Abstract
Histone protein post-translational modifications (PTMs) are significant for gene expression and DNA repair. Here we report the identification and validation of a new type of PTM in histones, lysine succinylation. The identified lysine succinylated histone peptides were verified by MS/MS of synthetic peptides, HPLC co-elution, and isotopic labeling. We identified 13, 7, 10, and 7 histone lysine succinylation sites in HeLa, mouse embryonic fibroblast, Drosophila S2, and Saccharomyces cerevisiae cells, respectively. We demonstrated that this histone PTM is present in all eukaryotic cells we examined. Mutagenesis of succinylation sites followed by functional assays implied that histone lysine succinylation can cause unique functional consequences. We also identified one and two histone lysine malonylation sites in HeLa and S. cerevisiae cells, respectively. Our results therefore increase potential combinatorial diversity of histone PTMs and suggest possible new connections between histone biology and metabolism.
Histones and p53 are among the proteins that are found to be most frequently modified (1, 2). Collective efforts from the research community identified at least 12 types of protein post-translational modifications (PTMs),1 most of which were identified by mass spectrometry (1, 3–5). In addition, the search for histone PTMs has not been exhausted and, not only novel sites, but also novel types of modifications continue to be discovered. For example, Hart and co-workers (4) recently identified O-GlcNAc modification as a new type of histone PTM. They demonstrated that O-GlcNAcylation is dynamically changed during mitosis and in response to heat shock.
Mounting evidence suggests that histone PTMs play a crucial regulatory role in diverse biological processes, such as cell differentiation and organismal development, and that aberrant modification of histones contributes to diseases, including cancer (6, 7). Thus, understanding this epigenetic process and its roles in cellular physiology and diseases demands a comprehensive understanding of all possible histone modifications.
At least two major mechanisms are thought to be associated with contributions of histone PTMs to dynamic chromatin-templated processes (1, 6). First, histone PTMs can directly modulate the packaging of chromatin by altering chemical structures of histones or internucleosomal interactions, through a change of the net charge, hydrogen bonding, size, or hydrophobicity in substrate PTM residues. A modified chromatin therefore in turn regulates the access of DNA-binding proteins, such as transcription factors. For example, neutralization of positive charges of lysine residues has been shown to disrupt interactions between positively charged lysine side chain and negatively charged DNA. Second, histone PTMs regulate chromatin structure and function by recruiting PTM-specific binding proteins (also called “readers”), which recognize modified histones via specialized structural folds, such as bromo, chromo, and plant homeo domain (PHD) domains (8–10). Conversely, histone PTMs can also function by inhibiting the interaction of specific binders with chromatin.
The remarkable regulatory potential of histone marks has been well illustrated in histone lysine acetylation and lysine methylation. Modifications at different locations (in the residues of histones) are involved in either activation or repression of gene expression. Acetylation versus methylation at the same histone site can be associated with very different transcriptional programs (11). Interestingly, lysine methylation exists in three forms: mono-, di-, and tri-methylation. Subtle chemical differences in these modifications may lead to very different outcomes. Different forms of lysine methylation can be enriched in different parts of chromatin (heterochromatin or euchromatin) (12). They can also be associated with different transcriptional regulatory elements of human genome. For example, histone H3K4 monomethylation specifically marks gene promoters, whereas H3K4 trimethylation is primarily associated with enhancers (13).
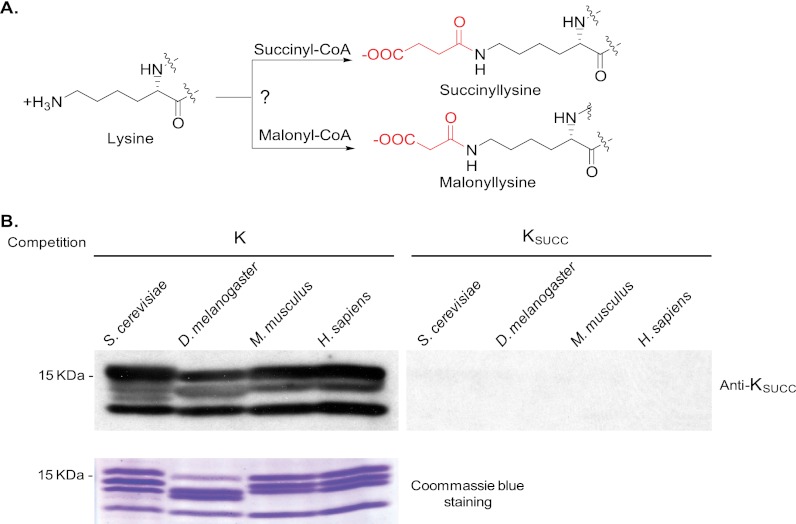
We recently discovered two types of novel PTMs called lysine succinylation and lysine malonylation in non-histone proteins (see Fig. 1A) (14, 15). In this study, we report that lysine succinylation and lysine malonylation are new types of histone PTMs. Our preliminary studies in Saccharomyces cerevisiae suggest lysine succinylation and malonylation in histones might have functional consequences.
