Abstract
Rather than simply being protein degradation products, peptides have proven to be important bioactive molecules. Bioactive peptides act as hormones, neurotransmitters and antimicrobial agents in vivo. The dysregulation of bioactive peptide signaling is also known to be involved in disease, and targeting peptide hormone pathways has been successful strategy in the development of novel therapeutics. The importance of bioactive peptides in biology has spurred research to elucidate the function and regulation of these molecules. Classical methods for peptide analysis have relied on targeted immunoassays, but certain scientific questions necessitated a broader and more detailed view of the peptidome–all the peptides in a cell, tissue or organism. In this review we discuss how peptidomics has emerged to fill this need through the application of advanced liquid chromatography-tandem mass spectrometry (LC-MS/MS) methods that provide unique insights into peptide activity and regulation.
Bioactive Peptide Action, Production and Signaling
Organisms seamlessly integrate numerous classes of molecules, including bioactive peptides, into biochemical pathways that enable all the processes needed for life (1-3). Insulin and glucagon, for example, are two of several well-known pancreatic peptide hormones that partake in hormonal regulation of physiological glucose metabolism (4, 5). Neuropeptides such as substance P (6, 7) and the enkephalins (8, 9) signal within the central nervous system (CNS) and mediate behavioral processes. There are also more than 20 known antimicrobial peptides (AMPs) (10) that have important roles in the innate immune system. The defensin family of antimicrobial intestinal peptides thwart infection by serving as endogenous antibiotics (11) and more recent experiments have also shown that the defensins regulate the intestinal microbiome (12), all the bacteria present in the gut, to extend the physiological role of these peptides even further.
These examples highlight the broad influence of bioactive peptides and demonstrate why these pathways are of interest. Moreover, the discovery of these peptides has begun to impact medicine with the development of approved drug liraglutide (13), a glucagon like peptide 1 (GLP-1) mimetic, and AMP mimics that are promising antibiotic agents (14). Many more examples can be presented as bioactive peptides can be found in many biological niches and play many roles. Conotoxins, for example, are a group of neurotoxic peptides produced by the venomous marine cone snail that are potential therapeutics (15-18). Here, we primarily focus on the application of peptidomics to problems in mammalian biology, the exception being the work described on honeybee neuropeptidomics, but we recognize that peptidomics has the potential to impact many different areas of biology.
The Production and Signaling of Bioactive Peptides
Though bioactive peptides have diverse functions and sequences, many of these peptides have analogous biochemical mechanisms that control their production, regulation and signaling (19) (Fig. 1). Bioactive peptide synthesis commences with the expression of a biologically inactive preprohormone that is typically 100-350 amino acids long (19). A signal peptide at the N-terminus (the “pre” in preprohormone) directs the peptide into the secretory pathway. The signal peptide is cleaved shortly after the peptide enters the lumen of the ER to afford the prohormone. Prohormones are then shuttled from the endoplasmic reticulum (ER) to the Golgi apparatus (Golgi). In the Golgi the prohormone begins its conversion into a mature bioactive peptide through proteolysis by a group of serine proteases called the prohormone convertases (PCs) (20, 21) (Fig. 1). This processing begins in the Golgi and continues into the secretory vesicles. Some peptides undergo additional proteolytic processing and post-translational modifications (acetylation, sulfation and amidation) to produce the active form (Fig. 2) (22, 23).
Fig. 1.

The key steps in the production and regulation of bioactive peptides. A prepropeptide is produced from mRNA encoding bioactive peptides. This peptide enters the secretory pathway through the ER and Golgi, before being packaged into secretory vesicles. In the trans Glogi as well as the secretory vesicles the prohormone encounters subtilisin-like proteases, called prohormone convertases, that process the prohormone to generate a mature form of the bioactive peptide. During this maturation process these peptides can also obtain additional posttranslational modifications, such as C-terminal amidation and N-terminal acetylation. Stimulation of cells can lead to secretion of the peptides through fusion of the vesicles with the plasma membrane. Once released the peptides can bind to receptors, typically G-protein coupled receptors (GPCRs) or receptor tyrosine kinases (RTKs), to elicit a cellular or physiological response. Once released, peptides can also undergo proteolysis to regulate their activity.
Fig. 2.

Posttranslational modifications found on bioactive peptides. Some modifications, such as N-terminal acetylation or C-terminal amidation, are common, while others, such as serine octanoylation are rare.
Peptides are released from cells into the extracellular milieu by fusion of secretory vesicles with the plasma membrane (2, 19) (Fig. 1). The secretion of a bioactive peptide is regulated by an external stimulus that causes the acute release of the peptide when needed (e.g. glucose release of insulin) (24, 25). After release, bioactive peptides travel to a target cell or tissue to bind their cognate membrane receptors, which initiates intracellular signal transduction and a biological response (26). The principle receptors involved in bioactive peptide signaling include seven transmembrane G-protein coupled receptors (GPCRs) (27) and receptor tyrosine kinases (RTKs) (28). Peptide signaling is terminated by removal of the peptide through renal clearance (29) or proteolyic inactivation of a peptide (30) (Fig. 1). By understanding these general pathways for a few peptides it has made it easier to discovery and characterization of new bioactive peptides, receptors and peptidases.
Bioactive Peptide Discovery
The methods used to investigate bioactive peptides have evolved as new techniques are introduced into biology. Classical methods for bioactive peptide discovery rely on bioassays that enable the biochemical purification of bioactive peptides from tissues by identifying fractions with a desired bioactivity (31). By performing multiple rounds of purification bioactive peptides could be purified and subsequently identified. Many important peptides were discovered through this type of approach including insulin (32), glucagon (33, 34), angiotensin II and the endorphins (35-37). Modern variants of this approach have replaced a cellular or physiological phenotypic bioassay with a targeted receptor-based assay to identify bioactive peptides and characterize the receptor too. The identification of GPCR ligands (38-40), for example, led to the discovery of sleep regulating orexins (41) and prolactin-releasing peptide (42) to demonstrate the effectiveness of this strategy in discovering new bioactive peptides.
To circumvent the need for bioassays, Tatemoto and Mutt developed an alternative approach bioactive peptide discovery that relies on the biochemical enrichment of peptides with a C-terminal amide (43-48), which are commonly found on bioactive peptides that are part of the secretory pathway (19). Oxidative cleavage of a C-terminal glycine by peptidyl alpha-amidating monooxygenase (PAM) (49-51) produces the C-terminal amide-containing peptides. C-terminal amides are thought to protect bioactive peptides from carboxypeptidase-mediated degradation and in some cases are required for bioactivity (52). Tatemoto and Mutt developed a biochemical method that enabled them to purify peptides with C-terminal amides from complex samples. This approach was highly successful and led to the discovery of neuropeptide Y (46), peptide histidine isoleucine (PHI(1-27)) (45) and galanin (48), which are now known to regulate a variety of physiological functions (3).
Lastly, bioactive peptides have also been discovered through messenger RNAs (mRNAs) that revealed a coding sequence containing PC-cleavage sites. Alternative splicing of the calcitonin gene affords two different mRNA products (53). Analysis of these splice forms revealed that one of the mRNAs encodes for the calcitonin peptide while the alternatively spliced form was predicted to encode a unknown peptide which was aptly named the calcitonin-gene related peptide (CGRP) (54). This predication was validated in subsequent experiments that demonstrated that CGRP was present in tissues (55, 56). Physiological experiments identified CGRP as a potent vasodilator (57). Most importantly, CGRP signaling was recently been linked the onset of migraines (58, 59) and this insight led to the development of CGRP-receptor antagonists a new class of anti-migraine drugs in clinical trials (60, 61). CGRP exemplifies the basic as well as biomedical impact of bioactive peptide discovery.
Bioactive Peptide Function
Pharmacology and genetics have been used to characterize the physiological functions for many peptides (9, 12). For instance, injection of glucagon like peptide 1 (GLP-1) reduces blood glucose levels to clearly define a role for GLP-1 in blood glucose homeostasis (62). Using pharmacology to ascertain peptide function is limited due to the poor bioavailability of most peptides (63). This is a particular problem in the study of neuropeptides since peptides do not readily cross the blood-brain barrier. Pharmacological studies with small-molecule receptor agonists (or antagonists) of neuropeptide receptors provide an alternative approach to understand the physiological function of a particular bioactive peptide (37, 61, 64). The analgesic natural product alkaloid morphine, for example, is an agonist for the opioid receptors to indicate that the endorphins, natural peptide agonists of the opioid receptors, are involved in pain sensation (37).
Genetic approaches have also proven to be a more general approach for studying bioactive peptide function in vivo (22, 65-67). For example, the role of the peptide YY (PYY), an intestinal hormone, in body mass regulation and feeding behavior was established by knocking this peptide out in a mouse model (68). Mice lacking PYY were obese and ate more than their wildtype counterparts, which demonstrates that PYY is a peripheral signal that is able to promote satiety (the feeling of fullness). These experiments also revealed that mice that ate protein, but not fat or carbohydrate, released more PYY and this finding provides a new model to explain why proteins are better appetite suppressants than fat or carbohydrate.
Knockout studies of peptides are often difficult, however, because a single gene sometimes produces more than one bioactive peptide (69-71). For example, the glucagon gene controls the production of glucagon, GLP-1 and GLP-2 (70, 71). As a result, knocking out the glucagon gene would result in loss of GLP-1 and GLP-2 as well. Glucagon and GLP-1 do have unique receptors though and knocking out the peptide receptors instead of the peptides enables the physiological functions of the bioactive peptide pathways to be assessed (27, 72, 73). To study glucagon signaling, for example, glucagon receptor knockout mice were generated and these mice showed improved glucose tolerance relative to their wildtype counterparts (74). The application of pharmacology and genetics has provided valuable information to further our understanding of the cellular and physiological roles of peptides and their receptors (Table 1).
Table 1.
Known peptide hormones and their physiological processes.
| Peptide | Physiological Process |
|---|---|
| insulin | glucose metabolism |
| angiotensin | blood pressure |
| oxytocin | social behavior |
| enkephalins | pain |
| ghrelin | appetite |
| orexins | sleep |
Proteolytic Regulation of Bioactive Peptides
An approach for controlling endogenous peptide signaling in medicine is to target the proteases and peptidases that regulate these pathways with small-molecule inhibitors (30, 75). A prototype example of the success of this approach is the inhibition of the angiotensin converting enzyme (ACE), a well studied exopeptidase, in the treatment of hypertension and cardiovascular disorders (75). ACE is responsible for the cleavage and activation of the angiotensin I peptide to generate angiotensin II, a ligand for the angiotensin receptor and a potent vasoconstrictor that causes hypertension. Understanding this mechanism led to the development of small-molecule ACE inhibitors as antihypertensive drugs that function by preventing the activation of angiotensin I. More recently, a similar strategy has successfully been employed in the development of anti-diabetic drugs that are inhibitors of dipeptidyl peptidase 4 (DPP4) activity.(30)
Bioactive peptides are often enzymatically regulated after release in ways that can regulate their activity. The insulinotropic hormone GLP-1 is released from the gut after a meal (30) and activates its receptors in the pancreas where it stimulates insulin biosynthesis and secretion (76). Detailed studies of GLP-1 signaling revealed that GLP-1 has a very short half-life in blood due to N-terminal processing of this peptide by a peptidase called DPP4 (77, 78). DPP4 removes an N-terminal dipeptide to inactivate GLP-1. Importantly, this pathway has been harnessed in the development of a new class of anti-diabetic drugs (30, 79) that inhibit DPP4. By blocking DPP4 activity endogenous GLP-1 levels are increased leading to higher levels of insulin and improved glucose tolerance (Fig. 3). The development of ACE and DPP4 inhibitors underscores the importance of bioactive peptides in basic research and medicine.
Fig. 3.

Peptide hormone signaling regulates physiology and can be harnessed for therapeutic gain. DPP4 inhibitors extend the half-life of the bioactive form of the insulinotropic hormone GLP-1. In doing so these inhibitors increase insulin secretion and improve physiological glucose tolerance, which is of tremendous value in the treatment of diabetes.
The Case for Peptidomics
The importance of peptides supports the need for better methods to analyze these molecules in a biological setting. Classical methods for peptide analysis rely mostly on immunoassay approaches that require specific antibodies to recognize the peptide of interest (80, 81). Immunoassays are valuable because they provide a means to detect the peptides from a variety of biological samples (80, 82, 83). While effective, immunoassays do not provide a global analysis, require the time-consuming generation of antibodies, and can sometimes suffer from antibody cross-reactivity with other peptides (82). For example, Jankowski, et al. discovered a novel vasoconstrictive peptides angiotensin A (Ang A) by mass spectrometry and showed that this peptide cross-reacts with antibodies against angiotensin II (AngII) (82). More specifically, cross-reactivity is a particular problem when studying peptide proteolysis since shorter or longer peptides may still contain the antibody epitope making it difficult to measure the peptide of interest. Peptidomics approaches can overcome the challenges associated with immunoassays because liquid chromatography-tandem mass spectrometry (LC-MS/MS) provides a specific and global analysis of peptidome. These attributes enable new types of experiments to elucidate important features of bioactive peptide regulation.
