Abstract
Nucleocytoplasmic transport occurs through the nuclear pore complex (NPC), which in yeast is a ∼50 MDa complex consisting of ∼30 different proteins. Small molecules can freely exchange through the NPC, but macromolecules larger than ∼40 kDa must be aided across by transport factors, most of which belong to a related family of proteins termed karyopherins (Kaps). These transport factors bind to the disordered phenylalanine-glycine (FG) repeat domains in a family of NPC proteins termed FG nups, and this specific binding allows the transport factors to cross the NPC. However, we still know little in terms of the molecular and kinetic details regarding how this binding translates to selective passage of transport factors across the NPC. Here we show that the specific interactions between Kaps and FG nups are strongly modulated by the presence of a cellular milieu whose proteins appear to act as very weak competitors that nevertheless collectively can reduce Kap/FG nup affinities by several orders of magnitude. Without such modulation, the avidities between Kaps and FG nups measured in vitro are too tight to be compatible with the rapid transport kinetics observed in vivo. We modeled the multivalent interactions between the disordered repeat binding sites in the FG nups and multiple cognate binding sites on Kap, showing that they should indeed be sensitive to even weakly binding competitors; the introduction of such competition reduces the availability of these binding sites, dramatically lowering the avidity of their specific interactions and allowing rapid nuclear transport.
Eukaryotic cells segregate their genetic material with a double-layered nuclear envelope, which protects the genomic DNA of the cell and enables highly regulated gene expression but also creates a barrier that a wide range of biomolecules must cross to maintain cell viability. All known transport in and out of the nucleus occurs through nuclear pore complexes (NPCs)1 embedded in the nuclear envelope. Although NPCs can be permeable to molecules below ∼9 nm in diameter (1), macromolecules and macromolecular complexes are typically actively carried across by transport factors. One family of such transport factors is the karyopherins (Kaps). Kaps can shuttle through the NPC on their own (2) or laden with cargo to be imported or exported (3–12) (reviewed in Refs. 13–15). Kaps interact with their cargo in a RanGTP-dependent manner, binding and releasing transported molecules on the appropriate side of the nuclear envelope (3–12) (reviewed in Refs. 13–15). The selectively permeable barrier of the NPC is formed by a family of proteins containing characteristic disordered phenylalanine-glycine (FG) repeats, termed FG nups (reviewed in Refs. 16–20), which surround and line the central channel of the NPC (21, 22) and which have been the focus of much study (2, 9, 13, 17, 23–28). These proteins interact specifically with Kaps and other transport factors, through multivalent interactions between the FG repeats and multiple binding sites along each transport factor (29–47). It has recently been shown in artificial nanoporous NPC mimics that, as long suspected, the interaction between transport factors and FG repeats is sufficient for transport (48–51); thus, a key to understanding transport through the NPC lies in understanding the interactions between FG nups and transport factors.
The FG repeat regions of FG nups have no stable structure and thus, like other such intrinsically disordered proteins (IDPs), are refractory to many conventional structure-based approaches (52–55). IDPs are extremely prevalent in eukaryotes: ∼30% of the eukaryotic proteome is predicted to contain significant intrinsically disordered domains (56). Much has been learned about these IDPs (recently reviewed in Refs. 52 and 57–61), which are often involved in interactions with multiple partners that require high specificity and low affinity (i.e. fast off-rates), such as those involved in signaling (52). Thus, this family of proteins is well suited for the job of gating transport across the NPC, a process that requires numerous rapid yet specific interactions. IDPs, however, are often difficult to purify (37) and can be very sensitive to buffer conditions (62). Thus, connecting in vitro observations to in vivo behavior and the function of IDPs is not straightforward.
