Abstract
Protein phosphorylation-dephosphorylation events play a primary role in regulation of almost all aspects of cell function including signal transduction, cell cycle, or apoptosis. Thus far, T cell phosphoproteomics have focused on analysis of phosphotyrosine residues, and little is known about the role of serine/threonine phosphorylation in early activation of the T cell receptor (TCR). Therefore, we performed a quantitative mass spectrometry-based analysis of the global phosphoproteome of human primary T cells in response to 5 min of TCR activation with anti-CD3 antibody. Combining immunoprecipitation with an antiphosphotyrosine antibody, titanium dioxide phosphopeptide enrichment, isobaric tag for the relative and absolute quantitation methodology, and strong cation exchange separation, we were able to identify 2814 phosphopeptides. These unique sites were employed to investigate the site-specific phosphorylation dynamics. Five hundred and seventeen phosphorylation sites showed TCR-responsive changes. We found that upon 5 min of stimulation of the TCR, specific serine and threonine kinase motifs are overrepresented in the set of responsive phosphorylation sites. These phosphorylation events targeted proteins with many different activities and are present in different subcellular locations. Many of these proteins are involved in intracellular signaling cascades related mainly to cytoskeletal reorganization and regulation of small GTPase-mediated signal transduction, probably involved in the formation of the immune synapse.
T lymphocytes are able to recognize specific antigenic peptides presented by molecules of the major histocompatibility complex on the surface of other cell types. This interaction is mediated by a dimeric specialized molecule called T cell receptor (TCR),1 which is part of a larger membrane complex in association with CD3 γ, δ, ε, and ζ chains. The binding between TCR and the major histocompatibility complex-antigen is of relatively low affinity, and it is stabilized by the association with co-receptors (CD4 or CD8). All of these molecules in turn recruit, via their intracellular domains, different polypeptides to carry out signal transduction. In addition to antigen recognition, coactivation by CD28 is required to trigger full activation of the T cell, which expresses then different cell surface molecules and releases soluble mediators (cytokines) that promote changes in the activity of different target cell types (1).
During the TCR-major histocompatibility complex-antigen recognition, T cells undergo considerable membrane and cytoskeletal rearrangements that lead to the formation of the immunological synapse (IS). During this maturation, precise molecular reorganizations occur at the interface between T cells and an antigen presenting cell. Cell motility, polarization, and receptor relocalization events are dependent on the lymphocyte cytoskeleton and are necessary for the maturation of the IS. TCR, co-receptors, intracellular signaling molecules, and adhesion receptors polarize to the IS and form small aggregates known as microclusters (2, 3), processes all dependent on functional microtubule and actin cytoskeleton. This results in the stabilization and functional maturation of the signaling complexes.
Protein phosphorylation is a major regulatory process in most intracellular signaling pathways (4). Signal transduction from the TCR is known to be dependent on the initial steps of several cytosolic tyrosine kinases (Lck, Fyn, and ZAP-70) and membrane proteins with tyrosine phosphatase activity (CD45). The intracellular signaling events follow engagement of the TCR including activation of different kinase cascades (PKC, MAPK, phosphoinositide 3-kinase, and PAK) (5–7). Important progress focused on elucidation of the roles and kinetics of early TCR-responsive tyrosine phosphorylation events during T cell activation has occurred. These studies have relied on the availability of highly specific antibodies that recognize phosphorylated tyrosine residues, making the detection of these phosphorylation events by flow cytometry or immunoblot easy (8–10). Recently, the use of MS coupled to phosphopeptide enrichment techniques has expanded the scope of these analysis by permitting the simultaneous detection and quantitation of hundreds or even thousands of phosphorylation sites in a sample, thus providing a broader, system wide view of the biological processes involved. Mass spectrometric mapping of tyrosine phosphorylation sites during TCR stimulation (11, 12) has provided important insights into the mechanism and connectivity of different pathways during early T cell activation, but fewer serine and threonine phosphorylation events have been characterized in the context of TCR signaling, despite their large number compared with tyrosine phosphorylation events. However, the complexity of the T cell serine and threonine phosphoproteome is starting to be recognized, and it seems now obvious that measuring the dynamics among the population of Ser and Thr phosphorylated residues will be critical for gaining a full understanding of T cell activation. Some recent studies have used a proteomic approach to address this issue using different lines of T lymphocytes (13, 14). Using P14 cytotoxic T lymphocytes, Navarro et al. (13) identified 2081 Ser and Thr phosphopeptides and found that 450 of them changed their abundance after 1 h of TCR stimulation. Proteins identified in this subset were involved in RNA post-translational modification, protein synthesis, cell death, gene transcription, and polymerization of actin. Another large scale quantitative phosphoproteomics experiment in Jurkat cells (14) reported the identification of thousands of phosphorylated sites, from which ∼600 tyrosine, serine, and threonine sites were up- or down-regulated in response to TCR activation for different times (5 and 15 min for Tyr; 15 or 60 min for Ser/Thr), and concluded that the scope of phosphorylation in response to TCR stimulation is widespread, and the proteins targeted were involved in all of the significant phenomena associated with T cell activation.
As observed by traditional biochemistry techniques, protein phosphorylation events following TCR activation maximize in a few minutes (as early as 1–2 min) for tyrosine phosphorylation, and some time later, at around 20 min, for most serine and threonine phosphorylated polypeptides. However, the precise timing and dynamics differ for particular proteins. We decided to study the phosphorylation changes occurring at a still early stage of the activation of the signaling cascades (5 min), between the very quick initial steps, and the well characterized events after 20 min of activation, because there is still a need to understand very early (5 min) Ser/Thr phosphorylation events in human primary T cells.
Although Jurkat cells are a very commonly accepted model to study TCR signaling (10, 11, 14), we have chosen here primary cells, because Jurkat cells lack specific signaling proteins as phosphatase and tensin homolog (8, 15), which could result in important differences in the phosphorylation profiles after TCR stimulation. Because of the usage of primary cells, isobaric mass tags (iTRAQ) (16, 17) were employed to measure differences in the phosphorylation profile after TCR stimulation.
To expand our study of protein phosphorylation changes in T cells at these early activation times, we focused our efforts not only on the analysis of tyrosine phosphorylation events employing immunoaffinity purification using P-Tyr-100 antibodies but also on serine and threonine phosphorylation using affinity chromatography based on TiO2. Overall, we identified and used for quantitation 2814 unique phosphorylated peptides (48 phosphorylated in Tyr and 2767 in Ser/Thr). Five hundred and seventeen of these phosphorylated sites (from 477 phosphopeptides) showed TCR-responsive changes. In-depth analysis of all the TCR-responsive phosphorylation sites resulted in the identification of 91 previously unreported phosphorylation sites. We show that TCR-responsive phosphorylation changes at these early times occur in proteins with many different activities and in different subcellular locations, with a large group of proteins involved in cytoskeletal reorganization, giving important clues about how the cytoskeleton interacts with plasma membrane in early TCR activation. To our knowledge, this data set is the broadest from human primary lymphocytes where changes in the intracellular levels of serine and threonine phosphorylation events are measured after 5 min of TCR stimulation.
EXPERIMENTAL PROCEDURES
Peripheral Blood Lymphocyte Isolation and Stimulation with anti-CD3
Peripheral blood lymphocytes were enriched by dilution of buffy coats (Blood Centers of the Pacific, San Francisco, CA) in PBS and centrifugation over a Ficoll-Paque (GE Healthcare) density gradient following standard procedures. The cells in the interface were collected and washed in PBS. After centrifugation at 1700 rpm for 10 min, supernatants were discarded, and each pellet was resuspended in RPMI 1640 culture medium with 10% FCS. The cells were left in culture for 2 h at 37 °C in the presence of CO2 to remove the adherent monocytes. Lymphocytes were recovered in the supernatant and resuspended at a concentration of 5 × 106 cells/ml in RPMI 1640 with 10% FCS.
Stimulation with anti-CD3 (clone HIT3a; BD Pharmingen, San Jose, CA) was performed as followed. For 5 min, 1 × 109 cells were treated with HIT3a primary antibody at a concentration of 1 μg/ml at 37 °C in presence of CO2 (incubator). Unstimulated cells were subjected to the same process as stimulated ones, but instead of antibody, PBS was added. Stimulated and unstimulated cells were centrifuged at 1200 rpm for 10 min and 2 ml of TRIzol reagent (Invitrogen) was added to each pellet and stored at −80 °C.
Cell Lysis, Reduction/Alkylation, and In-solution Digestion
1 × 109 cell pellets were thawed and resuspended in 22 ml of TRIzol reagent. Briefly, 0.2 ml of chloroform was added per 1 ml of TRIzol. The aqueous phase containing the RNA was removed, whereas the interface and the phenol phase containing DNA and proteins were kept. The removal of the DNA was done by precipitation with 100% ethanol (0.3 ml per ml of TRIzol). The supernatant was collected, and the proteins were isolated by precipitation with isopropyl alcohol, 1.5 ml of isopropanol/ml of TRIzol. Protein pellets were resuspended in 25 mm ammonium bicarbonate buffer containing 6 m guanidine HCl and 1 mm of NaF as phosphatase inhibitors. The amount of protein was checked by BCA protein assay kit (Pierce), and ∼30 mg of protein was recovered per sample. Next, 2 mm tris(2-carboxyethyl)phosphine was added to each sample, and they were incubated for 1 h at 56 °C to reduce cysteine side chains. Iodoacetamide was then added to the samples to a 4.2 mm concentration, and they were incubated 45 min in the dark at 21 °C to alkylate those cysteine side chains. The samples were then diluted 6-fold with 25 mm ammonium bicarbonate to reduce guanidine HCl concentration to 1 m, and 2% (w/w) modified trypsin (Promega, Madison, WI) was added. The pH was adjusted to 8.0 with 250 mm ammonium bicarbonate, and the samples were incubated 12 h at 37 °C. Digestion efficiency was checked by analyzing by LC-MS/MS aliquots of the digests containing 0.5 μg of the initial protein amount, desalted by using u-C18-ZipTips (Waters) according to the manufacturer's instructions.
Desalting of Digested Proteins
The digests were desalted using a MAX-RP Sep Pak® classic C18 cartridge (Waters) following the manufacturer's protocol. Briefly, the cartridges were conditioned with 70% acetonitrile (MeCN) and 0.1% TFA and then washed twice with 10 ml of 0.4% TFA in water. Next, the samples previously acidified were loaded onto the column, and the adsorbed material was washed 10–20 times the volume of the resin with 0.4% TFA in water. The peptides were eluted in three times with 0.5 ml of 70% MeCN, 0.1% TFA. The solvent was evaporated to dryness under vacuum, and the peptides were resuspended in the corresponding buffer to perform the phosphopeptide enrichment.