Peptidomics Workflow, Instrumentation and Analysis
Peptidomics refers to any method that provides a broad view of the entire peptide pool in a biological sample, the peptidome (84, 85). Modern peptidomics approaches rely heavily on LC-MS/MS to provide sensitive detection and identification of peptides from biological samples. The two predominant types of mass spectrometers used in peptidomics studies are quadrupole time-of-flight (Q-TOF) and ion trap (IT) instruments (86). These instruments enable quantitation using several approaches and provide the tandem mass spectra that are necessary to identify the peptide sequence. The key steps in every peptidomics experiment are peptide isolation, peptide fractionation and processing, mass spectrometry, and data analysis (Fig. 4). Svensson and colleagues provided an early example of an effective peptidomics workflow during their discovery of neuropeptides (84). Protease inhibitors are not effective enough to prevent peptide degradation so Svensson and coworkers used focused microwave radiation of rodent brains (84, 87, 88) to ablate proteolytic activity by heat denaturing all proteins prior to peptide isolation. Heating tissues prior to isolation of the peptidome has become a standard process in peptidomics even though different heating methods are used (e.g. ex-vivo heating of tissues by microwave (89) or boiling (90-92)).
Fig. 4.

The key steps in any peptidomics experiment include sample preparation and peptide isolation, processing of the sample (including isotopic labeling), and finally detection and data analysis. While these steps vary in subtle ways from experiment-to-experiment the overall workflow is consistent for most peptidomics experiments.
Following microwave irradiation the brain tissue was homogenized in slightly acidic buffer and centrifuged to remove any insoluble debris. Next, Svensson and colleagues fractionated their sample by passing it through a molecular weight cutoff (MWCO) filter (10 kilodalton) to separate the peptides from larger proteins (84). MWCO filters are used in almost every peptidomics experiment to enrich the peptidome. Other fractionation schemes can also be included to provide different views of the peptidome, including enzymatic digestion (e.g. trypsin), strong cation exchange (SCX) and isoelectric focusing (IEF) (92) (Fig. 4). The use of additional peptide fractionation improves the peptidome coverage. In one example a 10-fold increase in coverage was obtained through the addition of an SCX step into the workflow. The isolated neuropeptides were then subjected to reverse phase LC-MS/MS using an electrospray ionization (ESI) Q-TOF-MS. The identification of peptides is carried out using SEQUEST (93) or Mascot (94) software packages.
Using this workflow Svensson and colleagues identified greater than 500 neuropeptides (84). Some of these peptides are known bioactive neuropeptides, including substance P, beta-endorphin, and neurotensin, which validated the methodology. There were also a number of novel neuropeptides. Many of these novel peptides were derivatives of known neuropeptides, including longer or shorter peptide fragments and post-translationally modified forms. The common PTMs observed in this data set included acetylation, phosphorylation, and pyroglutamination (Fig. 2). The characterization of these novel neuropeptides highlights the value of peptidomics to obtain information that is inaccesible by other approaches. More generally, Svensson, et. al. demonstrated how to obtain quality peptidomics data and their approach has been emulated in all subsequent peptidomics studies.
Using Peptidomics to Discover Bioactive Peptides
Yamaguchi and colleagues developed a peptidomics strategy to identify new bioactive peptides by detecting peptides that are secreted by TT cells, a human thyroid cell line (95). By using peptidomics to detect amidated peptides in the conditioned media of TT cells they were able to detect known bioactive peptides, such as CGRP (53, 57). In addition, these experiments detected two novel C-terminally amidated peptides derived from the VGF gene called neuroendocrine regulatory peptides-1 and -2 (NERP-1 and -2). Subsequent histological studies revealed that NERP-1 and NERP-2 are present in vivo and that they co-localized in the rat brain (supraoptic nucleus and paraventricular nucleus) with vasopressin, a multifunctional peptide hormone (96-101). This led to the hypothesis that the NERPs and vasopressin may interact. This idea was validated when pharmacological delivery of the NERPs suppressed vasopressin release in a dose dependent fashion to demonstrate a physiological activity for these peptides (95) (Fig. 5). In this case, the detection of C-terminal amides using peptidomics led to the discovery of two novel bioactive peptides and provided a peptidomics version of the Tatemoto and Mutt approach (44). Sasaki, et. al. generalized this approach further to include peptides that lack C-terminal amides by performing cell based assays to determine which of the peptides they detected were able to induce intracellular calcium release, which is a hallmark of bioactive peptides (102).
Fig. 5.

Discovery of novel bioactive peptides using structural features of the peptide as a guide. A number of bioactive peptides contain C-terminal amides, which result from enzymatic production in the secretory pathway. Analysis of peptides secreted by TT cells using a peptidomics approach led to the discovery of the neuroendocrine regulatory peptides (NERPs). These peptides were later shown to co-localize with arginine vasopressin (AVP), a hormone involved in blood pressure, and physiological studies demonstrated that NERPs control AVP release in vivo. In doing so, this approach demonstrates the utility of peptidomics in accelerating the discovery of novel bioactive peptides.
In a more focused search Osaki and Minamino developed a peptidomics strategy to discover novel mammalian antimicrobial peptides (103). They took advantage of the fact that antimicrobial peptides are typically highly basic peptides—containing multiple lysines and arginines—and developed a cation exchange fractionation method that specifically enriched basic peptides. Treatment of QGP-1 cells, a human pancreatic endocrine cell line, with forskolin and carbachol promotes the secretion of intracellular peptides into the media. Secreted peptides were enriched by cation exchange, fractionated and desalted by HPLC, and analyzed by matrix assisted laser desorption ionization (MALDI)-MS to reveal a novel peptide fragment of insulin-like growth factor-binding protein 5 (IGFBP-5) (104, 105).
IGFBP-5 is a known secreted protein, but its proteolysis and antimicrobial activity had not been reported before. The peptide fragment of IGFBP-5 identified in this study was referred to as antimicrobial peptide-IGFBP-5 (AMP-IFGBP-5) (103). Synthesis of the AMP-IGFBP-5 enabled the antimicrobial activity of this peptide to be tested. AMP-IGFBP-5 had an IC50 of ~ 1 μM with E.coli K12 and was on par or better than the established antimicrobial peptides cathelicidin (106) and β-defensin-2 (107). Additional experiments revealed that this peptide was active against a number of Gram-negative and Gram-positive bacteria, as well as the fungus Pichia pastoris GS115. In addition, AMP-IGFBP-5 is present in the brain and gut of rats, suggesting a potential physiological role for this peptide in the innate immune system (Fig. 6).
Fig. 6.

Peptidomics of highly basic peptides from QGP-1 cells revealed the production of a series of IFGBP-5 peptides. While this protein was predicted to be secreted, it had not previously been hypothesized to produce bioactive peptides. The structure of the antimicrobial-IFGBP-5 (AMP-IFGBP-5) was highly charged and reminiscent of known defensin antimicrobial peptides. Subsequent antimicrobial and antifungal assays, demonstrated that AMP-IFGBP-5 has antimicrobial and antifungal activity equal to or better than that of defensins. Lastly, AMP-IFGBP-5 was present in the gut, which suggests that this peptide might play a role in innate immunity or regulation of the microbiome.
These examples highlight the utility of peptidomics to accelerate the discovery of novel bioactive peptides by enabling the detection and identification of these peptides. Most of these peptides would undetectable using standard molecular biology techniques used to identify proteins (e.g. silver stained SDS page gels or proteomics) (108, 109). In addition, immunoassays are not a discovery based approach because they require structural knowledge of the peptides first so that antibodies can be generated (110). Characterizing these new peptides these studies also indicate new functions for precursor proteins that generate these peptides, such as VGF (111) and IGFBP-5 (104, 105). As additional assays are integrated with peptidomics approaches the number of bioactive peptides discovered is bound to increase.
Quantitative Peptidomics
The above examples take advantage of the ability of peptidomics to identify peptides within complex mixtures (112). In some cases, however, it is important to quantify peptides between two samples to correlate changes in peptide levels with changes in a phenotype or genotype (89-92, 113-115). Quantitative peptidomics methods enable differences in relative peptide concentrations to be determined and can be carried out using isotopic labeling methods (113, 114, 116, 117) or a label-free approach (90-92, 118-121) (Fig. 7).
Fig. 7.
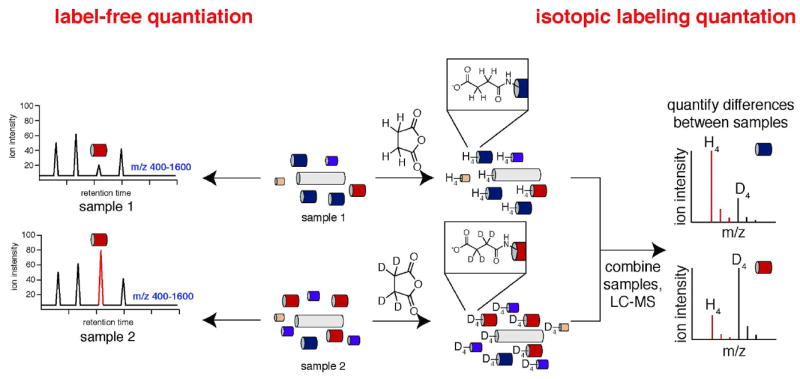
There are two primary approaches used for quantitative proteomics: stable isotope labeling methods and label-free approaches. Isotopic labeling relies on the chemical labeling of the peptidome using stable isotope variants of the same reagent. Analysis of the MS for each peptide should reveal a heavy and light labeled version of the peptide and the ratio of these peptides enables quantitation. By contrast, in label-free approaches samples are run sequentially and the peak intensities are used to determine changes in the concentration of the peptide between two samples.
In one example of an isotope labeling approach a light and heavy form (e.g. deuterium or carbon-13) of a chemical labeling reagent, such as succinic anhydride, is used to differentially label two peptidome samples. Succinic anhydride and the deuterated version of succinic anhydride, d4-succinic anhydride, are readily available. Labeling of two different peptidomes with succinic anhydride and d4-succinic anhydride results in a mass difference of 4 daltons between the same peptide in each sample (113) (Fig. 7). After separate labeling reactions the two samples are mixed and analyzed during in same LC-MS/MS experiment. Differences in relative peptide concentrations are measured by comparing the ratio of the light labeled peptide to the heavy labeled peptide in the mass spectrum (Fig. 7). This approach enables the relative abundance of peptides between different samples to be determined, and additional labels can be created if more samples are to be included. By contrast label-free approaches simply rely on the ion intensity, absolute signal at the MS detector, between two samples to provide relative changes in peptide levels between two samples (92).
Application of Quantitative Peptidomics to Behavioral Studies
Using an isotopic labeling strategy, Sweedler and colleagues investigated changes in neuropeptides in the honeybee brain during behavioral assays (113). Honeybees were chosen as the model system because they show amazingly sophisticated behavior (122, 123). They have approximately 100 neuropeptides, and in this report they were able to quantitatively measure about 50 of those peptides. Quantitation was accomplished using a chemical labeling approach with succinic anhydride and d4-succinic anhydride to differentially label the peptidomes in a series of pair-wise comparisons. Changes in neuropeptide levels were measured as honeybees were foraging for food (124). Four categories were created based on whether the honeybees preferred pollen (P) or nectar (N) as a food source, or whether the honeybees were isolated when they arrived (A) or departed (D) from the food (Fig. 8).
Fig. 8.
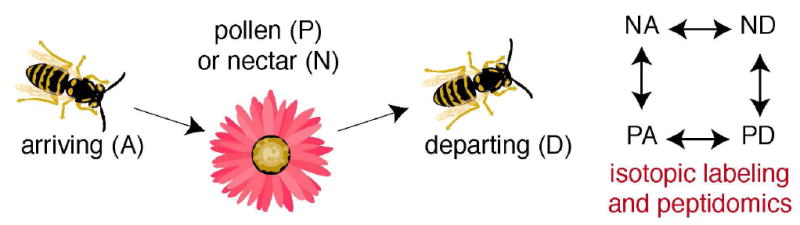
Neuropeptidomics of honeybees as they arrive (A) or depart (D) a food source (pollen (P) or nectar (N)) demonstrated that there are specific neuropeptides that are associated with food gathering in honeybees. By correlating changes in these peptide levels with phenotypes associated with forging, these studies provided new insights into the functions of these peptides, and revealed the ability of peptidomics to correlate changes in peptide levels with complex behaviors.