Transport through the NPC occurs rapidly, with ∼1000 transport events per pore per second (23) and individual Kap/cargo complexes crossing the NPC in <30 ms (63, 64). Even large mRNPs can cross the central channel of the NPC in 5–20 ms (65). During this time, individual Kaps may interact with as many as 40 FG nups (13, 23). Therefore, interactions between Kaps and FG nups are expected to be very weak, with rapid on-off rates and predicted affinities on the order of micromolar to millimolar (13, 23). Crystal structures (31–34, 66) and molecular dynamic simulations (41, 42, 67) indicate that these interactions occur via the phenylalanine side chains of FG nups inserting into hydrophobic pockets on the surface of Kaps. It is suggested that the multiple binding sites on each Kap are appropriately spaced to enable binding from multiple phenylalanines from the same FG repeat region (41, 42, 67). Measurements typically probe the avidity resulting from the combined binding strengths of these multiple simultaneous interaction sites rather than the affinity between individual phenylalanines and the Kap binding pockets.
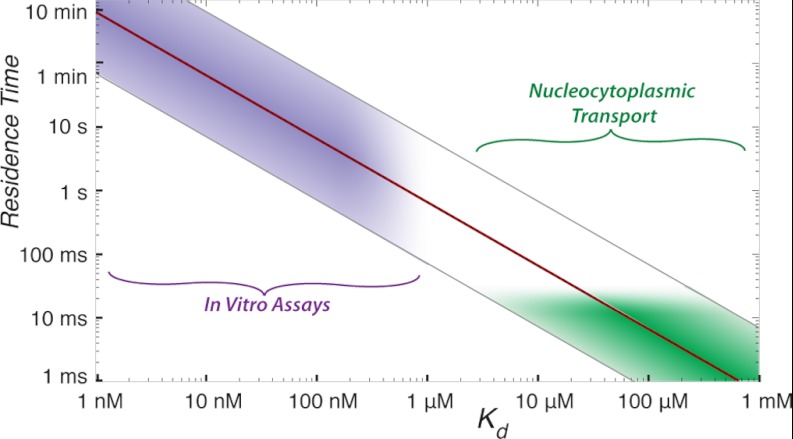
Unfortunately, there exists a fundamental problem in our understanding of nucleocytoplasmic transport. Reported Kap/FG nup affinities, measured in vitro, range from <51 pm (37) to ∼2 μm (68), with an average reported affinity of 270 nm (36, 37, 68, 69). These measured affinities are all too tight to explain the rapid transport through NPCs observed in vivo for two reasons. First, yeast contains 14 Kaps and other transport factors, at a total concentration of ∼20 μm (68, 70–72). These Kap concentrations are several hundred fold higher than the measured in vitro affinities between Kaps and FG nups, which would mean that FG domains in living cells are completely saturated with transport factors, a situation not seen in vivo (73). Second, given a diffusion-limited on-rate, an interaction with a 1 nm Kd would exhibit an off-rate of ∼0.1 s−1, which is many orders of magnitude too slow to explain the ∼30-ms transport times observed in vivo (23, 63, 64) (Fig. 1). Thus, the in vitro measurements reported in the literature seem incompatible with transport rates seen in vivo.
Fig. 1.
Current in vitro measurements are inconsistent with nucleocytoplasmic transport. Relationship between Kd and half-life of a binary complex (red line). Physiological on-rates for protein-protein interactions usually range from 104 to 106 m−1 s−1 (gray lines flanking red line). However, the measurements of Kd between Kaps and FG nups typically fall in the low nanomolar range, giving a range half-lives (purple region) that are inconsistent with half-lives expected for known transport rates of ∼30 ms (green region).
Here, we sought to investigate this issue. We first sought to determine whether the tight binding affinities measured were dependent on the assay chosen or on the particular subset of proteins studied. We therefore systematically surveyed binding strengths between three different Kaps and ten FG nups from the model organism, Saccharomyces cerevisiae (yeast) using two distinct binding assays: a bead binding and an overlay assay. Our results indicated that in isolation, Kap/FG nup interactions are indeed generally very tight. However, we also show a surprising dependence of nucleocytoplasmic transport kinetics on environmental factors not previously experimentally studied.
