Abstract
Objective
To extend recent conceptual and methodological advances in disparities research to include the incorporation of genomic information in analyses of racial/ethnic disparities in health care and health outcomes.
Data Sources
Published literature on human genetic variation, the role of genetics in disease and response to treatment, and methodological developments in disparities research.
Study Design
We present a conceptual framework for incorporating genomic information into the Institute of Medicine definition of racial/ethnic disparities in health care, identify key concepts used in disparities research that can be informed by genomics research, and illustrate the incorporation of genomic information into current methods using the example of HER-2 mutations guiding care for breast cancer.
Principal Findings
Genomic information has not yet been incorporated into disparities research, though it has direct relevance to concepts of race/ethnicity, health status, appropriate care, and socioeconomic status. The HER-2 example demonstrates how available genetic information can be incorporated into current disparities methods to reduce selection bias and measurement error. Advances in health information infrastructure may soon make standardized genetic information more available to health services researchers.
Conclusion
Genomic information can refine measurement of racial/ethnic disparities in health care and health outcomes and should be included wherever possible in disparities research.
Keywords: Health economics, social determinants of health, racial/ethnic differences in health and health care, personalized medicine, genomics
Despite the national commitment to eliminate health disparities (U.S. Department of Health and Human Services 2007; Institute of Medicine 2011), significant disparities persist in the quality of care received by racial/ethnic groups and subsequent health outcomes (Institute of Medicine 2003). In 2003, the Institute of Medicine (IOM) defined a health care disparity as “racial or ethnic differences in the quality of health care that are not due to patients' clinical needs and preferences, and appropriateness of intervention” (Institute of Medicine 2003). In measuring health care disparities, most studies focus on the coefficient of the race/ethnicity variable after adjusting for a range of covariates, but this approach does not acknowledge that racial/ethnic differences are mediated by—not independent of—socioeconomic status (McGuire et al. 1994; Agency for Healthcare Research and Quality 2011; Cook et al. current issue). Drawing from methods used in the economics literature on wage discrimination, McGuire, Cook, and others have advocated a methodological approach based on the IOM definition of health care disparity in which disparities are decomposed to estimate the contribution of socioeconomic status to racial/ethnic disparities in health care (McGuire et al. 1994; Cook et al. 2008). Similar methods have begun to appear in the health outcomes literature as well (Crown 2008). At the same time, advances in genomics research, through which patterns in the human genome are examined to identify variants or regions of the genome important to disease etiology or treatment response (National Human Genome Research Institute 2010), are increasing our understanding of the role of genetics in disease risk and treatment response (Manolio et al. 2008), and even the ways in which socioeconomic status can mediate genetic risk through gene–environment interactions (GEI) and epigenetic effects (Olden et al. 1978). A growing number of clinical guidelines, particularly in oncological care, now include practice recommendations based on a patient's genetic status (Amstutz and Carleton 2011). This new knowledge has not yet been widely incorporated into disparities research.
This paper addresses the implications of emerging genomic information for disparities research. We argue that genetic information is an important omitted variable in health services research generally and disparities research in particular, and one that will become even more important as time goes on. Genomic data can help to differentiate geographical ancestry from self-identified race; provide additional information about health status, appropriate care, and expected clinical outcomes; and shed new light on the importance of certain socioeconomic factors (e.g., greater exposure to stress) as critical control variables due to their role in amplifying genetic risk of disease. The inclusion of genetic variables in racial/ethnic disparities analyses, where possible, can thus help reduce measurement error and reduce bias in estimates of treatment effects, and allow for a better understanding of the sources of health disparities between self-identified racial/ethnic groups in our society.
We begin with a discussion of the intersection of emerging genomics research and four key concepts in disparities research: race, health status, clinical appropriateness, and socioeconomic status. We then present a conceptual framework showing how genomic information can be incorporated into disparities research, using the case of HER-2 status among women with breast cancer. Finally, recognizing that genomic information has yet to become widely available in health services data, we discuss statistical methods that test and help to control for measurement error due to the omission of variables that capture clinically relevant genetic heterogeneity.
Genomics Research Expanding Key Concepts Used in Disparities Research
Genomics research is expanding or refining several key concepts used in disparities research and in health services research more generally. Figure illustrates the IOM definition of racial/ethnic health care disparities, measures commonly used to operationalize this definition (Cook et al. current issue), and how specific measures can be extended or refined using genomic information. Below, we address the intersection of genomics research with four concepts central to disparities research: race, health status, clinical appropriateness, and socioeconomic status.
Figure 1.
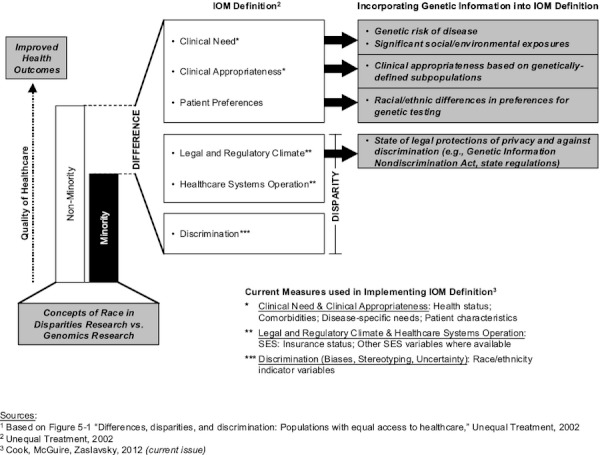
Incorporating Genomic Information into the IOM Definition of Health Care Disparities1
What Is “Race”?
Foundational to any discussion of disparities research is the meaning of “race.” The concept of race has been employed in very different ways in the fields of health services research and genomics research. Disparities research has been dominated by the use of the racial/ethnic categories institutionalized by the Office of Management and Budget (OMB) in 1997—the same categories used in the U.S. Census—to assess differences in the quality of care received across groups (Office of Management and Budget 2007). The National Institutes of Health (NIH) regulation requiring all federally funded researchers to report study participants based on the OMB categories was not based on presumed biological differences (National Institutes of Health 1993), but rather by concerns for distributive justice (e.g., equal access to the benefits and burdens of clinical research) (National Commission for the Protection of Human Subjects of Biomedical and Behavioral Research 1999; McCarthy 2006; Shields et al. 2010). The NIH requirement that researchers collect and report their study participants using these categories fostered the use of these same categories in data analysis. Thus, in most early genetics studies, the OMB racial/ethnic categories were used as a proxy for human genetic heterogeneity (i.e., human genetic variation or population structure). Human genetic variation, however, is a “continuous variable” that represents patterns of human mating and migration. These patterns can be mapped spatially (Parra 2004) and are often categorized according to geographical ancestry (Tishkoff and Verrelli 2009). Self-identified race variables have often been used as a proxy for geographical ancestry (e.g., black for “African ancestry”; white for “European ancestry”), although both geneticists and social scientists have argued that self-identified racial/ethnic categories are poor proxies for capturing human genetic heterogeneity (Royal and Dunston 2011; Shields et al. 2010).
In time, the field of human population genetics devised more precise ways to control for underlying population structure that have shown the limitations of the OMB racial/ethnic categories as a useful proxy for genetically meaningful groups. Methods for admixture mapping use data from the entire genome to empirically group individuals according to geographical ancestry based on their genetic data (Patterson et al. 2006). While empirical measures of geographical ancestry may be correlated with self-identified OMB racial/ethnic categories, OMB categories do not begin to capture the biological diversity of even the most basic geographical groupings. For example, a recent study showed that genetically-determined African ancestry among persons in the United States identifying as African American ranged from 1 to 99 percent (Bryc et al. 2011). Similar analyses have shown the extraordinary genetic diversity in European populations previously thought to be fairly homogeneous “whites” (Novembre et al. 2008).