Enrichment of Phosphorylated Peptides Using Titanium Dioxide
Titanium dioxide enrichment was performed using an AKTA Purifier (GE Healthcare) using 5-μm TiO2 beads (GL Sciences, Tokyo, Japan) (18, 19) in-house packed into a 2.0-mm × 2-cm analytical guard column (Upchurch Scientific, Oak Harbor, WA). Tryptic digests (30 mg) were resuspended in 1.75 ml of 35% MeCN, 200 mm sodium chloride, and 0.4% TFA and divided into seven aliquots of 250 μl, each containing 4.2 mg of material. Phosphopeptide enrichment was performed separately in each of these aliquots. Aliquots were loaded onto the TiO2 column at a flow rate of 2 ml/min. The column was then washed for 2 min with 35% MeCN, 200 mm NaCl, and 0.4% TFA to remove nonphosphorylated peptides. Phosphopeptides were eluted from the column using 1 m KH2PO4, pH 3.0, at a flow rate of 0.5 ml/min for 30 min directly onto an on-line coupled C18 macrotrap peptide column (Michrom Bioresources, Auburn, CA). This column was washed with 5% MeCN, 0.1% TFA for 14 min, and the adsorbed material was eluted in 400 μl of 50% MeCN, 0.1% TFA at a flow rate of 0.25 ml/min. Small fractions (1/200) of the eluates were analyzed by LC-MS/MS. The data were searched, allowing phosphorylation in serine, threonine, and tyrosine as variable modifications, to assess the enrichment in phosphopeptides on the eluates. 80% of all peptides identified were phosphorylated. Eluates of the nonstimulated samples were pulled together, and the same was done for the stimulated fractions; peptide amounts in the eluates were estimated based on absorbance at 280 nm, using a nanodrop system (Thermo Scientific). Both samples where then solvent evaporated in a speed vac system and stored at −20 °C until iTRAQ labeling.
iTRAQ Labeling
iTRAQ® labeling (AB Sciex) was performed as follows: 200 μg of each sample (stimulated and nonstimulated) were resuspended in 80 μl of iTRAQ dissolution buffer. The samples were then split in two equal parts to label 100 μg of peptide/iTRAQ channel. iTRAQ labeling reagents were reconstituted in 70 μl of ethanol, and two different isobaric reagents were used per condition, leaving the labeling as follows: unstimulated cells were labeled with iTRAQ tags 114 and 116, and stimulated cells were labeled with iTRAQ tags 115 and 117. The labeling reaction was performed for 1 h at 21 °C. An aliquot of each labeling reaction was then examined by LC-MS/MS and searched, allowing iTRAQ as a variable modification to confirm that at least 99% of all peptides identified showed iTRAQ labeling (20). Another aliquot containing a combination 1:1:1:1 of the four labeled samples was analyzed by LC-MS/MS to confirm that total peptide levels were similar in the four labeling reactions. The four labeling reactions were then combined, desalted using a Sep Pak as described earlier, solvent-evaporated in a speed vac system, and stored at −20 °C until immunoaffinity purification was performed.
Immunoaffinity Purification of Tyrosine-phosphorylated Peptides
Phosphotyrosine containing peptides were enriched from the iTRAQ-labeled TiO2 eluates using phosphotyrosine mouse monoclonal antibody (P-Tyr-100) coupled to protein G-agarose beads (Phosphoscan Kit, Cell Signaling Technology, Danvers, MA) (21). Peptides were dissolved in 1.4 ml of immunoaffinity purification buffer (50 mm MOPS, pH 7.2, 10 mm sodium phosphate, 50 mm NaCl). After verifying that the pH was neutral, the sample was incubated with phosphotyrosine antibody beads, adding 80 μl of slurry containing 40 μl of bead volume. The sample was incubated for 30 min at 4 °C with gentle rotation. The mixture was centrifuged at 1500 × g for 1 min, and the supernatant was collected and kept for analysis of serine and threonine phosphorylation. The beads were washed three times with 1 ml of immunoaffinity purification buffer and two times with 1 ml of water. Retained peptides were eluted twice with 0.15% TFA, vacuum dried, and desalted using C18 ZipTip at basic pH (22). The peptides were then analyzed by Nano-Lc-ESI-Qq-TOF tandem MS on a QSTAR Elite mass spectrometer (Applied Biosystems/MDS Sciex, Foster City, CA).
Strong Cation Exchange Chromatography
The supernatant collected from the immunoaffinity purification, consisting mainly of serine and threonine phosphopeptides, was desalted using a SepPak, solvent-evaporated, and resuspended in 300 μl of 5 mm KH2PO4, 30% MeCN, pH 2.7 (Buffer A). Strong cation exchange chromatography was performed on an ÄKTA purifier (GE Healthcare). Sample in buffer A was loaded in the column, a Tricorn 5/200 (GE Healthcare) packed in house with a 5-μm 300 Å pore polysulfoethyl A resin (Western Analytical, Lake Elsinore, CA). Buffer B consisted of 5 mm KH2PO4, 350 mm KCl, 30% MeCN at pH 2.7 (23). The chromatographic separation was performed at 0.35 ml/min with a gradient that went from 2% to 72% B during 52 min and from 72% to 100% B in 10 min. A total of 58 fractions were collected, desalted using μC18 ZipTips (Millipore), and analyzed by Nano-Lc-ESI-Qq-TOF tandem MS on a QSTAR Elite (Applied Biosystems/MDS Sciex).
Western Blotting for Phosphoproteins
Anti-CD3 lymphocyte lysates, stimulated for 5 min and unstimulated, were prepared by resuspension in TRIzol reagent, as mentioned above, and precipitated proteins were resuspended in gel loading buffer containing 200 mm Tris-HCl, pH 6.8, 8% SDS, 20% v/v glycerol, 5% 2-mercaptoethanol, and 0.01% bromphenol blue. Equal amounts of protein (20–50 μg) were separated by 4–20% gradient Tris-HCl gel (Bio-Rad) by SDS-PAGE (Bio-Rad), and then transferred onto a PVDF (Thermo Scientific, Rockford, IL). The membrane was blocked overnight at 4 °C in blocking buffer (25 mm Tris, pH 7.5, 150 mm NaCl, 0.1% Tween 20, 1% BSA) and then incubated with the indicated primary antibody in the same blocking buffer at 22 °C for 1 h. Mouse anti-human phosphotyrosine (P-Tyr-100), rabbit anti-human phospho-Akt (Ser-473), rabbit anti-human phospho-p44/42 MAPK (ERK1/2) (Thr-202/Tyr-204), rabbit anti-human phospho-stathmin (Ser-16), rabbit anti-human phospho-PAK1 (Ser-199/204)/PAK2 (Ser-192/197), rabbit anti-human phospho-p90RSK (Ser-380), rabbit anti-human phospho-GSK3α/β (Ser-21/9), and rabbit anti-human GAPDH antibodies (Cell Signaling Technology, Danvers, MA) were used as primary antibodies (1/1000 dilution). The membrane was washed three times for 10 min at 22 °C with TBS (25 mm Tris, pH 7.5, 150 mm NaCl, 0.1% Tween 20). Next, the membrane was incubated with secondary anti-rabbit IgG or anti-mouse IgG antibody directly conjugated to horseradish peroxidase (1/5000 dilution; Bio-Rad, GE Healthcare) for 1 h in blocking buffer and washed six times for 5 min with TBS and once for 5 min with PBS (25 mm Tris, pH 7.5, 150 mm NaCl). The blots were developed with chemiluminiscence ECL Kit (GE Healthcare).
Nano-LC-ESI-Qq-TOF Tandem Mass Spectrometry Analysis
Peptides were loaded onto a 75-μm × 150-mm reverse phase C18 PepMap column (Dionex, LC Packings, San Francisco, CA) to be separated using an Agilent 1100 series HPLC system equipped with an auto sampler (Agilent Technologies, Palo Alto, CA). A flow rate of 300 nl/min was used with a 2-h MeCN gradient (3–32%) in 0.1% formic acid. The LC eluate was coupled to a nano-ion spray source attached to a QSTAR Elite mass spectrometer (Applied Biosystems/MDS Sciex). Peptides were analyzed in positive ion mode. MS spectra were acquired between 350 and 1500 m/z for 0.4 s. For each MS spectrum, the two most intense multiple charged peaks were selected for collision-induced dissociation. Per precursor ion selected, two MS/MS were taken; the first one was acquired between 180 and 1500 m/z for 2.5 s with resolution set to low and an automatically collision-induced dissociation energy based upon peptide charge and m/z ratio. The second MS/MS was acquired between 112 and 119 m/z for 2.5 s with a resolution set to unit and a constant voltage of 65 v, to maximize generation of iTRAQ reporter ions. A dynamic exclusion window was applied which prevented the same m/z from being selected for 1 min after its acquisition. Typical performance characteristics were 10,000 resolution with 30 ppm mass measurement accuracy in both MS and MS/MS spectra.
Peptide and Protein Identification
QSTAR Elite data was analyzed with Analyst QS software (Applied Biosystems/MDS Sciex, Foster City, CA) and MS/MS centroid peak lists were generated using the Mascot.dll script. Peak lists were searched against the UniProtKB Homo sapiens database as of August 10th 2010 (containing 192.290 entries), using Protein Prospector version 5.8 (http://prospector.ucsf.edu) with the following parameters. Trypsin was the enzyme selected, and up to three missed cleavages were allowed. Carbamidomethylation of cysteine residues and iTRAQ labeling of lysine residues and N terminus of the protein were allowed as fixed modifications. N-Acetylation of the N terminus of the protein, loss of protein N-terminal methionine, pyroglutamate formation from peptide N-terminal glutamines, oxidation of methionine, and phosphorylation of serine, threonine, and tyrosine were allowed as variable modifications. A first search was done using a peptide tolerance for QSTAR data in MS and MS/MS mode of 150 ppm and 0.2 Da, respectively. Taking into account the systematic error from the first search, a second search was done with a MS mass tolerance of 60 ppm. Proteins were considered positively identified when at least one peptide with a Protein Prospector peptide expectation value lower than 0.1 was identified. The false positive rate was estimated by searching the data using a concatenated database that contains the original UniProtKB database, as well as a version of each original entry where the sequence has been randomized. Unique peptides were only considered; peptides common to several proteins were not used for quantitative analysis. Sequence ID and phosphorylation sites were manually confirmed for all peptides that showed significant changes in levels after stimulation. Sites described here were compared with lists of currently reported phosphorylation sites on PhosphositePlus (http://www.phosphosite.org) and Uniprot DB (http://www.uniprot.org).
Quantitation
Relative quantitation of peptide abundance was performed via calculation of the raw area of peaks corresponding to the four different iTRAQ labels, 114.1, 115.1, 116.1, and 117.1 m/z present in all MS/MS spectra. Areas were determined by Protein Prospector by using isotope correction values supplied by the vendor for these batches of the reagent. If a peptide with the same charge state had several MS/MS spectra, just the best scoring one was used for quantitation. Only MS/MS spectra with iTRAQ peak areas greater than 30 were selected. Areas of the iTRAQ reporter ions representing technical replicates of the same sample were used to calculate significance thresholds for the analysis, as described under “Results”.
To measure the relative abundance of a given phosphopeptide, the average of iTRAQ areas in the unstimulated sample was divided by the average of iTRAQ areas in the stimulated sample from that same MS/MS spectrum. The lower the ratio, the more abundant that phosphopeptide was in the stimulated sample.
