Abstract
Escherichia coli is the most important etiological agent of urinary tract infections (UTIs). Unlike uropathogenic E. coli, which causes symptomatic infections, asymptomatic bacteriuria (ABU) E. coli strains typically lack essential virulence factors and colonize the bladder in the absence of symptoms. While ABU E. coli can persist in the bladder for long periods of time, little is known about the genetic determinants required for its growth and fitness in urine. To identify such genes, we have employed a transposon mutagenesis approach using the prototypic ABU E. coli strain 83972 and the clinical ABU E. coli strain VR89. Six genes involved in the biosynthesis of various amino acids and nucleobases were identified (carB, argE, argC, purA, metE, and ilvC), and site-specific mutants were subsequently constructed in E. coli 83972 and E. coli VR89 for each of these genes. In all cases, these mutants exhibited reduced growth rates and final cell densities in human urine. The growth defects could be complemented in trans as well as by supplementation with the appropriate amino acid or nucleobase. When assessed in vivo in a mouse model, E. coli 83972carAB and 83972argC showed a significantly reduced competitive advantage in the bladder and/or kidney during coinoculation experiments with the parent strain, whereas 83972metE and 83972ilvC did not. Taken together, our data have identified several biosynthesis pathways as new important fitness factors associated with the growth of ABU E. coli in human urine.
INTRODUCTION
Urinary tract infections (UTIs) are among the most common bacterial infections in humans, affecting millions of people across the globe. It is estimated that there are more than 10 million cases of UTIs in Western Europe alone every year. UTIs also account for 25 to 40% of all nosocomial infections, making these infections an important medical and financial burden on our health care systems (17). Urinary tract infections affect primarily women; 40 to 50% of adult women will experience at least one UTI episode during their lifetime (8, 33). A UTI usually starts as a bladder infection but can, depending on the bacterial strain, ascend to the kidneys and may ultimately result in renal failure or dissemination to the bloodstream. The most important etiological agent of UTIs is Escherichia coli, which is associated with more than 80% of all such infections (34).
UTIs are classified into the following disease categories according to the focal point and severity of the infection: bacteriuria (the urine), cystitis (the bladder), pyelonephritis (the kidneys), and urosepsis (the blood). E. coli strains that cause cystitis and pyelonephritis have been studied extensively and are referred to as uropathogenic E. coli (UPEC). A subset of UPEC strains also cause urosepsis, and together with strains that cause neonatal meningitis and respiratory infection, these strains are collectively referred to as extraintestinal pathogenic E. coli (ExPEC) (18). In addition to well-documented symptomatic infections, many UTIs are asymptomatic. E. coli is also the major cause of asymptomatic bacteriuria (ABU); E. coli ABU strains cause few or no symptoms in the infected individual and establish bacteriuria in a commensal-like state. Despite the high prevalence of ABU, the molecular mechanisms associated with the ability of E. coli to establish this condition are not well understood. Previous studies have shown that many ABU E. coli strains fail to express key virulence factors normally associated with pathogenic UPEC strains, suggesting that this may constitute a mechanism of adaptation to long-term asymptomatic colonization of the human bladder (27, 32, 41, 40).
E. coli 83972 represents the prototypic ABU strain; it was originally isolated from a young Swedish girl who carried it asymptomatically for at least 3 years (4, 22). E. coli 83972 is well adapted for growth in the human urinary tract, where it can establish long-term bacteriuria (4, 15, 35). At the genome level, E. coli 83972 closely resembles the well-characterized and highly virulent UPEC isolate CFT073, suggesting that they may have a common ancestral origin (13, 19, 39). E. coli 83972 most likely represents a degenerated uropathogen which has lost the ability to produce a range of important virulence factors. It cannot adhere to bladder epithelial cells due to a lack of functional fimbriae and it does not activate host immune defenses during infection (20, 28). Nevertheless, E. coli 83972 grows well in human urine, with doubling times in the order of 25 to 45 min (12, 14, 27, 29). E. coli 83972 can also outcompete virulent UPEC strains in human urine and in the mouse bladder (29)—a phenotypic property that supports its use in the prophylactic treatment of patients who are refractory to conventional UTI therapy (5, 15). Extensive trials have suggested that deliberate colonization of the bladder with E. coli 83972 can prevent bacterial and fungal uropathogens from colonizing the urinary tract (5, 36–38). One common feature of E. coli 83972 and other ABU E. coli strains is the production of iron-chelating siderophores (23, 41, 42). Indeed, E. coli 83972 produces enterobactin, salmochelin, aerobactin, and yersiniabactin (42), and the genes responsible for the biosynthesis and transport of these molecules are highly expressed during growth in the iron-limited conditions of the human urinary tract (26).
Arguably, the ability to grow in human urine must be one of the prerequisites for ABU E. coli strains to establish long-term colonization of the human bladder. Bacterial metabolic processes are clearly important in relation to the persistence of E. coli strains in the urinary tract, with E. coli 83972 being an ideal strain for such studies, allowing for delineation between fitness and virulence. In this respect, we have previously shown, using transcriptomics, that a number of genes are upregulated in E. coli 83972 during growth in vivo in human urine compared to growth on laboratory medium (26). Here we have utilized E. coli 83972 and an additional ABU clinical isolate (VR89) to identify novel genes required for growth in human urine. Using a transposon mutagenesis approach, we show that genes involved in the biosynthesis of arginine (argC, argE, and carAB), methionine (metE), valine and leucine (ilvC), isoleucine (ilvA and ilvC), and adenine (purA) are important factors associated with the growth and fitness of ABU E. coli in human urine.
MATERIALS AND METHODS
Bacterial strains and growth medium.
The strains used in this study are listed in Table 1. All strains were grown at 37°C on solid or in liquid modified Luria-Bertani (LB) medium or in pooled sterile filtered urine collected from at least three healthy female volunteers with no history of UTI or antibiotic use within the last 2 months. The urine was pooled, filter sterilized, and stored at 4°C until use (within 1 to 2 days). Antibiotics were added when required at the following concentrations: gentamicin, 5 μg/ml; kanamycin, 25 μg/ml; chloramphenicol, 16 μg/ml; ampicillin, 100 μg/ml; and nalidixic acid, 50 μg/ml. Amino acid and nucleobase supplements were added at the following concentrations: uracil, 50 μg/ml; arginine, 100 μg/ml; adenine, 50 μg/ml; isoleucine, 100 μg/ml; and valine, 100 μg/ml.
Table 1.