EXPERIMENTAL PROCEDURES
Assay Design: The Bead Binding Assay
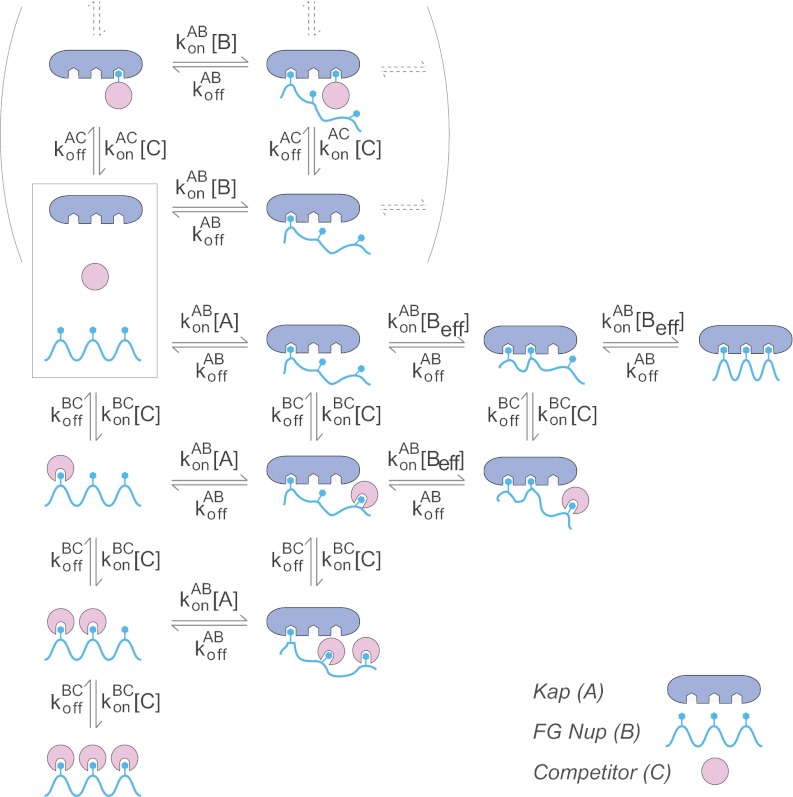
We chose to study the simplest possible transport reaction consisting of the fewest components necessary to support specific nucleocytoplasmic transport. Because transport factors have been shown to readily cross NPCs and artificial NPC mimics on their own (74–77), any kinetic explanation of nuclear transport must be able to account for the facilitated translocation of empty transport factors through a channel filled with FG repeats, without invoking any kind of modulation by cargo or Ran. We therefore chose to study the binding of unladen Kaps to FG nups. We designed our main assay (Fig. 2) to recapitulate many features of the interactions between Kaps and FG nups in the context of the NPC and explored different environments trying to best mimic the actual conditions faced by these proteins in the NPC.
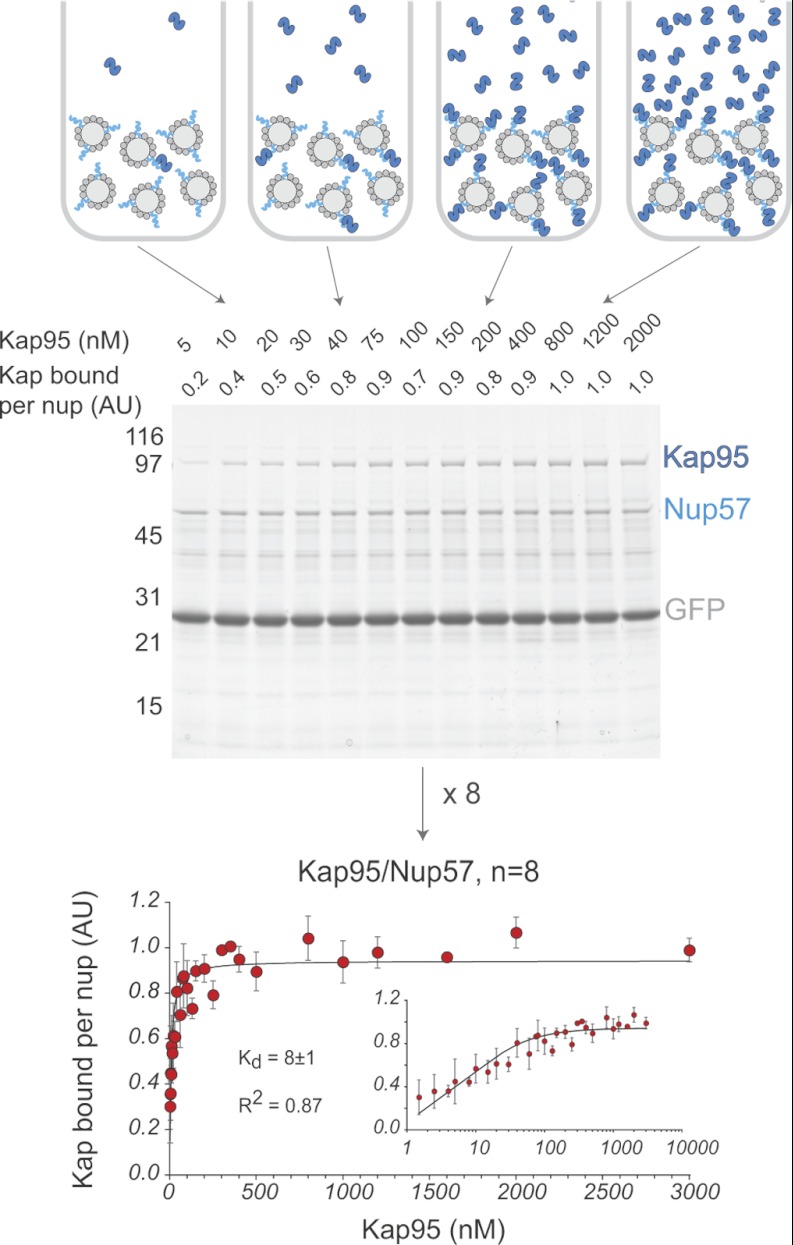
Fig. 2.
Measurement of Kap/FG nup affinities in vitro. Top panel, schematic of the bead binding assay. Dynabeads TALON (light gray) are saturated with His-tagged FG nups (turquoise) and GFP (dark gray). The beads are then incubated with increasing concentrations of Kap (dark blue). Middle panel, protein eluted from the beads is run on an SDS-PAGE gel and stained with Coomassie Blue. Band intensities are quantified by densitometry. Quantification is shown on the top of the gel. Molecular mass markers (in kilodaltons) are shown to the left. Bottom panel, the normalized and averaged data from eight measurements of the Kap95/Nup57 interaction are plotted (error bars showing standard deviation), and the best fit as calculated by Sigma Plot using Equation 1 is shown as a black line. The inset shows the data on a log/linear scale.