Combinations of these categories led to four different sample sets for peptidomics: nectar arriving (NA), nectar departing (ND), pollen arriving (PA), and pollen departing (PD) samples (Fig. 8). Peptidomics analysis compared NA to ND, NA to PA, ND to PD, and PA to PD. Of the 50 peptides measured in these studies, 8 were significantly different in one of the four pairwise comparisons. The changes were distributed between nectar (N) and pollen (P) foragers, as well as differences between arriving (A) and departing (D) honeybees. Together these changes were distinct enough to categorize the four different groups using the peptide signatures and canonical discriminant analysis (125). Brockmann and colleagues pushed the frontiers of peptidomics by making measurements with small amounts of tissue and demonstrating that the honeybee neuropeptidome could be used to correlate neuropeptides with behavior. This work sets the stage to subsequently study these peptides an their functions in honeybees or other related organisms.
Quantitative Peptidomics to Elucidate Peptidases and Proteases Substrates
One major question for every enzyme is the identification of natural substrates (126). For most peptidases and proteases, the identification of candidate substrates is a two-step process. Traditionally, the substrate specificity of the peptidase or protease is determined in vitro using synthetic peptide libraries (127, 128). Based on the enzyme specificity, the next step is to look for candidate endogenous substrates and then test these natural substrates in vitro (129). Finally, in the best case scenario, the activity of the enzyme is perturbed via genetics or chemical inhibition and the endogenous levels of the substrate are evaluated to determine the physiological relevance of the prediction (77).
While occasionally successful, this strategy often leads to many false positives because in vitro experiments are poor predictors of in vivo substrate utilization due to a number of factors that cannot be recapitulated in vitro (91, 130). First, in vitro experiments cannot account for the distribution of enzymes and substrates within cells and tissues. Therefore, enzyme-substrate relationships that are determined in vitro might not occur in vivo because the two molecules do not have the opportunity to interact. Next, assays with pure enzymes do not factor in the relative contributions of other enzymes in the proteome. As a consequence, even if an enzyme and substrate interact in vitro, other enzymes may be responsible for processing the peptide in vivo (126). Lastly, the presence of competitive substrates in vivo can prevent an enzyme from cleaving an in vitro substrate in vivo (131). Together these cases highlight the difficulty in trying to predict endogenous peptidase-substrate relationships from in vitro experiments (91).
Peptidomics offers an elegant solution to this problem by enabling the substrates of peptidases to be measured directly in cells and tissues (90-92, 114, 115, 132, 133). In doing so, substrate discovery using peptidomics takes into account all of the aforementioned challenges presented by in vitro experiments. These peptidomics experiments rely on the fact that perturbation of the activity of a peptidase–either by using genetics to knock it out or pharmacological inhibition–results in different amounts of substrates and products in cells and tissues. Comparison of samples with differences in peptidase activity using quantitative peptidomics can identify these endogenous substrates and products (Fig. 9). The identification of peptidase-regulated peptides can help explain the biochemical, cellular, and physiological functions of the peptidase. We provide several examples below that attempt to target particular enzymes involved in peptide biosynthesis, degradation, and signaling.
Fig. 9.

Identification of endogenous substrates of peptidases relies on peptidomics. While there are subtle differences in the methods for quantitation and data analysis the overall workflow for these experiments is similar. In this approach, comparison of the peptidomes of mice that differ in the activity of a particular peptidase/protease can reveal peptides regulated by the enzyme. These peptides will include substrates and/or products of the enzyme, which in turn, can be used to infer biochemical and biological function of the peptidase/protease.
Prohormone Convertases
Fricker and colleagues pioneered efforts to identify substrates and products of the PCs using peptidomics (134). As mentioned, PCs are the enzymes responsible for making the primary cut in the prohormone peptide in the secretory pathway that eventually leads to the production of the mature peptide (19). The PCs cleave prohormones at dibasic sites (e.g. KK, KR, RR), a very distinctive sequence that has been used to predict the presence of peptide hormones within longer genes. The PCs are a family of subtilsin-like serine proteases with seven members in mammals (135). PC1/3 and PC2 represent the majority of the PC activity in neuroendocrine cells and other tissues (135). For example, PC2 is involved in the production of a number of neuropeptides as well as insulin and glucagon in the pancreas.
Mice lacking PC2 show impaired neuropeptide processing by radioimmunoassasys (RIAs) that measured the levels of full-length peptide (136, 137). Like all immunoassays, RIAs suffer from issues with cross reactivity (82), and this problem may be exacerbated in peptide processing studies where incompletely processed peptides are very similar in structure, which would favor cross reactivity. As a result, Fricker and colleagues decided to employ a quantitative peptidomics approach to study PC2. In these experiments hypothalamic peptidomes from wildtype (PC2+/+) and PC2-null (PC2−/−) mice were compared by peptidomics to identify specific substrates of PC2 using an isotopic labeling method (134). The substrates would be expected to be elevated in the knockout mice, while the products would be elevated in the wildtype mice.
A number of neuropeptides were identified in this study including fragments of cocaine and amphetamine regulated transcript, chromogranins, preprotachykinins, provassopressin, and secretogranin (Table 2). The levels of the peptides detected in the PC2−/− sample were greatly reduced in comparison to the PC2+/+ sample. This data supports a general role for PC2 in the production of many neuropeptides in the hypothalamus. In addition, a number of protein fragments, most likely from the breakdown of proteins within the cell, were also detected in these samples. These fragments are not part of the secretory pathway and the levels of these fragments did not change, demonstrating the specificity of PC2 for secreted hormones. Finally, this specificity supports the notion that the methodology is capturing changes that are occurring in vivo, since any processing that occurred during the sample preparation would target secretory and non-secretory peptides equally. Fricker and colleagues also utilized their data to determine that aromatic residues or proline are preferred by PC2 at the P1’ and P2’ positions. In total, this work provided a physiological view of PC2 biochemistry and a template for comparative peptidomics studies that identify peptides that are regulated by peptidases and proteases.
Table 2.
Prohormone convertase 2 regulated peptides identified by peptidomics.
| Precursor | PC2−/−/PC2+/+ |
|---|---|
| CART(33-52) | < 0.05 |
| chromogranin A (392-402) | < 0.05 |
| proenkephalin(219-229) | < 0.05 |
| VGF(487-507) | < 0.05 |
| alpha-MSH | < 0.1 |
Prolyl Peptidases
One of the benefits of peptidomics is that it is unbiased and can be used to discover novel endogenous substrates for peptidases, which can help define the cellular and/or physiological function of peptidases (77). Examples of this are found in the analysis of members of the prolyl peptidase family. The prolyl peptidases are a family of serine peptidases that cleave peptides preferentially on the C-terminal side of proline residues (i.e. proline is strongly preferred at the P1 position). The prolyl peptidase family is composed of prolyl endopeptidases and dipeptidyl peptidases, which preferentially cut at proline residues at the penultimate position of the N-terminus (i.e. H2N-XP-peptide) (78).
Prolyl endopeptidase (PREP) is the founding member of the prolyl peptidase family (138, 139). Interest in this protein was derived from its unique selectivity for proline. Initial in vitro assays with PREP identified candidate substrates for the enzyme, such as vasopressin (139, 140), which led to new hypotheses about PREP function. Vasopressin was previously linked to memory performance and led to the development of selective PREP inhibitors, such as S17092 (141, 142), as potential anti-amnesic compounds. Interestingly, while S17092 inhibitors showed improved cognitive function in monkeys (143) and humans (144), PREP inhibition did not regulate physiological levels of vasopressin highlighting the difficulty in using in vitro experiments to predict endogenous substrates (140, 145).
To gain a deeper insight in the biochemistry of PREP in vivo, two groups utilized comparative peptidomics to investigate PREP substrates in the nervous system using selective pharmacological inhibition of PREP. The first study utilized the commercially available inhibitor, Z-ProProlinal, to study PREP activity in a rat model (133). In these experiments quantitation was performed using an isotopic labeling strategy termed iTRAQ (146). Using this approach, Tenorio-Laranga and colleagues identified a number of novel Prep substrates in the brains of inhibitor-treated rats. These were proline-containing peptides elevated in the inhibitor-treated samples versus the untreated samples. These peptides were not explicitly tested as Prep substrates, but fit into the known substrate profile for the enzyme. On the basis of their peptidomics data, Tenorio-Laranga and coworkers concluded that Prep is involved in the catabolism of peptides in the CNS (133).
Nolte and colleagues utilized the PREP inhibitor S17092 to study the impact of PREP on the CNS peptidome of mice (90). In this case, quantitation was performed using a label-free approach. The substrates identified in this study raised important questions about the PREP substrate specificity that were then studied in greater detail. Specifically, peptidomics was able to provide a possible explanation of why PREP had evolved a preference for cutting shorter peptides. One of the PREP substrates identified was a fragment of the bioactive peptide CGRP, CGRP(20-37), which was elevated in the S17092-treated sample (Fig. 10). The full length CGRP(1-37) was not a substrate, even though it contains the very same cut site. These data demonstrate that Prep uses sequence and length specificity to cleave a subset of proline-containing peptides in the nervous system.
Fig. 10.
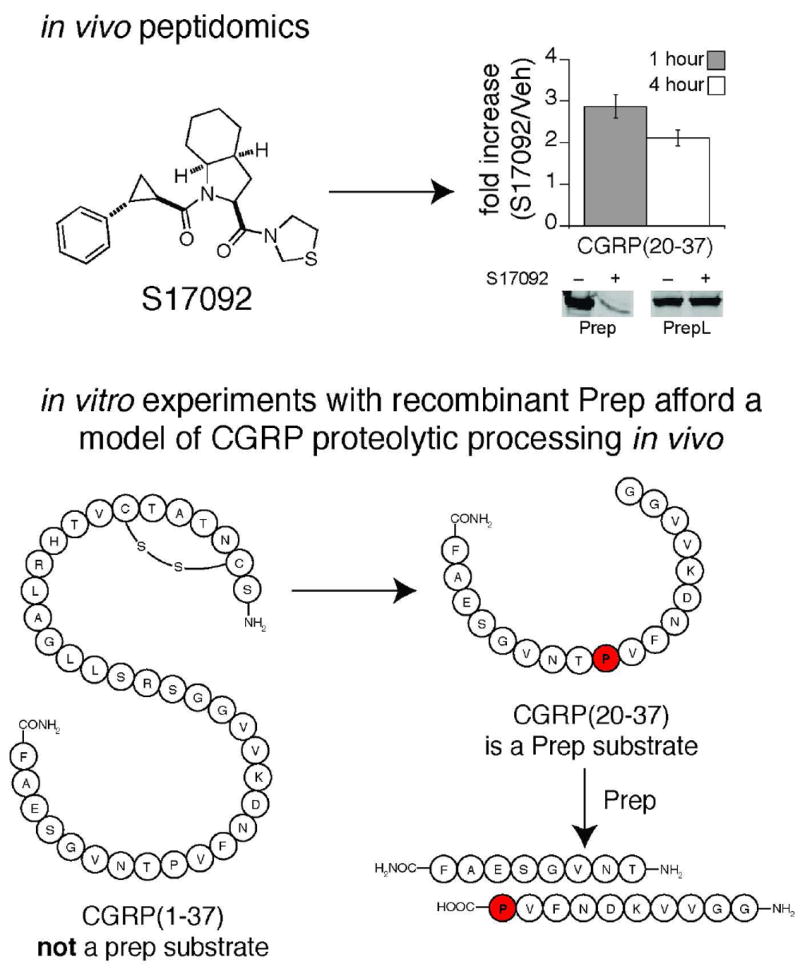
Peptidomics of Prep revealed that Prep regulates endogenous levels of CGRP(20-37). Follow up in vitro experiments demonstrated the Prep is able to process the shorter CGRP(20-37) but not the longer, full-length, peptide CGRP(1-37). This length preference enables Prep to participate in the catabolism of CGRP without regulating the bioactive form of the molecule (i.e., CGRP(1-37)), even though the cut sites are identical within the two molecules. This data supports a model of CGRP proteolysis in the nervous system where the full-length CGRP is processed by unknown enzymes followed by Prep processing of the proline-containing CGRP(20-37) fragment.
