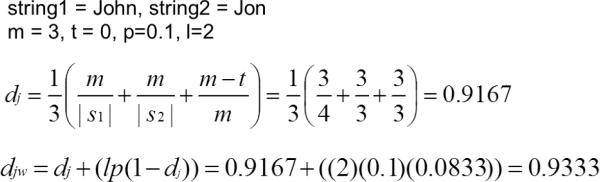Abstract
Record linkage is the task of identifying records from disparate data sources that refer to the same entity. It is an integral component of data processing in distributed settings, where the integration of information from multiple sources can prevent duplication and enrich overall data quality, thus enabling more detailed and correct analysis. Privacy-preserving record linkage (PPRL) is a variant of the task in which data owners wish to perform linkage without revealing identifiers associated with the records. This task is desirable in various domains, including healthcare, where it may not be possible to reveal patient identity due to confidentiality requirements, and in business, where it could be disadvantageous to divulge customers' identities. To perform PPRL, it is necessary to apply string comparators that function in the privacy-preserving space. A number of privacy-preserving string comparators (PPSCs) have been proposed, but little research has compared them in the context of a real record linkage application. This paper performs a principled and comprehensive evaluation of six PPSCs in terms of three key properties: 1) correctness of record linkage predictions, 2) computational complexity, and 3) security. We utilize a real publicly-available dataset, derived from the North Carolina voter registration database, to evaluate the tradeoffs between the aforementioned properties. Among our results, we find that PPSCs that partition, encode, and compare strings yield highly accurate record linkage results. However, as a tradeoff, we observe that such PPSCs are less secure than those that map and compare strings in a reduced dimensional space.
Keywords: privacy, record linkage, approximate matching, string comparison
1. Introduction
Information fusion is a process in which disparate, but related, pieces of data are combined in a meaningful way. The process of record linkage corresponds to a class of information fusion in which records2 held by multiple data owners are compared to identify and aggregate records referring to the same entity [1]. Record linkage is a crucial first step in data processing due to the fact that it enriches data quality, prevents duplication, and ultimately enables more accurate analysis [2]. Record linkage, however, is not always possible due to various societal concerns, as well as regulatory constraints, with respect to personal privacy [3]. As a result, the process of privacy-preserving record linkage (PPRL) has been established as a variant of the task in which the data owners can perform record linkage without revealing identifying information about the entities [4].
1.1. Applications and Opportunities
PPRL can enhance or enable novel record linkage applications in many different domains. One particular domain of interest is the biomedical realm, where the sharing of patient information is critical for research purposes [5]. The U.S. National Institutes of Health (NIH), for instance, requires that patient information used in federally funded studies must be shared for reuse in a “de-identified” form [6]. In other words, such data must not be shared with personal identifiers. Now, imagine that a life science researcher wishes to conduct a study on the relationship between two distinct biomedical attributes (e.g., DNA sequence and disease status) and intends to use such shared data. It is possible for patients to visit multiple healthcare providers, where their data is collected and applied in distinct studies [7]. If a patient's information is duplicated, queries issued across disparate submissions would result in overestimated correlations. On the other hand, if a patient's data is fragmented, such that a particular attribute (e.g., DNA sequence) is submitted from one institution and a separate attribute (e.g., disease status) is submitted by another, the query results would be underestimates. In this context, PPRL is necessary to mitigate bias in such statistical analyses without disclosing patient's identities3.
A second application of interest for PPRL is in counter-terrorism efforts. Currently, when a terrorist suspect is specified by an intelligence agency, record linkage is applied to identify records from multiple data owners to detect aliases or combine information about the individual to learn about their actions or co-conspirators [9]. More recently, from an international perspective, the European Union started to share information about inbound travelers (i.e., Passenger Name Records) to U.S. officials for inspection against terrorist “watchlists” [10]. These actions have met significant opposition due to concerns over personal privacy and autonomy (e.g., [11]). However, the adoption of PPRL could alleviate this tension because it eliminates the need to reveal the actual identities of suspects or travelers until a match on a watchlist is satisfied (i.e., only records referring to the suspect should be revealed) [12].
1.2. Overview of Record Linkage
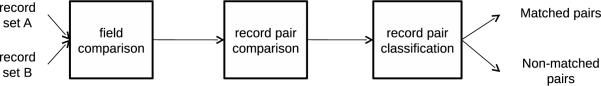
Record linkage is a multi-step process as depicted in Figure 1. The first step is field comparison, in which each field in each record pair is compared. In the record pair comparison step, the field similarities are converted into a single similarity score for each record pair. In the record pair classification step, the pairs are partitioned and classified into either the set of matches (e.g., record pairs that do refer to the same individual) or the set of non-matches (e.g., record pairs that do not refer to the same individual).
Figure 1.
Steps required in record linkage.
The focus of this study is the field comparison step. Further details of the other steps of the record linkage process can be found in Section 3.2.
1.3. Motivation for this Study
Within this paper, we consider the linkage of records from two data owners, which we refer to as Alice and Bob. Privacy-preserving string comparison is applied within the context of the following PPRL protocol.
First, Alice and Bob exchange the information required to encode records.
Next, Alice and Bob send encoded records to a third party Charlie.
Finally, the third party executes the linkage protocol.
We adopt the third party model based on the observation that such parties exist in real world privacy-preserving data sharing environments (e.g., [13]). In addition, in many cases, by using such parties, it is possible to create more efficient protocols [4].
A number of privacy-preserving string comparison techniques have been proposed in the literature, which begs the question, “Which is the best method”? It should first be recognized that privacy-preserving string comparators (PPSCs) are generally designed to compare encoded versions of an entity's identifying values. Unfortunately, many of these approaches function only when identifying values are recorded consistently and without error across disparate databases (e.g., [14, 15, 16, 17, 18]). This neglects the fact that variation (e.g., nicknames) or typographical error can corrupt personal identifiers [19]. In such cases, the application of equivalence-based models can result in subpar record linkage results. For example, the hashed4 value of “Jon” appears equally distant from the hashed values of “John” and “Sampson”5 [20].
In recognition of this limitation, various approximate comparators have been developed to compute string similarity, some of which have been translated into PPSCs (e.g., [21, 22]). In contrast to a measure of binary field similarity (i.e., exact matching), which results in a field similarity in [0,1], a measure of approximate field similarity provides a continuous measure of field similarity in the range [0,…,1]. Distance and similarity are inversely related (i.e., strings with a distance of 0 have a similarity of 1). While there is clear evidence (e.g., [23, 24]) that accounting for string similarity can improve record linkage results, it is unclear which PPSC should be selected for PPRL applications.
We recognize that several reviews of PPSCs have been conducted [25, 26, 27, 28], but they lack either the breadth (i.e., range of methods considered) or the depth necessary (i.e., a quantitative evaluation of the correctness, security, and computational complexity) to compare approaches on a common scale. For instance, Elmagarmid and colleagues [25] presented a comprehensive survey of record linkage techniques, but did not consider the privacy issues and the related PPRL techniques. Meanwhile, Trepetin [26] performed a brief review of PPSCs in association with the introduction of a new PPSC, but the review focused more on a qualitative discussion of computational complexity, rather than record linkage correctness and security of the various approaches. Another paper [27] provides an overview of privacy-preserving record linkage, current approaches, and open research questions, but again does not provide a formal, quantified analysis of existing approaches. In another study, Verykios and colleagues [28] performed a principled review of PPSCs, but focused on string comparison, rather than string comparison within the context of record linkage. Moreover, the latter study also neglected a formal security analysis of the PPSCs.
1.4. Contributions
Given the current state of affairs, in order for data owners to perform PPRL, they must sift through a variety of approaches that have not been directly compared to one another. Therefore, there is a need for a comprehensive evaluation of existing PPSCs that places the comparators on a level playing field. Our research makes the following contributions:
Taxonomy: We perform a comprehensive literature review of PPSCs. In doing so, we derive a taxonomy of existing approaches, which partitions PPSCs into distinct classes of techniques. Taxonomizing the approaches allows for analysis of features that are common to each class of methods.
Framework: We introduce a PPRL framework that is suitable for PPSCs that measure approximate field similarity, rather than field equivalence alone. Notably, this is an aspect of the broader PPRL problem which has been neglected.
Common Criteria: We define several quantifiable measures to compare PPSCs on common scales. These measures enable data managers to investigate the tradeoffs between PPSCs on three critical axes, namely 1) correctness in record linkage, 2) computational complexity as measured by running time, and 3) security.
Evaluation: We systematically investigate the relationship between six classes of PPSCs using a publicly available dataset. The fields present in the dataset consist of personal identifiers and demographics that have been proposed as key attributes for record linkage (e.g., personal name and residential address).
The remainder of the paper is organized as follows. In Section 2, we introduce a taxonomy of PPSCs. In Section 3, we present an overview of general record linkage frameworks and the specific methodologies used in this work. This section also presents the dataset utilized in our evaluation. In Section 4, we present details of the parameters used for each comparator in addition to the evaluation metrics. Then in Section 5, the correctness, computational complexity, and security metrics are reported for each comparator. Section 6 provides a discussion of factors affecting the performance of each comparator, limitations, and future work. Finally, we conclude the work in Section 7.
2. Background and Related Work
In this section, we introduce a taxonomy for privacy-preserving field comparators. Please note that only the privacy-preserving field comparators proposed in previous works are considered. This is in contrast to other aspects of the record linkage process, such as the record pair comparison and record pair classification steps. Holding constant the other steps in the record linkage process allows for the evaluation of multiple field comparison methods in an otherwise equivalent record linkage framework. This enables us to generalize the strengths and weaknesses of each class of methods.
A review of the literature surrounding privacy-preserving field comparators revealed the six broad categories shown in Table 1. The PPSCs empirically evaluated in this paper are denoted in italics. Several noteworthy privacy-preserving comparators more suitable for numerical fields [29, 30, 31] were considered, but were excluded from this study because our goal was to evaluate string, rather than numerical, comparators.
Table 1.