Fig. 1.
Detection of lysine succinylation in core histones of different species. A, chemical structure and hypothesized mechanism for lysine succinylation. B, Western blot analysis (top panel) of core histones from S. cerevisiae, D. melanogaster, M. musculus, and H. sapiens cells with or without competition of a peptide library bearing a fixed unmodified lysine (K) or succinylated lysine (KSUCC). The bottom panel shows the loading control.
EXPERIMENTAL PROCEDURES
Materials
All chemicals, unless otherwise indicated, were of the highest purity available or analytical grade purchased from Sigma-Aldrich. 2,2,3,3-D4-succinic acid was purchased from Cambridge Isotope Laboratories (Andover, MA). Dulbecco's modified Eagle's medium and YPD medium were purchased from Fisher. Schneider's Drosophila medium was purchased from Invitrogen. HeLa and mouse embryonic fibroblast cells were obtained from the ATCC (Manassas, VA). All of the synthetic peptides used in this study were synthesized through customer synthesis using N-(9-fluorenyl)methoxycarbonyl-Lys (mono-tert-butyl succinate)-OH.
Cell Culture and in Vivo Isotopic Labeling
HeLa and mouse embryonic fibroblast cells were grown to 95% confluence in high glucose (4.5 g/liter) Dulbecco's modified Eagle's medium (with glutamine and sodium pyruvate) containing 10% fetal bovine serum and 1% penicillin-streptomycin at 37 °C with 95% air and 5% CO2. Drosophila S2 cells were grown in Schneider's Drosophila medium containing 10% heat-inactivated fetal bovine serum at 26 °C until the cell density reached 1 × 107 cells/ml. Yeast (BY4741) cells were grown in YPD medium at 30 °C for 16 h with shaking at 230–270 rpm until A600 reached 2.4. For isotopic labeling, HeLa cells were grown in Dulbecco's modified Eagle's medium with 10% fetal bovine serum, 1% penicillin-streptomycin, and 50 mm of sodium D4-succinate for 24 h until 95% confluence. Extraction of the histones followed the acid extraction method described previously (16).
Peptide Sample Preparation and Affinity Enrichment
Chemical propionylation of histone extracts was performed using a procedure previously reported with slight modifications (17). Briefly, 3 mg of histone extracts were dissolved in 50 μl of 100 mm ammonium bicarbonate buffer (pH 8.0) and added with 300 μl of propionic anhydride in 300 μl of methanol. Ammonium hydroxide was added to adjust the solution pH to ∼8.0. After incubation at 51 °C for 20 min, the mixture was dried in a SpeedVac. In-solution histone digestion was carried out as previously reported (18). Enrichment of lysine succinylated and malonylated peptides from tryptic digest of histones, with or without in vitro propionylation, by peptide immunoprecipitation with pan anti-succinyllysine and anti-malonyllysine antibodies (PTM Biolabs Inc., Chicago, IL), was carried out as described previously (18).
HPLC/MS/MS Analysis
Peptide samples were analyzed by a NanoLC-1D plus HPLC system (Eksigent Technologies, Dublin, CA) coupled to an LTQ Orbitrap mass spectrometer (ThermoFisher Scientific, San Jose, CA) as described previously (14). The peptides were eluted from a home-made capillary Jupiter C12 column (10-cm length × 75-μm inner diameter, 4-μm particle size, 90 Å pore diameter; Phenomenex, St. Torrance, CA) with a 2-h gradient of 2% to 80% HPLC solvent B (0.1% formic acid in acetonitrile, v/v) in solvent A at a flow rate of 200 nl/min. High resolution full scan MS spectra (from m/z 350 to 1800) acquired in the Orbitrap with resolution r = 60,000 at m/z 400 was followed by MS/MS fragmentation of the 20 most intense ions in the linear ion trap analyzer with collisionally activated dissociation energy of 35%. Verification of lysine succinylated peptides by HPLC/MS/MS analysis of synthetic peptides was used the same method as described previously (14). Briefly, the affinity-enriched histone succinyllysine peptide, its synthetic counterpart, and their mixture were analyzed by nano-HPLC/MS/MS, respectively.