Strains and plasmids used in the study
| E. coli strain or plasmid | Relevant characteristicsa | Reference |
|---|---|---|
| E. coli strains | ||
| 83972 | OR:K5:H−, ABU isolate | 4 |
| 83972Nal | 83972, nalidixic acid resistant | This study |
| 83972Amp | 83972, ampicillin resistant | 7 |
| VR89 | ABU isolate | 27 |
| 83972ΔcarAB | This study | |
| 83972ΔargE | This study | |
| 83972ΔargC | This study | |
| 83972ΔpurA | This study | |
| 83972ΔmetE | This study | |
| 83972ΔilvA | This study | |
| 83972ΔilvC | This study | |
| 83972carAB::kan | This study | |
| 83972argE::kan | This study | |
| 83972argC::kan | This study | |
| 83972purA::kan | This study | |
| 83972metE::kan | This study | |
| 83972ilvA::kan | This study | |
| 83972ilvC::kan | This study | |
| VR89carAB::kan | This study | |
| VR89argE::kan | This study | |
| VR89argC::kan | This study | |
| VR89purA::kan | This study | |
| VR89metE::kan | This study | |
| VR89ilvC::kan | This study | |
| Plasmids | ||
| pACYC184 | Cloning vector, Camr | |
| pVR9 | carAB cloned into pACYC184, Camr | This study |
| pVR10 | argE cloned into pACYC184, Camr | This study |
| pVR11 | argC cloned into pACYC184, Camr | This study |
| pVR12 | purA cloned into pACYC184, Camr | This study |
| pVR13 | metE cloned into pACYC184, Camr | This study |
| pVR15 | ilvA cloned into pACYC184, Camr | This study |
| pVR16 | ilvC cloned into pACYC184, Camr | This study |
Camr, chloramphenicol resistance.
Growth experiments.
Monocultures were grown in pooled human urine in 96-well microtiter plates inoculated to an optical density at 600 nm (OD600) of 0.01 under shaking conditions (300 rpm on an orbital shaker) at 37°C. OD600 readings were acquired at 30-min intervals until stationary phase was reached. Each experiment included two to four replicates and was repeated at least three times. For competition experiments, an ampicillin-resistant variant of E. coli 83972 (E. coli 83972AMP) was used instead of the wild-type E. coli 83972; E. coli 83972AMP displayed a growth rate identical to that of wild-type E. coli 83972 under all conditions tested. Mixed cultures were inoculated into pooled human urine using a starting ratio of 1:1 and an initial OD600 of 0.05. Samples from each culture were extracted at the start of the cultivation, diluted, and cultured on LB plates with and without ampicillin to confirm the starting ratios of CFU. The cultures were then grown overnight at 37°C and 300 rpm on an orbital shaker. Samples were extracted after 16 h, diluted, and cultured on LB plates with and without ampicillin. Each competition experiment was performed in duplicate and repeated at least three times.
Transposon mutagenesis.
Transposon mutant libraries were constructed using the mariner-based transposon system carried on plasmid pBT20 (21). Bacterial suspensions of the donor (E. coli SM10-λpir containing pBT20) and the recipients (E. coli 83972NalR and E. coli VR89NalR) were recovered from overnight plates and resuspended in LB broth. These suspensions were adjusted to OD600s of 40 and 20, respectively, and mixed in a 1:1 ratio. Aliquots of 50 μl were spotted on a dry LB agar plate and incubated at 37°C for 6 h. The mating mixtures were subsequently recovered from the plate, resuspended in LB, and cultured on LB agar plates containing gentamicin and nalidixic acid. From these plates, the individual colonies were inoculated into microtiter plates containing LB broth or sterile filtered human urine. The plates were incubated statically overnight at 37°C and subsequently screened for mutants that could grow in LB broth but not in human urine. The insertion sites were determined using a semirandom PCR method (9).
Recombinant DNA techniques.
Specific mutants of E. coli 83972 and E. coli VR89 were constructed using the λ-red recombinase-based gene inactivation system as previously described (6). The primers are listed in Table 2. Following amplification, the PCR products were transformed into E. coli 83972 and VR89 carrying the helper plasmid pKD46 and the transformations were plated on selective medium. Deletion mutants were verified by PCR (primers are listed in Table 2). The kanamycin resistance cassette was subsequently removed by use of the flippase-encoding plasmid pCP20 (6). The plasmids pVR9-13 and pVR15-16 were constructed by amplifying the relevant genes from CFT073 using the primers listed in Table 2. PCR fragments were digested with BamHI and SalI or SphI and cloned into pACYC184. The resulting seven plasmids were transformed into the mutants to enable complementation.
Table 2.
Primers used in the study
| Primer purpose and name | Sequence |
|---|---|
| Construction of deletion mutants | |
| carAB forward | GAGTGAATATTCTCTGGAGGGTGTTTTGATTAAGTCAGCGTAATGTGTAGGCTGGAGCTGCTTCG |
| carAB reverse | CAGGGTGGTGTCGTAATGCACTTTATATTGCAGCGCACTGCCATATGAATATCCTCCTTA |
| argE forward | GGGTGATCAGTTCGCGGGTGGGCTTGATAAACCTCGTCTCTAATGTGTAGGCTGGAGCTGCTTCG |
| argE reverse | TACCGCCATTTATCGAGATTTACCGTGCTCTGATTGCCACATATGAATATCCTCCTTA |
| argC forward | TTCAGCGCAAAGCAATGATGCGGGAAAGTTAATCTCCGATTAATGTGTAGGCTGGAGCTGCTTC |
| argC reverse | GCACCGCTTGTGCCGCCGCGCCTTTCAGTAAGTTGTCTTCCATATGAATATCCTCCTTA |
| purA forward | GACTGAACGGGCTAAATATGTTGTACGCTACCAGGGCGGTTAATGTGTAGGCTGGAGCTGCTTC |
| purA reverse | ACGCGTCGAACGGGTCGCGCAGAATCATGGTTTCAGTACCATATGAATATCCTCCTTA |
| metE forward | TATTGGGCGGGGAACTCCACGCGTGAAGAACTGCTGTAATGTGTAGGCTGGAGCTGCTTCG |
| metE reverse | GGCCAGCCGCGCGTTTTCAGGCCACAGTCCGGGTTGACCCCATATGAATATCCTCCTTA |
| ilvA forward | AAGAGCGGTACTACGTGCACCGGTTTACGAGGCGGCGCAGGTCTAATAGGCTGGAGCTGCTTCGAA |
| ilvA reverse | GGTCGCCAAGTTCGAACGCCGCCAGTACGCGCCCGTAGTCCATATGAATATCCTCCTTA |
| ilvC forward | CAGGGTAAAAAAGTAGTCATCGTCGGCTGTGGCGCACAGTAATGTGTAGGCTGGAGCTGCTTCG |
| ilvC reverse | AGCCGCGCAGTTTCTTACCTACCTGCTCAATCGCATGGCATATGAATATCCTCCTTA |
| Verification of deletion mutants | |
| dapA | TGCGAGATGTGCTTGATCT |
| carB | CATCGGCATTCAGCGCCAT |
| yijP | GTTTAGCCTGGACTTCTGTG |
| argB | ACGCCACCCAGTTTGATAA |
| yjeT | TGAATTCGACAATCTGGCTG |
| yjeB | TCACTCCACCAGCAATAATT |
| yigM | GTCAACCTGATACTTCGCTA |
| metE | TGCACCATGTTCGCCAGTG |
| ilvC | TAACCCGCAACAGCAATACG |
| k1, pKD4 (reference) | CAGTCATAGCCGAATAGCCT |
| k2, pKD4 (reference) | CGGTGCCCTGAATGAACTGC |
| kt, pKD4 (reference) | CGGCCACAGTCGATGAATCC |
| Complementation | |
| carAB forward | CGGCGGATCCGCTAACAGGAGGAATTAACCGTGAGTGAATATTCT |
| carAB reverse | GCGCGTCGACTTATTTGATCTGCGC |
| argE forward | CGGCGGATCCGCTAACAGGAGGAATTAACCATGAATATTGATACT |
| argE reverse | GCGCGCATGCTTAATGCCAGCAAAA |
| argC forward | CGGCGGATCCGCTAACAGGAGGAATTAACCATGTTGAATACGCTG |
| argC reverse | GCGCGTCGACTTAAATAAGAGACTG |
| purA forward | CGGCGGATCCGCTAACAGGAGGAATTAACCATGGGTAACAACGTC |
| purA reverse | GGCGGTCGACTTACGCGTCGAACGG |
| metE forward | CGGCGGATCCGCTAACAGGAGGAATTAACCATGACAATTCTTAAT |
| metE reverse | GCCGGTCGACTCATCCCCGACGCAA |
| ilvA forward | CGGCGGATCCGCTAACAGGAGGAATTAACCATGATGGCAGACTCA |
| ilvA reverse | GCGCGTCGACTTAACCCGCCAAAAA |
| ilvC forward | CGGCGGATCCGCTAACAGGAGGAATTAACCATGGCTAACTACTTC |
| ilvC reverse | GCGCGTCGACTTAACCCGCAACAGC |
Mouse model of UTI.
