Abstract
Controlled production of reactive oxygen species leads to reversible oxidation of protein tyrosine phosphatases (PTPs) and has emerged as an important tier of regulation over phosphorylation-dependent signal transduction. We present a modified cysteinyl-labeling assay that detects reversible oxidation of members of each of the different PTP subclasses. Here, we describe the methods for enriching reversibly oxidized PTPs from complex protein extracts, illustrating the procedure in IMR90 fibroblasts.
INTRODUCTION
“All things are poison and nothing is without poison, only the dose permits something not to be poisonous”
-Paracelsus
Fifty years of investigation of the role of reactive oxygen species (ROS) has shed light on the manner by which cells use finely tuned redox signaling networks to regulate cellular homeostasis under physiological or disease states (1, 2). ROS, such as hydrogen peroxide (H2O2), are now established regulators of several cellular and physiological functions, including cell adhesion, cell migration, autophagy, senescence, and vascular tone (2–4). Protein phosphorylation is a reversible, dynamic process in which the net phosphorylation of a target substrate reflects the coordinated activity of the kinases that phosphorylate it and the protein phosphatases that catalyze the dephosphorylation reaction. In response to signals, the controlled, localized production of ROS exerts an additional tier of control by regulating the phosphoryl hydrolysis activity of protein tyrosine phosphatase (PTP) family members (5, 6). It is the architecture of the PTP catalytic center, particularly the conserved signature motif [His-Cys-(X)5-Arg-(Ser/Thr), where X is any amino acid] located at the base of the active site cleft (7), which creates an acidic environment that lowers the pKa (where Ka is the acid dissociation constant) of the catalytic cysteinyl residue. Hence, the side chain of the conserved cysteinyl residue is in a negatively charged thiolate form, making it a good nucleophile at neutral pH and making it particularly sensitive to cellular oxidants. Thus, oxidation of the catalytic residue of specific members of the PTP family abrogates their activity and facilitates phosphorylation-dependent signaling. However, PTP inactivation is transient: Upon oxidation, the formation of cyclic sulphenamides, for classical PTPs (8, 9), and intramolecular disulfide bonds with a vicinal cysteinyl residue, for dual-specificity PTPs (10), favors reversible inactivation over sulfinic or sulfonic acid states, which are largely irreversible. In addition, cysteinyl S-nitrosylation (11) and S-glutathionylation (12) have also been proposed to protect PTPs from irreversible oxidation and promote the reduction of the active-site cysteinyl residue. Lastly, PTPs are reduced back to their active form and specifically perform phosphoryl hydrolysis to terminate signaling.
We developed an assay that captures PTPs that have been reversibly oxidized in vivo. This protocol for enriching reversibly oxidized PTPs from complex protein extracts is a detailed description of the cysteinyl-labeling assay (13). This technique is challenging in several aspects. For example, first and foremost it is critical to avoid oxidation of proteins after cell lysis. Here, we provide detailed discussion of key aspects of the method to help an investigator overcome potential technical difficulties that may be encountered while performing this assay.
The cysteinyl-labeling assay uses an approach similar to the acyl-biotinyl exchange chemistry for identifying protein palmitoylation (14) and the biotin-switch technique for identifying S-nitrosylated proteins (15). It takes advantage of the unique, low pKa of the active-site cysteine residue that is a characteristic of members of the PTP family. The three-step method consists of (i) an alkylation step performed under anaerobic conditions to modify irreversibly the active-site cysteine of PTPs that were not modified by second messenger ROS molecules in vivo, (ii) a reactivation step in which active-site cysteines of PTPs that were reversibly oxidized (and protected in the first step) are reduced to their thiolate ion state by a reducing agent, and (iii) a modification step, in which the reduced Cys is modified by labeling with a biotinylated sulfhydryl-reactive probe at low pH. Hence, the biotinylated proteins generated in this assay are proteins with a low pKa cysteine residue that was reversibly oxidized upon acute or sustained generation of ROS molecules. We have used this technique successfully to enrich reversibly oxidized PTPs (13). In light of the ability of this assay to register all PTP subtypes, it should offer an approach to profiling of the entire ROS-regulated PTPome in a wide array of signaling paradigms.