Data Analysis and Validation
The mass spectrometric data analysis was performed by MASCOT search engine (v2.1; Matrix Science, London, UK). Peak lists were generated by extract_msn.exe software (v5.0; Thermo Scientific). For protein identification, the data from human, mouse, Drosophila, and yeast were searched against International Protein Index (IPI) human protein database (v 3.70, 87069 sequences), IPI mouse protein database (v 3.74,56860 sequences), UniProtKB Drosophila melanogaster protein database (taxonomy: 7227; 17,526 sequences), and Saccharomyces protein database (YeastORF, 6717 sequences), respectively. Significance threshold (p < 0.05) and Ions score cut-off (0) were used for protein identification. The identified proteins from each species were compiled into a new database for PTM analysis. The identified protein lists were included in supplemental Table 1. The search criteria were: mass error for parent ion mass was set as ±10 ppm and for fragment ion as ±0.5 Da. Enzyme was specified as trypsin with six missing cleavages. Methionine oxidation, lysine acetylation, lysine malonylation (K +86.00039 Da), and lysine succinylation (K +100.01604 Da) were specified as variable modifications. For the propionylated histone sample, lysine propionylation was also specified as a variable modification. All of the peptide identifications with Mascot ion score above 20 were manually verified based on their precursor MS and product MS/MS data.
Yeast Experiment
The yeast strain used to test silencing phenotypes for histone H2A and H2B is JDY187 (similar to JDY23 and a derivative of GFY167, MATα his3Δ200 leu2Δ1 trp1Δ63 lys2Δ0 ura3-167 met15Δ0 ade2::his RDN1::Ty1-MET15 TELV::ADE2 hta2-htb2::HygMX4 hta1-htb1::NatMX4 pJD78 [CEN URA3 HTA2-HTB2]). All other phenotypes were tested in JDY142 (similar to JDY92 and a derivative of S288C, MATα his3Δ200 leu2Δ0 lys2Δ0 trp1Δ63 ura3Δ0 met15Δ0 hta2-htb2::HygMX4 hta1-htb1::G418 pJD78 [CEN URA3 HTA2-HTB2]). The histone H2A and histone H2B mutants were generated by gene synthesis and integrated at endogenous HTA1-HTB1 locus. The phenotypic assays were described previously (19). Except for the Glu substitutions, phenotypes for all other histone H3 and H4 mutants were extracted from a previous study (19).
RESULTS
Initial Identification of Histone Lysine Succinylation
We hypothesize the existence of histone lysine succinylation in eukaryotic cells based on the fact that all the major PTMs are present in histones, including but not limited to phosphorylation, acetylation, methylation, ubiquitination, and O-GlcNAc modification. To test for the existence of lysine succinylation in core histones, we carried out Western blotting analysis of core histones from four eukaryotic species using an anti-succinyllysine antibody. The experiment detected lysine succinylation signals from the core histones of all four species tested (Fig. 1B). The signals can be efficiently competed away by a succinyllysine peptide library bearing a fixed succinyllysine at the seventh residue but not its corresponding unmodified peptide library. The specificity of the succinyllysine signals was demonstrated by competition experiments using a succinyllysine peptide library and dot-spot assay (14). These results indicate that lysine succinylation is found in histones and could represent an evolutionarily conserved histone mark in eukaryotic cells.
Lysine Succinylation Sites in Core Histones
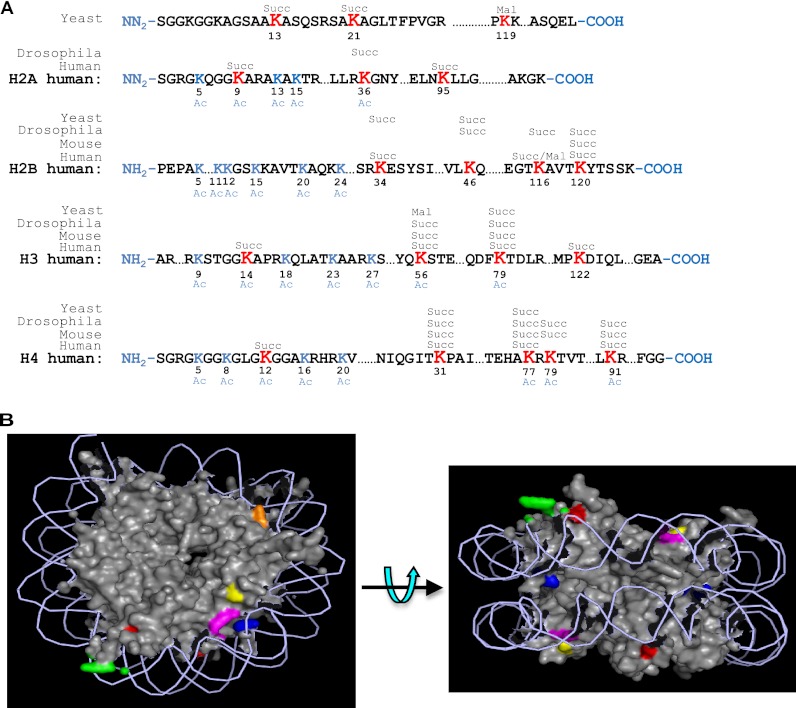
To identify succinyllysine sites, we extracted core histones from HeLa cells using a procedure described previously (16). The core histones were digested in solution, with or without chemical propionylation, and subjected to affinity enrichment using anti-succinyllysine antibody as reported (18). Lysine succinylated peptides were analyzed by HPLC/MS/MS analysis and protein sequence alignment to identify succinyllysine sites in histones. The succinyllysine residues can be identified based on a mass shift of + 100 Da at the lysine residue. The experiments led to the identification of 13 sites in HeLa core histones (Fig. 2A). Using the same experimental procedure, we also identified 7, 10, and 7 succinylation sites in histones extracted from cells of yeast, Drosophila, and mouse, respectively. The critical MS/MS spectra were manually verified to ensure the high quality of the peptide identification and are included in the text and supplemental materials for readers' reference (Figs. 3 and 4 and supplemental Figs. S4–S6).