Female C3H/HeJ mice (8 to 10 weeks) were purchased from the Animal Resources Centre, Western Australia, and housed in sterile cages with ad libitum access to sterile water. The mouse model of UTI with competitive mixed infection was performed as previously described (1). Mice were inoculated with a mixture of 2.5 × 108 CFU of E. coli 83972AMP and 2.5 × 108 CFU of each E. coli 83972 mutant, both grown for 20 h in LB medium. At 48 h after inoculation, urine was collected from each mouse and then mice were euthanized and the bladders and kidneys were excised. Bacterial numbers in the mouse urine, bladders, and kidneys were determined by plating on selective LB agar plates and by colony counts. The mixed strains were differentiated by their antibiotic resistance with E. coli 83972AMP resistant to ampicillin and the E. coli 83972 mutants resistant to kanamycin. Competitive fitness indices were calculated by dividing the CFU/ml or CFU/0.1 g for each E. coli 83972 mutant by that of E. coli 83972AMP. Differences in colonization between E. coli 83972AMP and each of the mutants were tested using the Wilcoxon matched-pairs signed-rank test. All experiments were carried out in strict accordance with the recommendations in the Animal Care and Protection Act (25a) and the Australian Code of Practice for the Care and Use of Animals for Scientific Purposes (23a). Approval for mouse infection studies was obtained from the University of Queensland Animal Ethics Committee (SCMB/471/09/NHMRC [NF]).
Microarray analysis.
The microarray results of the amino acid biosynthesis genes were extracted from a previously published transcriptomic analysis of E. coli 83972 during growth in pooled human urine and from urine specimens collected from three patients deliberately colonized with E. coli 83972 (26). GeneChip E. coli Genome 2.0 arrays (Affymetrix) were used for hybridization of the labeled cDNA. Three chips were hybridized with samples grown in three individual flasks in MOPS (morpholinepropanesulfonic acid) medium (including 0.2% glucose and 0.02% Casamino acids), three chips were hybridized with samples from cells grown in pooled human urine in three individual flasks, and nine chips were hybridized with samples obtained in triplicate from three individual patients; no pooling of samples was performed. The arrays were normalized, and fold changes during growth in urine compared to planktonic growth in MOPS were calculated.
Microarray data accession number.
The supporting microarray data have been deposited in ArrayExpress (http://www.ebi.ac.uk/arrayexpress/) with accession number E-MEXP-584.
RESULTS
Identification of mutants deficient in urine growth.
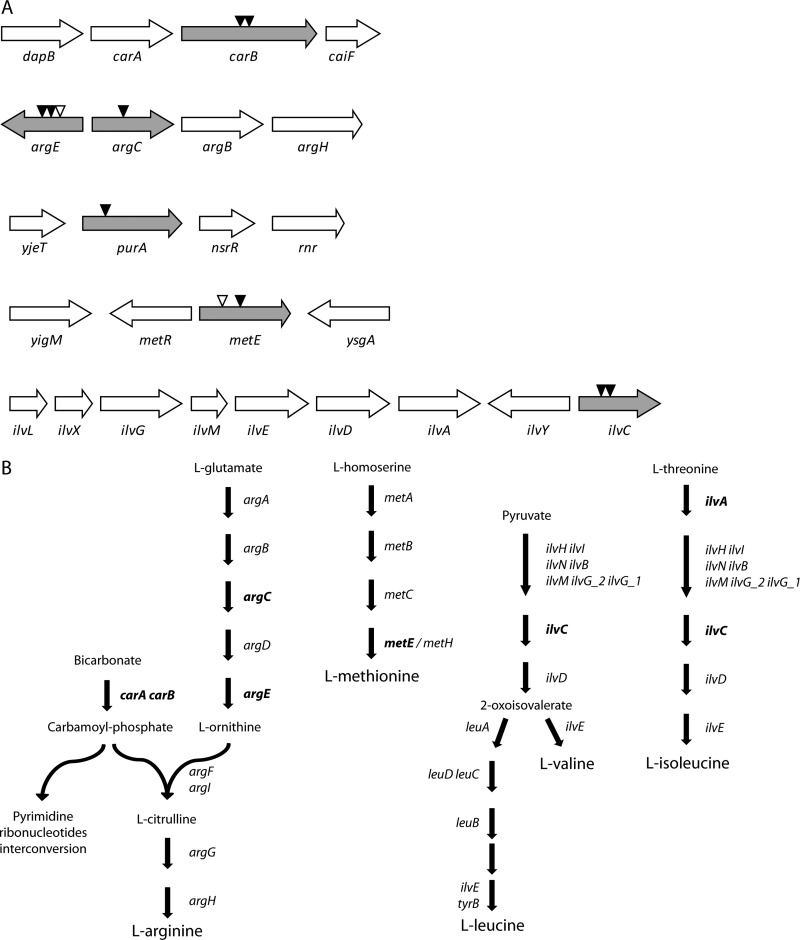
A transposon mutagenesis approach was used to identify genes that are important for the growth of two well-characterized ABU E. coli strains in human urine, i.e., the prototypic E. coli 83972 and E. coli VR89. A total of 960 and 1,152 transposon mutants of E. coli strains 83972 and VR89, respectively, were screened based on the following criteria: attenuated growth in sterile filtered pooled human urine and normal growth in LB broth. In total, 11 mutants that displayed a >90% reduction in final cell density following overnight growth in pooled human urine were identified. In order to identify the genes of interest, the location of the transposon insertions was determined by semirandom PCR. As illustrated in Fig. 1A, the insertions were located in genes related to amino acid, purine, and pyrimidine biosynthesis. Insertions were identified within carB, argE, argC, purA, metE, and ilvC. The carAB operon encodes the carbamoyl-phosphate synthetase, an essential enzyme in the biosynthesis of arginine and an important component of the pyrimidine metabolism (Fig. 1B). The metE gene product is required for the terminal step of methionine biosynthesis (Fig. 1B). The ilvC gene product is involved in one of the last common steps in the isoleucine-valine biosynthesis pathways (Fig. 1B). The purA gene encodes adenylosuccinate synthase, which catalyzes the first committed step toward the de novo synthesis of adenine. Notably, four of the six genes with insertions were identified in several mutants independently and two of the six insertions, i.e., argE and metE, were found in both strain backgrounds (Fig. 1A). argC has previously been shown to be important for the growth and virulence of a UPEC strain (31).