Given the well-documented and unacceptable gaps in the quality of health care received by self-identified white and minority patients in the U.S. health care system, disparities measured according to self-identified race/ethnicity will continue to be powerful policy levers for working toward equity, but these categories should not be considered adequate proxies for human genetic variation. Health services researchers will rarely have data available that include empirical measures of geographical ancestry. When available, these data could provide additional information regarding health status insofar as they capture as yet undiscovered, clinically relevant genetic variants that vary according to geographical ancestry, although the effect size of such variants will likely be modest compared to the contribution of social and environmental factors in explaining disparities (Hirschhorn and Gajdos 2005). In the vast majority of cases, however, data on geographical ancestry will not be available, and the best that can be done is to take great care in specifying precisely what is and is not being captured in the self-identified racial/ethnic variables being used.
Health Status
Patients' clinical need has been measured primarily as health status, using whatever general or disease-specific measures might be available. Recent advances in genomics research, most notably genome-wide association studies, have identified numerous genetic variants associated with increased disease risk, thereby identifying new subsets of patients likely to have more intensive future clinical needs. For example, seven genes account for approximately 10 percent of variation in diabetes risk (Voight et al. 2003). Thirty genetic variants account for close to 10 percent of inter-individual variation in HDL, LDL, and triglyceride levels indicative of cardiovascular disease (Kathiresan et al. 2008). BRCA1/2 mutations increase women's lifetime risk of breast cancer from 12 percent in the general population to about 80 percent (King, Marks, and Mandell 2009). Three genetic variants have been shown to account for a 2–4-fold increase in risk for developing age-related macular degeneration (Maller et al. 2006). Multiple variants have also been found to more than double the risk for nondiabetic end-stage renal disease, and these variants account for much of the excess risk among persons of African versus European ancestry (Kao et al. 2007). As data on genetic disease risk become validated and available to health services researchers, they should be included as individual-level measures in disparities research models to control for underlying differences in general or disease-specific health status, as appropriate.
Once individual mutations or constellations of variants have been shown to be reliable indicators of increased disease risk, they should be included in disparities analyses whenever possible as additional measures of health status in order to reduce measurement error. Some of these risk variants will vary in frequency among populations characterized according to geographical ancestry, and thus they will have varying degrees of correlation with the OMB racial/ethnic categories. A subset of these will have substantial clinical relevance. The 8q24 variant, for example, confers significantly increased risk of prostate cancer and is found predominantly in men of African ancestry (Freedman et al. 2008). However, in the proposed framework, we advocate the inclusion of genetic markers of health status as a means of reducing measurement error, not as proxies for race/ethnicity. The magnitude of the reduction in bias and improvements in precision as a result of incorporating genomic markers of health status will vary widely by phenotype and the degree to which the variant (or constellation of variants) is correlated with either the OMB race/ethnicity categories or outcome variables. Research results to date indicate that genetic variation will typically account for a small proportion of variance in disease risk and health outcomes among OMB-defined racial/ethnic groups, although in many cases genetic variation will be sufficiently informative to meaningfully reduce measurement error.
Clinical Appropriateness
Beyond controlling for health status, or clinical need more generally, genomics research is also beginning to generate information that can be used to match patients to optimal treatments (Beitelshees and Veenstra 2012). Genetically-tailored treatments are most widely established in cancer care, where it has now become standard practice to match patients to appropriate therapy according to their specific tumor biology (i.e., somatic mutations) (Garman, Nevins, and Potti 2011). Guidelines now recommend, for example, that women with breast cancer whose tumors are HER-2 positive receive the medication trastuzumab (Herceptin) to reduce risk of recurrence (Piccart-Gebhart et al. 2004). Approximately 10 percent of breast cancer patients have tumors that respond to trastuzumab; but among patients with HER-2 positive cancers, this figure is as high as 50 percent (Chang 2010). Use of trastuzumab among HER-2 negative women, by contrast, results in little clinical gain while exposing them to potential adverse effects of the drug (e.g., heart damage) (Romond et al. 2005). As another example, certain non-small cell lung cancers over-express the epidermal growth factor receptor (EGFR) (Paez et al. 2011). Among this subset of lung cancer patients, gefitinib (Iressa) inhibits this growth factor receptor and significantly improves survival (Kris et al. 2003). Some academic medical centers now routinely assess EGFR expression among all non-small cell lung cancer patients (Hayden 2009).
A growing number of examples outside oncology exist as well. Genetic variants that code for cytochrome P450 (CYP450) enzymes (i.e., CYP2C9, CYP2C19, and CYP2D6), which determine how fast an individual can metabolize certain medications (e.g., certain beta-blockers, tricyclic antidepressants, and anticoagulants), are being used to classify patients' metabolizing profiles and adjust dosing accordingly (Tomalik-Scharte et al. 2010). This type of genetic information is also being used to guide treatment decisions for thrombosis and HIV/AIDS (Mallal et al. 2010; Schwarz et al. 2004).
As with genetic measures of health status, once a specific genetic status is proven to characterize a clinically relevant subset of patients who would especially benefit from (or be harmed by) a given treatment, this information should be incorporated into health services research and disparities research whenever possible. Again, these clinically relevant variants will rarely be equally distributed across all human beings, and will more often differ in frequency among persons of differing geographical ancestry. Thus, the degree of correlation between genetic markers important to refining assessment of appropriate care and the OMB racial/ethnic categories used in disparities research will vary, depending on the variant(s) pertinent to the study at hand. To the extent that genomic data are important for guiding treatment choices, their omission in a given analysis will introduce a correlation between the treatment variable and the error term in the equation and will result in biased estimates of treatment effects.
Socioeconomic Status
In disparities research, socioeconomic status is conceptualized as mediating racial/ethnic disparities through health systems. However, a growing body of research shows how socioeconomic status also mediates health status through GEI and epigenetic effects, where environmental exposures that track with socioeconomic status are found to amplify genetic risk of disease (Hunter 2005; Jirtle and Skinner 2009a). GEI research, for example, investigates how complex relationships between genes and environmental stressors produce differential disease risk. A specific genetic variant may be common across all human populations but only deleteriously affect a biological process in the presence of a particular environmental exposure. For example, carriers of CD14 mutations are more susceptible to experiencing decreased lung function when exposed to airborne endotoxins (Smit et al. 2007). Poor diet (Low and Tai 2003) and air pollution (Moysich et al. 2009; Park et al. 1997) interact with specific genes to increase risk for breast cancer and cardiovascular disease. As genomics research identifies specific environmental exposures that are mediated by socioeconomic status and amplify genetic risk of disease, the inclusion of such exposure information, if available, would further reduce measurement error in disparities analyses. As a growing number of studies link air pollution to compounded genetic risk for decreased lung function (Ege et al. 2012), for instance, one can imagine geographic information systems (GIS)-linked air pollution exposure data becoming an expected covariate in future studies investigating asthma or chronic obstructive pulmonary disorder disparities.
A Methodological Framework for Incorporating Genomic Information into Health Disparities Research
Among the many intersections between current conceptual and methodological frameworks for examining racial/ethnic health disparities and genomic information, genomic information can be immediately applied to three areas: (a) accounting for previously unmeasured genetic variation that affects health status or treatment response; (b) refining measurement of “appropriate care”; and (c) incorporating genetic information indicating likely benefit from a given treatment into the assessment of health outcomes. We illustrate these areas using the example of HER-2 status guiding breast cancer care.