Tagore and colleagues (91, 92) extended peptidomics to study another prolyl peptidase, DPP4, in the kidney. While the role of DPP4 in plasma GLP-1 regulation was established (77) the function of this enzyme in other tissues, including the kidney, was less clear. Peptidomics comparison of wildtype (DPP4+/+) and DPP4 null (DPP4−/−) samples identified a number of DPP4 substrates elevated in the DPP4−/− samples. One interesting question that emerged from these studies was an attempt to explain how the penultimate proline containing substrates of DPP4 are generated in vivo. The data suggested that peptides are cleaved first by aminopeptidases until a penultimate proline is encountered. Penultimate proline peptides are not aminopeptidase substrates and are then released and cleaved by DPP4. In this model, aminopeptidase and DPP4 activities form a biochemical pathway that is responsible for the N-terminal degradation of proline-containing peptides (Fig. 11). This model was tested by adding peptides with internal prolines to tissue lysates with and without aminopeptidase activity. In the absence of aminopeptidase activity the production of penultimate proline containing peptides was reduced, which demonstrated a role for kidney aminopeptidase activity in the production of DPP4 substrates (91, 92). In total, these studies highlight the value of peptidomics in providing detailed information about peptidases in vivo.
Fig. 11.
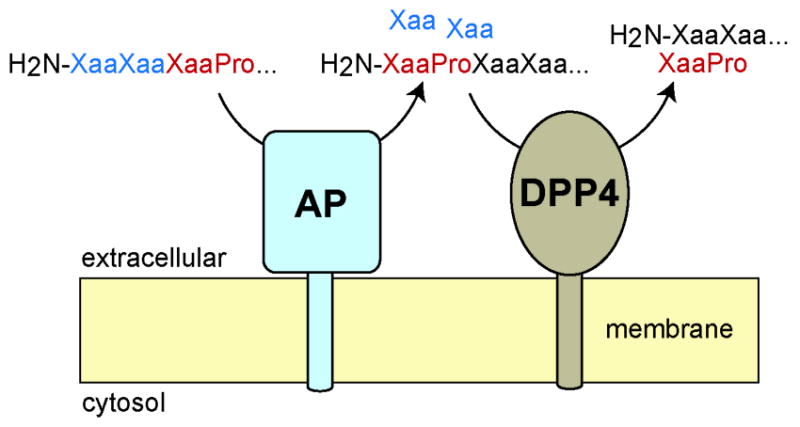
Peptidomics revealed an unappreciated physiological pathway for the renal catabolism of proline containing peptides that interlinks aminopeptidase (AP) and dipeptidyl peptidase 4 (DPP4) activities. These experiments demonstrate the value of peptidomics in understanding peptide processing and characterizing the biochemical and physiological functions of enzymes.
A Peptidomics Strategy to Elucidate the Proteolytic Pathways that Process Bioacitve Peptides
As mentioned, the regulation of bioactive peptides by peptidases is of basic interest and can also be used to develop novel therapeutics (e.g. DPP4 and ACE inhibitors). Despite the interest in these pathways we currently lack information about the proteolytic regulation of most bioactive peptides due to the lack of a general approach for elucidating these pathways in vivo. Specifically, any approach that can identify physiologically relevant peptide fragments can reveal the proteolytic pathways that process bioactive peptides and accelerate the discovery of the peptidases responsible for peptide processing. Tinoco, Kim, and colleagues developed a general peptidomics-based strategy to elucidate the endogenous pathways that regulate bioactive peptides coupling in vitro assays and in vivo peptidomics measurements of endogenous peptides (131) (Fig. 12).
Fig. 12.

A peptidomics-based approach to identify physiologically relevant proteolytic pathways that process peptide hormones. In this approach, peptidomics identification of fragments of bioactive peptides enables physiological-relevant proteolytic pathways to be identified. Application of this approach to the peptide hormone PHI(1-27) identified a previously unappreciated pathway for the C-terminal proteolysis of this peptide. Interestingly, this pathway was also shown to regulate the activity of PHI(1-27) in a cell-based glucose-stimulated insulin secretion (GSIS) assay.
This peptidomics-based approach was applied to investigate the proteolysis of PHI(1-27) (147), an intestinal peptide hormone which has been linked to a number of biological functions, including prolactin secretion (148) and glucose stimulated insulin secretion (GSIS) (149). First, PHI(1-27) was incubated with intestinal lysates and this revealed the production of PHI(3-27) and PHI(1-22) as the predominant fragments suggesting potential N- and C-terminus specific cut sites. In vitro experiments using chemical inhibitors demonstrated that the N-terminal processing is regulated by DPP4. Peptidomics experiments revealed the existence of PHI(1-22) but not PHI(3-27) in the intestine (150). The absence of PHI(3-27) in vivo was explained by the presence of competitive DPP4 substrates which can attenuate DPP4 processing of PHI(1-27). This hypothesis was supported by in vitro experiments with recombinant DPP4 where the presence of additional DPP4 substrates greatly slowed PHI(1-27) processing.
Lastly, the functional impact of the C-terminal proteolysis of PHI(1-27) was assessed using a GSIS assay (151) with mouse pancreatic β islets. In these experiments PHI(1-27) was active and able to promote insulin secretion, as reported, but PHI(1-22) was inactive to indicate that proteolysis of PHI(1-27) in vivo ablates the bioactivity of this peptide. Together this data demonstrates the utility of peptidomics in characterizing the endogenous proteolytic pathways that regulate bioactive peptide hormones. More generally, by coupling these experiments to improved strategies for peptidase discovery and bioassays, such as GSIS, additional enzymes and pathways that regulate important physiological pathways will be discovered.
Conclusions and Future Directions
As a whole, the application of LC-MS peptidomics has had a major impact on redefining the outlook of the peptide field. Advancements in peptidomics workflows and analytical tools have enabled improved strategies for identifying bioactive peptides and characterizing their functions (92). Peptidomics also helps to study peptidases and molecular pathways that regulate peptides in general (89-92, 114, 115, 131, 132). Because peptides are such powerful regulators of physiology, this research will impact our basic understanding of peptide and enzyme function in vivo, and may eventually provide novel insights that will be of benefit in the development of therapeutics like protease/peptidase inhibitors. Moreover, these same approaches can be extended from proteases to other classes of enzymes, such as kinases, that may modify bioactive peptides as well.
Looking forward there appears to be a number of opportunities for peptidomics to make an impact in biology and medicine. In the last decade, biologically active peptides encoded in short open reading frames (sORFs) have been serendipitously identified in mammals (152, 153), insects (154) and bacteria (155). These peptides encoded in sORFs, which lack signaling sequences and are released directly into the cytoplasm (156). Humanin was the first sORF derived peptide to be identified through a screen to identify genes that inhibit apoptosis and it exhibits neuroprotective properties (152, 157) by direct binding to proapoptotic proteins in the mitochondria. More recently, peptides encoded by the polished rice (pri) sORF gene have been shown to function as transcription factors that affect epidermal differentiation in Drosophila (158), suggesting that such peptides may be more ubiquitous than suspected. Improvements in the endogenous identification of sORF encoded peptides can be made by integrating genomic approaches with peptidomics (159-161).
Furthermore, as we improve our ability to deliver peptides in vivo the search for natural bioactive peptides is likely to ramp up. Natural product discovery was based on the fact that small-molecule drugs could be found by scouring nature’s stockpile of biologically active chemicals (162). Similar approaches for peptides have been hindered because peptides were traditionally considered to be poor drugs (163). The development of biologics aims to overcome many of these issues (164-166). Drugs such as liraglutide (13) and pramlintide (167) are peptides that are derived from GLP-1 and amylin, respectively. The emergence of these peptide drugs and improved methods for intracellular delivery of bioactive peptides (168, 169) and proteins (170) suggests a bright future for bioactive peptide medicines. Much of this will be predicated on the discovery of new bioactive peptides and will be accelerated by the inclusion of peptidomics.
Acknowledgments
This work was supported by the Mary Fieser Postdoctoral Fellowship (A.D.T.), Searle Scholar Award (A.S.), Burroughs Wellcome Fund Career Award in the Biomedical Sciences (A.S.), and National Institutes of Health Grant 1DP2OD002374 (A.S.).
Glossary
- LC-MS/MS
liquid chromatography-tandem mass spectrometry
- ER
endoplasmic reticulum
- Golgi
Golgi apparatus
- AMP
antimicrobial peptide
- GLP
glucagon like peptide
- GPCR
G-protein coupled receptor
- RTK
receptor tyrosine kinase
- PAM
peptidyl alpha-amidating monooxygenase
- PHI(1-27)
peptide histidine isoleucine
- mRNAs
messenger RNAs
- CGRP
calcitonin gene-related peptide
- PYY
peptide YY
- ACE
angiotensin converting enzyme
- DPP4
dipeptidyl peptidase 4
- Ang A
angiotensin A
- AngII
angiotensin II
- Q-TOF
quadrupole time-of-flight
- IT
ion trap
- MWCO
molecular weight cutoff
- SCX
strong cation exchange
- IEF
isoelectric focusing
- ESI
electrospray ionization
- NERP
neuroendocrine regulatory peptide
- MALDI
matrix assisted laser desorption ionization
- IGFBP-5
insulin-like growth factor-binding protein 5
- AMP-IFGBP-5
antimicrobial peptide-IGFBP-5
- PC
prohormone convertases
- PREP
prolyl endopeptidase
- iTRAQ
Isobaric tags for relative and absolute quantitation
- CNS
central nervous system
- GSIS
glucose stimulated insulin secretion
- sORF
short open reading frames
- PRI
polished rice
References
- 1.Berg JM, Tymoczko JL, Stryer L. Biochemistry. 6. W.H. Freeman; New York: 2007. [Google Scholar]
- 2.Gardner DG, Shoback DM, Greenspan FS. Greenspan’s basic & clinical endocrinology. 8. McGraw-Hill Medical; New York: 2007. [Google Scholar]
- 3.Kastin AJ. Handbook of biologically active peptides. Academic Press; Amsterdam; Boston: 2006. [Google Scholar]
- 4.Brady MJ, Saltiel AR. Insulin and Glucagon. John Wiley & Sons, Ltd; 2001. [Google Scholar]
- 5.Aronoff SL, Berkowitz K, Shreiner B, Want L. Glucose metabolism and regulation: Beyond insulin and glucagon. Diabetes Spectrum. 2004;17:183–190. [Google Scholar]
- 6.De Felipe C, Herrero JF, O’Brien JA, Palmer JA, Doyle CA, Smith AJ, Laird JM, Belmonte C, Cervero F, Hunt SP. Altered nociception, analgesia and aggression in mice lacking the receptor for substance P. Nature. 1998;392:394–397. doi: 10.1038/32904. [DOI] [PubMed] [Google Scholar]
- 7.Cao YQ, Mantyh PW, Carlson EJ, Gillespie AM, Epstein CJ, Basbaum AI. Primary afferent tachykinins are required to experience moderate to intense pain. Nature. 1998;392:390–394. doi: 10.1038/32897. [DOI] [PubMed] [Google Scholar]
- 8.Chang KJ, Cuatrecasas P. Multiple opiate receptors. Enkephalins and morphine bind to receptors of different specificity. J Biol Chem. 1979;254:2610–2618. [PubMed] [Google Scholar]
- 9.Kieffer BL. Opioids: first lessons from knockout mice. Trends in pharmacological sciences. 1999;20:19–26. doi: 10.1016/s0165-6147(98)01279-6. [DOI] [PubMed] [Google Scholar]