Fig 1.
Growth mutants identified by transposon mutagenesis. (A) Identified insertion sites of the mariner transposon in the two E. coli strains 83972 (black triangles) and VR89 (white triangles). The illustrated gene order and orientations refer to the genomic context of E. coli MG1655. (B) Biosynthesis pathways for the amino acids arginine, methionine, leucine, valine, and isoleucine. The gene(s) encoding the enzyme responsible for each step is indicated. Genes that were mutated (knocked out) in E. coli strain 83972 are indicated in boldface.
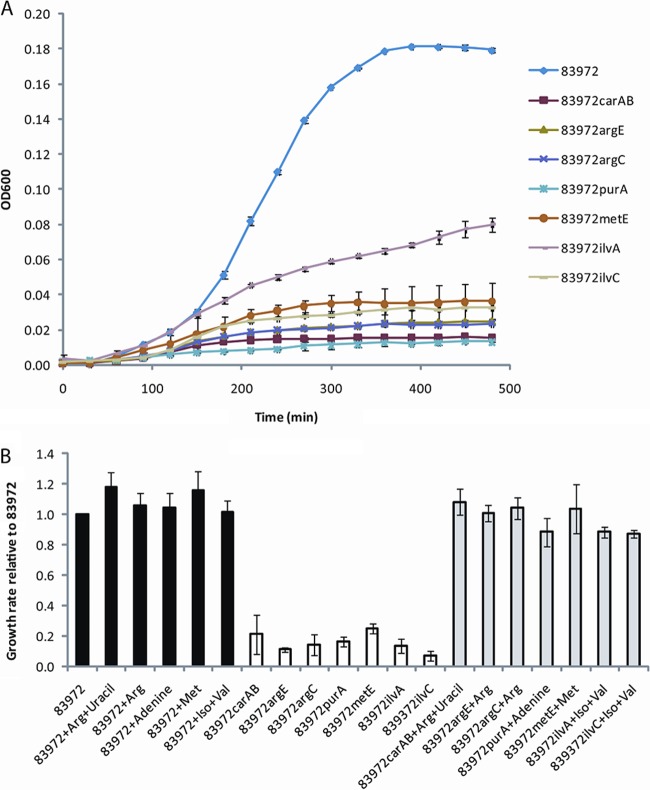
Characteristics of mutants deficient in urine growth.
To confirm the results obtained by the transposon mutagenesis screen, deletion mutants were constructed for each individual gene by λ-red-mediated homologous recombination. Six deletion mutants were made in each strain background (83972 and VR89) to validate the results. The resulting mutants were subsequently assayed for growth in sterile filtered pooled human urine. All of the mutants grew poorly in human urine (Fig. 2A). The six mutants in E. coli 83972 possessed a significantly reduced growth rate (7 to 25% of that observed for E. coli 83972) and achieved a significantly reduced final cell density (8 to 34% of that observed for E. coli 83972) (Fig. 2B and 3). Similar results were obtained in E. coli VR89 (data not shown).
Fig 2.
Growth of E. coli 83972 and its isogenic mutants in human urine. (A) The curves are shown as means of four replicates, and error bars indicate standard deviations (σn−1). (B) Growth rates of E. coli 83972 in human urine with and without addition of amino acids and/or nucleobases (black) and growth rates of its mutants in urine without (white) and with (gray) addition of amino acids and/or nucleobases. Growth rates are displayed as means of triplicates of independent experiments relative to the growth rate of E. coli 83972 grown in human urine with no addition of amino acids. Error bars indicate standard errors.
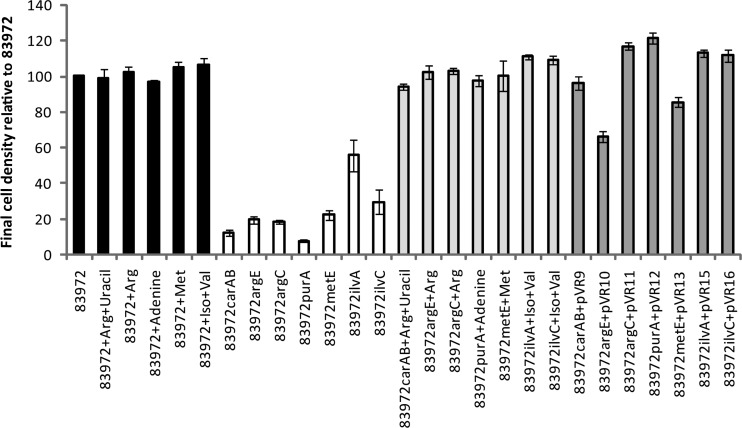
Fig 3.
Final cell density of E. coli 83972 and its isogenic mutants in human urine supplemented with the appropriate amino acids and/or nucleobases or complemented with plasmid expressing the defunct gene. Black bars, wild-type E. coli 83972 with and without addition of amino acids and/or nucleobases; white bars, isogenic mutants; light gray bars, isogenic mutants with addition of amino acids and/or nucleobases; dark gray bars, isogenic mutants complemented with plasmids expressing genes corresponding to the defunct gene. Growth characteristics of the isogenic mutants and the corresponding mutant expressing the control vector pACYC184 were identical. Values are means of at least triplicates, and error bars indicate standard errors.
IlvA catalyzes the first step in the isoleucine pathway and is not important for biosynthesis of valine or leucine. Therefore, it was expected that an ilvA mutant would be less attenuated for growth in urine than an ilvC mutant. In order to test this hypothesis, we constructed an ilvA mutant in E. coli 83972 and assessed its growth characteristics together with the ilvC mutant. As expected, when grown in urine, the ilvA mutant grew better than the ilvC mutant but was still significantly attenuated compared with the growth of the wild-type strain (Fig. 2A). The final cell density achieved by the ilvA mutant was significantly higher than those of the other mutants and represented approximately 50% of that observed for the wild-type strain (Fig. 3).
Mutants deficient in urine growth can be complemented in trans.
Each mutant generated was complemented with a plasmid expressing its respective nonfunctional gene. The final cell density following growth in pooled human urine was restored to wild-type levels for the carAB, argC, purA, metE, ilvA, and ilvC mutants (range of 86 to 121% of the final cell density of E. coli 83972) (Fig. 3). The argE mutant reached a final cell density that was 65% of that of E. coli 83972.
Mutants deficient in urine growth can be rescued by supplementation of amino acids.
In order to assess whether the various mutants could be rescued from their growth deficiency by end product complementation, each mutant was grown in the presence of the amino acids or nucleobases for which it was auxotrophic. While the growth of E. coli 83972 was not significantly altered by the presence of the supplements (growth rates and final cell densities were 97 to 118% of those for growth in urine without supplements), the growth of the mutants was fully restored by the addition of amino acids and/or nucleobases. The growth rates of the mutants were restored to 88 to 108% of that of the wild type (Fig. 2B), while the final cell densities were 94 to 111% of that of the wild type (Fig. 3). This demonstrates that these amino acids and nucleobases are limiting in human urine and that the biosynthesis pathways of arginine, methionine, valine, uracil, adenine, and isoleucine are essential for efficient growth of E. coli 83972 in human urine.