Phosphorylation Motif Analysis
To analyze the predicted consensus phosphorylation site motifs present in our set of TCR-responsive peptides, Motif-X algorithm (24) was run. Parameters used were: sequence window of ± 6 amino acids around the identified phosphorylated site, significance of 0.000001, and an occurrence of 35. The queried kinase motifs were from those reported in the Human Protein Reference Database phosphorylation database (25) and those described by Kinexus (www.kinexus.ca) as protein Ser/Thr kinase consensus phosphorylation site specificity.
We also performed a manual analysis of the enrichment we observed in the sequences of TCR-responsive peptides for some selected motifs for particular kinases compared with their presence in all protein sequences in the Human Uniprot database. Enrichment factors were calculated as reported previously (26).
RESULTS
Quantitative Analysis of Changes in Protein Phosphorylation Following Activation of Primary Human T Cells
To analyze the changes in protein phosphorylation in T lymphocytes following TCR activation, we used human primary lymphocytes isolated from leukocyte-rich buffy coats. Cells in culture were activated with 1 μg/ml anti-CD3 for 5 min. Fig. 1 shows the typical changes in the tyrosine phosphorylation profile we observe under these conditions by Western blot using P-Tyr-100 antiphosphotyrosine antibodies. To compare the relative occupancy of phosphorylated residues (tyrosine and serine/threonine) in proteins extracted from resting and cells that were stimulated for 5 min, we used a mass spectrometry-based iTRAQ approach. The work flow of the experiment is depicted in Fig. 2. Briefly, the cells were lysed, and proteins were TRIzol-extracted and digested with trypsin (see “Experimental Procedures”). Digests were enriched in phosphopeptides by affinity chromatography using TiO2 columns. Equal amounts (as estimated by absorbance at 280 nm) of the eluates of the TiO2 column from unstimulated or stimulated T cells were labeled with iTRAQ reagents. To be able to determine significance thresholds for the changes in abundance levels from the iTRAQ ratio distributions of the peptides, two iTRAQ channels were used per sample: Two separate aliquots of peptides from control (unstimulated) cells were labeled with either 114 or 116 iTRAQ tags, and two other separate aliquots of peptides from stimulated cells were labeled with either 115 or 117 iTRAQ tags. Subsequently the four differentially labeled samples were combined. To further enrich phosphotyrosine-containing peptides, immunoprecipitation using P-Tyr-100 was carried out. This fraction was analyzed separately by MS. The supernatant of the immunoprecipitation, mainly phosphoserine- and phosphothreonine-containing peptides, was subjected to additional separation by strong cation exchange chromatography to reduce complexity of the fractions to be analyzed by mass spectrometry. A total of 2883 phosphopeptides from 1372 proteins were identified; of these peptides, 2836 were phosphorylated on serine or threonine, and 48 were phosphorylated on tyrosine. The false discovery rate of peptide identification was below 0.8%, as estimated from searches against a randomized database.
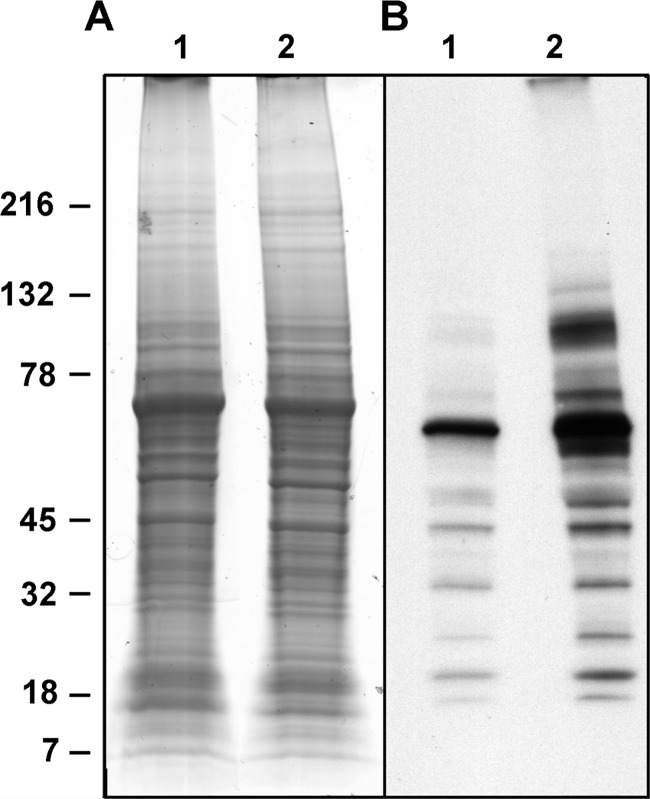
Fig. 1.
Global tyrosine phosphorylation changes in T lymphocytes upon stimulation with anti-CD3 antibody. The cells were incubated in control medium (lanes 1) or in the presence of 1 μg/ml anti-CD3 antibody for 5 min (lanes 2) and then lysed. Aliquots of protein extracted from total cells lysates were subjected to SDS-PAGE and stained with colloidal Coomassie (A). Identical amounts of sample were loaded in a second gel and immunoblotted with P-Tyr-100 antibody (B).
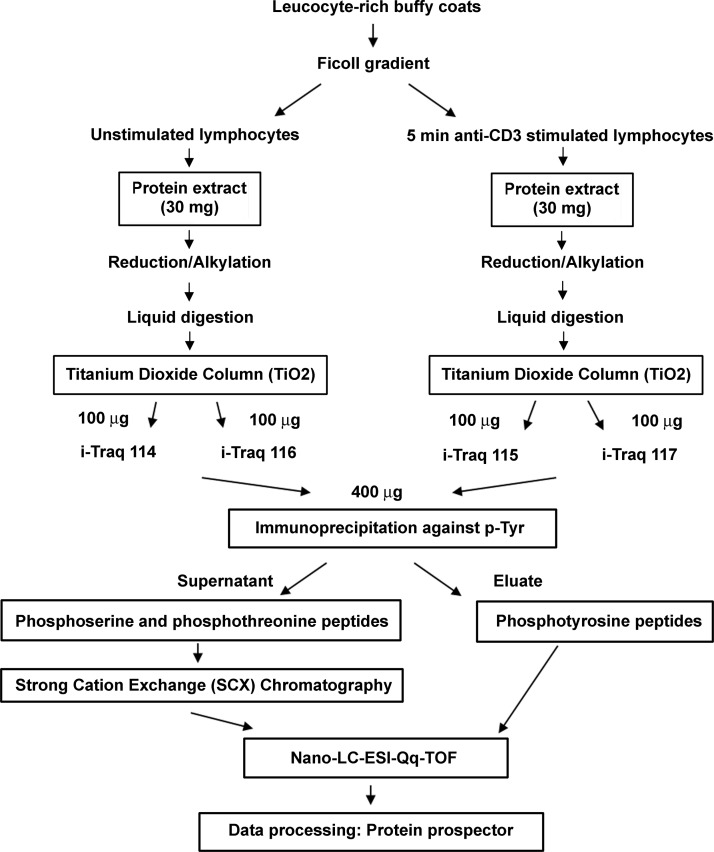
Fig. 2.
Experimental workflow. The different steps of sample preparation and analysis (cell isolation, protein extraction, phosphoenrichment, iTRAQ labeling, and LC-MS/MS analysis) are summarized in this diagram.
Quantitative data for the phosphorylated peptides were generated using the peak areas of the iTRAQ reporter ions. First we analyzed the experimental dispersion of the system using the technical replicates we included by labeling the control and stimulated samples with two iTRAQ channels each. As we can see from the plots representing the peak area in one iTRAQ channel versus the second channel corresponding to the same sample (control or stimulated cells) for each given peptide, peak areas of the two technical replicates per sample were very consistent (see supplemental Fig. 1). We excluded from downstream analysis peptides for which the coefficient of variation between the peak area values for the two iTRAQ channels for the same sample were greater than 30%. 2814 phosphopeptides (48 phosphorylated in Tyr and 2767 in Ser or Thr) were then used for quantization. For comparison between the control and stimulated cells, the averages of the areas of the 114 and 116 ions versus the 115 and 117 ions were considered. Logarithms to base 10 of the ratio of these two values are plotted in Fig. 3 (panels iii), for both the phosphotyrosine containing set (Fig. 3B, panel iii) and the phosphoserine/phosphothreonine set (Fig. 3A, panel iii). For the Ser/Thr phosphorylation set, the histogram shows a broad distribution with mode close to 0. Fig. 3 (panels i and ii) also shows the distribution of iTRAQ ratios for both channels in the control sample (ratio 114/116) and in the stimulated sample (ratio 115/117). Because in this case both iTRAQ channels represent the same sample and they were mixed in 1:1 ratio, dispersion in these plots must be due to stochastic fluctuations in the intensities of the iTRAQ reporter ions, so it represents the random dispersion of the ratios in technical replicates. We are considering significant changes in the log10 ratios of the control versus stimulated samples that exceed three times the value of the mode of the distribution (0) plus/minus the standard deviation observed in the distribution of log10 ratios of the technical replicate of the control sample (ratio 114/116) (±0.22, equivalent to a 1.6-fold change).
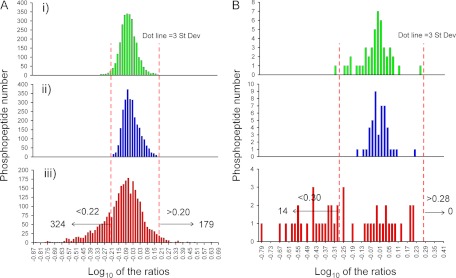
Fig. 3.
Distribution of relative levels of phosphopeptides in control versus stimulated T lymphocytes. Histograms showing logarithm base 10 of the ratios between the indicated iTRAQ reporter ions were constructed for the phosphoserine/threonine containing peptides (A) and phosphotyrosine peptides (B). Technical replicates of the control versus control sample (114/116 ions) (panels i) and stimulated versus stimulated sample (115/117 ions) (panels ii) represent stochastic variation of reporter ion intensities during analysis. Log10 ratios of the average values for the control sample (114 and 116) and the stimulated sample (115 and 117) are in panels iii). Three times the biggest value of the standard deviation of the log10 ratios in distributions in panels i and ii was selected as a threshold for significant changes in relative levels in panel iii (0.21, equivalent to 1.6-fold change in A; 0.295, equivalent to a 1.8-fold changes in B). The mode of the distribution in A, panel iii, is −0.01, so phosphopeptides with a logarithm of area ratios in A, panel iii, smaller than −0.22 are considered up-regulated, and those with a logarithm greater than 0.2 down-regulated after TCR stimulation. In B, panel iii, phosphopeptides with values smaller than −0.31 are considered up-regulated, and those with values greater than 0.28 down-regulated after TCR stimulation. In total, 517 phosphorylation sites were TCR-regulated after 5 min of CD3 stimulation.
The same analysis was done for the phosphotyrosine containing set. In this case, histograms of log10 ratios for technical replicates show a similar distribution centered at 0. The log10 ratio of the control versus stimulated values (Fig. 3B, panel iii), however, is clearly shifted toward negative values. Because these peptides come from the same pool as the phosphoserine/phosphothreonine peptides, and they were clearly showing that the mixing of the labeled tryptic digests of control and stimulated cells was done in a 1 to 1 ratio, the shift in the log10 values for most of the phosphotyrosine containing peptides quantified must have a real biological origin. Again, we are considering as significant the changes in the log10 ratios of the control versus stimulated samples that exceed three times the standard deviation observed in the distribution of log10 ratios of the technical replicate of the control sample (ratio 114/116) (±0.295, equivalent to a 1.8-fold change).