Fig. 2.
Identification of succinyllysine residues in histone proteins. A, illustration of identified histone succinylated sites from S. cerevisiae, D. melanogaster, M. musculus, and H. sapiens. Succinylation sites are shown in bold red type. Three malonyllysine sites identified in yeast and HeLa cells are also indicated. Reported acetylation sites are shown in blue type. B, position of succinylated residues in yeast nucleosome. The crystal structure is from the Protein Data Bank (code 1ID3) and is shown as spheres. DNA is shown as lines. The succinylated residues are labeled in color: yellow, H3K79; orange, H4K31; purple, H4K77; green, H2AK13; red, H2AK21; and blue, H2BK37.
Fig. 3.
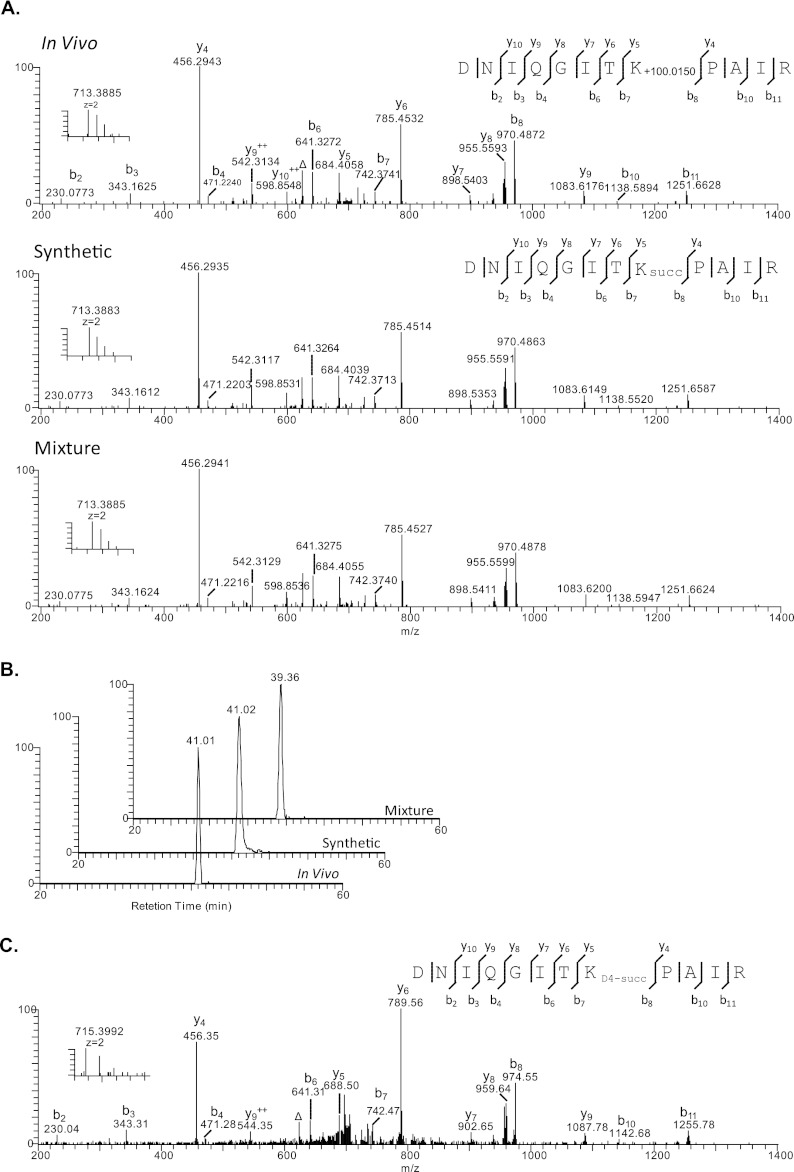
HPLC and mass spectrometric verification of histone succinylation at H4K31 (DNIQGITK+100.0150 PAIR). A, high resolution MS/MS spectra of H4K31 succinylation peptide (DNIQGITK+100.0150 PAIR) from affinity-enriched HeLa histone extract using anti-succinyllysine pan antibody (top), the synthetic DNIQGITKsuccPAIR (middle), and the mixture of them (bottom). The insets show the precursor ions. The label Δ designates b or y ions with water and/or ammonia loss. B, extracted ion chromatograms of the in vivo derived H4K31 peptide (top), its synthetic counterpart (middle), and their mixture (bottom). C, MS and MS/MS spectrum of D4-succinyllysine peptide.