MATERIALS
Anti-biotin–horseradish peroxidase (HRP) antibody (Cat. no. 7075, Cell Signaling Technology)
Antibodies against specific PTPs, such as RPTPα (Cat no. 07-472, Upstate), SHP- 2 (Cat. no. sc-280, Santa Cruz) or PTP1B (FG6)
Aprotinin (Cat. no. 11388, United States Biochemical)
Bond-Breaker tris(2-carboxyethyl)phosphine (TCEP) solution (Cat. no. 77720, Thermo Scientific)
Bovine serum albumin (Cat. no 03117057001, Roche)
Bromophenol blue (Cat. no. B-6131, Sigma Aldrich)
Catalase Aspergillus niger (Cat. no. 219261, Calbiochem)
Cultured cells, such as IMR90 fibroblasts
EGTA (Cat. no. E-0396, Sigma Aldrich)
Enhanced chemiluminescence (ECL) (Cat. no. RPN2106, GE Healthcare)
EZ-Link iodoacetyl-PEG2-biotin (Cat no. 21334, Thermo Scientific)
Glass beads, 3 mm (Cat. no. 11-312A, Fisher Scientific)
Glycerol (Cat. no. M778-07, J. T. Baker)
Glycine (Cat. no. G7126, Sigma Aldrich)
Iodoacetic acid (IAA) (Cat. no. 35603, Thermo Scientific)
Leupeptin (Cat. no. L6543, Invitrogen)
β-Mercaptoethanol (Cat. no. M-3148, Sigma Aldrich)
Methanol (Cat. no. 9070-03, J. T. Baker)
Nonfat dry milk (Carnation)
Sodium acetate (Cat. no. S-7545, Sigma Aldrich)
Sodium chloride (Cat. no. X190, Amresco)
Sodium docecyl sulfate (SDS) (Cat. no. L-6026, Sigma Aldrich)
Streptavidin-Sepharose (Cat. no. 17-5113-01, GE Healthcare)
Sucrose (Cat. no. S9378, Sigma Aldrich)
Surfact-Amps Nonidet P-40 (Cat. no. 28324, Thermo Scientific)
Superoxide dismutase (SOD) bovine liver (Cat. no. 574593, Calbiochem)
Tween-20 (Cat. no. P1379, Sigma Aldrich)
Tris-HCl (Cat. no. 0497, Amresco)
Zeba desalt spin columns, 2 ml (Cat no. 89889, Thermo Scientific)
EQUIPMENT
Amber 1.5 ml microcentrifuge tubes (Cat. no. 05-408-134, Fisher Scientific)
Büchner flask (50 ml, 125 ml)
Cell culture dishes 100 mm
Cell lifter (Cat. no. 3008, Corning.)
Centrifuge
Electrophoresis and blotting apparatus
Hoffman open-side tubing clamp (Fisher Scientific)
Hypoxic glove box (COY Laboratory Products.)
Oxygen electrode (DO-166, Lazar.)
Rotator
Temperature-controlled shaker
RECIPES
Recipe 1: Lysis Buffer Part A
| Sodium Acetate 50 mM | |
| Sodium Chloride 150 mM | |
| Glycerol, 10% (vol/vol) (v/v) | |
| Degassed distilled deionized H2O (ddH2O) | |
| Prepare 90 ml. Refer to the “Instructions” section for details of preparation. |
Recipe 2: Degassed Lysis Buffer
Lysis Buffer (Part A)
Nonoxidized NP-40 Solution (Surfact-Amps Nonidet P-40), 1% (v/v)
Prepare 100 ml. Refer to the “Instructions” section for details of preparation.
Recipe 3: IAA Stock Solution
Prepare 100 μl of 1 M IAA in ethanol immediately before use. Protect from light by preparing in an amber 1.5-ml centrifuge tube.
Note: To minimize light exposure and air oxidation, IAA powder should be aliquoted in amber 1.5-ml centrifuge tubes when the bottle is first opened.
Recipe 4: SOD Stock
250 U/μl in dH2O
Prepare in the necessary volume to achieve activity indicated, aliquot 25 μl per tube into 1.5-ml amber tubes, and store at −20°C.
Note: The specific activity varies for each batch of SOD.
Recipe 5: Catalase Stock
125 U/μl in dH2O
Prepare in the necessary volume to achieve activity indicated, aliquot 25 μl per tube into 1.5-ml amber tubes, and store at −20°C.
Note: The specific activity varies for each batch of catalase.
Recipe 6: IAA-Supplemented Degassed Lysis Buffer
Degassed Lysis Buffer (Recipe 2)
IAA Stock (Recipe 3)
SOD Stock (Recipe 4)
Catalase Stock (Recipe 5)
Aprotinin
Leupeptin
Prepare 10 ml. Refer to the “Instructions” section for details of preparation.
Recipe 7: Degassed Lysis Buffer Without IAA
Degassed Lysis Buffer (Recipe 2)
SOD Stock (Recipe 4)
Catalase Stock (Recipe 5)
Aprotinin
Leupeptin
Prepare 2 ml. Refer to the “Instructions” section for details of preparation.
Recipe 8: Iodoacetyl Polyethylene Glycol (IAP)-Biotin Stock
Prepare a 25 mM solution in ddH2O and store at −20°C.
Recipe 9: Phosphate-Buffered Saline (PBS)
NaCl, 137 mM
KCl, 2.7 mM
Na2HPO4O, 10 mM
KH2PO4O, 2 mM
ddH2O
Prepare 1 liter and adjust pH to 7.5.
Recipe 10: 4× Laemmli Buffer
Tris-HCl, 50 mM pH 6.8
SDS, 2.5%
β-Mercaptoethanol, 500 mM
Sucrose, 300 mM
EGTA, 1.0 mM
Bromophenol Blue, 0.008%
ddH2O
Prepare 25 ml and aliquot in 1.5 ml tubes.