Phosphopeptides showing more than a 1.6-fold level change in the phosphoserine/phosphothreonine set, or 1.8-fold changes in abundance in the phosphotyrosine set contain thus TCR-responsive phosphorylation sites. In total, 517 of the phosphosites (from 477 phosphopeptides, see supplemental Table 1) showed TCR-responsive changes; 338 were up-regulated (14 phosphotyrosine and 324 phosphoserine/phosphothreonine-containing peptides), and 179 were down-regulated, all in the phosphoserine/threonine set (see supplemental Table 1).
Analysis of all the TCR-responsive phosphorylation sites resulted in the identification of 91 new sites or 18% of the responsive ones (supplemental Table 2). Sequence identification and assignment of the phosphorylation site for all the TCR-responsive phosphopeptides were confirmed by manual inspection of the MS/MS spectra (see spectra in supplemental MS/MS data).
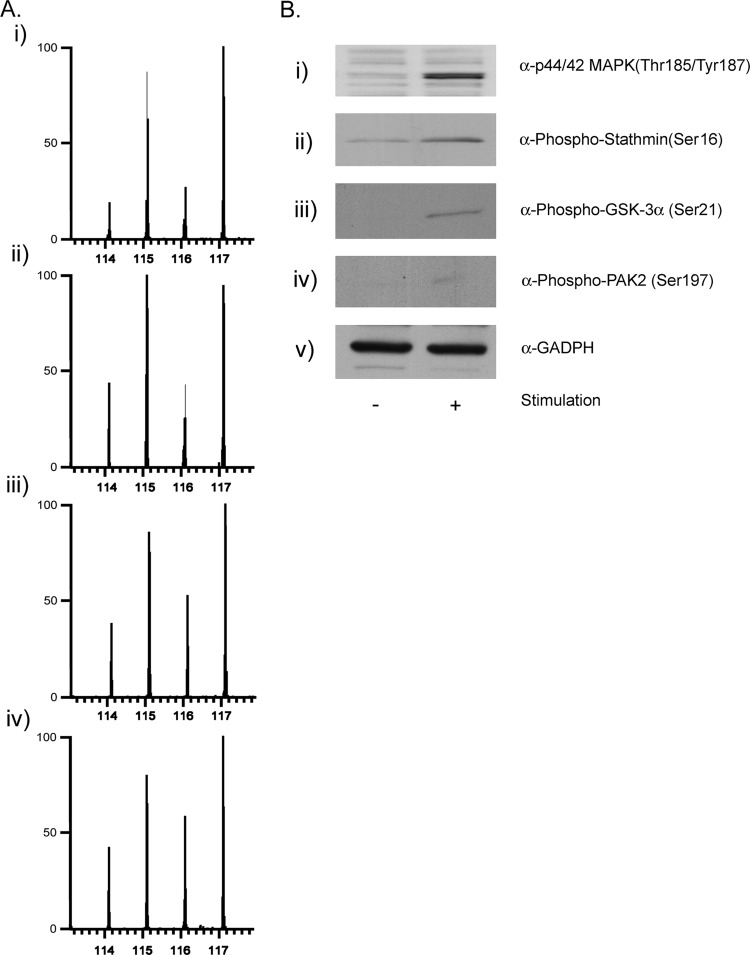
We have confirmed changes in relative levels between control and 5-min stimulated samples for some of the identified TCR-responsive phosphorylation sites by Western blot, to further validate the analysis of the iTRAQ data (Fig. 4B). Not many antibodies are commercially available against the phosphorylated forms of the proteins found in this study. We have performed immunoblots against (i) phospho-ERK 2 (doubly phosphorylated on Thr-185 and Tyr-187), (ii) phospho-stathmin on Ser-16, (iii) phospho-GSK-3α on Ser-21, and (iv) phospho-PAK 2 on Ser-197. Fig. 4A shows the ion intensities of the reporter ions of MS/MS spectra of phosphopeptides that contain the above mentioned phosphorylation sites in the same set of proteins (i to iv). We conclude that, at least for this small subset of proteins that we have tested, the relative changes inferred from the immunoblots and form the MS/MS data are in good agreement.
Fig. 4.
Correlation between the changes in relative levels of phosphorylation after TCR stimulation observed by immunoblot and by MS. In A, MS/MS spectra showing iTRAQ reporter ions generated from precursors with m/z 612.774+ (panel i), 616.963+ (panel ii), 700.834+ (panel iii), and 767.053+ (panel iv), which correspond to peptides spanning residues Val-173–Arg-191, Ala-15–Arg-27, Thr-7–Lys-27, Ser-197–Lys-216 on ERK1/2, stathmin, GSK3, and PAK, respectively, and phosphorylated in Thr-185/Tyr-187, Ser-16, Ser-21, and Ser-197. In B, 5 min anti-CD3 stimulated and unstimulated lymphocytes were lysed, protein was extracted, and the aliquots were subjected to SDS-PAGE and immunoblotted using antibodies against the specific phosphorylated sequences in ERK1/2 (panel i), stathmin (panel ii), GSK3 (panel iii), and PAK 2 (panel iv) or against GAPDH (panel v).
Bioinformatic Analysis of the TCR-responsive Proteins
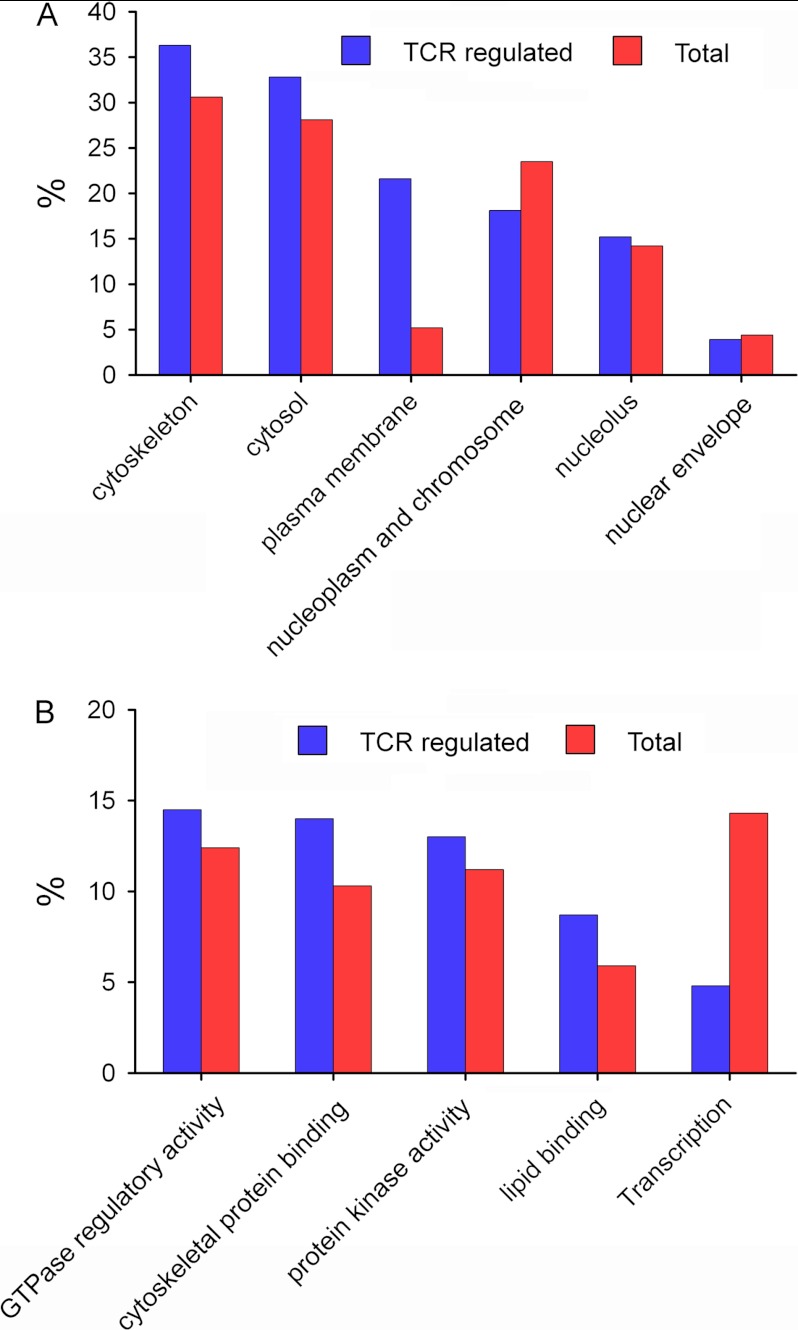
All 517 identified TCR-responsive phosphorylation sites belong to a set of 334 proteins. We have analyzed this set of phosphoproteins involved in the TCR signaling pathways using DAVID Bioinformatics Resources 6.7 (http://david.abcc.ncifcrf.gov/) to obtain their cellular localization and molecular function (Fig. 5). Most of these proteins are located in the cytoskeleton, cytosol, plasma membrane, and nucleoplasm/chromosomes. We have performed a parallel analysis for the whole set of proteins identified by MS. The results indicated that in proteins containing TCR-responsive phosphorylation sites, some categories seem overrepresented, particularly plasma membrane proteins. Most of these plasma membrane proteins are also associated to cytoskeleton according to their Gene Ontology annotations. In the same way, molecular functions (Fig. 5B) are very diverse, but a striking number have GTPase regulatory activity, cytoskeletal protein binding, kinase activity, and lipid-binding domains. We have also observed how phosphorylation on proteins associated with transcription processes highly decreased after TCR stimulation.
Fig. 5.
Distribution of functional categories of the identified proteins. Cellular localization and molecular function of the 334 TCR-responsive proteins and all identified proteins are represented in A and B, respectively. The categories were distributed using DAVID bioinformatics resources (http://david.abcc.ncifcrf.gov/).
