Abstract
Autophagy is used by eukaryotes in bulk cellular material recycling and in immunity to intracellular pathogens. We evaluated the role of macrophage autophagy in the response to Cryptococcus neoformans and Candida albicans, two important opportunistic fungal pathogens. The autophagosome marker LC3 (microtubule-associated protein 1 light chain 3 alpha) was present in most macrophage vacuoles containing C. albicans. In contrast, LC3 was found in only a few vacuoles containing C. neoformans previously opsonized with antibody but never after complement-mediated phagocytosis. Disruption of host autophagy in vitro by RNA interference against ATG5 (autophagy-related 5) decreased the phagocytosis of C. albicans and the fungistatic activity of J774.16 macrophage-like cells against both fungi, independent of the opsonin used. ATG5-knockout bone marrow-derived macrophages (BMMs) also had decreased fungistatic activity against C. neoformans when activated. In contrast, nonactivated ATG5-knockout BMMs actually restricted C. neoformans growth more efficiently, suggesting that macrophage autophagy plays different roles against C. neoformans, depending on the macrophage type and activation. Interference with autophagy in J774.16 cells also decreased nonlytic exocytosis of C. neoformans, increased interleukin-6 secretion, and decreased gamma interferon-induced protein 10 secretion. Mice with a conditionally knocked out ATG5 gene in myeloid cells showed increased susceptibility to intravenous C. albicans infection. In contrast, these mice manifested no increased susceptibility to C. neoformans, as measured by survival, but had fewer alternatively activated macrophages and less inflammation in the lungs after intratracheal infection than control mice. These results demonstrate the complex roles of macrophage autophagy in restricting intracellular parasitism by fungi and reveal connections with nonlytic exocytosis, humoral immunity, and cytokine signaling.
INTRODUCTION
Cryptococcus neoformans and Candida albicans are two of the most important human-pathogenic fungi. C. neoformans is a soil saprophyte with a worldwide distribution, whereas C. albicans is a commensal in the human digestive tract and skin. Despite these differences, these two microorganisms are common causes of severe systemic infections in the vast population of immunocompromised individuals that results from the AIDS pandemic, hematologic malignancies, and therapies created by modern medicine, such as transplantation, intravenous catheters, and immunosuppressive drugs.
C. neoformans is unique among the medically important fungi for bearing a polysaccharide capsule, which is its major virulence attribute (50). Cryptococcosis most commonly presents as a meningoencephalitis that is fatal when untreated and that kills about 20% of the patients, despite treatment with amphotericin B and flucytosine (34). Moreover, these drugs lead to acute renal failure in 49% to 65% of the patients (5), poignant reminders that antifungal therapy is far from optimal. The global burden of cryptococcosis exceeds 1 million cases a year, of whom about 600,000 die (33). The initial infection with C. neoformans, usually subclinical, is believed to commonly occur in early childhood (11) due to inhalation of desiccated yeast cells and spores. Upon deposition in the lung, fungal particles are phagocytosed by alveolar macrophages and the yeast cells can survive within mature phagosomes. The host response leads to the formation of granulomas containing the intracellular or extracellular yeast, which herald the state of latency (8, 21).
C. albicans is a commensal in most humans but can cause a wide range of diseases, especially in hosts with impaired immunity (19). The most common ones are superficial infections of the oropharyngeal, esophageal, or vaginal mucosa and skin. In the presence of risk factors such as immunosuppression, use of antibacterial therapy, major abdominal surgery, or placement of venous and urinary catheters, C. albicans cells can breach epithelial barriers and reach the bloodstream, from where they disseminate to cause invasive candidiasis with a mortality of 30 to 40% (9). Macrophages are among the most important immune cells in controlling C. albicans cells in tissues and the bloodstream, both because they phagocytose and kill the fungus and because they secrete proinflammatory cytokines (47).
Autophagy is a mechanism by which cells recycle cytoplasmic material and organelles. It is conserved through all Eukarya and has been shown to play important roles in homeostasis and pathogenesis (26). Several forms of autophagy are recognized, but only macroautophagy will be dealt with in this report. This process results in the sequestration of intracellular material inside a double-membrane vesicle named an autophagosome. The formation of these vesicles requires two ubiquitin-like conjugates, one formed by ATG5 (autophagy-related 5) and ATG12 and the other formed by LC3 (microtubule-associated protein 1 light chain 3 alpha) and phosphatidylethanolamine (20). After their formation, autophagosomes fuse with lysosomes, resulting in the digestion and recycling of nutrients (26).
In addition to its role in nutritional homeostasis, autophagy has recently been established as a mechanism of innate immunity against viruses, bacteria, and protozoa (6). The formation of autophagosomes around pathogens found in the cytoplasm, such as Sindbis virus (22) or Listeria monocytogenes (39), leads to their clearance by lysosomal degradation. It also allows the cell to overcome pathogen-mediated inhibition of phagosomal maturation, as demonstrated by the gamma interferon (IFN-γ)-dependent, autophagy-mediated clearance of Mycobacterium tuberculosis (12). A recent study found that autophagy is also involved in the interaction between C. neoformans and macrophages (37). Screening an RNA interference library, the autophagy genes Atg2, Atg5, and Atg9 were each identified to be necessary for C. neoformans phagocytosis and replication in and escape from Drosophila S2 cells and murine macrophage cell lines.
Given that cryptococcosis and invasive candidiasis are intimately related to the host immune status, it is important to understand how the host interacts with and controls these fungi. In this study, we addressed the role of macrophage autophagy in this interaction, showing that it participates in multiple steps of the in vitro and in vivo immune response against C. neoformans and C. albicans.
MATERIALS AND METHODS
Strains and reagents.
C. neoformans var. grubii (strain H99, serotype A) and C. albicans (strain SC5314) cultures were grown in Sabouraud dextrose broth (Difco) at 37°C with agitation at 150 rpm for 1 to 2 days. J774.16 murine macrophage-like cells were obtained from ATCC and cultivated in feeding medium (Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum, 10% NCTC 109 medium, and 1% nonessential amino acids) at 37°C with 10% CO2. pEGFP-LC3 was a kind gift from Beth Levine (Dallas, TX), obtained under permission from Noboru Mizushima (Tokyo, Japan). IgG1 murine monoclonal antibody (MAb) to cryptococcal capsular polysaccharide 18B7 (3) was purified from hybridoma culture supernatants. 1,1-Dioctadecyl-3,3,3,3-tetramethylindocarbocyanine perchlorate (DiI; Vybrant DiI cell labeling solution; Invitrogen) was used at a final concentration of 5 μg/ml.
Animals.
C57BL/6 mice (6- to 8-week-old females) obtained from the National Cancer Institute (NCI) were used to obtain primary macrophages by peritoneal lavage. Mice with myelocyte-specific ATG5 conditional knockout have been described (52) and were obtained as breeding pairs from Herbert Virgin (St. Louis, MO). The conditional-knockout and control mice were obtained by crossing two animals homozygous for a flox-flanked ATG5 exon (ATG5flox) (13); one of the mice was also heterozygous for the Cre recombinase expressed under the control of the lysozyme gene (Lyz Cre+). Pups were genotyped by PCR as previously described (51). As the two strains used to generate the conditional-knockout and control mice were from two different backgrounds (129S and C57BL/6), the resulting pups were not isogenic, and thus, all experiments were performed with littermates. Bone marrow-derived macrophages (BMMs) were obtained by flushing bone marrow cells from 6- to 8-week-old male or female mice and differentiating them in Dulbecco's modified Eagle's medium supplemented with 20% L929 supernatant for 7 days. Macrophages were assayed at days 6 to 7.
Experiments with mice in this study were carried out in strict observance of Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC) and National Institutes of Health (NIH) guidelines. All experiments were approved by the Albert Einstein College of Medicine Institutional Animal Care and Use Committee (IACUC).
ATG5 knockdown by shRNA lentiviral transduction.
RNA interference was used to reduce expression of ATG5, a gene that is essential for autophagy (27). J774.16 cells were stably transduced with lentiviral vectors encoding five different small hairpin RNAs (shRNAs) that target ATG5 and one scrambled shRNA without targets in the murine genome. Transduced cells were then selected and maintained in medium supplemented with 5 μg/ml puromycin. Knockdown efficiency was assessed by immunoblotting using rabbit polyclonal antibody to murine ATG5 (Santa Cruz Biotechnology), followed by detection with horseradish peroxidase and chemiluminescence (Thermo Scientific). Sample loading was controlled by stripping the membranes and reprobing with mouse monoclonal antibody to β-actin. To obtain optimal ATG5-knockdown efficiency, individual transduced cells were cloned by limiting dilution, and those ones with the lowest ATG5 expression were selected.