Taxonomy of privacy-preserving field comparators. The comparators evaluated in this paper are denoted in italics.
| Equivalence Testing n-gram Methods | Exact Matching [14, 15, 16, 17, 18] |
| Number of Common Bigrams [21] | |
| Bloom Filter [32] | |
| Trigrams [33] | |
| Reference Space Embedding | Public Reference Space Embedding [34] |
| Private Reference Space Embedding [35] | |
| Dissimilarity Matrix [30] | |
| Teamwork | Edit Similarity [36] |
| Secret Sharing [29] | |
| Euclidean Distance [31] | |
| Phonetic Filtering Guess & Encode Errors | Phonetic Filter [37] |
| Random Introduction of Errors [22, 38] | |
| Systematic Introduction of Errors [39] |
2.1. Exact Matching Methods
As alluded to, many PPSC protocols use variations of an “encode-and-compare” model in which identifiers are encoded and compared for equality [14, 15, 16, 17, 18]. When a plaintext string (i.e., human readable) is encoded, it is transformed into a form known as an encoding.
Comparators based on equivalence testing do not take into account the notion of similarity between the strings. This means that strings very similar in the plaintext space are generally very dissimilar in the encoded space [20]. Our hypothesis is that leveraging similarity will improve record linkage accuracy. To gauge the degree of improvement achieved through the use of approximate string comparators, we will use the Exact Matching comparator as a baseline.
2.2. n-gram Methods
In the n-gram class of PPSCs, strings are broken into the n-grams that compose the string, where n is an integer that can range from 1 to the length of the string. n-grams are usually padded on both ends with n − 1 blank spaces. to properly account for the first and last letters. For example, the 2-grams, also called bigrams, associated with the name “John” are “J”, “Jo”, “oh”, “hn”, and “n”.
2.2.1. Number of common bigrams
Churches and Christen proposed an approximate PPSC based on determining the number of bigrams shared in common between strings [21]. However, previous evaluations have already demonstrated this comparator did not perform well in record linkage and is too computationally intense to be practical [25, 26]. Therefore, we do not evaluate this comparator in this study.
2.2.2. Trigrams
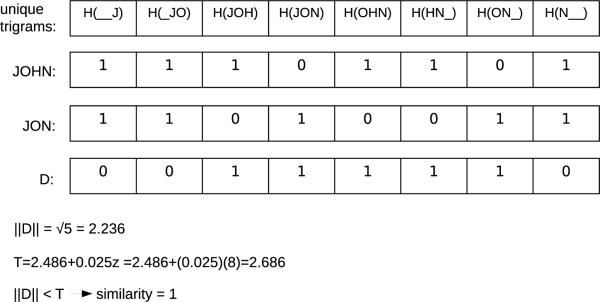
The comparator in [33] uses the set of encoded 3-grams, or trigrams, associated with the string. Each string is encoded as a sparse binary vector where each cell in the vector corresponds to a trigram. The length of the vector is the number of possible trigrams for the given alphabet. Each cell contains the count of the number of times the trigram corresponding to the cell appears in the given string. For example, “0” in a cell indicates the trigram to which the cell corresponds is not present in the given string whereas a “1” in a cell indicates the trigram to which the cell corresponds appears once in the given string. When two strings are compared, a difference vector D is calculated (see Figure 2) where each cell represents the difference in the number of times a trigram appears in each string. To determine the similarity of two strings, a threshold T is used6:
| (1) |
where z is the number of unique trigrams contained among the set of trigrams belonging to both strings. If the magnitude of the difference vector D is less than the threshold T, the strings are assigned a similarity score of 1 (i.e., the strings are completely similar); otherwise, the strings are assigned a similarity score of 0 (i.e., the strings are completely dissimilar).
Figure 2.
An example of the Trigrams comparator. H represents a hash function. The vector D contains the difference in the count of trigrams found in each string. For ease of representation, only trigrams present in at least one of the sample strings are shown in this figure. Assume that the value “JOHN' is held by Alice and the value “JON' by Bob. The magnitude of the vector is less than the threshold, so the similarity is determined to be 1.
2.2.3. Bloom Filter
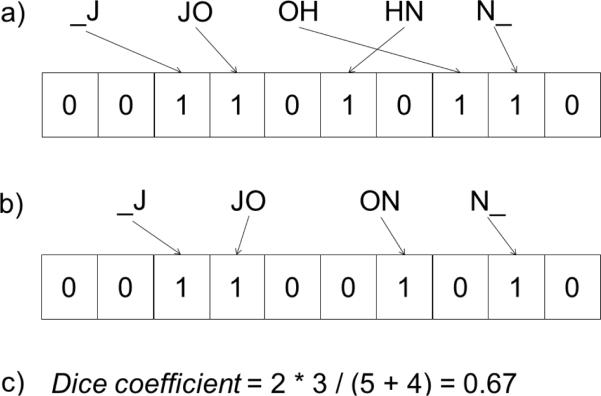
This comparator encodes a string by hashing the n-grams of the string into a Bloom filter [32]. A Bloom filter is a data structure commonly used to determine set membership or to compare sets of stored items [40, 41, 42]. It is represented as a bit array where all bits are initialized to 0. When an item is stored in the Bloom filter, the bits corresponding to the item are set to 1. When an additional item is stored in that Bloom filter, a bit set to 1 by the previous item can be “hit” again by the newly added item. In this case, the value of the bit remains at 1. This means that false positives, but not false negatives, are possible. The Bloom Filter approach in [32] generates an encoding for each string by hashing the bigrams associated with the string into a Bloom filter using q different hash functions. When Alice and Bob compare their strings, they agree upon the number of bits that compose the Bloom filter (i.e., its length), the number q of hash functions to apply, and which hash functions to utilize. Alice then encodes her string by hashing its corresponding bigrams into a new Bloom filter using all of the q agreed upon hash functions. Bob encodes his string similarly. To determine the approximated similarity of their strings, the corresponding Bloom filters are compared using a set-based similarity measure, such as the Dice coefficient:
| (2) |
where α and β are the Bloom filters containing the hashed bigrams of Alice's and Bob's encoded strings, respectively. An example is shown in Figure 3.
Figure 3.
An example of the Bloom filter comparator where each Bloom filter has 10 bits and a single hash function is used to hash each bigram into the filter. The Dice coefficient (Equation 2) is used to determine the similarity of the Bloom filters. The number of intersecting bits in the Bloom filters is 3, the number of bits set in Bloom filter a is 5, and the number of bits set in Bloom filter b is 4, yielding a similarity of 0.67.
2.3. Embedding Methods
The comparators in the Embedding class use a set of reference strings to define an embedding space. Alice and Bob embed a string by calculating the distance (as determined by some distance function7) between their string and the reference sets, resulting in a vector of distances. To compare the strings, Alice and Bob compare the distance vectors associated with each of their strings. The idea behind this approach is that if the strings are similar, they will also be of similar distance in the reference space.
Embedding approaches have been proposed that utilize either publicly available [34] or privately held [35] reference spaces. In the latter approach, Alice and Bob generate the reference space, which is kept secret from Charlie. We select this comparator for evaluation because a privately held reference space provides greater security than a publicly available reference space. For simplicity, this comparator will henceforth be referred to as Embedding.
The Embedding approach [35] is based on the use of the Sparse Map variant of Lipschitz embeddings. The general idea is that the embedding space is defined by several reference sets, each of which contain randomly generated reference strings. When a string is embedded in this space, its distance to each of those reference sets is measured. The distance from a string s to a reference set is the minimal distance between s and each string composing the reference set. The embedding of a string is therefore a vector of distances, where each element j denotes the distance from the string to reference set j. To compare strings, the Euclidean distance of their embeddings is calculated. A greedy resampling heuristic is used to limit the computational complexity by reducing the dimensionality of the embeddings such that, rather using all possible reference sets, only a subset of the most information-rich reference sets are used. A proper explanation is beyond the scope of this paper, but we refer the reader to [35] for the Embedding comparator and [43] for Sparse Map.
2.4. Teamwork Methods
The comparators in the teamwork class are so-called because they require that Alice and Bob interactively cooperate throughout the protocol to compare their strings.
2.4.1. Edit Similarity
The Levenshtein edit distance between two strings is the minimal number of insertions, deletions, and substitutions required to transform one string into the other [44]. A dynamic programming algorithm is often used to compute edit distance such that the minimal edit distance between the strings is iteratively calculated. For example, if the edit distance between strings of lenth d and e is calculated, a matrix of size d × e is set up. At the completion of the algorithm, the matrix cell M[d][e] represents the minimal edit distance between the two strings. Further details can be found in [45].
Attalah and colleagues present a comparator for securely calculating edit distance without revealing either string [36]. In this protocol, the standard dynamic programming algorithm for edit distance is applied with the exception that the matrix and its values are split between Alice and Bob. Alice and Bob fill in the cells of their respective matrices using a protocol that allows them to determine the optimal method for calculating matrix values, without revealing any information to one another. At the completion of the protocol, Alice and Bob encode and send the value in the final cell of the matrix to the third party Charlie. Charlie sums the values he received from Alice and Bob to determine the final edit distance.
While the original method [36] is edit distance, a more informative variant of this metric, edit similarity, was selected for evaluation in this manuscript. The difference between edit distance and edit similarity is discussed in Section 4.1.6.
2.5. Phonetic Filtering Methods
Phonetic filters are used to generate a phonetic representation of a string. This representation can overcome various errors, such typos and the use of nicknames. For example, “John” and “Jon” can be mapped to the same phonetic encoding (see Figure 4). Multiple phonetic encoding strategies, such as Soundex [37], Metaphone [46], and NYSIIS [47] have been used in record linkage applications.
Figure 4.
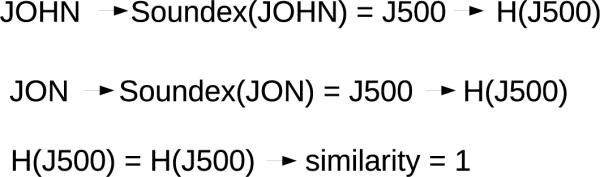
An example of the Phonetic Filter comparator. The Soundex code for each string is hashed and checked for equality. The hashed Soundex codes are equivalent, so the similarity of the strings is 1.