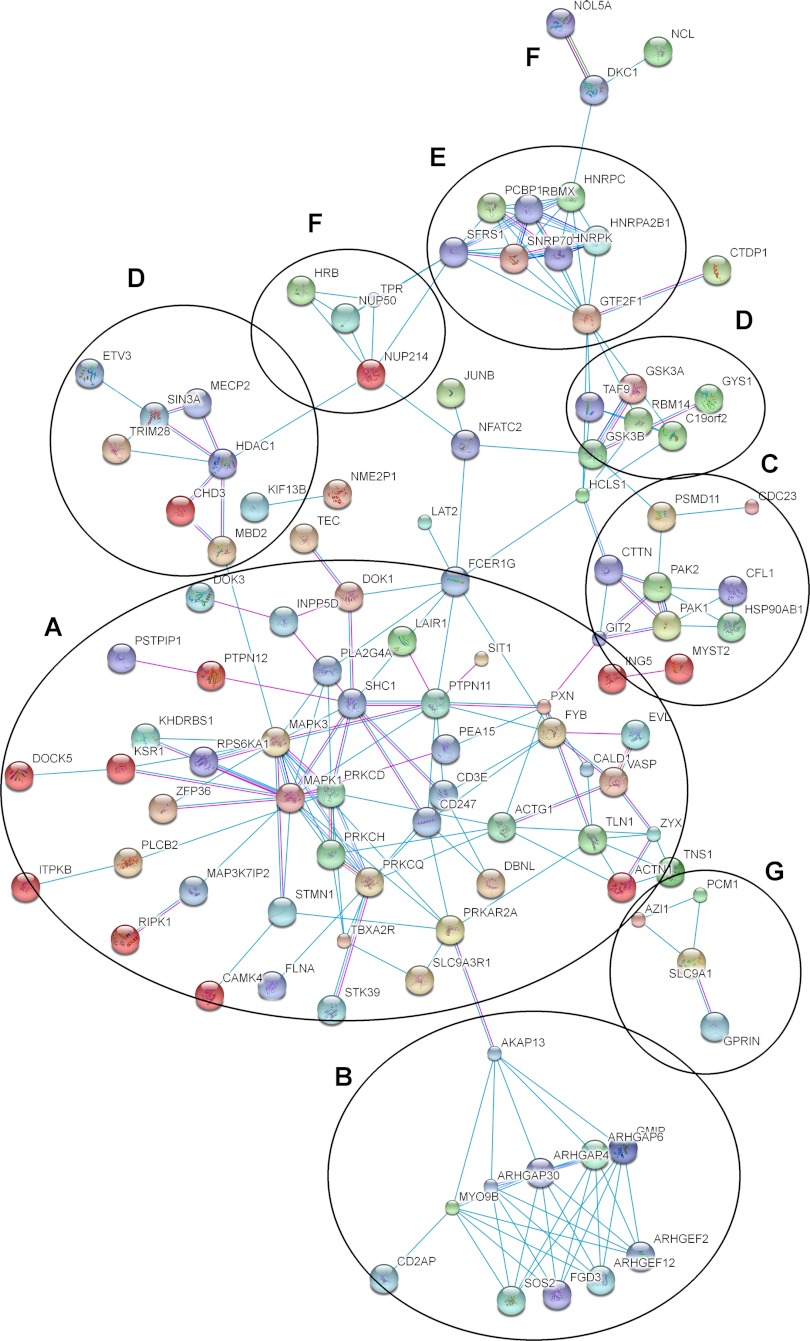
Next, we organized TCR-regulated phosphoproteins into protein-protein interaction networks. We have used STRING (Search Tool for the Retrieval of Interacting Genes/Protein, http://string-db.org/) to find interactions between them based on previously described experimental evidences. We obtained by plotting a high confidence evidence network (score > 0.7) seven different clusters (Fig. 6). The largest cluster contains proteins interacting directly with the TCR complex or the main reported pathways like CD3ζ, CD3ε, NFAT (nuclear factor of activated T cells), ERK1/2, STMN1 (stathmin), and EVL (enabled/vasodilator-stimulated phosphoprotein-like protein) and also in cytoskeleton regulation as talin, zixin, and actin (Fig. 6A). Several Rho-GTPase proteins formed another defined cluster where they are all involved in cytoskeleton regulation and cytoskeletal polarity between T cell and APC (antigen-presenting cell) (27, 28) (string83.embl.de) (Fig. 6B). Cortactin, cofilin-1, and p21-activated protein kinase 1/2 (PAK1/2) interact among them and formed another cluster also involved in cytoskeleton organization (Fig. 6C) (8, 29). Other clusters can also be seen involved in regulation of transcription (either initiation or repression) (Fig. 6D), pre-mRNA processing (Fig. 6E), trafficking through nucleus (Fig. 6F), and microtubule and centrosome assembly (Fig. 6G) (30, 31).
Fig. 6.
Interaction networks for some groups of TCR-responsive proteins. Different clusters of interacting proteins were identified using STRING to obtain a high confidence evidence network. Connecting lines show reported interaction based on experimental evidence (blue lines) or databases (pink lines). A, proteins interacting directly with the TCR complex or the main reported pathways; B, Rho-GTPases, C, cytoskeletal regulators; D, regulation of transcription; E, pre-mRNA processing; F, trafficking through nucleus; G, microtubule and centrosome assembly.
Analysis of the Kinase-specific Motives in TCR-responsive Phosphopeptides
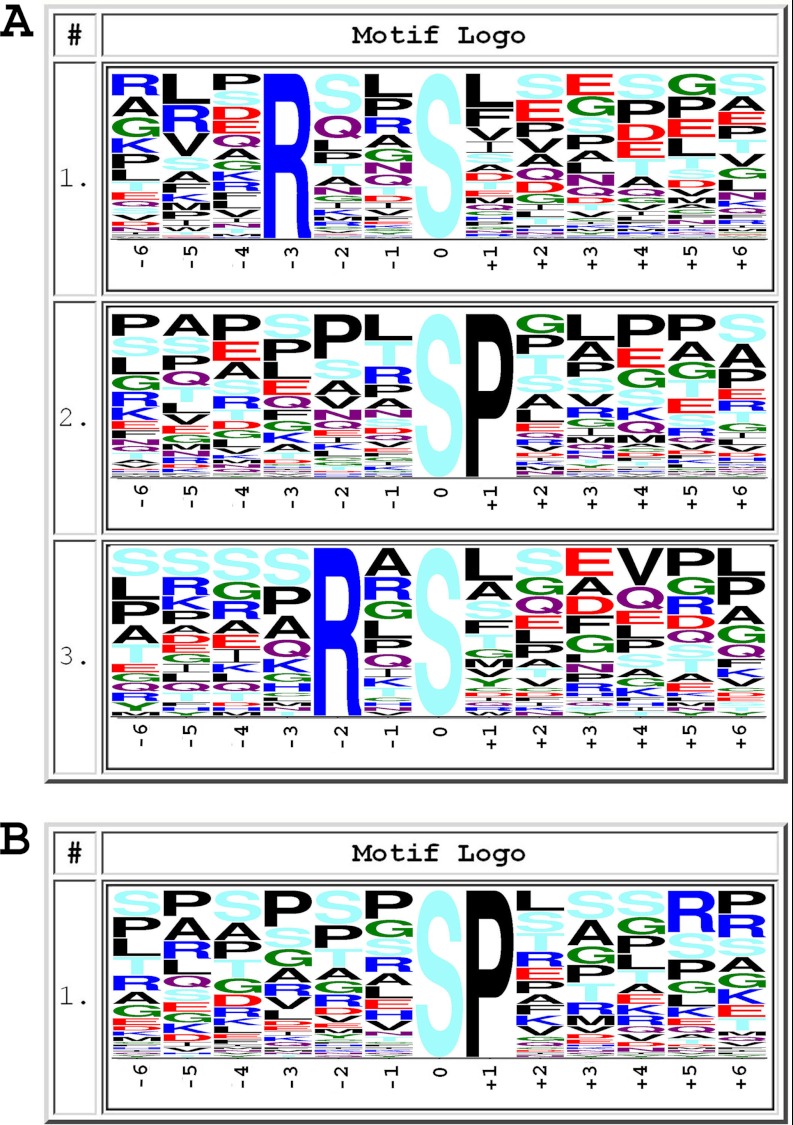
Kinase specificity is defined by the amino acid sequences surrounding the phosphorylation sites. We have performed an analysis of the predicted consensus phosphorylation site motifs present in our set of TCR-responsive peptides. We employed the Motif-X algorithm (24) by using a sequence window of ± 6 amino acids around the identified phosphorylated site. Three motifs were identified in the set of phosphopeptides up-regulated after TCR stimulation: RXX(pS/pT), (pS/pT)P, and RX(pS/pT); and just one, (pS/pT)P, was identified in the set of down-regulated phosphopeptides (Fig. 7). According to the Human Protein Reference Database, the consensus phosphorylation site motif RXX(pS/pT) is specific for kinases such as CaMK1, 2, and 4; PKC; and Akt. Motif RX(pS/pT) is specific for PKC, PKA, and Aurora-A kinase, and the proline-directed kinase motif (pS/pT)P is specific for ERK1/2, GSK3, and cyclin-dependent kinase.
Fig. 7.
Frequency plots of residues surrounding serine, threonine, or tyrosine phosphorylation sites. The height of the residues represents the frequency with which they occur at the respective positions. The color of the residues represents their physicochemical properties. The frequency plots were generated with motif X algorithm. A shows motifs found in up-regulated TCR-responsive phosphopeptides, and B shows those found in down-regulated phosphopeptides.
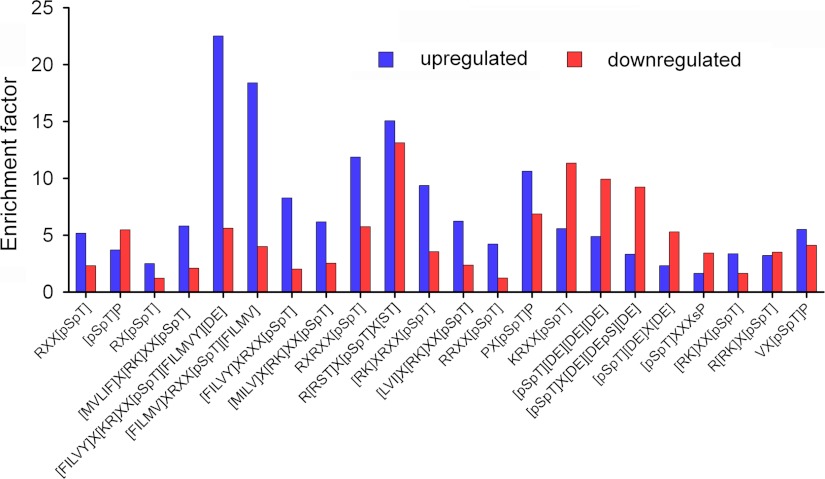
We have also performed a manual analysis of the enrichment we observed in the sequences of TCR-responsive peptides for some selected motifs for particular kinases (26), compared with their presence in all protein sequences in the Human Uniprot database (Fig. 8 and supplemental Table 4). Consensus motifs for CaMK, p90RSK, PKD, and ERK1/2 are enriched in the subset of phosphopeptides up-regulated after TCR stimulation, whereas motifs for PKA, cell division control protein 2 homolog, casein kinase 2 are enriched in the pool of down-regulated phosphopeptides.
Fig. 8.