Macrophage-fungus interaction assays. (i) C. neoformans.
Two different assays were used to study the interaction between C. neoformans and macrophages. For the killing assay, described in detail elsewhere (29), shRNA-transduced J774.16 cells were plated on 96-well tissue culture plates at a density of 2.5 × 104 cells per well in feeding medium supplemented with or without 100 U/ml recombinant IFN-γ (Roche) and 500 ng/ml lipopolysaccharide (LPS; Sigma-Aldrich). After 24 h, these cells were infected with C. neoformans cells opsonized either with MAb 18B7 (10 μg/ml) to the polysaccharide capsule of C. neoformans or with mouse serum complement (Pel-Freez) at an effector-to-target cell ratio of 1:1. After 24 h of infection, J774.16 cells were lysed with distilled water and the number of viable C. neoformans cells was determined by counting of the CFU on Sabouraud dextrose agar plates after incubation at 30°C for 2 days. The second assay measured phagocytosis kinetics and was performed according to a recently described method (31). Briefly, J774.16 cells and C. neoformans were independently labeled with the fluorescent dyes 9-H-(1,3-dichloro-9,9-dimethylacridin-2-one-7-yl)-succinimidyl ester (DDAO-SE) and 5-chloromethylfluorescein diacetate (CMFDA), respectively. After phagocytosis, the cells were incubated with the cell wall stain Uvitex 2B (Polysciences Inc.), which stains only fungi that have not been phagocytosed, and 7-amino-actinomycin D (7-AAD), which stains dead macrophages. The killing assays were repeated at least four times with at least three samples per condition each time, whereas the phagocytosis assay was done once in triplicate.
(ii) C. albicans.
shRNA-transduced cells were plated at 105 cells per well in an 8-chamber polystyrene tissue culture glass slide (Becton Dickinson) and grown overnight before use in the phagocytosis assay. C. albicans cells were collected after 24 h of growth and washed 3× in phosphate-buffered saline (PBS). Yeast cells were added to the macrophage monolayer at an effector/target cell ratio of 1:5, and the suspension was incubated at 37°C for 30 min. Cocultures were then washed with Hank's buffered saline solution (HBSS; pH 7.2), and the slides were stained with 0.01% acridine orange (Sigma-Aldrich) for 45 s by the method of Pruzanski and Saito (36). The slides were gently washed with HBSS and stained for 45 s with 0.05% crystal violet (Sigma-Aldrich) dissolved in 0.15 M NaCl. Finally, the slides were rinsed 3 times with PBS, mounted on microscope coverslips, and sealed at the edge with nail polish. The phagocytic index was determined by a fluorescence microscope (Axiovert 200 M inverted microscope; Zeiss). For each experiment, the macrophages in 5 fields in each well were counted, and at least 100 macrophages were analyzed in each well. The phagocytic index was the ratio of the number of intracellular yeast cells to the number of macrophages counted. Colony counts were made to determine the number of viable C. albicans yeast cells after phagocytosis. For CFU determination, shRNA-transduced and control J774.16 macrophages were infected with C. albicans cells as described above. After 30 min of incubation, macrophages were lysed by forcibly pulling the culture through a 26-gauge needle 5 times. The lysates were serially diluted and plated on Sabouraud dextrose agar at 37°C. CFU determinations were made after 48 h. Controls also consisted of yeast cells grown without macrophages. All tests were repeated at least three times.
Immunofluorescence microscopy.
J774.16 cells or primary peritoneal macrophages were plated on poly-l-lysine-coated 35-mm glass-bottom plates (MatTek) and allowed to adhere for 24 h. The cell monolayers were then infected with C. neoformans or C. albicans at effector-to-target cell ratios of 1:1 or 1:10, and the cells were coincubated for different times between 15 min and 24 h. At the desired time interval, the cells were fixed and permeabilized with methanol at −20°C for 10 min and stained with rabbit polyclonal antibody to LC3, followed by a fluorescein-conjugated secondary. In some experiments, Uvitex 2B was used as well to highlight the fungal cells. After staining, the coverslips were sealed in mounting medium (0.1 M propyl gallate in PBS with 50% glycerol) and imaged under one of three different microscopes: an Olympus microscope equipped with a ×100 1.3-numerical-aperture (NA) objective for regular epifluorescence, a Zeiss Axioskop 200 M microscope equipped with a ×63 1.4-NA objective for regular epifluorescence and deconvolution microscopy, and a Leica SP2 laser-scanning confocal microscope for confocal microscopy. The LC3 immunofluorescence microscopy experiments were repeated numerous times, with similar results.
Live cell imaging.
J774.16 cells were transiently transfected with pEGFP-LC3 using Lipofectamine 2000 according to the manufacturer's instructions. C. neoformans cells were collected from an overnight culture, washed twice in PBS, and resuspended at 107 cells/ml in PBS containing 5 μM DDAO-SE or 5-(and-6)-chloromethyl seminaphthorhodafluor acetate (CM-SNARF 1). The labeling reaction was carried out overnight on an orbital shaker at 37°C. At approximately 24 h after transfection, J774.16 cells were transferred to a glass-bottom 35-mm dish (MatTek) and incubated for 24 h in Earle's balanced salts solution (EBSS) or feeding medium supplemented with 100 U/ml IFN-γ and 0.5 μg/ml LPS. The plates were then transferred to a heated-stage Perkin-Elmer UltraView RS-3 spinning-disk confocal microscope in an atmosphere of humidified 5% CO2. Labeled C. neoformans (10:1 yeast/macrophage ratio) and 10 μg/ml MAb 18B7 were added immediately before imaging started. In some experiments, the macrophages were loaded with the lysosomal marker 10-kDa dextran-Texas Red (Invitrogen) between transfection with pEGFP-LC3 and infection with C. neoformans. The live-imaging experiment was repeated at least three times.
Image analysis.
Epifluorescence images were analyzed with AxioVision (Zeiss), ImageJ, and Photoshop software. Epifluorescence z stacks were deconvolved with a constrained iterative algorithm with AxioVision. Spinning-disk confocal image sets were manipulated on ImageJ and Photoshop software. Images were collected in color on an Olympus microscope; all other images were collected in gray scale and pseudocolored later. All spinning-disk confocal images were enhanced for contrast and brightness to highlight the weak signal.
Transmission electron microscopy.
J774.16 cells were grown on 6-well tissue culture dishes for 24 h and infected with C. neoformans at a 1:1 effector-to-target cell ratio in the presence of 10 μg/ml 18B7. After 12 h, the cells were fixed with 2.5% glutaraldehyde in 0.1 M sodium cacodylate buffer, postfixed with 1% osmium tetroxide followed by 2% uranyl acetate, dehydrated through a graded series of ethanol, and embedded in LX112 resin (LADD Research Industries, Burlington, VT). Ultrathin sections were cut on a Reichert Ultracut UCT ultramicrotome, stained with uranyl acetate followed by lead citrate, and viewed on a JEOL 1200EX transmission electron microscope at 80 kV.
Quantification of nonlytic exocytosis by flow cytometry.
Nonlytic exocytosis of C. neoformans was quantified using a flow cytometric method as described previously (31). Briefly, J774.16 and C. neoformans cells were each independently labeled with the cell tracking dyes DDAO-SE and CMFDA, respectively, and coincubated for 2 h. The monolayer was then stained with Uvitex 2B and 7-AAD, followed by flow cytometric sorting of macrophages with internalized fungi. The sorted cells were incubated for 24 h and then analyzed by flow cytometry again to determine the percentage of macrophages that underwent nonlytic exocytosis. The results from multiple experiments were pooled for statistical analysis, as done before (2, 24, 29).
Murine survival experiments. (i) C. neoformans.