Accounting for Previously Unmeasured Genetic Variation
To date, the growing knowledge about genetic variation and the role of genetics in predicting an individual's risk of disease or response to a given treatment has rarely been incorporated into disparities research methods. Determining the best methods for addressing the issue of previously unobserved genetic influences that may be correlated with both treatment and outcomes is a current challenge for the health services research field. As increasing amounts of evidence-based, clinically relevant genetic information becomes available, the incorporation of such data into analyses of health disparities can be expected to reduce measurement error and selection bias, and improve the value of racial/ethnic disparities research.
We describe below an analytic framework that incorporates genetic variables in disparities research in anticipation of the growing availability of genomic data, recognizing that many of the data needed to implement the proposed framework are not yet widely available to health services researchers. Central to this framework is the acknowledgment that there is clinically relevant genetic heterogeneity within and between the OMB-defined racial/ethnic groups. Ideally, one would want to control for genetic variability within and between self-identified OMB racial/ethnic groups with robust markers of geographical ancestry, as well as known markers relevant to health status and disease risk. This would address the selection bias problems (i.e., bias introduced by the omission of variables that are correlated with both treatment and patient outcomes) associated with unobserved heterogeneity in treatment selection and response. In addition to including genetic variables in our framework, we utilize, within each racial/ethnic group, the decomposition methods that have been used to assess racial/ethnic disparities in health care in the recent literature (Cook et al. current issue). Finally, these methods are extended to incorporate racial/ethnic disparities in nonrandom selection (matching patients to treatments based on their genetic profile) into treatment and heterogeneity of treatment response (due to one's genetic make-up) within treatment categories. The resulting analytic framework combines statistical methods from both disparities research and outcomes research, and incorporates genetic information to enable more robust analyses of racial/ethnic disparities in health care utilization and outcomes.
Any number of factors can undermine the reliability of statistical inferences drawn from observational data. Common problems in observational data analysis include omitted variables, measurement error, joint causation, and unobserved factors (e.g., positive HER-2 status) that are correlated with both treatment selection and patient outcomes. The use of race/ethnicity categories as proxies for underlying genetic variation is a measurement error problem because of the imprecision of the OMB race categories and the wide genetic variation found within these racial categories (Novembre et al. 2008; Bryc et al. 2011). As a result, disease risk among genetic subgroups within a given OMB racial/ethnic category would be expected to vary. The potential clinical benefit to be derived from a given treatment can also vary dramatically according to genotype. Thus, genetic variation found within the OMB racial/ethnic categories has implications both for errors in treatment selection (i.e., appropriate care) as well as observed variation in treatment response and health outcomes. Selection bias in treatment choice and variation in treatment response can be reduced by identifying clinically relevant genetic subgroups. As genetic data become increasingly available, the inclusion of these variables will enable researchers to explain a greater proportion of disease burden and outcomes.
Some variables that are important predictors of health care utilization (e.g., health status, socioeconomic status) can be readily observed by researchers, but others (e.g., family history, genetic traits) may not be observed. The inability to include such variables can lead to a special type of measurement error known as omitted variable bias. The most direct solution is to include the missing data when possible. However, when this is not possible, researchers can use statistical methods that test and correct for selection bias in treatment and outcomes. To illustrate these points more concretely, consider the example of breast cancer treatment. A basic framework is provided in Equations (1–4).
Refining Measurement of “Appropriate Care”
| (1) |
| (2) |
Equations (1) and (2) represent the clinician's diagnostic and treatment process. YB and YW represent the prescribed treatments for blacks and whites, respectively. As discussed above, the OMB categories are a blunt instrument for classifying disease risk and controlling for heterogeneity of treatment selection. White women are known to be at higher risk for developing breast cancer than black women, but black women are diagnosed at later stages of disease and have higher mortality (Jemal et al. 2008). However, once diagnosed, the differential risk of disease may have limited utility in guiding treatment; matching women to the best treatment to optimize outcomes is the priority. As detailed above, within each racial category, information about HER-2 status has become a critical predictor of treatment selection. Thus, genetic testing for HER-2 receptor status should help to improve access to appropriate care for women with breast cancer, regardless of race. To the extent that black women experience poorer access to appropriate treatments for breast cancer, testing for HER-2 status may reduce racial disparities in breast cancer treatment. Using the counterfactual approach (online Supporting Information Appendix SA2), it would be possible to evaluate racial disparities in treatment with trastuzumab under the assumption that black women had the same mean HER-2 status and other characteristics as white women.
While the example of HER-2 is useful in illustrating the value of matching patients to treatment based on their mutation status, it simultaneously highlights a limitation of currently available genomic data of particular relevance to disparities researchers. Though roughly the same proportion of African American and white women will have a negative HER-2 status (Lund et al. 2006; Swede et al. 2007), black women with HER-2 negative status will be far more likely to have triple negative cancer (which is a much more aggressive form of breast cancer) (Carey et al. 2010; Cleator, Heller, and Coombes 2006). The vast majority of genomic studies in breast cancer, however—as with all conditions—have been conducted in European ancestry populations (Haga 2007), and so the genetics research needed to fully understand the etiology and treatment of triple negative cancer has not yet been performed. Large scale genomic studies of women of African ancestry with breast cancer are only now being undertaken (Hutter et al. 2011).
The racial disparities literature has largely focused on differential access to appropriate care by race. In the following sections, the racial disparities framework is extended to include an assessment of disparities in patient outcomes that may arise from disparities in care.
Incorporating Genomic Information into the Assessment of Health Outcomes
| (3) |
| (4) |
Equations (3) and (4) illustrate predicted outcomes OB and OW contingent upon observed patient characteristics XB and XW and treatment choice YB and YW for blacks and whites, respectively. One would expect that the use of HER-2 positive gene expression results to guide treatment selection should also lead to improvements in treatment outcomes within each racial category. By reducing within-group variation, the ability to measure true between group differences will be improved.
Once again, Equations (3) and (4) could be used to implement the traditional counterfactual approach to assess racial/ethnic differences in treatment outcomes. The counterfactual approach would enable researchers to examine whether differential outcomes by race/ethnicity persist after controlling for the same rate of treatment with trastuzumab, as well as other patient characteristics such as health status and socioeconomic status.
Discussion
Advances in genomics research are generating new insight into the role of genetics in disease risk and treatment response. The pace of discovery is impressive, promising an increasing number of clinically relevant genetic markers that will become validated as evidence-based indicators of health status or clinical need and used to guide medical care. Due to patterns of human migration, these genetic markers often vary in frequency according to geographical ancestry, which in turn is often correlated with, but not to be confused with, groups based on self-identified race/ethnicity. The complex interplay of human genetic diversity and the OMB categories of race/ethnicity commonly deployed in disparities research present distinctive challenges. To incorporate emerging genomic data into cutting-edge disparities research, several key concepts and measures, including race, health status, clinical appropriateness, and socioeconomic status, must be refined or expanded. Furthermore, appropriate genomic variables that account for population structure, health status, and appropriate care should be included wherever possible to reduce measurement error in disparities research models. Realizing the potential value of genomic information to disparities research will depend on addressing key normative and practical issues.
First and foremost, standards for evaluating the scientific reliability of emerging genomic information are needed. When is the scientific evidence strong enough to warrant including genetic information in disparities studies? In general, the scientific process for identifying control variables in any health services research is a function of evidence from the literature. In this sense, the threshold for inclusion of genomic variables in disparities studies is no different. The emerging field of personalized medicine, however, has been slow to embrace evidence-based medicine (Khoury et al. 2011), and the pace of research has made it difficult to reach consensus regarding the clinical utility of many genetic markers. Currently, the Centers for Disease Control's Evaluation of Genomic Applications in Practice and Prevention Initiative (Teutsch et al. 2011) is the premier source of independent, evidence-based assessment of the clinical utility and reliability of genomic applications. The U.S. Preventive Services Task Force (Agency for Healthcare Research and Quality 2011), Cochrane Collaboration (Sivell et al. 2005), and Secretary's Advisory Committee on Genetics, Health and Society (National Institutes of Health 2012) have also published reports assessing the validity and reliability of specific genetic applications, as have professional organizations recommending the use of specific mutations to guide treatment (Carlson et al. 2009; Levin et al. 2011; Lynch 2007; Burtness et al. 2010; Greenberg et al. 2006). When deciding whether mutations associated with disease risk or treatment response should be included in disparities research, their use in clinical practice and inclusion in evidence-based guidelines are reasonable evidentiary thresholds for health services researchers to consider.
