Abstract
To understand how the chromosomal passenger complex ensures chromosomal stability, it is crucial to identify its substrates and to find ways to specifically inhibit the enzymatic core of the complex, Aurora B. We therefore developed a chemical genetic approach to selectively inhibit human Aurora B. By mutating the gatekeeper residue Leu-154 in the kinase active site, the ATP-binding pocket was enlarged, but kinase function was severely disrupted. A unique second site suppressor mutation was identified that rescued kinase activity in the Leu-154 mutant and allowed the accommodation of bulky N6-substituted adenine analogs. Using this analog-sensitive Aurora B kinase, we found that retention of the chromosomal passenger complex at the centromere depends on Aurora B kinase activity. Furthermore, analog-sensitive Aurora B was able to use bulky ATPγS analogs and could thiophosphorylate multiple proteins in cell extracts. Utilizing an unbiased approach for kinase substrate mapping, we identified several novel substrates of Aurora B, including the nucleosomal-binding protein HMGN2. We confirmed that HMGN2 is a bona fide Aurora B substrate in vivo and show that its dynamic association to chromatin is controlled by Aurora B.
Faithful chromosome segregation requires that the duplicated sister chromatids bi-orient on the mitotic spindle and that anaphase onset does not start before this is accomplished for all chromosomes. The chromosomal passenger complex is essential for this because it specifically destabilizes incorrectly attached spindle microtubules from the kinetochores of the chromosomes and acts on the mitotic checkpoint that inhibits the anaphase-promoting complex/cyclosome until all chromosomes have acquired the correct bipolar attachments (1). In addition, this complex is important for cytoplasmic division and may have additional functions outside mitosis, such as DNA damage repair in G2 (2) and the epigenetic silencing of gene expression (3). Although it is accepted that the CPC1 is essential for proper cell division, its potential functions outside mitosis are only beginning to be uncovered. To reveal new in vivo functions of the CPC and to understand how this complex is capable of fulfilling all of these different functions, it is important to specifically and completely inhibit the enzymatic core of the complex (Aurora B) without affecting the stability of the other CPC subunits (INCENP, borealin, and survivin).
Current approaches to inhibit Aurora B (small interfering RNA and small molecule inhibitors) are important research tools, but they do suffer from variations in the level of protein knockdown or kinase inhibition (4). In particular, the presence of two other Aurora kinases (A and C) with a high degree of homology to Aurora B makes it particularly challenging to identify small molecules that selectively inhibit Aurora B (5). Because of the high level of active site homology, finding an inhibitor concentration that completely inhibits the kinase of interest in cells without affecting any other kinase is nearly impossible. Hence, using the current approaches to target Aurora B makes it difficult to unequivocally resolve the in vivo functions of the CPC and may complicate the assignment of true Aurora B substrates. We have therefore developed a chemical-genetic system that allows specific Aurora B inhibition and direct substrate identification.
Chemical genetics refers to a strategy where a kinase is genetically engineered to render it capable of utilizing non-natural ATP analogs to be preferentially utilized as substrates and additionally to be sensitive to unique inhibition by cell-permeable ATP analogs (6, 7). This so-called analog-sensitive kinase harbors a specific mutation in the ATP-binding pocket that changes a bulky amino acid (i.e. methionine, leucine, phenylalanine, or threonine) into a small amino acid (glycine or alanine). Mutation of this “gatekeeper” residue enlarges the ATP-binding pocket, allowing it to accommodate bulky side chains of ATP analogs and making it susceptible to cell-permeable derivatives of the Src inhibitor PP1 (PP1 inhibitors) (8). Approximately 30% of kinases lose their catalytic activity after mutation of the gatekeeper residue, but functionality can be restored by introduction of one or more second site suppressor mutations (9). Catalytic activity is critical when attempting to map direct kinase substrates in an unbiased manner (10).
Human Aurora B turned out to be one of the kinases that did not tolerate mutation of the gatekeeper residue (Leu-154) and for which mutation of the predicted second sites failed to restore functionality. We here describe the identification of a unique second site suppressor mutation that restored activity of the Aurora B gatekeeper mutants and that made the kinase susceptible to inhibition by PP1 analogs. Using these analog-sensitive Aurora B mutants, we demonstrate that retention of the CPC at the centromere depends on Aurora B kinase activity. We also show that the active Aurora B is capable of using bulky ATPγS analogs to thiophosphorylate multiple proteins in complex cell extracts, including a number of known Aurora B substrates. Because this approach is not biased with respect to known consensus sites or for particular functional categories of putative substrates, it is particularly useful for identifying novel direct substrates. Indeed, we found a number of potential novel Aurora B phosphorylation sites on previously reported substrates, as well as novel substrates of the kinase including the nucleosomal-binding protein HMGN2.
EXPERIMENTAL PROCEDURES
Mutagenesis and Cloning
Aurora BL154A and Aurora BL154G were generated by site-directed mutagenesis (QuikChange; Agilent Technologies, Wilmington DE) using the following primers: forward GGAGGATCTACTTGATTGCAGAGTATGCCCCCCGCGC and reverse CCGCGGGGGGCATACTCTGCAATCAAGTCGATCCTCC for L154A and forward GGAGGATCTACTTGATTGGAGAGTATGCCCCCCGCGG and reverse CCGCGGGGGGCATACTCTCCAATCAAGTAGATCCTCC for L154G, and FLAG-Aurora Bwt (pCR3; Invitrogen) and GST-Aurora Bwt (pGEX-4T-1; GE Healthcare) as templates. Aurora BL154A/H250Y and Aurora BL154G/H250Y were generated from the FLAG-Aurora BL154A (pCR3), FLAG-Aurora BL154G (pCR3; Invitrogen), plasmids by mutagenesis PCR using forward GATTGAGGGGCGCATGTACAATGAGAAGGTGGATC and reverse GATCCACCTTCTCATTGTACATGCGCCCCTCAATC as primers. The Aurora BL154A/H250Y and Aurora BL154G/H250Y PCR products were digested with EcoRI and ligated into the enhanced green fluorescent protein-C2 (Clontech), pCR3-vsv (11), pGEX-4T-1, and pFastBac (Invitrogen) vectors, to introduce N-terminal GFP, vesicular stomatitis virus, or GST tags, respectively, or for expression in Sf9 insect cells. Full-length HMGN2 was generated by PCR using the following primers forward ATTGAATTCCCCAAGAGAAAGGCTGAAG and reverse ATTCTCGAGGGGCCCTCACTTGGCATCTCCAGC. The PCR product was ligated into the pJET1.2/blunt cloning vector (Fermentas by Thermo Scientific, St. Leon-Rot, Germany). The vector was digested with EcoRI and ApaI, and HMGN2 was ligated into enhanced green fluorescent protein C2. All of the constructs were verified by DNA sequencing.
Cell Culture and Transfection
U2OS (human osteosarcoma cell line) and HeLa (human cervical cancer cell line) cells were cultured in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 6% fetal calf serum (Invitrogen), 1 mm ultraglutamine (Lonza, Cologne, Germany), and streptomycin/penicillin (Invitrogen). The cell lines were maintained in 5% CO2 at 37 °C. All cDNA transfections were performed using the standard calcium phosphate transfection protocol.
Purification of Recombinant Proteins
The Aurora B/pGEX-4T-1 vectors were introduced into BL21 bacteria, and the transformants were plated on LB-agar plates containing ampicillin and chloramphenicol (both from Sigma-Aldrich). Protein expression was induced for 4 h at 30 °C by the addition of 1 mm isopropyl β-d-thiogalactopyranoside (Sigma-Aldrich). After induction, bacteria pellets were lysed in lysis buffer containing 10 mm EGTA, 10 mm EDTA, 0.1% Tween 20, 250 mm NaCl, 5 mm DTT, 0.325 mg/ml lysosome, and protease inhibitors (CompleteTM; Roche Applied Science). The cells were sonicated and centrifuged at 38,724 relative centrifugal force (rcf) for 30 min at 4 °C. Supernatants were coupled to glutathione-Sepharose 4B beads (Amersham Biosciences), and proteins were eluted in buffer containing 50 mm Tris (pH 8.0), 20 mm reduced glutathione and 75 mm KCl. The GST-Aurora B/His-INCENP complexes were purified from baculovirus-infected SF9 cells. Protein expression was induced for 4 days at 27 °C. SF9 were pelleted and lysed in buffer containing: 50 mm Tris (pH 8.0), 400 mm NaCl, 1.0% Nonidet P-40, 0.5% sodium deoxycholate, 20 mm glycerol-2-phosphate, 0.3 mm NaVO3, 1 μg/ml leupeptin, 1 μg/ml pepstatin, 1 μg/ml aprotinin, and 1 mm PMSF (all from Sigma-Aldrich). The cells were sonicated and centrifuged at 15,870 relative centrifugal force (rcf) for 20 min at 4 °C. Supernatants were coupled to nickel-nitrilotriacetic acid-agarose beads (Qiagen) and washed twice in buffer containing 100 mm Na2HPO4, 100 mm NaH2PO4, 300 mm NaCl, 10% glycerol, and 20 mm imidazole. Protein complexes were eluted in buffer containing 250 mm imidazole.