- 10.Lai YP, Gallo RL. AMPed up immunity: how antimicrobial peptides have multiple roles in immune defense. Trends in Immunology. 2009;30:131–141. doi: 10.1016/j.it.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Selsted ME, Ouellette AJ. Mammalian defensins in the antimicrobial immune response. Nature immunology. 2005;6:551–557. doi: 10.1038/ni1206. [DOI] [PubMed] [Google Scholar]
- 12.Salzman NH, Hung K, Haribhai D, Chu H, Karlsson-Sjoberg J, Amir E, Teggatz P, Barman M, Hayward M, Eastwood D, Stoel M, Zhou Y, Sodergren E, Weinstock GM, Bevins CL, Williams CB, Bos NA. Enteric defensins are essential regulators of intestinal microbial ecology. Nature immunology. 2010;11:76–83. doi: 10.1038/ni.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Buse JB, Sesti G, Schmidt WE, Montanya E, Chang CT, Xu Y, Blonde L, Rosenstock J. Switching to once-daily liraglutide from twice-daily exenatide further improves glycemic control in patients with type 2 diabetes using oral agents. Diabetes Care. 2010;33:1300–1303. doi: 10.2337/dc09-2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chongsiriwatana NP, Patch JA, Czyzewski AM, Dohm MT, Ivankin A, Gidalevitz D, Zuckermann RN, Barron AE. Peptoids that mimic the structure, function, and mechanism of helical antimicrobial peptides. Proc Natl Acad Sci U S A. 2008;105:2794–2799. doi: 10.1073/pnas.0708254105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kelley WP, Schulz JR, Jakubowski JA, Gilly WF, Sweedler JV. Two Toxins from Conus striatus That Individually Induce Tetanic Paralysis†. Biochemistry. 2006;45:14212–14222. doi: 10.1021/bi061485s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gilly WF, Richmond TA, Duda TF, Jr, Elliger C, Lebaric Z, Schulz J, Bingham JP, Sweedler JV. A diverse family of novel peptide toxins from an unusual cone snail, Conus californicus. J Exp Biol. 2011;214:147–161. doi: 10.1242/jeb.046086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lewis RJ. Conotoxin venom peptide therapeutics. Adv Exp Med Biol. 2009;655:44–48. doi: 10.1007/978-1-4419-1132-2_5. [DOI] [PubMed] [Google Scholar]
- 18.Twede VD, Miljanich G, Olivera BM, Bulaj G. Neuroprotective and cardioprotective conopeptides: an emerging class of drug leads. Curr Opin Drug Discov Devel. 2009;12:231–239. [PMC free article] [PubMed] [Google Scholar]
- 19.Strand FL. Neuropeptides : regulators of physiological processes. MIT Press; Cambridge, Mass: 1999. [Google Scholar]
- 20.Xu H, Shields D. Prohormone processing in the trans-Golgi network: endoproteolytic cleavage of prosomatostatin and formation of nascent secretory vesicles in permeabilized cells. J Cell Biol. 1993;122:1169–1184. doi: 10.1083/jcb.122.6.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jung LJ, Scheller RH. Peptide processing and targeting in the neuronal secretory pathway. Science. 1991;251:1330–1335. doi: 10.1126/science.2003219. [DOI] [PubMed] [Google Scholar]
- 22.Yang J, Brown MS, Liang G, Grishin NV, Goldstein JL. Identification of the acyltransferase that octanoylates ghrelin, an appetite-stimulating peptide hormone. Cell. 2008;132:387–396. doi: 10.1016/j.cell.2008.01.017. [DOI] [PubMed] [Google Scholar]
- 23.Gigoux V, Escrieut C, Silvente-Poirot S, Maigret B, Gouilleux L, Fehrentz JA, Gully D, Moroder L, Vaysse N, Fourmy D. Met-195 of the cholecystokinin-A receptor interacts with the sulfated tyrosine of cholecystokinin and is crucial for receptor transition to high affinity state. J Biol Chem. 1998;273:14380–14386. doi: 10.1074/jbc.273.23.14380. [DOI] [PubMed] [Google Scholar]
- 24.Hohmeier HE, Mulder H, Chen G, Henkel-Rieger R, Prentki M, Newgard CB. Isolation of INS-1-derived cell lines with robust ATP-sensitive K+ channel-dependent and -independent glucose-stimulated insulin secretion. Diabetes. 2000;49:424–430. doi: 10.2337/diabetes.49.3.424. [DOI] [PubMed] [Google Scholar]
- 25.Moynihan KA, Grimm AA, Plueger MM, Bernal-Mizrachi E, Ford E, Cras-Meneur C, Permutt MA, Imai S. Increased dosage of mammalian Sir2 in pancreatic beta cells enhances glucose-stimulated insulin secretion in mice. Cell Metab. 2005;2:105–117. doi: 10.1016/j.cmet.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 26.Matsumoto H, Noguchi J, Horikoshi Y, Kawamata Y, Kitada C, Hinuma S, Onda H, Nishimura O, Fujino M. Stimulation of prolactin release by prolactin-releasing peptide in rats. Biochemical and biophysical research communications. 1999;259:321–324. doi: 10.1006/bbrc.1999.0789. [DOI] [PubMed] [Google Scholar]
- 27.Hansotia T, Baggio LL, Delmeire D, Hinke SA, Yamada Y, Tsukiyama K, Seino Y, Holst JJ, Schuit F, Drucker DJ. Double incretin receptor knockout (DIRKO) mice reveal an essential role for the enteroinsular axis in transducing the glucoregulatory actions of DPP-IV inhibitors. Diabetes. 2004;53:1326–1335. doi: 10.2337/diabetes.53.5.1326. [DOI] [PubMed] [Google Scholar]
- 28.Hotamisligil GS, Peraldi P, Budavari A, Ellis R, White MF, Spiegelman BM. IRS-1-mediated inhibition of insulin receptor tyrosine kinase activity in TNF-alpha- and obesity-induced insulin resistance. Science. 1996;271:665–668. doi: 10.1126/science.271.5249.665. [DOI] [PubMed] [Google Scholar]
- 29.Bennett HP, McMartin C. Peptide hormones and their analogues: distribution, clearance from the circulation, and inactivation in vivo. Pharmacol Rev. 1978;30:247–292. [PubMed] [Google Scholar]
- 30.Thornberry NA, Weber AE. Discovery of JANUVIA (Sitagliptin), a selective dipeptidyl peptidase IV inhibitor for the treatment of type 2 diabetes. Curr Top Med Chem. 2007;7:557–568. doi: 10.2174/156802607780091028. [DOI] [PubMed] [Google Scholar]
- 31.Ozawa A, Lindberg I, Roth B, Kroeze WK. Deorphanization of Novel Peptides and Their Receptors. Aaps Journal. 2010;12:378–384. doi: 10.1208/s12248-010-9198-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rosenfeld L. Insulin: discovery and controversy. Clin Chem. 2002;48:2270–2288. [PubMed] [Google Scholar]
- 33.Kimball CP, Murlin JR. AQUEOUS EXTRACTS OF PANCREAS. Journal of Biological Chemistry. 1923;58:337–346. [Google Scholar]
- 34.Staub A, Sinn L, Behrens OK. Purification and crystallization of glucagon. J Biol Chem. 1955;214:619–632. [PubMed] [Google Scholar]
- 35.Hughes J. Isolation of an endogenous compound from brain with pharmacological properties similar to morphine. Brain Research. 1975;88:295–308. doi: 10.1016/0006-8993(75)90391-1. [DOI] [PubMed] [Google Scholar]
- 36.Hughes J, Smith TW, Kosterlitz HW, Fothergill LA, Morgan BA, Morris HR. Identification of 2 related pentapeptides from brain with potenet opiate agonist activity. Nature. 1975;258:577–579. doi: 10.1038/258577a0. [DOI] [PubMed] [Google Scholar]
- 37.Brownstein MJ. A brief history of opiates, opioid peptides, and opioid receptors. Proc Natl Acad Sci U S A. 1993;90:5391–5393. doi: 10.1073/pnas.90.12.5391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kotarsky K, Niclas EN. Reverse pharmacology and the deorphanization of 7TM receptors. Drug Discovery Today: Technologies. 2004;1:99–104. doi: 10.1016/j.ddtec.2004.07.003. [DOI] [PubMed] [Google Scholar]
- 39.Civelli O. GPCR deorphanizations: the novel, the known and the unexpected transmitters. Trends in pharmacological sciences. 2005;26:15–19. doi: 10.1016/j.tips.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 40.Nelson CP, Challiss RAJ. “Phenotypic” pharmacology: The influence of cellular environment on G protein-coupled receptor antagonist and inverse agonist pharmacology. Biochemical Pharmacology. 2007;73:737–751. doi: 10.1016/j.bcp.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 41.Sakurai T, Amemiya A, Ishii M, Matsuzaki I, Chemelli RM, Tanaka H, Williams SC, Richardson JA, Kozlowski GP, Wilson S, Arch JR, Buckingham RE, Haynes AC, Carr SA, Annan RS, McNulty DE, Liu WS, Terrett JA, Elshourbagy NA, Bergsma DJ, Yanagisawa M. Orexins and orexin receptors: a family of hypothalamic neuropeptides and G protein-coupled receptors that regulate feeding behavior. Cell. 1998;92:573–585. doi: 10.1016/s0092-8674(00)80949-6. [DOI] [PubMed] [Google Scholar]
- 42.Hinuma S, Habata Y, Fujii R, Kawamata Y, Hosoya M, Fukusumi S, Kitada C, Masuo Y, Asano T, Matsumoto H, Sekiguchi M, Kurokawa T, Nishimura O, Onda H, Fujino M. A prolactin-releasing peptide in the brain. Nature. 1998;393:272–276. doi: 10.1038/30515. [DOI] [PubMed] [Google Scholar]
- 43.Tatemoto K, Mutt V. Chemical determination of polypeptide hormones. Proc Natl Acad Sci U S A. 1978;75:4115–4119. doi: 10.1073/pnas.75.9.4115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tatemoto K, Mutt V. Isolation of two novel candidate hormones using a chemical method for finding naturally occurring polypeptides. Nature. 1980;285:417–418. doi: 10.1038/285417a0. [DOI] [PubMed] [Google Scholar]
- 45.Tatemoto K, Mutt V. Isolation and characterization of the intestinal peptide porcine PHI (PHI-27), a new member of the glucagon--secretin family. Proc Natl Acad Sci U S A. 1981;78:6603–6607. doi: 10.1073/pnas.78.11.6603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tatemoto K, Carlquist M, Mutt V. Neuropeptide Y--a novel brain peptide with structural similarities to peptide YY and pancreatic polypeptide. Nature. 1982;296:659–660. doi: 10.1038/296659a0. [DOI] [PubMed] [Google Scholar]
- 47.McDonald TJ, Jornvall H, Tatemoto K, Mutt V. Identification and characterization of variant forms of the gastrin-releasing peptide (GRP) FEBS letters. 1983;156:349–356. doi: 10.1016/0014-5793(83)80527-4. [DOI] [PubMed] [Google Scholar]
- 48.Tatemoto K, Rokaeus A, Jornvall H, McDonald TJ, Mutt V. Galanin - a novel biologically active peptide from porcine intestine. FEBS letters. 1983;164:124–128. doi: 10.1016/0014-5793(83)80033-7. [DOI] [PubMed] [Google Scholar]
- 49.Bradbury AF, Smyth DG. Peptide amidation. Trends in biochemical sciences. 1991;16:112–115. doi: 10.1016/0968-0004(91)90044-v. [DOI] [PubMed] [Google Scholar]
- 50.Evans JP, Blackburn NJ, Klinman JP. The catalytic role of the copper ligand H172 of peptidylglycine alpha-hydroxylating monooxygenase: a kinetic study of the H172A mutant. Biochemistry. 2006;45:15419–15429. doi: 10.1021/bi061734c. [DOI] [PubMed] [Google Scholar]
- 51.Chufan EE, De M, Eipper BA, Mains RE, Amzel LM. Amidation of bioactive peptides: the structure of the lyase domain of the amidating enzyme. Structure. 2009;17:965–973. doi: 10.1016/j.str.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Merkler DJ. C-terminal amidated peptides: production by the in vitro enzymatic amidation of glycine-extended peptides and the importance of the amide to bioactivity. Enzyme and microbial technology. 1994;16:450–456. doi: 10.1016/0141-0229(94)90014-0. [DOI] [PubMed] [Google Scholar]
- 53.Amara SG, Jonas V, Rosenfeld MG, Ong ES, Evans RM. Alternative RNA processing in calcitonin gene expression generates mRNAs encoding different polypeptide products. Nature. 1982;298:240–244. doi: 10.1038/298240a0. [DOI] [PubMed] [Google Scholar]