In vivo, Kaps freely move about the nucleus and cytoplasm, whereas one end of each FG nup is anchored to the inner surface of the central channel of the NPC. To mimic this surface attachment of FG nups, as our main assay, we used a binding assay in which Kaps were free in solution, whereas His-tagged FG nups were bound to the surface of magnetic beads. Excess His-tagged GFP was bound to the beads along with the FG nup to ensure that all His-binding sites on the beads were occupied. We incubated the immobilized FG nup and GFP with at least a 3-fold molar excess of Kap (to ensure, as in vivo, the Kap is never limiting), over a subnanomolar to micromolar range of Kap concentrations. To simulate the well studied interactions involved in transport of unladen Kaps (78), neither cargo nor Ran was present in these binding reactions. After the binding reaction reached equilibrium, the beads were washed very quickly to remove unbound material, and bound proteins were eluted. The elutions were run on an SDS-PAGE gel, and Coomassie Blue-stained bands corresponding to the Kap and FG Nup were quantified by densitometry. Each Kap band was normalized to the corresponding FG nup band, and the results (Kapbound, in arbitrary units) were plotted against Kap concentration ([Kap], in nm) (Fig. 2). To combine results from multiple repeats, each curve was normalized to 100% maximum binding, and the normalized intensities were averaged across all repeats. The normalized, averaged curve was then fit to determine a measure of avidity (Kd) using Equation 1 (79),
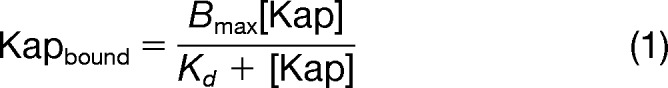 |
where Kapbound is the normalized Coomassie intensity of the Kap band at each concentration of Kap; Bmax is the value to which the curve saturates, the maximum normalized Coomassie intensity of the Kap band; [Kap] is the concentration of Kap; and Kd is the resulting avidity.