Fig. 4.
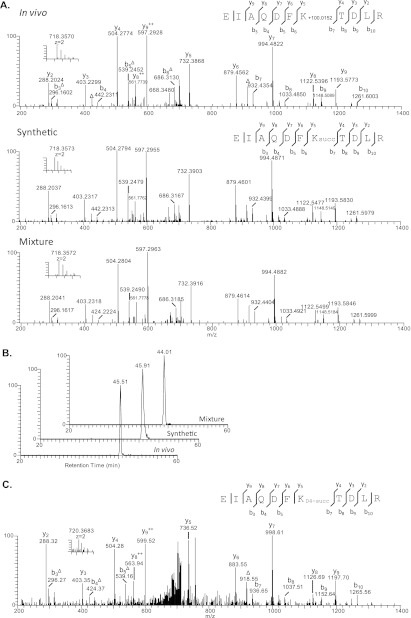
HPLC and mass spectrometric verification of histone succinylation at H4K31 (EIAQDFK+100.0152TDLR). A, high resolution MS/MS spectra of in vivo H3K79 succinylation peptide (EIAQDFK+100.0152TDLR) from affinity-enriched HeLa histone extract (top), the synthetic EIAQDFKsuccTDLR (middle), and the mixture of them (bottom). B, extracted ion chromatograms of the in vivo derived H3K79 peptide (top), its synthetic counterpart (middle), and their mixture (bottom). C, MS and MS/MS spectrum of D4-succinyllysine peptide.
Confirmation of Histone Succinyllysine Peptides by MS/MS and HPLC Co-elution
We are reasonably confident that the identified mass shift of + 100 Da is caused by lysine succinylation, because the histone succinyllysine peptides were affinity-purified before MS/MS analysis. Because lysine succinylation is a relatively new PTM, it is desirable to confirm the structure of the identified peptides to ensure that the derived mass shifts of + 100 Da is caused by lysine succinylation. MS/MS and HPLC co-elution are gold standards for verifying peptide identification. Toward this goal, we chemically synthesized three representative succinyllysine peptides: DNIQGITKsuccPAIR, EIAQDFKsuccTDLR, and TVTAMDVVYALKsuccR. We then carried out pair-wise MS/MS and co-elution experiments between the synthetic peptides and their in vivo counterparts, respectively.
Our result showed that the high resolution MS/MS fragmentation patterns of in vivo DNIQGITK+100.0150PAIR peptide, the synthetic DNIQGITKsuccPAIR peptide, and their mixture were almost identical (Fig. 3A). Furthermore, the mixture of the in vivo DNIQGITK+100.0150PAIR peptide and the synthetic succinyllysine counterpart showed a single co-eluted peak in the HPLC chromatogram, indicating that the detected + 100.0150 mass shift was caused by a succinyl group (Fig. 3B). Using the same method, we also confirmed the peptide identification for EIAQDFKsuccTDLR, TVTAMDVVYALKsuccR (Fig. 4 and supplemental Fig. S1).
Isotopic Labeling of Histone Succinyllysine Peptides
To further establish the presence of lysine succinylation in core histones, we carried out in vivo isotopic labeling followed by HPLC/MS/MS analysis of histone peptides as described previously (14). In this experiment, we labeled HeLa cells with isotopic succinate (sodium 2,2,3,3-D4-succinate) for 24 h. The core histones were extracted and analyzed using the above described procedure. D4-Labeled succinyllysine was identified in 11 histone peptides (Figs. 3C and 4C and supplemental Fig. S3), suggesting that histone lysine succinylation can be labeled in similar fashion to histone lysine acetylation, presumably by using a succinyl coenzyme A precursor (20).