- 54.Fisher LA, Kikkawa DO, Rivier JE, Amara SG, Evans RM, Rosenfeld MG, Vale WW, Brown MR. Stimulation of noradrenergic sympathetic outflow by calcitonin gene-related peptide. Nature. 1983;305:534–536. doi: 10.1038/305534a0. [DOI] [PubMed] [Google Scholar]
- 55.Rosenfeld MG, Mermod JJ, Amara SG, Swanson LW, Sawchenko PE, Rivier J, Vale WW, Evans RM. Production of a novel neuropeptide encoded by the calcitonin gene via tissue-specific RNA processing. Nature. 1983;304:129–135. doi: 10.1038/304129a0. [DOI] [PubMed] [Google Scholar]
- 56.Gibson SJ, Polak JM, Bloom SR, Sabate IM, Mulderry PM, Ghatei MA, McGregor GP, Morrison JF, Kelly JS, Evans RM, et al. Calcitonin gene-related peptide immunoreactivity in the spinal cord of man and of eight other species. The Journal of neuroscience : the official journal of the Society for Neuroscience. 1984;4:3101–3111. doi: 10.1523/JNEUROSCI.04-12-03101.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Brain SD, Williams TJ, Tippins JR, Morris HR, MacIntyre I. Calcitonin gene-related peptide is a potent vasodilator. Nature. 1985;313:54–56. doi: 10.1038/313054a0. [DOI] [PubMed] [Google Scholar]
- 58.Durham PL. CGRP-receptor antagonists--a fresh approach to migraine therapy? The New England journal of medicine. 2004;350:1073–1075. doi: 10.1056/NEJMp048016. [DOI] [PubMed] [Google Scholar]
- 59.Ashina M, Bendtsen L, Jensen R, Schifter S, Olesen J. Evidence for increased plasma levels of calcitonin gene-related peptide in migraine outside of attacks. Pain. 2000;86:133–138. doi: 10.1016/s0304-3959(00)00232-3. [DOI] [PubMed] [Google Scholar]
- 60.Olesen J, Diener HC, Husstedt IW, Goadsby PJ, Hall D, Meier U, Pollentier S, Lesko LM. Calcitonin gene-related peptide receptor antagonist BIBN 4096 BS for the acute treatment of migraine. The New England journal of medicine. 2004;350:1104–1110. doi: 10.1056/NEJMoa030505. [DOI] [PubMed] [Google Scholar]
- 61.Ho TW, Mannix LK, Fan X, Assaid C, Furtek C, Jones CJ, Lines CR, Rapoport AM. Randomized controlled trial of an oral CGRP receptor antagonist, MK-0974, in acute treatment of migraine. Neurology. 2008;70:1304–1312. doi: 10.1212/01.WNL.0000286940.29755.61. [DOI] [PubMed] [Google Scholar]
- 62.Xiao Q, Giguere J, Parisien M, Jeng W, St-Pierre SA, Brubaker PL, Wheeler MB. Biological activities of glucagon-like peptide-1 analogues in vitro and in vivo. Biochemistry. 2001;40:2860–2869. doi: 10.1021/bi0014498. [DOI] [PubMed] [Google Scholar]
- 63.Egleton RD, Davis TP. Bioavailability and transport of peptides and peptide drugs into the brain. Peptides. 1997;18:1431–1439. doi: 10.1016/s0196-9781(97)00242-8. [DOI] [PubMed] [Google Scholar]
- 64.Roth BL, Baner K, Westkaemper R, Siebert D, Rice KC, Steinberg S, Ernsberger P, Rothman RB. Salvinorin A: a potent naturally occurring nonnitrogenous kappa opioid selective agonist. Proc Natl Acad Sci U S A. 2002;99:11934–11939. doi: 10.1073/pnas.182234399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Miyawaki K, Yamada Y, Yano H, Niwa H, Ban N, Ihara Y, Kubota A, Fujimoto S, Kajikawa M, Kuroe A, Tsuda K, Hashimoto H, Yamashita T, Jomori T, Tashiro F, Miyazaki J, Seino Y. Glucose intolerance caused by a defect in the entero-insular axis: a study in gastric inhibitory polypeptide receptor knockout mice. Proc Natl Acad Sci U S A. 1999;96:14843–14847. doi: 10.1073/pnas.96.26.14843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hampton LL, Ladenheim EE, Akeson M, Way JM, Weber HC, Sutliff VE, Jensen RT, Wine LJ, Arnheiter H, Battey JF. Loss of bombesin-induced feeding suppression in gastrin-releasing peptide receptor-deficient mice. Proc Natl Acad Sci U S A. 1998;95:3188–3192. doi: 10.1073/pnas.95.6.3188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Weber HC. Regulation and signaling of human bombesin receptors and their biological effects. Current opinion in endocrinology, diabetes, and obesity. 2009;16:66–71. doi: 10.1097/med.0b013e32831cf5aa. [DOI] [PubMed] [Google Scholar]
- 68.Batterham RL, Heffron H, Kapoor S, Chivers JE, Chandarana K, Herzog H, Le Roux CW, Thomas EL, Bell JD, Withers DJ. Critical role for peptide YY in protein-mediated satiation and body-weight regulation. Cell Metab. 2006;4:223–233. doi: 10.1016/j.cmet.2006.08.001. [DOI] [PubMed] [Google Scholar]
- 69.Colwell CS, Michel S, Itri J, Rodriguez W, Tam J, Lelievre V, Hu Z, Liu X, Waschek JA. Disrupted circadian rhythms in VIP- and PHI-deficient mice. American journal of physiology Regulatory, integrative and comparative physiology. 2003;285:R939–949. doi: 10.1152/ajpregu.00200.2003. [DOI] [PubMed] [Google Scholar]
- 70.Orskov C, Holst JJ, Knuhtsen S, Baldissera FG, Poulsen SS, Nielsen OV. Glucagon-like peptides GLP-1 and GLP-2, predicted products of the glucagon gene, are secreted separately from pig small intestine but not pancreas. Endocrinology. 1986;119:1467–1475. doi: 10.1210/endo-119-4-1467. [DOI] [PubMed] [Google Scholar]
- 71.Kieffer TJ, Habener JF. The glucagon-like peptides. Endocrine reviews. 1999;20:876–913. doi: 10.1210/edrv.20.6.0385. [DOI] [PubMed] [Google Scholar]
- 72.Sun YG, Chen ZF. A gastrin-releasing peptide receptor mediates the itch sensation in the spinal cord. Nature. 2007;448:700–703. doi: 10.1038/nature06029. [DOI] [PubMed] [Google Scholar]
- 73.Vuguin PM, Kedees MH, Cui L, Guz Y, Gelling RW, Nejathaim M, Charron MJ, Teitelman G. Ablation of the glucagon receptor gene increases fetal lethality and produces alterations in islet development and maturation. Endocrinology. 2006;147:3995–4006. doi: 10.1210/en.2005-1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gelling RW, Du XQ, Dichmann DS, Romer J, Huang H, Cui L, Obici S, Tang B, Holst JJ, Fledelius C, Johansen PB, Rossetti L, Jelicks LA, Serup P, Nishimura E, Charron MJ. Lower blood glucose, hyperglucagonemia, and pancreatic alpha cell hyperplasia in glucagon receptor knockout mice. Proc Natl Acad Sci U S A. 2003;100:1438–1443. doi: 10.1073/pnas.0237106100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Patchett AA, Harris E, Tristram EW, Wyvratt MJ, Wu MT, Taub D, Peterson ER, Ikeler TJ, ten Broeke J, Payne LG, Ondeyka DL, Thorsett ED, Greenlee WJ, Lohr NS, Hoffsommer RD, Joshua H, Ruyle WV, Rothrock JW, Aster SD, Maycock AL, Robinson FM, Hirschmann R, Sweet CS, Ulm EH, Gross DM, Vassil TC, Stone CA. A new class of angiotensin-converting enzyme inhibitors. Nature. 1980;288:280–283. doi: 10.1038/288280a0. [DOI] [PubMed] [Google Scholar]
- 76.Kjems LL, Holst JJ, Volund A, Madsbad S. The influence of GLP-1 on glucose-stimulated insulin secretion: effects on beta-cell sensitivity in type 2 and nondiabetic subjects. Diabetes. 2003;52:380–386. doi: 10.2337/diabetes.52.2.380. [DOI] [PubMed] [Google Scholar]
- 77.Marguet D, Baggio L, Kobayashi T, Bernard AM, Pierres M, Nielsen PF, Ribel U, Watanabe T, Drucker DJ, Wagtmann N. Enhanced insulin secretion and improved glucose tolerance in mice lacking CD26. Proc Natl Acad Sci U S A. 2000;97:6874–6879. doi: 10.1073/pnas.120069197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rosenblum JS, Kozarich JW. Prolyl peptidases: a serine protease subfamily with high potential for drug discovery. Curr Opin Chem Biol. 2003;7:496–504. doi: 10.1016/s1367-5931(03)00084-x. [DOI] [PubMed] [Google Scholar]
- 79.Augeri DJ, Robl JA, Betebenner DA, Magnin DR, Khanna A, Robertson JG, Wang A, Simpkins LM, Taunk P, Huang Q, Han SP, Abboa-Offei B, Cap M, Xin L, Tao L, Tozzo E, Welzel GE, Egan DM, Marcinkeviciene J, Chang SY, Biller SA, Kirby MS, Parker RA, Hamann LG. Discovery and preclinical profile of Saxagliptin (BMS-477118): a highly potent, long-acting, orally active dipeptidyl peptidase IV inhibitor for the treatment of type 2 diabetes. Journal of medicinal chemistry. 2005;48:5025–5037. doi: 10.1021/jm050261p. [DOI] [PubMed] [Google Scholar]
- 80.Deacon CF, Holst JJ. Immunoassays for the incretin hormones GIP and GLP-1. Best practice & research Clinical endocrinology & metabolism. 2009;23:425–432. doi: 10.1016/j.beem.2009.03.006. [DOI] [PubMed] [Google Scholar]
- 81.Bidlingmaier M, Freda PU. Measurement of human growth hormone by immunoassays: current status, unsolved problems and clinical consequences. Growth hormone & IGF research : official journal of the Growth Hormone Research Society and the International IGF Research Society. 2010;20:19–25. doi: 10.1016/j.ghir.2009.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jankowski V, Vanholder R, van der Giet M, Tolle M, Karadogan S, Gobom J, Furkert J, Oksche A, Krause E, Tran TN, Tepel M, Schuchardt M, Schluter H, Wiedon A, Beyermann M, Bader M, Todiras M, Zidek W, Jankowski J. Mass-spectrometric identification of a novel angiotensin peptide in human plasma. Arterioscler Thromb Vasc Biol. 2007;27:297–302. doi: 10.1161/01.ATV.0000253889.09765.5f. [DOI] [PubMed] [Google Scholar]
- 83.Deacon CF, Johnsen AH, Holst JJ. Degradation of glucagon-like peptide-1 by human plasma in vitro yields an N-terminally truncated peptide that is a major endogenous metabolite in vivo. J Clin Endocrinol Metab. 1995;80:952–957. doi: 10.1210/jcem.80.3.7883856. [DOI] [PubMed] [Google Scholar]
- 84.Svensson M, Skold K, Svenningsson P, Andren PE. Peptidomics-based discovery of novel neuropeptides. J Proteome Res. 2003;2:213–219. doi: 10.1021/pr020010u. [DOI] [PubMed] [Google Scholar]
- 85.Hummon AB, Richmond TA, Verleyen P, Baggerman G, Huybrechts J, Ewing MA, Vierstraete E, Rodriguez-Zas SL, Schoofs L, Robinson GE, Sweedler JV. From the genome to the proteome: uncovering peptides in the Apis brain. Science. 2006;314:647–649. doi: 10.1126/science.1124128. [DOI] [PubMed] [Google Scholar]
- 86.Domon B, Aebersold R. Review - Mass spectrometry and protein analysis. Science. 2006;312:212–217. doi: 10.1126/science.1124619. [DOI] [PubMed] [Google Scholar]
- 87.Skold K, Svensson M, Norrman M, Sjogren B, Svenningsson P, Andren PE. The significance of biochemical and molecular sample integrity in brain proteomics and peptidomics: stathmin 2-20 and peptides as sample quality indicators. Proteomics. 2007;7:4445–4456. doi: 10.1002/pmic.200700142. [DOI] [PubMed] [Google Scholar]
- 88.Parkin MC, Wei H, O’Callaghan JP, Kennedy RT. Sample-dependent effects on the neuropeptidome detected in rat brain tissue preparations by capillary liquid chromatography with tandem mass spectrometry. Analytical chemistry. 2005;77:6331–6338. doi: 10.1021/ac050712d. [DOI] [PubMed] [Google Scholar]
- 89.Che FY, Lim J, Pan H, Biswas R, Fricker LD. Quantitative neuropeptidomics of microwave-irradiated mouse brain and pituitary. Molecular & cellular proteomics : MCP. 2005;4:1391–1405. doi: 10.1074/mcp.T500010-MCP200. [DOI] [PubMed] [Google Scholar]