2.5.1. Phonetic Filter
Karakasidis and Verykios suggest using the Soundex phonetic filter to transform a string into its phonetic representation and then encoding the phonetic representation [37]. If the encoded phonetic representations match, the strings are given a similarity score of 1; otherwise, they are given a similarity score of 0.
2.6. Guess & Encode Errors Methods
When strings are encoded and compared, small differences in the plaintext strings can result in very different ciphertexts. To overcome the small differences that can be created through typographical, spelling, or other errors in strings, a class of of approaches have been proposed that attempt to preemptively generate errors that might be associated with a string. Rather than compare a single pair of strings, the sets of error-riddled relatives of the initial strings are compared [22, 38, 39]. When Alice and Bob compare their strings, if any of the error-riddled relatives of Alice's string match any of the error-riddled relatives of Bob's string, the strings are declared a match. However, these comparators result in many false positives with weak matching precision and require unreasonable storage [26].
3. Materials and methods
The previous section presented a taxonomy of existing PPSCs. However, string comparison is only one part of the larger record linkage process. In this section, we provide a broad overview of the steps involved in record linkage: field comparison, record pair comparison, and record pair classification. This section also provides the details of the algorithm used in the record pair comparison step and the process used for record pair classification. Finally, the dataset and implementation details are provided.
3.1. Notation
For this work, we assume each record is comprised of k fields that are useful for linkage purposes. For example, first name, last name, and gender are fields that could be included in each record. A denotes a set of records, a indicates a record within set A, and a[i] refers to the value of the ith field in record a, where i ∈ {1,…,k}. We B, b, and b[i], to represent a set of records. The goal of record linkage is to correctly classify all record pairs 〈a, b〉, into the class M (match) or the class U (non-match). In traditional forms of record linkage, a third intermediate class may by included, in which record pairs are set aside for clerical review by a human who will determine the true match status. However, in the context of PPRL, clerical review clearly compromises privacy and, thus, this third, intermediate class is excluded.
3.2. Overview of Record Linkage
As briefly described in Section 1.2 and depicted in Figure 1, record linkage involves multiple steps. The first step is field comparison in which each field in each record pair is compared, resulting in a vector, called γ, of size k. Each cell in the vector, γ〈a,b〉[i], indicates the similarity of field i in record pair 〈a, b〉. In the record pair comparison step, the k-sized γ similarity vector for each record pair is converted into a single similarity score for the record pair. In the record pair classification step, the pairs are partitioned and classified into either the set M or U. The specific instantiations of these steps in this study are as follows:
Field comparison: Six experimental string comparators (Section 4.1) are used for field comparison. Additionally, a non-PPSC [23] shown to work well in record linkage [48, 49] is used as a reference standard.
Record pair comparison: A modification (Section 3.3.2) to a widely adopted algorithm (Section 3.3.1) that allows for continuous field similarity scores is used for record pair comparison.
Record pair classification: During this step, the record pairs are classified into the class M or U based on their similarity scores. Further details are provided in Section 3.4.
Throughout this work we hold constant the methods for both record pair comparison and classification so that we can clearly evaluate the field comparison step. The following subsections provide details of the comparison and classification steps as they were applied. Information regarding the implementation of the field comparators is presented in Section 4.
3.3. Record pair comparison
The Fellegi-Sunter (FS) algorithm typically works with binary field similarity scores, which are converted into a single similarity score for each record pair. This algorithm can be modified to allow for the incorporation of continuous field similarity scores. Both of these methods are described below and are used for record pair comparison in this study.
3.3.1. Fellegi-Sunter
The binary field comparison method is generally used with FS, so the γ vector is filled in as follows:
| (3) |
where “==” indicates complete agreement. The m and u vectors represent the conditional probability of field agreement given match status. These vectors are computed once per record linkage and are calculated over all record pairs:
| (4) |
| (5) |
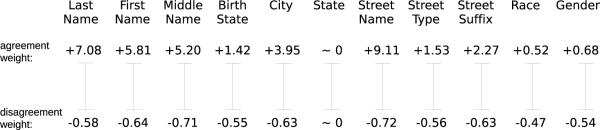
where “==” indicates complete agreement. In this evaluation we know the true match status for the test datasets, so we calculate the conditional probabilities exactly. This best case scenario allows clear evaluation of the string comparators without the hindrance of errors due to estimation. As this technique is used for all comparators, it places them on a level playing field and does not affect the comparisons provided8. The conditional match probabilities are then applied to calculate a vector of agreement weights, wa, and disagreement weights, wd, for each field i = 1, …, k:
| (6) |
| (7) |
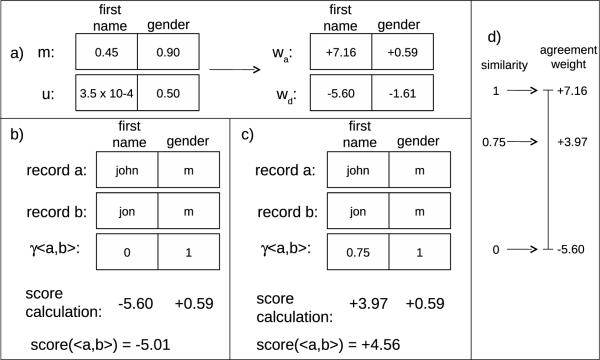
To recap, the conditional probabilities of agreement given match status are calculated for each field. These conditional probabilities are then used to calculate an agreement and disagreement weight for each field. For an example, see Figure 5, part a.
Figure 5.
Record linkage with the FS algorithm. a) Match parameters associated with the dataset. b) Calculation of the record pair score for the record pair when binary matching is used for the field comparison step and traditional FS is used for score calculation. c) Calculation of the record pair score for the record pair when Edit Similarity is used for the field comparison and the Winkler modification to FS is used for score calculation. d) Demonstration of the numerical scale laid out by the agreement and disagreement weights for the first name field.
The next step is to apply these per-field agreement and disagreement weights to calculate a total score for each record pair 〈a, b〉. This score is calculated for each record pair according to Equation 8.
| (8) |
Therefore, if field i agrees, the agreement weight associated with field i, wa[i], will be added to the total score for the record pair; if field i disagrees, the disagreement weight associated with field i, wd[i], will be added to the total score for the record pair (Figure 5, part b).
3.3.2. Winkler modification for continuous Fellegi-Sunter
The original FS algorithm is designed to work with only binary field comparison metrics (i.e., fields either completely agree or disagree). To incorporate approximate field comparators that detect similarity in addition to agreement/disagreement, the FS algorithm must be modified. Winkler introduced a modification, that incorporates continuous field comparators, which yields a continuous score in the range [0,…,1] to capture the similarity of strings, rather than binary agreement or disagreement [23]. Under the modification, the conditional probabilities, agreement weights, and disagreement weights are calculated in the manner previously described. However, the agreement weights now lay out a scale onto which the field comparison scores are mapped. A field similarity of 0 maps to the value of the disagreement weight, and a field similarity of 1 maps to the value of the agreement weight. Intermediate field similarities map to intermediate score weights. For example, if the field similarity score is 0.75, then the score at the 75th percentile on the scale laid out by the agreement and disagreement weights is selected. For an example, see Figure 5, parts c and d. More formally, the score calculated according to this method is given as follows:
| (9) |
3.4. Record pair classification
The record pair comparison step assigns a similarity score to each record pair. We use the following approach to classify all record pairs into M and U based on their similarity scores. Each record file A studied in this paper contains 1,000 records and is linked to a record file B that also contains 1,000 records. Each record in A must be compared to each record in B, which results in 1, 000×1, 000 = 106 record pairs that must be classified into the sets M and U. Each record in A has a record in B that refers to the same individual, so we know 1,000 of the record pairs, or 0.001%, are true matches. Therefore, we classify the 1,000 record pairs having the highest scores as M and all other record pairs as U9.
3.5. Dataset creation
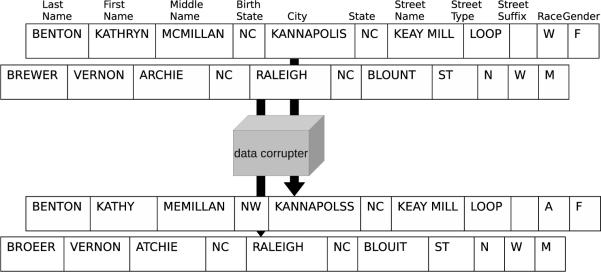
To perform our evaluation with a large set of records, we downloaded the publicly-available North Carolina voter registration (NCVR) files [55], which contain 6,190,504 individual records. From these records, we randomly selected (without replacement) 100 datasets, each containing 1,000 records, which we refer to as A1, …, A100. To generate datasets B1, …, B100 to which we link the aforementioned sets, we implemented a “data corrupter” based on the research of Pudjijono and Christen [56]. The corrupter introduced optical character recognition errors (e.g., S swapped for 8), phonetic errors (e.g., ph swapped for f), and typographic errors, such as insertions, deletions, transpositions, and substitutions. The probability with which the errors are introduced was chosen to be consistent with the error rates seen in real datasets [56].
In addition to character-level errors, we extended the data corrupter by introducing token-level errors. These errors were introduced at frequencies estimated to be encountered in real record linkage datasets. Nicknames were substituted for full names (and vice-versa) with probability 0.15, addresses are changed to represent an individual moving with probability 0.1, last names were changed with probability 0.1 for females (due to the common practice of taking the husband's last name), 0.01 for males, and last names were hyphenated with probability 0.01. The nicknames were based on the Massmind Nicknames Database [57], the last names were based on the 2000 U.S. Census names dataset [58], and the addresses were selected from a subset of the NCVR file not used in the record linkage datasets. Figure 6 provides an example of two records and their corrupted counterparts.
Figure 6.
An example of two records and their corrupted counterparts.
In each case, the fields “street direction”, “race”, and “gender” are compared with exact matching rather than approximate matching. Each of these fields consists of a single letter, so approximate matching is not possible.