The practical challenge for health services researchers trying to operationalize clinical guidelines based on genomic information lies in the lack of research infrastructure. The genomic information necessary to define denominator populations is not readily available in claims data or even electronic medical records. Germline (e.g., BRCA1/2) or somatic (e.g., HER-2) mutation status, which is used to determine appropriate treatment, is often only available in medical charts or other data sources (e.g., cancer registries). Few datasets outside cancer include genomic information. Even within fully automated electronic health records (EHRs), mutation status is rarely entered electronically (Wilke et al. 2010). This data gap limits the feasibility of mounting large-scale studies assessing disparities in receipt of guideline-recommended care where genomic information is used to define the applicable patient populations.
Some health plans and health systems have begun collecting genetic information on patients' genetic status for select variants key to directing care, particularly in the areas of cancer and cardiovascular disease. The VA Hypertension Primary Care Longitudinal cohort, for example, has been incorporating genetic markers into EHRs since 2003 (Salem et al. 2005; Puppala et al. 2007). Recently, Kaiser Permanente was funded to link genetic and clinical data for up to 500,000 enrollees to assess the efficacy and toxicity of drugs, as well as the effect of different environmental exposures across different genetic profiles (Thomson et al. 2011; Wilke et al. 2010). The Kaiser Permanente Research Program on Genes, Environment and Health has now genotyped the DNA and the length of chromosome tips of more than 100,000 individuals. This program is merging these data with patients' EHRs, providing a valuable, integrated database. Such data resources will become more common in the coming years (Kaiser Permanente 2009b). Other initiatives to incorporate genomic data into EHR systems include the Harvard University/Partners Healthcare system i2b2, the Vanderbilt BioVu44 database, and the multi-center, National Human Genome Research Institute-funded eMERGE (electronic Medical Records and Genomics Network) (Kohane 2008).
The infrastructure to facilitate the routine incorporation of genomic data into EHRs in the future is key to making use of the added power of genetic information to understand variation in disease risk and treatment outcomes among different populations, and to craft interventions most likely to ensure optimal outcomes for all. Federal initiatives determined to accelerate adoption of EHRs will likely be a major force in developing the infrastructure that will begin capturing clinically relevant genetic information and making it available to health services researchers. Adoption rates of EHRs remains low nationally, at approximately 13 percent among physicians and 8 percent among hospitals (DesRoches et al. 2010; Jha et al. 2009), with safety net providers facing particular barriers to adoption (Shields et al. 2008; Jha et al. 2001). However, large providers and health systems are already beginning to transform their information systems by merging data (including genomic data) previously collected only in research into EHR formats (Massachusetts General Hospital 2009; Wilke et al. 2010).
As the first generation of genetic testing data becomes available to health services researchers, these data should be interpreted with care. Concerns about privacy and genetic discrimination may result in biased samples of patients for whom germline genetic test results are available, thus limiting generalizability. Patient preferences for testing play a significant role in determining whether a patient receives genetic testing (Bruno et al. 2011). Importantly, patient preferences for genetic testing have been shown to differ according to patients' race and socioeconomic status, with minority patients less likely to be aware of and less likely to believe in the benefits of testing (Peters, Rose, and Armstrong 2007; Halbert, Kessler, and Mitchell 2009). In reality, patients are rarely equipped to make informed decisions about their health care preferences (O'Connor et al. 2010; Peters et al. 2004). The complex nature of genetic information may make it more difficult for patients to understand available information and may lead to heightened anxieties about how genetic information will be used. The passage of the Genetic Information Nondiscrimination Act of 2008 may allay these fears over time.
The field of genomics is growing at an exponential rate, and new technological capacity has yielded results that have eclipsed findings from only a few years ago. As a result, an increasing wealth of clinically relevant, validated genomic data will become available to health services researchers for use in disparities research models in the coming years. Although we believe social, environmental, and political factors will continue to account for the lion's share of racial/ethnic disparities in the United States, we also believe that genetic data will be valuable in accounting for unobserved differences in health status and treatment heterogeneity due to genetic variation that may contribute to underlying prevalence of illness, treatment selection, and treatment response. As genomics research continues to identify clinically relevant patient subpopulations and knowledge of the complex factors that predict disease and treatment outcomes increases, we anticipate a gradual shift from statistical models with extremely large variability in selecting appropriate treatments and in predicting health outcomes toward models that explain an ever growing proportion of inter-individual variation. This added analytical power, in turn, has the potential to increase our understanding of the various sources of racial/ethnic disparities in health care and health outcomes, and generate new interventions aimed at reducing disparities.
Acknowledgments
Joint Acknowledgment/Disclosure Statement: The authors thank Tom McGuire, Wylie Burke, Mehdi Najafzadeh, and John Ayanian for comments on earlier drafts of this manuscript. Carly Hudelson, Marcelo Cerullo, and Anna Boonin Schachter also deserve special thanks for their superb research assistance. This work was conducted with support from Harvard Catalyst | The Harvard Clinical and Translational Science Center (NIH Award UL1 RR 025758 and financial contributions from Harvard University and its affiliated academic health care centers). The content is solely the responsibility of the authors and does not necessarily represent the official views of Harvard Catalyst, Harvard University and its affiliated academic health care centers, the National Center for Research Resources, or the National Institutes of Health. This work was also supported by a grant from the National Human Genome Research Institute R01 HG003475-03 (A. Shields) and the OptumInsight Fellowship Program (W. Crown).
Disclosures: None.
Disclaimers: None.
SUPPORTING INFORMATION
Additional supporting information may be found in the online version of this article:
Appendix SA1: Author Matrix.
Appendix SA2: Decomposition Methods, the Counterfactual, and Sample Selection Bias Models.
Please note: Wiley-Blackwell is not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Agency for Healthcare Research and Quality. 2010 National Healthcare Disparities Report. Rockville, MD: Agency for Healthcare Research and Quality; 2011. [Google Scholar]
- Agency for Healthcare Research and Quality. “ U.S. Preventive Services Task Force”. 2012. [accessed on February 1, 2012]. Available at http://www.ahrq.gov/clinic/uspstfix.htm.
