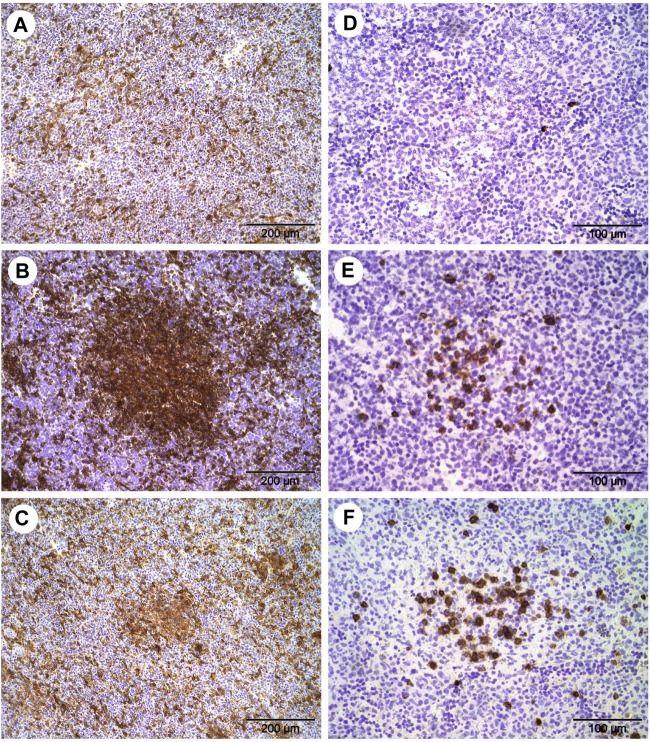
The challenge strains were found to spread in CAM and internal organs in the course of infection. Characteristically, inclusions and foci of infected tissue cells were more abundant in C. psittaci-infected embryos than in samples from the C. abortus experiments (Fig. 2). The first inclusions of C. psittaci were detectable by immunohistochemistry from the first day postinfection in the ectodermic epithelium of the CAM. Larger foci of C. psittaci-infected cells were observed in the mesenchymal part of the CAM from 3 dpi onwards (Fig. 2A), as well as in liver, spleen (Fig. 2B and C), and bursa of Fabricius from 4 dpi onwards. In heart and intestine, C. psittaci was only sporadically detected between 6 dpi and 8 dpi, whereas it was never seen in stomach and brain tissue. C. abortus bacteria were consistently found in lower numbers in the respective embryonic tissues (Fig. 2D to F). In particular, C. abortus counts were significantly lower in CAM (at 4, 6, and 8 dpi), liver (at 4 dpi), and spleen (at 4 and 8 dpi) (data not shown).
Fig 2.
(A to F) Images of immunohistochemical staining of Chlamydia (brown) and cell nuclei (blue) in CAM and embryonic liver and spleen at 8 dpi. (A) C. psittaci in CAM. (B) C. psittaci in liver. (C) C. psittaci in spleen. (D) C. abortus in CAM (Ec, ectoderm; Me, mesenchyme; En, endoderm). (E) C. abortus in liver. (F) C. abortus in spleen. (G and H) Electron microscopic images of intracellular C. psittaci in CAM of the chicken embryos. Panels G and H show 5 dpi and 6 dpi, respectively. Arrowhead, elementary body; long arrow, reticulate body; short arrow, intermediate body; asterisk, cell division of RBs; M, mitochondria.
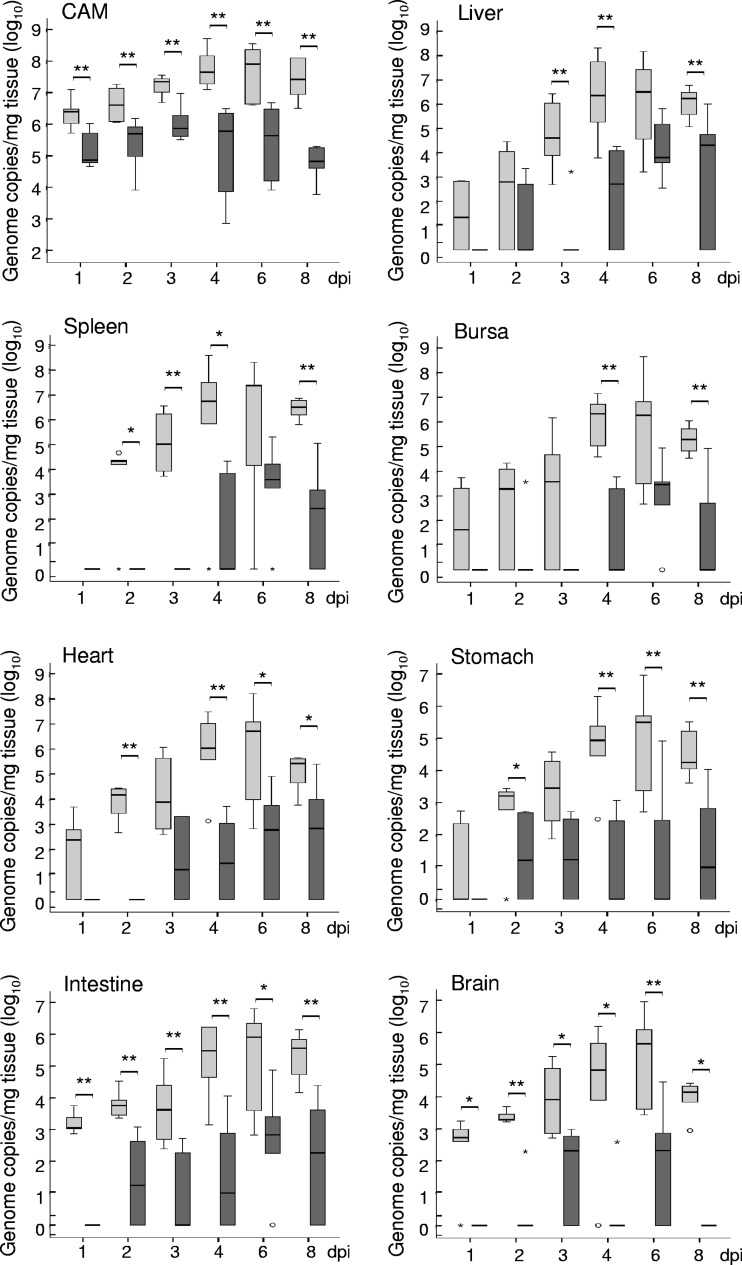
Using quantitative real-time PCR, genomic DNA of the challenge strains was detected in all organs examined, i.e., CAM, liver, spleen, bursa of Fabricius, stomach, intestine, heart, and brain (Fig. 3). Tissue samples from C. psittaci-infected embryos showed generally higher DNA contents than their counterparts from the C. abortus experiment.