Selected kinase motifs present in TCR-responsive phosphopeptides. We represent the enrichment factors for different motives in the sets of up- and down-regulated phosphopeptides (letters inside brackets indicate possible alternative amino acids in that position). Enrichment factors (supplemental Table 4) were calculated by taking into account the number of times the consensus phosphorylation site motifs appeared randomly in the database and the number of TCR-responsive phosphopeptides containing that motif (see “Experimental Procedures” for details). Motifs in the x axis correspond to the following kinases: CaMK1, (F/I/L/M/V)XRXX(pS/pT)(F/I/L/M/V); CaMK2, RXX(pS/pT); (M/V/L/I/F)X(R/K)XX(pS/pT) or (F/I/L/V/Y)X(K/R)XX(pS/pT)(F/I/L/M/V/Y)(D/E); CaMK4, RXX(pS/pT) or (F/I/L/V/Y)XRXX(pS/pT); PKA, RXX(pS/pT), RX(pS/pT), KRXX(pS/pT), or R(R/K)X(pS/pT); PKC, RXX(pS/pT), RX(pS/pT), or (R/K)XX(pS/pT); ERK1/2, (pS/pT)P, PX(pS/pT)P, or VX(pS/pT)P; GSK3, (pS/pT)P, PX(pS/pT)P, or (pS/pT)XXXsP; aurora A, RX(pS/pT); Chk1, (M/I/L/V)X(R/K)XX(pS/pT); Akt, RXX(pS/pT), RXRXX(pS/pT), or R(R/S/T/)X(pS/pT)X(S/T); p90RSK, RXRXX(pS/pT) or (R/K)XRXX(pS/pT); PKD, (L/V/I)X(R/K)XX(pS/pT); Zip kinase, RRXX(pS/pT); cyclin-dependent kinase 5, (pS/pT)P or PX(pS/pT)P; cell division control protein 2 homolog, KRXX(pS/pT); and casein kinase 2, (pS/pT)(D/E)(D/E)(D/E), (pS/pT)X(D/E)(D/E/pS)(D/E), or (pS/pT)(D/E)X(D/E).
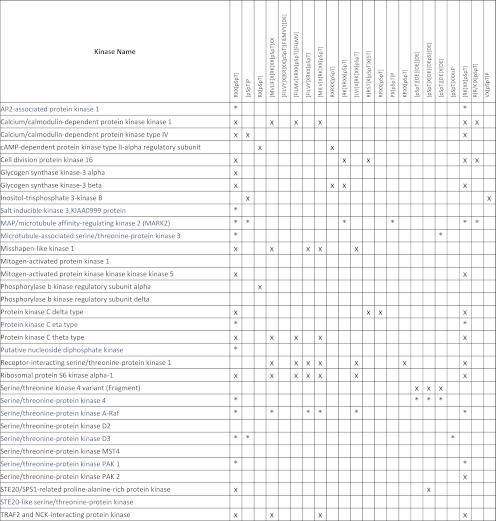
From the 517 TCR-responsive phosphosites, we found 33 belonging to protein kinases; 11 of these phosphosites are down-regulated and 22 up-regulated upon TCR stimulation. We have searched for kinase-specific motifs in this subset of peptides (Table II). Most of these protein kinases (except peptides from δ subunit of phosphorylase b kinase, ERK2, serine threonine protein kinase D2, and serine threonine protein kinase MST4) had at least one phosphopeptide containing one of the kinase-specific phosphorylation motives analyzed here, as shown in Table II. The majority of the TCR-responsive phosphopeptides found in protein kinases could be a product of the activity of several different kinases (see sequences in supplemental Table 1). For example, ribosomal protein s6 kinase α1 (p90RSK) could be phosphorylated on Ser-380 (in the sequence QLFRGFsFVATGL) by CaMK2, 1, and 4; Chk1; PKD; or PKC. In contrast, TCR-responsive phosphorylation sites identified in other kinases are much more specific, like Ser-58 in PAK2 (in the sequence PRHKIIsIFSGTE), between all the motifs considered here, only fits with PKC-directed motifs. In some cases different sites in the same kinase show opposite changes upon TCR activation: mitogen-activated protein/microtubule affinity regulating kinase 2 is identified with one up-regulated phosphopeptide (Ser-376) with a p90RSK-specific motif and one down-regulated phosphopeptide with a GSK3 specific motif (Ser-456).
Table II. Kinase-specific motifs found in the TCR responsive phosphopeptides in protein kinases.
In blue and with an asterisk we represent the kinase-specific motifs found in downregulated phosphopeptides present in the sequences of protein kinases. Upregulated ones are represented in black. See legend of Figure 8 for kinase specificity for these motives.
DISCUSSION
Different methods for the activation of T cells in culture using anti-CD3 antibodies have been developed and used by different groups. In most cases, they used antibodies attached to the plastic surface of the culture dishes (32, 33). Some other studies add the soluble antibody to the medium that contains the cells (3, 34, 35). We tested both protocols for efficiency of the activation of the signaling cascades (as assessed by Western blot using an anti-phosphotyrosine antibody), using either culture plates precoated with anti-CD3 (using antibody solutions 5 or 10 μg/ml) or soluble antibody added to the cells (at concentrations of 1 and 2 μg/ml). In our hands, activation was more efficient, as suggested by the intensity and pattern of immunoreactive bands, using the antibody in solution, (data do not shown). The results were similar using 1 or 2 μg/ml, so we decided to carry out our experiments adding anti-CD3 antibody to the culture medium at 1 μg/ml.
The large scale, unbiased analysis of the changes in T cell phosphorylation levels performed here establishes that at 5 min after TCR stimulation, widespread serine and threonine phosphorylation and dephosphorylation is taking place. Five minutes following TCR activation the main wave of tyrosine phosphorylation, which includes the very initial steps of the signaling cascades, is already fading, and serine/threonine phosphorylation signaling events are becoming predominant. We see how these events affect proteins with many different activities and in different subcellular locations. At this time point we still can observe in our data set increased tyrosine phosphorylation in some of the proteins that represent the first steps of the signaling pathway, like CD3 ζ chain. We do not detect tyrosine phosphorylation of some of the major tyrosine kinase substrates immediately downstream of the TCR that are observed shortly after TRC stimulation, like LAT (linker of activated T cells), SLP-76, PLCγ, Vav, or ZAP-70, perhaps because of the kinetics of the experimental system; as we mention above, tyrosine phosphorylation events peak at an earlier time, within 1–2 min after TCR activation, and the resulting phosphorylated forms of these proteins can be only transiently detected. During TCR activation, tyrosine phosphorylation of CD3 ζ chain is followed by recruitment of the tyrosine kinases Lck, Fyn, and ZAP-70 that phosphorylate the transmembrane adaptor protein LAT, which relocates to microclusters during TCR activation. Unexpectedly, we observe here how LAT gets dephosphorylated in Ser-135 upon TCR stimulation. How phosphorylation in this residue affects LAT activity is unknown, but it possibly adds a new control mechanism over its activity complementary to its phosphorylation by ZAP-70.
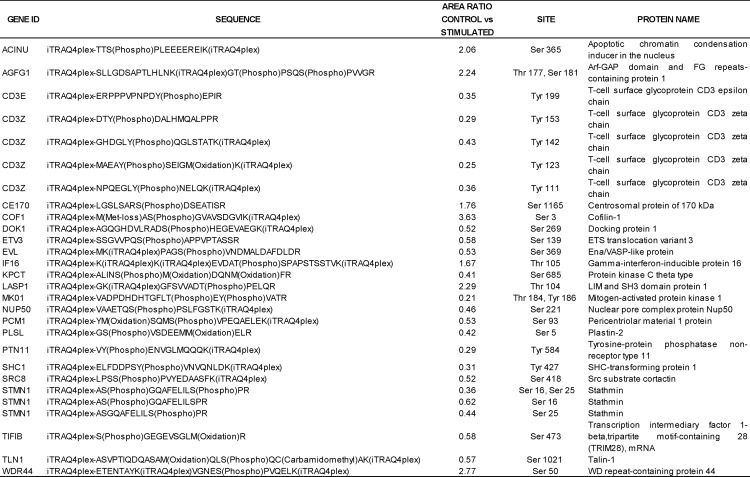
Indeed, a number of the proteins we have identified as TCR-responsive have been previously described to participate in TCR-modulated signaling pathways. From all of the 517 TCR-responsive phosphorylation sites identified in this study, 29 were previously characterized as TCR-responsive sites in the literature (Table I) (13, 14). For example, TCR stimulation generated dephosphorylation of the actin-binding proteins cofilin-1 on Ser-3 and LIM and SH3 domain protein 1 on Thr-104 and induced phosphorylation of others, as docking protein 1, which modulates integrin activation by competing with talin-1. We also observe an increase in phosphorylation on talin-1 Ser-1021 (Table I) (14, 36, 37). We have found that many other sites, previously unknown to be TCR-responsive, do change phosphorylation levels on proteins previously known to play defined roles in TCR signaling (supplemental Table 3). An example is the above mentioned Ser-135 in LAT. We have found 101 of these additional sites. From these sites, seven have never been described as a phosphorylation site on the corresponding protein; examples of this are Ser-886 in neuroblast differentiation-associated protein, Ser-394 and Thr-2209 on filamin-A, and Ser-135 on drebrin-like protein, which is a key component of the immunological synapse that regulates T cell activation by bridging TCRs and the actin cytoskeleton to gene activation and endocytic processes (38).
Table I. TCR responsive phosphorylation sites identified in this study and previously known to be involved in TCR signaling.
Phosphorylation sites are labeled in the sequence of the peptides.
The time frame we have chosen in this experiment to analyze the changes in the phosphoproteome of T cells (5 min) overlap in time with extensive cytoskeletal reorganization in the immune synapse. Dynamic rearrangement of the actin and microtubule cytoskeletons is essential for TCR signaling (2). Following the interaction between T cell and APC, a number of proteins (TCR complexes and signaling molecules such as ZAP-70 (ζ chain-associated protein kinase of 70 kDa), LAT, GRB2 (growth factor receptor-bound protein-2), GRAP2 (GRB2-related adaptor protein-2), and SLP-76 (SRC homology-2 domain-containing leukocyte protein of 76 kDa)) (3, 39) aggregate into microclusters at the periphery of the immune synapse, and then they move centripetally toward central areas in which signal extinction occurs (40). This movement of signaling microclusters is controlled by actin cytoskeleton reorganizations; between 5 and 15 min after TCR-APC interaction, accumulation of F-actin in these areas is elevated, and there is active actin polymerization (41).