- Amstutz U, Carleton BC. “Pharmacogenetic Testing: Time for Clinical Practice Guidelines”. Clinical Pharmacology and Therapeutics. 2011;89((6)):924–7. doi: 10.1038/clpt.2011.18. [DOI] [PubMed] [Google Scholar]
- Beitelshees AL, Veenstra DL. “Evolving Research and Stakeholder Perspectives on Pharmacogenomics”. Journal of the American Medical Association. 2011;306((11)):1252–3. doi: 10.1001/jama.2011.1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruno M, Digennaro M, Tommasi S, Stea B, Danese T, Schittulli F, Paradiso A. “Attitude Towards Genetic Testing for Breast Cancer Susceptibility: A Comparison of Affected and Unaffected Women”. European Journal of Cancer Care. 2010;19((3)):360–8. doi: 10.1111/j.1365-2354.2009.01067.x. [DOI] [PubMed] [Google Scholar]
- Bryc K, Auton A, Nelson MR, Oksenberg JR, Hauser SL, Williams S, Froment A, Bodo JM, Wambebe C, Tishkoff SA, Bustamante CD. “Genome-Wide Patterns of Population Structure and Admixture in West Africans and African Americans”. Proceedings of the National Academy of Sciences. 2010;107((2)):786–91. doi: 10.1073/pnas.0909559107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burtness B, Anadkat M, Basti S, Hughes M, Lacouture ME, McClure JS, Myskowski PL, Paul J, Perlis CS, Saltz L, Spencer S. “NCCN Task Force Report: Management of Dermatologic and Other Toxicities Associated with EGFR Inhibition in Patients with Cancer”. Journal of the National Comprehensive Cancer Network. 2009;7(suppl 1):S5–S21. doi: 10.6004/jnccn.2009.0074. quiz S22-24-S5-21; quiz S22-24. [DOI] [PubMed] [Google Scholar]
- Carey LA, Perou CM, Livasy CA, Dressler LG, Cowan D, Conway K, Karaca G, Troester MA, Tse CK, Edmiston S, Deming SL, Geradts J, Cheang MCU, Nielsen TO, Moorman PG, Earp HS, Millikan RC. “Race, Breast Cancer Subtypes, and Survival in the Carolina Breast Cancer Study”. Journal of the American Medical Association. 2006;295((21)):2492–2502. doi: 10.1001/jama.295.21.2492. [DOI] [PubMed] [Google Scholar]
- Carlson RW, Moench SJ, Hammond ME, Perez EA, Burstein HJ, Allred DC, Vogel CL, Goldstein LJ, Somlo G, Gradishar WJ, Hudis CA, Jahanzeb M, Stark A, Wolff AC, Press MF, Winer EP, Paik S, Ljung BM, the NCCN HER2 Testing in Breast Cancer Task Force “HER2 Testing in Breast Cancer: NCCN Task Force Report and Recommendations”. Journal of the National Comprehensive Cancer Network. 2006;4:S1–S22. [PubMed] [Google Scholar]
- Chang HR. “Trastuzumab-Based Neoadjuvant Therapy in Patients with HER2-Positive Breast Cancer”. Cancer. 2010;116((12)):2856–2867. doi: 10.1002/cncr.25120. [DOI] [PubMed] [Google Scholar]
- Cleator S, Heller W, Coombes RC. “Triple-Negative Breast Cancer: Therapeutic Options”. Lancet Oncology. 2007;8((3)):235–244. doi: 10.1016/S1470-2045(07)70074-8. [DOI] [PubMed] [Google Scholar]
- Cook BL, McGuire TG, Meara E, Zaslavsky AM. “Adjusting for Health Status in Non-Linear Models of Health Care Disparities”. Health Services and Outcomes Research Methodology. 2008;9((1)):1–21. doi: 10.1007/s10742-008-0039-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook BL, McGuire TG, Zaslavsky AM. “Measuring Racial/Ethnic Disparities in Health Care: Methods and Practical Issues”. Health Services Research. 2012 doi: 10.1111/j.1475-6773.2012.01387.x. doi: 10.1111/j.1475-6773.2012.01413.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crown WH. “There's a Reason They Call Them Dummy Variables: A Note on the Use of Structural Equation Techniques in Comparative Effectiveness Research”. PharmacoEconomics. 2010;28((10)):947–955. doi: 10.2165/11537750-000000000-00000. [DOI] [PubMed] [Google Scholar]
- DesRoches CM, Campbell EG, Rao SR, Donelan K, Ferris TG, Jha A, Kaushal R, Levy DE, Rosenbaum S, Shields AE, Blumenthal D. “Electronic Health Records in Ambulatory Care–A National Survey of Physicians”. New England Journal of Medicine. 2008;359((1)):50–60. doi: 10.1056/NEJMsa0802005. [DOI] [PubMed] [Google Scholar]
- Ege MJ, Strachan DP, Cookson WOCM, Moffatt MF, Gut I, Lathrop M, Kabesch M, Genuneit J, Büchele G, Sozanska B, Boznanski A, Cullinan P, Horak E, Bieli C, Braun-Fahrländer C, Heederik D, von Mutius E. “Gene-Environment Interaction for Childhood Asthma and Exposure to Farming in Central Europe”. Journal of Allergy and Clinical Immunology. 2011;127((1)):144.e1–4. doi: 10.1016/j.jaci.2010.09.041. [DOI] [PubMed] [Google Scholar]
- Freedman ML, Haiman CA, Patterson N, McDonald GJ, Tandon A, Waliszewska A, Penney K, Steen RG, Ardlie K, John EM, Oakley-Girvan I, Whittemore AS, Cooney KA, Ingles SA, Altshuler D, Henderson BE, Reich D. “Admixture Mapping Identifies 8q24 as a Prostate Cancer Risk Locus in African-American men”. Proceedings of the National Academy of Sciences. 2006;103((38)):14068–68. doi: 10.1073/pnas.0605832103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garman KS, Nevins JR, Potti A. “Genomic Strategies for Personalized Cancer Therapy”. Human Molecular Genetics. 2007;16((R2):R226–R26. doi: 10.1093/hmg/ddm184. [DOI] [PubMed] [Google Scholar]
- Greenberg PL, Rigsby CK, Stone RM, Deeg HJ, Gore SD, Millenson MM, Nimer SD, O'Donnell MR, Shami PJ, Kumar R. “NCCN Task Force: Transfusion and Iron Overload in Patients with Myelodysplastic Syndromes”. Journal of the National Comprehensive Cancer Network. 2009;7(suppl 9):S1–S16. doi: 10.6004/jnccn.2009.0082. [DOI] [PubMed] [Google Scholar]
- Haga SB. “Impact of Limited Population Diversity of Genome-Wide Association Studies”. Genetics in Medicine. 2010;12((2)):81–4. doi: 10.1097/GIM.0b013e3181ca2bbf. [DOI] [PubMed] [Google Scholar]
- Halbert CH, Kessler LJ, Mitchell E. “Genetic Testing for Inherited Breast Cancer Risk in African Americans”. Cancer Investigation. 2005;23((4)):285–295. doi: 10.1081/cnv-58819. [DOI] [PubMed] [Google Scholar]
- Hayden EC. “Personalized Cancer Therapy Gets Closer”. Nature. 2009;458((7235)):131–2. doi: 10.1038/458131a. [DOI] [PubMed] [Google Scholar]
- Hirschhorn JN, Gajdos ZKZ. “Genome-Wide Association Studies: Results from the First Few Years and Potential Implications for Clinical Medicine”. Annual Review of Medicine. 2011;62((1)):11–24. doi: 10.1146/annurev.med.091708.162036. [DOI] [PubMed] [Google Scholar]
- H.R. 493–110th Congress. “Genetic Information Nondiscrimination Act of 2008, H.R. 493, 110th Congress, 2nd Session”. 2008.