Fig 3.
Quantification by real-time PCR of C. psittaci (light gray bars) and C. abortus (dark gray bars) in CAM and internal organs of challenged embryonated chicken eggs. Chlamydiae were quantified by including decimal dilutions of a genome copy standard. The final load is given in genome copies per 1 mg tissue. Medians are significantly different (*, P ≤ 0.05; **, P ≤ 0.01) between C. psittaci- and C. abortus-infected tissues (calculated by Mann-Whitney U test).
Chlamydial DNA was detected in CAM samples from both experimental series at all time points. While the highest rate of 1.1 × 108 C. psittaci genome copies per mg of infected tissue was found at 6 dpi, the maximum load of C. abortus in CAM was 8.0 × 105 copies/mg at 3 dpi.
Internal organs of the embryo harbored lower specific amounts of the agent than CAM. C. psittaci reached its maximum copy numbers at 4 dpi or 6 dpi with a subsequent decrease in all organs (Fig. 3). C. abortus was detected later and in smaller amounts using PCR. Mock-infected embryos always tested negative.
The presence of elementary bodies (EBs), reticulate bodies (RBs), and intermediate bodies in the electron micrographs (Fig. 2G and H) indicates that the bacteria went through complete developmental cycles after inoculation. The largest amounts of C. psittaci inclusions, which were typically localized close to cell nuclei or mitochondria, were found in CAM at 5 dpi and 6 dpi. At 5 dpi, the RBs were still clearly outnumbering EBs. Mature RBs showed evidence of fission at 6 dpi. Subsequently, at 7 and 8 dpi, apoptotic cells and bodies alongside vacuolated cells and multilaminar bodies were emerging in the CAM cells (data not shown).
To demonstrate the challenge strains' viability in the infected tissue, cell cultures of reisolates from CAM and liver were examined using confocal laser scanning microscopy (see Supplement S2 in the supplemental material). C. psittaci was recovered from avian CAM and liver tissue at 4 and 8 dpi, but C. abortus-infected tissue contained smaller amounts of the agent (recovered from CAM at 4 and 8 dpi and from liver only at 8 dpi).
Immune cell influx in CAM and embryonic organs.
In the control embryos, macrophages were observed in liver and intestine from ED 11 onwards and in CAM and heart from ED 13 onwards. High numbers of macrophages were found in spleen (from ED 13 onwards) and bursa of Fabricius compared to the other organs tested (from ED 12 onwards; data not shown). Generally, macrophages were evenly spread throughout the tissue in mock-infected embryos (Fig. 4A).
Fig 4.
Immunohistochemical staining of immune cells in spleen of chicken embryos after infection with C. psittaci and C. abortus at 8 dpi. Immune cells appear in brown, and cell nuclei appear in blue. (A) Staining of macrophages after mock infection. (B) Staining of macrophages in C. psittaci infection. (C) Staining of macrophages in C. abortus infection. (D) Staining of γδ T cells after mock infection. (E) Staining of γδ T cells in C. psittaci infection. (F) Staining of γδ T cells in C. abortus infection.
The data from infected embryonic tissue (Table 1) indicate a significant increase in macrophage numbers in both CAM and internal organs (P ≤ 0.05) compared to mock infection. Macrophage accumulations were found in both C. psittaci- and C. abortus-infected tissue (Fig. 4B and C). C. psittaci-infected embryos had significantly higher macrophage numbers in CAM than their C. abortus-infected counterparts.
Table 1.
Fold increases of the number of macrophages and γδ T cells in different tissues from Chlamydia-infected chicken embryos
| Cells | dpi | Fold increase of immune cells per mm2a |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CAM |
Liver |
Spleen |
Bursa |
Heart |
Intestine |
||||||||
| Cps | Cab | Cps | Cab | Cps | Cab | Cps | Cab | Cps | Cab | Cps | Cab | ||
| Macrophages | 1 | NC | NC | 1.7AC | 1 | ND | ND | ND | ND | NC | ND | 1 | ND |
| 2 | 10AC | NC | 1.6C | 1 | ND | ND | 6.3A | ND | NC | ND | 1 | ND | |
| 3 | 15.2AC | 2.2 | 1.8AC | 1 | 2.2AC | 1.1 | 1 | ND | 1 | ND | 1 | ND | |
| 4 | 188AC | 38B | 3.2AC | 1.4 | 3AC | 1.8B | 1.5A | ND | 6.3A | ND | 1.2 | ND | |
| 6 | 431AC | 156B | 9.6A | 12.7B | 2.5A | 3.2B | 2.5A | ND | 8.8A | ND | 2.1A | ND | |
| 8 | 82.3A | 75B | 33.8AC | 9.5B | 4.9AC | 2.8B | 2A | ND | 13.5A | ND | 5.4A | ND | |
| γδ T cells | 4 | NC | NC | NC | NC | NC | NC | NC | ND | NC | ND | NC | ND |
| 6 | NC | NC | 6 | NC | 6A | 6 | NC | ND | NC | ND | NC | ND | |
| 8 | 6A | 6B | 3.3A | 3.3B | 6.3A | 11.3B | 1 | ND | NC | ND | 1 | ND | |
Four embryos per dpi; 5 regions of interest counted. Letters beside values indicate significant difference (P ≤ 0.05; A, mock versus C. psittaci; B, mock versus C. abortus; C, C. psittaci versus C. abortus) after C. psittaci infection or C. abortus infection as calculated by Mann-Whitney U test. Significances refer to the original values. Cps, C. psittaci; Cab, C. abortus. NC, no cells found. ND, not determined because of small organ size (C. psittaci) or low infection rate (C. abortus).
γδ T cells were detected in liver, spleen (Fig. 4D), bursa of Fabricius, and intestine of mock-infected chicken embryos at ED 18 but never in CAM. Following infection with C. psittaci and C. abortus, γδ T cells were observed in CAM at 8 dpi (P ≤ 0.05) (Table 1). Additionally, γδ T cells were found in significantly higher numbers than in controls in liver at 8 dpi and in spleen at 6 dpi and 8 dpi (Fig. 4E and F). There were no significant differences between the two chlamydial strains (Table 1). In contrast, αβ T cells were not observed in infected tissues or in controls.
mRNA expression of essential chlamydial genes.