For intraperitoneal infection, 500 μl of C. neoformans suspension containing 105 to 107 cells of strain H99 cells in PBS was injected in the peritoneal cavity. For intratracheal infections, mice were anesthetized with 100 mg/kg of body weight ketamine and 10 mg/kg of body weight xylazine. An incision was made on the neck, and 50 μl of C. neoformans suspensions containing 2.5 × 104 to 106 C. neoformans strain H99 in PBS was injected into the trachea using a 250-μl glass syringe and a 26-gauge needle. The incision was closed using veterinary adhesive (Vetbond; 3M). For survival studies, infected mice were inspected daily and euthanized by CO2 inhalation in case of distress. Each survival experiment had at least 10 littermate mice of the same sex and age per group.
(ii) C. albicans.
For survival studies, mice were infected by inoculation of 5 × 105 C. albicans cells in the tail vein and inspected daily. The experiment was performed with 10 littermate female mice (age, 6 to 8 weeks) per group.
Lung and brain fungal burden.
ATG5-conditional-knockout and control littermate mice were infected intratracheally with 105 C. neoformans strain H99 cells and euthanized 3, 7, and 14 days later. Their lungs and brains were removed and homogenized in sterile PBS. The homogenates were serially diluted and plated on Sabouraud agar. After incubation for 2 days at 30°C, the number of colonies was counted and multiplied by the dilution to determine the number of CFU. There were five female mice aged 6 to 8 weeks per group per time interval.
Quantification of cytokines.
Samples were prepared for cytokine quantification using a multiplexed microsphere assay (Luminex, Austin, TX) and a kit with reagents for 22 cytokines and chemokines: granulocyte colony-stimulating factor, granulocyte-macrophage colony-stimulating factor, IFN-γ, interleukin-10 (IL-10), IL-12 (p70), IL-13, IL-15, IL-17, IL-1α, IL-1β, IL-2, IL-4, IL-5, IL-6, IL-7, IL-9, IFN-γ-induced protein 10 (IP-10), keratinocyte-derived cytokine, monocyte chemoattractant protein 1, macrophage inflammatory protein 1α (MIP-1α), RANTES, and tumor necrosis factor alpha (TNF-α) (Millipore, Billerica, MA). The first sample to be quantified was prepared by plating the control and ATG5 shRNA-transduced J774.16 cells on petri dishes and infecting with C. neoformans strain H99 opsonized with 10 μg/ml 18B7 for 24 h at a 1:10 macrophage/C. neoformans ratio. The supernatant was then collected, centrifuged, filtered, supplemented with a cocktail of protease inhibitors (Roche), and frozen until analyzed. The analysis was performed with supernatants from three independent experiments. The other samples were lung homogenates from the day 14 fungal burden assay described above; immediately after plating for determination of the numbers of CFU, a cocktail of protease inhibitors was added to the homogenates and they were frozen until analyzed. The analysis was performed with lung homogenates from five mice per group.
Histology.
ATG5-conditional-knockout and control mice were euthanized by CO2 inhalation and exsanguination at 3, 7, and 14 days after intratracheal infection with 105 C. neoformans strain H99 cells. Five littermate females aged 6 to 8 weeks per group per time point were used. The lungs, spleen, liver, and brain were dissected, fixed in 10% neutral formalin, routinely processed to paraffin, sectioned (5 μm), and stained with hematoxylin-eosin. Samples for immunohistochemistry (IHC) were sectioned to a thickness of 5 μm and deparaffinized in xylene followed by graded alcohols. Antigen retrieval was performed in 10 mM sodium citrate buffer at pH 6.0, heated to 96°C, for 30 min. Endogenous peroxidase activity was blocked using 0.3% hydrogen peroxide in water. The sections were stained by routine IHC methods with the primary antibody to Ym-1 (R&D Systems, Minneapolis, MN), followed by horseradish peroxidase-conjugated mouse IgG secondary antibody. The antibody binding was detected using an avidin-biotin-horseradish peroxidase system, followed by the use of diaminobenzidine as the chromogen. All immunostained sections were lightly counterstained with hematoxylin. Both hematoxylin-eosin and IHC slides were evaluated semiquantitatively by a board-certified veterinary pathologist.
Statistical analysis.
The results of the in vitro experiments with macrophages and the fungal burden and cytokine quantification assays were compared by two-way analysis of variance (ANOVA) using GraphPad Prism software (GraphPad Software Inc., La Jolla, CA). Nonlytic exocytosis results were compared by two-tailed chi-square test with Yate's correction (GraphPad Software Inc.). Survival in murine infection studies was compared by log-rank (Mantel-Cox) test using GraphPad Prism software. Results from multiple survival studies were evaluated with Cox proportional hazards regression using IBM SPSS software.
RESULTS
Reduction in autophagy alters antifungal activity of macrophages.
To determine whether autophagy is involved in macrophage immunity against fungi, we reduced ATG5 expression with RNA interference. This was achieved with stable transduction of lentiviral vectors in J774.16 cells with shRNAs that targeted different regions of the ATG5 gene. J774.16 clones transduced with three of these shRNAs, with ATG5 expression levels of 25% to 63% compared with the control (Fig. 1A), were infected with C. albicans or C. neoformans to assess phagocytosis and killing of the pathogens. We found no significant effect of ATG5 knockdown in phagocytosis of C. neoformans after 2 h of coincubation (Fig. 1B) but observed significantly decreased phagocytosis of C. albicans after 30 min of coincubation (Fig. 1C). A series of CFU experiments was then done to evaluate C. neoformans killing. At 24 h after incubation, ATG5 knockdown in J774.16 cells resulted in a dose-dependent decrease of fungistatic activity against both antibody (Ab)- and complement-opsonized C. neoformans (Fig. 1D and E). At 2 h of coincubation, there was no significant difference in the number of CFU independent of the activation status (data not shown). The C. albicans killing assays could not be done with long coincubation times because filamentation made it impossible to count fungal cells. However, after 30 min of coincubation we observed a significant reduction in the ability of J774.16 cells to kill C. albicans (Fig. 1F).
Fig 1.
Lack of ATG5 decreases phagocytosis and intracellular killing of C. neoformans and C. albicans. (A) Two different shRNA sequences targeting murine ATG5 were transduced into J774.16 cells (clones 30 and 31), as was a scrambled shRNA control (C−). After selection with puromycin and cloning, J774.16 clones were evaluated by immunoblotting for ATG5 and β-actin expression. The proportion of ATG5 expression in comparison with the scrambled shRNA control was calculated by densitometry, and the two clones with the highest knockdown efficiency were selected for further experiments. (B) Antibody-opsonized C. neoformans phagocytosis after 2 h of coincubation using ATG5 shRNA-transfected J774.16 cells. The y axis represents the percentage of total macrophages with internalized C. neoformans cells. Bars represent the means of three independent experiments. (C) C. albicans phagocytosis after 30 min of coincubation with ATG5 shRNA J774.16 cells. The y axis represents the phagocytic index, calculated by dividing the number of internalized fungal cells by the total number of macrophages. Bars represent the means of five independent experiments. (D and E) C. neoformans killing assays after 24 h of incubation with ATG5 shRNA-transfected J774.16 cells. Monoclonal antibody and serum complement were used as opsonins in panels D and E, respectively. Bars represent the means and SEMs of 4 to 16 independent experiments. (F) C. albicans killing assay after 30 min of coincubation with ATG5 shRNA-transfected J774.16 cells. Bars represent the means and SEMs of 3 independent samples. (G and H) Secretion of IL-6 and IP-10 by shRNA-transfected cells. Each clone was incubated for 24 h with or without antibody-opsonized C. neoformans, and the supernatants were used for cytokine quantification. Bars represent the means and SEMs of 3 independent samples. *, P < 0.05 on the Bonferroni posttest that followed ANOVA analyses.
To confirm the results obtained with J774.16 cells with primary cells, BMMs were isolated from ATG5-conditional-knockout and littermate control mice. These macrophages efficiently restricted growth of Ab- and complement-opsonized C. neoformans, and their anticryptococcal activity was enhanced by IFN-γ and LPS stimulation. In contrast to the result obtained with ATG5 shRNA J774.16 cells, nonactivated BMMs showed better killing of C. neoformans when ATG5 was knocked out. Upon activation, however, a higher number of CFU was recovered from ATG5-knockout macrophages infected with complement-opsonized but not those infected with Ab-opsonized C. neoformans (see Fig. S1 in the supplemental material).