- Hunter DJ. “Gene-Environment Interactions in Human Diseases”. Nature Reviews Genetics. 2005;6:287–98. doi: 10.1038/nrg1578. [DOI] [PubMed] [Google Scholar]
- Hutter CM, Young AM, Ochs-Balcom HM, Carty CL, Wang T, Chen CTL, Rohan TE, Kooperberg C, Peters U. “Replication of Breast Cancer GWAS Susceptibility Loci in the Women's Health Initiative African American SHARe Study”. Cancer Epidemiology Biomarkers & Prevention. 2011;20((9)):1950–59. doi: 10.1158/1055-9965.EPI-11-0524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Institute of Medicine. Crossing the Quality Chasm: A New Health System for the 21st Century. Washington, DC: National Academy Press; 2001. [PubMed] [Google Scholar]
- Institute of Medicine. Unequal Treatment: Confronting Racial and Ethnic Disparities in Health Care. Washington, DC: The National Academies Press; 2003. [PubMed] [Google Scholar]
- Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. “Cancer Statistics, 2009”. CA: A Cancer Journal for Clinicians. 2009;59((4)):225–49. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- Jha AK, DesRoches CM, Campbell EG, Donelan K, Rao SR, Ferris TG, Shields A, Rosenbaum S, Blumenthal D. “Use of Electronic Health Records in US Hospitals”. New England Journal of Medicine. 2009a;360((16)):1628–38. doi: 10.1056/NEJMsa0900592. [DOI] [PubMed] [Google Scholar]
- Jha AK, DesRoches CM, Shields AE, Miralles PD, Zheng J, Rosenbaum S, Campbell EG. “Evidence of an Emerging Digital Divide among Hospitals That Care for the Poor”. Health Affairs. 2009b;28((6)):w1160–w70. doi: 10.1377/hlthaff.28.6.w1160. [DOI] [PubMed] [Google Scholar]
- Jirtle RL, Skinner MK. “Environmental Epigenomics and Disease Susceptibility”. Nature Reviews Genetics. 2007;8((4)):253–62. doi: 10.1038/nrg2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser Permanente. “DNA of 100,000 Kaiser Permanente Members Genotyped in 15 Months, Creating Novel Resource for Research”. Division of Research. 2011 [accessed on November 2, 2011]. Available at: http://researchnews.kaiser.org/?p=1811. [Google Scholar]
- Kao WHL, Klag MJ, Meoni LA, Reich D, Berthier-Schaad Y, Li M, Coresh J, Patterson N, Tandon A, Powe NR, Fink NE, Sadler JH, Weir MR, Abboud HE, Adler SG, Divers J, Iyengar SK, Freedman BI, Kimmel PL, Knowler WC, Kohn OF, Kramp K, Leehey DJ, Nicholas SB, Pahl MV, Schelling JR, Sedor JR, Thornley-Brown D, Winkler CA, Smith MW, Parekh RS. “MYH9 Is Associated with Nondiabetic End-Stage Renal Disease in African Americans”. Nature Genetics. 2008;40((10)):1185–92. doi: 10.1038/ng.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kathiresan S, Willer CJ, Peloso GM, Demissie S, Musunuru K, Schadt EE, Kaplan L, Bennett D, Li Y, Tanaka T, Voight BF, Bonnycastle LL, Jackson AU, Crawford G, Surti A, Guiducci C, Burtt NP, Parish S, Clarke R, Zelenika D, Kubalanza KA, Morken MA, Scott LJ, Stringham HM, Galan P, Swift AJ, Kuusisto J, Bergman RN, Sundvall J, Laakso M, Ferrucci L, Scheet P, Sanna S, Uda M, Yang Q, Lunetta KL, Dupuis J, de Bakker PIW, O'Donnell CJ, Chambers JC, Kooner JS, Hercberg S, Meneton P, Lakatta EG, Scuteri A, Schlessinger D, Tuomilehto J, Collins FS, Groop L, Altshuler D, Collins R, Lathrop GM, Melander O, Salomaa V, Peltonen L, Orho-Melander M, Ordovas JM, Boehnke M, Abecasis GR, Mohlke KL, Cupples LA. “Common Variants at 30 Loci Contribute to Polygenic Dyslipidemia”. Nature Genetics. 2009;41((1)):56–65. doi: 10.1038/ng.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoury MJ, Berg A, Coates R, Evans J, Teutsch SM, Bradley LA. “The Evidence Dilemma in Genomic Medicine”. Health Affairs. 2008;27((6)):1600–11. doi: 10.1377/hlthaff.27.6.1600. [DOI] [PubMed] [Google Scholar]
- King M-C, Marks JH, Mandell JB. “Breast and Ovarian Cancer Risks due to Inherited Mutations in BRCA1 and BRCA2”. Science (New York, N.Y.) 2003;302((5645)):643–46. doi: 10.1126/science.1088759. [DOI] [PubMed] [Google Scholar]
- Kohane IS. “Using Electronic Health Records to Drive Discovery in Disease Genomics”. Nature Reviews Genetics. 2011;12((6)):417–28. doi: 10.1038/nrg2999. [DOI] [PubMed] [Google Scholar]
- Kris MG, Natale RB, Herbst RS, Lynch TJ, Prager D, Belani CP, Schiller JH, Kelly K, Spiridonidis H, Sandler A, Albain KS, Cella D, Wolf MK, Averbuch SD, Ochs JJ, Kay AC. “Efficacy of Gefitinib, an Inhibitor of the Epidermal Growth Factor Receptor Tyrosine Kinase, in Symptomatic Patients with Non–Small Cell Lung Cancer”. Journal of the American Medical Association. 2003;290((16)):2149–58. doi: 10.1001/jama.290.16.2149. [DOI] [PubMed] [Google Scholar]
- Levin B, Barthel JS, Burt RW, David DS, Ford JM, Giardiello FM, Gruber SB, Halverson AL, Hamilton S, Kohlmann W, Ludwig KA, Lynch PM, Marino C, Martin EW, Jr, Mayer RJ, Pasche B, Pirruccello SJ, Rajput A, Rao MS, Shike M, Steinbach G, Terdiman JP, Weinberg D, Winawer SJ. “Colorectal Cancer Screening Clinical Practice Guidelines”. Journal of the National Comprehensive Cancer Network. 2006;4((4)):384–420. doi: 10.6004/jnccn.2006.0033. [DOI] [PubMed] [Google Scholar]
- Low YL, Tai ES. “Understanding Diet-Gene Interactions: Lessons from Studying Nutrigenomics and Cardiovascular Disease”. Mutation Research. 2007;622((1-2):7–13. doi: 10.1016/j.mrfmmm.2007.01.015. [DOI] [PubMed] [Google Scholar]
- Lund MJ, Butler EN, Hair BY, Ward KC, Andrews JH, Oprea-Ilies G, Bayakly AR, O'Regan RM, Vertino PM, Eley JW. “Age/Race Differences in HER2 Testing and in Incidence Rates for Breast Cancer Triple Subtypes: A Population-Based Study and First Report”. Cancer. 2010;116((11)):2549–59. doi: 10.1002/cncr.25016. [DOI] [PubMed] [Google Scholar]
- Lynch PM. “Current Approaches in Familial Colorectal Cancer: A Clinical Perspective”. Journal of the National Comprehensive Cancer Network. 2006;4((4)):421–30. doi: 10.6004/jnccn.2006.0034. [DOI] [PubMed] [Google Scholar]
- Mallal S, Phillips E, Carosi G, Molina JM, Workman C, Tomažič J, Jägel-Guedes E, Rugina S, Kozyrev O, Cid JF, Hay P, Nolan D, Hughes S, Hughes SHughesA, Ryan S, Fitch N, Thorborn D, Benbow A. “HLA-B* 5701 Screening for Hypersensitivity to Abacavir”. New England Journal of Medicine. 2008;358((6)):568–79. doi: 10.1056/NEJMoa0706135. [DOI] [PubMed] [Google Scholar]
- Maller J, George S, Purcell S, Fagerness J, Altshuler D, Daly MJ, Seddon JM. “Common Variation in Three Genes, Including a Noncoding Variant in CFH, Strongly Influences Risk of age-Related Macular Degeneration”. Nature Genetics. 2006;38((9)):1055–59. doi: 10.1038/ng1873. [DOI] [PubMed] [Google Scholar]
- Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, McCarthy MI, Ramos EM, Cardon LR, Chakravarti A, Guttmacher AE, Kong A, Kruglyak L, Mardis E, Rotimi CN, Slatkin M, Valle D, Whittemore AS, Boehnke M, Clark AG, Eichler EE, Gibson G, Haines JL, Mackay TF, McCarroll SA, Visscher PM. “Finding the Missing Heritability of Complex Diseases”. Nature. 2009;461((7265)):747–53. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massachusetts General Hospital. Massachusetts General Hospital Cancer Center Opens Molecular Pathology Lab to Genetically Profile all Patient Tumors. Boston: News, Massachusetts General Hospital; 2009. [Google Scholar]
- McCarthy CR. “Historical Background of Clinical Trials Involving Women and Minorities”. Academic Medicine. 1994;69((9)):695–98. doi: 10.1097/00001888-199409000-00002. [DOI] [PubMed] [Google Scholar]
- McGuire TG, Alegria M, Cook BL, Wells KB, Zaslavsky AM. “Implementing the Institute of Medicine Definition of Disparities: An Application to Mental Health Care”. Health Services Research. 2006;41((5)):1979–2005. doi: 10.1111/j.1475-6773.2006.00583.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moysich KB, Shields PG, Freudenheim JL, Schisterman EF, Vena JE, Kostyniak P, Greizerstein H, Marshall JR, Graham S, Ambrosone CB. “Polychlorinated Biphenyls, Cytochrome P4501A1 Polymorphism, and Postmenopausal Breast Cancer Risk”. Cancer Epidemiology, Biomarkers & Prevention, A Publication of the American Association for Cancer Research, Cosponsored by the American Society of Preventive Oncology. 1999;8((1)):41–44. [PubMed] [Google Scholar]
- National Commission for the Protection of Human Subjects of Biomedical and Behavioral Research. The Belmont Report: Ethical Principles and Guidelines for the Protection of Human Subjects of Research. Washington, DC: Government Printing Office; 1978. [PubMed] [Google Scholar]
- National Human Genome Research Institute. “ Genetic and Genomic Science”. 2010. [accessed on March 12, 2012]. Available at http://www.genome.gov/19016904.