When data was missing, it received a weight of 0 and therefore did not factor into the record linkage. This practice was also used in [59].
3.6. Implementation Details
All methods were implemented in Perl, a language known for its string handling capabilities. The running time for PPRL computation was measured using the Time∷HiRes package. The datasets were stored as flat text files. Experiments were run on a 2.5 GHz quad core PC with 4GB of memory.
4. Experimental Design
4.1. PPSCs Evaluated and Parameter Settings
We selected a subset of the aforementioned PPSCs for evaluation in this manuscript. PPSCs that have performed poorly in previous evaluations were not considered [21, 22, 39]. To comprehensively evaluate the range of techniques, a comparator was selected from each of the categories described above, with the exception of the “Guess & Encode” methods, which have been shown to perform poorly in a previous record linkage study [26]. The PPSCs selected, and the parameters invoked, are discussed in Sections 4.1.1 through 4.1.6 below. In all cases, we attempted to use the parameters suggested in the original articles. Where this was not possible or parameters were not suggested, this is explicitly stated.
4.1.1. Exact Matching
For the exact match comparator, each identifier is hashed (by the widely-used SHA-1 hash function) and compared. The output of the comparison is 1 if the hashed strings match exactly, and 0 otherwise. In practice, each string should be concatenated with a random string known as “salt” to prevent a dictionary attack; i.e., an exhaustive hashing of plaintexts to determine which encoded value corresponds to each plaintext value [60].
4.1.2. Bloom Filter
The Bloom Filter comparator was evaluated because it showed promising results in a preliminary evaluation [61]. As recommended in the original paper [32], we use a Bloom filter of length 1,000 bits and use 30 hash functions, all variations of SHA-1, to hash each bigram into the Bloom filter. Strings are padded with spaces on both ends in bigram creation as mentioned earlier.
4.1.3. Trigrams
The Trigrams [33] comparator was favorably reviewed [28] and was therefore selected for evaluation in this work. Each string was padded on both ends with two spaces in trigram creation. SHA-1, in conjunction with “salt”, was again applied to hash each trigram. A space-efficient implementation was used where each string was represented as a bag of trigrams and their associated counts. This is in contrast to representing each string as a very sparse vector of all possible trigrams where the value for the cell associated with each trigram is the number of times the trigram occurs in a given string.
4.1.4. Embedding
The Embedding approach described in [35] was selected to represent the “Reference Space Embedding” class due to the ability of a privately held reference space to provide greater security. With respect to the parameters for this comparator, we attempted to follow the recommendations of the authors of [35] as much as possible. As such, twenty strings were used in generating sixteen reference sets. For each field, the length of the reference strings is the average length of strings in the field. With respect to the greedy resampling heuristic, 10% of random pairs were sampled (without replacement) as recommended in [43]. To determine the optimal number of coordinates in the final embeddings, we systematically tested all values in the range [2,…,16]. We found that using 9 coordinates produced the best record linkage results in a small test set, and was therefore selected as the dimensionality of the final embeddings. Details can be found in the Appendix.
Euclidean distance was used to determine the similarity between embeddings. For each field, the Euclidean distance was normalized by the largest Euclidean distance seen to normalize the similarities into the range [0,…,1].
4.1.5. Phonetic Filter
In the Phonetic Filter comparator, strings were transformed by the Soundex phonetic filter, hashed, and tested for equivalence. In practice, salting should be used to protect against a dictionary attack. Perl's Text∷Soundex package was used.
4.1.6. Edit Similarity
The Edit Similarity approach in [36] was selected for evaluation due to its rigorous treatment of privacy protection. We draw attention to the distinction between distance and similarity. Distance is in the range [0,…,∞], where lower distance indicates strings are alike. Similarity, on the other hand, is in the range [0,…,1], where 1 indicates strings are completely alike and 0 indicates the strings are completely unalike. Distance can be converted to similarity as follows:
| (10) |
While the original comparator, as presented, returns the edit distance between two strings, we believe that edit similarity is a more informative measure because it incorporates string length. For example, an edit distance of three between the strings “Bob” and “Jan” is very different from an edit distance of three between the strings “Catherine” and “Katerina”. The edit distance protocol described above can be modified to report edit similarity when Alice and Bob divide their final answers (i.e., the last cell in their matrices) by the maximum string length. Note, this does not reveal any additional information because Alice and Bob already know the string lengths as these are revealed in the protocol.
4.2. Reference Standard String comparator
To compare the PPSCs to a standard, we use the Jaro-Winkler distance, a non-privacy-preserving string comparator that, while highly dependent on heuristics, has been shown to work very well in record linkage [23, 48, 49]. Winkler introduced a modification to the Jaro distance that incorporated the observation that errors are more likely to occur at the end of a string, rather than the beginning [23]. The modified comparator therefore places greater emphasis on the characters of the beginning of the string in determining string similarity. The Jaro-Winkler string comparator is defined as:
| (11) |
where dj is the Jaro distance for strings s1 and s2, l is the length of the prefix common to both strings (up to 4 characters), and p is a scaling factor. The default value for p is 0.1. The Jaro distance [51] is defined as:
| (12) |
where m is the number of matching characters and t is the number of transpositions. A transposition is the swapping of two adjacent letters (for example, “John” and “Jonh”). Two characters c1 and c2 are considered matching if they are no further than – 1 characters apart. An example is shown in Figure 7.
Figure 7.
An example of the Jaro-Winkler distance between the strings “JOHN” and “JON”.
4.3. Evaluation Metrics
We evaluate each string comparator along three axes: 1) correctness in record linkage, 2) security (the extent to which the strings are protected in the linkage process), and 3) computational complexity as determined by the running time.
4.3.1. Correctness in Record Linkage
To evaluate correctness in record linkage, we examine the ability of each string comparator to accurately classify record pairs into the sets M and U. As mentioned earlier, only 1,000 of the possible record pairs are true matches. The number of true negatives (999,000) so dominates the number of true positives that some measures, such as specificity, were not very informative. Therefore, to focus on the 0.001% of record pairs that are true matches, the True Positive (TP) rate10 is examined for each comparator. The TP rate is defined as:
| (13) |
where TP is the number of true positives and FP is the number of false positives. The TP rate reports the proportion of record pairs predicted to be matches that are, in fact, true matches. A comparator that perfectly classifies the record pairs has a TP rate of 1.
4.3.2. Computational Complexity
We use the running time to evaluate the computational complexity of each comparator. In this paper, a record linkage method that requires more than one day to process the linkage of two record files, each containing 1,000 records, is considered computationally infeasible for real world use. We make this assumption because the datasets used in this work are relatively small compared to typical real world record linkage datasets. Additionally, the running time increases quadratically with the number of records11. Therefore, comparators that take more than one day to run on these small datasets would take much longer to run on larger datasets.
4.3.3. Security
We next study the extent to which each comparator protects the security of the records. Recall that in the protocol presented in Section 1.3, data owners Alice and Bob rely on a third party Charlie to perform the record linkage. Specifically, Alice and Bob first exchange the information required to encode their records (which varies for each comparator). This information can be viewed as a “key” which is then used to encode the records. Charlie receives the encoded records and performs record linkage without the key. To conduct a security analysis of each comparator, we make the following standard assumptions with respect to the knowledge and the behavior of all parties in the above protocol:
All parties strictly follow the protocol in data operations and communications. This means that Alice and Bob always encode the records using the shared encoding key. They will only send the encoded record to Charlie. None of the parties will maliciously change the content of the records or the linkage results.
Charlie sees the encoded records, but not the original records.
Charlie does not know the encoding key.
Under these assumptions, the security analysis of the PPSCs focuses on how much information Charlie can acquire from the encodings to infer the original record. Since each field is independently encoded in all comparators, we pick one field (e.g., last name) to illustrate our metric for evaluating security. Here we use the mutual information entropy, a standard practice in security evaluations, to quantify the dependence of the plaintext (original field) and the encoding [20].
We use the dataset shown in Table 2 as a running example throughout this section to illustrate the entropy calculations. Notice that there are two people with the last name “ADAMS” in the population. Also note that both the name “SMITH” and “SMYTH” map to the same phonetic encoding.
Table 2.
Sample plaintexts and encodings for a population of size 5. The Phonetic Filter encodings are used as an example and SHA represents a hash function.
| Plaintexts (x ∈ X) | Encodings (y ∈ Y) |
|---|---|
| ADAMS | SHA(A352) |
| ADAMS | SHA(A352) |
| JOHNSON | SHA(J525) |
| SMITH | SHA(S530) |
| SMYTH | SHA(S530) |
Formally, let random variable X denote the original information at the selected field, which takes values x1 …xn. Let P(xi) be the probability mass function of outcome xi. The entropy (H)of X is then defined as:
| (14) |
In our specific case, X is a variable whose sample space is all plaintext last names present in the entire NCVR database, and x refers to each specific name. In the sample dataset provided in Table 2, H(X) is calculated as shown in Table 3. In this example, H(X) = 0.579.
Table 3.
Calculation of H(X) for the sample population. The contribution to H(X) is defined in Equation 14.
| Unique Plaintexts (x ∈ X) | P(x) | Contribution to H(X) |
|---|---|---|
| ADAMS | 0.4 | 0.159 |
| JOHNSON | 0.2 | 0.140 |
| SMITH | 0.2 | 0.140 |
| SMYTH | 0.2 | 0.140 |
| Sum | 1.0 | 0.579 |
Similarly, we consider random variable Y as the encoded form of this selected field, which takes values y1 …ym and let P(y) be its probability mass function. Then the entropy (H) of Y can be similarly defined as: . In our specific case, Y is a variable whose sample space is the encodings corresponding to each last name, and y refers to each specific encoding. For example, in the Phonetic Filter comparator, the encoded form is a hashed version of a phonetic encoding. In the sample dataset provided in Table 2, H(Y) is calculated as shown in Table 4. In this example, H(Y) = 0.458.
Table 4.
