Abstract
Salmonella enterica subsp. I serovar Enteritidis exhibits type III secretion system 2 (TTSS2)-dependent early colonization and inflammation kinetics faster than those of closely related S. enterica serovar Typhimurium. To investigate the accelerated TTSS-2-dependent pathogenic potential of S. Enteritidis, we focused on its genome. Results of a previously published comparative genomic study revealed the presence of mutually exclusive genes in both serovars. In this study, we investigated the roles of six S. Enteritidis-specific genes in vivo by using differential fluorescence induction (DFI) through putative gene-specific promoters. The promoter construct associated with the gene locus SEN1140 induced green fluorescent protein (GFP) expression in the gut lumen, lamina propria, mesenteric lymph nodes, and related systemic organs. To further investigate the potential role of SEN1140, we compared a SEN1140 deletion mutant with S. Typhimurium in a TTSS1-deficient background. Interestingly, the S. Enteritidis mutant lacking SEN1140 did not show the unique TTSS-2-dependent early colonization and inflammation kinetic phenotype of S. Typhimurium. Consistent with this result, complementation of SEN1140 restored the TTSS-2-dependent accelerated inflammatory potential of S. Enteritidis. This report presents a suitable screening strategy that uses a combination of DFI, fluorescence-activated cell sorting, quantitative PCR, and wild-type isogenic tagged-strain techniques to explore the unique roles of S. Enteritidis-specific genes in bacterial pathogenesis.
INTRODUCTION
Salmonella enterica bacteria are pathogenic organisms that cause an array of diseases. A few of the members of this species show host specificity, while others cause diseases in a wide range of hosts (10). Two of the main serovars of S. enterica, which cause gastroenteritis in humans and systemic and lethal infections in genetically susceptible mice, are S. enterica serovars Enteritidis and Typhimurium (2). These pathogenic bacteria and the diseases they cause are major public health concerns in many countries. Studies have shown that the vast majority of Salmonella virulence factors and associated pathogenicities result from multiple pathogen-host interactions. The deletion of known virulence determinants could be advantageous for identifying and analyzing novel interactions or virulence factors involved in bacterial adaptiveness. Indeed, this technique has facilitated the identification of various SPI-1 effector proteins (18, 29, 45) and the characterization of the roles of SPI-1, SPI-2 (19, 20), and many other Salmonella virulence factors. Surprisingly, rare cases of human Salmonella infection with SPI-1-deficient strains have been reported (22) and the prevalence of such mutants has also been identified in animal husbandry (35). Thus, mutants lacking major known virulence determinants are also of importance for the study of health and disease. The mode and mechanism of S. Typhimurium pathogenesis have been extensively studied, and recently, many studies have been focusing on the comparison of the infection profiles of S. Typhimurium and S. Enteritidis, as the genomes of both of these serovars have been sequenced and made available (27, 44). The comparative analysis of both genomes shows the presence of many sequences in S. Enteritidis that are not present in the chromosome of S. Typhimurium. These gene sequences originated primarily from prophages or genomic islands that were acquired through horizontal gene transfer and are designated S. Enteritidis-specific “regions of differentiation.” Presumably, differences between the contents of the genomes of the two serovars might also reflect differences between their infection profiles.
S. Enteritidis-specific gene sequences represent approximately 2% of the total size of the S. Enteritidis genome, and many of these sequences are still not annotated. We have previously reported that S. Enteritidis has accelerated invasion kinetics and shows early SPI-1-independent inflammation, compared with SPI-1-deficient S. Typhimurium, in specific-pathogen-free, streptomycin-pretreated C57BL/6 mice (43). Thus, it would be interesting to identify the genetic factors in S. Enteritidis that contribute to its early infection process. It has been recently reported that S. Enteritidis infection involves additional genes that are absent from S. Typhimurium (3). A comparative analysis of S. Enteritidis and S. Typhimurium shows 98% similarity in their genome sequences (44), suggesting that the extra gene elements present in S. Enteritidis (2% of the genome) are most likely associated with its accelerated inflammation kinetics. Gene disruption, followed by in vitro or in vivo analysis of the mutant generated, is an established conventional way of identifying the roles of genes in the infection process, but this method is laborious when used to explore multiple mutations. Here we present a method that uses a simple screen for the identification of S. Enteritidis-specific promoters and their corresponding genes that might contribute to the accelerated infection process of S. Enteritidis. The screen represents a promising combination of bioinformatic analysis, differential fluorescence induction (DFI), wild-type isogenic tagged-strain (WITS), fluorescence-activated cell sorting (FACS), and quantitative real-time PCR (qRT-PCR) techniques to evaluate the role of S. Enteritidis gene products. This combinatorial method is based on the in vivo expression of green fluorescent protein (GFP) in differentially tagged wild-type isogenic strains using an S. Enteritidis-specific promoter cloned into a promoterless GFP plasmid. The spatiotemporal endogenous promoter-driven expression of GFP in different host tissues might indicate the involvement of its respective gene(s) in various host-pathogen interactions in vivo. Subsequently, a small mutant library of selected genes could be created on the basis of the promoters screened instead of developing an exhaustive mutant library to conduct cumbersome in vivo analyses of individual mutants. The techniques proposed in this study are superior to RNA sequencing, as this approach works precisely even in a background of numerous other bacterial species and high loads of host-derived cells.
In the present study, our findings demonstrated that the involvement of an uncharacterized S. Enteritidis gene, SEN1140, in the invasion process resulted in accelerated SPI-1-independent inflammation. SEN1140 was likely acquired through genetic transfer from a prophage to the S. Enteritidis genome and encodes a hypothetical phage protein whose function is not yet known. It is well known that prophages contain additional cargo genes called morons or lysogenic conversion genes that play insignificant roles in the phage life cycle. Interestingly, these morons get incorporated into the chromosomes of bacteria infected with moron-bearing prophages (5). The morons in many pathogenic bacteria have been studied and shown to encode proven or suspected virulence factors; however, S. Enteritidis P125109 has been poorly studied in this regard. The results of this study suggest a role for a moron element such as SEN1140 in S. Enteritidis virulence and pathogenesis.
MATERIALS AND METHODS
Bacterial strains and growth condition.
Bacterial strains (Table 1) were grown for 12 h at 37°C in Luria-Bertani (LB) medium supplemented with 0.3 M NaCl, diluted 1:20 in fresh LB medium, and subcultured for another 4 h under mild aeration until an optical density of 0.6 was obtained. The bacteria were washed in ice-cold phosphate-buffered saline (PBS), and 5 × 107 CFU were suspended in 50 μl cold PBS for use in the in vivo experiments. Prior to the start of the in vivo experiments, all strains were tested for growth attenuation for 16 h in 10 ml of culture medium at 37°C at 150 rpm under aerated conditions.
Table 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant genotype and/or phenotype | Background or resistance | Reference |
|---|---|---|---|
| Strains | |||
| M1511 | TTSS-1− (invC::aphT); Smr Kmr | M1525 | 43 |
| M1525 | S. Enteritidis 125109 wild type; Smr | Wild type | 42 |
| SB566 | TTSS-1− (invC::aphT); Smr Kmr | SB300 | 38 |
| Z290 | SPI-1-deficient SEN1140 mutant (invC::aphT SEN1140::cat); Smr Kmr Cmr | M1511 | This study |
| Z291 | SEN1140 mutant (SEN1140::cat); Smr Cmr | M1525 | This study |
| Z292 | Z290 complemented with SEN1140 through pCH112; Ampr | Z290 | This study |
| Plasmids | |||
| pM968 | Promoterless GFPmut2 in promoterless pBAD24 vector | Ampr | 41 |
| pM975 | bla PssaG GFPmut2 plasmid with oripMB1 | Ampr | 28 |
| pM973 | bla PssaH GFPmut2 plasmid with oripMB1 | Ampr | 40 |
| pCH112 | hilA ORF cloned into PBAD/myc-His; oripBR322 | Ampr | 28 |
| pKD3 | bla FRT cat FRT PS1 PS2 oriR6K | Cmpr | 25 |
| pKD46 | bla PBAD gam bet exo pSC101 oriTS | Ampr | 25 |
| pM2152 | 450 bp upstream (P0999) of SEN0999 cloned into pM968 between BamHI and XbaI | Ampr | This study |
| pM2153 | 500 bp upstream (P4250) of SEN4250 cloned into pM968 between BamHI and XbaI | Ampr | This study |
| pM2154 | 550 bp upstream (P1013) of SEN1013 cloned into pM968 between BamHI and XbaI | Ampr | This study |
| pM2155 | 750 bp upstream (P1140) to SEN1140 cloned into pM968 between BamHI and XbaI | Ampr | This study |
| pM2156 | 650 bp upstream (P1975) to SEN1975 cloned into pM968 between BamHI and XbaI | Ampr | This study |
| pM2157 | 700 bp upstream (P1994) to SEN1994 cloned into pM968 between BamHI and XbaI | Ampr | This study |
| pM2158 | SEN1140 ORF with 750-bp upstream region cloned into pCH112 between NcoI and XbaI; oripBR322 | Ampr | This study |
Ethics statement.