Phenotypic Analysis of Mutations on Succinylated Residues in Budding Yeast
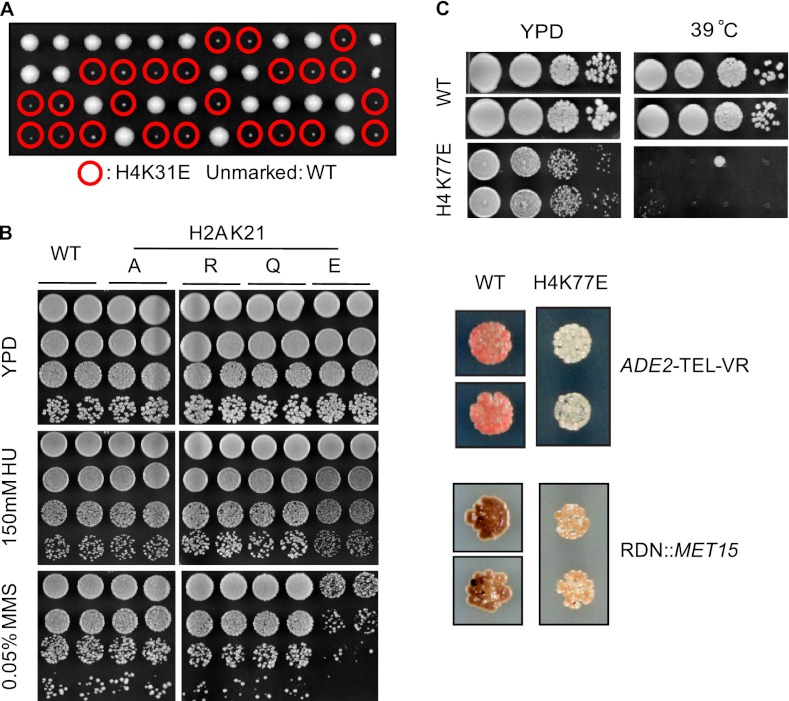
To gain some insight into the biological function of lysine succinylation in core histones, we mutated the modified residue to alanine and arginine to prevent succinylation and to glutamic acid to mimic constitutively succinylated lysine. Of the six residues, we found that the Glu substitution, but not Ala or other substitutions on histone H4K31, significantly reduces cell viability (Fig. 5A). Cells bearing Glu substitutions on other sites had no obvious effect on cell growth. After testing under various conditions as described before (19), we found no significant difference for all mutations on histone H2AK13 and H2BK37 compared with wild type. In addition, all of the mutations on histone H3K79 lose silencing at telomeres and rDNA. No distinction was observed among the Glu substitution and other mutations. However, we found that histone H2AK21E, but not K21A or K21R, was sensitive to methyl methanesulfonate (Fig. 5B), suggesting a potentially deleterious effect of succinylation of this residue. In addition, we identified several unique phenotypes of histone H4K77E mutation (Fig. 5C). It causes a loss of silencing at both telomere and rDNA, with a more profound effect on telomeric silencing. On the other hand, there is no silencing defect in a K77R mutant, whereas the K77A mutant loses silencing at telomere but has slightly increased rDNA silencing (19). Furthermore, the K77E substitution becomes temperature-sensitive at 37 and 39 °C, which is not observed in the other mutants tested.
Fig. 5.
Phenotypic analysis of mutations on succinylated residues in budding yeast. A, H4K31E mutant has severe growth defect. Circled in red are the spores with K31E mutants. The picture was taken after 4 days of incubation at 30 °C. B, histone H2AK21E mutant is specifically sensitive to methyl methanesulfonate. Two independent colonies containing the corresponding mutations were tested on either YPD or YPD-containing hydroxyurea (HU) or methyl methanesulfonate (MMS). The picture was taken after 3 days of incubation at 30 °C. C, histone H4K77E mutant is temperature-sensitive and loses silencing at both rDNA and telomere.
Identification of Histone Lysine Malonylation and Phenotypic Analysis of the Kmal Sites in Budding Yeast
Lysine malonylation has been reported recently in both bacteria and mammalian cells (15). However, it is not known whether lysine malonylation exists in histone proteins. By using affinity enrichment with pan anti-malonyllysine antibody and mass spectrometry, we identified one and two malonyllysine sites in histones from HeLa and S. cerevisiae cells, respectively (Fig. 2A). The malonyllysine peptides can be unambiguously identified based upon the characteristic neutral loss of CO2 peaks in their MS/MS spectra (15) and their high resolution precursor ion masses (supplemental Fig. S7 and S8).
To probe the potential function of lysine malonylation in budding yeast, we mutated both of the Kmal sites (histone H2AK119 and histone H3K56) to glutamic acid to mimic constitutive modification and analyzed the phenotypes of these mutations as described before (19). We did not detect significant change of fitness for the H2AK119 mutant compared with wild type. However, the H3K56E mutant reduced cell viability in yeast, as shown in both plasmid shuffling assay and tetrad analyses (supplemental Fig. S9).
DISCUSSION
In this study, seven sites of Ksucc in yeast were identified, of which two sites are on histone H2A, two on histone H2B, two on histone H4, and one on histone H3, respectively. Interestingly, none of these sites are within the N-terminal tail, a region heavily modified in other ways. In fact, two of the succinylated sites (H2AK13 and H2BK37) are located right at the end of its N-terminal tail (Fig. 2B), where the histones make close contact with DNA. Of the six succinylated sites, except H3K79, all of them are somewhat near to the double-stranded DNA. The position of these succinylated residues and the nature of lysine succinylation suggest that it may interfere with the interaction between histones and the negatively charged DNA. Evidently, Glu substitution of histone H4K31, which is located close to the dyad axis of nucleosome, causes a dramatic reduction in cell viability. In addition, it is worth notice that none of the sites are buried within the nucleosome, allowing potential access to “writers” and the “readers.” Among the succinylated lysines, H3K79 is also well known to be methylated. Methylation on H3K79 is carried out by Dot1p in S. cerevisiae and is critical for gene activation (21). Identification of succinylation on this residue raises the possibility that its function may be affected by this “new” modification.