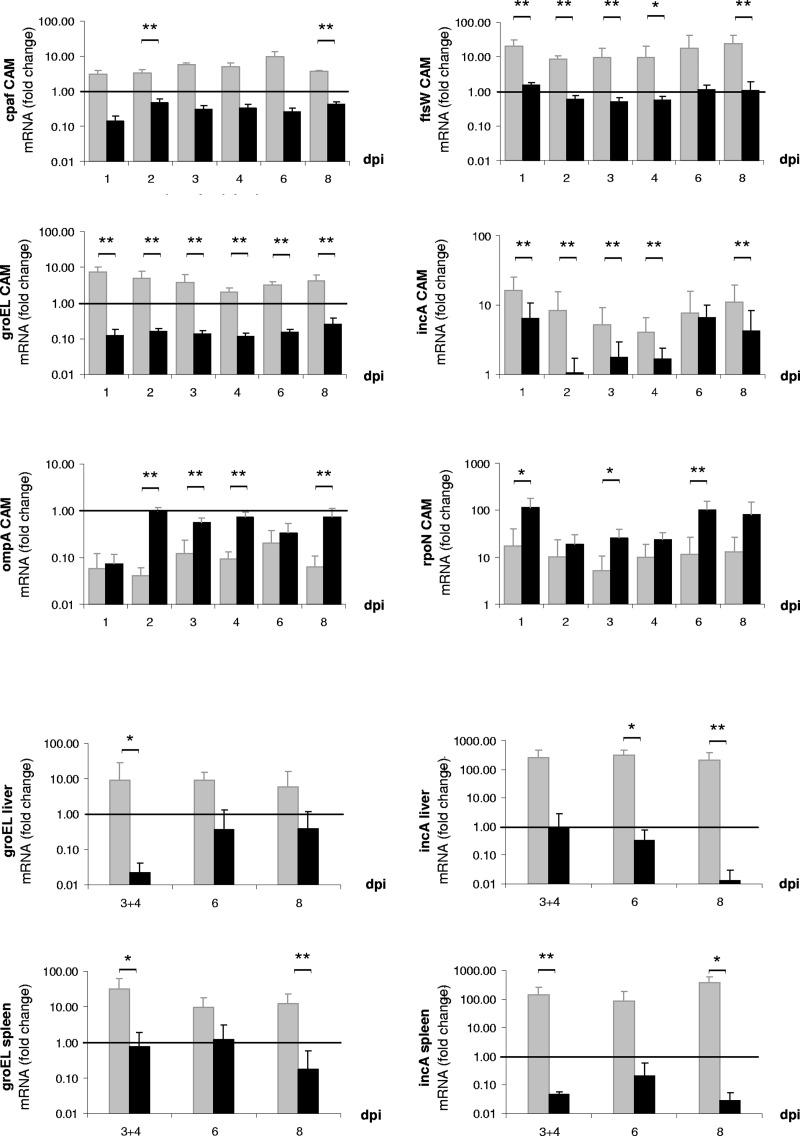
Quantification of transcripts for selected chlamydial genes in embryonic CAM, liver, and spleen revealed generally higher transcription levels in C. psittaci than in C. abortus (Fig. 5). For instance, in CAM, mRNA expression rates of the gene encoding the protease CPAF were increased in C. psittaci infection from 1 dpi onwards, and significant differences were found at 2 dpi and 8 dpi (both P ≤ 0.01). In the case of the chlamydial stress response gene groEL, the effect was even more pronounced, with significant differences between C. psittaci and C. abortus occurring at all time points (P ≤ 0.01). Similar distinctions were observed for the genes encoding chlamydial inclusion membrane protein IncA and cell division factor FtsW in CAM samples. Downregulation of cpaf and groEL in C. abortus contrasted to upregulation in C. psittaci.
Fig 5.
mRNA expression levels of virulence-associated genes of C. psittaci (gray bars) and C. abortus (black bars) in tissue of CAM and embryonic liver and spleen. Data are presented as fold changes compared to chlamydial inocula prior to administration. Means are significantly different (*, P ≤ 0.05; **, P ≤ 0.01) between C. psittaci and C. abortus (calculated by Mann-Whitney U test).
In the case of the chlamydial RNA polymerase factor RpoN, both Chlamydia species responded to infection by upregulation. However, C. abortus showed significantly higher rates of mRNA expression of rpoN than C. psittaci at 1 dpi, 3 dpi, and 6 dpi.
The mRNA expression patterns of groEL and incA in liver and spleen largely resembled the findings from CAM. In these organs, no upregulation of cpaf, ftsW, and rpoN was found for C. psittaci and C. abortus (data not shown). The gene of the chlamydial major outer membrane protein A, ompA, tended toward downregulation in CAM (Fig. 5), liver, and spleen in both C. psittaci- and C. abortus-infected embryos (data for liver and spleen not shown).
Transcription of immune-related genes.