Calculation of H(Y) for the sample population. The contribution to H(Y) is defined in Equation 14.
| Unique Encodings (y ∈ Y) | P(y) | Contribution to H(Y) |
|---|---|---|
| SHA(A352) | 0.4 | 0.159 |
| SHA(J525) | 0.2 | 0.140 |
| SHA(S530) | 0.4 | 0.159 |
| Sum | 1.0 | 0.458 |
Let each pair of outcomes (xi, yj)(i.e., the encoded form of xi is yj) occur with probability P(xi, yj). The joint entropy of variables X and Y is then defined as:
| (15) |
In our specific case, the joint entropy is calculated by examining the unique (plaintext, encoding) pairs. This is acheived by concatenating the plaintexts and encodings as shown in Table 5. In this example, H(X,Y) = 0.203.
Table 5.
Calculation of H(X,Y) for the sample population. The contribution to H(X,Y) is defined in Equation 15.
| Unique (x,y) pairs | P(x,y) | P(x) | P(y) | Contribution to H(X,Y) |
|---|---|---|---|---|
| ADAMS_SHA(A352) | 0.4 | 0.4 | 0.4 | 0.064 |
| JOHNSON_SHA(J525) | 0.2 | 0.2 | 0.2 | 0.028 |
| SMITH_SHA(S530) | 0.2 | 0.2 | 0.4 | 0.056 |
| SMYTH_SHA(S530) | 0.2 | 0.2 | 0.4 | 0.056 |
| Sum | 1.0 | 0.203 |
The mutual information (MI) of variables X and Y is defined as:
| (16) |
Therefore, the MI with respect to the sample population show in Table 2 is 0.579 + 0.458 − 0.203 = 0.833. Based on information theory, the mutual information measures the dependence of the two variables. Thus, if an encoding scheme is secure, the plaintext and the ciphertext should be independent of one another (i.e., X and Y are independent), then MI(X, Y) = 0. In general, lower MI indicates higher independence between the plaintext and the ciphertext and thus greater encoding security12.
5. Results
In this section, we examine the results with respect 1) correctness in record linkage, 2) computational complexity, and 3) security.
5.1. Correctness in Record Linkage
To ensure that the agreement and disagreement weights associated with each field are in line with intuition, Figure 8 depicts the weights from a randomly selected 〈Ai, Bi〉 linkage. As expected, the agreement weights are always higher than the disagreement weights, which means that the score for a record pair increases when its fields agree and decreases when its fields do not agree. Also as expected, information-rich fields, such as last name, are more important than information-poor fields, such as gender, as indicated by the high agreement weight for last name. While first name is still important, it is not as discriminatory as last name, as indicated by the lower agreement weight. This agrees with intuition as there are 274,790 unique last names as compared to 187,743 unique first names in the NCVR database, indicating that the field last name contains more information.
Figure 8.
Example of the agreement and disagreement weights for each field from a sample dataset.
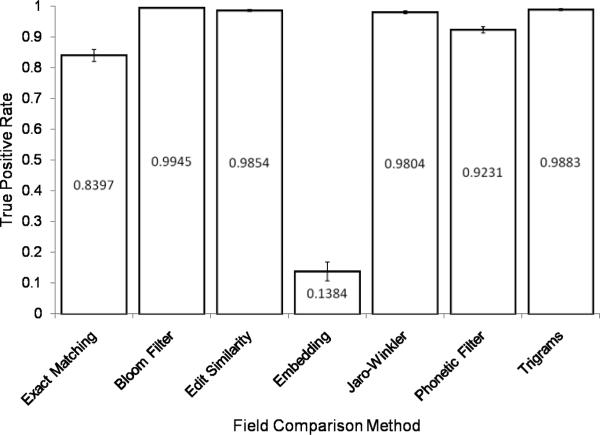
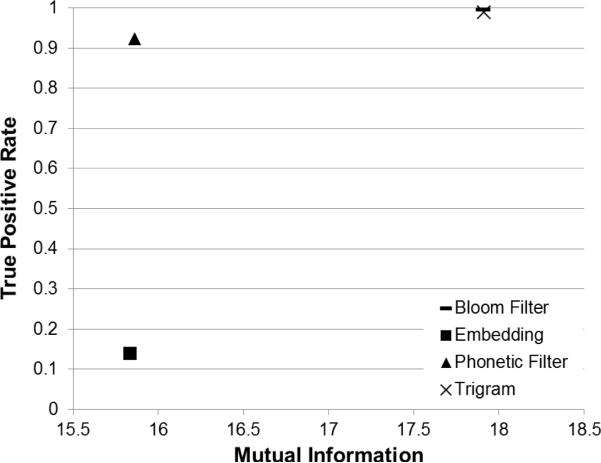
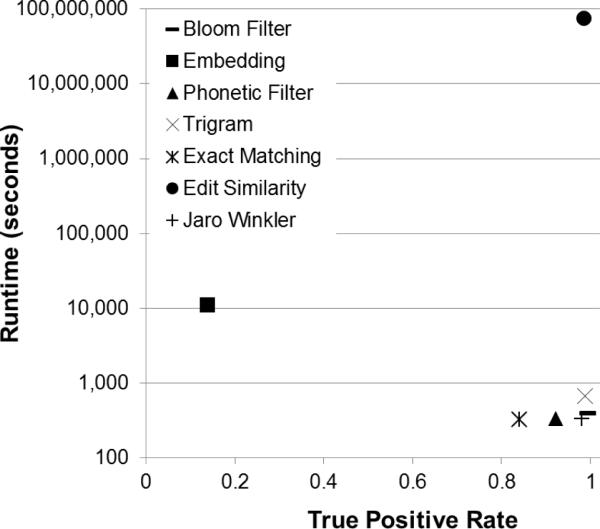
In Figure 9, the TP rate13 is reported for each string comparator. The Bloom Filter comparator performs best (TP rate = 0.9945) followed closely by the Trigrams (TP rate = 0.9883) and Edit Similarity (TP rate = 0.9854) comparators. The Embedding comparator does not perform as well as the others and achieves only a TP rate of 0.1384. Of note, all of the approximate PPSCs, with the exception of the Embedding comparator, outperform the baseline, traditional PPSC Exact Matching (TP rate = 0.8397). Also of note, several of the approximate PPSCs outperform the reference standard, Jaro-Winkler (TP rate = 0.9804).
Figure 9.
True Positive Rate. The values reported are the averages over the 100 pairs of record files. Standard deviation error bars are shown.
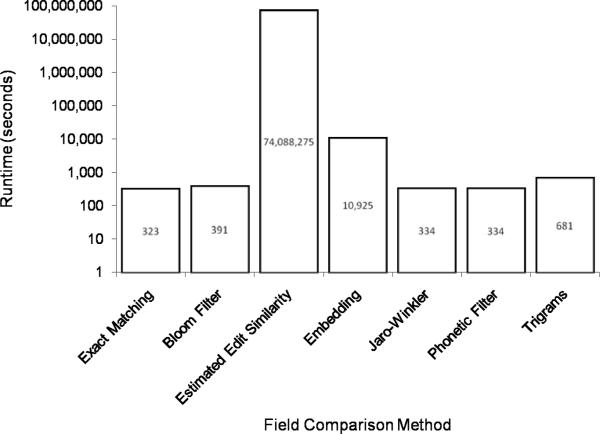
5.2. Computational Complexity
It is also important to examine which comparators are computationally feasible for real record linkage applications. Figure 10 provides the running time of each field comparison method for comparing the fields in 106 record pairs. Edit Similarity required too much time to complete, so a smaller record linkage with files containing 10 records each (resulting in 100 pairs) was performed. The resulting running time, 74.09 ± 13.43 seconds, was then extrapolated to provide an estimate of the time required to compare the fields in 106 record pairs. In practice, this would take over two years to run and is clearly infeasible. Additionally, a time-consuming part of the protocol (specifically, an interactive cryptographic technique known as “1-out-of-n oblivious transfer”) was not included in this implementation and would only serve to further increase the running time14.
Figure 10.
Field Comparison Running times. The values reported are the averages, in log scale, over the 100 pairs of record files. Standard deviation error bars are shown.
The running time of the Embedding comparator was an order of magnitude longer than the other comparators. The Exact Matching, Jaro-Winkler, and Phonetic Filter comparators all have comparable running times at around 330 seconds. The Bloom Filter is slightly slower at nearly 400 seconds and the Trigrams comparator trails at nearly 700 seconds.
5.3. Security
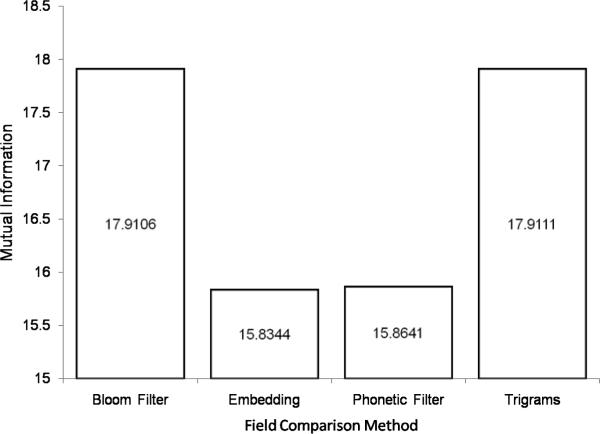
We examine the MI between the plaintext and encoded form of the last names associated with each field comparator. These results are shown in Figure 11. The order of security as indicated by low MI is Embedding > Phonetic Filter >> Bloom Filter > Trigrams which agrees with intuition (see qualitative discussion below)15. Further details of the calculations are provided in Table 6. For reference, H(Plaintext) = 8.9586 and there are 274,790 unique last names in the NCVR database.
Figure 11.
The Mutual Information between the plain texts and encoded plain texts associated with each field comparison comparator.
Table 6.