The appropriate committee (Kantonales Veterinäramt, Zürich, Switzerland; license number ZH201/2007) approved all animal work. All animals were maintained in strict accordance with the good animal practice guidelines of the relevant national and/or welfare bodies.
Mouse infection experiment.
The animal experiments were performed in compliance with all relevant federal guidelines and the institutional policies of ETH Zürich. The infection experiments were performed in individual ventilated cages as described previously (4). All C57BL/6 mice were pathogen free and bred at the Rodent Center HCI, ETH Zürich. Mice were pretreated intragastrically with 50 mg of streptomycin and infected 24 h later with 5 × 107 CFU (oral gavage) of the corresponding bacterial strain. When needed for bacterial coinfection experiments, freshly grown cultures of bacterial strains were mixed in equal proportions and a dose of 5 × 107 CFU of a mixed inoculum was administered to streptomycin-pretreated C57BL/6 mice. Fecal samples were collected at 12 and 24 h for estimation of lipocalin 2 as a marker of early inflammation. Bacterial loads in the cecum, mesenteric lymph nodes (MLN), liver, and spleen were determined upon plating of the homogenates on MacConkey agar plates supplemented with appropriate antibiotics. The tissues were collected in an optimum cutting temperature (OCT) compound (Sakura Finetek Inc.) for cryosectioning, snap-frozen in liquid nitrogen, and stored at −80°C. For fluorescent imaging and GFP counting, the tissues were collected in a 4% paraformaldehyde (PFA) solution (in PBS) and processed separately.
Preparation of WITS of S. Enteritidis.
S. Enteritidis P125109 wild-type strain M1525 was tagged with specific barcode sequences to generate WITS (14). Insertion of tags into the S. Enteritidis genome was achieved by a conventional phage transduction method. Briefly, a small aliquot of the recipient wild-type S. Enteritidis strain (M1525) grown overnight was transduced with phage lysate obtained from individual donor WITS for 15 min at 37°C. The transduced colonies were selected on LB agar plates supplemented with kanamycin (50 μg/ml) and screened for the integration of individual tags into the chromosome by PCR using individual WITS tag-specific primers in combination with the ydgA-Fw common primer (Table 2).
Table 2.
Primers used in this study
| Primer | Sequence (5′–3′) |
|---|---|
| p999-Fw | GGGTCTAGA AATGAACATAAAAAAAATTC |
| p999-Rw | GCCGGATCCAATAACCTCTATATATAATC |
| p4250-Fw | GTTTCTAGAATATCGGATACTGCTGCACT |
| p4250-Rw | GCGGGATCCACTCCATTTAAAAAGCTAAT |
| p1013-Fw | GGGTCTAGATTGCTCATAAAATGATATCA |
| p1013-Rw | GGCGGATCC ATGTTTTCCTGAAGTGTGTA |
| p1975-Fw | GCGTCTAGA AGTTTCCAATATATCATTTG |
| p1975-Rw | GCGGGATCCCAGATTAACTCCCTGAGATA |
| p1994-Fw | GCGTCTAGAAGTTTCAAAAGTTATGATCA |
| p1994-Rw | CGGGGATCCATGTTAGGTTAATGTTAGTT |
| p1140-Fw | AATTCTAGA CAGCAGCTCATCGTGGCGAC |
| p1140-Rw | GGCGGATCC TTCCTATTGCTGATTAATAC |
| pM968-Fw | TCCGCTTACAGACAAGCTGTG |
| pM968-Rw | TACATAACCTTCGGGCATGGC |
| WITS-1 | ACGACACCACTCCACACCTA |
| WITS-2 | ACCCGCAATACCAACAACTC |
| WITS-11 | ATCCCACACACTCGATCTCA |
| WITS-13 | GCTAAAGACACCCCTCACTCA |
| WITS-17 | TCACCAGCCCACCCCCTCA |
| WITS-19 | GCACTATCCAGCCCCATAAC |
| WITS-21 | ACAACCACCGATCACTCTCC |
| ydgA-Fw | GGCTGTCCGCAATGGGTC |
| SEN1140-Fw | GCAAGCCTGCTGAGGAACGGGATTTTTGTATTAATCAGCAATAGGAATGTGTAGGCTGGAGCTGCTTC |
| SEN1140-Rw | GATGTGTGGCAATTCTTACCGAGGGTGTTGAGCAATCCTCTCTAAAATATGAATATCCTCCTTAGTT |
| SEN1140-comp-Fw | TTA ACC ATG GAG CAG CTC ATC GTG GCG AC |
| SEN1140-comp-Rw | GTT CTC TAG ACT ATT TCT GCG AGG CTA TAT |
Plasmid cloning.
The genomes of S. Enteritidis and S. Typhimurium have already been compared (44). For this study, six S. Enteritidis-specific genes were selected on the basis of their putative encoded protein domains. Putative promoters for the selected S. Enteritidis-specific genes were PCR amplified from the genomic DNA of S. Enteritidis P125109 using promoter-specific primer pairs (Table 2) as described earlier. Briefly, the PCR mixture contained 1× reaction buffer, 0.5 mM deoxynucleoside triphosphate mix, 1.25 mM magnesium chloride, 0.5 μM promoter-specific forward and reverse primers, 1 ng of S. Enteritidis P125109 genomic template DNA, and 1 U of Pfu DNA polymerase. The PCR products were obtained after 30 PCR cycles of 95°C for 1 min, followed by 53°C for 45 s and 72°C for 1 min. The amplicons were gel eluted and column purified. The fragments were introduced between the BamHI and XbaI sites of promoterless GFPmut2 plasmid pM968 to obtain recombinant plasmids (Table 1) and transformed into individual WITS of S. Enteritidis P125109 (wild-type) strain M1525. The transformants were selected on LB agar plates supplemented with ampicillin (100 μg/ml) and kanamycin (50 μg/ml). Transformants were screened by colony PCR; the clones were subjected to sequencing of plasmid DNA for confirmation of the incorporation of selected promoter regions into plasmid pM968 using primer pM968-Fw (Table 2). These manipulations generated the following combinations of chromosomal tags with plasmid constructs in strain M1525: WITS1-pM2152, WITS2-pM2153, WITS11-pM2154, WITS13-pM2156, WITS19-pM2157, and WITS21-pM2155.
Sample preparations for FACS analysis.
To examine GFP expression through the control of the individually cloned promoters, the recombinant plasmid-bearing WITS of S. Enteritidis (M1525) were grown overnight in LB medium supplemented with 0.3 M NaCl and appropriate antibiotics. Strain M1525 with the WITS17 tag and plasmid pM975 (ssaG promoter in pM968) was used as the positive control for this set of experiment. The cultures were subcultured to an optical density of 0.6 and subsequently mixed in equal proportions (1:1:1:1:1:1:1) representing equal numbers of CFU of all of the strains in a mixed pool. Approximately 107 CFU of the mixed-inoculum pool was administered orally to a group of streptomycin-pretreated C57BL/6 mice; the cecal contents were collected from each mouse at 2 days postinfection (p.i.) and diluted in 500 μl of PBS. The cecal suspensions were FACS sorted to analyze the GFP expression in the bacterial strains.