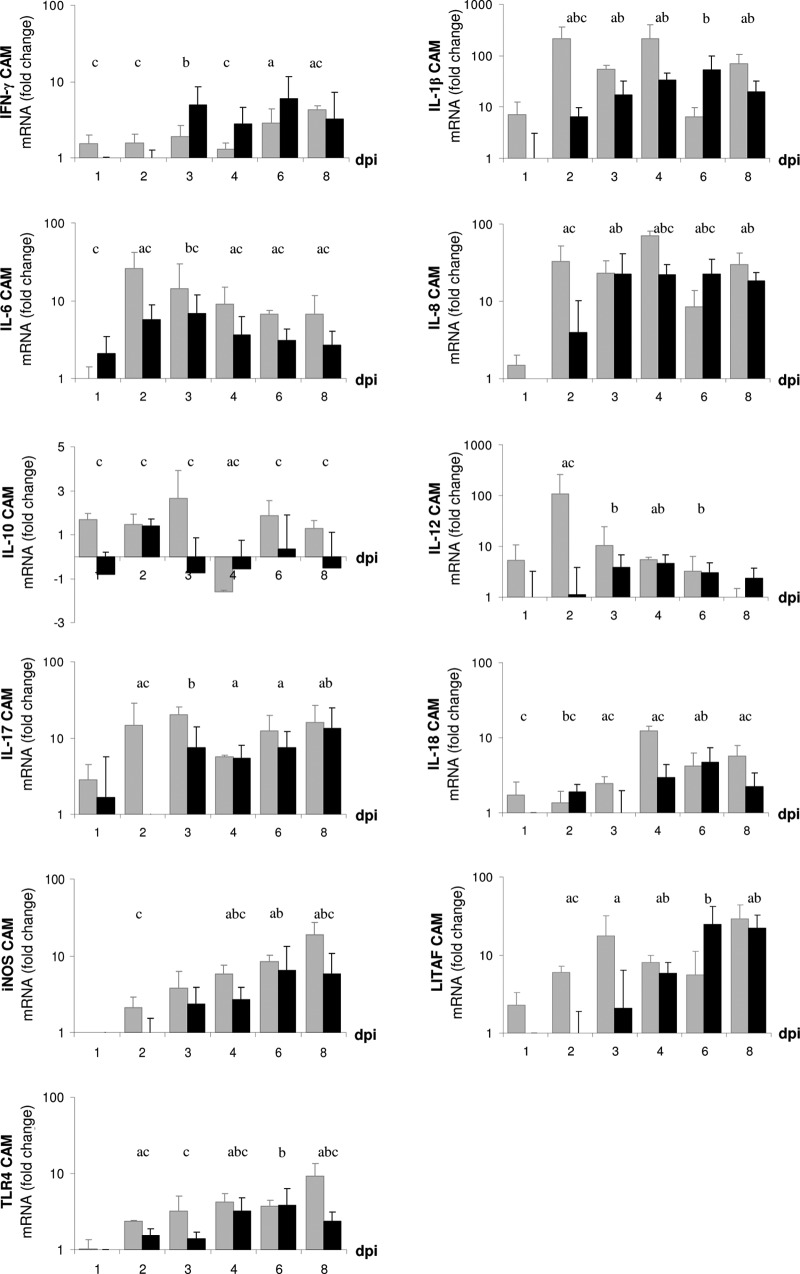
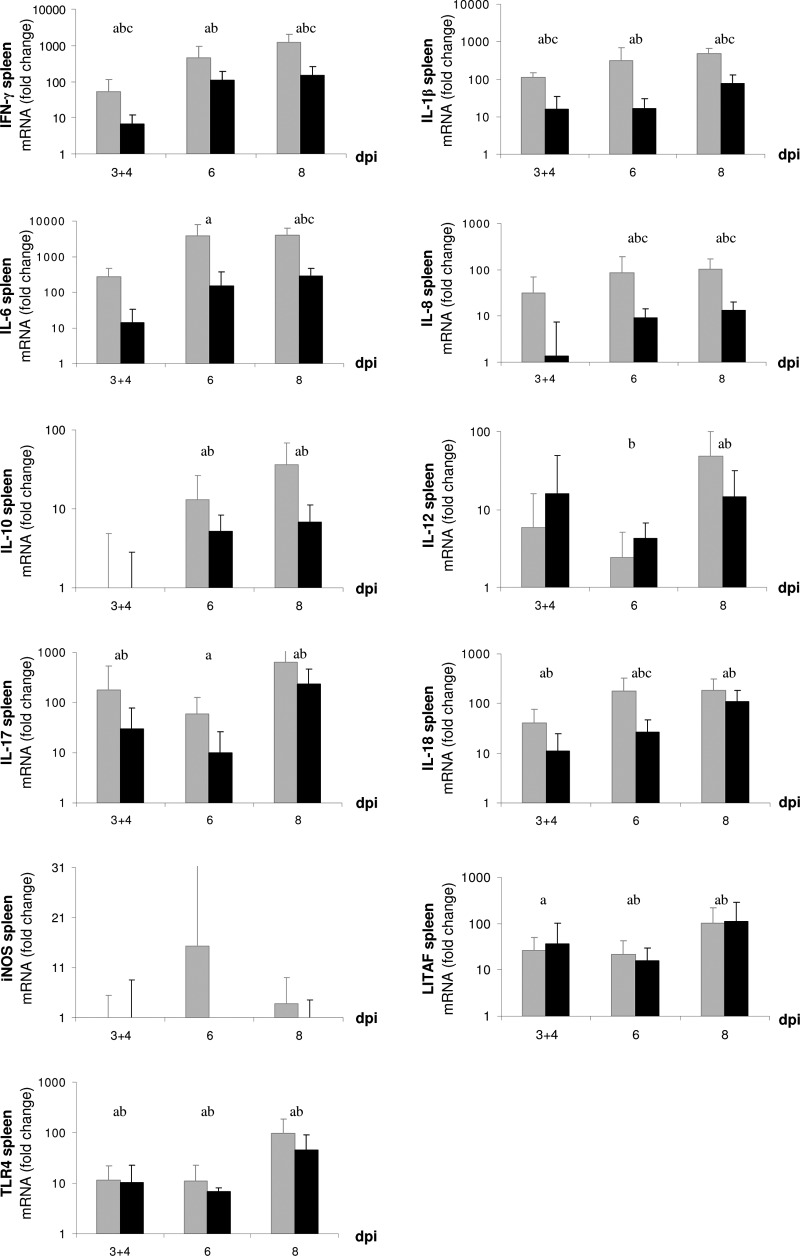
Examination by quantitative real-time RT-PCR of tissue samples from CAM (1 dpi to 8 dpi) and liver and spleen (3 dpi to 8 dpi) showed that mRNA expression of several immune mediators underwent dynamic changes in the course of both infection experiments. Expression of the inflammatory mediators iNOS (with the exception of spleen), IL-1β, IL-6, IL-8, LITAF, and IL-17, as well as IFN-γ, IL-12, IL-18, and TLR4, was significantly upregulated in CAM (Fig. 6), liver (see Supplement S3 in the supplemental material), and spleen (Fig. 7) of C. psittaci- and C. abortus-infected embryos. In general, the response pattern was similar for both strains, but C. psittaci induced a more rapid reaction than C. abortus. The expression of some genes (those for iNOS, IL-6, IL-8, and TLR4) was significantly higher during C. psittaci infection than during C. abortus infection, whereas only marginal and nonsignificant differences both in CAM and organs have been observed with others (those for IFN-γ, IL-12, IL-17, and LITAF) from 3 dpi onwards. Expression rates of IL-4 and IL-10 were only moderately increased in CAM throughout both experiments (data not shown).
Fig 6.
Relative quantification of mRNA expression of immune-related proteins in CAM of embryonated chicken eggs challenged with C. psittaci (gray bars) and C. abortus (black bars) compared to mock infection. Means are significantly different (P ≤ 0.05). a, C. psittaci versus mock; b, C. abortus versus mock; c, C. psittaci versus C. abortus.