We observe in this study how the most abundant group of TCR-regulated phosphoproteins after 5 min of activation is associated to cytoskeleton; this set of proteins should therefore be informative of the molecular mechanisms regulating the cytoskeletal reorganization dynamics in the IS, to control TCR signaling. Formation of membrane structures resembling IS has been observed when anti-CD3 bound to glass surfaces has been used to stimulate T cells, in an antibody-specific fashion (39). Anti-CD3 stimulation thus seems to be able to trigger cytoskeletal rearrangements associated to IS formation. In a time frame (5 min) coincident with the time observed for the cytoskeletal rearrangements observed when T cells are presented anti-CD3 bound to glass surfaces (39) or during IS formation in APC-T cell systems, we see extensive modifications in cytoskeletal proteins, which suggests that these events could also participate in the formation of the IS. At the molecular level, we see an increase of phosphorylation on actin itself (Ser-239 on β-actin), and several regulatory proteins. Actin cytoskeleton controls B cell receptor dynamics at the plasma membrane through membrane-cytoskeleton linkers (42); it is unknown how this happens in the TCR. We see in our data how talin-1, which acts as a membrane-cytoskeleton linker, increases its phosphorylation levels in residues Ser-1021, Ser-1201, and Ser-446 as a result of TCR activation. This protein has been shown to interact with adhesion molecules of the integrin family and cytoskeletal components in a Rap1-dependent manner (43), and it is localized on the peripheral zone of the immunological synapse (44). Based on the sequences that we observe that surround the phosphorylated residues, talin-1 could be a substrate of protein kinases like PKC and CAMK2 and 4, which we observe to be responsive to TCR stimulation in our experiment (supplemental Table 1). Several other actin-binding proteins, which could be involved in the movement of those microclusters, show changes in phosphorylation; as an example, CapZ-interacting protein (dephosphorylated at Ser-1164) regulates the ability of F-actin capping protein to remodel actin filament assembly. We observe dephosphorylation of particular residues in a considerable number of proteins related to the actin cytoskeleton: phosphatase and actin regulator 2, myosin 18A, paxillin, myosin 9 (phosphorylated on Ser-1713 and dephosphorylated on Ser-1942), dematin (increased in phosphorylation in 5 sites: Ser-92, Ser-96, Ser-105, Ser-113, and Ser-333 and dephosphorylated in either Ser-16 or Ser-18), LIM, SH3 protein, CAP-Gly domain-containing linker protein, and cofilin-1 (supplemental Table 1). So both protein kinase and phosphate activities triggered after 5 min of TCR engagement act over the TCR-responsive set of phosphoproteins in the actin cytoskeleton. Inhibitory phosphorylation sites, as occurring in the case of myosin IIA (45) impair signaling microclusters movement and TCR signaling, but once the TCR is triggered, some specific phosphatase(s) act(s) over these phosphorylation sites, releasing their inhibitory effect, as a necessary step for the microclusters relocation to the synapse. We observe changes in the phosphorylation status of particular residues in several different regulatory phosphatase subunits (protein phosphatase 1 regulatory (inhibitor) subunit 11, protein phosphatase 1 regulatory (inhibitor) subunit 12A (PPP1R12A), protein phosphatase inhibitor 2, putative uncharacterized protein PSTPIP1, proline-serine-threonine phosphatase-interacting protein 1, RNA polymerase II subunit A C-terminal domain phosphatase, tyrosine-protein phosphatase nonreceptor type 11 (PTPN11), and protein tyrosine phosphatase, nonreceptor type 12 (PTPN12). Which of them, if any, are related to the dephosphorylation events observed at this time during TCR activation remains a challenging task for future works.
The processes occurring in the immune synapse also involve reorganization of the microtubules. The microtubule organizing center needs to reorient and contact the plasma membrane at the TCR-APC contact site, and it requires the effect of protein kinases located in the microclusters, like tyrosine kinases Lck and Fyn and other kinases as PKC ε, δ, and θ (46, 47). We observe how several PKC isoforms alter their phosphorylation levels at 5 min. PKC θ gets tyrosine- and serine-phosphorylated in several sites (Tyr-313, Thr-141, Ser-306, Ser-685, and Ser-695) in our data. PKCε is dephosphorylated in Ser-674. Other proteins are also required for microtubule organizing center polarization, and we can see them here changing phosphorylation levels: PDLIM1 (PDZ and LIM domain protein 1), which increases its phosphorylation in Ser-206 (newly identified, see supplemental Table 2) and which gets dephosphorylated in Ser-129; LAT interaction proteins, like the adapter protein GRB2-associated binding protein 3, which is seen to be increased in phosphorylation at Ser-345; and its interacting protein SHC1 (phosphorylated on Tyr-427). Some important regulatory proteins as the earlier mentioned tyrosine protein phosphatase nonreceptor type 11 (PTN11) are able to bind GRB2-associated binding protein 3. Microtubule organizing center recruitment is regulated by interactions between tubulin and molecular motors like dynein, responsible for transporting cellular cargo along the microtubules (48), which shows increased phosphorylation levels in our data set.
Rac/Rho GTPases are inactive in quiescent cells but become activated with TCR stimulation in response to increased intracellular diacylglicerol levels. GTPases are important regulators of the cytoskeleton. Some of them are localized in the microclusters, like CDC42 or Rap1. They are likely to be the key for actin and microtubules cytoskeleton interplay in the immunological synapse conditioning its shape and stability. We can see in the list of TCR-responsive phosphoproteins several regulators of its activity: residues on CDC42 Rho GTPase effector protein, Ras GTPase-activating-like protein IQGAP2, and Rap1 GTPase-activating protein 2 when present increased phosphorylation levels. As for the kinases responsible for these modifications, there could be several candidates according to the motives that surround the phosphorylated residues. PKC might be responsible for Ser-564 phosphorylation of Rap1 GTPase-activating protein 2, and GSK3 or ERK might be responsible for Ser-45 and Ser-612/613. A possible kinase responsible for IQGAP2 phosphorylation is p90RSK. We observe increased phosphorylation on PKD2; PKD has been described to control the activity and plasma membrane localization of the GTPase Rap1 (49).
A set of GAP and GEF also shows changes in their phosphorylation levels (Rho GTPase-activating proteins 4, 6, 25, and 30, Rho guanine nucleotide exchange factors 2 and 12, and CDC42 effector protein 3). Several of these are able to interact with A kinase (PRKA) anchor protein 13 (nononcogenic Rho GTPase-specific GTP exchange factor, AKAP13). AKAP13 has been described to function as scaffolding proteins to coordinate a Rho signaling pathway and, in addition, function as a protein kinase A-anchoring protein (50, 51). We can observe increased phosphorylation in AKAP13 and its interacting protein cAMP-dependent protein kinase, regulatory, type IIα (PRKAR2A). This could provide a link for the regulation of cytoskeleton by cAMP levels via Rac/Rho GTPases during TCR responses. PKA is anchored to the TCR by the ezrin-myosin family (52) and works as a potent negative regulator of T cell immune function. It phosphorylates the Src kinase Csk, which upon activation inhibits Lck and Fyn maintaining the T cell homeostasis, but cAMP could also regulate the cytoskeleton rearrangements on the IS by this action through the Rac/Rho GTPases.
Changes in phosphorylation levels at 5 min after TCR stimulation affect even proteins involved in nuclear processes related to gene expression, transcription factors (we see key regulators of cytokine gene expression in T cells like NFAT I and β-catenin (53, 54) decreasing its phosphorylation in inhibitory residues upon TCR triggering), transcriptional regulators like histone deacetylases (Ser-393 in histone deacetylase 1) and methyl-CpG binding proteins. Proteins involved in mRNA processing and splicing present changes in their phosphorylation state at this early time, which is in agreement with what has been observed for later times (13), but our results show that changes in the regulatory post-translational modifications in this set of proteins are in fact much earlier, and the nuclear reprogramming of the T cell starts a few minutes after TCR stimulation.
Many additional experiments will be necessary to explore all the possible consequences of the changes in the T cell phosphoproteome shown here to happen after TCR stimulation. Changes in protein phosphorylation collected in large scale experiments like this one represent global views of the cellular state and so can be invaluable to design targeted experiments to elucidate the patterns of the transfer of information inside the cell. The exact role of the changes in phosphorylation on the sites that are responsive to TCR stimulation in T cells remains a task for future research.
Acknowledgments
We thank Jonathan Trinidad for guidance with phosphopeptide enrichment and strong cation exchange chromatography, Kati Medzihradszky for advice regarding confirmation of some phosphorylated sites, and David Maltby for assistance in the running of the mass spectrometer.
Footnotes
* This work was supported by National Center of Research Resources Grants P41RR001614 and RR012961. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- TCR
- T cell receptor
- IS
- immunological synapse
- iTRAQ
- isobaric tag for the relative and absolute quantitation
- LC-MS/MS
- liquid chromatography tandem mass spectrometry
- PBL
- peripheral blood lymphocytes.
REFERENCES
- 1. Bridgeman J. S., Sewell A. K., Miles J. J., Price D. A., Cole D. K. (2012) Structural and biophysical determinants of αβ T-cell antigen recognition. Immunology 135, 9–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lasserre R., Alcover A. (2010) Cytoskeletal cross-talk in the control of T cell antigen receptor signaling. FEBS Lett. 584, 4845–4850 [DOI] [PubMed] [Google Scholar]
- 3. Yokosuka T., Sakata-Sogawa K., Kobayashi W., Hiroshima M., Hashimoto-Tane A., Tokunaga M., Dustin M. L., Saito T. (2005) Newly generated T cell receptor microclusters initiate and sustain T cell activation by recruitment of Zap70 and SLP-76. Nat. Immunol. 6, 1253–1262 [DOI] [PubMed] [Google Scholar]
- 4. Pawson T., Scott J. D. (2005) Protein phosphorylation in signaling: 50 years and counting. Trends Biochem. Sci. 30, 286–290 [DOI] [PubMed] [Google Scholar]
- 5. van der Merwe P. A., Dushek O. (2011) Mechanisms for T cell receptor triggering. Nat. Rev. Immunol. 11, 47–55 [DOI] [PubMed] [Google Scholar]
- 6. Pearce L. R., Komander D., Alessi D. R.(2010) The nuts and bolts of AGC protein kinases. Nat. Rev. Mol. Cell Biol. 11, 9–22 [DOI] [PubMed] [Google Scholar]
- 7. Finlay D., Cantrell D. (2011) The coordination of T-cell function by serine/threonine kinases. Cold Spring Harb. Perspect. Biol. 3, a002261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Houtman J. C., Houghtling R. A., Barda-Saad M., Toda Y., Samelson L. E. (2005) Early phosphorylation kinetics of proteins involved in proximal TCR-mediated signaling pathways. J. Immunol. 175, 2449–2458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhu M., Janssen E., Zhang W. (2003) Minimal requirement of tyrosine residues on linker for activation of T cells in T cell activation and thymocyte development. J. Immunol. 170, 325–333 [DOI] [PubMed] [Google Scholar]