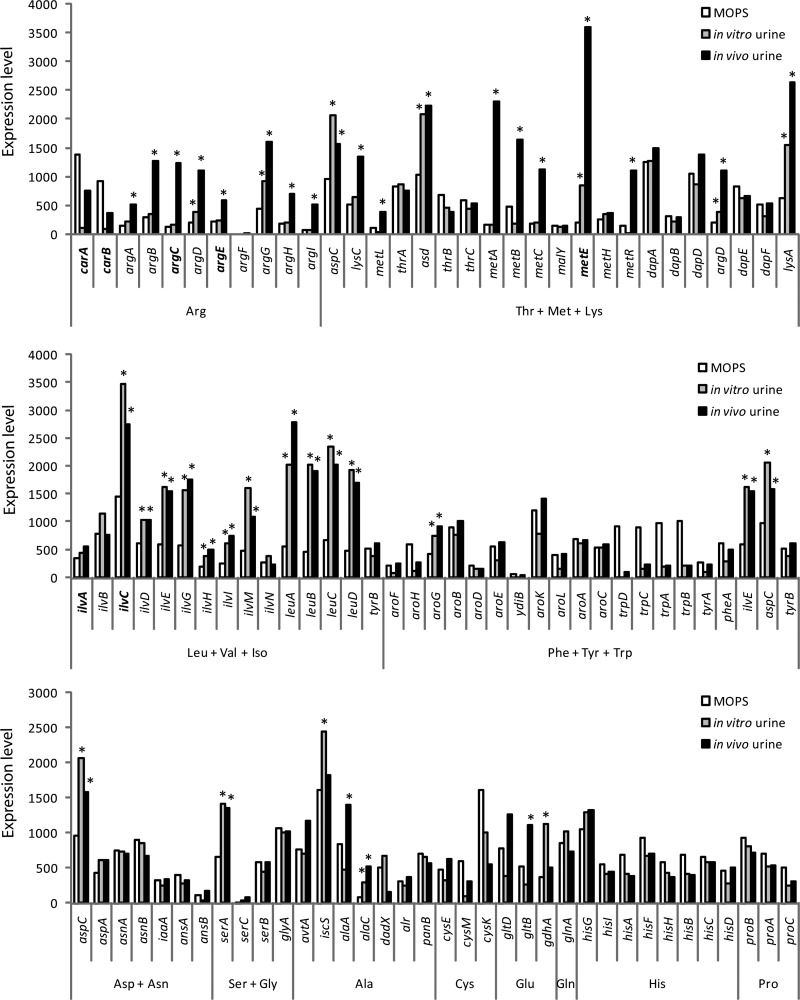
Global gene expression analysis of genes involved in amino acid biosynthesis.
We have previously examined the global gene expression profile of E. coli 83972 during growth in urine—both in vitro and in vivo (in human volunteers) (26). The expression profile of E. coli 83972 obtained during growth in urine was compared with that during exponential growth in minimal medium (MOPS-glucose including 0.02% Casamino acids). The expression levels of the genes involved in the biosynthesis of amino acids are displayed in Fig. 4. Of the six genes identified in our transposon mutagenesis screen, four were significantly upregulated during growth in urine compared to their levels during growth in minimal medium, i.e., argC, argE, metE, and ilvC were all induced 1.7- to 17-fold in vivo (Fig. 4). Indeed, the metE transcript was the 9th highest expressed in vivo of all 8,716 transcripts in the global gene expression analysis of E. coli 83972. It should also be noted that less than 5% of all 8,716 transcripts had absolute expression levels above 1,000, revealing that many amino acid biosynthesis genes were highly expressed in general. Of the 10 highest expressed amino acid-related genes in vivo, five were involved in biosynthesis of leucine, valine, and isoleucine and four were involved in biosynthesis of threonine, methionine, and lysine (Fig. 4). In contrast, genes involved in biosynthesis of phenylalanine, tyrosine, and tryptophan were among the lowest expressed of all amino acid-related genes in urine (9 of the 13 lowest among the 98 genes displayed in Fig. 4).
Fig 4.
Expression levels of E. coli 83972 in minimal medium (MOPS) and human urine (in vitro and in vivo) of all genes involved in the biosynthesis of amino acids, as determined by microarray analysis. Genes that were mutated in E. coli 83972 are indicated in boldface. Asterisks indicate expression levels in human urine that were significantly higher than those in MOPS (P < 0.05).
Interestingly, the mutations that significantly reduced growth in human urine (Fig. 1) were found among the amino acid pathways displaying the highest proportion of significantly upregulated genes (Fig. 4). In the Arg, Leu+Val+Iso, and Thr+Met+Lys pathways, 73%, 73%, and 52% of the genes, respectively, displayed significant upregulation in urine. Other pathways displaying a high proportion of induced genes were the Glu and Ala pathways (67% and 43%, respectively), while the remaining pathways contained a significantly lower proportion of induced genes (0 to 25%). Although the numbers of genes represented in the Glu and Ala pathways are comparably low, this may indicate that the biosynthesis of glutamate and alanine might also be important for growth in human urine. Taken together, the gene expression data support the growth profiles observed for our amino acid mutants and suggest that these amino acids are limiting in human urine.
Competition in pooled human urine.
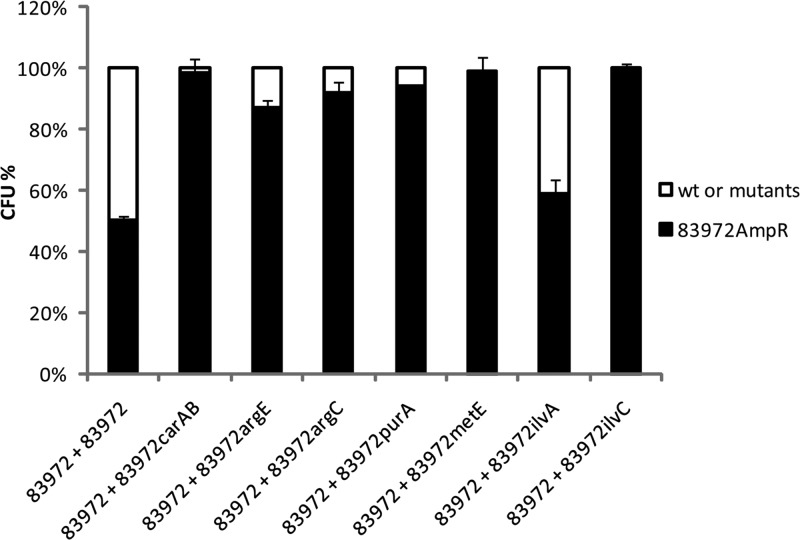
The severely attenuated growth phenotype displayed by the mutants suggested that they would be outcompeted by the parent E. coli 83972 strain during mixed growth in pooled human urine. To investigate this, we pitted an ampicillin-resistant version of E. coli 83972 (E. coli 83972AMP) against wild-type E. coli 83972 (control) and each mutant using a 1:1 inoculation ratio. The number of CFU of each strain was then enumerated after 16 h of growth. The growth of E. coli strains 83972 and 83972AMP was identical, indicating the suitability of E. coli 83972AMP for these assays. In the mixed competition assays, six of the seven mutants were significantly outcompeted by E. coli 83972AMP, which constituted 83 to 100% of the final population after 16 h of growth (Student's paired t test, P < 0.01) (Fig. 5). The exception was E. coli 83972ilvA, which was not outcompeted by E. coli 83972AMP.