Recipe 11: Electrophoresis Running Buffer
Tris-Base, 25 mM pH 8.3
Glycine, 192 mM
SDS, 1%
Prepare 4 liters and store at room temperature.
Recipe 12: Transfer Buffer
Tris-Base, 25 mM
Glycine, 192 mM
Methanol, 5%
Prepare 4 liters and store at 4°C.
Recipe 13: Tris-Buffered Saline Tween (TBST)
Tris-HCl, 25 mM pH 7.5
NaCl, 150 mM
Tween-20 0.05% (v/v)
Prepare 4 liters and store at room temperature.
Recipe 14: 5% Nonfat Milk Blocking Solution
Nonfat Milk Powder, 5% (weight/vol) (w/v)
TBST (Recipe 13)
Prepare 100 ml.
Recipe 15: Primary Antibody Dilution Buffer
Bovine Serum Albumin, 1% (w/v)
TBST (Recipe 13)
Prepare 100 ml and store at −20°C.
INSTRUCTIONS
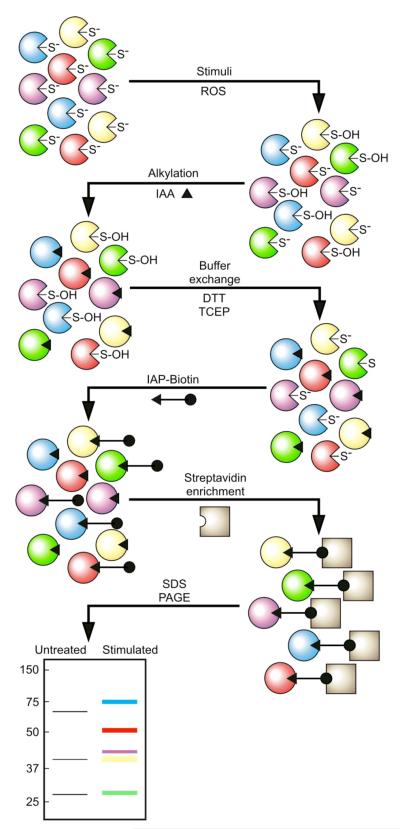
This three-step method is designed to label reversibly oxidized PTPs (Fig. 1). The mildly acidic lysis buffer in the first step is supplemented with iodoacetic acid to eliminate those PTPs that remain in a reduced state after a stimulus. However, PTPs that are reversibly oxidized on their catalytic cysteine residue are protected from this alkylation step. The second step consists of a buffer exchange to eliminate the alkylating agent from the lysates and reduce the oxidized catalytic cysteines back to their active state. Lastly, the restored reactive catalytic cysteinyl residues are labeled with a biotinylated sulfhydryl-reactive compound under mildly acidic conditions, and the PTPs are purified by affinity precipitation. The low pH of the lysis buffer is designed to keep the restored reactive PTP catalytic cysteine as a thiolate ion and to favor its labeling with sulfhydryl-reactive reagents over less-reactive cysteine, histidine, and methionine residues.
Fig. 1.
Profiling of reversible PTP oxidation. The catalytic Cys residue of members of the PTP family of enzymes is present as a thiolate under basal cellular conditions. This feature makes it a good nucleophile at neutral pH and allows it to react with phospho–amino acid residues, ROS, or certain sulhydryl-reactive compounds. After a stimulus that generates localized production of ROS, the active-site Cys residue of those specific PTPs that encounter the ROS becomes oxidized, and PTP activity is abrogated. The pool of reversibly oxidized PTPs is represented by a sulfenic acid (S-OH). In the first step of the assay, the active-site Cys residue of those PTPs that were not modified by second messenger ROS molecules in vivo is modified irreversibly by alkylation under anaerobic conditions in vitro. In the second step, the active-site Cys residues of PTPs that were reversibly oxidized and thus protected from alkylation in the first step are reduced. DTT indicates dithiothreitol and TCEP indicates tris(2-carboxyethyl)phosphine. The third step consists of a modification in which the reduced Cys residue is modified by a biotinylated sulfhydryl-reactive probe under low-pH labeling conditions. Probe-labeled PTPs are then captured on streptavidin beads, enriched, and subjected to SDS-PAGE protein fractionation, transferred onto a nitrocellulose membrane, and detected with an antibody against biotin or a PTP-specific antibody.
It is important to note that a rapid cellular lysis in a hypoxic environment, together with use of a thoroughly degassed lysis buffer, appear to be critical steps leading to specificity of labeling and sensitivity of detection. In addition, we found that healthy, low passage cells worked best for measuring inducible PTP oxidation.
Preparation of Lysis Buffers
To prevent the spontaneous oxidation of PTPs by oxygen in the air that is dissolved in the buffer solutions, it is essential to degas all lysis buffers thoroughly before use. The IAA-supplemented degassed lysis buffer (recipe 6) and the degassed lysis buffer without IAA (recipe 7) should be prepared in a hypoxic glove box station equilibrated with 100% argon.