- National Institutes of Health. “ National Institutes of Health Revitalization Act: Subtitle B, Clinical Research Equity Regarding Women and Minorities”. 1993. 42 U.S.C.: National Institutes of Health Revitalization Act.
- National Institutes of Health. “ Secretary's Advisory Committee on Genetics, Health, and Society”. 2012. [accessed on March 12, 2012]. Available at http://oba.od.nih.gov/SACGHS/sacghs_home.html.
- Novembre J, Johnson T, Bryc K, Kutalik Z, Boyko AR, Auton A, Indap A, King KS, Bergmann S, Nelson MR, Stephens M, Bustamante CD. “Genes Mirror Geography within Europe”. Nature. 2008;456((7218)):98–101. doi: 10.1038/nature07331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor AM, Wennberg JE, Legare F, Llewellyn-Thomas HA, Moulton BW, Sepucha KR, Sodano AG, King JS. “Toward the ‘Tipping Point’: Decision Aids and Informed Patient Choice”. Health Affairs. 2007;26((3)):716–25. doi: 10.1377/hlthaff.26.3.716. [DOI] [PubMed] [Google Scholar]
- Office of Management and Budget. Recommendations from the Interagency Committee for the Review of the Racial and Ethnic Standards to the Office of Management and Budget Concerning Changes to the Standards for the Classification of Federal Data on Race and Ethnicity. Washington, DC: Federal Register; 1997. pp. 36873–946. [Google Scholar]
- Olden K, Freudenberg N, Dowd J, Shields AE. “Discovering how Environmental Exposures Alter Genes Could Lead to New Treatments for Chronic Illnesses”. Health Affairs. 2011;30((5)):833–41. doi: 10.1377/hlthaff.2011.0078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paez JG, Jänne PA, Lee JC, Tracy S, Greulich H, Gabriel S, Herman P, Kaye FJ, Lindeman N, Boggon TJ, Naoki K, Sasaki H, Fujii Y, Eck MJ, Sellers WR, Johnson BE, Meyerson M. “EGFR Mutations in Lung Cancer: Correlation with Clinical Response to Gefitinib Therapy”. Science. 2004;304((5676)):1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- Park SK, O'Neill MS, Wright RO, Hu H, Vokonas PS, Sparrow D, Suh H, Schwartz J. “HFE Genotype, Particulate Air Pollution, and Heart Rate Variability: A Gene-Environment Interaction”. Circulation. 2006;114((25)):2798–805. doi: 10.1161/CIRCULATIONAHA.106.643197. [DOI] [PubMed] [Google Scholar]
- Parra EJ. “Human Pigmentation Variation: Evolution, Genetic Basis, and Implications for Public Health”. American Journal of Physical Anthropology. 2007;134((S45):85–105. doi: 10.1002/ajpa.20727. [DOI] [PubMed] [Google Scholar]
- Patterson N, Hattangadi N, Lane B, Lohmueller KE, Hafler DA, Oksenberg JR, Hauser SL, Smith MW, O'Brien SJ, Altshuler D, Daly MJ, Reich D. “Methods for High-Density Admixture Mapping of Disease Genes”. American Journal of Human Genetics. 2004;74((5)):979–1000. doi: 10.1086/420871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters N, Rose A, Armstrong K. “The Association between Race and Attitudes about Predictive Genetic Testing”. Cancer Epidemiology Biomarkers & Prevention. 2004;13((3)):361–5. [PubMed] [Google Scholar]
- Peters E, Hibbard J, Slovic P, Dieckmann N. “Numeracy Skill and the Communication, Comprehension, and Use of Risk-Benefit Information”. Health Affairs. 2007;26((3)):741–48. doi: 10.1377/hlthaff.26.3.741. [DOI] [PubMed] [Google Scholar]
- Piccart-Gebhart MJ, Procter M, Leyland-Jones B, Goldhirsch A, Untch M, Smith I, Gianni L, Baselga J, Bell R, Jackisch C, Cameron D, Dowsett M, Barrios CH, Steger G, Huang CS, Andersson M, Inbar M, Lichinitser M, Láng I, Nitz U, Iwata H, Thomssen C, Lohrisch C, Suter TM, Rüschoff J, Suto T, Greatorex V, Ward C, Straehle CC, McFadden E, Dolci MS, Gelber RD, Herceptin Adjuvant (HERA) Trial Study Team “Trastuzumab after Adjuvant Chemotherapy in HER2-Positive Breast Cancer”. New England Journal of Medicine. 2005;353((16)):1659–72. doi: 10.1056/NEJMoa052306. [DOI] [PubMed] [Google Scholar]
- Puppala S, Coletta DK, Schneider J, Hu SL, Farook VS, Dyer TD, Arya R, Blangero J, Duggirala R, DeFronzo RA, Jenkinson CP. “Genome-Wide Linkage Screen for Systolic Blood Pressure in the Veterans Administration Genetic Epidemiology Study (VAGES) of Mexican-Americans and Confirmation of a Major Susceptibility Locus on Chromosome 6q14. 1”. Human Heredity. 2011;71((1)):1–10. doi: 10.1159/000323143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romond EH, Perez EA, Bryant J, Suman VJ, Geyer CE, Jr, Davidson NE, Tan-Chiu E, Martino S, Paik S, Kaufman PA, Swain SM, Pisansky TM, Fehrenbacher L, Kutteh LA, Vogel G, Visscher DW, Yothers G, Jenkins RB, Brown AM, Dakhil SR, Mamounas EP, Lingle WL, Klein PM, Ingle JN, Wolmark N. “Trastuzumab Plus Adjuvant Chemotherapy for Operable HER2-Positive Breast Cancer”. New England Journal of Medicine. 2005;353((16)):1673–84. doi: 10.1056/NEJMoa052122. [DOI] [PubMed] [Google Scholar]
- Royal CDM, Dunston GM. “Changing the Paradigm from ‘Race’ to Human Genome Variation”. Nature Genetics. 2004;36(suppl 11):S5–7. doi: 10.1038/ng1454. [DOI] [PubMed] [Google Scholar]