In short, the effect of autophagy on macrophage antifungal activity depended on the opsonin, macrophage type, and activation state. In J774.16 cells, autophagy knockdown decreased phagocytosis and killing of C. albicans and fungistasis of both complement- and antibody-opsonized C. neoformans. In nonactivated BMMs, autophagy knockout increased fungistasis of C. neoformans, whereas in activated BMMs, autophagy knockout decreased antifungal activity.
Autophagy knockdown alters the cytokine response to C. neoformans.
Our next step was to study the role of autophagy on secretion of cytokines, both on macrophages alone and on macrophages infected with C. neoformans. The shRNA-transduced J774.16 cells were cultivated with or without antibody-opsonized C. neoformans for 24 h, and the supernatants were collected to quantify 22 different cytokines and chemokines (see Table S1 in the supplemental material). J774.16 cells transduced with two different shRNAs targeting ATG5 secreted more IL-6 after infection with C. neoformans (Fig. 1G). An opposite trend was observed with the chemokine IP-10 (CXCL-10), which was significantly reduced in the supernatants of ATG5 shRNA cells, both with and without C. neoformans infection (Fig. 1H).
Phagosomes containing fungi have the autophagy-specific marker LC3.
After observing that autophagy contributed to fungistasis and cytokine secretion, we studied the mechanisms that could be involved in this process. Initially, we infected both J774.16 cells and primary macrophages with C. neoformans or C. albicans and performed immunofluorescence against LC3, a specific autophagosome marker (18). These experiments revealed that after Ab-mediated phagocytosis, some of the C. neoformans phagosomes became autophagosomes in J774.16 cells (Fig. 2A) and in primary macrophages (see Fig. S2A in the supplemental material). A control experiment performed with J774.16 cells in which the LC3 antibody was omitted (Fig. 2B) showed that the LC3-positive phagosome was not an artifact caused by direct binding of the secondary antibody. Kinetic experiments revealed that autophagosome formation could be observed at as early as 30 min and as late as 24 h after infection but was most frequently observed at about 12 h after infection (data not shown). At 12 h after infection with Ab-opsonized C. neoformans, 36 of 44 (82%) macrophages with internalized C. neoformans contained the fungi in LC3-positive phagosomes. Of note, we did not observe LC3-positive phagosomes in 54 of the J774.16 cells infected with complement-opsonized C. neoformans at 12 h postinfection (Fig. 2C).
Fig 2.
Phagosomes containing C. neoformans acquire LC3. (A) LC3 localization in J774.16 cells infected with antibody-opsonized C. neoformans for 12 h. (B) Negative control done in parallel with the images shown in panel C. The samples were processed and imaged identically, except for the exclusion of the LC3 antibody. (C) Effect of the opsonin on LC3 localization in J774.16 cells infected with C. neoformans for 12 h. (D) Montage of time-lapse images of EGFP-LC3-tranfected J774.16 cells infected with C. neoformans. From the top left, each frame represents a single slice through the center of the cell, with an interval between each image of 24 min. The complete time-lapse series is included in Movie S1 in the supplemental material. (A to C) z-stack projections of stacks collected on a Leica SP2 point-scanning microscope. LC3 (green) was labeled by immunofluorescence, and the cell wall (blue) was labeled with Uvitex 2B. DIC, differential interference contrast. Bars, 10 μm.
To understand how the autophagosome was formed, J774.16 cells were transiently transfected with enhanced green fluorescent protein (EGFP)-tagged LC3 for live imaging of C. neoformans phagocytosis. These movies confirmed with a different methodology that autophagosomes formed around C. neoformans hours after infection and suggested that LC3 was recruited by sequential fusion of small autophagosomes with the C. neoformans vacuole (Fig. 2D; see Movie S1 in the supplemental material). In addition, inclusion of the endosomal marker 10-kDa dextran-Texas Red suggested that this structure was formed after fusion of lysosomes with the C. neoformans vacuole and that the small autophagosomes that fused were not autophagolysosomes (see Fig. S2B in the supplemental material).
Transmission electron microscopy was used to search for ultrastructural hallmarks of autophagy such as vesicles with double membranes. We imaged J774.16 cells 12 h after infection with C. neoformans, as this was the time when autophagosomes were more frequent (see Fig. S2C in the supplemental material). Additionally, we examined archival transmission electron microscopy images collected from the lungs of C. neoformans-infected mice from a previous study (10) (data not shown). We did not observe autophagosome hallmarks such as double membranes surrounding fungal cells or undigested cytosolic contents inside the C. neoformans vacuole in any of these experiments. Instead, consistent with the live-imaging results, we observed double-membrane vesicles in close proximity to and fusing with the C. neoformans vacuole.
In contrast, in J774.16 cells infected with C. albicans for 2 h, most fungal cells were contained in LC3-positive vacuoles (Fig. 3A). The autophagosome marker could be found surrounding both yeast and hyphal forms of the fungus. Two negative-control experiments confirmed that the observed signal came from macrophage LC3 surrounding the fungal cells (Fig. 3B). Later experiments showed that LC3 is recruited to phagosomes containing C. albicans at all time points tested between 15 min and 24 h, with occasional images suggesting that even C. albicans cells that were not fully phagocytosed were already LC3 positive (see Fig. S3A in the supplemental material). Surprisingly, we observed that some C. albicans cells that were clearly not internalized or even in close proximity to macrophages were nevertheless LC3 positive (see Fig. S3B in the supplemental material).
Fig 3.
Phagosomes containing C. albicans acquire LC3. (A) LC3 localization in J774.16 cells infected with C. albicans for 2 h. (B) Negative controls done in parallel with the images shown in panel A. The images show samples that were processed and imaged exactly like those in panel A, but with two differences. On the left-side images, the LC3 antibody was omitted to test if the secondary antibody recognized nonspecifically any macrophage structure. The right-side images show C. albicans cells that were incubated without macrophages and stained with the LC3 and secondary antibodies to test if the antibody recognized C. albicans. The controls show autofluorescent cells but not the bright LC3 layer surrounding the internalized C. albicans. LC3 (green) was labeled by immunofluorescence, and all images were collected by epifluorescence on a Zeiss microscope. Bars, 10 μm.
Thus, in C. albicans-infected macrophages, LC3 is recruited to the membranes of most phagosomes shortly after phagocytosis. In contrast, in C. neoformans-infected macrophages, the recruitment of LC3 takes a longer time, happens in only a fraction of the infected macrophages, and depends on the opsonin, as only antibody-opsonized fungi end up in LC3-positive vacuoles.
Autophagy knockdown reduces C. neoformans nonlytic exocytosis.
During the epifluorescence and live-imaging experiments, we observed C. neoformans cells that were not inside macrophages but were surrounded by a layer of LC3-containing material after infection of both J774.16 cells (see Fig. S4A in the supplemental material) and primary murine macrophages (data not shown). To ascertain the validity and reproducibility of this observation, we infected J774.16 cells with antibody-opsonized C. neoformans in a tissue culture dish, washed the noningested fungi after 2 h, and 24 h after infection collected the supernatant, which should be enriched with fungal cells that had been exocytosed. Half of these cells were fixed in cold methanol, and both fixed and nonfixed samples were stained with LC3 antibody, showing that LC3 surrounded cells in both samples. Additionally, some cells were also stained with the lipophilic probe DiI, previously shown to stain lipid bilayers outside the C. neoformans cell wall (30). None of the LC3 layers in either fixed or nonfixed cells stained with DiI (data not shown), suggesting that those structures contained the LC3 protein but not a lipid bilayer. To exclude the possibility of nonspecific binding of antibodies to C. neoformans, an aliquot of the fungal cells that were used for infection was stained and imaged under identical conditions, without any observable fluorescence surrounding the cells (see Fig. S4B in the supplemental material). A similar observation is apparent at the end of a movie in which we captured the autophagosome formation (see Fig. S4C and Movie S2 in the supplemental material): after accumulation of EGFP-LC3 around the C. neoformans cells, they were expelled from the cytoplasm of the macrophage to the medium. This movie probably does not depict a genuine nonlytic exocytosis event, though, because the J774.16 cell appeared to be dead at the end, likely due to phototoxicity and incubation in EBSS with no serum or nutrients. Together with our serendipitous observations during the LC3 immunolocalization studies, this led us to hypothesize that autophagy was involved in nonlytic exocytosis, and we examined whether autophagy knockdown altered the rate of nonlytic exocytosis. Using flow cytometry to measure nonlytic exocytosis as we recently described (31), two of the ATG5-knockdown J774.16 strains had 10% to 20% decreased rates of nonlytic exocytosis of C. neoformans compared to the control cells (Table 1).