Boil 200 ml of ddH2O for about 1 hour in a glass bottle containing glass beads to help degassing and close hermetically. Allow the buffer to cool to room temperature.
-
Gently pour 75 ml of degassed ddH2O into a 125-ml Büchner flask; add 10 ml glycerol, sodium acetate (final concentration 50 mM), and sodium chloride (final concentration 150 mM); and mix under vacuum (<5 to 10 mm Hg to avoid evaporation) for a few hours to overnight. Add further degassed ddH2O to a final volume of 90 ml. This is Lysis Buffer Part A (Recipe 1).
Note: We usually perform the mixing under vacuum overnight.
-
Add 10 ml of a 10% Nonoxidized NP-40 Solution to Lysis Buffer Part A (Recipe 1) to obtain a final volume of 100 ml and adjust the pH to 5.5 with 0.1 N HCl.
Note: Nonoxidized NP-40 is used to avoid potential postlysis oxidation.
-
Transfer 20 ml of the pH 5.5 buffer to a 50-ml Büchner flask and degas for 20 min on ice and 45 to 60 min at room temperature at 35 mm Hg using a vacuum pump. This is Degassed Lysis Buffer (Recipe 2).
Note: (i) At this point transfer all required solutions and reagents to the hypoxic glove box. (ii) The flask containing the degassed lysis buffer (Recipe 2) should be kept on ice in the argon-filled hypoxic glove box until needed further.
-
Measure pO2 level of an aliquot of the Degassed Lysis Buffer 2 with oxygen electrode according to the manufacturer’s instructions inside the hypoxic glove box.
Note: The pO2 of the lysis buffer varies between 0.35 and 0.50 parts per million (ppm), which minimizes postlysis oxidation. If pO2 is too high, continue to degas. This step only needs to be performed the first time a vacuum pump set up is used to determine length of time required for complete degassing.
Pour 10 ml of Degassed Lysis Buffer (Recipe 2) in a 15-ml foil-covered tube. Keep on ice.
-
Immediately before use, prepare IAA–Supplemented Lysis Buffer (Recipe 6) by adding freshly prepared IAA (Recipe 3) (final concentration 10 mM), SOD (Recipe 4) (final concentration, 125 U/ml), catalase (Recipe 5) (final concentration, 250 U/ml), and leupeptin and aprotinin (final concentration for each, 5 μg/ml).
Note: Reserve 2 ml of Degassed Lysis Buffer (Recipe 2) to prepare Degassed Lysis Buffer Without IAA (Recipe 7) to use for the positive-control samples.
Cellular Extraction
This part of the procedure involves extraction of the proteins under anaerobic conditions to prevent oxidation during sample preparation from cells that have been subjected to a stimulation or treatment and from unstimulated control cells. Examples of stimuli that could be used for the experimental samples include exposing the cells to hydrogen peroxide, phorbol 12-myristate 13-acetate, insulin, platelet-derived growth factor-BB, epidermal growth factor, endothelin, angiotensin, or hypoxia.
One key control that should be included is processing one sample of unstimulated cells without IAA in the lysis buffer (Recipe 7). This control validates the efficiency of the biotinylated probe. All thiols that are reactive at pH 5.5 should be labeled by the alkylating-biotinylated probe in the absence of IAA.
Assemble the items necessary for cell lysis and prepare immediately before use the IAA-Supplemented Degassed Lysis Buffer (Recipe 6) and the Degassed Lysis Buffer without IAA (Recipe 7) in a hypoxic glove box station equilibrated with 100% argon.
Bring in the cells through the airlock.
-
Carefully remove all of the media from the cells and rapidly add 600 μl (for a 10-mm culture dish) of ice-cold IAA-Supplemented Degassed Lysis Buffer (Recipe 6).
Note: Use Degassed Lysis Buffer Without IAA (Recipe 7) for the positive-control samples in order to maximize the IAP labeling.
Gently remove the cells using a cell lifter and immediately transfer to an amber 1.5-ml centrifuge tube.
-
Incubate on a temperature-controlled shaker for 1 hour at 25°C to permit alkylation. Protect from light.
Note: From this step on, the lysate can be manipulated outside the argon-filled chamber.
Centrifuge the alkylated protein extract at 14,000g for 10 min at 4°C.
Determine the protein content of the supernatant with the Bradford assay.
Biotinylation of Reversibly Oxidized Cysteinyl Residues
The removal of the alkylating agent at this step allows for the subsequent reactivation of the reversibly oxidized PTPs by TCEP. The architecture of the signature motif, His-Cys-(X)5-Arg(Ser/Thr), confers an unusually low pKa (4.5 to 5.5) on the invariant catalytic cysteinyl residue of PTP family members. Hence, performing the labeling with the biotinylated, sufhydryl-reactive probe at pH 5.5 favors the reaction with the thiolate ions of the PTP catalytic cysteine in comparison to the protonated cysteines present in other proteins. As a negative control, one sample should not be exposed to TCEP, which serves as an indicator of the efficiency of the alkylation that occurred in the cellular lysis process. All thiols that are reactive at pH 5.5 should be alkylated during cellular lysis and should not be labeled by the alkylating probe in the second step if the TCEP-reducing agent is omitted at the reactivation step.