Immunoprecipitation and in Vitro Kinase Reactions
U2OS cells were transiently transfected with FLAG- or GFP-tagged cDNA constructs. The cells were treated with nocodazole (250 μg/ml) for 18 h, and mitotic cells were collected by shake-off. Mitotic cells were lysed as described previously (12), and cleared lysates were added to protein A/G beads coupled to anti-GFP or anti-FLAG (M2; Sigma-Aldrich). Immunoprecipitated or recombinant kinases were included into a 25-μl reaction containing kinase buffer (10 mm MgCl2, 25 mm HEPES, pH 7.5, 25 mm β-glycerophosphate, 0.5 mm DTT and 0.5 mm vanadate), 0.2 mg/ml histone H3 (Roche Diagnostics), and 100 μm ATP with or without 2 μm NA-PP1 (Calbiochem/Merck Chemicals). For thiophosphorylation assays, different concentrations of ATPγS or bulky ATPγS analogs (Biolog, Hayward, CA) were used. After 30 min at 30 °C, the reactions were stopped by the addition of 25 μl of 2× sample buffer. Thiophosphorylated samples were subsequently incubated with 2.5 mm p-nitrobenzyl mesylate (Epitomics, Burlingame, CA). This thiol-specific alkylating agent generates a bio-orthogonal tiophosphate ester that is recognized by a thiophosphate ester-specific antibody (Epitomics) (13).
Western Blotting
Protein samples were separated by SDS-PAGE and transferred to nitrocellulose membrane by standard procedures. Membranes were blocked with 4% milk in Tris-buffered saline containing 1% Tween 20 (TBST) prior to incubation with the following primary antibodies in 4% milk-TBST: mouse anti-GST (GE Healthcare), rabbit anti-Ser(P)-10 histone H3 (Upstate Biotechnology, Charlottesville, VA), mouse anti-FLAG (Campro Scientific, Berlin, Germany), rabbit anti-GFP (gift of The Netherlands Cancer Institute), mouse anti-Aurora B (BD Biosciences, Breda, The Netherlands), mouse anti-vesicular stomatitis virus (Sigma-Aldrich), rabbit anti-thiophosphate ester (Epitomics), or anti-Ser(P)-25/29 HMGN2 (Acris Antibodies, Herford, Germany). After extensive washing in TBST, the blots were incubated with horseradish peroxidase-conjugated goat anti-mouse or goat anti-rabbit antibodies (DAKO, Glostrup, Denmark) in 4% milk-TBST. Immunocomplexes were visualized using the ECL chemiluminescence detection kit from Amersham Biosciences (GE Healthcare).
FACS
The cells were cultured and transfected as described above. Thymidine (2.5 μm; Sigma) was added to block the cells in G1/S phase. The cells were released in medium containing 1 μm pacilitaxel (Sigma-Aldrich) plus DMSO or different concentrations of PP1 analogs. After 18 h, the cells were harvested and fixed in 70% ethanol. The cells were washed two times in PBS with 0.05% Tween 20 and incubated with mouse anti-MPM2 (Upstate Biotechnology) as primary antibody and goat anti-mouse Cy5 (Jackson ImmunoResearch, West Grove, PA) as secondary antibody. The cells were subsequently incubated with staining solution containing propidium iodide and RNase and analyzed by flow cytometry (FACSCalibur; BD Sciences).
Immunofluorescence Microscopy
The cells were cultured on coverslips and transfected as described above. After a thymidine block, the cells were released in medium containing 20 μm S-trityl-l-cysteine (Sigma-Aldrich) plus either DMSO or 2 μm NA-PP1 for 14 h. The coverslips were fixed in 4% PFA, followed by ice-cold methanol. After blocking in Dulbecco's PBS plus 3% BSA, coverslips were incubated overnight at 4 °C with the following antibodies: Human anti-CREST (Cortex Biochem, San Leandro CA), mouse anti-Aurora B (BD Biosciences), and rabbit anti-Survivin (R & D Systems, Abingdon, Oxon, UK). For secondary antibodies, goat anti-human Alexa-647, goat anti-mouse Alexa-488, and goat anti-rabbit Alexa-568 (all from Molecular Probes by Invitrogen) were used, and DNA was stained with 1 μg/ml 4′,6′-diamino-2-phenylindole. Images were acquired with a Zeiss LSM 510 Meta confocal microscope with 63× 1.4 N.A. objective. The images were analyzed with Image J. For HMGN2 immunofluorescence, a brief PEM/Triton 0.2% pre-extraction was performed prior to fixation with 4% PFA, and rabbit anti-HMGN2 polyclonal antibody (Acris Antibodies) was used as primary antibody.
Isolation of Thiophosphorylated Peptides
Covalent capture of thiophosphorylated substrate proteins was preformed essentially as described (14) except for the following modifications. The labeled HeLa cell lysates were denatured by adding 60% by volume solid urea, 1 m tris(2-carboxyethyl)phosphine to 10 mm and incubating at 55 °C for 1 h. Proteins were then digested by diluting the urea to 2 m by the addition of 100 mm NH4HCO3 (pH 8), adding additional tris(2-carboxyethyl)phosphine to 10 mm final, 0.5 m EDTA to 1 mm, and trypsin (Promega, Madison, WI) 1:20 by weight. The labeled lysates were digested for 16 h at 37 °C, acidified to 0.5% TFA, and desalted by using a Sep-Pak C18 column (Waters, Milford, MA) eluting into 1 ml of 50% acetonitrile, 0.1% TFA. The desalted peptides were dried by using a speed vacuum to 40 μl. The pH of the peptides was adjusted by adding 40 μl of 200 mm HEPES (pH 7.0), 75 μl acetonitrile and brought to pH 7.0 by the addition of 10% NaOH. The peptide solution was then added to 100 μl of iodoacetyl beads (Pierce) pre-equilibriated with 200 mm HEPES (pH 7.0) and incubated with end-over-end rotation at room temperature in the dark for 16 h. The beads were then added to small disposable columns, washed with H2O, 5 m NaCl, 50% acetonitrile, 5% formic acid, and 10 mm DTT, followed by elution with 100 μl and 200 μl (300 μl total) 1 mg/ml oxone (Sigma-Aldrich), desalted, and concentrated on a 10-μl ZipTip (Millipore, Billerica, MA) eluting into 60 μl of total volume. The resulting phosphopeptides were concentrated to 10 μl and analyzed by LC-MS/MS.
Mass Spectrometry
The phosphopeptides resulting from oxidation-promoted hydrolysis and concentration on a ZipTip were concentrated to 10 μl and separated using a 2–50% acetonitrile gradient on a NanoAcquity (Waters) reversed phase 75 micron capillary ultra performance liquid chromatography column on-line to an LTQ Velos Orbitrap mass spectrometer (Thermo-Fisher, Waltham MA) by both electron transfer dissociation and higher energy C-trap dissociation analysis. Peak lists were generated by using PAVA software (UCSF Release 2011) and searched by batch tag on Protein Prospector software (UCSF Version 5.3.1) for phosphopeptides with the following conditions: SwissProt.2011.07.06 (20,237 protein entries, 3,618,677 enzyme fragments) for Homo sapiens or SwissProt.2011.07.06.random.concat (40,474 protein entries, 7,224,371 enzyme fragments) for H. sapiens; precursor charge range, 2,3,4, monoisotopic masses; parent mass tolerance, 20 ppm; fragment mass tolerance, 20 ppm for higher energy C-trap dissociation and 0.5 Da for electron transfer dissociation; digest, trypsin; nonspecific, 0 termini; maximum missed cleavages, 2; constant modes, 0; expectation calculation method, linear tail fit; variable modes, acetyl (protein N-terminal), acetyl + oxidation (protein N-terminal M) Gln → pyro-Glu (N-terminal Q) Met-loss (protein N-terminal M) Met-loss + acetyl (protein N-terminal M) oxidation (M) phospho(S/T). Using the random.concat decoy database, we determined that the false discovery rate was 1% or less during most searches using the following parameters: minimum score protein, 22; minimum score peptide, 15; maximum E value protein, 0.01; maximum E value peptide, 0.05; DB peptide, variable mods, protein mods, SLIP Threshold 6 (15). Therefore we utilized search compare with the same parameters during all subsequent searches when not searching against the decoy database as we compared at least six samples, and therefore the error rate would be negligible (a multiple of the 1% false discovery rate). Phosphosites where the SLIP score is below 6 are reported as ambiguous. Phosphosites that were identified in at least two experiments in the plus INCENP/AurBL154G/H250Y samples (at least two biological replicates were performed per sample) and not in the negative controls (by extracted ion chromatogram) were reported as putative Aurora B substrates.
Viewing Annotated Spectra of Results
Accompanying this document as supplemental information are two zipped peak list files and one results file (tab-delimited text files). These files allow all of the search results to be freely viewed using a spectral viewer; e.g. MS-Viewer (part of the public Protein Prospector website). This software is already on the public website found at this address: http://prospector.ucsf.edu/prospector/cgi-bin/msform.cgi?form=msviewer. The results can be viewed by uploading peak list and results files accompanying this document. Alternatively, the results can be accessed using the links below (where we have already uploaded the results and set up the display parameters).