Table 1.
ATG5 shRNA decreases nonlytic exocytosis of C. neoformans
| Clone | No. of cells |
% exocytosis | P valuea | |
|---|---|---|---|---|
| Total | Exocytosed | |||
| Clone 31 | 86,116 | 41,445 | 48 | <0.001 |
| C−b | 102,459 | 61,123 | 60 | |
| Clone 30 | 80,568 | 49,671 | 62 | <0.001 |
| C− | 54,582 | 37,581 | 69 | |
P values were calculated by Fisher's exact test with the Yates correction. Results were pooled from two independent experiments with clone 30 and three independent experiments with clone 31.
C−, scrambled shRNA control.
Infection of mice with conditional knockout of macrophage autophagy.
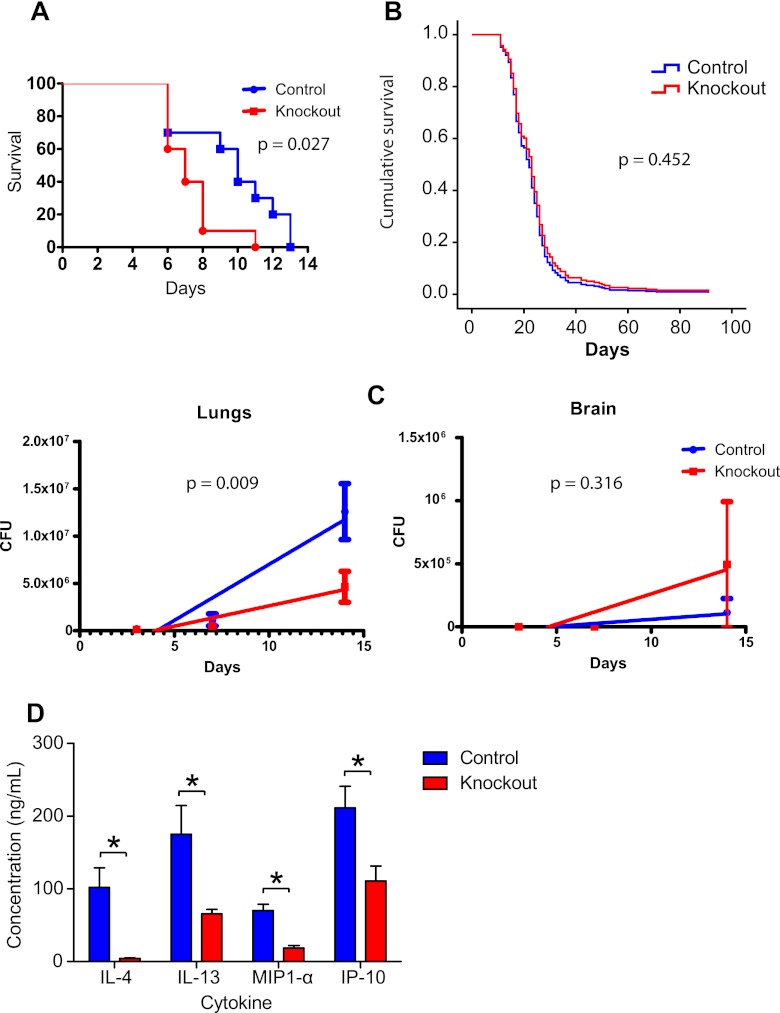
To evaluate the in vivo significance of the results that we achieved with in vitro studies, we used murine models of cryptococcosis and systemic candidiasis with conditional knockout of ATG5 in macrophages. These knockout mice died faster than controls when infected intravenously with C. albicans (Fig. 4A). Experiments with C. neoformans were done using two different routes of infection (intraperitoneal and intratracheal), different inocula (from 2.5 × 104 to 106 C. neoformans cells per mouse), and mice at different ranges of age and sex, but no statistically significant differences were found (see Fig. S5 in the supplemental material). Even when the results of all C. neoformans survival experiments were combined using Cox proportional hazards regression, there was no significant difference in survival between the two strains (Fig. 4B).
Fig 4.
ATG5-conditional-knockout increases murine mortality due to candidiasis but not cryptococcosis. (A) Mouse survival experiment after intravenous C. albicans infection. Each group had 10 mice. (B) Cox proportional hazards regression combining the results of seven survival experiments with mice infected with C. neoformans using different routes and inocula. No statistically significant difference in survival was observed in any individual experiment (see Fig. S5 in the supplemental material) or when the data from all animals (84 per group) were combined. (C) Lung and brain fungal burdens of ATG5-conditional-knockout mice infected intratracheally with 105 C. neoformans cells. Samples were collected from five mice per group at each time interval (3, 7, and 14 days after infection). (D) Lung cytokine quantification. The four cytokines with statistically significant differences in concentration in the lung homogenates are shown. Bars represent means and SEMs. *, P < 0.05 in the Bonferroni posttest that followed a two-way ANOVA.
To further investigate the reason why there was no difference in murine survival despite the fact that autophagy-negative macrophages were less able to contain C. neoformans, we measured fungal burden, cytokine expression, and tissue inflammation in infected mice. We chose the intratracheal route with an inoculum of 105 C. neoformans cells; under these conditions, mice start dying 16 days after infection, so the evaluations were done at 3, 7, and 14 days. C. neoformans burdens at the primary infection site, lungs, and at the target organ, brain, were quantified by CFU enumeration. At all three times evaluated, the numbers of CFU in the lungs were lower in the ATG5-conditional-knockout mice, and this difference was statistically significant when analyzed by linear regression (Fig. 4C). On the other hand, the brain fungal burden showed no statistically significant difference (P > 0.05). On day 14, in addition to CFU counting, we used the lung homogenates to quantify cytokines and chemokines (see Table S2 in the supplemental material). ATG5-conditional-knockout mice had lower concentrations of IL-4, IL-13, MIP-1α, and IP-10 in the lung homogenates than control mice (Fig. 4D).
We also evaluated the histology of cryptococcal intratracheal infection in ATG5-conditional-knockout and control mice (Fig. 5A to C). In both groups of mice, spleens and livers had no notable findings. The brains had lesions only at day 14. The most striking findings were in the lungs of infected mice, with a marked difference between ATG5-conditional-knockout and control mice. Differences in the lung lesions on day 3 were limited, although only 2 of 5 ATG5-conditional-knockout mice had pyogranulomatous inflammation with intralesional C. neoformans, whereas 4 of 5 ATG5 control mice had such lesions. On day 7, small tan-red lesions appeared in the lungs of most mice from both groups upon gross inspection. While 5 of 5 ATG5-conditional-knockout mice but only 3 of 5 control mice had pyogranulomatous inflammation, the inflammation in the knockout mice was less severe and contained notably fewer neutrophils than that in the control animals. On day 14, the gross pulmonary lesions in the controls grew to large translucent nodules, whereas the small tan-red lesions in ATG5-conditional-knockout mice were mostly unchanged (Fig. 5D). Histologically, the amount of airspace free of inflammation and/or C. neoformans cells in the ATG5-conditional-knockout animals was, on average, nearly double that in the control mice (48% versus 28%, respectively). Control mice, therefore, had more significant pulmonary disease than the ATG5-conditional-knockout mice. Control mice also had greater perivascular lymphocytic cuffing (moderate), generally a reflection of the severity of acute lung inflammation, than ATG5-conditional-knockout animals (minimal).
Fig 5.
ATG5 conditional knockout alters the murine immune response to intratracheal C. neoformans infection. (A to C) Histology of the lungs of ATG5-conditional-knockout and control mice 3, 7, and 14 days after intratracheal infection with 105 C. neoformans cells. Hematoxylin-eosin-stained sections were evaluated semiquantitatively by a veterinary pathologist. Five mice were used per group per time interval. (A) The arrow points to a reactive vessel. (B) The arrows point to infiltrated neutrophils with dense polymorphic nuclei and scant cytoplasm, whereas the arrowheads indicate infiltrated macrophages with round nuclei and large cytoplasm. (C) The arrowheads point to alveoli that are not occupied by C. neoformans or inflammatory cells in the lungs of conditional-knockout mice. (D) Macroscopic appearance of the lungs from 3 ATG5-conditional-knockout mice and 3 control mice infected with C. neoformans for 14 days showing large translucent nodules in the controls and smaller tan-red nodules in the ATG5-conditional-knockout animals. (E) IHC localization of Ym-1, a marker of alternatively activated macrophages, on lung sections from days 3 and 14. The arrows point to clusters of alternatively activated macrophages on the lungs of control mice infected with C. neoformans for 3 days. Bars, 500 μm (images at ×2.5 and ×4 magnifications) and 50 μm (images at ×40 magnification).