This part of the procedure also includes steps for minimizing nonspecific interactions. Nonspecific interactions with the streptavidin-Sepharose beads are minimized by preclearing the desalted sample with the streptavidin-Sepharose beads before reducing the oxidized PTPs and addition of IAP-Biotin.
Loosen the cap of a desalting column, twist off the column’s bottom, and place desalting column in a 15-ml conical tube.
Centrifuge column at 1000g for 2 min to remove storage solution.
Equilibrate the column by applying 1 ml of Degassed Lysis Buffer (Recipe 2) three times.
Place desalting column in a clean 15-ml conical tube.
Slowly apply the appropriate volume of alkylated lysate containing 1 mg of protein to the center of the column.
Elute the IAA-free sample by centrifuging at 1000g for 2 min and transfer the sample to an amber 1.5-ml centrifuge tube.
Transfer the desired volume of streptavidin-Sepharose bead slurry (20 μl per sample) to a microcentrifuge tube and add 1 ml of PBS (Recipe 9).
Spin at 3000g for 30 s and remove the PBS.
Wash two more times with 1 ml of PBS, centrifuging to pellet the beads between washes.
Resuspend the beads in cold Lysis Buffer (Recipe 2) to obtain a 50% slurry (the same as the initial volume).
Add 20 μl of the prepared streptavidin-Sepharose slurry to the microcentrifuge tube containing the IAA-free (desalted) sample and incubate on a rotator at 4°C for 30 min.
Spin at 3000g for 2 min at 4°C and transfer the supernatant containing the oxidized PTPs to a fresh amber 1.5 ml microcentrifuge tube.
-
Reduce the oxidized PTPs by adding TCEP (final concentration 1 mM) and incubate on a temperature-controlled shaker for 30 min at 25°C. Protect from light.
Note: Prepare one sample without TCEP treatment as a negative control.
14. Label the reactivated PTPs by adding IAP-Biotin (Recipe 8) to a final concentration of 5 mM and incubate on a temperature-controlled shaker for 1 hour at 25°C.
Enrichment and Detection of Biotinylated PTPs
Add 25 μl of 50% streptavidin-Sepharose/PBS slurry to the precleared and biotinylated sample and incubate for 16 hours at 4°C on a rotating wheel.
-
Centrifuge (3000g, 2 min, 4°C) and wash the beads three times with 1 ml PBS, centrifuging after each wash and discarding the supernatant.
Note: Stringent washes can also be performed to disrupt protein-protein interactions because streptavidin-biotin interactions are extremely strong. We have used buffers containing 0.5 M KCl, 0.5 M NaCl, and 0.2 M glycine (pH 2) successfully.
Resuspend the beads (25 to 40 μl) in 20 μl 4× Laemmli Sample Buffer (Recipe 10) and heat at 95°C for 90 s.
Centrifuge the beads in sample buffer at 12,000g for 1 min at room temperature.
Separate the proteins in the samples on a 10% (w/v) polyacrylamide SDS–polyacrylamide gel electrophoresis (PAGE) and transfer electrophoretically onto nitrocellulose membranes at 100 V and ~4°C for 90 min.
Block the nitrocellulose membrane in 5% Nonfat Milk Blocking Solution (Recipe 14) for 1 hour at room temperature.
-
Incubate the nitrocellulose membrane overnight at 4°C with PTP-specific antibodies or biotin-HRP antibodies diluted in Primary Antibody Dilution Buffer (Recipe 15).
Note: Antibodies should be used at the recommended concentrations, or, if not characterized for immunoblotting, then appropriate concentrations will have to be determined empirically.
Wash the membranes three times for 10 min with TBST (Recipe 13).
-
If using PTP-specific antibodies, then incubate the membrane for 2 hours with peroxidase-conjugated secondary antibodies diluted 1:10000 in 5% Nonfat Milk Blocking Solution (Recipe 14) and then wash the membranes three times for 10 min with TBST (Recipe 13).
Note: This step is only necessary if PTP-specific antibodies are used. If the samples are detected with biotin-HRP antibodies, then this step is unnecessary because the biotin-HRP antibodies are already conjugated to horseradish peroxidase.
Visualize the immunoreactive bands with ECL solutions according to the manufacturer’s protocol.
TROUBLESHOOTING
Success in detecting reversible oxidation of PTPs with the cysteinyl-labeling assay relies on a thorough preparation of the lysis buffers, the rapidity of execution once the lysis buffers are prepared, analysis of healthy cells, and performing the appropriate controls. We perform the following controls to validate the assay. We prepare one positive-control sample by extracting the cells in lysis buffer lacking IAA, which validates the efficiency of the biotinylation. We prepare an IAA-treated sample without TCEP treatment, which serves as a negative control measuring the efficiency of the alkylation occurring in the cellular lysis process.