FACS analysis.
S. Enteritidis (wild-type) strain M1525 harboring the promoterless plasmid pM968 was used as the negative control for FACS analysis using a BD Biosciences FACSAria system. The side scatter (SSC-A) on the y axis and GFP fluorescence (FL1-A GFP) on the x axis were used to gate on single bacterial cells. A total of 105 cells for each GFP-positive or GFP-negative fraction were collected from each sample suspension and enriched overnight in LB medium supplemented with ampicillin (100 μg/ml) and kanamycin (50 μg/ml). The genomic DNA was isolated from each bacterial culture and subjected to qRT-PCR using WITS-specific primers in combination with the ydgA-specific common primer to determine the WITS copy numbers in a bacterial population. The relative WITS tag proportion directly represented the ratio of the individual recombinant plasmid present with respect to the specific WITS in the FACS-sorted samples.
Analysis of GFP-expressing Salmonella in the lamina propria.
Streptomycin-pretreated C57BL/6 mice were infected with GFP reporter plasmid (pM973)-bearing Salmonella M1511, Z290, and SB566 mutant strains. Cecal tissues were collected at day 2 p.i., incubated overnight in a 4% PFA solution (in PBS) at 4°C, incubated for 8 h in 20% sucrose (in PBS) at 4°C, and then snap-frozen in OCT compound (Sakura Finetek Inc.). Cryosections 20 μm thick were obtained on a clean glass slide and air dried for a minimum of 4 h. Subsequently, the cryosections were blocked in the dark for 1 h in 10% goat serum prepared in PBS. The GFP-expressing recombinant plasmid-bearing S. Enteritidis bacteria were enumerated in the mucosa as described earlier. Briefly, the sections were stained with anti-Armenian hamster αCD54 antibody (clone 3E2; BD Biosciences) for 1 h and then incubated in a mixture of phalliodin-A647 (FluoProbes) (1:100), anti-Armenian hamster–Cy3 (Jackson ImmunoResearch Laboratories; 1:200) antibodies, and 4′,6-diamidino-2-phenylindole (DAPI) stain (Sigma-Aldrich; 1:1,000). After incubation, the sections were washed repeatedly with PBS and mounted with Vectashield (Vector Laboratories). Six to nine sections per cecum sample were analyzed for the enumeration of GPF-expressing bacteria in the lamina propria.
Chromosomal gene disruption and plasmid-based complementation.
Chromosomal deletion of the SEN1140 gene was done by using the standard lambda-red recombinase system with a pKD3 template plasmid (9, 23). The phage lysate of the mutant developed was prepared, and the stable double mutant strains Z290 and Z291 were obtained by transducing the mutation into recipient strains M1511 (S. Enteritidis invC::aphT) and M1525 (S. Enteritidis, wild type), respectively. For complementation of the SEN1140 gene, plasmid pM2158 was constructed by manipulating plasmid pCH112 (28). Briefly, the SEN1140 gene along with its 750-bp upstream sequence was PCR amplified using primers SEN1140-comp-Fw and SEN1140-comp-Rw (Table 2). The resulting amplicon was digested with nucleases NcoI and XbaI. The hilA open reading frame (ORF) between the NcoI and XbaI sites in plasmid pCH112 was replaced with the SEN1140 ORF and its corresponding 750-bp upstream promoter sequence to obtain plasmid pM2158. Subsequent introduction of the recombinant plasmid (pM2158) into mutant strain Z290 to rescue the function of SEN1140 resulted in complemented strain Z292. Plasmid pM2158 also contained a PBAD promoter, which remained inactive in the absence of arabinose; however, the plasmid could express SEN1140 under the control of its cloned promoter.
Histopathological evaluation.
Segments of the ileum, cecum, and colon were fixed and embedded in OCT compound (Sakura Finetek Inc.), snap-frozen in liquid nitrogen, and stored at −80°C. The cryosections (5 μm) were obtained on glass slides and stained with hematoxylin and eosin (H&E) after drying for at least 2 h at room temperature. The stained histopathological sections were evaluated on the basis of a previously described scoring system for the quantitative analysis of cecal inflammation (4, 43). The H&E-stained sections (5 μm) were scored independently on the basis of the following criteria.
(i) Submucosal edema.
Submucosal edema was observed at ×100 magnification for the distance between the tunica muscularis and lamina mucosalis mucosae. The feature was scored as follows: no significant pathological changes in the submucosal features, score 0; mild edema when the submucosa accounts for <50% of the diameter of the entire intestinal wall from the tunica muscularis to the epithelium, score 1; moderate edema when the submucosa covers nearly 50 to 80% of the intestinal diameter, score 2; significant edema when the submucosa covers more than 80% of the diameter of the entire intestinal wall, score 3.
(ii) PMN cell infiltration of the lamina propria.
Polymorphonuclear (PMN) cells in the lamina propria were enumerated at ×400 magnification, and the average number of PMN cells/high-power field (HPF) was graded by using the following criteria: 0 to 5 PMN cells/HPF was assigned a score of 0, 5 to 20 PMN cells/HPF was assigned a score of 1, 21 to 60 PMN cells/HPF was assigned a score of 2, 61 to 100 PMN cells/HPF was assigned a score of 3, and more than 100 PMN cells/HPF was assigned a score of 4.
(iii) Goblet cells.
Nearly 10 different regions of the cecal epithelium were observed at ×400 magnification to determine the average number of goblet cells per HPF. The appearance of more than 28 goblet cells/HPF was scored 0, considering that they are a normal feature of specific-pathogen-free mice and the disappearance of goblet cells would increase the score. A score of 1 was assigned for 11 to 28 cells/HPF, a score of 2 was assigned for 1 to 10 cells/HPF, and a score of 3 represented <1 goblet cell or the complete disappearance of goblet cells from the cecum as observed in the cryosections.
(iv) Epithelial integrity.
Epithelial integrity was observed at ×400 magnification, and a score of 0 was assigned when there were no detectable pathological changes in the cecal epithelial layer. Epithelial desquamation was categorized grade 1, and erosion of the epithelial layer characterized by a gap of 1 to 10 epithelial cells/lesion was considered grade 2. However, a gap of more than 10 epithelial cells/lesion is a characteristic of epithelial ulceration, which was categorized as grade 3.
The independent scores for submucosal edema, PMN cell infiltration, goblet cells, and the epithelial integrity of each tissue sample were averaged and combined. The combined pathological scores ranged from 0 to 13 arbitrary units covering the following inflammation levels: intact intestine without any sign of inflammation, a pathoscore of 0; minimal sign of inflammation (which is commonly found in the ceca of specific-pathogen-free mice and generally not considered a pathological feature), a pathoscore of 1 or 2; slight inflammation (a minimal sign of tissue pathology), a pathoscore of 3 or 4; moderate inflammation, a pathoscore of 5 to 8; significant inflammation, a pathoscore of 9 to 13.
Lipocalin 2 estimation.
Lipocalin 2 was measured using an enzyme-linked immunosorbent assay (ELISA) of 12- and 24-h fecal samples using a commercially available kit (R&D Systems). Briefly, for the early detection of cecal inflammation at an intermittent time point, fecal samples were collected from the mice at 12 and 24 h. The samples were weighed and suspended in 500 μl of sterile PBS. For the lipocalin 2 ELISA, the wells were coated with an anti-lipocalin 2 capture antibody (diluted 1:200 in PBS), washed with ELISA wash buffer (0.05% Tween 20 in PBS [PBST]), and blocked with 2% bovine serum albumin solution in PBS. The diluted fecal samples and lipocalin 2 standards were added to the respective wells and incubated overnight at 4°C. The wells were washed with wash buffer; subsequently, 100 μl of biotin-labeled anti-lipocalin 2 antibody diluted 1:200 in blocking solution was added to each well and incubated at room temperature for 1 h. Horseradish peroxidase-labeled streptavidin (Biolegend) was diluted 1:1,000 in PBS and added to each well after washing with wash buffer. The wells were developed with 100 μl of substrate buffer containing 2,2′-azino-bis(3-ethylbenzothiazoline-6-sulfonic acid) (ABTS; 0.01%) and hydrogen peroxide (0.1%). The absorbance at 405 nm was measured on an ELISA plate reader, and the lipocalin 2 values were expressed as nanograms of lipocalin 2 per gram of feces.