Together, the results of the experimental cryptococcosis models suggested that knocking out ATG5 in macrophages could alter the balance of classical and alternative activation of macrophages. To test this hypothesis further, lung sections taken from mice infected 3 and 14 days before were used for immunohistochemistry with an antibody to Ym-1, a chitinase family protein that is specifically produced by macrophages activated by Th2 cytokines (38). On day 3, several foci of Ym-1-positive cells were visible in the lungs from control mice, whereas foci were rare in the ATG5-conditional-knockout mice. On day 14, there was no difference between the two groups of mice, with both presenting intense labeling throughout the lung (Fig. 5E).
In conclusion, infection of ATG5-conditional-knockout mice with C. albicans results in decreased survival, whereas C. neoformans infections result in no difference in survival. However, ATG5-conditional-knockout mice have decreased lung fungal burden, decreased lung cytokines (IL-4, IL-13, MIP-1α and IP-10), less intense pyogranulomatous pneumonia, and delayed alternative activation of macrophages.
DISCUSSION
The importance of autophagy in the response against intracellular pathogens has been recognized by studies that have used a broad variety of viral, bacterial, and protozoan pathogens to uncover the diverse roles of autophagy mechanisms as effectors and regulators of innate and adaptive immunity (6). Our results indicate the involvement of autophagy mechanisms in cellular and host defense against the fungal pathogens C. neoformans and C. albicans, with the caveat that their effects appear to be dependent on the fungal species, macrophage type, opsonin, and activation state. The key experiments described in this report are summarized in Table S3 in the supplemental material.
Our first experiment established the involvement of autophagy by disrupting the autophagic machinery, followed by evaluating its effects on the ability of macrophages to phagocytose fungi and kill or restrict their intracellular growth. In vitro experiments with J774.16 cells were done using RNA interference instead of pharmacological autophagy inhibitors because these drugs could also inhibit the pathogen's own autophagic machinery, which is dispensable for C. albicans survival in macrophages (32) but necessary for C. neoformans virulence (16). Infection of J774.16 cells transduced with shRNA revealed that control cells efficiently phagocytosed and killed C. albicans, whereas those with decreased autophagy were less able to ingest and control the pathogen. These results suggest that the macrophage response against C. albicans is mediated in part by autophagy. The experiments with C. neoformans, on the other hand, had more complex results. With J774.16 cells, interference with autophagy did not alter phagocytosis but significantly decreased fungistatic activity. This result contrasts with the previous report of an approximately 25% reduction in intracellular replication of C. neoformans in macrophages with siRNA against ATG5 (37), perhaps due to differences in the macrophage cell line used (J774.16 versus RAW264.7), in the methodology used to measure macrophage antifungal activity (killing assay versus fluconazole protection assay), or in the multiplicity of infection (1:1 versus 1:10, macrophages/C. neoformans cells). With BMMs, the effect of ATG5 knockout in fungistatic activity was dependent on the activation of macrophages. Nonactivated ATG5-knockout BMMs were less permissive to fungal growth, as described for RAW264.7 cells. In contrast, when the BMMs were activated by IFN-γ and LPS, the lack of ATG5 made them more permissive to fungal growth. These results suggest that macrophage activation might increase antifungal activity by inducing autophagy, as has been described with M. tuberculosis (12).
Having shown that autophagy has a function in the response of macrophages to C. neoformans and C. albicans, we localized the autophagy marker LC3 on infected J774.16 cells. We observed that in some of the J774.16 cells containing C. neoformans, the fungal phagosome contained LC3. These LC3-positive C. neoformans vacuoles were observed at as early as 30 min after infection but were more easily observed at later times of infection. Qin et al. also observed LC3-positive phagosomes containing C. neoformans in 40 to 60% of infected J774.A1 cells; however, they reported a faster kinetics, with a peak of LC3 positivity 3 h after infection, somewhat before the acquisition of the lysosomal marker LAMP-1 (37). Our data with the lysosomal marker dextran-Texas Red suggest that LC3-positive vacuoles are formed after phagolysosomal fusion. Transmission electron microscopy and live imaging with EGFP-tagged LC3 confirmed that this marker was acquired by sequential fusion with small LC3-positive vesicles and not by a de novo formation of a double-membrane autophagosome. In contrast to what was observed with C. neoformans, most C. albicans phagosomes were LC3 positive shortly after and apparently even during phagocytosis. A similar pattern of fast LC3 recruitment was reported for phagocytosis of zymosan particles (40). This work also showed that LC3 recruitment was mediated by recognition of the zymosan by Toll-like receptor 2 (TLR2), a pattern recognition receptor that recognizes C. albicans cell wall components (48). Our results thus suggest that LC3 recruitment to the C. albicans phagosome might be mediated by recognition of pathogen-associated molecular patterns leading to increased clearance of the fungus. TLR2, however, is probably of little importance in C. neoformans infection because the polysaccharide capsule masks recognition of cell wall components (28, 49). Consequently, the differential recognition by TLR2 could explain why phagosomes containing C. albicans acquire LC3 immediately, whereas the ones containing C. neoformans do not.
As C. neoformans is not phagocytosed by macrophages unless an opsonin is present, we were able to dissect the effects of autophagy on pathogens that were internalized by antibody- or complement-mediated phagocytosis. In C. neoformans killing assays using both types of autophagy-deficient macrophages, we observed that the effect of autophagy knockdown on fungistatic activity was more pronounced upon complement opsonization than in the presence of antibody. However, we never observed LC3-positive C. neoformans-containing vacuoles in multiple experiments in which the fungi were opsonized with complement. Murine IgG1, such as the one used in this work, mediates phagocytosis of C. neoformans mostly via Fcγ receptor III (41) or complement receptor 3 (CR3), even in the absence of complement (44). In contrast, capsule-bound complement fragment C3b mostly interacts with CR3 (4). These receptors have different downstream effectors, so the absence of LC3 in phagosomes containing complement-opsonized C. neoformans and in a fraction of those containing antibody-opsonized fungi suggests an inhibitory signal from CR3 engagement with regard to the recruitment of the autophagy machinery.
Taken together, these results show that in vitro macrophage autophagy is important in immunity against C. neoformans and C. albicans. For some pathogens that escape into the cytoplasm, autophagy works by pathogen encasement inside membranes that can then be fused with lysosomes for degradation (22, 39). C. neoformans and C. albicans, however, remain inside a vacuole. For other pathogens, such as Mycobacterium tuberculosis (12), encasing the bacterium inside autophagosomes helps the host cell overcome pathogen-driven blockage of phagosomal maturation, acidification, and fusion with lysosomes. This must not be the case here, as the C. neoformans vacuole matures rapidly (21) and maturation should be complete by the time that fusion with autophagosomes starts. Instead, fusion with autophagosomes could deliver a toxic payload, as described for M. tuberculosis (1). In that work, the authors purified autophagosomal contents and detected mycobactericidal peptides derived from digested ubiquitin. Recent studies with M. tuberculosis expanded the notion of autophagosomal delivery of antimicrobial peptides, which are derived from ribosomal protein S30 (also known as ubiquicidin) and polyubiquitinated proteins targeted to autophagosomes by the adaptor p62 (35). One of the antimicrobial peptides described by these authors as being active against M. tuberculosis, ubiquicidin, has also been shown to affect C. albicans (46). It is thus feasible that autophagy inhibits antibody-opsonized C. neoformans by engulfing and digesting intracellular material that is then delivered to the fungal vacuole, leading to the accumulation of antifungal peptides. An additional possibility is that the antifungal mechanism affected by interference with ATG5 is autophagosome independent, such as that described for Toxoplasma gondii (51). Lack of macrophage ATG5 impaired IFN-γ-mediated recruitment of the Irga6 GTPase to the parasitophorous vacuole, resulting in decreased immunity to T. gondii. This effect happened in spite of the fact that no LC3 was recruited to the parasitophorous vacuole, suggesting a possible explanation for our observations with complement-opsonized C. neoformans.