Aurora B-as_ETD: http://prospector2.ucsf.edu/prospector/cgi-bin/mssearch.cgi?report_title=MS-Viewer&search_key=fddk75hfbx&search_name=msviewer.
Aurora B-as_HCD: http://prospector2.ucsf.edu/prospector/cgi-bin/mssearch.cgi?report_title=MS-Viewer&search_key=kcwtffdeo6&search_name=msviewer.
RESULTS
Human Aurora B Does Not Tolerate Mutation of the Leucine 154 Gatekeeper
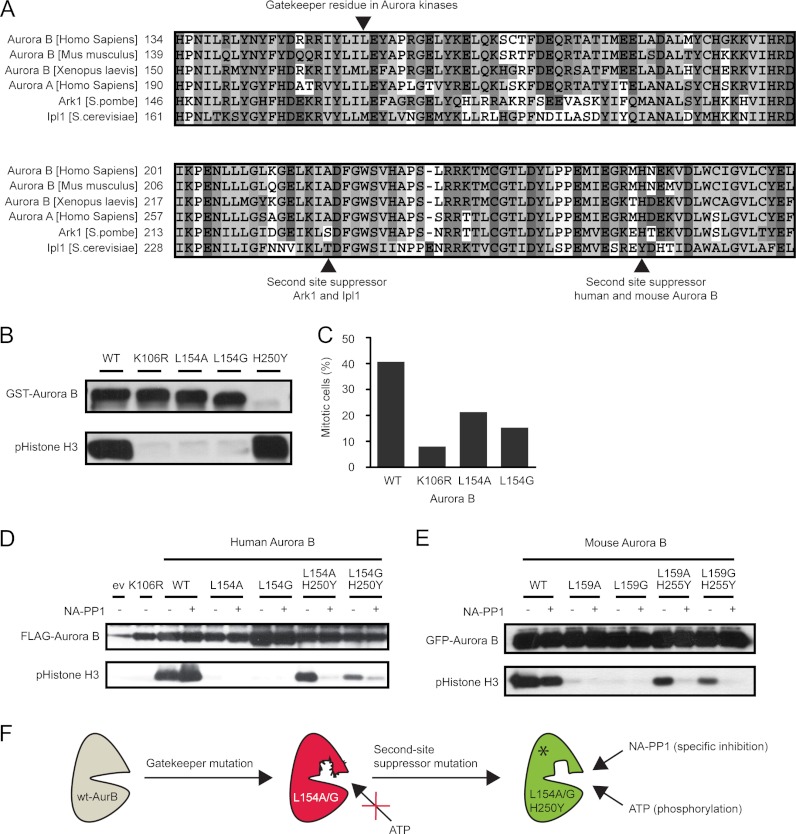
Generation of an analog-sensitive kinase requires engineering of the ATP-binding pocket by mutation of a bulky amino acid (the so-called gatekeeper) into a small amino acid (glycine or alanine), to accommodate bulky side chains of ATP analogs, making it susceptible to cell-permeable derivatives of the Src inhibitor PP1 (PP1 inhibitors) (7, 8). The gatekeeper of human Aurora B is leucine 154 (Fig. 1A) (16), and this hydrophobic residue was mutated into the smaller alanine or glycine residues to alter the shape of the ATP-binding pocket. Recombinant Aurora Bwt readily phosphorylated Ser-10 of histone H3 in vitro, but both Aurora B gatekeeper mutants failed to do so, similar to a known kinase-dead Aurora BK106R mutant (Fig. 1B). In cells, Aurora B resides in a complex with INCENP, borealin, and survivin (the chromosomal passenger complex, CPC), and the binding of Aurora B to the C-terminal IN-box of INCENP is required for full Aurora B kinase activity (17–19). To test whether the gatekeeper mutants were active when in complex with the other CPC members, FLAG-tagged Aurora BL154A and Aurora BL154G mutants were expressed in U2OS cells, immunoprecipitated from mitotic cell extracts, and the protein complexes were subjected to in vitro kinase reactions. Also under these conditions only wild-type Aurora B displayed kinase activity, whereas the kinase-dead and gatekeeper mutants were inactive (Fig. 1D). Thus, mutation of the L154 gatekeeper residue is not tolerated in human Aurora B.
Fig. 1.
The H250Y mutation rescues kinase activity of Aurora B gatekeeper mutants. A, multiple sequence alignment of the Aurora kinases in different species. The gatekeeper residue and the second site suppressor mutation found in Ark1 and Ipl1 as well as the newly identified second site suppressor mutation for human and mouse Aurora B are indicated. B, in vitro kinase assay with the indicated recombinant GST-tagged Aurora B proteins and recombinant histone H3 as substrate. Because one master mix of kinase reaction buffer plus histone H3 was used, each reaction was supplemented with the same amount of substrate. Phosphorylation of histone H3 was detected with an antibody specific for phosphoserine 10. Note that even a small amount of Aurora BH250Y results in massive phosphorylation of histone H3. C, mitotic index of paclitaxel-treated U2OS cells transfected with cDNAs encoding the indicated Aurora B mutants. All three mutants are dominant negative over the endogenous kinase and therefore reduce the mitotic delay induced by paclitaxel. D and E, FLAG-tagged human Aurora B mutants (D) and GFP-tagged mouse Aurora B mutants (E) were immunoprecipitated from mitotic U2OS cells. Kinase reactions were performed in the presence of ATP and with or without NA-PP1 (2 μm). Histone H3 was used as substrate. F, the H250Y mutation rescues kinase activity of Aurora B gatekeeper mutants. The double mutants are inhibited by NA-PP1.
Identification of a Unique Second Site Suppressor Mutation for the Human Aurora B Gatekeeper Mutants
Approximately 30% of kinases do not tolerate gatekeeper mutations, and we showed that Aurora B falls within this group. For several intolerant kinases, second site suppressor mutations were identified that could rescue kinase activity (9). For the yeast homologs of Aurora B (Ipl1 in Saccharomyces cerevisiae and Ark1 in Schizosaccharomyces pombe), such a second site suppressor mutation was identified (20, 21), and we therefore expected to find the second site suppressor residue for human Aurora B via amino acid sequence alignment. In the Ipl1-as and Ark1-as mutants, Thr-244 and Ser-229, respectively, were changed into glycine or alanine (20, 21). Surprisingly, mammalian Aurora B and Aurora A already carry an alanine at that position (Fig. 1A), indicating that mammalian Auroras require a different second site suppressor mutation. Recently four Aurora B mutations were identified in cell lines with acquired resistance for the small molecule Aurora B inhibitor ZM447439 (22). Three of these mutations were located in the catalytic active site, whereas the fourth, H250Y, was in close proximity with the activation loop and strongly enhanced the Aurora B kinase activity (22) (Fig. 1B). Interestingly, multiple sequence alignment showed that Ipl1 has a tyrosine residue at this position (Fig. 1A). To test whether the H250Y mutation could rescue kinase activity of the Aurora B gatekeeper mutants, we engineered the Aurora BL154A/H250Y and Aurora BL154G/H250Y double mutants by site-directed mutagenesis. FLAG-tagged mutant proteins were expressed in U2OS cells and immunoprecipitated from mitotic cell extracts, and kinase activity of the protein complexes against histone H3 was tested in vitro. Strikingly, the Aurora BL154A/H250Y mutant phosphorylated histone H3 similar to wild-type Aurora B, but unlike Aurora Bwt, its activity was inhibited by the bulky PP1 analog NA-PP1 (Fig. 1, D and F). The Aurora BL154G/H250Y mutant displayed lower kinase activity than the Aurora BL154A/H250Y mutant, but its activity was further inhibited by NA-PP1 (Fig. 1, D and F). This shows that the H250Y mutation acts as a second site suppressor and rescues the kinase activity of the Aurora B gatekeeper mutants (Fig. 1F). Of note, both mutants needed an intact CPC (or at least INCENP) to be active, because bacterially expressed recombinant Aurora B mutants were not active (data not shown). Importantly, we found exactly the same results for mouse Aurora B where loss of kinase activity caused by mutation of the Leu-159 gatekeeper residue into alanine was rescued by mutation of His-255 into tyrosine (Fig. 1E).