RESULTS
DFI coupled with qPCR identified promoter P1140 as overrepresented in the mouse gut.
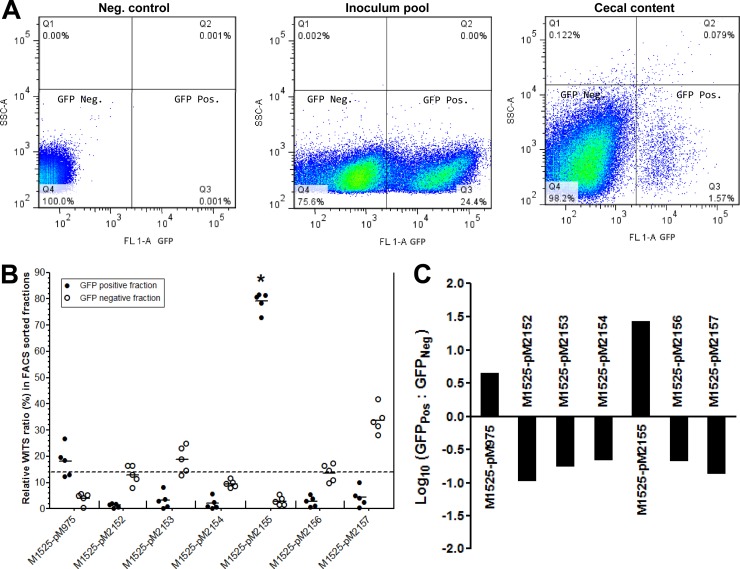
The streptomycin-pretreated C57BL/6 mice were coinfected with differentially tagged wild-type S. Enteritidis (M1525) strains containing GFP reporter plasmid constructs. These plasmids harbored putative promoter elements for unique S. Enteritidis genes (see Fig. S1 in the supplemental material) cloned upstream of the GFP ORF between the XbaI and BamHI sites. We proposed that GFP would be expressed through these plasmid constructs under the control of the cloned putative promoters. The analysis was limited to seven different promoters at a time, as there were only seven unique M1525 WITS (numbered WITS1, -2, -11, -13, -17, -19, and -21). The analysis included strain M1525 carrying intracellular GFP reporter plasmid pM975 as a control strain. Thus, the scheme allowed the analysis of six other putative promoters of S. Enteritidis. Initially, the genomic DNA isolated from the inoculum pool was subjected to relative quantification of the WITS tags using tag-specific primers (Table 2) by qPCR. All seven tags were present in equal ratios in the inoculum pool, indicating the presence of an equal population of differentially tagged S. Enteritidis strains in the inoculum (Fig. 1A). The relative equal ratio of WITS tags was also observed in the DNA obtained from the fecal samples of mice at day 1 p.i., indicating that all of the strains had the same opportunity for in vivo colonization; this result ruled out stomach acid-related bottlenecks and reiterated inoculum proportions (Fig. 1B). Subsequently, the cecal samples obtained at day 2 p.i. were FACS sorted on the basis of their GFP expression profile using DFI. The GFP-positive fraction obtained represented a net of 24% of the GFP-expressing bacterial population (Fig. 2A). However, at day 2 p.i., the promoter construct pM2155 (upstream region of SEN1140; P1140) was statistically predominant (P < 0.0001) in the GFP-positive fraction, accounting for 76 to 78% of the GFP-expressing population (Fig. 2B). Alternatively, the ratio of GFP-positive to GFP-negative fractions (∼80-fold) was statistically significant only for S. Enteritidis strains harboring P1140 of all of the promoters tested. Furthermore, marginal GFP expression was observed for the M1525 strain containing pM975 (Fig. 2B and C and 3A), which could be attributed to phagocytosis by luminally recruited PMN leukocytes and the subsequent release of Salmonella back into the lumen after destruction of the immune cell. Thus, the activation of the putative promoter P1140 through the promoter construct pM2155 in the mouse gut could suggest novel roles for SEN1140 in S. Enteritidis interactions with the host, possibly influencing subsequent inflammation.
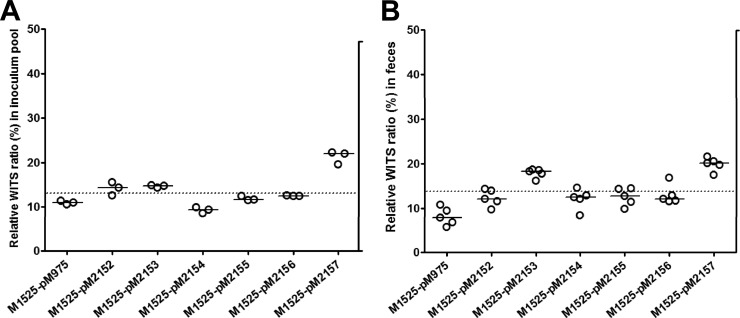
Fig 1.
Recombinant promoter construct-bearing Salmonella strains colonize equally in vivo. Freshly grown cultures of seven differentially WITS of S. Enteritidis wild-type strain M1525 harboring different promoter constructs integrated into a GFP reporter plasmid were mixed in equal proportions (1:1:1:1:1:1:1) to create a mixed-inoculum pool. (A) Individual WITS tags from three independent replicates of the inoculum pool were quantified by qPCR. All WITS showed an average equal proportion of 11 to 19% of the inoculum pool. (B) At day 1 p.i., all seven strains were found to have colonized well and to be shed in equal ratios in the feces. All strains showed corresponding relative WITS ratios of about 14.28%, which represents the theoretical average WITS ratio (shown by the dashed line) of individual WITS in a pool of seven strains.
Fig 2.
The GFP-positive fraction was represented mainly by one promoter construct. A mixed-inoculum pool (1:1:1:1:1:1:1) of WITS of S. Enteritidis having specific promoter constructs was used to infected a group of five streptomycin-pretreated C57BL/6 mice, whose cecal contents were analyzed at day 2 p.i. (A) Cecum samples were collected and suspended in 500 μl PBST solution. The suspension was diluted 1:100, passed through a 50-μm sieve, and FACS sorted to collect GFPPos and GFPNeg bacterial fractions from the third and fourth quadrants, respectively, during FACS sorting. The FACS was calibrated with a negative-control strain bearing plasmid pM968. (B) Relative WITS counts of GFPPos (●) and GFPNeg (○) fractions of cecal contents. Collected GFP fractions were enriched in LB medium supplemented with an appropriate antibiotic. Genomic DNA was isolated from the culture, and qPCR was performed for WITS. Most of the GFPPos fraction was represented by strain M1525/pM2155. *, P < 0.001 (Kruskal-Wallis test). (C) Statistics of FACS-sorted GFPPos and GFPNeg fractions obtained from cecal contents on a logarithmic scale showing higher expression of GFP through promoter construct pM2155 than the positive control pM975 in the mouse cecum.
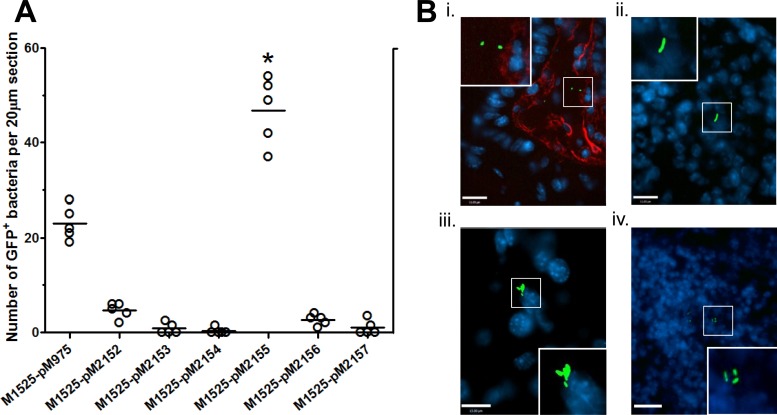
Fig 3.