- 10. Abraham R. T., Weiss A. (2004) Jurkat T cells and development of the T-cell receptor signaling paradigm. Nat. Rev. Immunol. 4, 301–308 [DOI] [PubMed] [Google Scholar]
- 11. Salomon A. R., Ficarro S. B., Brill L. M., Brinker A., Phung Q. T., Ericson C., Sauer K., Brock A., Horn D. M., Schultz P. G., Peters E. C. (2003) Profiling of tyrosine phosphorylation pathways in human cells using mass spectrometry. Proc. Natl. Acad. Sci. U.S.A. 100, 443–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nguyen V., Cao L., Lin J. T., Hung N., Ritz A., Yu K., Jianu R., Ulin S. P., Raphael B. J., Laidlaw D. H., Brossay L., Salomon A. R. (2009) A new approach for quantitative phosphoproteomic dissection of signaling pathways applied to T cell receptor activation. Mol. Cell. Proteomics 8, 2418–2431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Navarro M. N., Goebel J., Feijoo-Carnero C., Morrice N., Cantrell D. A. (2011) Phosphoproteomic analysis reveals an intrinsic pathway for the regulation of histone deacetylase 7 that controls the function of cytotoxic T lymphocytes. Nat. Immunol. 12, 352–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mayya V., Lundgren. D. H., Hwang S. I., Rezaul K., Wu L., Eng J. K., Rodionov V., Han D. K. (2009) Quantitative phosphoproteomic analysis of T cell receptor signaling reveals system-wide modulation of protein-protein interactions. Sci. Signal. 2, ra46. [DOI] [PubMed] [Google Scholar]
- 15. Carrascal M., Ovelleiro D., Casas V., Gay M., Abian J. (2008) Phosphorylation analysis of primary human T lymphocytes using sequential IMAC and titanium oxide enrichment. J. Proteome Res. 7, 5167–5176 [DOI] [PubMed] [Google Scholar]
- 16. Iwai L. K., Benoist C., Mathis D., White F. M. (2010) Quantitative phosphoproteomic analysis of T cell receptor signaling in diabetes prone and resistant mice. J. Proteome Res. 9, 3135–3145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Moulder R., Lönnberg T., Elo L. L., Filén J. J., Rainio E., Corthals G., Oresic M., Nyman T. A., Aittokallio T., Lahesmaa R. (2010) Quantitative proteomics analysis of the nuclear fraction of human CD4+ cells in the early phases of IL-4-induced Th2 differentiation. Mol. Cell. Proteomics 9, 1937–1953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pinkse M. W., Uitto P. M., Hilhorst M. J., Ooms B., Heck A. J. (2004) Selective isolation at the femtomole level of phosphopeptides from proteolytic digests using 2D-NanoLC-ESI-MS/MS and titanium oxide precolumns. Anal. Chem. 76, 3935–3943 [DOI] [PubMed] [Google Scholar]
- 19. Larsen M. R., Thingholm T. E., Jensen O. N., Roepstorff P., Jørgensen T. J. (2005) Highly selective enrichment of phosphorylated peptides from peptide mixtures using titanium dioxide microcolumns. Mol. Cell. Proteomics 4, 873–886 [DOI] [PubMed] [Google Scholar]
- 20. Trinidad J. C., Specht C. G., Thalhammer A., Schoepfer R., Burlingame A. L. (2006) Comprehensive identification of phosphorylation sites in postsynaptic density preparations. Mol. Cell. Proteomics 5, 914–922 [DOI] [PubMed] [Google Scholar]
- 21. Rush J., Moritz A., Lee K. A., Guo A., Goss V. L., Spek E. J., Zhang H., Zha X. M., Polakiewicz R. D., Comb M. J. (2005) Immunoaffinity profiling of tyrosine phosphorylation in cancer cells. Nat. Biotechnol. 23, 94–101 [DOI] [PubMed] [Google Scholar]
- 22. Gilar M., Olivova P., Daly A. E., Gebler J. C. (2005) Two-dimensional separation of peptides using RP-RP-HPLC system with different pH in first and second separation dimensions. J. Sep. Sci. 28, 1694–1703 [DOI] [PubMed] [Google Scholar]
- 23. Villén J., Gygi S. P. (2008) The SCX/IMAC enrichment approach for global phosphorylation analysis by mass spectrometry. Nat. Protoc. 3, 1630–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Schwartz D., Gygi S. P. (2005) An iterative statistical approach to the identification of protein phosphorylation motifs from large-scale data sets. Nat. Biotechnol. 23, 1391–1398 [DOI] [PubMed] [Google Scholar]
- 25. Amanchy R., Periaswamy B., Mathivanan S., Reddy R., Tattikota S. G., Pandey A. (2007) A curated compendium of phosphorylation motifs. Nat. Biotechnol. 25, 285–286 [DOI] [PubMed] [Google Scholar]
- 26. Biarc J., Chalkley R. J., Burlingame A. L., Bradshaw R. A. (2011) The induction of serine/threonine protein phosphorylations by a PDGFR/TrkA chimera in stably transfected PC12 cells. Mol. Cell. Proteomics 10.1074/mcp.M111.013375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Birkenfeld J., Nalbant P., Yoon S. H., Bokoch G. M. (2008) Cellular functions of GEF-H1, a microtubule-regulated Rho-GEF: Is altered GEF-H1 activity a crucial determinant of disease pathogenesis? Trends Cell Biol. 18, 210–219 [DOI] [PubMed] [Google Scholar]
- 28. van Duijn T. J., Anthony E. C., Hensbergen P. J., Deelder A. M., Hordijk P. L. (2010) Rac1 recruits the adapter protein CMS/CD2AP to cell-cell contacts. J. Biol. Chem. 285, 20137–20146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ammer A. G., Weed S. A. (2008) Cortactin branches out: Roles in regulating protrusive actin dynamics. Cell Motil. Cytoskeleton 65, 687–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ambrose J. C., Wasteneys G. O. (2008) CLASP modulates microtubule-cortex interaction during self-organization of acentrosomal microtubules. Mol. Biol. Cell 19, 4730–4737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Didier C., Merdes A., Gairin J. E., Jabrane-Ferrat N. (2008) Inhibition of proteasome activity impairs centrosome-dependent microtubule nucleation and organization. Mol. Biol. Cell 19, 1220–1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tse H. M., Thayer T. C., Steele C., Cuda C. M., Morel L., Piganelli J. D., Mathews C. E. (2010) NADPH oxidase deficiency regulates Th lineage commitment and modulates autoimmunity. J. Immunol. 185, 5247–5258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Goldberg M. R., Luknar-Gabor N., Zadik-Mnuhin G., Koch P., Tovbin J., Katz Y. (2007) Synergy between LPS and immobilized anti-human CD3ε mAb for activation of cord blood CD3+ T cells. Int. Immunol. 19, 99–103 [DOI] [PubMed] [Google Scholar]
- 34. Shan X., Balakir R., Criado G., Wood J. S., Seminario M. C., Madrenas J., Wange R. L. (2001) Zap-70-independent Ca2+ mobilization and Erk activation in Jurkat T cells in response to T-cell antigen receptor ligation. Mol. Cell. Biol. 21, 7137–7149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rebeaud F., Hailfinger S., Posevitz-Fejfar A., Tapernoux M., Moser R., Rueda D., Gaide O., Guzzardi M., Iancu E. M., Rufer N., Fasel N., Thome M. (2008) The proteolytic activity of the paracaspase MALT1 is key in T cell activation. Nat. Immunol. 9, 272–281 [DOI] [PubMed] [Google Scholar]
- 36. Oxley C. L., Anthis N. J., Lowe E. D.., Vakonakis I., Campbell I. D., Wegener K. L. (2008) An integrin phosphorylation switch: The effect of β3 integrin tail phosphorylation on Dok1 and talin binding. J. Biol. Chem. 283, 5420–5426 [DOI] [PubMed] [Google Scholar]
- 37. Das V., Nal B., Roumier A., Meas-Yedid V., Zimmer C., Olivo-Marin J. C., Roux P., Ferrier P., Dautry-Varsat A., Alcover A. (2002) Membrane-cytoskeleton interactions during the formation of the immunological synapse and subsequent T-cell activation. Immunol. Rev. 189, 123–135 [DOI] [PubMed] [Google Scholar]
- 38. Le Bras S., Foucault I., Foussat A., Brignone C., Acuto O., Deckert M. (2004) Recruitment of the actin-binding protein HIP-55 to the immunological synapse regulates T cell receptor signaling and endocytosis. J. Biol. Chem. 279, 15550–15560 [DOI] [PubMed] [Google Scholar]
- 39. Bunnell S. C., Kapoor V., Trible R. P., Zhang W., Samelson L. E. (2001) Dynamic actin polymerization drives T cell receptor-induced spreading: A role for the signal transduction adaptor LAT. Immunity 14, 315–329 [DOI] [PubMed] [Google Scholar]
- 40. Varma R., Campi G., Yokosuka T., Saito T., Dustin M. L. (2006) T cell receptor-proximal signals are sustained in peripheral microclusters and terminated in the central supramolecular activation cluster. Immunity 25, 117–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Alarcón B., Mestre D., Martínez-Martín N. (2011) The immunological synapse: A cause or consequence of T-cell receptor triggering? Immunology 133, 420–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Treanor B., Depoil D., Gonzalez-Granja A., Barral P., Weber M., Dushek O., Bruckbauer A., Batista F. D. (2010) The membrane skeleton controls diffusion dynamics and signaling through the B cell receptor. Immunity 32, 187–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Reicher B., Barda-Saad M. (2010) Multiple pathways leading from the T-cell antigen receptor to the actin cytoskeleton network. FEBS Lett. 584, 4858–4864 [DOI] [PubMed] [Google Scholar]
- 44. Critchley D. R. (2000) Focal adhesions: The cytoskeletal connection. Curr. Opin. Cell Biol. 12, 133–139 [DOI] [PubMed] [Google Scholar]
- 45. Ilani T., Vasiliver-Shamis G., Vardhana S., Bretscher A., Dustin M. L. (2009) T cell antigen receptor signaling and immunological synapse stability require myosin IIA. Nat. Immunol. 10, 531–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Cunningham N. R., Hinchcliff E. M., Kutyavin V. I., Beck T., Reid W. A., Punt J. A. (2011) GSK3-mediated instability of tubulin polymers is responsible for the failure of immature CD4+CD8+ thymocytes to polarize their MTOC in response to TCR stimulation. Int. Immunol. 23, 693–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kuhné M. R., Lin J., Yablonski D., Mollenauer M. N., Ehrlich L. I., Huppa J., Davis M. M., Weiss A. (2003) Linker for activation of T cells, zeta-associated protein-70, and Src homology 2 domain-containing leukocyte protein-76 are required for TCR-induced microtubule-organizing center polarization. J. Immunol. 171, 860–866 [DOI] [PubMed] [Google Scholar]
- 48. Dustin M. L., Chakraborty A. K., Shaw A. S. (2010) Understanding the structure and function of the immunological synapse. Cold Spring Harb. Perspect Biol. 2, a002311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Medeiros R. B., Dickey D. M., Chung H., Quale A. C., Nagarajan L. R., Billadeau D. D., Shimizu Y. (2005) Protein kinase D1 and the β1 integrin cytoplasmic domain control β1 integrin function via regulation of Rap1 activation. Immunity 23, 213–226 [DOI] [PubMed] [Google Scholar]
- 50. Smith F. D., Langeberg L. K., Cellurale C., Pawson T., Morrison D. K., Davis R. J., Scott J. D. (2010) AKAP-Lbc enhances cyclic AMP control of the ERK1/2 cascade. Nat. Cell Biol. 12, 1242–1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Michel J. J., Scott J. D. (2002) AKAP mediated signal transduction. Annu. Rev. Pharmacol. Toxicol. 42, 235–257 [DOI] [PubMed] [Google Scholar]
- 52. Mosenden R., Taskén K. (2011) Cyclic AMP-mediated immune regulation–overview of mechanisms of action in T cells. Cell Signal. 23, 1009–1016 [DOI] [PubMed] [Google Scholar]
- 53. Lovatt M., Bijlmakers M. J. (2010) Stabilisation of β-catenin downstream of T cell receptor signalling. PLoS One 5, e12794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Chung E. J., Hwang S. G., Nguyen P., Lee S., Kim J. S., Kim J. W., Henkart P. A., Bottaro D. P., Soon L., Bonvini P., Lee S. J., Karp J. E., Oh H. J., Rubin J. S., Trepel J. B. (2002) Regulation of leukemic cell adhesion, proliferation, and survival by β-catenin. Blood 100, 982–990 [DOI] [PubMed] [Google Scholar]