Fig 7.
Relative quantification of mRNA expression of immune mediators in embryonic spleen after infection with C. psittaci (gray bars) and C. abortus (black bars) compared to mock infection. Means are significantly different (P ≤ 0.05). a, C. psittaci versus mock; b, C. abortus versus mock; c, C. psittaci versus C. abortus.
Spleen tissue (Fig. 7) showed a pronounced upregulation of most genes, in particular IL-6, IL-17, IL-18, IFN-γ, and TLR4. Significant differences between C. psittaci- and C. abortus-infected embryos were only found for mRNA expression levels of IL-1β (3 and 4 dpi, 8 dpi) and IL-8 (6 dpi, 8 dpi). The transcription rates of IL-6 and IL-18 were also statistically higher in C. psittaci-infected embryos at 8 dpi and 6 dpi, respectively. Furthermore, the C. psittaci infection induced significant upregulation of IFN-γ at 3 and 4 dpi and at 8 dpi, both in Chlamydia versus the control and C. psittaci versus C. abortus. IL-4 and IL-10 expression in spleen did not differ between C. psittaci and C. abortus infection. In both infection experiments, spleen was the only organ where no upregulation of iNOS was found.
In both experimental series, the data on regulation of immune-associated genes in liver closely resembled those from spleen, with the exception of iNOS, which was upregulated (see Supplement S3 in the supplemental material).
The mRNA expression of lymphotactin and MIP-1β was never significantly increased in CAM, liver, or spleen (data not shown). No mRNA of IL-15 was found.
Reference experiments (cell culture control).
To obtain a baseline for transcription of the selected genes in standard cell culture, we recorded quantitative growth curves for both strains in BGM cell culture and determined mRNA expression rates of the same gene panel in the course of in vitro infection. The main results were as follows. Although C. psittaci proliferated slightly better, the growth curves of the two agents were closer to each other than in the chicken embryo experiments (see Supplement S4 in the supplemental material). No significant differences in mRNA expression rates of the genes ftsW, incA, and rpoN were observed, while transcription of groEL, cpaf, and ompA differed between C. psittaci and C. abortus at individual time points (see Supplement S5 in the supplemental material). However, the behaviors of all six genes were clearly different from those in the in ovo experiments.
DISCUSSION
Dissemination of the pathogens.
To characterize the pathogens' dissemination, three different methods were used: immunohistochemistry, quantitative real-time PCR, and transmission electron microscopy. While all methods succeeded in detecting the infectious agents, PCR proved most sensitive as expected, and immunohistochemistry identified only samples containing more than 1,000 copies/mg tissue. Using transmission electron microscopy, we were able to demonstrate different intracellular proliferation stages of C. psittaci in CAM of the chicken embryo that resembled the typical morphologies evolving during the chlamydial developmental cycle (Fig. 2G and H).
For host infection, Chlamydia spp. are capable of entering a wide range of different cell types, e.g., endothelial and epithelial cells and macrophages (19, 37). The fact that chlamydial organisms were localized in all three layers of CAM and internal organs indicates a systemic dissemination of the bacteria in the present chicken embryo model. Moazed and colleagues (30) described a systemic distribution of C. pneumoniae via alveolar and peritoneal macrophages, as well as blood cells, in a murine infection model. While the dissemination mechanisms are not completely clear, it is conceivable that, in the present experiments, the chlamydiae have been transported by blood monocytes or lymphocytes from CAM to the embryonic organs.
Regulation of essential chlamydial genes.
As a mechanism of survival and evasion of elimination, chlamydial virulence-associated factors integrated in the inclusion membrane (39) or secreted into host cell cytoplasm (51) can modulate the host immune response. In the present study, we have examined six well-investigated essential chlamydial genes in order to characterize the pathogen's regulation in the course of infection. Notably, C. psittaci showed generally higher mRNA expression rates of the incA, ftsW, groEL, and cpaf genes than C. abortus in the infected chicken embryos.
The IncA protein, which localizes at the cytoplasmatic side of the inclusion membrane (39), interacts with cytosolic host proteins and forms long fibers used as cytosolic tracks extending from the inclusion (46). Its expression by C. abortus has not been shown in vivo so far, but a moderate mRNA expression in C. psittaci was shown in vitro by Goellner et al. (21). As we observed incA upregulation of C. psittaci in CAM and organs, it is conceivable that elevated expression levels of this protein would contribute to a stabilization of the inclusion where the bacteria are residing. Accordingly, the low numbers of C. abortus bacteria in the embryonic inner organs coincide with downregulation in liver and spleen, whereas in CAM its expression was upregulated but lower in C. abortus than in C. psittaci.
The state of regulation of the FtsW protein as part of the complex mediating cell division in C. trachomatis (20) and C. pneumoniae (9) can be an indicator of the pathogen's capability to replicate in the host. Interestingly, specific mRNA of ftsW was detected in C. psittaci-infected CAM at a significantly higher level than in its C. abortus counterpart at all time points (Fig. 5), which could explain the better proliferation of C. psittaci throughout the infection (Fig. 3). A study in HD11 avian monocytes/macrophages showed that cell division genes of C. psittaci, among them ftsW, were weakly expressed while chlamydial proliferation was also reduced (2). Other studies had demonstrated downregulation of the gene following exposure to IFN-γ (9, 21). In contrast, the significant upregulation of ftsW in the present model could suggest that, despite the onset of the embryonic immune response, the conditions were not as adverse to the pathogen as in the in vitro models.
The chlamydial heat shock protein GroEL (Hsp60) serves as a chaperone during intracellular development (56) and seems to be involved in the induction of immune response during human chlamydial infection, where it colocalizes with infiltrating macrophages (24). Correspondingly, groEL gene expression was significantly upregulated in CAM, liver, and spleen (Fig. 5) and coincided with macrophage influx in the course of infection with the potentially more virulent C. psittaci but not with the C. abortus strain.
The chlamydial protease-like activity factor (CPAF) was associated with inhibition of apoptosis and intracellular trafficking, as well as suppression of major histocompatibility complex class I (MHC-I)- and MHC-II-mediated antigen expression in Chlamydia-infected cells (55), but more functions have been attributed to it (14, 44). While current knowledge is mostly based on HeLa cell culture models using C. trachomatis (11, 40), data for C. psittaci and C. abortus are scarce. In the present in ovo study, a significantly increased mRNA expression level of cpaf was found in CAM during C. psittaci infection but not in C. abortus infection.