Besides phagocytosis, macrophages have other important activities in immunity to fungi. These phagocytes secrete cytokines and chemokines that attract other cells of the immune system and modulate their action. Autophagy has extensive links with cytokine signaling, being induced by proinflammatory cytokines such as IFN-γ, TNF-α, and IL-1 and inhibited by anti-inflammatory or Th2-type cytokines such as IL-4, IL-10, and IL-13. Autophagy also controls the production and secretion of IL-1, IL-18, TNF-α, and type I IFN (14). With this in mind, we tested the production of 22 cytokines and chemokines by ATG5 shRNA-transduced J774.16 cells both with and without infection with C. neoformans. Autophagy-deficient cells secreted more IL-6, a cytokine that has been shown to augment macrophage activity against C. neoformans (43). This result could be interpreted as an autocrine loop circumventing the decreased antifungal activity of the macrophage. Interference with ATG5 also decreased secretion of IP-10, which was observed in the two clones of shRNA-transduced J774.16 cells both with and without infection. IP-10 was also reduced in the lungs of ATG5-conditional-knockout mice 14 days after infection with C. neoformans. These two very different experimental situations nevertheless point to a specific role for ATG5 in secreting this chemokine. This result agrees with a previous report that treating the murine intestinal epithelial cell line Mode-K with the autophagy inhibitor 3-methyladenine decreased IP-10 secretion by inhibiting the secretion of vesicles containing the chemokine (15). IP-10 is a chemokine secreted by monocytes, fibroblasts, and epithelial and endothelial cells, among others, and increases chemotaxis of monocytes/macrophages and natural killer, dendritic, and T cells (23). It is also involved in orchestrating neutrophilic pneumonia (25), so the reduction in IP-10 could explain the decreased neutrophilic infiltrates that we observed in the ATG5-conditional-knockout mice.
We explored whether macrophage autophagy was involved in nonlytic exocytosis of C. neoformans from macrophages based on the serendipitous finding that some extracellular C. neoformans cells were covered in an LC3-positive layer. This observation suggested that these cells were once contained in an autophagosomal compartment and had retained LC3 upon their exit from the host cells. Control experiments demonstrated that this LC3 cover was derived from the macrophage, as the fungal protein was not recognized by the LC3 antibody. This observation resembles the mechanism of nonlytic release of poliovirus that hijacks cellular autophagosomes to assemble the machinery that mounts viral particles (42). Assembled virions are encased by autophagosomes which fuse with the cytoplasmic membrane, releasing viruses inside an LC3-positive vesicle without lysing the host cell (17). To test if this was the case, we stained exocytosed C. neoformans cells with LC3 antibody and the membrane probe DiI. However, none of the LC3-positive layers bound the probe, suggesting that only LC3 protein and not autophagosome membranes remained in exocytosed fungi. Subsequent measurements of nonlytic exocytosis in the ATG5 shRNA-transfected J774.16 cells showed that autophagy is indeed involved in this phenomenon. A similar finding was also reported by Qin et al. (37). Using a fluconazole protection assay to measure escape of C. neoformans cells from RAW264.7 macrophages, they have shown that transfection with siRNA against ATG5 decreased escape by about 25% (37). Autophagy is also involved in other instances of nonlytic release of lysosomal contents. In microglial cells, LC3-positive bacterium-containing phagolysosomes have been shown to fuse with the cytoplasmic membrane and release their contents into the medium (45). Moreover, ATG5-dependent fusion of lysosomes with the plasma membrane has also been observed in osteoclasts (7). Along with our data, these studies with other types of autophagy-dependent exocytosis may help provide an understanding of the mechanism by which C. neoformans nonlytic exocytosis occurs.
After demonstrating these effects of autophagy on the interactions of macrophages with two fungi, we used ATG5-conditional-knockout mice to study the physiological role of autophagy during experimental C. albicans and C. neoformans infections. In the first experiment, we infected these mice with the two fungi and compared their survival with that of the controls. With intravenous C. albicans infection, the results were consistent with those observed in vitro: median survival of 6 days for conditional-knockout mice and 10 days for controls. This result demonstrates that antifungal autophagy is also important in vivo. In contrast, multiple experiments with C. neoformans using different inocula and routes of infection in mice of different ages and sexes showed no difference in survival rates. This result is not very surprising, bearing in mind that the in vitro results point to a balance of decreased macrophage fungistatic activity on the one hand and decreased nonlytic exocytosis plus increased secretion of proinflammatory IL-6 on the other hand. To better understand how cryptococcosis developed in mice in which macrophages lack autophagy, we used these mice in fungal burden, histopathology, IHC, and cytokine dosing experiments. While the control mice developed an intense pneumonia with large neutrophilic infiltrates and early alternative activation of macrophages, the conditional-knockout mice developed a less intense pneumonia and later alternative activation. These results suggest that mice that lack autophagy mount an altogether different immune response to C. neoformans infection.
The results presented here suggest that macrophage autophagy is involved in several steps in the interaction of these cells with two different fungal pathogens, C. neoformans and C. albicans. This reinforces the importance of immunological autophagy, which is involved in the response to an ever increasing number of microbes. As autophagy is involved in various other neoplastic, neurodegenerative, and autoimmune diseases (26), it is a prime pharmacological target. Our work suggests that autophagy-modulating drugs could also be used to improve antifungal therapy.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Institutes of Health (grants HL059842, AI033774, AI033142, and AI052733 to A.C., AI52733 to J.D.N., and 1K22A1087817-01A1 to L.R.M.), the Center for AIDS Research at the Albert Einstein College of Medicine, an Irma T. Hirschl/Monique Weill-Caulier Trust Research award to J.D.N., a CAPES/Fulbright scholarship to P.A., and a LIU-Post-Faculty Research grant to L.R.M.
We thank Martha Feldmesser, Beth Levine, Ana Maria Cuervo, Noboru Mizushima, Herbert Virgin, and Dee Dao, all of whom provided invaluable materials or helpful insights. We also thank Rani Sellers at the Einstein Histotechnology and Comparative Pathology facility for help with the murine studies and critical reading of the manuscript, as well as personnel at the Analytical Imaging and Flow Cytometry core facilities for technical assistance.