Analog-sensitive Aurora B Mutants Are Inhibited by Low Concentrations of NA-PP1 in Cells
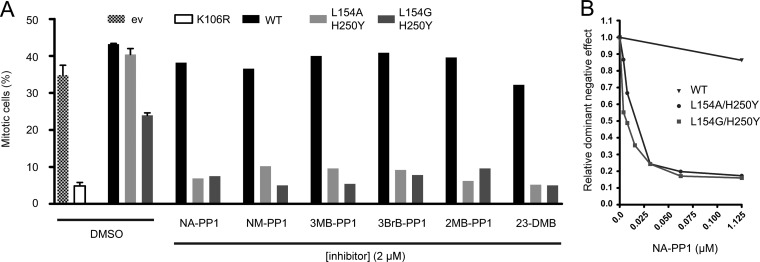
To study the function of Aurora BL154A/H250Y and Aurora BL154G/H250Y in cells, we analyzed the behavior of Aurora BL154A/H250Y and Aurora BL154G/H250Y expressing U2OS cells in paclitaxel. Paclitaxel is a microtubule-stabilizing drug that induces a mitotic delay because of the activity of the mitotic checkpoint (23). Inhibition of Aurora B in paclitaxel-treated cells compromises the mitotic checkpoint, and cells exit mitosis (24, 25). The mitotic index of cells treated with paclitaxel was therefore used as a measure of Aurora B kinase function. Aurora Bwt-expressing cells were arrested in mitosis after exposure to paclitaxel, and the addition of 2 μm NA-PP1 did not affect this mitotic arrest (Figs. 1C and 2). As expected, the mitotic index of Aurora BK106R-expressing cells was reduced to <5% (Figs. 1C and 2A), confirming that this kinase-dead mutant is dominant negative and that reduced Aurora B activity overrides the mitotic checkpoint when microtubules are stabilized by paclitaxel (11, 24, 26). Interestingly, although overexpression of the inactive Aurora BL154A and Aurora BL154G single mutants perturbed the response to paclitaxel (Fig. 1C), Aurora BL154A/H250Y-expressing cells reached a mitotic index of approximately 40%, similar to Aurora Bwt-expressing cells (Fig. 2A). However, the addition of NA-PP1 to the Aurora BL154A/H250Y expressing cells resulted in a dramatic reduction in the mitotic index, indicating that Aurora BL154A/H250Y only acts in a dominant negative fashion when inhibited by NA-PP1 (Fig. 2A). In line with its reduced activity (Fig. 1D), cells expressing Aurora BL154G/H250Y displayed a reduced mitotic index in paclitaxel (± 25%) compared with Aurora Bwt and Aurora BL154A/H250Y expressing cells. The addition of NA-PP1 further reduced the mitotic index, showing that Aurora BL154G/H250Y was also sensitive to NA-PP1 (Fig. 2). To test which PP1 inhibitor analog was the most potent inhibitor of Aurora B-as kinases, we tested six different PP1 inhibitors (NA-PP1, 1-(1,1-dimethylethyl)-3-(1-naphthalenylmethyl)-PP1, 1-(tert-butyl)-3-(3-methylbenzyl)-PP1, (1-(tert-butyl)-3-(3-bromobenzyl)-PP1, 1-(tert-butyl)-3-(2-methylbenzyl)-PP1, and (1-(tert-butyl)-3-(2,3-dimethylbenzyl)-PP1) against Aurora BL154A/H250Y- and Aurora BL154G/H250Y-expressing cells and determined the mitotic index after paclitaxel treatment (Fig. 2a and supplemental Fig. S1). All of these inhibitors inhibited the Aurora B-as mutants at a concentration of 2 μm, but titration of NA-PP1, 1-(1,1-dimethylethyl)-3-(1-naphthalenylmethyl)-PP1, and (1-(tert-butyl)-3-(2,3-dimethylbenzyl)-PP1 showed that both Aurora B-as mutants were most sensitive to NA-PP1 (supplemental Fig. S1). Further titration of this compound demonstrated that 0.06 μm of NA-PP1 was sufficient to compromise the response to paclitaxel because of efficient Aurora B kinase inhibition (Fig. 2b).
Fig. 2.
The analog-sensitive Aurora B mutants are inhibited by PP1 analogs in cells. A, U2OS cells expressing the indicated proteins were released from a thymidine-induced G1/S block into medium containing paclitaxel plus or minus the indicated PP1 analogs. Seventeen hours after release, the mitotic index was determined by propidium iodide/MPM2 monoclonal antibody labeling and FACS analysis. B, similar to A, but now different concentrations of NA-PP1 were added to the cells. For each transfected cell population, the mitotic index without PP1 inhibitor was set to 100%, and the relative reduction in mitotic index in the presence of PP1 inhibitor was determined.
Aurora B Kinase Activity Is Required to Retain CPC Localization at the Centromere
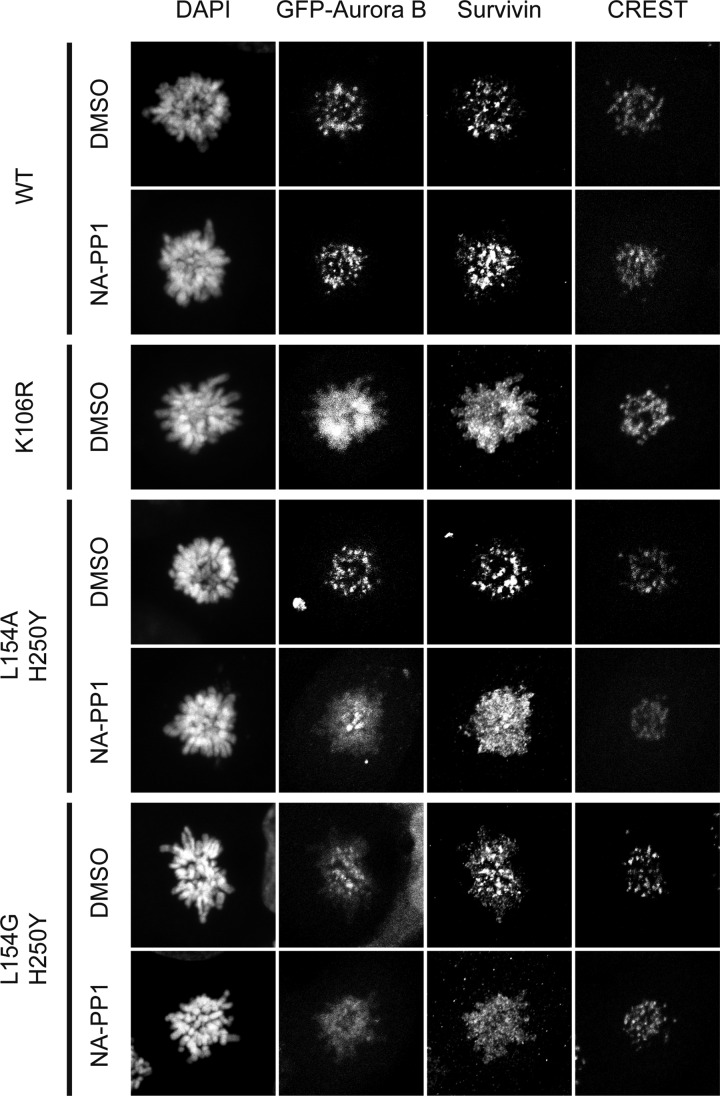
To further prove that the Aurora BL154A/H250Y and Aurora BL154G/H250Y analog-sensitive mutants behave as Aurora Bwt in the absence of cell-permeable ATP analogs, we analyzed their localization in mitosis. As expected, GFP-tagged Aurora Bwt co-localized with endogenous survivin to centromeres in prometaphase (Fig. 3) and to the central spindle in anaphase (not shown) (1). The addition of NA-PP1 did not affect this centromeric localization. The Aurora BK106R kinase-dead mutant failed to localize properly to centromeres and was displaced over the chromosomal arms (Fig. 3). This also resulted in the displacement of endogenous survivin (Fig. 3). This atypical localization of Aurora Bk106R was seen before, only after heavy overexpression of the kinase-dead protein (24). Interestingly, when we overexpressed GFP-tagged Aurora BL154A/H250Y, it localized normally to centromeres together with survivin (Fig. 3), yet when we inhibited its kinase activity with NA-PP1, both exogenous Aurora B and endogenous survivin displaced from the centromeres to the chromosomal arms (Fig. 3), confirming that Aurora B kinase activity is required for centromeric localization of CPC proteins (Aurora B and survivin) in mitosis (27). In line with this, the Aurora BL154G/H250Y mutant that is slightly less active than the L154A/H250Y mutant was partially displaced from the centromeres in the absence of NA-PP1 and completely displaced when NA-PP1 was present (Fig. 3). Based on this we conclude that centromeric retention of survivin and Aurora B requires Aurora B kinase activity.
Fig. 3.
Localization of the analog-sensitive Aurora B mutants. U2OS cells expressing the indicating GFP-tagged proteins were fixed 14 h after release from a thymidine-induced G1/S block into the Eg5 inhibitor S-trityl-l-cysteine (44) with or without NA-PP1 (2 μm). Centromeres were detected with human CREST antiserum and endogenous survivin with anti-survivin polyclonal antibody. DNA was counterstained with 4′,6′-diamino-2-phenylindole (DAPI). Note the displacement over the chromosomal arms when kinase activity is inhibited by NA-PP1.