In the cell culture controls, mRNA expression patterns of all six genes were clearly different from those of the in ovo experiments, thus indicating that the observed transcription patterns in ovo were indeed reflective of in vivo responses, probably to host defense, and not intrinsic to the individual strain.
Immunology.
The capability of different Chlamydia strains to induce cytokine and chemokine expression in vitro was reviewed by Rusconi and Greub (45). Although mouse models were suggested to elucidate the role of immune mediators in vivo (43), the immunological processes during infection with C. psittaci or C. abortus are not completely understood yet. In this context, our investigations have demonstrated for the first time that in ovo infection with Chlamydia induced a cellular influx, as well as cytokine and chemokine gene expression in the chicken embryo. Similar to the Chlamydia species' capability to induce an immune defense reaction in chicken embryos, a recent study on Aspergillus fumigatus also revealed elevated transcription of cytokines (22).
The present data allow the following conclusions: (i) the embryonic immune responses were characterized by an early and acute inflammatory reaction; (ii) both C. psittaci and C. abortus induced similar mRNA expression profiles for a range of immune-related genes in the organs of chicken embryos, which were significantly different to mock infection; (iii) the immune reaction in CAM, liver, and spleen emerged faster to C. psittaci than to C. abortus, and the latter correlated well with the distinct invasiveness and virulence of the two bacterial strains; and (iv) the general response patterns of immunologically important mediators were largely the same in both experiments. Besides, some immune genes had transcription levels that were significantly higher in C. psittaci-infected embryos than in C. abortus-infected embryos and vice versa. Interestingly, C. abortus proved capable of eliciting higher or similar transcription levels of a subset of cytokines (IL-8, LITAF, IL-1β, and IFN-γ in CAM and spleen) at some time points, although its load in those tissues was lower than that of C. psittaci (Fig. 6 and 7). We have no exhaustive explanation for the lack of correlation between bacterial load and inflammatory response intensity in the respective organs. On the one hand, if C. abortus were more proinflammatory, it should express particularly immunogenic antigens that could trigger a stronger response. However, there are no data from genome or proteome analysis to support this. On the other hand, C. psittaci is probably more efficient in actively suppressing the host immune response, which enables better dissemination in the host. For instance, upregulation of cpaf enabled chlamydiae to influence the host reaction in their favor (see below) (14, 55). Our general observation of higher rates of mRNA expression per bacterial cell in C. psittaci may also be supportive of the latter hypothesis, as are the present data on embryo survival (Fig. 1).
We showed for the first time an influx of macrophages and γδ T cells in chicken embryos during Chlamydia infection. Studies from other groups revealed activation of macrophages in vitro (1) and their ability to systemically transport chlamydiae in the body (30). We assume the same mechanisms having occurred in the present in ovo study, although further investigations are necessary. Chicken γδ T cells were believed to emigrate into spleen and intestine in three waves during embryonic development, starting at ED 15 (15). We have not found γδ T cells in the uninfected controls before ED 18, but our results demonstrate that the Chlamydia infection resulted in an earlier and more pronounced emergence of γδ T cells in embryonic organs.
The recognition of Chlamydia by TLR4 has often been described (7, 31), as has activation of macrophages by TLR4 via LPS-binding (48) and subsequent production of bactericidal nitric oxide (NO) (26). Production of NO by iNOS is important for suppression of intracellular pathogens (54). We found an early upregulation of the iNOS transcription after chlamydial infection of the embryos. The mRNA expression of iNOS was shown to be continuously rising in the course of infection with both C. psittaci and C. abortus (Fig. 6), which concurred with the increase of TLR4 expression rates. The increase of TLR4 and iNOS mRNA expression may indicate activation of macrophages following Chlamydia infection, although we cannot rule out that elevated macrophage numbers contributed to the present findings.
Both in ovo infections resulted in an inflammatory response characterized by the rapid expression of IL-1β, IL-8, and LITAF seen in CAM, liver, and spleen of the infected embryos. Transcription of IL-17, as a further regulator of inflammation, was induced in CAM and spleen. IL-1β can stimulate T cells to produce IL-17 (47). Similar to other T cells, the γδ T cells have been described as a source of IL-17 (38). In the chicken embryos, the occurrence of γδ T cells correlated with the late transcription of IL-17 in liver. In contrast, in CAM and spleen, the expression of IL-17 was detected before γδ T cells entered the scene. It might be possible that other embryonic cell types are also able to produce IL-17 in response to the infection.
IFN-γ plays an important role in the defense against C. trachomatis and C. pneumoniae in infected mice (43). Here we found a general upregulation of IFN-γ expression in both C. psittaci and C. abortus experiments, but the higher expression rate during C. psittaci infection in spleen and liver contrasted with the situation in CAM, where C. abortus infection caused higher upregulation, albeit at a considerably lower level (Fig. 6). This finding could indicate the embryo's attempt to control the infection at an early stage in CAM tissue, since IFN-γ as an essential mediator of the innate immune response is known to retard chlamydial growth and dissemination (21, 32).
The fact that we measured active IL-12 and IL-18 mRNA expression from 2 dpi onwards and subsequently increased transcription of IFN-γ might be taken as evidence for a coordinated production of IL-12, IL-18, and IFN-γ in chicken embryos. The elevated macrophage counts could suggest that IL-12 and IL-18 were produced by chicken embryonic macrophages in response to Chlamydia infection, thus inducing IFN-γ expression. Nevertheless, the cell type that secretes IFN-γ in Chlamydia-infected chicken embryos has yet to be determined. Notably, we never found any αβ T cells in the avian embryo. Even though our group recently described the ability of chicken γδ T cells to express IFN-γ mRNA (36), the final proof that γδ T cells identified in the present study actually account for the IFN-γ expression remains to be delivered.
In conclusion, the present study has revealed different virulence properties of the two zoonotic agents C. psittaci and C. abortus in chicken embryos. While the invasion capability of the two Chlamydia species differed, the elicited immunological response was largely similar but tended to be more rapid and intense in C. psittaci infection. The relatively higher virulence of C. psittaci correlated well with a higher transcription level for a range of different chlamydial proteins thought to be responsible for better survival, dissemination, and propagation. The results indicate a relevance of these essential factors for the differences in pathogenicity to avian hosts among the various chlamydial agents. The present in vivo data provide a basis for a better understanding of host-pathogen interactions in Chlamydia-infected organisms. The fact that C. psittaci has coped far better than C. abortus with the onset of the avian embryo's immune response by upregulating selected genes may be a key to understanding the mechanisms underlying host adaptation and etiopathology.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to David Longbottom (Moredun Institute, Scotland) for providing C. abortus strain S26/3. We thank Katrin Schlehahn and Simone Bettermann for excellent technical assistance and Roland Diller for help with the statistical analysis.