- 90.Nolte WM, Tagore DM, Lane WS, Saghatelian A. Peptidomics of prolyl endopeptidase in the central nervous system. Biochemistry. 2009;48:11971–11981. doi: 10.1021/bi901637c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tagore DM, Nolte WM, Neveu JM, Rangel R, Guzman-Rojas L, Pasqualini R, Arap W, Lane WS, Saghatelian A. Peptidase substrates via global peptide profiling. Nat Chem Biol. 2009;5:23–25. doi: 10.1038/nchembio.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Tinoco AD, Tagore DM, Saghatelian A. Expanding the dipeptidyl peptidase 4-regulated peptidome via an optimized peptidomics platform. J Am Chem Soc. 2010;132:3819–3830. doi: 10.1021/ja909524e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sadygov RG, Cociorva D, Yates JR., 3rd Large-scale database searching using tandem mass spectra: looking up the answer in the back of the book. Nat Methods. 2004;1:195–202. doi: 10.1038/nmeth725. [DOI] [PubMed] [Google Scholar]
- 94.Elias JE, Haas W, Faherty BK, Gygi SP. Comparative evaluation of mass spectrometry platforms used in large-scale proteomics investigations. Nat Methods. 2005;2:667–675. doi: 10.1038/nmeth785. [DOI] [PubMed] [Google Scholar]
- 95.Yamaguchi H, Sasaki K, Satomi Y, Shimbara T, Kageyama H, Mondal MS, Toshinai K, Date Y, Gonzalez LJ, Shioda S, Takao T, Nakazato M, Minamino N. Peptidomic identification and biological validation of neuroendocrine regulatory peptide-1 and -2. J Biol Chem. 2007;282:26354–26360. doi: 10.1074/jbc.M701665200. [DOI] [PubMed] [Google Scholar]
- 96.Nielsen S, DiGiovanni SR, Christensen EI, Knepper MA, Harris HW. Cellular and subcellular immunolocalization of vasopressin-regulated water channel in rat kidney. Proc Natl Acad Sci U S A. 1993;90:11663–11667. doi: 10.1073/pnas.90.24.11663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Nielsen S, Chou CL, Marples D, Christensen EI, Kishore BK, Knepper MA. Vasopressin increases water permeability of kidney collecting duct by inducing translocation of aquaporin-CD water channels to plasma membrane. Proc Natl Acad Sci U S A. 1995;92:1013–1017. doi: 10.1073/pnas.92.4.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Robertson GL, Shelton RL, Athar S. The osmoregulation of vasopressin. Kidney Int. 1976;10:25–37. doi: 10.1038/ki.1976.76. [DOI] [PubMed] [Google Scholar]
- 99.Insel TR. The challenge of translation in social neuroscience: a review of oxytocin, vasopressin, and affiliative behavior. Neuron. 2010;65:768–779. doi: 10.1016/j.neuron.2010.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Rozen-Zvi B, Yahav D, Gheorghiade M, Korzets A, Leibovici L, Gafter U. Vasopressin receptor antagonists for the treatment of hyponatremia: systematic review and meta-analysis. Am J Kidney Dis. 2010;56:325–337. doi: 10.1053/j.ajkd.2010.01.013. [DOI] [PubMed] [Google Scholar]
- 101.Donaldson ZR, Young LJ. Oxytocin, vasopressin, and the neurogenetics of sociality. Science. 2008;322:900–904. doi: 10.1126/science.1158668. [DOI] [PubMed] [Google Scholar]
- 102.Sasaki K, Takahashi N, Satoh M, Yamasaki M, Minamino N. A peptidomics strategy for discovering endogenous bioactive peptides. Journal of proteome research. 2010;9:5047–5052. doi: 10.1021/pr1003455. [DOI] [PubMed] [Google Scholar]
- 103.Osaki T, Sasaki K, Minamino N. Peptidomics-Based Discovery of an Antimicrobial Peptide Derived from Insulin-Like Growth Factor-Binding Protein 5. Journal of proteome research. 2011;10:1870–1880. doi: 10.1021/pr101114a. [DOI] [PubMed] [Google Scholar]
- 104.Jones JI, Gockerman A, Busby WH, Jr, Camacho-Hubner C, Clemmons DR. Extracellular matrix contains insulin-like growth factor binding protein-5: potentiation of the effects of IGF-I. J Cell Biol. 1993;121:679–687. doi: 10.1083/jcb.121.3.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Mukherjee A, Wilson EM, Rotwein P. Insulin-like growth factor (IGF) binding protein-5 blocks skeletal muscle differentiation by inhibiting IGF actions. Mol Endocrinol. 2008;22:206–215. doi: 10.1210/me.2007-0336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sorensen OE, Follin P, Johnsen AH, Calafat J, Tjabringa GS, Hiemstra PS, Borregaard N. Human cathelicidin, hCAP-18, is processed to the antimicrobial peptide LL-37 by extracellular cleavage with proteinase 3. Blood. 2001;97:3951–3959. doi: 10.1182/blood.v97.12.3951. [DOI] [PubMed] [Google Scholar]
- 107.Schroder JM, Harder J. Human beta-defensin-2. Int J Biochem Cell Biol. 1999;31:645–651. doi: 10.1016/s1357-2725(99)00013-8. [DOI] [PubMed] [Google Scholar]
- 108.Gharahdaghi F, Weinberg CR, Meagher DA, Imai BS, Mische SM. Mass spectrometric identification of proteins from silver-stained polyacrylamide gel: a method for the removal of silver ions to enhance sensitivity. Electrophoresis. 1999;20:601–605. doi: 10.1002/(SICI)1522-2683(19990301)20:3<601::AID-ELPS601>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 109.Link AJ, Eng J, Schieltz DM, Carmack E, Mize GJ, Morris DR, Garvik BM, Yates JR., 3rd Direct analysis of protein complexes using mass spectrometry. Nat Biotechnol. 1999;17:676–682. doi: 10.1038/10890. [DOI] [PubMed] [Google Scholar]
- 110.Kishi T, Grass L, Soosaipillai A, Shimizu-Okabe C, Diamandis EP. Human kallikrein 8: immunoassay development and identification in tissue extracts and biological fluids. Clin Chem. 2003;49:87–96. doi: 10.1373/49.1.87. [DOI] [PubMed] [Google Scholar]
- 111.Hahm S, Mizuno TM, Wu TJ, Wisor JP, Priest CA, Kozak CA, Boozer CN, Peng B, McEvoy RC, Good P, Kelley KA, Takahashi JS, Pintar JE, Roberts JL, Mobbs CV, Salton SR. Targeted deletion of the Vgf gene indicates that the encoded secretory peptide precursor plays a novel role in the regulation of energy balance. Neuron. 1999;23:537–548. doi: 10.1016/s0896-6273(00)80806-5. [DOI] [PubMed] [Google Scholar]
- 112.Chan WC, White PD. Fmoc solid phase peptide synthesis : a practical approach. Vol. 222. Oxford University Press; New York: 2000. [Google Scholar]
- 113.Brockmann A, Annangudi SP, Richmond TA, Ament SA, Xie F, Southey BR, Rodriguez-Zas SR, Robinson GE, Sweedler JV. Quantitative peptidomics reveal brain peptide signatures of behavior. Proc Natl Acad Sci U S A. 2009;106:2383–2388. doi: 10.1073/pnas.0813021106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Che FY, Biswas R, Fricker LD. Relative quantitation of peptides in wild-type and Cpe(fat/fat) mouse pituitary using stable isotopic tags and mass spectrometry. Journal of mass spectrometry : JMS. 2005;40:227–237. doi: 10.1002/jms.742. [DOI] [PubMed] [Google Scholar]
- 115.Che FY, Fricker LD. Quantitation of neuropeptides in Cpe(fat)/Cpe(fat) mice using differential isotopic tags and mass spectrometry. Anal Chem. 2002;74:3190–3198. doi: 10.1021/ac015681a. [DOI] [PubMed] [Google Scholar]
- 116.Morano C, Zhang X, Fricker LD. Multiple isotopic labels for quantitative mass spectrometry. Analytical chemistry. 2008;80:9298–9309. doi: 10.1021/ac801654h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ramos-Ortolaza DL, Bushlin I, Abul-Husn N, Annangudi SP, Sweedler J, Devi LA. Quantitative neuroproteomics of the synapse. Methods in molecular biology. 2010;615:227–246. doi: 10.1007/978-1-60761-535-4_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Nanni P, Levander F, Roda G, Caponi A, James P, Roda A. A label-free nano-liquid chromatography-mass spectrometry approach for quantitative serum peptidomics in Crohn’s disease patients. Journal of chromatography. B, Analytical technologies in the biomedical and life sciences. 2009;877:3127–3136. doi: 10.1016/j.jchromb.2009.08.003. [DOI] [PubMed] [Google Scholar]
- 119.Scholz B, Skold K, Kultima K, Fernandez C, Waldemarson S, Savitski MM, Soderquist M, Boren M, Stella R, Andren P, Zubarev R, James P. Impact of temperature dependent sampling procedures in proteomics and peptidomics - a characterization of the liver and pancreas post mortem degradome. Molecular & cellular proteomics : MCP. 2011;10 doi: 10.1074/mcp.M900229-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Quintana LF, Campistol JM, Alcolea MP, Banon-Maneus E, Sol-Gonzalez A, Cutillas PR. Application of label-free quantitative peptidomics for the identification of urinary biomarkers of kidney chronic allograft dysfunction. Molecular & cellular proteomics : MCP. 2009;8:1658–1673. doi: 10.1074/mcp.M900059-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Menzel C, Guillou V, Kellmann M, Khamenya V, Juergens M, Schulz-Knappe P. High-throughput biomarker discovery and identification by mass spectrometry. Combinatorial chemistry & high throughput screening. 2005;8:743–755. doi: 10.2174/138620705774962373. [DOI] [PubMed] [Google Scholar]
- 122.Menzel R, Muller U. Learning and memory in honeybees: from behavior to neural substrates. Annu Rev Neurosci. 1996;19:379–404. doi: 10.1146/annurev.ne.19.030196.002115. [DOI] [PubMed] [Google Scholar]
- 123.Whitfield CW, Cziko AM, Robinson GE. Gene expression profiles in the brain predict behavior in individual honey bees. Science. 2003;302:296–299. doi: 10.1126/science.1086807. [DOI] [PubMed] [Google Scholar]
- 124.Ruppell O, Pankiw T, Page RE., Jr Pleiotropy, epistasis and new QTL: the genetic architecture of honey bee foraging behavior. J Hered. 2004;95:481–491. doi: 10.1093/jhered/esh072. [DOI] [PubMed] [Google Scholar]
- 125.McLachlan GJ. Discriminant analysis and statistical pattern recognition. Wiley; New York: 1992. [Google Scholar]
- 126.Saghatelian A, Trauger SA, Want EJ, Hawkins EG, Siuzdak G, Cravatt BF. Assignment of endogenous substrates to enzymes by global metabolite profiling. Biochemistry. 2004;43:14332–14339. doi: 10.1021/bi0480335. [DOI] [PubMed] [Google Scholar]
- 127.Backes BJ, Harris JL, Leonetti F, Craik CS, Ellman JA. Synthesis of positional-scanning libraries of fluorogenic peptide substrates to define the extended substrate specificity of plasmin and thrombin. Nat Biotechnol. 2000;18:187–193. doi: 10.1038/72642. [DOI] [PubMed] [Google Scholar]
- 128.Leiting B, Pryor KD, Wu JK, Marsilio F, Patel RA, Craik CS, Ellman JA, Cummings RT, Thornberry NA. Catalytic properties and inhibition of proline-specific dipeptidyl peptidases II, IV and VII. Biochem J. 2003;371:525–532. doi: 10.1042/BJ20021643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Mentlein R, Gallwitz B, Schmidt WE. Dipeptidyl-peptidase IV hydrolyses gastric inhibitory polypeptide, glucagon-like peptide-1(7-36)amide, peptide histidine methionine and is responsible for their degradation in human serum. European journal of biochemistry / FEBS. 1993;214:829–835. doi: 10.1111/j.1432-1033.1993.tb17986.x. [DOI] [PubMed] [Google Scholar]
- 130.Saghatelian A, Cravatt BF. Assignment of protein function in the postgenomic era. Nature chemical biology. 2005;1:130–142. doi: 10.1038/nchembio0805-130. [DOI] [PubMed] [Google Scholar]