Analog-sensitive Aurora B Mutant Can Thiophosphorylate Multiple Proteins in Cell Extracts
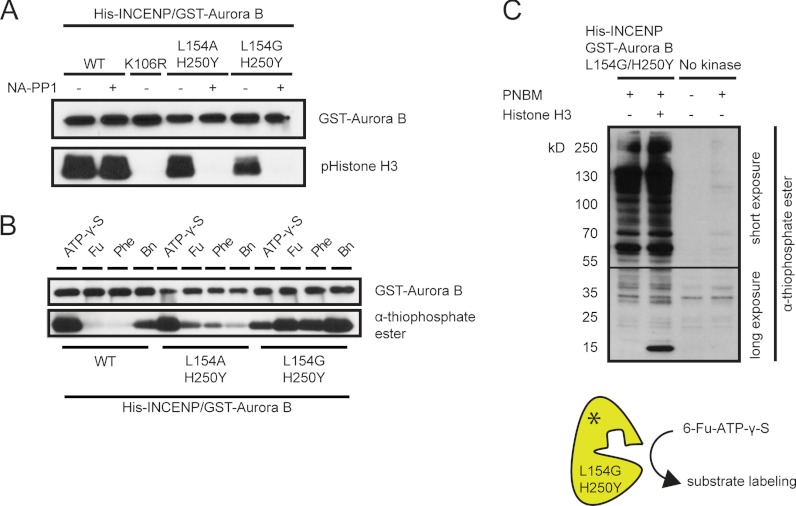
To selectively label and isolate Aurora B substrates from cell extracts, one has to use bulky ATPγS analogs to thiophosphorylate target proteins. However, to achieve this, it is crucial to obtain sufficient amounts of active recombinant kinase that can utilize these bulky ATPγS analogs. We therefore isolated recombinant Aurora B in complex with His-INCENP from insect (Sf9) cells and tested whether this recombinant protein complex phosphorylated histone H3 in vitro. Similar to our previous results with the immunoprecipitated proteins (Fig. 1D), we found that the recombinant INCENP/Aurora Bwt and INCENP/Aurora BL154A/H250Y protein complexes readily phosphorylated histone H3 in vitro, whereas phosphorylation by INCENP/Aurora BL154G/H250Y was reduced (Fig. 4A). Again, NA-PP1 only inhibited the recombinant analog-sensitive Aurora B mutants and not Aurora Bwt.
Fig. 4.
The Aurora B analog-sensitive mutant thiophosphorylates multiple proteins in cell extracts. A, in vitro kinase assay with recombinant His-INCENP/GST-tagged Aurora B, co-purified from SF9 cells, in the presence of ATP and with or without NA-PP1 (2 μm). Histone H3 was used as a substrate. B, in vitro kinase assay with recombinant His-INCENP/GST-tagged Aurora B (wild-type or analog-sensitive mutants) in the presence of ATPγS, N6-furfuryl-ATPγS (Fu), N6-phenylethyl-ATPγS (Phe), or N6-benzyl-ATPγS (Bn). Thiophosphorylated histone H3 was detected with a thiophosphate ester epitope-specific antibody (13). C, in vitro kinase assay with His-INCENP/GST-Aurora BL154G/H250Y in the presence of N6-furfuryl-ATPγS. 50 μg of a mitotic HeLa cell lysate was used as input, and histone H3 was spiked into this lysate to serve as positive control. For the lower part of the blot (containing proteins with molecular masses of 15–50 kDa), a longer exposure (5 min instead of 15 s for the short exposure) is shown.
Remarkably, however, when we tested three different bulky ATPγS analogs, we found that INCENP/Aurora BL154G/H250Y, but not INCENP/Aurora BL154A/H250Y and INCENP/Aurora Bwt efficiently thiophosphorylated histone H3 in the presence of all three bulky ATPγS analogs (Fig. 4B). Because INCENP/Aurora Bwt showed some reactivity with N6-benzyl-ATPγS and thiophosphorylation of histone H3 by INCENP/Aurora BL154G/H250Y was more efficient with N6-furfuryl-ATPγS than with N6-phenylethyl-ATPγS (Fig. 4B), we choose N6-furfuryl-ATPγS to perform a kinase reaction in a whole cell extract prepared from mitotic HeLa cells. As shown in Fig. 4C, INCENP/Aurora BL154G/H250Y could still thiophosphorylate recombinant histone H3 in the presence of 50 μg of total cell extract and more importantly, we detected multiple thiophosphorylated proteins in the extract in which recombinant His-INCENP/Aurora BL154G/H250Y was present that were not present in the negative control, indicating that these proteins were thiophosphorylated by INCENP/Aurora BL154G/H250Y.
Identification of Aurora Substrates and Their Phosphorylation Sites in Cell Extracts
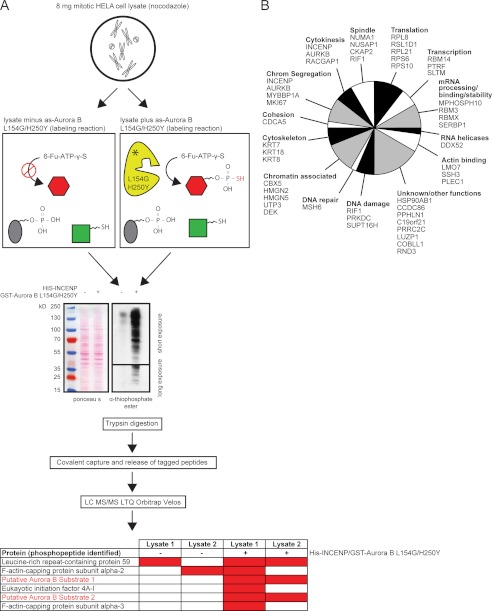
Next, we performed two independent kinase reactions with 4 μg of recombinant His-INCENP/Aurora BL154G/H250Y complex and 4 mg of mitotic cell extract or with 4 mg of mitotic cell extract without the addition of the recombinant protein complex (negative control) (Fig. 5A). Thiophosphorylated peptides were isolated according to the covalent capture and release methodology described by Blethrow et al. (10) and Hertz et al. (14), and peptides were analyzed by mass spectrometry. The phosphopeptides were then compared between the samples, and only peptides that were never identified in the negative control samples but were present with INCENP/Aurora BL154G/H250Y were predicted to be putative Aurora B substrates (Fig. 5A). In total 114 phosphorylated peptides were identified in the plus His-INCENP/Aurora BL154G/H250Y samples, revealing 92 unique phosphosites in a total of 58 proteins (Table I and supplemental Table S1). When we only considered the phosphosites with a SLIP score threshold of 6 or singly present in the peptide (15) (n = 70; Table I and supplemental Table S1), we found in 57 of the 70 phosphosites (81%) an arginine at position −2, in line with the previously predicted Aurora consensus motif (K/R)X(S/T)(I/L/V) (28) (where X represents any residue) and in line with a recent peptide-library screen by Alexander et al. (29) (Table I and supplemental Table S2). Similar to that work, we also found a preference for hydrophobic residues in the +1 (36/57 sites) or +2 position (14/57 sites) (Table I and supplemental Table S2). Moreover, in line with the published finding that a proline at position +1 is not tolerated by Aurora kinases (29), we did not find peptides with an arginine at −2 and a proline at +1. In the 13 non-RX(S/T) sites, we found in two cases an arginine or a lysine at −1 and in three cases an arginine at −3, in each case combined with an hydrophobic residue in position +1 or +2, a situation that may also be considered as Aurora-specific (30). For five of the eight remaining non-RX(S/T) sites, an adjacent RX(S/T) motif was not present in the purified peptide, and these sites could thus represent novel unexpected Aurora sites (supplemental Table S2). However, given the fact that one of these sites is a validated Cdk1 site in INCENP (Thr-59) (31, 32), we deem this possibility unlikely.
Fig. 5.
Identification of putative Aurora B substrates by covalent capture and release of thiolabeled peptides. A, set-up of Aurora B substrate screen using the covalent capture-and-release method. B, potential Aurora B substrates containing the RX(S/T) consensus motif (including ambiguous sites). Substrates were only listed if thiophosphorylated peptides were found in two independent experiments.
Table I. Overview of the 92 thiophosphorylated sites found in all 114 peptides (peptides were found in one or two experiments) corresponding to 58 proteins.
| Number of phosphosites | Site |
||
|---|---|---|---|
| SLIP score of ≥6 or singly present in peptide | Ambiguous | ||
| Total | 92 | 70 | 22 |
| RX(S/T)Φ | 43 | 36 | 7 |
| RX(S/T)XΦ | 18 | 14 | 4 |
| RX(S/T)XX | 7 | 7 | 0 |
| Non-RX(S/T) | 24 | 13 | 11 |
Importantly, whereas Thr-232 of Aurora B was most likely missed because of the nearby cysteine (Cys-235) that can form a thioether linkage with the iodoacetyl-agarose beads and is therefore not liberated by oxidation-promoted hydrolysis (10), within the RX(S/T) group we found back a number of known Aurora B substrates and sites, such as INCENP Ser-893; Ser-894 (18, 19), Myb-binding protein 1A/MYBBP1A Ser-1303 (33); and vimentin/VIM Ser-73 (34) (supplemental Table S2). Moreover, we here confirm that the novel site in INCENP (Ser-72) is indeed an Aurora site. A recombinant protein of the N terminus of INCENP (amino acids 1–80) was generated and used as substrate in an in vitro kinase reaction with the recombinant INCENP/Aurora Bwt complex. Indeed, INCENP 1–80 was readily phosphorylated in vitro, but its phosphorylation was significantly reduced when Ser-72 was mutated into alanine (Fig. 6A). Overall, the thiophosphate labeling approach revealed candidate Aurora B substrates involved in a wide range of cellular functions such as chromosome segregation, cytokinesis, spindle formation, chromatin remodeling, DNA repair, the DNA damage response, mRNA processing, and transcription and translation (Fig. 5B). Moreover, we identified at least 20 potential Aurora B substrates that were not found in a recent screen that combined quantitative phosphoproteomics with Aurora kinase inhibition using small molecules (30) (Table II).