This study was financially supported by the Federal Ministry of Higher Education and Research (BMBF) of Germany under grant 0314108, “PET/CT for imaging of infection and inflammation: chlamydial infections.” The funding sources had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Published ahead of print 11 June 2012
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1. Bas S, et al. 2008. The proinflammatory cytokine response to Chlamydia trachomatis elementary bodies in human macrophages is partly mediated by a lipoprotein, the macrophage infectivity potentiator, through TLR2/TLR1/TLR6 and CD14. J. Immunol. 180:1158–1168 [DOI] [PubMed] [Google Scholar]
- 2. Beeckman DS, Vanrompay DC. 2010. Biology and intracellular pathogenesis of high or low virulent Chlamydophila psittaci strains in chicken macrophages. Vet. Microbiol. 141:342–353 [DOI] [PubMed] [Google Scholar]
- 3. Beeckman DS, Vanrompay DC. 2009. Zoonotic Chlamydophila psittaci infections from a clinical perspective. Clin. Microbiol. Infect. 15:11–17 [DOI] [PubMed] [Google Scholar]
- 4. Berndt A, Methner U. 2001. Gamma/delta T cell response of chickens after oral administration of attenuated and non-attenuated Salmonella typhimurium strains. Vet. Immunol. Immunopathol. 78:143–161 [DOI] [PubMed] [Google Scholar]
- 5. Berndt A, et al. 2007. Chicken cecum immune response to Salmonella enterica serovars of different levels of invasiveness. Infect. Immun. 75:5993–6007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Betts HJ, Wolf K, Fields KA. 2009. Effector protein modulation of host cells: examples in the Chlamydia spp. arsenal. Curr. Opin. Microbiol. 12:81–87 [DOI] [PubMed] [Google Scholar]
- 7. Bulut Y, et al. 2009. Chlamydial heat shock protein 60 induces acute pulmonary inflammation in mice via the Toll-like receptor 4- and MyD88-dependent pathway. Infect. Immun. 77:2683–2690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Burnet FM, Rountree PM. 1935. Psittacosis in the developing egg. J. Pathol. 40:471–481 [Google Scholar]
- 9. Byrne GI, et al. 2001. Chlamydia pneumoniae expresses genes required for DNA replication but not cytokinesis during persistent infection of HEp-2 cells. Infect. Immun. 69:5423–5429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Carvajal BG, Methner U, Pieper J, Berndt A. 2008. Effects of Salmonella enterica serovar Enteritidis on cellular recruitment and cytokine gene expression in caecum of vaccinated chickens. Vaccine 26:5423–5433 [DOI] [PubMed] [Google Scholar]
- 11. Chen D, et al. 2010. Autoprocessing and self-activation of the secreted protease CPAF in Chlamydia-infected cells. Microb. Pathog. 49:164–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Coers J, Starnbach MN, Howard JC. 2009. Modeling infectious disease in mice: co-adaptation and the role of host-specific IFNgamma responses. PLoS Pathog. 5:e1000333 doi:10.1371/journal.ppat.1000333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Donachie WD. 1993. The cell cycle of Escherichia coli. Annu. Rev. Microbiol. 47:199–230 [DOI] [PubMed] [Google Scholar]
- 14. Dong F, Su H, Huang Y, Zhong Y, Zhong G. 2004. Cleavage of host keratin 8 by a Chlamydia-secreted protease. Infect. Immun. 72:3863–3868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dunon D, Cooper MD, Imhof BA. 1993. Migration patterns of thymus-derived gamma delta T cells during chicken development. Eur. J. Immunol. 23:2545–2550 [DOI] [PubMed] [Google Scholar]
- 16. Ehricht R, Slickers P, Goellner S, Hotzel H, Sachse K. 2006. Optimized DNA microarray assay allows detection and genotyping of single PCR-amplifiable target copies. Mol. Cell Probes 20:60–63 [DOI] [PubMed] [Google Scholar]
- 17. Eissenberg LG, Wyrick PB, Davis CH, Rumpp JW. 1983. Chlamydia psittaci elementary body envelopes: ingestion and inhibition of phagolysosome fusion. Infect. Immun. 40:741–751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fields KA, Hackstadt T. 2002. The chlamydial inclusion: escape from the endocytic pathway. Annu. Rev. Cell Dev. Biol. 18:221–245 [DOI] [PubMed] [Google Scholar]
- 19. Gaydos CA, Summersgill JT, Sahney NN, Ramirez JA, Quinn TC. 1996. Replication of Chlamydia pneumoniae in vitro in human macrophages, endothelial cells, and aortic artery smooth muscle cells. Infect. Immun. 64:1614–1620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gérard HC, et al. 2001. Expression of Chlamydia trachomatis genes encoding products required for DNA synthesis and cell division during active versus persistent infection. Mol. Microbiol. 41:731–741 [DOI] [PubMed] [Google Scholar]
- 21. Goellner S, et al. 2006. Transcriptional response patterns of Chlamydophila psittaci in different in vitro models of persistent infection. Infect. Immun. 74:4801–4808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jacobsen ID, et al. 2010. Embryonated eggs as an alternative infection model to investigate Aspergillus fumigatus virulence. Infect. Immun. 78:2995–3006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kaleta EF, Taday EM. 2003. Avian host range of Chlamydophila spp. based on isolation, antigen detection and serology. Avian Pathol. 32:435–461 [DOI] [PubMed] [Google Scholar]
- 24. Kol A, Bourcier T, Lichtman AH, Libby P. 1999. Chlamydial and human heat shock protein 60s activate human vascular endothelium, smooth muscle cells, and macrophages. J. Clin. Invest. 103:571–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Longbottom D, Coulter LJ. 2003. Animal chlamydioses and zoonotic implications. J. Comp. Pathol. 128:217–244 [DOI] [PubMed] [Google Scholar]
- 26. MacMicking J, Xie QW, Nathan C. 1997. Nitric oxide and macrophage function. Annu. Rev. Immunol. 15:323–350 [DOI] [PubMed] [Google Scholar]