Entropy Analysis
| PPSC | H(Y) | H(Plaintext,Y) | MI(Plaintext,Y) | # Unique Encodings |
|---|---|---|---|---|
| Bloom Filter | 8.9581 | 0.0060 | 17.9106 | 274,430 |
| Embedding | 6.9008 | 0.0250 | 15.8344 | 55,097 |
| Phonetic Filter | 6.9214 | 0.0159 | 15.8641 | 5,815 |
| Trigram | 8.9586 | 0.0060 | 17.9111 | 274,790 |
At first glance, it appears unusual that Embedding and Phonetic Filter are similar in entropy while the number of unique encodings associated with each comparator is quite different. However, this can be explained by considering the distribution of probabilities associated with each of those unique encodings. The probabilities within the Phonetic Filter are relatively evenly distributed resulting in a higher entropy per encoding, whereas the probabilities within the Embedding are relatively skewed resulting in a lower entropy per encoding.
As mentioned earlier, Edit Similarity is the most secure method, with respect to what is revealed by the encodings, because the method does involve encoding the strings. Therefore, Charlie cannot learn anything by examining the string encodings as he never sees string encodings. Similarly, Exact Matching is not included in this analysis as each plaintext value maps to a unique encoding, and the mutual information is therefore 0.
5.4. Summary of Results
The tradeoffs between correctness, computational complexity, and security are visualized in Figures 12, 13, and 14.
Figure 12.
The tradeoffs between correctness as measured by TP rate, and security, as measured by MI.
Figure 13.
The tradeoffs between computational complexity (in log scale), as measured by running time, and security, as measured by MI.
Figure 14.
The tradeoffs between correctness, as measured by TP rate, and computational complexity (in log scale), as measured by the running time.
Figure 12 shows that the highly accurate comparators, (Bloom Filter and Trigrams), tend to be less secure. The Embedding comparator is more secure but not as accurate. An exception is the Phonetic Filter comparator which is both highly accurate and secure.
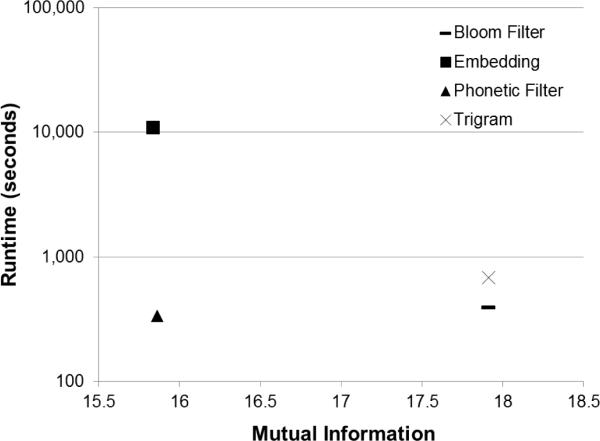
Figure 13 shows the general trend that the faster comparators are not as secure. Conversely, the Embedding comparator is very secure, but is also very slow. The Phonetic Filter comparator is an exception to this trend as it is both secure and fast as compared to the other comparators.
The tradeoff between correctness and computational complexity is more complex, as shown in Figure 14. The Edit Similarity comparator is highly accurate yet is orders of magnitudes slower than the other metrics. Many of the other comparators achieve both high accuracy and expedient running times.
6. Discussion
In this section, we provide a summary of the results in addition to a discussion of the results with respect to each of the three axes evaluated. Finally, we present additional considerations and limitations of this work.
6.1. Correctness in Record Linkage
The Exact Matching comparator achieves a TP rate of 0.8397. We believe it is able to correctly classify some record pairs as the large number of fields (i.e., eleven) allows for error correction. When there are many fields, even if several fields do not agree exactly, the number of additional fields that do agree increases the record pair's similarity score are enough to achieve a correct classification. We hypothesize that when fewer fields are included in each record, the ability to perform approximate string comparison will become even more important. This is because there will be fewer additional fields to increase the record pair's similarity score and “push it over” the classifying line. However, this same error correction affects each of the PPSCs equally, and the results are therefore still comparable.
Perhaps surprisingly, several of the PPSCs (Bloom Filter, Trigrams, and Edit Similarity) achieved higher TP rates than the non-privacy-preserving reference standard, Jaro-Winkler. One possible reason for this is that the data corrupter did not introduce errors in a way that Jaro-Winkler expects; the corrupter introduced errors uniformly throughout the string whereas Jaro-Winkler expects to see fewer errors at the beginning. This may have handicapped Jaro-Winkler, and it may perform better on datasets that contain fewer errors at the beginning of strings. Additionally, Jaro-Winkler is particularly well-suited for personal names, and in these experiments it was applied to non-name strings, such as cities and street names.
The Embedding approach did not perform as well as other PPSCs. One reason for this may be the use of edit distance, rather than edit similarity, as the distance function. Another possible reason may be the use of randomly generated reference strings. If the reference strings were chosen more systematically such that they better cover the comparison space, the results might improve. While testing on a small sample indicated that using nine reference sets to define the embedding space is optimal (see Appendix for further details), further calibration of this parameter may yield better results.
Although the Bloom Filter already performs very well, further improvements may be realized if the parameters (i.e., filter length and the number of hash functions used) were tailored to the expected length of the strings in each field. For example, far fewer bits are set to one when the bigrams of the two-digit state code are hashed into a Bloom filter than when the bigrams of the longer field, street address, are hashed.
The Phonetic Filter is limited in that only the first several characters of a string are integrated into its encoding. In long or multi-token strings, such as “Lori Beth” (double name) or “Windy Hills” (street address), often the first token is the only one captured by this approach.
In nearly all cases (with the exception of the Embedding approach), approximate field comparison provided greater accuracy than exact matching, confirming our hypothesis stated in Section 2.1.
6.2. Computational Complexity
While Edit Similarity provides excellent security and high correctness, its computational complexity renders it unreasonable. The other comparators perform the relatively expensive encoding process once per record (2,000 times in the case of linking two 1,000-record files), and then perform an inexpensive record pair comparison process for each record pair (106 times). However, Edit Similarity does not include an up-front encoding process, so the expensive edit distance matrix calculation must be performed for each record pair16. Since the number of record pairs is quadratic with respect to the number of input records, the running time of this approach quickly becomes infeasible. Additionally, this comparator relies on an expensive form of encryption. Specifically, this protocol incorporates a secure minimum finding protocol, which requires six expensive cryptographic operations for each cell of the matrix. An additional expensive operation included in this protocol, 1-out-of-n oblivious transfer, was not implemented but would serve to further increase the running time.
The Embedding approach involves multiple steps. Generating the initial string embeddings and record pair comparison are relatively fast steps. However, the greedy resampling heuristic, during which the reference sets used are whittled down from all reference sets to a subset of “good” information-rich reference sets, is computationally expensive. If reference strings are selected with care so as to use information-rich reference strings (as opposed to randomly generated reference strings), the time-intensive greedy resampling step could be eliminated and the correctness of results may improve.
A tradeoff between the running time and the storage space exists with the Trigrams. While we used a space-efficient implementation, a vectorized implementation would yield reduced running time.
6.3. Security
Edit Similarity is the most secure method in terms of the information revealed by the encodings – since this method does not involve revealing an encoded form of the information, no information is revealed by the encoding.
As indicated by the MI values, Embedding and Phonetic Filter are the most secure and approximately equivalent. Little information is revealed by Embedding because Charlie is unaware of the strings defining the reference space; thus, the embeddings provide little information. Each phonetically filtered string is a Soundex code of the form letter, number, number, number resulting in 26,000 possible Soundex codes. In practice, the 274,790 unique last names in the NCVR database were mapped to only 5,815 phonetic filter encodings, resulting in information loss. Therefore, the Phonetic Filter approach also does not provide much information.
The Bloom Filter and Trigrams approaches are the least secure as indicated by MI. In both of these comparators, salt is used to prevent a dictionary attack. However, these comparators are still subject to frequency analysis. The frequency distribution of n-grams is not as well-defined as the frequency distribution of full names; however, an attacker may still be able to use this information to determine the values associated with some n-grams. The Trigrams comparator additionally reveals the length of each string to Charlie. He can derive this information based on the number of trigrams associated with each string17. The Bloom Filter comparator does not directly reveal the length of each string due to the fact that false positives and “crowding” in the Bloom filter can occur, but it likely provides Charlie with the ability to bound the string length based on the number of bits set to one.
The goal of this security analysis was to evaluate all of the PPSCs in a common, quantified way. As such, the analysis is focused on answering the question “Given only the ciphertexts, how much can Charlie learn about the plaintexts?” Another question that could be asked is “Given external data sources, such as the distribution of plaintexts and ciphertexts in the population, how much can Charlie learn about the plaintexts?” We leave this analysis to future work. In practice, an attacker could mount different attacks specific to the unique features of each PPSC. Therefore, additional security analyses specific to these unique features could also be conducted.
6.4. Additional Considerations
Several aspects of the record linkage process require further research prior to adoption in real PPRL applications.
First of all, our experiments were performed with relatively small datasets, but record linkage is often performed on a much larger scale. In such cases, a technique called blocking is used to make the task computationally feasible in which records are sorted into bins with other similar records [62]. For example, records may be “blocked” into bins based on the first letter of last name. Comparisons could then be made only to records in the same bin, which would greatly reduce the number of record pairs that must be compared. It is reasonable to consider each of the 1,000-record files analyzed in our experiments as representative of a single block in a larger record linkage. We recognize that certain privacy-preserving blocking methods exist (e.g., [63, 64, 65]); however, it has yet to be determined if such methods are practical for real world application18.
Second, the goal of record linkage is to use the information available within records to determine which records refer to the same entity. Therefore in practice, fields used in record linkage may be numerical, rather than just string-based. Some of the PPSCs considered are more extendable to numerical fields than others. We believe Bloom Filter, Exact Matching, and Trigrams should work equally well on numerical fields. If the distance function and embedding space were selected specifically for numerical fields, Embedding would also be applicable for numerical fields. The Jaro-Winkler and Phonetic Filter approaches are specifically tuned to work for strings and are not applicable to numerical fields.