- 131.Tinoco AD, Kim YG, Tagore DM, Wiwczar J, Lane WS, Danial NN, Saghatelian A. A Peptidomics Strategy To Elucidate the Proteolytic Pathways That Inactivate Peptide Hormones. Biochemistry. 2011;50:2213–2222. doi: 10.1021/bi2000033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Che FY, Yan L, Li H, Mzhavia N, Devi LA, Fricker LD. Identification of peptides from brain and pituitary of Cpe(fat)/Cpe(fat) mice. Proc Natl Acad Sci U S A. 2001;98:9971–9976. doi: 10.1073/pnas.161542198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Tenorio-Laranga J, Valero ML, Mannisto PT, Sanchez del Pino M, Garcia-Horsman JA. Combination of snap freezing, differential pH two-dimensional reverse-phase high-performance liquid chromatography, and iTRAQ technology for the peptidomic analysis of the effect of prolyl oligopeptidase inhibition in the rat brain. Anal Biochem. 2009;393:80–87. doi: 10.1016/j.ab.2009.06.019. [DOI] [PubMed] [Google Scholar]
- 134.Pan H, Che FY, Peng B, Steiner DF, Pintar JE, Fricker LD. The role of prohormone convertase-2 in hypothalamic neuropeptide processing: a quantitative neuropeptidomic study. J Neurochem. 2006;98:1763–1777. doi: 10.1111/j.1471-4159.2006.04067.x. [DOI] [PubMed] [Google Scholar]
- 135.Steiner DF. The proprotein convertases. Curr Opin Chem Biol. 1998;2:31–39. doi: 10.1016/s1367-5931(98)80033-1. [DOI] [PubMed] [Google Scholar]
- 136.Allen RG, Peng B, Pellegrino MJ, Miller ED, Grandy DK, Lundblad JR, Washburn CL, Pintar JE. Altered processing of pro-orphanin FQ/nociceptin and pro-opiomelanocortin-derived peptides in the brains of mice expressing defective prohormone convertase 2. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2001;21:5864–5870. doi: 10.1523/JNEUROSCI.21-16-05864.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Berman Y, Mzhavia N, Polonskaia A, Furuta M, Steiner DF, Pintar JE, Devi LA. Defective prodynorphin processing in mice lacking prohormone convertase PC2. Journal of neurochemistry. 2000;75:1763–1770. doi: 10.1046/j.1471-4159.2000.0751763.x. [DOI] [PubMed] [Google Scholar]
- 138.Walter R, Shlank H, Glass JD, Schwartz IL, Kerenyi TD. Leucylglycinamide released from oxytocin by human uterine enzyme. Science. 1971;173:827–829. doi: 10.1126/science.173.3999.827. [DOI] [PubMed] [Google Scholar]
- 139.Toide K, Iwamoto Y, Fujiwara T, Abe H. JTP-4819: a novel prolyl endopeptidase inhibitor with potential as a cognitive enhancer. J Pharmacol Exp Ther. 1995;274:1370–1378. [PubMed] [Google Scholar]
- 140.Toide K, Okamiya K, Iwamoto Y, Kato T. Effect of a novel prolyl endopeptidase inhibitor, JTP-4819, on prolyl endopeptidase activity and substance P- and arginine-vasopressin-like immunoreactivity in the brains of aged rats. J Neurochem. 1995;65:234–240. doi: 10.1046/j.1471-4159.1995.65010234.x. [DOI] [PubMed] [Google Scholar]
- 141.Cavasin MA, Rhaleb NE, Yang XP, Carretero OA. Prolyl oligopeptidase is involved in release of the antifibrotic peptide Ac-SDKP. Hypertension. 2004;43:1140–1145. doi: 10.1161/01.HYP.0000126172.01673.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Barelli H, Petit A, Hirsch E, Wilk S, De Nanteuil G, Morain P, Checler F. S 17092-1, a highly potent, specific and cell permeant inhibitor of human proline endopeptidase. Biochem Biophys Res Commun. 1999;257:657–661. doi: 10.1006/bbrc.1999.0366. [DOI] [PubMed] [Google Scholar]
- 143.Schneider JS, Giardiniere M, Morain P. Effects of the prolyl endopeptidase inhibitor S 17092 on cognitive deficits in chronic low dose MPTP-treated monkeys. Neuropsychopharmacology. 2002;26:176–182. doi: 10.1016/S0893-133X(01)00307-4. [DOI] [PubMed] [Google Scholar]
- 144.Morain P, Lestage P, De Nanteuil G, Jochemsen R, Robin JL, Guez D, Boyer PA. S 17092: a prolyl endopeptidase inhibitor as a potential therapeutic drug for memory impairment. Preclinical and clinical studies. CNS Drug Rev. 2002;8:31–52. doi: 10.1111/j.1527-3458.2002.tb00214.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Bellemere G, Vaudry H, Morain P, Jegou S. Effect of prolyl endopeptidase inhibition on arginine-vasopressin and thyrotrophin-releasing hormone catabolism in the rat brain. J Neuroendocrinol. 2005;17:306–313. doi: 10.1111/j.1365-2826.2005.01308.x. [DOI] [PubMed] [Google Scholar]
- 146.Wiese S, Reidegeld KA, Meyer HE, Warscheid B. Protein labeling by iTRAQ: a new tool for quantitative mass spectrometry in proteome research. Proteomics. 2007;7:340–350. doi: 10.1002/pmic.200600422. [DOI] [PubMed] [Google Scholar]
- 147.Tatemoto K, Mutt V. Isolation and characterization of the intestinal peptide porcine PHI (PHI-27), a new member of the glucagon--secretin family. Proc Natl Acad Sci U S A. 1981;78:6603–6607. doi: 10.1073/pnas.78.11.6603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Werner S, Hulting AL, Hokfelt T, Eneroth P, Tatemoto K, Mutt V, Maroder L, Wunsch E. Effect of the peptide PHI-27 on prolactin release in vitro. Neuroendocrinology. 1983;37:476–478. doi: 10.1159/000123598. [DOI] [PubMed] [Google Scholar]
- 149.Szecowka J, Tatemoto K, Mutt V, Efendic S. Interaction of a newly isolated intestinal polypeptide (PHI) with glucose and arginine to effect the secretion of insulin and glucagon. Life Sci. 1980;26:435–438. doi: 10.1016/0024-3205(80)90162-9. [DOI] [PubMed] [Google Scholar]
- 150.Menschaert G, Vandekerckhove TTM, Baggerman G, Schoofs L, Luyten W, Van Criekinge W. Peptidomics Coming of Age: A Review of Contributions from a Bioinformatics Angle. Journal of proteome research. 2010;9:2051–2061. doi: 10.1021/pr900929m. [DOI] [PubMed] [Google Scholar]
- 151.Danial NN, Walensky LD, Zhang CY, Choi CS, Fisher JK, Molina AJ, Datta SR, Pitter KL, Bird GH, Wikstrom JD, Deeney JT, Robertson K, Morash J, Kulkarni A, Neschen S, Kim S, Greenberg ME, Corkey BE, Shirihai OS, Shulman GI, Lowell BB, Korsmeyer SJ. Dual role of proapoptotic BAD in insulin secretion and beta cell survival. Nat Med. 2008;14:144–153. doi: 10.1038/nm1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Hashimoto Y, Ito Y, Niikura T, Shao Z, Hata M, Oyama F, Nishimoto I. Mechanisms of neuroprotection by a novel rescue factor humanin from Swedish mutant amyloid precursor protein. Biochemical and Biophysical Research Communications. 2001;283:460–468. doi: 10.1006/bbrc.2001.4765. [DOI] [PubMed] [Google Scholar]
- 153.Guo B, Zhai D, Cabezas E, Welsh K, Nouraini S, Satterthwait AC, Reed JC. Humanin peptide suppresses apoptosis by interfering with Bax activation. Nature. 2003;423:456–461. doi: 10.1038/nature01627. [DOI] [PubMed] [Google Scholar]
- 154.Hashimoto Y, Kondo T, Kageyama Y. Lilliputians get into the limelight: novel class of small peptide genes in morphogenesis. Dev Growth Differ. 2008;50(Suppl 1):S269–276. doi: 10.1111/j.1440-169X.2008.00994.x. [DOI] [PubMed] [Google Scholar]
- 155.Wadler CS, Vanderpool CK. A dual function for a bacterial small RNA: SgrS performs base pairing-dependent regulation and encodes a functional polypeptide. Proc Natl Acad Sci U S A. 2007;104:20454–20459. doi: 10.1073/pnas.0708102104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Hashimoto Y, Kondo T, Kageyama Y. Lilliputians get into the limelight: Novel class of small peptide genes in morphogenesis. Development Growth & Differentiation. 2008;50:S269–S276. doi: 10.1111/j.1440-169X.2008.00994.x. [DOI] [PubMed] [Google Scholar]
- 157.Tajima H, Niikura T, Hashimoto Y, Ito Y, Kita Y, Terashita K, Yamazaki K, Koto A, Aiso S, Nishimoto I. Evidence for in vivo production of Humanin peptide, a neuroprotective factor against Alzheimer’s disease-related insults. Neuroscience Letters. 2002;324:227–231. doi: 10.1016/s0304-3940(02)00199-4. [DOI] [PubMed] [Google Scholar]
- 158.Kondo T, Plaza S, Zanet J, Benrabah E, Valenti P, Hashimoto Y, Kobayashi S, Payre F, Kageyama Y. Small Peptides Switch the Transcriptional Activity of Shavenbaby During Drosophila Embryogenesis. Science. 2010;329:336–339. doi: 10.1126/science.1188158. [DOI] [PubMed] [Google Scholar]
- 159.Castellana NE, Payne SH, Shen ZX, Stanke M, Bafna V, Briggs SP. Discovery and revision of Arabidopsis genes by proteogenomics. Proc Natl Acad Sci U S A. 2008;105:21034–21038. doi: 10.1073/pnas.0811066106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Washietl S, Findeiss S, Muller SA, Kalkhof S, von Bergen M, Hofacker IL, Stadler PF, Goldman N. RNAcode: Robust discrimination of coding and noncoding regions in comparative sequence data. Rna-a Publication of the Rna Society. 2011;17:578–594. doi: 10.1261/rna.2536111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Yang XH, Tschaplinski TJ, Hurst GB, Jawdy S, Abraham PE, Lankford PK, Adams RM, Shah MB, Hettich RL, Lindquist E, Kalluri UC, Gunter LE, Pennacchio C, Tuskan GA. Discovery and annotation of small proteins using genomics, proteomics, and computational approaches. Genome Research. 2011;21:634–641. doi: 10.1101/gr.109280.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Koehn FE, Carter GT. The evolving role of natural products in drug discovery. Nat Rev Drug Discov. 2005;4:206–220. doi: 10.1038/nrd1657. [DOI] [PubMed] [Google Scholar]
- 163.Adessi C, Soto C. Converting a peptide into a drug: strategies to improve stability and bioavailability. Curr Med Chem. 2002;9:963–978. doi: 10.2174/0929867024606731. [DOI] [PubMed] [Google Scholar]
- 164.Gartlehner G, Hansen RA, Jonas BL, Thieda P, Lohr KN. The comparative efficacy and safety of biologics for the treatment of rheumatoid arthritis: a systematic review and metaanalysis. J Rheumatol. 2006;33:2398–2408. [PubMed] [Google Scholar]
- 165.Bostrom J, Yu SF, Kan D, Appleton BA, Lee CV, Billeci K, Man W, Peale F, Ross S, Wiesmann C, Fuh G. Variants of the antibody herceptin that interact with HER2 and VEGF at the antigen binding site. Science. 2009;323:1610–1614. doi: 10.1126/science.1165480. [DOI] [PubMed] [Google Scholar]
- 166.Jazirehi AR, Bonavida B. Cellular and molecular signal transduction pathways modulated by rituximab (rituxan, anti-CD20 mAb) in non-Hodgkin’s lymphoma: implications in chemosensitization and therapeutic intervention. Oncogene. 2005;24:2121–2143. doi: 10.1038/sj.onc.1208349. [DOI] [PubMed] [Google Scholar]
- 167.Hollander PA, Levy P, Fineman MS, Maggs DG, Shen LZ, Strobel SA, Weyer C, Kolterman OG. Pramlintide as an adjunct to insulin therapy improves long-term glycemic and weight control in patients with type 2 diabetes: a 1-year randomized controlled trial. Diabetes Care. 2003;26:784–790. doi: 10.2337/diacare.26.3.784. [DOI] [PubMed] [Google Scholar]
- 168.Moellering RE, Cornejo M, Davis TN, Del Bianco C, Aster JC, Blacklow SC, Kung AL, Gilliland DG, Verdine GL, Bradner JE. Direct inhibition of the NOTCH transcription factor complex. Nature. 2009;462:182–188. doi: 10.1038/nature08543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Walensky LD, Kung AL, Escher I, Malia TJ, Barbuto S, Wright RD, Wagner G, Verdine GL, Korsmeyer SJ. Activation of apoptosis in vivo by a hydrocarbon-stapled BH3 helix. Science. 2004;305:1466–1470. doi: 10.1126/science.1099191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Cronican JJ, Thompson DB, Beier KT, McNaughton BR, Cepko CL, Liu DR. Potent delivery of functional proteins into Mammalian cells in vitro and in vivo using a supercharged protein. ACS Chem Biol. 2010;5:747–752. doi: 10.1021/cb1001153. [DOI] [PMC free article] [PubMed] [Google Scholar]