Fig. 6.
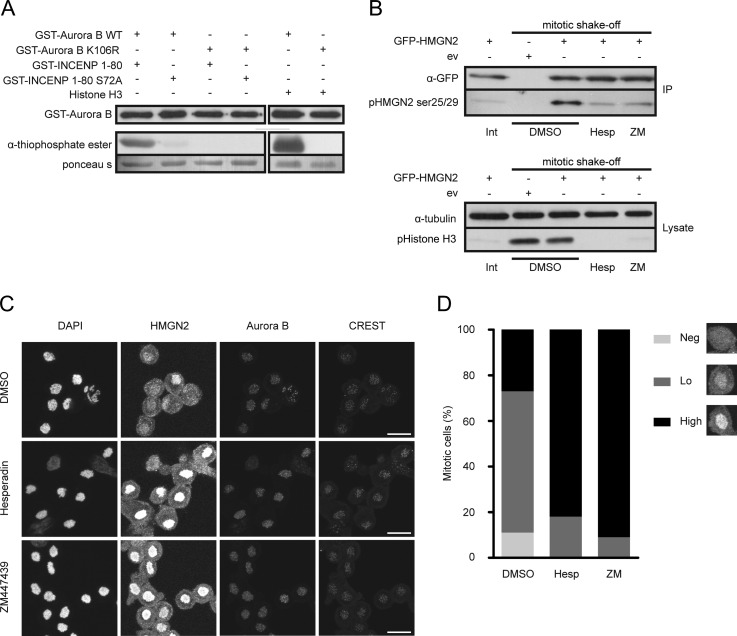
Validation of potential novel Aurora B phosphorylation sites and substrate. A, in vitro kinase assay with recombinant His-INCENP/GST-tagged Aurora B (wild-type and kinase-dead), in the presence of ATPγS. Recombinant GST-INCENP 1–80, GST-INCENP 1–80S72A, or histone H3 was added as substrate. B, U2OS cells were transfected with either empty GFP vector (ev) or a plasmid encoding GFP-HMGN2. Mitotic cells were collected by mitotic shake-off, and the remaining adherent cells were used as interphase cell input. The overexpressed proteins were immunoprecipitated from the interphase or mitotic cell lysates with an anti-GFP antibody. The precipitated proteins were separated by SDS-PAGE, and Western blots were probed with an antibody specific for phosphorylated Ser-25/Ser-29 in HMGN2 (middle panel) and subsequently reprobed with an anti-GFP antibody (upper panel). Western blots of whole cell extracts were probed with an antibody specific for phosphorylated histone H3 (Ser-10, lower panel). C and D, U2OS cells were released into nocodazole in the presence or absence of the indicated Aurora B inhibitors. HMGN2, Aurora B, and centromeres (CREST) were visualized with specific antibodies. Images of fields of mitotic cells were captured by a Zeiss LSM microscope (C), and the fraction of mitotic cells with no, low, or high levels of HMGN2 on mitotic chromosomes was quantified (D). For each condition >100 cells were counted. Scale bar = 20 micrometer.
Table II. Aurora B substrates unique to this screen.
The criteria for inclusion include phosphopeptide(s) found in two independent experiments, RX(S/T) motif, E-score ≤ 10E-4, SLIP score ≥ 6, and substrate not found by Kettenbach et al. (30).
| Name | Full name | Phosphosite | Site found in other phosphoproteome database? (www.phosphosite.org) | Reported function |
|---|---|---|---|---|
| DEK | DEK | Thr-67 | No | Chromatin associated |
| UTP3 | Something about silencing protein 10 | Ser-462 | No | Chromatin associated |
| HMGN2 | Non-histone chromosomal protein HMG-17 | Ser-29 | Yes | Chromatin associated |
| HMGN5 | High mobility group nucleosome-binding domain-containing protein 5 | Ser-20, Ser-24 | Yes | Chromatin associated |
| KRT8 | Keratin, type II cytoskeletal 8 | Thr-6, Ser-34 | No/Yes | Cytoskeleton |
| KRT7 | Keratin, type II cytoskeletal 7 | Ser-27 | No | Cytoskeleton |
| RIF1 | Telomere-associated protein RIF1 | Ser-2205 | Yes | Spindle/DNA damage |
| PRKDC | DNA-dependent protein kinase catalytic subunit | Ser-511 | Yes | DNA damage |
| RPS10 | 40 S ribosomal protein S10 | Thr-118 | No | Translation |
| RPL21 | 60 S ribosomal protein L21 | Ser-104 | Yes | Translation |
| RPL8 | 60 S ribosomal protein L8 | Ser-130 | No | Translation |
| PTRF | Polymerase I and transcript release factor | Ser-300 | Yes | Transcription |
| SSH3 | Protein phosphatase Slingshot homolog 3 | Ser-37 | Yes | Actin binding |
| RACGAP1 | Rac GTPase-activating protein 1 | Thr-249 | Yes | Cytokinesis |
| MPHOSPH10 | M phase phosphoprotein 10 | Thr-332 | No | mRNA processing |
| HSP90AB1 | Heat shock protein HSP 90-β | Ser-452 | Yes | Unknown |
| COBLL1 | Cordon-bleu protein-like 1 | Ser-955 | No | Unknown |
| PPHLN1 | Periphilin-1 | Ser-110 | Yes | Unknown |
| PRRC2C/BAT2D1 | BAT2 domain-containing protein 1 | Ser-1013 | No | Unknown |
| C19orf21 | Uncharacterized protein C19orf21 | Ser-348 | No | Unknown |
HMGN2 Is a Mitotic Substrate of Aurora B
To further validate our approach, we next asked whether the high mobility group nucleosomal binding protein 2 (HMGN2) we identified as a potential Aurora B substrate (Table II and Fig. 5B) could be validated as a bona fide in vivo substrate of the kinase. Because HMGN2 is highly phosphorylated in mitosis (35), we tested whether Aurora B kinase activity was responsible for its mitotic phosphorylation. Mitotic HeLa cells were treated with two different small molecule inhibitors (hesperadin and ZM447439), both shown to inhibit Aurora B significantly better than Aurora A (24, 25), and GFP-tagged HMGN2 was immunoprecipitated from these extracts. The mitotic phosphorylation of Ser-25/Ser-29 in HMGN2 was clearly inhibited by hesperadin and ZM447439, as was the phosphorylation of histone H3 at Ser-10, a well known substrate of Aurora B (Fig. 6B). Interestingly, HMGN2 binds to chromatin in interphase, but the protein dissociates from chromatin when cells enter mitosis (supplemental Fig. S2) (35). Both Ser-25 and Ser-29 lie within the nucleosomal binding domain of HMGN2 and an HMGN2 S25E/S29E mutant no longer binds to nucleosomes in vitro (36). We therefore tested whether the mitotic dissociation of HMGN2 was mediated by Aurora B. Indeed when we let cells enter into mitosis in the presence of hesperadin or ZM447439, we observed a dramatic increase in the number of mitotic cells with enhanced chromosomal localization of HMGN2 (Fig. 6, C and D, and supplemental Fig. S2). In line with this, HMGN2 appears to reassociate with chromatin in anaphase when Aurora B translocates from the centromeres to the central spindle (supplemental Fig. S2). We have thus identified HMGN2 as a novel mitotic target of Aurora B whose dynamic chromatin association is under control of the kinase.
DISCUSSION
We successfully generated an important new research tool for the functional analysis of human Aurora B, a kinase essential for genomic stability. In contrast to a recent study that identified L138V in Aurora B as a second site suppressor mutation based on modeling using the published crystal structure of Aurora A bound to ADP (37), the here described H250Y mutation that restored activity of the Aurora B gatekeeper mutants (L154A/G) was found by combining orthologous Aurora sequence alignment and reported information on Aurora B mutations that enhanced kinase activity (38). This could thus be considered as an alternative strategy to find second site suppressor mutations for kinases intolerant to gatekeeper mutations.
The unique analog-sensitive mutant of Aurora B is functional in cells and inhibited by low concentrations of the PP1 analog NA-PP1, opening up the possibility to stably replace endogenous Aurora B kinase for this mutant and generate cell lines in which Aurora B can be selectively inhibited. Moreover, the Aurora BL154G/H250Y double mutant is capable of using bulky ATP analogs to thiophosphorylate proteins in cell extracts. This resulted in the isolation of multiple unique phosphopeptides of which 71% corresponded to either a RX(S/T)Φ or a RX(S/T)XΦ motif (where Φ stands for a hydrophobic residue).