Fig 5.
Competition experiment in human urine cultures between E. coli 83972 and its mutants, mixed 1:1 at the starting point. After 16 h of growth under hydrodynamic conditions at 37°C, the number of CFU of each strain in the mixed biofilm population was determined. The data shown are the means of three independent experiments in different batches of urine, and error bars indicate standard errors. wt, wild type.
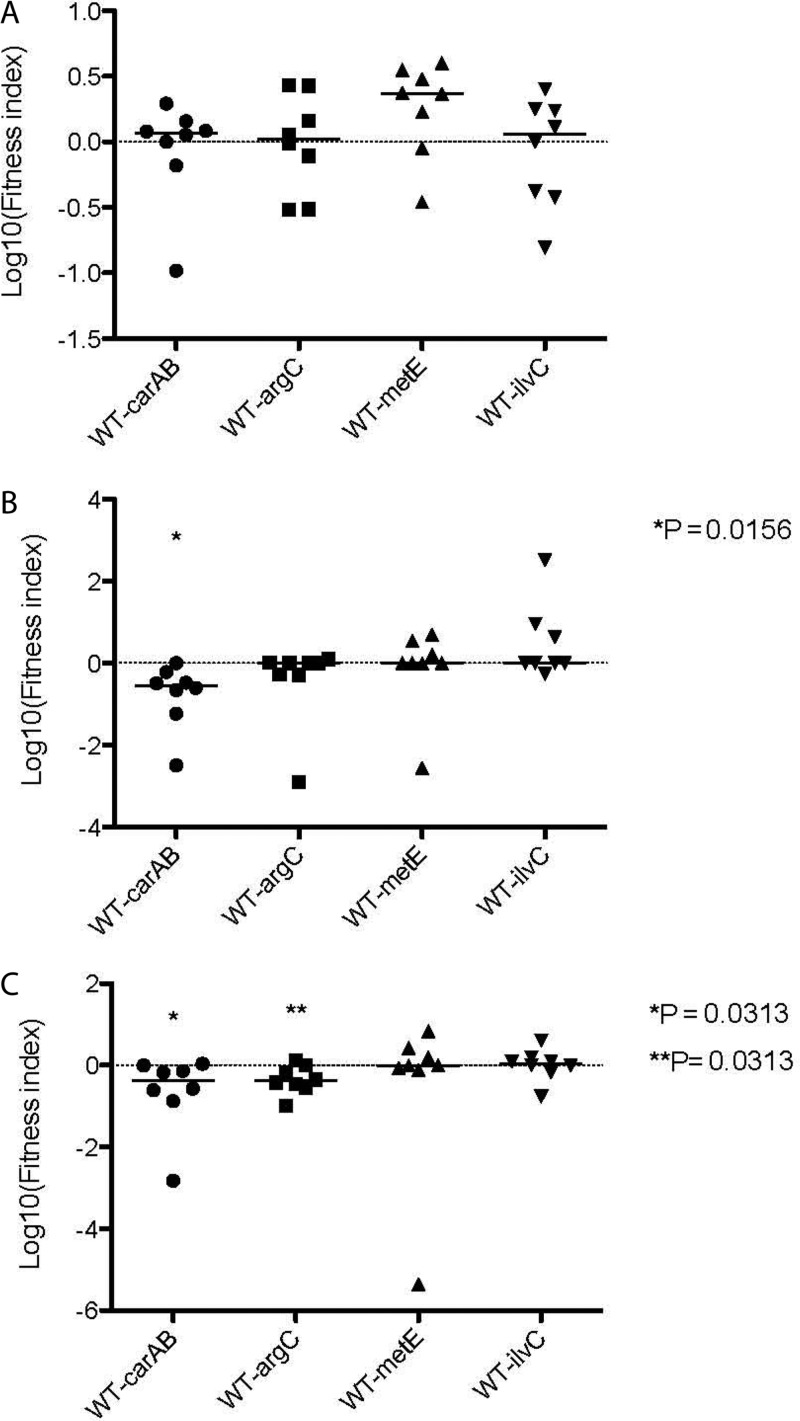
Contribution of genes required for growth in human urine to colonization of the mouse urinary tract.
To assess the role of the genes identified in our transposon mutagenesis screen on colonization of the mouse urinary tract, we examined the ability of the wild-type and mutant strains to survive in competitive-colonization experiments. Mice were coinoculated with wild-type and mutant strains (viz., 83972carAB, 83972argC, 83972metE, and 83972ilvC) in a 1:1 ratio. In the mouse model, E. coli 83972carAB was significantly outcompeted by E. coli 83972 in the bladder (P = 0.0156) and kidney (P = 0.0313) of infected mice (Fig. 6). Of the remaining mutants, only the E. coli 83972argC strain exhibited a colonization defect; this strain was significantly outcompeted by E. coli 83972 in the kidney (P = 0.0313) of infected mice. Interestingly, no difference in total bacterial counts was observed between E. coli 83972 and any of the mutants in the urine of infected mice (Fig. 6A).
Fig 6.
Competitive colonization of mouse urine, bladder, and kidneys. E. coli 83972AMP and each mutant were mixed at a 1:1 ratio and inoculated into a group of eight mice. Urine (A), bladders (B), and kidneys (C) were collected 48 h after inoculation and processed for quantitative colony counts. Asterisks indicate significant differences in colonization.
DISCUSSION
Our knowledge of the factors that are involved in the virulence of UPEC has advanced significantly over the last few decades. We currently have a relatively good understanding of the repertoire of adhesins, invasins, and toxins that are necessary for uropathogenicity (24). However, our knowledge of the physiological factors required for growth and survival of E. coli in human urine, and in the urinary tract, is limited. Relatively few reports have addressed this important issue compared to the number of studies on virulence. Nevertheless, the importance of metabolic processes in the persistence and colonization of the urinary tract by bacterial strains is increasingly appreciated (2, 3). Urine is a complex growth medium. It contains significant amounts of urea and creatinine but is iron limiting. Urine has also been reported to contain essential amino acids, purines, and pyrimidines (25). However, it is unclear if these are available in concentrations that are sufficient to support growth in the absence of de novo growth factors for synthesis. Previous reports have suggested that the biosynthesis of arginine and guanine is important for growth and virulence of uropathogens (31). In addition, a study by Hull and Hull suggested a requirement for glutamine, serine, leucine, methionine, phenylalanine, proline, and uracil (16).
Transcriptomic analyses of E. coli growing in human urine from patients and in vitro have provided valuable information about the nutritional requirement of UTI strains (11, 26, 29). In order to understand how ABU strains grow and persist in the urinary tract and to complement our transcriptomic data, we analyzed transposon mutant libraries of E. coli ABU strains 83972 and VR89. Screening for mutants deficient in urine growth identified six genes required for efficient growth in human urine. The mutants were defunct in genes involved in the biosynthesis of arginine (carB, argC, and argE), methionine (metE), valine, isoleucine, and leucine (ilvC), as well as adenine and uracil (carB and purA). Two genes, i.e., argE and metE, were identified independently in both strain backgrounds. All six mutants, in both E. coli strain 83972 and VR89, had severely affected growth rates (7 to 25% of that of the wild type) and reached a significantly reduced final cell density (8 to 34% of that of the wild type) compared with those of their isogenic parent strain. These six mutants were also easily outcompeted by the wild-type strain during competition experiments. Amino acid or nucleobase supplementation readily rescued the growth deficiency of the mutants, demonstrating that these growth factors are limiting in human urine.