False Positive Signals
High background labeling may be observed when oxidation occurs during the cellular lysis step. The most common causes leading to postlysis oxidation are (i) the IAA has not been freshly prepared, (ii) inadequate degassing of the lysis buffer, (iii) slow execution when lysing the cells, (iv) exposure of samples to light during the alkylation step, (v) the reaction has not been conducted under mildly acidic conditions, and (vi) processing unhealthy, stressed cells. We have observed an increasing amount of background oxidation when the cell lines have been passaged too many times. Also, some cancer cell lines have a high production of ROS leading to an increased background of reversibly oxidized PTPs (16), which can make it challenging to detect changes in the amount of stimulus-induced reversible oxidation in such backgrounds.
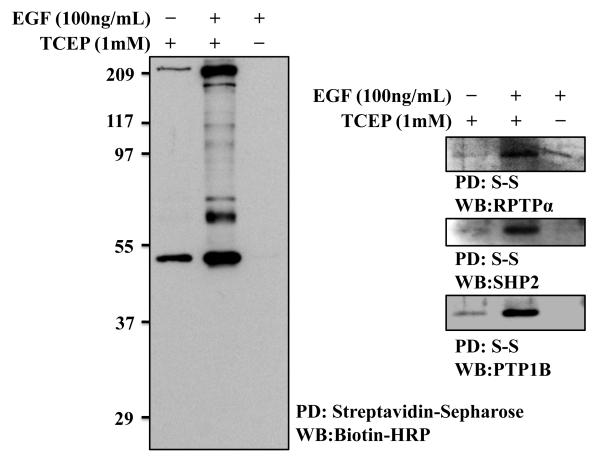
Although not empirically tested to determine whether this has an effect, we culture the cells in low-glucose medium [Dulbecco’s minimum essential medium (DMEM) low glucose (Cat. no. 11885, Invitrogen)], and we use medium without phenol red when serum starving [DMEM low glucose without phenol red (Cat. no. D5921, Sigma Aldrich)]. Figure 2 show representative experiment of serum-deprived IMR90 cells exposed to epidermal growth factor (EGF).
Fig. 2.
Detection of EGF–dependent reversible oxidation of PTPs in IMR90 cells. IMR90 fibroblasts were cultured to confluency in low-glucose DMEM supplemented with 10% fetal bovine serum and nonessential amino acids. On the day before the experiment, cells were serum-deprived in low-glucose DMEM without phenol red for a total of 16 hours. Healthy cells were then incubated with 100 ng/ml EGF for 2 min and transferred to an argon-equilibrated hypoxic chamber through the airlock, and the cysteinyl-labeling assay was performed. Proteins were detected with biotin-HRP, RPTPα, SHP-2, or PTP1B (FG6) antibodies. Proteins were then detected with HRP-conjugated secondary antibodies, and the immunoreactive bands were visualized by ECL. Reversible oxidation of RPTPα, SHP-2, and PTP1B was observed upon 2 min of EGF stimulation in IMR90 cells. IAP labeling detected the reversible oxidation of PTPs specifically because no immunoreactive band was visualized when TCEP was omitted from the reducing step. Probing for biotinylated PTPs with biotin-HRP revealed an increase in staining of several bands, confirming the successful enrichment of multiple reversibly oxidized proteins. PD, Pull-down; WB, Western Blot.
Low Sensitivity
Sensitivity issues are usually the result of an inadequate labeling by the sulfhydryl-reactive probe. This can occur when (i) the alkylating agent is inefficiently removed during the desalting procedure, (ii) reduction of the reversibly oxidized PTPs is ineffective, (iii) the reaction has not been conducted under mildly acidic conditions, (iv) when the sulfhydryl reactive probe has been mishandled (for example, exposed to light or subjected to free-thaw cycles), or (v) insufficient boiling of the sample in Laemmli sample buffer.
RELATED TECHNIQUES
Similar protocols for identifying reversibly oxidized PTPs exist, which use neutral pH and SDS to denature proteins from the cellular extract and to allow the alkylating agents to gain access to hidden sulfhydryl-reactive Cys residues (17, 18). However, these denaturing conditions disrupt the active-site architecture (and low pKa), thus making the method less specific for PTPs. Another approach avoided denaturants but focused on measuring the decrease in labeling with the biotinylated thiol-reactive probe that accompanied oxidation, which limits the dynamic range of the assay (18).
Another affinity-based approach uses antibodies raised against the terminally and irreversibly oxidized (that is sulfonic acid, Cys-SO3H) Val-His-Cys-Ser-Ala-Gly peptide from the PTP signature motif. This strategy has potential to be applied to proteomic screening of reversibly oxidized PTPs (19). However, the in vitro terminal oxidation of all reversibly oxidized PTPs, which is essential for antibody recognition, would not allow distinction between reversibly oxidized PTPs and the terminally oxidized PTPs that are abundant in cancer cells (16). Furthermore, it is not clear whether this antibody detects all members of the PTP family.