Thus far we identified only highly abundant proteins present in a nonfractionated cell extract as potential Aurora B substrates, but we expect to find more (less abundant) Aurora B substrates when using fractionated cell extracts, enriched for certain subcellular compartments, as input for the kinase reactions (10). Based on the recent screen by Kettenbach et al. (30) that combined quantitative phosphoproteomics with Aurora kinase inhibition using small molecules, our candidates represent only a small portion of the Aurora B substrates. However, the use of chemical inhibitors may affect multiple downstream pathways and nonspecific pathways because of off target kinase inhibition, whereas our screen will identify only direct Aurora B substrates. Both screens may thus be nicely complementary in finding relevant Aurora B targets. Indeed, we identified at least 20 potential Aurora B substrates that were not uncovered by the Kettenbach screen (30) (Table II). Importantly, we found RACGAP1, which is a validated substrate of Aurora B (34, 39), and for HMGN2, we demonstrated in this study that it is a novel mitotic target of Aurora B. Moreover, 22 of the 40 recovered RX(S/T) Aurora phosphosites were also found in other phosphoproteomic data sets (www.phosphosite.org) (40), indicating that the majority of these sites are also phosphorylated in vivo.
Advantages of the chemical genetic approach are that only direct targets of the kinase are labeled and that we can use different functionally relevant subcellular fractions as input, such as purified chromosome fractions and isolated midbodies, to enable more comprehensive coverage of less abundant Aurora substrates. However, the challenge will be to verify whether the identified sites are also in vivo phosphorylated and to establish whether the sites are indeed specific for Aurora B. Because the recombinant kinase is no longer confined to its normal cellular locations and Aurora B and A tend to phosphorylate overlapping peptide sequences (29), it is expected that both Aurora B and A substrates will be found. Indeed, 5 of the 17 phospho-sites that overlapped with the screen by Kettenbach et al. (30) were clustered as Aurora A specific sites in that study, whereas 8 of 17 were clustered as Aurora B specific and 4 of 20 were clustered as Aurora ambiguous (the latter meaning that the site is mostly likely phosphorylated by both Aurora A and B). Thus, combining the advantages of different substrate screens together with functional validation experiments, will build a complete picture of the physiologically relevant Aurora B substrates.
To prove that the in vitro thiophosphate labeling approach could indeed identify novel in vivo Aurora B substrates, we selected HMGN2 as a candidate for further validation. HMGN2 belongs to a group of non-histone chromosomal proteins (HMGN1–5) that bind to nucleosomes and modulate the structure and function of chromatin. HMGN2 reduces the compaction of chromatin fibers most likely to facilitate gene expression (41) and promotes repair of UV-induced DNA lesions (42). HMGN2 is highly phosphorylated in mitosis, and this phosphorylation coincides with the dissociation of the protein from mitotic chromosomes (35). We showed that Aurora B kinase is responsible for its mitotic phosphorylation and dissociation from chromosomes. Interestingly, the Aurora B phosphorylation sites (Ser-25 and Ser-29) lie within the RRSARLSA core of the nucleosomal binding domain that is conserved between all the members of this protein family (41), and we therefore predict that all HMGN proteins will be Aurora B targets. Indeed, Ser-20 and Ser-24 of HMGN5 were also identified as potential Aurora sites in our screen (Table I, Fig. 5B, and supplemental Table S1), and similar to HMGN2 also HMGN5 dissociates from mitotic chromosomes (43). Currently, we do not know why the HMGN proteins have to dissociate from chromatin when cells enter mitosis. Because they reduce chromatin compaction, their mitotic dissociation might contribute to full chromatin condensation in mitosis promoting proper chromosome segregation in anaphase.
In conclusion, we have generated a powerful new tool to inhibit Aurora B kinase activity and to identify downstream targets of the kinase. The fact that we uncovered candidate substrates involved in a wide range of cellular functions could indicate that Aurora B may have multiple functions outside mitosis or that the putative substrates have additional functions during cell division.
Acknowledgments
We thank Prof. R. H. Medema for critical reading of the manuscript.
Footnotes
* This work was supported by National Institutes of Health Grants NCRR RR015804 and NCRR RR001614; by the Howard Hughes Medical Institute; by Dutch Cancer Society Grant UU 2009-4311 and Netherlands Organization for Scientific Research Grant Vidi 917.66.332 (to S. M. A. L., R. C. C. H., and M. J. M. V.); a predoctoral fellowship from Genentech and a Rapid Response Innovation Award from the MJFF (to N. T. H.); and National Institutes of Health Grant R01EB001987 and additional funds form the Howard Hughes Medical Institute (to N. T. H. and K. M. S.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- as
- analog sensitive
- CPC
- chromosomal passenger complex
- HMGN
- high mobility group nucleosome-binding domain-containing protein
- INCENP
- inner centromere protein
- NA-PP1
- 1-(1,1-dimethylethyl)-3-(1-naphthalenyl)-PP1
- PP1
- 1H-pyrazolo[3,4-d]pyrimidin-4-amine
- SLIP
- site location in peptide.
REFERENCES
- 1. Ruchaud S., Carmena M., Earnshaw W. C. (2007) Chromosomal passengers: Conducting cell division. Nat. Rev. Mol. Cell Biol. 8, 798–812 [DOI] [PubMed] [Google Scholar]
- 2. Monaco L., Kolthur-Seetharam U., Loury R., Murcia J. M., de Murcia G., Sassone-Corsi P. (2005) Inhibition of Aurora-B kinase activity by poly(ADP-ribosyl)ation in response to DNA damage. Proc. Natl. Acad. Sci. U.S.A. 102, 14244–14248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sabbattini P., Canzonetta C., Sjoberg M., Nikic S., Georgiou A., Kemball-Cook G., Auner H. W., Dillon N. (2007) A novel role for the Aurora B kinase in epigenetic marking of silent chromatin in differentiated postmitotic cells. EMBO J. 26, 4657–4669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Weiss W. A., Taylor S. S., Shokat K. M. (2007) Recognizing and exploiting differences between RNAi and small-molecule inhibitors. Nat. Chem. Biol. 3, 739–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lens S. M., Voest E. E., Medema R. H. (2010) Shared and separate functions of polo-like kinases and Aurora kinases in cancer. Nat. Rev. Cancer 10, 825–841 [DOI] [PubMed] [Google Scholar]
- 6. Shah K., Liu Y., Deirmengian C., Shokat K. M. (1997) Engineering unnatural nucleotide specificity for Rous sarcoma virus tyrosine kinase to uniquely label its direct substrates. Proc. Natl. Acad. Sci. U.S.A. 94, 3565–3570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bishop A. C., Ubersax J. A., Petsch D. T., Matheos D. P., Gray N. S., Blethrow J., Shimizu E., Tsien J. Z., Schultz P. G., Rose M. D., Wood J. L., Morgan D. O., Shokat K. M. (2000) A chemical switch for inhibitor-sensitive alleles of any protein kinase. Nature 407, 395–401 [DOI] [PubMed] [Google Scholar]
- 8. Bishop A., Buzko O., Heyeck-Dumas S., Jung I., Kraybill B., Liu Y., Shah K., Ulrich S., Witucki L., Yang F., Zhang C., Shokat K. M. (2000) Unnatural ligands for engineered proteins: New tools for chemical genetics. Annu. Rev. Biophys. Biomol. Struct. 29, 577–606 [DOI] [PubMed] [Google Scholar]
- 9. Zhang C., Kenski D. M., Paulson J. L., Bonshtien A., Sessa G., Cross J. V., Templeton D. J., Shokat K. M. (2005) A second-site suppressor strategy for chemical genetic analysis of diverse protein kinases. Nat. Methods 2, 435–441 [DOI] [PubMed] [Google Scholar]