- Salem RM, Pandey B, Richard E, Fung MM, Garcia EP, Brophy VH, Schork NJ, O'Connor DT, Bhatnagar V. “The VA Hypertension Primary Care Longitudinal Cohort: Electronic Medical Records in the Post-Genomic era”. Health Informatics Journal. 2005;16((4)):274–74. doi: 10.1177/1460458210380527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz UI, Ritchie MD, Bradford Y, Li C, Dudek SM, Frye-Anderson A, Kim RB, Roden DM, Stein CM. “Genetic Determinants of Response to Warfarin during Initial Anticoagulation”. New England Journal of Medicine. 2008;358((10)):999–1008. doi: 10.1056/NEJMoa0708078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shields AE, Fortun M, Hammonds EM, King PA, Lerman C, Rapp R, Sullivan PF. “The use of Race Variables in Genetic Studies of Complex Traits and the Goal of Reducing Health Disparities: A Transdisciplinary Perspective”. American Psychologist. 2005;60((1)):77–103. doi: 10.1037/0003-066X.60.1.77. [DOI] [PubMed] [Google Scholar]
- Shields AE, Shin P, Leu MG, Levy DE, Betancourt RM, Hawkins D, Proser M. “Adoption of Health Information Technology in Community Health Centers: Results of a National Survey”. Health Affairs. 2007;26((5)):1373–83. doi: 10.1377/hlthaff.26.5.1373. [DOI] [PubMed] [Google Scholar]
- Sivell S, Iredale R, Gray J, Coles B. “Cancer Genetic Risk Assessment for Individuals at Risk of Familial Breast Cancer”. Cochrane Database of Systematic Reviews (Online) 2007;2:CD003721. doi: 10.1002/14651858.CD003721.pub2. [DOI] [PubMed] [Google Scholar]
- Smit LAM, Heederik D, Doekes G, Koppelman GH, Bottema RWB, Postma DS, Wouters IM. “Endotoxin Exposure, CD14 and Wheeze among Farmers: A Gene-Environment Interaction”. Occupational and Environmental Medicine. 2011;68((11)):826–31. doi: 10.1136/oem.2010.060038. [DOI] [PubMed] [Google Scholar]
- Swede H, Gregorio DI, Tannenbaum SH, Brockmeyer JA, Ambrosone C, Wilson LL, Pensa MA, Gonsalves L, Stevens RG, Runowicz CD. “Prevalence and Prognostic Role of Triple-Negative Breast Cancer by Race: A Surveillance Study”. Clinical Breast Cancer. 2011;11((5)):332–41. doi: 10.1016/j.clbc.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teutsch SM, Bradley LA, Palomaki GE, Haddow JE, Piper M, Calonge N, Dotson WD, Douglas MP, Berg AO. “The Evaluation of Genomic Applications in Practice and Prevention (EGAPP) Initiative: Methods of the EGAPP Working Group”. Genetics in Medicine. 2009;11((1)):3–3. doi: 10.1097/GIM.0b013e318184137c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson J, Garry D, Poncz M, Center S, Torok-Storb B, Friedman A, Cooke J, Institutes JDG, San Francisco DS, Hatzopoulos A. “Kaiser Permanente & UCSF Receive NIH Award for Genetic Epidemiology Research”. Journal of Investigative Medicine. 2010;58((1)):6–9. [Google Scholar]
- Tishkoff SA, Verrelli BC. “Patterns of Human Genetic Diversity: Implications for Human Evolutionary History and Disease”. Annual Review of Genomics and Human Genetics. 2003;4:293–340. doi: 10.1146/annurev.genom.4.070802.110226. [DOI] [PubMed] [Google Scholar]
- Tomalik-Scharte D, Lazar A, Fuhr U, Kirchheiner J. “The Clinical Role of Genetic Polymorphisms in Drug-Metabolizing Enzymes”. Pharmacogenomics Journal. 2007;8((1)):4–15. doi: 10.1038/sj.tpj.6500462. [DOI] [PubMed] [Google Scholar]
- U.S. Department of Health and Human Services. Healthy People 2010: Understanding and Improving Health. Washington, DC: U.S. Government Printing Office; 2000. [Google Scholar]
- Voight BF, Scott LJ, Steinthorsdottir V, Morris AP, Dina C, Welch RP, Zeggini E, Huth C, Aulchenko YS, Thorleifsson G, McCulloch LJ, Ferreira T, Grallert H, Amin N, Wu G, Willer CJ, Raychaudhuri S, McCarroll SA, Langenberg C, Hofmann OM, Dupuis J, Qi L, Segre AV, van Hoek M, Navarro P, Ardlie K, Balkau B, Benediktsson R, Bennett AJ, Blagieva R, Boerwinkle E, Bonnycastle LL, Bostrom KB, Bravenboer B, Bumpstead S, Burtt NP, Charpentier G, Chines PS, Cornelis M, Couper DJ, Crawford G, Doney ASF, Elliott KS, Elliott AL, Erdos MR, Fox CS, Franklin CS, Ganser M, Gieger C, Grarup N, Green T, Griffin S, Groves CJ, Guiducci C, Hadjadj S, Hassanali N, Herder C, Isomaa B, Jackson AU, Johnson PRV, Jorgensen T, Kao WHL, Klopp N, Kong A, Kraft P, Kuusisto J, Lauritzen T, Li M, Lieverse A, Lindgren CM, Lyssenko V, Marre M, Meitinger T, Midthjell K, Morken MA, Narisu N, Nilsson P, Owen KR, Payne F, Perry JRB, Petersen A-K, Platou C, Proenca C, Prokopenko I, Rathmann W, Rayner NW, Robertson NR, Rocheleau G, Roden M, Sampson MJ, Saxena R, Shields BM, Shrader P, Sigurdsson G, Sparso T, Strassburger K, Stringham HM, Sun Q, Swift AJ, Thorand B, Tichet J, Tuomi T, van Dam RM, van Haeften TW, van Herpt T, van Vliet-Ostaptchouk JV, Walters GB, Weedon MN, Wijmenga C, Witteman J, Bergman RN, Cauchi S, Collins FS, Gloyn AL, Gyllensten U, Hansen T, Hide WA, Hitman GA, Hofman A, Hunter DJ, Hveem K, Laakso M, Mohlke KL, Morris AD, Palmer CNA, Pramstaller PP, Rudan I, Sijbrands E, Stein LD, Tuomilehto J, Uitterlinden A, Walker M, Wareham NJ, Watanabe RM, Abecasis GR, Boehm BO, Campbell H, Daly MJ, Hattersley AT, Hu FB, Meigs JB, Pankow JS, Pedersen O, Wichmann HE, Barroso I, Florez JC, Frayling TM, Groop L, Sladek R, Thorsteinsdottir U, Wilson JF, Illig T, Froguel P, van Duijn CM, Stefansson K, Altshuler D, Boehnke M, McCarthy MI. “Twelve Type 2 Diabetes Susceptibility Loci Identified through Large-Scale Association Analysis”. Nature Genetics. 2010;42((7)):579–89. doi: 10.1038/ng.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilke RA, Xu H, Denny JC, Roden DM, Krauss RM, McCarty CA, Davis RL, Skaar T, Lamba J, Savova G. “The Emerging Role of Electronic Medical Records in Pharmacogenomics”. Clinical Pharmacology and Therapeutics. 2011;89((3)):379–86. doi: 10.1038/clpt.2010.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