Direct sulfenic acid–labeling methods using dimedone or close derivatives have also been used to detect reversible oxidation of proteins in vivo (20–24). However, no reversibly oxidized PTPs have yet been identified from complex protein extracts with this approach.
NOTES AND REMARKS
In addition to IAP, we have also used an activity-based probe to label reversibly oxidized PTPs in angiomyolipoma cells. The α-bromobenzylphosphonate probe used in our previous study (13) is a suicide substrate for PTPs (25) and leads to the formation of a derivatized cysteinyl-phosphate enzyme intermediate when the reactivated active-site Cys residue reacts with the phosphorus center of the probe. This is similar to the intermediate formed between a PTP and its tyrosine-phosphorylated substrate. A second reaction mechanism has also been proposed to explain the labeling of PTPs by this probe (26). Use of this probe would be expected to enhance specificity for PTP family members compared with general alkylating agents. Overall, through the use of a biotinylated derivative of IAA under mildly acidic conditions, the cysteinyl-labeling assay described here has minimal labeling of proteins lacking a low pKa Cys residue. However, the development and improvement of specific probes would aid proteomic approaches to quantify the reversibly oxidized PTPome.
Acknowledgments
We thank members of the Tonks laboratory, T. C. Meng and M. Dagnell for helpful discussions and J. Duffy and C. Eberstark for assistance with figures.
Funding: Supported by NIH grants to N.K.T. (R01-GM55989). B.B. was the recipient of a postdoctoral fellowship from the Heart and Stroke Foundation of Canada and was supported by the Cold Spring Harbor Laboratory Association.
Footnotes
Competing interests: The authors declare that they have no competing interests.
References and Notes
- 1.Gruber J, Schaffer S, Halliwell B. The mitochondrial free radical theory of ageing—Where do we stand? Front. Biosci. 2008;13:6554–6579. doi: 10.2741/3174. [DOI] [PubMed] [Google Scholar]
- 2.Valko M, Leibfritz D, Moncol J, Cronin MT, Mazur M, Telser J. Free radicals and antioxidants in normal physiological functions and human disease. Int. J. Biochem. Cell Biol. 2007;39:44–84. doi: 10.1016/j.biocel.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 3.Storz P. Reactive oxygen species in tumor progression. Front. Biosci. 2005;10:1881–1896. doi: 10.2741/1667. [DOI] [PubMed] [Google Scholar]
- 4.Azad MB, Chen Y, Gibson SB. Regulation of autophagy by reactive oxygen species (ROS): Implications for cancer progression and treatment. Antioxid. Redox Signal. 2009;11:777–790. doi: 10.1089/ars.2008.2270. [DOI] [PubMed] [Google Scholar]
- 5.Juarez JC, Manuia M, Burnett ME, Betancourt O, Boivin B, Shaw DE, Tonks NK, Mazar AP, Doñate F. Superoxide dismutase 1 (SOD1) is essential for H2O2-mediated oxidation and inactivation of phosphatases in growth factor signaling. Proc. Natl. Acad. Sci. U.S.A. 2008;105:7147–7152. doi: 10.1073/pnas.0709451105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tonks NK. Protein tyrosine phosphatases: From genes, to function, to disease. Nat. Rev. Mol. Cell Biol. 2006;7:833–846. doi: 10.1038/nrm2039. [DOI] [PubMed] [Google Scholar]
- 7.Barford D, Flint AJ, Tonks NK. Crystal structure of human protein tyrosine phosphatase 1B. Science. 1994;263:1397–1404. [PubMed] [Google Scholar]
- 8.Salmeen A, Andersen JN, Myers MP, Meng TC, Hinks JA, Tonks NK, Barford D. Redox regulation of protein tyrosine phosphatase 1B involves a sulphenyl-amide intermediate. Nature. 2003;423:769–773. doi: 10.1038/nature01680. [DOI] [PubMed] [Google Scholar]
- 9.Yang J, Groen A, Lemeer S, Jans A, Slijper M, Roe SM, den Hertog J, Barford D. Reversible oxidation of the membrane distal domain of receptor PTPalpha is mediated by a cyclic sulfenamide. Biochemistry. 2007;46:709–719. doi: 10.1021/bi061546m. [DOI] [PubMed] [Google Scholar]
- 10.Salmeen A, Barford D. Functions and mechanisms of redox regulation of cysteine-based phosphatases. Antioxid. Redox Signal. 2005;7:560–577. doi: 10.1089/ars.2005.7.560. [DOI] [PubMed] [Google Scholar]