H3K56 is known to be acetylated, and acetylation on this residue is important for histone deposition (22). We identified H3K56 lysine malonylation in yeast and lysine succinylation in Drosophila, mouse, and human. Substitution of H3K56 with glutamic acid in yeast caused lethality (supplemental Fig. S9), whereas substitution of the site with alanine or arginine did not change yeast cell viability (19, 23). Because histone H3K56 localizes near the nucleosome entry site and is proximate to the DNA double helix, the change of charge from positive to negative at the site by malonylation might interfere with its interaction with DNA, causing the loss of cell viability.
Although we have not yet performed careful experiments to determine the stoichiometry of histone lysine succinylation and lysine malonylation at specific sites, our initial studies suggest that the overall stoichiometry of these two modifications are less abundant than histone lysine methylation and lysine acetylation, the two most abundant histone PTMs, and are in line with lysine butyrylation. In some cells, some histone lysine succinylation sites can be detected by HPLC/MS/MS, without the enrichment step and are more abundant than some low abundant histone acetyllysine and methyllysine sites (data not shown). Given the fact that histones are so abundant in the cells, low stoichiometry PTMs does not necessarily exclude the possibility that they have important biological functions. As an example, we recently demonstrated that some histone lysine butyrylation mark, a histone PTM with much lower stiochiometry than lysine acetylation, is associated with specific genomic regions, likely having important functions in transcriptional regulation (data not shown).
Although we cannot conclusively exclude the possibility that lysine malonylation and lysine succinylation is caused by chemical reaction, originating directly from malonyl- and succinyl-CoA, a few lines of evidence indicate that enzyme-catalyzed lysine succinylation and lysine malonylation exist in cells. First, in this study, we have identified unique Ksucc sites that have not been reported to either have lysine acetylation (H2AK95, H2BK116, and H2BK120) or any other PTMs (H2AK95). Second, half of the Ksucc sites we identified are highly conserved among species. Additionally, most of Ksucc sites are located in the globular domain and the C terminus instead of the N-terminal tails of histones, which are among the residues that easily access other chemicals for chemical reactions. Nevertheless, we argue that both enzyme-catalyzed histone PTMs and chemical reaction-induced histone PTMs may have significant biological consequences, because the removal of these modifications could be a critical event for cellular detoxicification process.
Lysine acetylation neutralizes the positive side chain of histone lysine residues, affecting chromatin structure and function by modulating interactions between histones and DNA and recruiting “histone code readers” (e.g. a bromo-domain protein). Within the nucleosome core particle, four lysine residues have direct contact with DNA, and another four of them span the major groove (24), implying a key role in their interactions with DNA through charge-charge interaction. Lysine succinylation and malonylation induce even more significant structural changes than lysine acetylation by changing a positively charged residue to a negative charge. In terms of charge state change, lysine succinylation and malonylation are of the same magnitude as phosphorylation, producing a two-charge shift in the substrate residues. Given the crucial role of histone lysine PTMs (e.g. lysine acetylation and methylation) in DNA-templated processes, such a dramatic structural disturbance is likely to have significant consequences for chromatin structure and function. Therefore, lysine succinylation and malonylation are likely to play important roles in histone structure and function.
Footnotes
* This work was supported by National Institutes of Health Roadmap Program Grants U54RR020389, RO1CA126832, and RO1DK082664 (to J. D. B. and Y. Z.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviation used is:
- PTM
- post-translational modification.