Choice of Proteins
We produced milligram quantities of full-length Kaps and FG nups by specific modifications of bacterial expression techniques, greatly increasing the yield of protein over purification of endogenous or even overexpressed protein from yeast. Bacterially expressed Kaps and FG nups have previously been shown to be functional and to recapitulate many of the key behaviors of the native proteins. For example, like the endogenous proteins, bacterially expressed FG nups are natively unfolded (24, 27, 28) and interact specifically with Kaps and other transport factors (2, 36, 68, 80–83). Indeed, these recombinant proteins have even been shown to mimic selective nuclear transport through a functionalized nanopore (48, 49). Correspondingly, all of the recombinant Kaps and FG nups used in this work were shown to be fully competent for specific binding (supplemental Figs. 1 and 2). We studied three of the best characterized Kaps: Kap95, Kap123, and Kap121 (8, 34, 37, 40, 43, 45, 68, 72, 83, 85–90). Unless otherwise noted, all FG nup constructs used in this study contained the full amino acid sequence, including the NPC attachment domain and the natively unfolded FG domain(s). All of the yeast FG nups were included in this survey except for Nup2, Nup116, and Nup159.
Recombinant Protein Expression and Purification
Plasmids encoding Kap95 in pGEX-2TK and Kap121 in pGEX-4T1 were a kind gift from John Aitchison (see supplemental Table 1 for a list of Kap plasmids). Kap123 was codon-optimized for expression in bacteria (GenScript) and cloned into the BamHI site of pGEX-Tev (a kind gift from Yuh Min Chook). These plasmids were transformed into BL21DE3 pLys or RIL cells and grown with the following protocol. Growth medium consisted of TB (12.5 g/L tryptone, 25.5 g/L yeast extract, 5.6 g/L glycerol) and TBB (11.5 g of KH2PO4 and 62.5 g of K2HPO4 per 500 ml). One colony of cells was inoculated into 27 ml of TB + 3 ml of TBB + antibiotics and incubated overnight shaking at 37 °C. The 30-ml culture was spun down, resuspended in 45 ml of TB + 5 ml of TBB + antibiotics and incubated shaking at 37 °C for 8 h. The 50-ml culture was spun down, resuspended into 1–2 ml of TB, and inoculated into 900 ml of TB + 100 ml of TBB + antibiotics. This 1-liter culture was incubated with shaking at 37 °C for 5 h. The cells were then induced with 1 mm isopropyl 1-thio-β-d-galactopyranoside and incubated for 16 h at 30 °C (for Kap95 and Kap121) or 37 °C (for Kap123). The cells were pelleted and stored at −80 °C until ready for use.
Cell pellets from 1 liter of culture were thawed and resuspended in 20 ml of TBT buffer (20 mm Hepes, pH 7.4, 110 mm KOAc, 2 mm MgCl2, 0.1% Tween 20, 10 μm CaCl2, 10 μm ZnCl2) plus 2 mm DTT, 18 mg/liter PMSF, and 0.4 mg/liter pepstatin A. The cells were lysed by passing several times through a Microfluidizer® (Microfluidics), and the lysate was clarified by spinning 90 min at 40,000 rpm (145,000 × gav) in a Ti50.2 rotor (Beckman) at 4 °C. The lysate was then filtered through a 0.22-μm filter and incubated with 25–50 ml of glutathione-Sepharose Fast Flow (GE Healthcare) or 10 ml of ProCatch Glutathione Resin (Miltenyi Biotech) for 2 h at 4 °C. The resin was then washed with five bed volumes of cold (4 °C) TBT + 1 mm DTT + 18 mg/liter PMSF + 0.4 mg/liter pepstatin A; once with five bed volumes of cold (4 °C) TBT + 0.5–1 m NaCl + 1 mm DTT + 18 mg/liter PMSF + 0.4 mg/liter pepstatin A; once with five bed volumes of room temperature TBT + 100 μm ATP + 1 mm DTT + 18 mg/liter PMSF + 0.4 mg/liter pepstatin A; and once with five bed volumes of cold (4 °C) TBT + 1 mm DTT (no protease inhibitors). For Kap95 and Kap121, the GST-Kaps were