Third, some of the comparators are more sensitive to the way in which information is partitioned into fields. For example, residential address can be represented as a single field, such as {Street Address}, or can be broken into more granular bits such as {Street Number}, {Street Name}, {Street Suffix}, and {Street Direction}. The Exact Matching approach will perform best when fields are partitioned into smaller-sized fields because this approach cannot leverage the similarity information within a field. The Phonetic Filter and Jaro-Winkler approaches also perform better when fields are represented at more granular levels, as both of these comparators rely on the tokens at the beginning of a string. The other approximate PPSCs are less sensitive to the way in which information is divided into fields, but only to a certain extent. If too many fields are combined, the FS matching algorithm loses the ability to distinguish between more and less informative fields. For example, in the extreme case, all fields could be concatenated into a single field. In this case, less informative fields, such as gender, cannot be distinguished from more important fields, such as name, and therefore the classification correctness is expected to decrease.
Fourth, one of the limitations of this research is that, though fields were selected to only contain strings, the inclusion of some numerical tokens was unavoidable. For example, although street number was excluded, sometimes the street name itself contains numbers (i.e., “Highway 194”). Although this was very rare and efforts were made to include only string fields, this may have provide a slight advantage to string comparators more tolerant of numerical fields. In future research, we intend to determine how to best handle fields with a mixture of strings and numeric information.
7. Conclusions
In this work, we provided a principled and comprehensive evaluation of the state-of-the-art privacy-preserving string comparators (PPSCs). The evaluation considered three axes critical to privacy-preserving record linkage (PPRL) applications: 1) correctness, 2) computational complexity, and 3) security. This research used a real dataset, evaluated the PPSCs on a common quantified space, and provided the information needed to support decisions in designing a PPRL protocol in the real world. Using our methods, and analysis, a data manager can clearly model the tradeoffs and choose the best string comparator based on his resource constraints. In summary, the Bloom Filter and Trigrams comparators were found to be the most correct. The Edit Similarity comparator was the most secure, followed by the Embedding and Phonetic Filter comparators. The Edit Similarity comparator proved to be computationally expensive, whereas the Phonetic Filter, Exact Match, and Bloom Filter comparators were the fastest PPSCs. In future work, we intend to research privacy-preserving blocking schemes to ensure the feasibility of conducting record linkage of large datasets.
A. Embedding Parameter Selection.
The Embedding comparator [35] evaluated in this paper uses the Sparse Map variant of Lipschitz embeddings. A detailed description of Sparse Map can be found in [43]. Rather than using all reference sets that compose the embedding space, Sparse Map employs a greedy resampling heuristic to whittle down to a subset containing the most informative strings. These reference sets serve as coordinates that define the embedding space. A recommended number of coordinates was not specified in [35]; therefore, we systematically evaluated all possible subset sizes from 2 coordinates up to the maximum number of coordinates. In this case, the maximum number of coordinates was determined to be 16 (as recommended in [35]). The results are shown in Figure 15. The highest true positive rate was achieved using subsets of size both 9 and 13. We selected the subset of minimal size and use 9 coordinates to define the embedding space used in this work.
Figure 15.
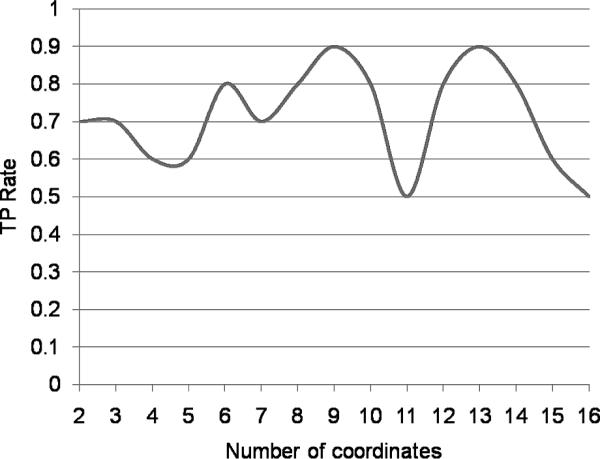
The TPR achieved for the Embedding comparator is dependent upon the number of reference sets used in estimating distance between strings.
Acknowledgements
This research was supported by grants R01-LM009989 and 2-T15LM07450-06 from the National Library of Medicine, National Institutes of Health and grants CNS-0845803, CNS-0964350, and the Team for Research in Ubiquitous Secure Technologies (TRUST) grant CCF-0424422 from the National Science Foundation. The authors wish to thank the anonymous referees for their diligent reviews and assistance in the preparation of this manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
In order for record linkage to be possible, the records must be drawn from the same population.
Statistical matching, a tangentially related technique that preserves the statistical inferential properties without requiring the correct assignment of matching record pairs [8], can also be applied in this query-based setting where the accuracy of only aggregated information is important.
A cryptographic hash function is a deterministic encoding function that converts a plaintext string into a fixed-length encoding.
This statement does not apply to the specific class of hash functions known as locality-sensitive hash functions.
This data-independent threshold was experimentally determined in [33] and was shown to work well in practice.
While any distance function can be used, selection of an appropriate, informative function is important for achieving accurate record linkage results, as stated in [35].
In practice, the Expectation Maximization (EM) algorithm can be used to estimate the conditional probabilities associated with each feature [50, 51, 52, 53]. EM can also be applied to estimate the proportion of matches anticipated among Alice and Bob's records, which provides guidance on where to set the classification boundary.
This method does not enforce a one-to-one mapping of records. In practice, this may be enforced through an exhaustive linear sum assignment procedure or a greedy matching heuristic. Futher details can be found in [54].
The TP rate is also referred to as precision in the computer science community.
With respect to the theoretical complexity, let k represent the number of fields in each record, and let n represent the number of records held by each dataholder. The theoretical complexity of record linkage is then O(kn2). However, we assume that k is much less than n, and therefore assume record linkage is O(n2).
Note that Edit Similarity is an exception to this analysis because records are not encoded in this protocol.
The way the classification boundary is set, as described in Section 3.4, means that the number of false positives (FP) is equivalent to the number of false negatives (FN). While recall is often reported in information retrieval experiments, it provides no additional information in the case where FP equals FN, and is therefore not reported.
The values in 5.1 Section were generated using an insecure, yet far quicker, version of edit similarity.
The >> symbol indicates “much greater than”.
However, a practical shortcut to reducing the computation time for this approach is to calculate the edit distance between all pairs of unique values only, rather than all record pairs.
The string length is generally the number of trigrams plus two when the strings are padded with two spaces on both ends.
Non-privacy-preserving blocking techniques have, however, been widely used in practice. A detailed empirical survey can be found in [66].
References
- [1].Torra V. Data Mining and Knowledge Discovery Handbook. Springer; 2005. Information fusion: Methods and aggregation operators; pp. 1005–1016. [Google Scholar]
- [2].Herzog T, Scheueren F, Winkler W. Data quality and record linkage techniques. Springer; 2007. [Google Scholar]
- [3].U.S. General Accounting Office . Record linkage and privacy: issues in creating new federal research and statistical information, Technical Report GAO-01-126SP. U.S. General Accounting Office; 2001. [Google Scholar]
- [4].Clifton C, Kantarcioglu M, Doan A, Schadow G, Vaidya J, Elmagarmid A, Suciu D. Privacy-preserving data integration and sharing. 9th ACM SIGMOD Workshop on Research Issues in Data Mining and Knowledge Discovery.pp. 19–26. [Google Scholar]
- [5].Safran C, Bloomrosen M, Hammond WE, Labkoff S, Markel-Fox S, Tang PC, Detmer DE. Toward a national framework for the secondary use of health data: An American Medical Informatics Association white paper. Journal of the American Medical Informatics Association. 2007;14:1–9. doi: 10.1197/jamia.M2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].U.S. National Institutes of Health Policy for sharing of data obtained in NIH supported or conducted genome-wide association studies, NOT-OD-07-088. 2007 [Google Scholar]
- [7].Malin B, Sweeney L. How (not) to protect genomic data privacy in a distributed network: using trail re-identification to evaluate and design anonymity protection systems. Journal of Biomedical Informatics. 2004;37:179–192. doi: 10.1016/j.jbi.2004.04.005. [DOI] [PubMed] [Google Scholar]
- [8].D'Orazio M, Zio MD, Scanu M. Statistical Matching: Theory and Practice (Wiley Series in Survey Methodology) John Wiley & Sons; 2006. [Google Scholar]
- [9].Committee on Technical and Privacy Dimensions of Information for Terrorism Prevention and Other National Goals. National Research Council . Protecting Individual Privacy in the Struggle Against Terrorists. The National Academies Press; Washington, DC: 2008. [Google Scholar]
- [10].Krouse W, Elias B. Technical Report RL33645. Congressional Research Service; 2009. Terrorist watchlist checks and air passenger pre-screening. [Google Scholar]
- [11].Fulton S. EU seeks privacy enforcement rights in US courts through diplomatic agreement. International Business Times; 2010. [Google Scholar]
- [12].Kushner D. Vegas 911. IEEE Spectrum. 2006;43:44–49. [Google Scholar]
- [13].Quantin C, Bouzelat H, Allaert F, Benhamiche A, Faivre J, Dusserre L. Automatic record hash coding and linkage for epidemiological follow-up data confidentiality. Methods of Information in Medicine. 1998:271–277. [PubMed] [Google Scholar]