- 10. Blethrow J. D., Glavy J. S., Morgan D. O., Shokat K. M. (2008) Covalent capture of kinase-specific phosphopeptides reveals Cdk1-cyclin B substrates. Proc. Natl. Acad. Sci. U.S.A. 105, 1442–1447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lens S. M., Wolthuis R. M., Klompmaker R., Kauw J., Agami R., Brummelkamp T., Kops G., Medema R. H. (2003) Survivin is required for a sustained spindle checkpoint arrest in response to lack of tension. EMBO J. 22, 2934–2947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Klein U. R., Nigg E. A., Gruneberg U. (2006) Centromere targeting of the chromosomal passenger complex requires a ternary subcomplex of Borealin, Survivin, and the N-terminal domain of INCENP. Mol. Biol. Cell 17, 2547–2558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Allen J. J., Li M., Brinkworth C. S., Paulson J. L., Wang D., Hübner A., Chou W. H., Davis R. J., Burlingame A. L., Messing R. O., Katayama C. D., Hedrick S. M., Shokat K. M. (2007) A semisynthetic epitope for kinase substrates. Nat. Methods 4, 511–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hertz T. N., Wang B. T., Allen J. J., Zhang C., Dar A. C., Burlingame A. L., Shokat K. M. (2010) Chemical genetic approach for kinase-substrate mapping by covalent capture of thiophosphopeptides and analysis by Mass Spectrometry, in Current Protocols in Chemical Biology, pp. 15–36, John Wiley and Sons, New York: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Baker P. R., Trinidad J. C., Chalkley R. J. (2011) Modification site localization scoring integrated into a search engine. Mol. Cell. Proteomics 10.1074/mcp.M111.008078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Blethrow J., Zhang C., Shokat K. M., Weiss E. L. (2004) Design and use of analog-sensitive protein kinases. Curr. Protoc. Mol. Biol., Chapter 18, Unit 18.11, John Wiley and Sons, New York: [DOI] [PubMed] [Google Scholar]
- 17. Bishop J. D., Schumacher J. M. (2002) Phosphorylation of the carboxyl terminus of inner centromere protein (INCENP) by the Aurora B kinase stimulates Aurora B kinase activity. J. Biol. Chem. 277, 27577–27580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Honda R., Körner R., Nigg E. A. (2003) Exploring the functional interactions between Aurora B, INCENP, and survivin in mitosis. Mol. Biol. Cell 14, 3325–3341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sessa F., Mapelli M., Ciferri C., Tarricone C., Areces L. B., Schneider T. R., Stukenberg P. T., Musacchio A. (2005) Mechanism of Aurora B activation by INCENP and inhibition by hesperadin. Mol. Cell 18, 379–391 [DOI] [PubMed] [Google Scholar]
- 20. Pinsky B. A., Kung C., Shokat K. M., Biggins S. (2006) The Ipl1-Aurora protein kinase activates the spindle checkpoint by creating unattached kinetochores. Nat. Cell Biol. 8, 78–83 [DOI] [PubMed] [Google Scholar]
- 21. Hauf S., Biswas A., Langegger M., Kawashima S. A., Tsukahara T., Watanabe Y. (2007) Aurora controls sister kinetochore mono-orientation and homolog bi-orientation in meiosis-I. EMBO J. 26, 4475–4486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Girdler F., Sessa F., Patercoli S., Villa F., Musacchio A., Taylor S. (2008) Molecular basis of drug resistance in Aurora kinases. Chem. Biol. 15, 552–562 [DOI] [PubMed] [Google Scholar]
- 23. Jordan M. A., Wilson L. (2004) Microtubules as a target for anticancer drugs. Nat. Rev. Cancer 4, 253–265 [DOI] [PubMed] [Google Scholar]
- 24. Ditchfield C., Johnson V. L., Tighe A., Ellston R., Haworth C., Johnson T., Mortlock A., Keen N., Taylor S. S. (2003) Aurora B couples chromosome alignment with anaphase by targeting BubR1, Mad2, and Cenp-E to kinetochores. J. Cell Biol. 161, 267–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hauf S., Cole R. W., LaTerra S., Zimmer C., Schnapp G., Walter R., Heckel A., van Meel J., Rieder C. L., Peters J. M. (2003) The small molecule hesperadin reveals a role for Aurora B in correcting kinetochore-microtubule attachment and in maintaining the spindle assembly checkpoint. J. Cell Biol. 161, 281–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Murata-Hori M., Wang Y. L. (2002) The kinase activity of Aurora B is required for kinetochore-microtubule interactions during mitosis. Curr. Biol. 12, 894–899 [DOI] [PubMed] [Google Scholar]
- 27. Wang F., Ulyanova N. P., van der Waal M. S., Patnaik D., Lens S. M., Higgins J. M. (2011) A positive feedback loop involving Haspin and Aurora B promotes CPC accumulation at centromeres in mitosis. Curr. Biol. 21, 1061–1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cheeseman I. M., Anderson S., Jwa M., Green E. M., Kang J., Yates J. R., 3rd, Chan C. S., Drubin D. G., Barnes G. (2002) Phospho-regulation of kinetochore-microtubule attachments by the Aurora kinase Ipl1p. Cell 111, 163–172 [DOI] [PubMed] [Google Scholar]
- 29. Alexander J., Lim D., Joughin B. A., Hegemann B., Hutchins J. R., Ehrenberger T., Ivins F., Sessa F., Hudecz O., Nigg E. A., Fry A. M., Musacchio A., Stukenberg P. T., Mechtler K., Peters J. M., Smerdon S. J., Yaffe M. B. (2011) Spatial exclusivity combined with positive and negative selection of phosphorylation motifs is the basis for context-dependent mitotic signaling. Sci. Signal. 4, ra42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kettenbach A. N., Schweppe D. K., Faherty B. K., Pechenick D., Pletnev A. A., Gerber S. A. (2011) Quantitative phosphoproteomics identifies substrates and functional modules of Aurora and polo-like kinase activities in mitotic cells. Sci. Signal. 4, rs5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Goto H., Kiyono T., Tomono Y., Kawajiri A., Urano T., Furukawa K., Nigg E. A., Inagaki M. (2006) Complex formation of Plk1 and INCENP required for metaphase-anaphase transition. Nat. Cell Biol. 8, 180–187 [DOI] [PubMed] [Google Scholar]
- 32. Hümmer S., Mayer T. U. (2009) Cdk1 negatively regulates midzone localization of the mitotic kinesin Mklp2 and the chromosomal passenger complex. Curr. Biol. 19, 607–612 [DOI] [PubMed] [Google Scholar]
- 33. Perrera C., Colombo R., Valsasina B., Carpinelli P., Troiani S., Modugno M., Gianellini L., Cappella P., Isacchi A., Moll J., Rusconi L. (2010) Identification of Myb-binding protein 1A (MYBBP1A) as a novel substrate for Aurora B kinase. J. Biol. Chem. 285, 11775–11785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Goto H., Yasui Y., Kawajiri A., Nigg E. A., Terada Y., Tatsuka M., Nagata K., Inagaki M. (2003) Aurora-B regulates the cleavage furrow-specific vimentin phosphorylation in the cytokinetic process. J. Biol. Chem. 278, 8526–8530 [DOI] [PubMed] [Google Scholar]
- 35. Cherukuri S., Hock R., Ueda T., Catez F., Rochman M., Bustin M. (2008) Cell cycle-dependent binding of HMGN proteins to chromatin. Mol. Biol. Cell 19, 1816–1824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ueda T., Catez F., Gerlitz G., Bustin M. (2008) Delineation of the protein module that anchors HMGN proteins to nucleosomes in the chromatin of living cells. Mol. Cell. Biol. 28, 2872–2883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Johnson E. O., Chang K. H., de Pablo Y., Ghosh S., Mehta R., Badve S., Shah K. (2011) PHLDA1 is a crucial negative regulator and effector of Aurora A kinase in breast cancer. J. Cell Sci. 124, 2711–2722 [DOI] [PubMed] [Google Scholar]
- 38. Girdler F., Gascoigne K. E., Eyers P. A., Hartmuth S., Crafter C., Foote K. M., Keen N. J., Taylor S. S. (2006) Validating Aurora B as an anti-cancer drug target. J. Cell Sci. 119, 3664–3675 [DOI] [PubMed] [Google Scholar]
- 39. Minoshima Y., Kawashima T., Hirose K., Tonozuka Y., Kawajiri A., Bao Y. C., Deng X., Tatsuka M., Narumiya S., May W. S., Jr., Nosaka T., Semba K., Inoue T., Satoh T., Inagaki M., Kitamura T. (2003) Phosphorylation by Aurora B converts MgcRacGAP to a RhoGAP during cytokinesis. Dev. Cell 4, 549–560 [DOI] [PubMed] [Google Scholar]
- 40. Hornbeck P. V., Chabra I., Kornhauser J. M., Skrzypek E., Zhang B. (2004) PhosphoSite: A bioinformatics resource dedicated to physiological protein phosphorylation. Proteomics 4, 1551–1561 [DOI] [PubMed] [Google Scholar]
- 41. Postnikov Y., Bustin M. (2010) Regulation of chromatin structure and function by HMGN proteins. Biochim. Biophys. Acta 1799, 62–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Subramanian M., Gonzalez R. W., Patil H., Ueda T., Lim J. H., Kraemer K. H., Bustin M., Bergel M. (2009) The nucleosome-binding protein HMGN2 modulates global genome repair. FEBS J. 276, 6646–6657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Rochman M., Postnikov Y., Correll S., Malicet C., Wincovitch S., Karpova T. S., McNally J. G., Wu X., Bubunenko N. A., Grigoryev S., Bustin M. (2009) The interaction of NSBP1/HMGN5 with nucleosomes in euchromatin counteracts linker histone-mediated chromatin compaction and modulates transcription. Mol. Cell 35, 642–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Debonis S., Skoufias D. A., Indorato R. L., Liger F., Marquet B., Laggner C., Joseph B., Kozielski F. (2008) Structure-activity relationship of S-trityl-l-cysteine analogues as inhibitors of the human mitotic kinesin Eg5. J. Med. Chem. 51, 1115–1125 [DOI] [PubMed] [Google Scholar]