To determine whether the metabolic genes identified through transposon mutagenesis were required for the growth and fitness of E. coli ABU strains during colonization of the urinary tract, four mutants were assayed in an ascending UTI mouse model, viz., E. coli 83972carAB, 83972argC, 83972metE, and 83972ilvC. In these experiments, there was no significant difference in the numbers of mutant and wild-type cells recovered from the urine of infected mice. These findings may result from differences in the composition of human and mouse urine. It is also possible that the continuous replenishment of nutrients in the urine of mice (unlike the in vitro batch culture), as well as growth in the proximity of epithelial cells and cellular debris, may contribute to these differences.
Some differences were observed with respect to bladder and kidney colonization of the E. coli mutants 83972argC and 83972carAB. The mutant strain E. coli 83972argC was impaired in the colonization of the kidneys, while E. coli 83972carAB was outcompeted in both the bladder and the kidneys. This correlates well with previous studies of an argC mutant of UPEC isolate CP9 which was also significantly impaired in colonization of the mouse kidneys but not in colonization of the bladder (31). We note that our study involved the use of Toll-like receptor 4 (TLR4)-deficient mice, a characteristic which likely accounts for the ability of E. coli 83972 to colonize the kidneys. Interestingly, a previous study showed that argG does not significantly impact the growth and persistence of UPEC strain CFT073 in the mouse UTI model (2). This may suggest that different steps in the arginine biosynthesis pathway have different effects on the fitness of UPEC strains in vivo.
Several studies have demonstrated that iron is an important limiting factor during growth in urine and that most E. coli strains that colonize the urinary tract, including ABU isolates, have numerous iron acquisition systems (10, 29). E. coli 83972 possesses functional yersiniabactin-, enterobactin-, aerobactin-, salmochelin-, and citrate-dependent iron transport systems (39, 42). Many of the genes involved in these systems are highly upregulated during growth in urine (14, 29). As many of these systems are functionally redundant with respect to iron acquisition, it was not unexpected that our mutagenesis screen did not identify any iron acquisition genes. Russo et al. found several urine-upregulated genes (i.e., iroN and ure1) that were not essential for growth in urine (30). This demonstrates the value of combining different experimental approaches, namely, mutagenesis and transcriptomics.
Although our libraries were not saturated, our findings indicate that the growth advantage displayed by E. coli 83972 compared with the growth of pathogenic strains is not related to unique biosynthetic pathways. Many of the same nutrients appear to be required for both pathogenic (3, 16, 30, 31) and asymptomatic (present study) isolates. This is interesting in terms of delineating between fitness per se and virulence. We note that the argC, ilvA, ilvC, and metE genes were also significantly upregulated (1.7- to 15-fold) in the pyelonephritis isolate CFT073 grown in human urine compared to their levels in minimal medium (data extracted from a previous microarray study [14]). This indicates that the genes identified as important for growth of the two ABU E. coli strains in the present study might also be important for growth and colonization of UPEC isolates.
Based on genomic sequencing, E. coli 83972 differs surprisingly little from highly pathogenic UTI strains, suggesting that genetic adaptation to commensalism through gene loss and point mutations—rather than distinct biosynthetic pathways—may explain its superior growth characteristics (28, 39). Many virulence factors, fimbriae in particular, are costly to produce. It remains to be determined if the loss of key virulence factors can increase fitness for growth in urine; this may very well be the case for E. coli 83972.
In conclusion, our results point to the classification of several biosynthesis pathways as bona fide bacterial fitness factors. Such findings may offer a novel target for the development of future antimicrobial agents. Compounds that specifically inhibit one or more metabolic pathways may have potential for the treatment of symptomatic urinary tract infections.
ACKNOWLEDGMENTS
We thank Birthe Bondo, Barbara Arnts, and Chelsea Stewart for excellent technical assistance.
This work was supported by a grant from Lundbeckfonden (grant no. R17-A1603). M.A.S. is supported by an ARC Future Fellowship (FT100100662) and a grant from the National Health and Medical Research Council of Australia (569676).
Footnotes
Published ahead of print 2 July 2012
REFERENCES
- 1. Allsopp LP, et al. 2010. UpaH is a newly identified autotransporter protein that contributes to biofilm formation and bladder colonization by uropathogenic Escherichia coli CFT073. Infect. Immun. 78:1659–1669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Alteri CJ, Mobley HL. 2012. Escherichia coli physiology and metabolism dictates adaptation to diverse host microenvironments. Curr. Opin. Microbiol. 15:3–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Alteri CJ, Smith SN, Mobley HL. 2009. Fitness of Escherichia coli during urinary tract infection requires gluconeogenesis and the TCA cycle. PLoS Pathog. 5:e1000448 doi:10.1371/journal.ppat.1000448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Andersson P, et al. 1991. Persistence of Escherichia coli bacteriuria is not determined by bacterial adherence. Infect. Immun. 59:2915–2921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Darouiche RO, et al. 2001. Pilot trial of bacterial interference for preventing urinary tract infection. Urology 58:339–344 [DOI] [PubMed] [Google Scholar]
- 6. Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ferrieres L, Hancock V, Klemm P. 2007. Biofilm exclusion of uropathogenic bacteria by selected asymptomatic bacteriuria Escherichia coli strains. Microbiology 153:1711–1719 [DOI] [PubMed] [Google Scholar]
- 8. Foxman B. 2002. Epidemiology of urinary tract infections: incidence, morbidity, and economic costs. Am. J. Med. 113(Suppl 1A):5S–13S [DOI] [PubMed] [Google Scholar]
- 9. Friedman L, Kolter R. 2004. Genes involved in matrix formation in Pseudomonas aeruginosa PA14 biofilms. Mol. Microbiol. 51:675–690 [DOI] [PubMed] [Google Scholar]
- 10. Garcia EC, Brumbaugh AR, Mobley HLT. 2011. Redundancy and specificity of Escherichia coli iron acquisition systems during urinary tract infection. Infect. Immun. 79:1225–1235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hagan EC, Lloyd AL, Rasko DA, Faerber GJ, Mobley HL. 2010. Escherichia coli global gene expression in urine from women with urinary tract infection. PLoS Pathog. 6:e1001187 doi:10.1371/journal.ppat.1001187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hancock V, Ferrieres L, Klemm P. 2007. Biofilm formation by asymptomatic and virulent urinary tract infectious Escherichia coli strains. FEMS Microbiol. Lett. 267:30–37 [DOI] [PubMed] [Google Scholar]