REFERENCES
- 1. Kouzarides T. (2007) Chromatin modifications and their function. Cell 128, 693–705 [DOI] [PubMed] [Google Scholar]
- 2. Dai C., Gu W. (2010) p53 post-translational modification: Deregulated in tumorigenesis. Trends Mol. Med. 16, 528–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Garcia B. A., Shabanowitz J., Hunt D. F. (2007) Characterization of histones and their post-translational modifications by mass spectrometry. Curr. Opin. Chem. Biol. 11, 66–73 [DOI] [PubMed] [Google Scholar]
- 4. Sakabe K., Wang Z., Hart G. W. (2010) β-N-acetylglucosamine (O-GlcNAc) is part of the histone code. Proc. Natl. Acad. Sci. U.S.A. 107, 19915–19920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kruse J. P., Gu W. (2008) SnapShot: p53 posttranslational modifications. Cell 133, 930-30.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ruthenburg A. J., Li H., Patel D. J., Allis C. D. (2007) Multivalent engagement of chromatin modifications by linked binding modules. Nat. Rev. Mol. Cell Biol. 8, 983–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Martin C., Zhang Y. (2007) Mechanisms of epigenetic inheritance. Curr. Opin. Cell Biol. 19, 266–272 [DOI] [PubMed] [Google Scholar]
- 8. Wysocka J., Swigut T., Xiao H., Milne T. A., Kwon S. Y., Landry J., Kauer M., Tackett A. J., Chait B. T., Badenhorst P., Wu C., Allis C. D. (2006) A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature 442, 86–90 [DOI] [PubMed] [Google Scholar]
- 9. Wysocka J., Swigut T., Milne T. A., Dou Y., Zhang X., Burlingame A. L., Roeder R. G., Brivanlou A. H., Allis C. D. (2005) WDR5 associates with histone H3 methylated at K4 and is essential for H3 K4 methylation and vertebrate development. Cell 121, 859–872 [DOI] [PubMed] [Google Scholar]
- 10. Zeng L., Zhou M. M. (2002) Bromodomain: An acetyl-lysine binding domain. FEBS Lett. 513, 124–128 [DOI] [PubMed] [Google Scholar]
- 11. Jenuwein T., Allis C. D. (2001) Translating the Histone Code. Science 293, 1074–1080 [DOI] [PubMed] [Google Scholar]
- 12. Martin C., Zhang Y. (2005) The diverse functions of histone lysine methylation. Nat. Rev. Mol. Cell Biol. 6, 838–849 [DOI] [PubMed] [Google Scholar]
- 13. Heintzman N. D., Stuart R. K., Hon G., Fu Y., Ching C. W., Hawkins R. D., Barrera L. O., Van Calcar S., Qu C., Ching K. A., Wang W., Weng Z., Green R. D., Crawford G. E., Ren B. (2007) Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 39, 311–318 [DOI] [PubMed] [Google Scholar]
- 14. Zhang Z., Tan M., Xie Z., Dai L., Chen Y., Zhao Y. (2011) Identification of lysine succinylation as a new post-translational modification. Nat. Chem. Biol. 7, 58–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Peng C., Lu Z., Xie Z., Cheng Z., Chen Y., Tan M., Luo H., Zhang Y., He W., Yang K., Zwaans B. M., Tishkoff D., Ho L., Lombard D., He T. C., Dai J., Verdin E., Ye Y., Zhao Y. (2011) The first identification of lysine malonylation substrates and its regulatory enzyme. Mol. Cell. Proteomics 10, 10.1074/mcp.M111.012658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Shechter D., Dormann H. L., Allis C. D., Hake S. B. (2007) Extraction, purification and analysis of histones. Nat. Protoc. 2, 1445–1457 [DOI] [PubMed] [Google Scholar]
- 17. Garcia B. A., Mollah S., Ueberheide B. M., Busby S. A., Muratore T. L., Shabanowitz J., Hunt D. F. (2007) Chemical derivatization of histones for facilitated analysis by mass spectrometry. Nat. Protoc 2, 933–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kim S. C., Sprung R., Chen Y., Xu Y., Ball H., Pei J., Cheng T., Kho Y., Xiao H., Xiao L., Grishin N. V., White M., Yang X. J., Zhao Y. (2006) Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol. Cell 23, 607–618 [DOI] [PubMed] [Google Scholar]
- 19. Dai J., Hyland E. M., Yuan D. S., Huang H., Bader J. S., Boeke J. D. (2008) Probing nucleosome function: A highly versatile library of synthetic histone H3 and H4 mutants. Cell 134, 1066–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. MacDonald M. J., Fahien L. A., Brown L. J., Hasan N. M., Buss J. D., Kendrick M. A. (2005) Perspective: Emerging evidence for signaling roles of mitochondrial anaplerotic products in insulin secretion. Am. J. Physiol. Endocrinol Metab. 288, E1–E15 [DOI] [PubMed] [Google Scholar]
- 21. van Leeuwen F., Gafken P. R., Gottschling D. E. (2002) Dot1p modulates silencing in yeast by methylation of the nucleosome core. Cell 109, 745–756 [DOI] [PubMed] [Google Scholar]
- 22. Xu F., Zhang K., Grunstein M. (2005) Acetylation in histone H3 globular domain regulates gene expression in yeast. Cell 121, 375–385 [DOI] [PubMed] [Google Scholar]
- 23. Nakanishi S., Sanderson B. W., Delventhal K. M., Bradford W. D., Staehling-Hampton K., Shilatifard A. (2008) A comprehensive library of histone mutants identifies nucleosomal residues required for H3K4 methylation. Nat. Struct. Mol. Biol. 15, 881–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Luger K., Richmond T. J. (1998) DNA binding within the nucleosome core. Curr. Opin. Struct. Biol. 8, 33–40 [DOI] [PubMed] [Google Scholar]