- [14].Berman JJ. Zero-check: A zero-knowledge protocol for reconciling patient identities across institutions. Archives of Pathology & Laboratory Medicine. 2004;128:344–346. doi: 10.5858/2004-128-344-ZAZPFR. [DOI] [PubMed] [Google Scholar]
- [15].Agrawal R, Evfimievski A, Srikant R. Information sharing across private databases. 2003 ACM SIGMOD International Conference on Management of Data; San Diego, CA. pp. 86–97. [Google Scholar]
- [16].Eycken EV, Haustermans K, Buntinx F, Ceuppens A, Weyler J, Wauters E, Oyen HV, Schaever MD, den Berge DV, Haelterman M. Evaluation of the encryption procedure and record linkage in the belgian national cancer registry. Archives of public health. 2000;58:281–294. [Google Scholar]
- [17].Quantin C, Bouzelat H, Allaert F, Benhamiche A, Faivre J, Dusserre L. How to ensure data security of an epidemiological follow-up:quality assessment of an anonymous record linkage procedure. International Journal of Medical Informatics. 1998;49:117–122. doi: 10.1016/s1386-5056(98)00019-7. [DOI] [PubMed] [Google Scholar]
- [18].Agrawal R, Asonov D, Srikant R. Enabling sovereign information sharing using web services. 2004 ACM SIGMOD international conference on Management of data.pp. 873–877. [Google Scholar]
- [19].Hernández MA, Stolfo SJ. Real-world data is dirty: Data cleansing and the merge/purge problem. Data Mining and Knowledge Discovery. 1998;2:9–37. [Google Scholar]
- [20].Stallings W. Pearson Education. 5th edition 2002. Cryptography and Network Security: Principles and Practice; p. 86. [Google Scholar]
- [21].Churches T, Christen P. Some methods for blindfolded record linkage. BMC Medical Informatics and Decision Making. 2004;4:9. doi: 10.1186/1472-6947-4-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Song DX, Wagner D, Perrig A. Practical techniques for searches on encrypted data. 2000 IEEE Symposium on Security and Privacy, b; Oakland, CA. 2000. p. 44. [Google Scholar]
- [23].Porter EH, Winkler WE. Research Report RR97/02. U.S. Census Bureau; 1997. Approximate string comparison and its effect on an advanced record linkage system. [Google Scholar]
- [24].Tromp M, Reitsma J, Ravelli A, Meray N, Bonsel G. Record linkage: making the most out of errors in linking variables. American Medical Informatics Association Annual Symposium; pp. 779–783. [PMC free article] [PubMed] [Google Scholar]
- [25].Elmagarmid A, Ipeirotis P, Verykios V. Duplicate record detection: A survey. IEEE Transactions on Knowledge and Data Engineering. 2007;19:1–16. [Google Scholar]
- [26].Trepetin S. Privacy-preserving string comparisons in record linkage systems: A review. Information Security Journal: A Global Perspective. 2008;17:253–266. [Google Scholar]
- [27].Hall R, Fienberg S. Privacy-preserving record linkage. In: Domingo-Ferrer J, Magkos E, editors. Privacy in Statistical Databases, volume 6344 of Lecture Notes in Computer Science. Springer; Berlin / Heidelberg: 2011. pp. 269–283. [Google Scholar]
- [28].Verykios VS, Karakasidis A, Mitrogiannis VK. Privacy preserving record linkage approaches. International Journal of Data Mining and Modelling and Management. 2009;1:206–221. [Google Scholar]
- [29].Kaya SV, Pedersen TB, Savas E, Saygin Y. Efficient Privacy Preserving Distributed Clustering Based on Secret Sharing, volume 4819 of Lecture Notes in Computer Science. Springer; pp. 280–291. [Google Scholar]
- [30].Inan A, Kaya S, Saygin Y, Savas E, Hintoglu A, Levi A. Privacy preserving clustering on horizontally partitioned data. Data and Knowledge Engineering. 2007;63:646–666. [Google Scholar]
- [31].Ravikumar P, Fienberg SE. A secure protocol for computing string distance metrics. IEEE Workshop on Privacy and Security Aspects of Data Mining; Brighton, England. pp. 40–46. [Google Scholar]
- [32].Schnell R, Bachteler T, Reiher J. Privacy-preserving record linkage using Bloom filters. BMC Medical Informatics and Decision Making. 2009;9:41. doi: 10.1186/1472-6947-9-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Hylton JA. Master's thesis. Massachusetts Institute of Technology; Cambridge, Massachusetts: 1996. Identifying and Merging Related Bibliographic Records. [Google Scholar]
- [34].Pang C, Gu L, Hansen D, Maeder A. Intelligent Patient Management. volume 189. Springer; Berlin / Heidelberg: 2009. Privacy-preserving fuzzy matching using a public reference table; pp. 71–89. [Google Scholar]
- [35].Scannapieco M, Figotin I, Bertino E, Elmagarmid A. Privacy preserving schema and data matching. ACM SIGMOD International Conference on Management of Data; Beijing, China. pp. 653–664. [Google Scholar]
- [36].Atallah M, Kerschbaum F, Du W. Secure and private sequence comparisons. ACM Workshop on Privacy in the Electronic Society; Washington, DC. pp. 39–44. [Google Scholar]
- [37].Karakasidis A, Verykios VS. Privacy preserving record linkage using phonetic codes. 4th Balkan Conference in Informatics; Thessaloniki, Greece. pp. 101–106. [Google Scholar]
- [38].Jonas J, Pattak PB, Litchko JP. Using advanced information technology to combat insider threats. Journal of Organizational Excellence. 2001;20:19–27. [Google Scholar]
- [39].Du W, Atallah MJ. Technical Report, CERIAS. Purdue University; 2001. Protocols for Secure Remote Database Access with Approximate Matching. [Google Scholar]
- [40].Bloom BH. Space/time trade-offs in hash coding with allowable errors. Commun. ACM. 1970;13:422–426. [Google Scholar]
- [41].Broder A, Mitzenmacher M. Network applications of Bloom filters: A survey. Internet Mathematics. 2002;1:636–646. [Google Scholar]
- [42].Jain N. Using Bloom filters to refine web search results. 7th International Workshop on Web Databases; Baltimore, MD. pp. 25–30. [Google Scholar]
- [43].Hjaltason G, Samet H. Properties of embedding methods for similarity searching in metric spaces. IEEE Transactions on Pattern Analysis and Machine Intelligence. 2003;25:530–549. [Google Scholar]
- [44].Levenshtein VI. Binary Codes Capable of Correcting Deletions, Insertions and Reversals. Soviet Physics Doklady. 1966;10:707. [Google Scholar]
- [45].Marzal A, Vidal E. Computation of normalized edit distance and applications. IEEE Transactions on Pattern Analysis and Machine Intelligence. 1993;15:926–932. [Google Scholar]
- [46].Binstock A, Rex J. Metaphone: A modern Soundex. In: Binstock A, Rex J, editors. Practical Algorithms for Programmers. Addison Wesley; 1995. [Google Scholar]
- [47].Grannis SJ, Overhage JM, McDonald CJ. Analysis of identifier performance using a deterministic linkage algorithm. American Medical Informatics Association Annual Symposium; San Antonio, TX. pp. 305–309. [PMC free article] [PubMed] [Google Scholar]
- [48].Cohen WW, Ravikumar P, Fienberg SE. A comparison of string distance metrics for name-matching tasks. IJCAI-03 Workshop on Information Integration; Acapulco, Mexico. pp. 73–78. [Google Scholar]
- [49].Grannis SJ, Overhage JM, McDonald C. Real world performance of approximate string comparators for use in patient matching. Medinfo; San Francisco, CA. pp. 43–47. [PubMed] [Google Scholar]
- [50].Grannis SJ, Overhage JM, Hui S, McDonald CJ. Analysis of a probabilistic record linkage technique without human review. American Medical Informatics Association Annual Symposium; Washington, DC. pp. 259–263. [PMC free article] [PubMed] [Google Scholar]
- [51].Jaro MA. UNIMATCH: a record linkage system. U.S. Census Bureau; Washington, DC: 1978. [Google Scholar]
- [52].Winkler WE. Research Report RR00/05. U.S. Census Bureau; 2000. Using the EM algorithm for weight computation in the Fellegi-Sunter model of record linkage. [Google Scholar]
- [53].Torra V, Domingo-Ferrer J. Information fusion in data mining. Springer; 2003. Record linkage methods for multidatabase data mining; pp. 101–132. [Google Scholar]
- [54].Lenz R. Measuring the disclosure protection of micro aggregated business microdata. An analysis taking as an example the german structure of costs survey. Journal of Official Statistics. 2006;22:681–710. [Google Scholar]
- [55]. [Last accessed June 2, 2010];North Carolina Voter Registration Database. ftp://www.app.sboe.state.nc.us/data,
- [56].Christen P, Pudjijono A. Accurate synthetic generation of realistic personal information. 13th Pacific-Asia Conference on Advances in Knowledge Discovery and Data Mining; Bangkok, Thailand. pp. 507–514. [Google Scholar]
- [57]. [Last accessed June 2, 2010];Massmind Nicknames Database. http://techref.massmind.org/techref/ecommerce/nicknames.htm,
- [58].U.S. Census Bureau [Last accessed June 2, 2010];Population Division. http://www.census.gov/genealogy/names/namesfiles.html,
- [59].Gomatam S, Carter R, Ariet M, Mitchell G. An empirical comparison of record linkage procedures. Statistics in Medicine. 2002;21:1485–1496. doi: 10.1002/sim.1147. [DOI] [PubMed] [Google Scholar]
- [60].Schneier B. Applied Cyptogography: Protocols, Algorithms and Source Code in C. Wiley; 1996. [Google Scholar]
- [61].Durham E, Xue Y, Kantarcioglu M, Malin B. Private medical record linkage with approximate matching. American Medical Informatics Association Annual Symposium; [PMC free article] [PubMed] [Google Scholar]
- [62].Baxter R, Christen P, Churches T. A comparison of fast blocking methods for record linkage. ACM SIGKDD Workshop on Data Cleaning, Record Linkage and Object Consolidation; Washington, DC. pp. 25–27. [Google Scholar]
- [63].Al-Lawati A, Lee D, McDaniel P. Blocking-aware private record linkage. International Workshop on Information Quality in Information Systems; Baltimore, MD. pp. 59–68. [Google Scholar]
- [64].Kantarcioglu M, Inan A, Jiang W, Malin B. Formal anonymity models for efficient privacy-preserving joins. Data Knowledge Engineering. 2009;68:1206–1223. [Google Scholar]
- [65].Inan A, Kantarcioglu M, Ghinita G, Bertino E. Private record matching using differential privacy. 13th International Conference on Extending Database Technology, EDBT '10, ACM; New York, NY, USA. 2010. pp. 123–134. [Google Scholar]
- [66].Lenz R, Vorgrimler D. fdzarbeitspapier nr. 4 edition 2005. Matching German turnover tax statistics. [Google Scholar]