- 11.Chen YY, Chu HM, Pan KT, Teng CH, Wang DL, Wang AH, Khoo KH, Meng TC. Cysteine S-nitrosylation protects protein-tyrosine phosphatase 1B against oxidation-induced permanent inactivation. J. Biol. Chem. 2008;283:35265–35272. doi: 10.1074/jbc.M805287200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barrett WC, DeGnore JP, Keng YF, Zhang ZY, Yim MB, Chock PB. Roles of superoxide radical anion in signal transduction mediated by reversible regulation of protein-tyrosine phosphatase 1B. J. Biol. Chem. 1999;274:34543–34546. doi: 10.1074/jbc.274.49.34543. [DOI] [PubMed] [Google Scholar]
- 13.Boivin B, Zhang S, Arbiser JL, Zhang ZY, Tonks NK. A modified cysteinyl-labeling assay reveals reversible oxidation of protein tyrosine phosphatases in angiomyolipoma cells. Proc. Natl. Acad. Sci. U.S.A. 2008;105:9959–9964. doi: 10.1073/pnas.0804336105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Drisdel RC, Alexander JK, Sayeed A, Green WN. Assays of protein palmitoylation. Methods. 2006;40:127–134. doi: 10.1016/j.ymeth.2006.04.015. [DOI] [PubMed] [Google Scholar]
- 15.Jaffrey SR, Snyder SH. The biotin switch method for the detection of S-nitrosylated proteins. Sci. STKE. 2001;86:pl1. doi: 10.1126/stke.2001.86.pl1. [DOI] [PubMed] [Google Scholar]
- 16.Lou YW, Chen YY, Hsu SF, Chen RK, Lee CL, Khoo KH, Tonks NK, Meng TC. Redox regulation of the protein tyrosine phosphatase PTP1B in cancer cells. FEBS J. 2008;275:69–88. doi: 10.1111/j.1742-4658.2007.06173.x. [DOI] [PubMed] [Google Scholar]
- 17.Kwon J, Lee SR, Yang KS, Ahn Y, Kim YJ, Stadtman ER, Rhee SG. Reversible oxidation and inactivation of the tumor suppressor PTEN in cells stimulated with peptide growth factors. Proc. Natl. Acad. Sci. U.S.A. 2004;101:16419–16424. doi: 10.1073/pnas.0407396101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kwon J, Qu CK, Maeng JS, Falahati R, Lee C, Williams MS. Receptor-stimulated oxidation of SHP-2 promotes T-cell adhesion through SLP-76-ADAP. EMBO J. 2005;24:2331–2341. doi: 10.1038/sj.emboj.7600706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Persson C, Sjöblom T, Groen A, Kappert K, Engström U, Hellman U, Heldin CH, den Hertog J, Ostman A. Preferential oxidation of the second phosphatase domain of receptor-like PTP-alpha revealed by an antibody against oxidized protein tyrosine phosphatases. Proc. Natl. Acad. Sci. U.S.A. 2004;101:1886–1891. doi: 10.1073/pnas.0304403101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Benitez LV, Allison WS. The inactivation of the acyl phosphatase activity catalyzed by the sulfenic acid form of glyceraldehyde 3-phosphate dehydrogenase by dimedone and olefins. J. Biol. Chem. 1974;249:6234–6243. [PubMed] [Google Scholar]
- 21.Ellis HR, Poole LB. Novel application of 7-chloro-4-nitrobenzo-2-oxa-1,3-diazole to identify cysteine sulfenic acid in the AhpC component of alkyl hydroperoxide reductase. Biochemistry. 1997;36:15013–15018. doi: 10.1021/bi972191x. [DOI] [PubMed] [Google Scholar]
- 22.Conway ME, Poole LB, Hutson SM. Roles for cysteine residues in the regulatory CXXC motif of human mitochondrial branched chain aminotransferase enzyme. Biochemistry. 2004;43:7356–7364. doi: 10.1021/bi0498050. [DOI] [PubMed] [Google Scholar]
- 23.Charles RL, Schröder E, May G, Free P, Gaffney PR, Wait R, Begum S, Heads RJ, Eaton P. Protein sulfenation as a redox sensor: Proteomics studies using a novel biotinylated dimedone analogue. Mol. Cell. Proteomics. 2007;6:1473–1484. doi: 10.1074/mcp.M700065-MCP200. [DOI] [PubMed] [Google Scholar]
- 24.Leonard SE, Reddie KG, Carroll KS. Mining the thiol proteome for sulfenic acid modifications reveals new targets for oxidation in cells. ACS Chem. Biol. 2009;4:783–799. doi: 10.1021/cb900105q. [DOI] [PubMed] [Google Scholar]
- 25.Kumar S, Zhou B, Liang F, Wang WQ, Huang Z, Zhang ZY. Activity-based probes for protein tyrosine phosphatases. Proc. Natl. Acad. Sci. U.S.A. 2004;101:7943–7948. doi: 10.1073/pnas.0402323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liang F, Kumar S, Zhang ZY. Proteomic approaches to studying protein tyrosine phosphatases. Mol. Biosyst. 2007;3:308–316. doi: 10.1039/b700704n. [DOI] [PubMed] [Google Scholar]
- Boivin B, Yang M, Tonks NK. Targeting the reversibly oxidized protein tyrosine phosphatase family. Sci. Signal. 2010;3:pl2. doi: 10.1126/scisignal.3137pl2. [DOI] [PMC free article] [PubMed] [Google Scholar]