Construct pM2155 is expressed at systemic sites in vivo. All of the constructs were tested in C57BL/6 for GFP expression at the systemic sites monitored after cecal colonization. (A) Cecal PFA sections were stained for immunofluorescence microscopy of the lamina propria. Shown are the counts of all of the GFP-expressing bacteria in the lamina propria. *, P < 0.05 (unpaired t test with Welch's correction). (B) Immunofluorescence staining of the lamina propria (i), spleen (ii), liver (iii), and MLN (iv) showing GFP-expressing strain M1525/pM2155. Bacteria expressing GFP at the host tissue sites are shown in the insets of all immunofluorescence images. Bars, 15 μm.
Wild-type S. Enteritidis with a P1140-harboring plasmid exhibits GFP expression in mouse secondary lymphoid organs.
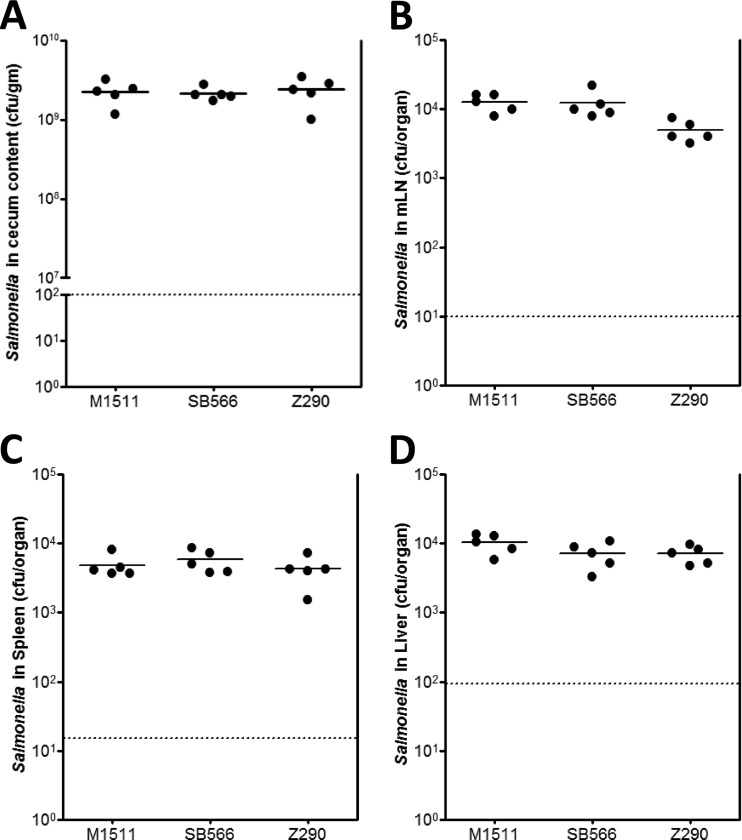
DFI revealed the activation of S. Enteritidis-specific putative promoter P1140 through strain M1525/pM2155 in the intestinal lumen as mentioned above. However, the gut colonization capacity in C57BL/6 mice pretreated with streptomycin was equivalent in mutant strain deficient in SEN1140 (strain Z290) and the parental M1525 strain (Fig. 4A). Therefore, we investigated the role of SEN1140 in secondary lymphoid tissues. Interestingly, the P1140 promoter in strain M1525/pM2155 was active in gut-associated lymphoid tissues, including the lamina propria (Fig. 3A), with a mean count of 46.80 (standard error of the mean, ±3.184)/20-μm section (6 sections/mouse; 5 mice). Qualitatively, the draining MLN and systemic sites, i.e., the spleen and liver, also showed GFP expression through pM2155 reflecting activation of putative promoter P1140 (Fig. 3B). Furthermore, P1140 activation was twice as high as that of positive control pM975, which was harbored by wild-type strain M1525, in the observed lamina propria tissues (P < 0.05). Overall, the activation of the putative P1140 promoter in the secondary lymphoid tissues might indicate the involvement of its downstream gene SEN1140 in multiple bacterial interactions inside the host. These interactions might also include virulence-specific roles of SEN1140 genes unique to S. Enteritidis. However, the identification of key factors involved in the accelerated inflammation kinetics of TTSS1-deficient S. Enteritidis remains a challenge.
Fig 4.
Strain Z290 colonizes efficiently in vivo. Different groups of streptomycin-pretreated C57BL/6 mice were infected with S. Enteritidis strain M1511 (SPI-1 deficient), S. Typhimurium strain SB566 (SPI-1 deficient), and S. enteritidis strain Z290 (SPI-1 deficient, SEN1140::cat). The cecum (A), MLN (B), spleen (C), and liver (D) were isolated at day 2 p.i., and the CFU in each organ were enumerated. P > 0.05 for Z290 versus the other strains.
SEN1140 contributes to TTSS2-dependent accelerated inflammation kinetics.
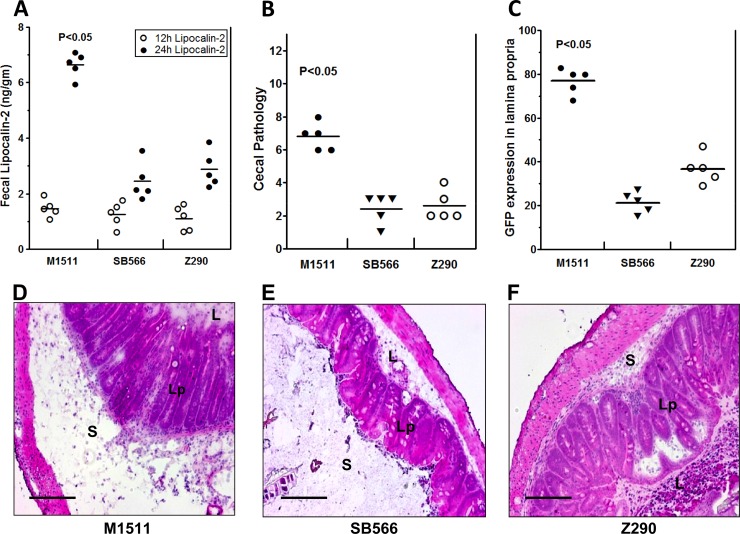
Our previous work comparing the virulence of S. Enteritidis and S. Typhimurium revealed significant differences in inflammation kinetics when invasion and subsequent inflammation were triggered via an alternative pathway in a TTSS2-dependent manner (43). However, the molecular mechanisms underlying this phenomenon remained largely unknown. In the present study, we have shown that the genetic deletion of SEN1140 in a TTSS1-deficient background results in differences between the inflammation kinetic profiles of these two serovars. Streptomycin-pretreated C57BL/6 mice were infected with the S. Enteritidis double mutant Z290 (invC::aphT SEN1140::cat); parallel and independent infections were conducted with S. Enteritidis parent strain M1511 (invC::aphT) and the S. Typhimurium equivalent SB566 (invC::aphT), respectively. All of these strains were carrying GFP reporter plasmid pM973 to facilitate fluorescence microscopic screening of the lamina propria. Upon infection, all strains colonized efficiently in vivo and exhibited comparable gut colonization levels (Fig. 4). However, TTSS1-deficient S. Enteritidis parent strain M1511 was able to induce early inflammation. The degree of inflammation was assessed indirectly through the fecal levels of lipocalin 2, which is one of the many possible biomarkers of intestinal inflammation, reflecting PMN cell infiltration of the lamina propria and the subsequent luminal secretion of glycoprotein. Initially, the mean lipocalin 2 concentration in strain Z290-infected mice was similar to the levels observed in mice infected with strains M1511 and SB566 at 12 h p.i.; however, significant differences were observed at 24 h p.i. (Fig. 5A). The mean lipocalin 2 concentration produced by strain Z290 infection was approximately 2.893 (standard deviation [SD], ±0.643) ng/g feces, whereas the concentration produced by strain M1511 infection was 6.637 (SD, ±0.445) ng/g feces (P < 0.05) at 24 h p.i. Notably, the lipocalin 2 level produced by strain Z290 infection was comparable to that produced by strain SB566 infection, i.e., 2.453 (SD, ±0.673) ng/g feces. Furthermore, the cecal pathology score was consistent with the lipocalin 2 concentrations (Fig. 5A) and reflected a pronounced difference in the degree of inflammation at day 2 p.i. (Fig. 5B). The early inflammation profile observed in the TTSS1-deficient S. Enteritidis (M1511) strain and its difference from the S. Typhimurium equivalent strain (SB566) were consistent with our previously published results (43). However, the degree of inflammation from strain Z290 (M1511, SEN1140::cat) was comparable to that from strain SB566. It became clear that the deletion of SEN1140 lowered the degree of inflammation expected from the SPI-1-deficient S. Enteritidis strain. This reduction in the inflammation profile could possibly be considered deferred inflammation kinetics. It was intriguing to observe that the deletion of a single gene, i.e., SEN1140, in a TTSS1-deficient background could explain the kinetic differences between these serovars, highlighting the previously unknown roles of S. Enteritidis-specific genes.