Footnotes
Published ahead of print 18 June 2012
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1. Alonso S, Pethe K, Russell DG, Purdy GE. 2007. Lysosomal killing of Mycobacterium mediated by ubiquitin-derived peptides is enhanced by autophagy. Proc. Natl. Acad. Sci. U. S. A. 104:6031–6036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Alvarez M, Casadevall A. 2006. Phagosome extrusion and host-cell survival after Cryptococcus neoformans phagocytosis by macrophages. Curr. Biol. 16:2161–2165 [DOI] [PubMed] [Google Scholar]
- 3. Casadevall A, et al. 1998. Characterization of a murine monoclonal antibody to Cryptococcus neoformans polysaccharide that is a candidate for human therapeutic studies. Antimicrob. Agents Chemother. 42:1437–1446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cross CE, Collins HL, Bancroft GJ. 1997. CR3-dependent phagocytosis by murine macrophages: different cytokines regulate ingestion of a defined CR3 ligand and complement-opsonized Cryptococcus neoformans. Immunology 91:289–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Deray G. 2002. Amphotericin B nephrotoxicity. J. Antimicrob. Chemother. 49(Suppl 1):37–41 [DOI] [PubMed] [Google Scholar]
- 6. Deretic V, Levine B. 2009. Autophagy, immunity, and microbial adaptations. Cell Host Microbe 5:527–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Deselm CJ, et al. 2011. Autophagy proteins regulate the secretory component of osteoclastic bone resorption. Dev. Cell 21:966–974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dromer F, Casadevall A, Perfect JR, Sorrell T. 2011. Cryptococcus neoformans: latency and disease. In Heitman J, Kozel TR, Kwon-Chung JK, Perfect JR, Casadevall A. (ed), Cryptococcus from human pathogen to model yeast. ASM Press, Washington, DC [Google Scholar]
- 9. Eggimann P, Bille J, Marchetti O. 2011. Diagnosis of invasive candidiasis in the ICU. Ann. Intensive Care 1:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Feldmesser M, Kress Y, Novikoff P, Casadevall A. 2000. Cryptococcus neoformans is a facultative intracellular pathogen in murine pulmonary infection. Infect. Immun. 68:4225–4237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Goldman DL, et al. 2001. Serologic evidence for Cryptococcus neoformans infection in early childhood. Pediatrics 107:E66 doi:10.1542/peds.107.5.e66 [DOI] [PubMed] [Google Scholar]
- 12. Gutierrez MG, et al. 2004. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell 119:753–766 [DOI] [PubMed] [Google Scholar]
- 13. Hara T, et al. 2006. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature 441:885–889 [DOI] [PubMed] [Google Scholar]
- 14. Harris J. 2011. Autophagy and cytokines. Cytokine 56:140–144 [DOI] [PubMed] [Google Scholar]
- 15. Hoermannsperger G, et al. 2009. Post-translational inhibition of IP-10 secretion in IEC by probiotic bacteria: impact on chronic inflammation. PLoS One 4:e4365 doi:10.1371/journal.pone.0004365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hu G, et al. 2008. PI3K signaling of autophagy is required for starvation tolerance and virulence of Cryptococcus neoformans. J. Clin. Invest. 118:1186–1197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jackson WT, et al. 2005. Subversion of cellular autophagosomal machinery by RNA viruses. PLoS Biol. 3:e156 doi:10.1371/journal.pbio.0030156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kabeya Y, et al. 2000. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 19:5720–5728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kim J, Sudbery P. 2011. Candida albicans, a major human fungal pathogen. J. Microbiol. 49:171–177 [DOI] [PubMed] [Google Scholar]
- 20. Levine B, Mizushima N, Virgin HW. 2011. Autophagy in immunity and inflammation. Nature 469:323–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Levitz SM, et al. 1999. Cryptococcus neoformans resides in an acidic phagolysosome of human macrophages. Infect. Immun. 67:885–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Liang XH, et al. 1998. Protection against fatal Sindbis virus encephalitis by beclin, a novel Bcl-2-interacting protein. J. Virol. 72:8586–8596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Liu M, et al. 2011. CXCL10/IP-10 in infectious diseases pathogenesis and potential therapeutic implications. Cytokine Growth Factor Rev. 22:121–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ma H, Croudace JE, Lammas DA, May RC. 2006. Expulsion of live pathogenic yeast by macrophages. Curr. Biol. 16:2156–2160 [DOI] [PubMed] [Google Scholar]
- 25. Michalec L, et al. 2002. CCL7 and CXCL10 orchestrate oxidative stress-induced neutrophilic lung inflammation. J. Immunol. 168:846–852 [DOI] [PubMed] [Google Scholar]
- 26. Mizushima N, Levine B, Cuervo AM, Klionsky DJ. 2008. Autophagy fights disease through cellular self-digestion. Nature 451:1069–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mizushima N, et al. 2001. Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J. Cell Biol. 152:657–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nakamura K, et al. 2006. Limited contribution of Toll-like receptor 2 and 4 to the host response to a fungal infectious pathogen, Cryptococcus neoformans. FEMS Immunol. Med. Microbiol. 47:148–154 [DOI] [PubMed] [Google Scholar]
- 29. Nicola AM, Casadevall A. 2012. In vitro measurement of phagocytosis and killing of Cryptococcus neoformans by macrophages. Methods Mol. Biol. 844:189–197 [DOI] [PubMed] [Google Scholar]
- 30. Nicola AM, Frases S, Casadevall A. 2009. Lipophilic dye staining of Cryptococcus neoformans extracellular vesicles and capsule. Eukaryot. Cell 8:1373–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nicola AM, Robertson EJ, Albuquerque P, Derengowski Lda S, Casadevall A. 2011. Nonlytic exocytosis of Cryptococcus neoformans from macrophages occurs in vivo and is influenced by phagosomal pH. mBio 2(4):e00167–11 doi:10.1128/mBio.00167-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Palmer GE, Kelly MN, Sturtevant JE. 2007. Autophagy in the pathogen Candida albicans. Microbiology 153:51–58 [DOI] [PubMed] [Google Scholar]
- 33. Park BJ, et al. 2009. Estimation of the current global burden of cryptococcal meningitis among persons living with HIV/AIDS. AIDS 23:525–530 [DOI] [PubMed] [Google Scholar]
- 34. Perfect JR, et al. 2010. Clinical practice guidelines for the management of cryptococcal disease: 2010 update by the Infectious Diseases Society of America. Clin. Infect. Dis. 50:291–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ponpuak M, et al. 2010. Delivery of cytosolic components by autophagic adaptor protein p62 endows autophagosomes with unique antimicrobial properties. Immunity 32:329–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pruzanski W, Saito S. 1988. Comparative study of phagocytosis and intracellular bactericidal activity of human monocytes and polymorphonuclear cells. Application of fluorochrome and extracellular quenching technique. Inflammation 12:87–97 [DOI] [PubMed] [Google Scholar]
- 37. Qin QM, et al. 2011. Functional analysis of host factors that mediate the intracellular lifestyle of Cryptococcus neoformans. PLoS Pathog. 7:e1002078 doi:10.1371/journal.ppat.1002078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Raes G, et al. 2002. Differential expression of FIZZ1 and Ym1 in alternatively versus classically activated macrophages. J. Leukoc. Biol. 71:597–602 [PubMed] [Google Scholar]
- 39. Rich KA, Burkett C, Webster P. 2003. Cytoplasmic bacteria can be targets for autophagy. Cell. Microbiol. 5:455–468 [DOI] [PubMed] [Google Scholar]
- 40. Sanjuan MA, et al. 2007. Toll-like receptor signalling in macrophages links the autophagy pathway to phagocytosis. Nature 450:1253–1257 [DOI] [PubMed] [Google Scholar]
- 41. Saylor CA, Dadachova E, Casadevall A. 2010. Murine IgG1 and IgG3 isotype switch variants promote phagocytosis of Cryptococcus neoformans through different receptors. J. Immunol. 184:336–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Schlegel A, Giddings TH, Jr, Ladinsky MS, Kirkegaard K. 1996. Cellular origin and ultrastructure of membranes induced during poliovirus infection. J. Virol. 70:6576–6588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Siddiqui AA, Shattock RJ, Harrison TS. 2006. Role of capsule and interleukin-6 in long-term immune control of Cryptococcus neoformans infection by specifically activated human peripheral blood mononuclear cells. Infect. Immun. 74:5302–5310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Taborda CP, Casadevall A. 2002. CR3 (CD11b/CD18) and CR4 (CD11c/CD18) are involved in complement-independent antibody-mediated phagocytosis of Cryptococcus neoformans. Immunity 16:791–802 [DOI] [PubMed] [Google Scholar]
- 45. Takenouchi T, et al. 2009. The activation of P2X7 receptor impairs lysosomal functions and stimulates the release of autophagolysosomes in microglial cells. J. Immunol. 182:2051–2062 [DOI] [PubMed] [Google Scholar]
- 46. Tollin M, et al. 2003. Antimicrobial peptides in the first line defence of human colon mucosa. Peptides 24:523–530 [DOI] [PubMed] [Google Scholar]
- 47. van de Veerdonk FL, Kullberg BJ, Netea MG. 2010. Pathogenesis of invasive candidiasis. Curr. Opin. Crit. Care 16:453–459 [DOI] [PubMed] [Google Scholar]
- 48. Villamon E, et al. 2004. Toll-like receptor-2 is essential in murine defenses against Candida albicans infections. Microbes Infect. 6:1–7 [DOI] [PubMed] [Google Scholar]
- 49. Yauch LE, Mansour MK, Shoham S, Rottman JB, Levitz SM. 2004. Involvement of CD14, Toll-like receptors 2 and 4, and MyD88 in the host response to the fungal pathogen Cryptococcus neoformans in vivo. Infect. Immun. 72:5373–5382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zaragoza O, et al. 2009. The capsule of the fungal pathogen Cryptococcus neoformans. Adv. Appl. Microbiol. 68:133–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zhao Z, et al. 2008. Autophagosome-independent essential function for the autophagy protein Atg5 in cellular immunity to intracellular pathogens. Cell Host Microbe 4:458–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zhao Z, et al. 2007. Coronavirus replication does not require the autophagy gene ATG5. Autophagy 3:581–585 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.