- 27. McClenaghan M, Herring AJ, Aitken ID. 1984. Comparison of Chlamydia psittaci isolates by DNA restriction endonuclease analysis. Infect. Immun. 45:384–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Miyairi I, Ramsey KH, Patton DL. 2010. Duration of untreated chlamydial genital infection and factors associated with clearance: review of animal studies. J. Infect. Dis. 201(Suppl 2):S96–S103 [DOI] [PubMed] [Google Scholar]
- 29. Miyairi I, et al. 2007. The p47 GTPases Iigp2 and Irgb10 regulate innate immunity and inflammation to murine Chlamydia psittaci infection. J. Immunol. 179:1814–1824 [DOI] [PubMed] [Google Scholar]
- 30. Moazed TC, Kuo CC, Grayston JT, Campbell LA. 1998. Evidence of systemic dissemination of Chlamydia pneumoniae via macrophages in the mouse. J. Infect. Dis. 177:1322–1325 [DOI] [PubMed] [Google Scholar]
- 31. Ohashi K, Burkart V, Flohe S, Kolb H. 2000. Cutting edge: heat shock protein 60 is a putative endogenous ligand of the Toll-like receptor-4 complex. J. Immunol. 164:558–561 [DOI] [PubMed] [Google Scholar]
- 32. Ouellette SP, et al. 2006. Global transcriptional upregulation in the absence of increased translation in Chlamydia during IFNgamma-mediated host cell tryptophan starvation. Mol. Microbiol. 62:1387–1401 [DOI] [PubMed] [Google Scholar]
- 33. Pantchev A, Sting R, Bauerfeind R, Tyczka J, Sachse K. 2010. Detection of all Chlamydophila and Chlamydia spp. of veterinary interest using species-specific real-time PCR assays. Comp. Immunol. Microbiol. Infect. Dis. 33:473–484 [DOI] [PubMed] [Google Scholar]
- 34. Pantchev A, Sting R, Bauerfeind R, Tyczka J, Sachse K. 2009. New real-time PCR tests for species-specific detection of Chlamydophila psittaci and Chlamydophila abortus from tissue samples. Vet. J. 181:145–150 [DOI] [PubMed] [Google Scholar]
- 35. Pfaffl MW. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pieper J, Methner U, Berndt A. 2008. Heterogeneity of avian gammadelta T cells. Vet. Immunol. Immunopathol. 124:241–252 [DOI] [PubMed] [Google Scholar]
- 37. Rasmussen SJ, et al. 1997. Secretion of proinflammatory cytokines by epithelial cells in response to Chlamydia infection suggests a central role for epithelial cells in chlamydial pathogenesis. J. Clin. Invest. 99:77–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Roark CL, Simonian PL, Fontenot AP, Born WK, O'Brien RL. 2008. Gammadelta T cells: an important source of IL-17. Curr. Opin. Immunol. 20:353–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rockey DD, Scidmore MA, Bannantine JP, Brown WJ. 2002. Proteins in the chlamydial inclusion membrane. Microbes Infect. 4:333–340 [DOI] [PubMed] [Google Scholar]
- 40. Rödel J, et al. 2012. Persistent Chlamydia trachomatis infection of HeLa cells mediates apoptosis resistance through a Chlamydia protease-like activity factor-independent mechanism and induces high mobility group box 1 release. Infect. Immun. 80:195–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rodolakis A, Yousef Mohamad K. 2010. Zoonotic potential of Chlamydophila. Vet. Microbiol. 140:382–391 [DOI] [PubMed] [Google Scholar]
- 42. Rohde G, Straube E, Essig A, Reinhold P, Sachse K. 2010. Chlamydial zoonoses. Dtsch. Arztebl. Int. 107:174–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Rottenberg ME, Gigliotti-Rothfuchs A, Wigzell H. 2002. The role of IFN-gamma in the outcome of chlamydial infection. Curr. Opin. Immunol. 14:444–451 [DOI] [PubMed] [Google Scholar]
- 44. Rupp J, et al. 2007. Chlamydia pneumoniae directly interferes with HIF-1alpha stabilization in human host cells. Cell Microbiol. 9:2181–2191 [DOI] [PubMed] [Google Scholar]
- 45. Rusconi B, Greub G. 2011. Chlamydiales and the innate immune response: friend or foe? FEMS Immunol. Med. Microbiol. 61:231–244 [DOI] [PubMed] [Google Scholar]
- 46. Suchland RJ, Rockey DD, Weeks SK, Alzhanov DT, Stamm WE. 2005. Development of secondary inclusions in cells infected by Chlamydia trachomatis. Infect. Immun. 73:3954–3962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sutton C, Brereton C, Keogh B, Mills KH, Lavelle EC. 2006. A crucial role for interleukin (IL)-1 in the induction of IL-17-producing T cells that mediate autoimmune encephalomyelitis. J. Exp. Med. 203:1685–1691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Takeda K. 2005. Toll-like receptor. Nihon Rinsho Meneki Gakkai Kaishi 28:309–317 (In Japanese.) [DOI] [PubMed] [Google Scholar]
- 49. Taylor GA, Feng CG, Sher A. 2007. Control of IFN-gamma-mediated host resistance to intracellular pathogens by immunity-related GTPases (p47 GTPases). Microbes Infect. 9:1644–1651 [DOI] [PubMed] [Google Scholar]
- 50. Valdes TI, Kreutzer D, Moussy F. 2002. The chick chorioallantoic membrane as a novel in vivo model for the testing of biomaterials. J. Biomed. Mater. Res. 62:273–282 [DOI] [PubMed] [Google Scholar]
- 51. Valdivia RH. 2008. Chlamydia effector proteins and new insights into chlamydial cellular microbiology. Curr. Opin. Microbiol. 11:53–59 [DOI] [PubMed] [Google Scholar]
- 52. Vanrompay D, Lyons JM, Morre SA. 2006. Animal models for the study of Chlamydia trachomatis infections in the female genital infection. Drugs Today (Barc.) 42(Suppl A):55–63 [PubMed] [Google Scholar]
- 53. Walder G, et al. 2003. Chlamydophila abortus pelvic inflammatory disease. Emerg. Infect. Dis. 9:1642–1644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Webb JL, Harvey MW, Holden DW, Evans TJ. 2001. Macrophage nitric oxide synthase associates with cortical actin but is not recruited to phagosomes. Infect. Immun. 69:6391–6400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zhong G. 2011. Chlamydia trachomatis secretion of proteases for manipulating host signaling pathways. Front. Microbiol. 2:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zügel U, Kaufmann SH. 1999. Role of heat shock proteins in protection from and pathogenesis of infectious diseases. Clin. Microbiol. Rev. 12:19–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.