Fig 5.
Deletion of the SEN1140 gene lowers inflammation in the absence of TTSS1. The mouse groups shown in Fig. 4 were also analyzed for secretion of lipocalin 2 as a marker of cecal inflammation. (A) Fecal samples were collected from each group of mice at 12 and 24 h p.i. and suspended in sterile PBS, and the lipocalin 2 concentrations in the supernatants of fecal samples were estimated by ELISA. (B) Cecum sections from each group of mice were examined to determine pathoscores. (C) PFA sections of mouse cecum were stained for immunofluorescence microscopy, and the number of CFU of each strain expressing GFP in the lamina propria was determined. (D to F) H&E-stained representative cecum sections from each group of mice showing induced cecal inflammation: D, S. Enteritidis strain M1511 (SPI-1 deficient); E, S. Typhimurium strain SB566 (SPI-1 deficient; F, S. Enteritidis strain Z290 (SPI-1 deficient, SEN1140::cat). L, lumen; Lp, lamina propria; S, submucosal edema. Bars, 200 μm.
In an independent experiment, we tested the differences in colonization and inflammation kinetics between SEN1140 single mutant Z291 and its parental S. Enteritidis wild-type strain, M1525. We observed efficient colonization by these strains in all of the organs assessed (see Fig. S3A in the supplemental material). The cecal inflammation profiles of strains M1525 and Z291 were found to be comparable (see Fig. S3B in the supplemental material). Further, we could not observe any significant difference between the competitive indexes (log10) of M1525 and Z291 upon the execution of an independent coinfection experiment (n = 7); both strains had equal access to systemic organs (see Fig. S3C in the supplemental material). These data suggest that the disruption of SEN1140 in an SPI-1-proficient strain could not address the differences in inflammation kinetics observed between M1511 and Z290 in previous experiments. When taken together, all of the data collectively suggest a possible role for SEN1140 in S. Enteritidis-induced cecal inflammation in a TTSS-2-dependent manner.
SEN1140 augments the mucosal tissue load in a TTSS2-dependent manner.
TTSS1-deficient S. Enteritidis demonstrated accelerated inflammation kinetics, as mentioned above; however, this phenotype could arise for several possible reasons, including efficient sampling of S. Enteritidis, faster replication, or increased resistance to host defenses, which result in a net increase in the S. Enteritidis mucosal tissue load compared with that of S. Typhimurium. Alternatively, the former could provoke a more sensitive cytokine response, resulting in accelerated inflammation despite similar colonization levels. However, a previous report confirmed that TTSS1-deficient S. Enteritidis produced a greater mucosal tissue load, i.e., 10-fold more CFU in the lamina propria on day 2 than did the S. Typhimurium equivalent (43). Therefore, we investigated whether the abrogated inflammation of Z290 results from differential mucosal colonization. Cecum tissues were analyzed by fluorescence microscopy (see Materials and Methods). As expected, strain M1511 reached a mean tissue density of 77.21 (SD, ±5.96) cells per tissue section, while that of the S. Typhimurium equivalent, SB566, was significantly lower at a mean of 21.20 (SD, ±4.76) cells per tissue section (Fig. 5C). Interestingly, strain Z290 had approximately 2-fold attenuated cecal lamina propria colonization compared with that of its parent strain, M1511, but was comparable to SB566. Thus, the inefficient lamina propria colonization of strain Z290 could further support its corresponding inflammation kinetics compared with strain M1511, which efficiently colonized the lamina propria and produced a high pathoscore at day 2 p.i. Furthermore, the complementation of SEN1140 in strain Z290 rescued the accelerated inflammation phenotype, which was comparable to that of M1511 at day 2 p.i. (P > 0.05) (see Fig. S2 in the supplemental material), confirming the role of SEN1140 in a TTSS2-dependent alternative pathway of S. Enteritidis-induced inflammation.
DISCUSSION
Since the beginning of evolution, bacteria have continuously adapted to survive under exposed conditions through the manipulation of their genome contents and protein expression profiles. In some cases, small changes in genome content led to the evolution of bacteria with altered phenotypes (6). In pathogenic bacteria, genetic changes might result in an altered in vivo phenotype and host adaptation. In some cases, this enhances the virulence and/or transmission of the bacterium (5, 33). Salmonella is a pathogenic organism that has undergone numerous changes in its genome content throughout the evolutionary process. Among other members of this species, S. Typhimurium (SL1344) and S. Enteritidis (P125109) have been sequenced and annotated. Although the average nucleotide sequence identity between the shared orthologs of these serovars was reported to be 98.98% (44), approximately 6.4% of the genes present in S. Enteritidis are specific to this group and not found in S. Typhimurium. Indeed, S. Enteritidis possesses additional gene sequences in its genome that are not present in the genome of S. Typhimurium or the closely related serovar S. Gallinarum (7, 44). The functions of most of the S. Enteritidis genes, those shared with extensively studied S. Typhimurium, are now know; however, the extra gene elements present in S. Enteritidis have yet to be explored for their functional importance. There are few reports addressing the roles of S. Enteritidis-specific gene elements in the invasiveness and pathogenicity of the serovar (39). The SEN1975 gene has been reported as a novel virulence factor of S. Enteritidis that modulates the host immune response, as it bears close homology with the mammalian Toll/interleukin-1 (IL-1) receptor family of proteins (30). Similarly, the deletion of prophage-like element ϕSE12 severely attenuates the strain and promotes protective immunity against S. Enteritidis (1), suggesting the involvement of this element in the enhancement of S. Enteritidis virulence.