- 13. Hancock V, Seshasayee AS, Ussery DW, Luscombe NM, Klemm P. 2008. Transcriptomics and adaptive genomics of the asymptomatic bacteriuria Escherichia coli strain 83972. Mol. Gen. Genet. 279:523–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hancock V, Vejborg RM, Klemm P. 2010. Functional genomics of probiotic Escherichia coli Nissle 1917 and 83972, and UPEC strain CFT073: comparison of transcriptomes, growth and biofilm formation. Mol. Genet. Genomics 284:437–454 [DOI] [PubMed] [Google Scholar]
- 15. Hull R, et al. 2000. Urinary tract infection prophylaxis using Escherichia coli 83972 in spinal cord injured patients. J. Urol. 163:872–877 [PubMed] [Google Scholar]
- 16. Hull RA, Hull SI. 1997. Nutritional requirements for growth of uropathogenic Escherichia coli in human urine. Infect. Immun. 65:1960–1961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Johansen TE, et al. 2006. Hospital acquired urinary tract infections in urology departments: pathogens, susceptibility and use of antibiotics. Data from the PEP and PEAP-studies. Int. J. Antimicrob. Agents 28(Suppl 1):S91–S107 [DOI] [PubMed] [Google Scholar]
- 18. Kaper JB, Nataro JP, Mobley HL. 2004. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2:123–140 [DOI] [PubMed] [Google Scholar]
- 19. Klemm P, Hancock V, Schembri MA. 2007. Mellowing out: adaptation to commensalism by Escherichia coli asymptomatic bacteriuria strain 83972. Infect. Immun. 75:3688–3695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Klemm P, Roos V, Ulett GC, Svanborg C, Schembri MA. 2006. Molecular characterization of the Escherichia coli asymptomatic bacteriuria strain 83972: the taming of a pathogen. Infect. Immun. 74:781–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kulasekara HD, et al. 2005. A novel two-component system controls the expression of Pseudomonas aeruginosa fimbrial cup genes. Mol. Microbiol. 55:368–380 [DOI] [PubMed] [Google Scholar]
- 22. Lindberg U, et al. 1975. Asymptomatic bacteriuria in schoolgirls. II. Differences in Escherichia coli causing asymptomatic bacteriuria. Acta Paediatr. Scand. 64:432–436 [DOI] [PubMed] [Google Scholar]
- 23. Mabbett AN, et al. 2009. Virulence properties of asymptomatic bacteriuria Escherichia coli. Int. J. Med. Microbiol. 299:53–63 [DOI] [PubMed] [Google Scholar]
- 23a. National Health and Medical Research Council 2004. Australian code of practice for the care and use of animals for scientific purposes, 7th ed National Health and Medical Research Council, Australia: http://www.nhmrc.gov.au/_files_nhmrc/publications/attachments/ea16.pdf [Google Scholar]
- 24. Nielubowicz GR, Mobley HL. 2010. Host-pathogen interactions in urinary tract infection. Nat. Rev. Urol. 7:430–441 [DOI] [PubMed] [Google Scholar]
- 25. Parvy PR, Bardet JI, Rabier DM, Kamoun PP. 1988. Age-related reference values for free amino acids in first morning urine specimens. Clin. Chem. 34:2092–2095 [PubMed] [Google Scholar]
- 25a. Queensland Parliamentary Counsel 2009. Animal Care and Protection 2001, reprint 3A. Office of the Queensland Parliamentary Counsel, Queensland, Australia: http://www.legislation.qld.gov.au/LEGISLTN/CURRENT/A/AnimalCaPrA01.pdf [Google Scholar]
- 26. Roos V, Klemm P. 2006. Global gene expression profiling of the asymptomatic bacteriuria Escherichia coli strain 83972 in the human urinary tract. Infect. Immun. 74:3565–3575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Roos V, Nielsen EM, Klemm P. 2006. Asymptomatic bacteriuria Escherichia coli strains: adhesins, growth and competition. FEMS Microbiol. Lett. 262:22–30 [DOI] [PubMed] [Google Scholar]
- 28. Roos V, Schembri MA, Ulett GC, Klemm P. 2006. Asymptomatic bacteriuria Escherichia coli strain 83972 carries mutations in the foc locus and is unable to express F1C fimbriae. Microbiology 152:1799–1806 [DOI] [PubMed] [Google Scholar]
- 29. Roos V, Ulett GC, Schembri MA, Klemm P. 2006. The asymptomatic bacteriuria Escherichia coli strain 83972 outcompetes uropathogenic E. coli strains in human urine. Infect. Immun. 74:615–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Russo TA, Carlino UB, Mong A, Jodush ST. 1999. Identification of genes in an extraintestinal isolate of Escherichia coli with increased expression after exposure to human urine. Infect. Immun. 67:5306–5314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Russo TA, Jodush ST, Brown JJ, Johnson JR. 1996. Identification of two previously unrecognized genes (guaA and argC) important for uropathogenesis. Mol. Microbiol. 22:217–229 [DOI] [PubMed] [Google Scholar]
- 32. Salvador E, et al. 2012. Comparison of asymptomatic bacteriuria Escherichia coli isolates from healthy individuals versus those from hospital patients shows that long-term bladder colonization selects for attenuated virulence phenotypes. Infect. Immun. 80:668–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Schappert SM. 1999. Ambulatory care visits to physician offices, hospital outpatient departments, and emergency departments: United States, 1997. Vital Health Stat. 13(143):1–39 [PubMed] [Google Scholar]
- 34. Stamm WE. 1991. Catheter-associated urinary tract infections: epidemiology, pathogenesis, and prevention. Am. J. Med. 91:65S–71S [DOI] [PubMed] [Google Scholar]
- 35. Sunden F, Hakansson L, Ljunggren E, Wullt B. 2010. Escherichia coli 83972 bacteriuria protects against recurrent lower urinary tract infections in patients with incomplete bladder emptying. J. Urol. 184:179–185 [DOI] [PubMed] [Google Scholar]
- 36. Sundén F, Håkansson L, Ljunggren E, Wullt B. 2006. Bacterial interference—is deliberate colonization with Escherichia coli 83972 an alternative treatment for patients with recurrent urinary tract infection? Int. J. Antimicrob. Agents 28(Suppl 1):S26–S29 [DOI] [PubMed] [Google Scholar]
- 37. Trautner BW, Darouiche RO, Hull RA, Hull S, Thornby JI. 2002. Pre-inoculation of urinary catheters with Escherichia coli 83972 inhibits catheter colonization by Enterococcus faecalis. J. Urol. 167:375–379 [PMC free article] [PubMed] [Google Scholar]
- 38. Trautner BW, Hull RA, Darouiche RO. 2003. Escherichia coli 83972 inhibits catheter adherence by a broad spectrum of uropathogens. Urology 61:1059–1062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Vejborg RM, Friis C, Hancock V, Schembri MA, Klemm P. 2010. A virulent parent with probiotic progeny: comparative genomics of Escherichia coli strains CFT073, Nissle 1917 and ABU 83972. Mol. Genet. Genomics 283:469–484 [DOI] [PubMed] [Google Scholar]
- 40. Vejborg RM, Hancock V, Schembri MA, Klemm P. 2011. Comparative genomics of Escherichia coli strains causing urinary tract infections. Appl. Environ. Microbiol. 77:3268–3278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Watts RE, et al. 2010. Escherichia coli isolates causing asymptomatic bacteriuria in catheterized and noncatheterized individuals possess similar virulence properties. J. Clin. Microbiol. 48:2449–2458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Watts RE, et al. 2012. Contribution of siderophore systems to growth and urinary tract colonization of asymptomatic bacteriuria Escherichia coli. Infect. Immun. 80:333–344 [DOI] [PMC free article] [PubMed] [Google Scholar]