S. Enteritidis and S. Typhimurium are identical in gene distribution and alignment in the key pathogenicity islands, SPI-1 and SPI-2, of their genomes. These serovars colonize host tissues equally during the course of infection. Surprisingly, in a comparison of these serovars in a TTSS1-deficient background, S. Enteritidis showed accelerated invasion and inflammation patterns via an alternative pathway in streptomycin-pretreated mice (43). The reported difference in the inflammation patterns was attributed to additional gene elements present in the genome of S. Enteritidis (43). However, the identification of virulence factors remains a technical challenge for many researchers and many have reported on modern methods of screening for genes expressed in vivo. Phenotypic study upon the deletion of specific genes is still an influential and widely accepted approach. Alternatively, screening for promoters could ultimately indicate the expression of a related gene (36). In this regard, one of the most widely practiced approaches is to develop a promoter trap library for analysis of the differential expression of the reporter protein under variable conditions at different host sites; this method seems more promising and convenient than other more expensive methods, such as microarray and transcriptome analysis (8, 46). In this study, the putative promoter regions of six distinct S. Enteritidis-specific genes were studied. These genes were chosen on the basis of their presence in the S. Enteritidis chromosome or relative similarity to some domains within mammalian proteins as assessed with SMART and Pfam tools (12, 24). Of the experimental promoters selected, the putative promoter upstream of SEN1140 induced the expression of GFP in vitro and in vivo. Often in highly sensitive and advanced screening systems, such as microarrays, genes that are expressed both in vitro and in vivo show reduced expression upon data analysis, indicating that these genes are constitutively expressed; however, this could be misleading. It has been reported that many constitutive genes are differentially expressed under various host conditions and in various infection types. In addition, the altered expression of a few constitutive genes was reported in a comparison of enteritis and typhoid models of infection (37). This provides an adequate platform for the selection of a widely expressive promoter associated with the SEN1140 gene for the assessment of its possible role in in vivo S. Enteritidis infection. The use of the WITS technique in this study mediated the indirect identification of individual promoter constructs and facilitated the quantification of exclusive promoter-bearing strains directly from in vitro cultures or in vivo cecal samples. The WITS technique has been developed for the design of phenotypically identical strains bearing different signature tags in noncoding regions of the chromosome (26). This technique has been demonstrated to facilitate a comprehensive view of the spatial and stochastic nature of the isogenic tagged strains within a host (14).
In this study, we demonstrated that the additional in-frame deletion of SEN1140 in an SPI-1-deficient S. Enteritidis strain (Z290) lowers the inflammation profile, compared with that of a TTSS1-deficient parental strain (M1511), in streptomycin-pretreated mice. The concentration of lipocalin 2 in the feces of infected mice directly reflected the severity of the inflammation. Lipocalin 2 is a Th17-induced antimicrobial peptide produced by host epithelial cells against luminal bacteria (15). The secretion of lipocalin 2 is a secondary antimicrobial response, whereas the primary response involves the recruitment of neutrophils to the site of inflammation, which is induced through the action of IL-13/IL-17. Lipocalin 2 inhibits the growth of intestinal bacteria by preventing their acquisition of iron, an essential nutrient element. It has been reported that Salmonella infection results in marked IL-17- and IL-22-dependent intestinal epithelial induction and luminal accumulation of the antimicrobial peptide lipocalin 2 (34). Determination of the concentration of the highly stable lipocalin 2 protein is convenient for evaluation of the severity of diarrheal inflammation using the cecal or fecal contents of an infected host.
The restoration of the inflammation rate through complementation of SEN1140 confirms its contribution to the accelerated inflammation kinetics of S. Enteritidis pathogenicity in vivo. SEN1140 encodes a hypothetical phage protein with an unknown function. SEN1140 homologs have been observed in S. Gallinarum/Pullorum (strain RKS5078), S. Dublin (strains CT_02021853 and 3246), and S. Gallinarum (strain 287/91); however, some S. enterica serovars, such as S. Typhimurium (strains DT104, SL1344, DT2, and D23580), S. Bongori (strain 12419), S. Hadar, and other related serovars, did not show the presence of any such homolog, as observed in the available online database of typed Salmonella strains. Analysis of the primary sequence showed the presence of a YxxL/I–(x)6–8–YxxL/I (x denotes any amino acid) consensus sequence for an immunoreceptor tyrosine-based activation motif (ITAM) between residues 40 and 60 and a consensus sequence (I/V/L/S)xYxx(L/V) for its inhibition motif (ITIM) between residues 155 and 162 of the SEN1140-encoded protein. These motifs have been reported to actively modulate immune responses by activating signaling cascades (3) or by counterbalancing certain immune processes (16, 17, 30, 31). Moreover, it has been established that the signals derived from ITAM-containing receptors specifically control interstitial neutrophil migration toward the site of bacterial infection (13).
The majority of S. Enteritidis-specific gene clusters range in size from >3 to >40 kb. These unique gene clusters are called regions of difference of S. Enteritidis, which are involved primarily in prophage-related functions (44). Studies utilizing Escherichia coli K-12 and Shigella models have shown that differences between the DNA contents of related bacterial genomes are attributable to differences between the DNA sequences of the associated prophages (6). In the S. Enteritidis genome, the SEN1140 gene is present in the ϕSE12 prophage element (44). Studies have indicated that some prophages contain additional cargo genes, termed morons, which are not required for the phage cycle. However, many morons from prophages in pathogenic bacteria might encode additional virulence factors that change the phenotype of the lysogen (6). Such moron elements are also present in the S. Enteritidis genome (6, 21, 32), contributing to its virulence and pathogenesis; therefore, the phage element SEN1140 could also be considered a moron that influences the virulence of S. Enteritidis. The identification of morons that encode fitness factors in several of the S. Typhimurium prophages has led to the hypothesis that the phage-mediated reassortment of virulence and fitness factors is a key driving force in the optimization of the Salmonella-host interaction and the emergence of new epidemic clones (11). Some of the well-studied examples of moron-encoded functions include superoxide dismutases, enzymes for lipopolysaccharide modifications, and bacterial effector proteins secreted into host cells through specialized type III secretion systems (21). The effector protein SopE represents another appropriate model of the involvement of immunomodulatory morons in bacterial pathogenesis (32). Thus, it would be interesting and essential to evaluate the rationale for the presence of such morons within the S. Enteritidis genome and to identify any domains in the proteins that these genetic elements, such as SEN1140, encode. This analysis will be extended to investigate the role of other S. Enteritidis-specific genes in invasion or virulence to obtain a better understanding of bacterial adaptiveness. We hypothesize that phage-encoded morons with immunomodulatory functions might facilitate the adaptation of Salmonella strains to new hosts and provide an interesting topic for future research.
Supplementary Material
ACKNOWLEDGMENTS
We extend our gratitude to Thomas Christian Weber and staff of the RCHCI, ETH Zürich, for facilitating and maintaining mice for in vivo experiments. We thank Neera Singh for her constructive comments on the preparation of the manuscript.
This work was supported by the UBS Optimus Foundation (Zürich, Switzerland) and by the Department of Biotechnology, Ministry of Science and Technology, New Delhi, India (grant BT/PR14489/Med/29/207/2010).
Footnotes
Published ahead of print 2 July 2012
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1. Araya DV, et al. 2010. Deletion of a prophage-like element causes attenuation of Salmonella enterica serovar Enteritidis and promotes protective immunity. Vaccine 28:5458–5466 [DOI] [PubMed] [Google Scholar]
- 2. Asahara T, et al. 2011. Protective effect of Lactobacillus casei strain Shirota against lethal infection with multi-drug resistant Salmonella enterica serovar Typhimurium DT104 in mice. J. Appl. Microbiol. 110:163–173 [DOI] [PubMed] [Google Scholar]
- 3. Barrow AD, Trowsdale J. 2006. You say ITAM and I say ITIM, let's call the whole thing off: the ambiguity of immunoreceptor signalling. Eur. J. Immunol. 36:1646–1653 [DOI] [PubMed] [Google Scholar]
- 4. Barthel M, et al. 2003. Pretreatment of mice with streptomycin provides a Salmonella enterica serovar Typhimurium colitis model that allows analysis of both pathogen and host. Infect. Immun. 71:2839–2858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bäumler AJ, Tsolis RM, Ficht TA, Adams LG. 1998. Evolution of host adaptation in Salmonella enterica. Infect. Immun. 66:4579–4587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Brüssow H, Canchaya C, Hardt WD. 2004. Phages and the evolution of bacterial pathogens: from genomic rearrangements to lysogenic conversion. Microbiol. Mol. Biol. Rev. 68:560–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Carver TJ, et al. 2005. ACT: the Artemis comparison tool. Bioinformatics 21:3422–3423 [DOI] [PubMed] [Google Scholar]
- 8. Chiang HI, et al. 2008. Gene expression profiling in chicken heterophils with Salmonella enteritidis stimulation using a chicken 44 K Agilent microarray. BMC Genomics 9:526 doi:10.1186/1471-2164-9-526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Eswarappa SM, et al. 2008. Differentially evolved genes of Salmonella pathogenicity island: insights into the mechanism of host specificity in Salmonella. PLoS One 3(12):e3829 doi:10.1371/journal.pone.0003829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Figueroa-Bossi N, Uzzau S, Maloriol D, Bossi L. 2001. Variable assortment of prophages provides a transferable repertoire of pathogenic determinants in Salmonella. Mol. Microbiol. 39(2):260–271 [DOI] [PubMed] [Google Scholar]
- 12. Finn RD, et al. 2010. The Pfam protein families database. Nucleic Acids Res. 38:D211–D222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Graham DB, et al. 2009. ITAM signaling by Vav family rho guanine nucleotide exchange factors regulates interstitial transit rates of neutrophils in vivo. PLoS One 4(2):e4652 doi:10.1371/journal.pone.0004652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Grant AJ, et al. 2008. Modelling within—host spatiotemporal dynamics of invasive bacterial disease. PLoS Biol. 6(4):e74 doi:10.1371/journal.pbio.0060074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Griffin AJ, McSorley SJ. 2011. Development of protective immunity to Salmonella, a mucosal pathogen with a systemic agenda. Mucosal Immunol. 4:371–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hamerman JA, Lanier LL. 2006. Inhibition of immune responses by ITAM-bearing receptors. Science STKE 320:1–7 [DOI] [PubMed] [Google Scholar]
- 17. Hamerman JA, Tchao NK, Lowell CA, Lanier LL. 2005. Enhanced Toll-like receptor responses in the absence of signaling adaptor DAP12. Nat. Immunol. 6:579–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hapfelmeier S, et al. 2004. Role of the Salmonella pathogenicity island 1 effector proteins SipA, SopB, SopE, and SopE2 in Salmonella enterica subspecies 1 serovar Typhimurium colitis in streptomycin-pretreated mice. Infect. Immun. 72:795–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hapfelmeier S, et al. 2008. Microbe sampling by mucosal dendritic cells is a discrete, MyD88-independent step in DeltainvG S. Typhimurium colitis. J. Exp. Med. 205:437–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hapfelmeier S, et al. 2005. The Salmonella pathogenicity island (SPI)-1 and SPI-2 type III secretion systems allow Salmonella serovar Typhimurium to trigger colitis via MyD88-dependent and MyD88- independent mechanisms. J. Immunol. 174:1675–1685 [DOI] [PubMed] [Google Scholar]
- 21. Hendrix RW, Lawrence JG, Hatfull GF, Casjens S. 2000. The origins and ongoing evolution of viruses. Trends Microbiol. 8:504–508 [DOI] [PubMed] [Google Scholar]
- 22. Hu Q, et al. 2008. Salmonella enterica serovar Senftenberg human clinical isolates lacking SPI-1. J. Clin. Microbiol. 46:1330–1336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lesic B, Rahme LG. 2008. Use of the lambda Red recombinase system to rapidly generate mutants in Pseudomonas aeruginosa. BMC Mol. Biol. 9:20 doi:10.1186/1471-2199-9-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Letunic I, Doerks T, Bork P. 2009. SMART 6: recent updates and new developments. Nucleic Acids Res. 37:D229–D232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lostroh CP, Bajaj V, Lee CA. 2000. The cis requirements for transcriptional activation by HilA, a virulence determinant encoded on SPI-1. Mol. Microbiol. 37:300–315 [DOI] [PubMed] [Google Scholar]
- 26. Mastroeni P, Grant A, Restif O, Maskell D. 2009. A dynamic view of the spread and intracellular distribution of Salmonella enterica. Nat. Rev. Microbiol. 7:73–80 [DOI] [PubMed] [Google Scholar]
- 27. McClelland M, et al. 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413:852–856 [DOI] [PubMed] [Google Scholar]
- 28. Misselwitz B, et al. 2011. RNAi screen of Salmonella invasion shows role of COPI in membrane targeting of cholesterol and Cdc42. Mol. Syst. Biol. 7:474 doi:10.1038/msb.2011.7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Müller AJ, et al. 2009. The S. Typhimurium effector SopE induces caspase-1 activation in stromal cells to initiate gut inflammation. Cell Host Microbe 6:125–136 [DOI] [PubMed] [Google Scholar]
- 30. Newman RM, Salunkhe P, Godzik A, Reed JC. 2006. Identification and characterization of a novel bacterial virulence factor that shares homology with mammalian Toll/interleukin-1 receptor family proteins. Infect. Immun. 74:594–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pasquier B, et al. 2005. Identification of Fcα RI as an inhibitory receptor that controls inflammation: dual role of FcRγ ITAM. Immunity 22:31–42 [DOI] [PubMed] [Google Scholar]
- 32. Pelludat C, Mirold S, Hardt WD. 2003. The SopEPhi phage integrates into the ssrA gene of Salmonella enterica serovar Typhimurium A36 and is closely related to the Fels-2 prophage. J. Bacteriol. 185:5182–5191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rabsch W, et al. 2002. Salmonella enterica serotype Typhimurium and its host-adapted variants. Infect. Immun. 70:2249–2255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Raffatellu M, et al. 2009. Lipocalin-2 resistance confers an advantage to Salmonella enterica serotype Typhimurium for growth and survival in the inflamed intestine. Cell Host. Microbe 5:476–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rahn K, et al. 1992. Amplification of an invA gene sequence of Salmonella Typhimurium by polymerase chain reaction as a specific method of detection of Salmonella. Mol. Cell. Probes 6:271–279 [DOI] [PubMed] [Google Scholar]
- 36. Rediers H, Rainey PB, Vanderleyden J, De Mot R. 2005. Unraveling the secret lives of bacteria: use of in vivo expression technology and differential fluorescence induction promoter traps as tools for exploring niche-specific gene expression. Microbiol. Mol. Biol. Rev. 69:217–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rollenhagen C, Bumann D. 2006. Salmonella enterica highly expressed genes are disease specific. Infect. Immun. 74:1649–1660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Schlumberger MC, et al. 2007. Two newly identified SipA domains (F1, F2) steer effector protein localization and contribute to Salmonella host cell manipulation. Mol. Microbiol. 65:741–760 [DOI] [PubMed] [Google Scholar]
- 39. Silva CA, et al. 2012. Infection of mice by Salmonella enterica serovar Enteritidis involves additional genes that are absent in the genome of serovar Typhimurium. Infect. Immun. 80:839–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Songhet P, et al. 2011. Stromal IFN-γR-signaling modulates goblet cell function during Salmonella Typhimurium infection. PLoS One 6(7):e22459 doi:10.1371/journal.pone.0022459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Stecher B, et al. 2004. Flagella and chemotaxis are required for efficient induction of Salmonella enterica serovar Typhimurium colitis in streptomycin-pretreated mice. Infect. Immun. 72:4138–4150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Suar M, et al. 2006. Virulence of broad- and narrow-host-range Salmonella enterica serovars in the streptomycin-pretreated mouse model. Infect. Immun. 74:632–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Suar M, et al. 2009. Accelerated type III secretion system 2-dependent enteropathogenesis by a Salmonella enterica serovar enteritidis PT4/6 strain. Infect. Immun. 77:3569–3577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Thomson NR, et al. 2008. Comparative genome analysis of Salmonella Enteritidis PT4 and Salmonella Gallinarum 287/91 provides insights into evolutionary and host adaptation pathways. Genome Res. 18:1624–1637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tsolis RM, Adams LG, Ficht TA, Bäumler AJ. 1999. Contribution of Salmonella typhimurium virulence factors to diarrheal disease in calves. Infect. Immun. 67:4879–4885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wang S, et al. 2010. Transcriptomic responses of Salmonella enterica serovars Enteritidis and Typhimurium to chlorine-based oxidative stress. Appl. Environ. Microbiol. 76:5013–5024 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.