eluted from the glutathione-Sepharose resin with 10 mm reduced glutathione (Sigma) in 100 mm Tris, pH 7.4. Buffer was changed by dialysis into TBT without glutathione, and the GST was cleaved off using the thrombin cleavage capture kit (Novagen) following the manufacturer's instructions. Kap123 was eluted by on-resin overnight cleavage of the GST using Tev protease (purified as a His-tagged fusion (91)) in 20 mm Tris, pH 7.5, 150 mm NaCl, 20 mm B-mercaptoethanol + 0.1 mm EDTA + 20% glycerol at 4 °C. Flow-through containing cleaved Kap123 and His-Tev was collected and incubated with TALON-Sepharose to remove the His-Tev. The final Kap123 was dialyzed into TBT. Kaps were stored at −80 °C for long term storage or at 4 °C for <1 month. Kaps were centrifuged for 2 h at 55,000 rpm (112,000 × gav) in a TLA55 rotor (Beckman) at 4 °C immediately prior to use to ensure that no microaggregates were present.
The genes encoding FG nups were amplified from either genomic DNA or a plasmid containing the gene and inserted into vectors of the pET family. The His6 tag was positioned on the structured anchor end of the FG nups (92). The plasmid encoding the non-FG domain of Nup100 (amino acids 571–960) was a kind gift from Josef Franke. These plasmids were then transformed into BL21DE3 Gold cells, which were found to give the best expression with minimal proteolysis of the expressed proteins. Expression conditions were determined for each individual protein, varying induction times from 1–16 h and induction temperatures from 22–37 °C. Some FG nups cleaved in vivo at all of the expression conditions tested. However, many of these nups expressed full-length when coexpressed with Kap95. Nups in a kanamycin-resistant plasmid were coexpressed with Kap95 in pGEX-2TK, whereas nups in an ampicillin-resistant plasmid were coexpressed with Kap95 in pET24. After the appropriate induction, the cells were harvested and stored at −80 °C until ready for purification (see supplemental Table 1 for FG nup plasmids and supplemental Table 2 for FG nup expression conditions).
Cell pellets were defrosted and resuspended in four volumes of lysis buffer (50 mm NaH2PO4, pH 8, 300 mm NaCl) plus 8 m urea + 18 mg/liter PMSF + 0.4 mg/liter pepstatin A. The urea was necessary to keep the FG nups soluble and should not interfere with their function because the FG domains are natively unfolded, and we directly tested that the non-FG portion of Nup100 does not directly interact with Kap95 (supplemental Fig. 5) (24). The cells were lysed by passing several times through a Microfluidizer® (Microfluidics). The lysine mimic 6-amino-n-caproic-acid (Sigma) was added to a final concentration of 5 mg/ml to minimize proteolysis, and cell debris was spun down in a Ti50.2 Rotor (Beckman), 40,000 rpm at room temperature. The supernatant was put through a 0.22-μm filter, and the clarified lysate was incubated for >2 h at room temperature with TALON-Sepharose resin (Clontech). The resin was washed with more than five bed volumes of lysis buffer + 0.8 m urea + 18 mg/liter PMSF + 0.4 mg/liter pepstatin A, and protein was eluted in lysis buffer + 0.8 m urea + 18 mg/liter PMSF + 0.4 mg/liter pepstatin A + 150 mm imidazole. Glycerol was added to 10%, and the enriched FG nups were stored at −80 °C until ready for use.
Preparation of His-depleted Escherichia coli and Yeast Lysate
1 liter of BL21 DE3 RIL cells with were grown in LB to an A600 of 0.8 and incubated for 16 h at 37 °C. 1 liter of BL21 DE3 RIL cells containing the plasmid Kap95 in pET24a (to express untagged Kap95) were grown in LB to an A600 of 0.8, induced with 1 mm isopropyl 1-thio-β-d-galactopyranoside, and incubated for 4 h at 30 °C. The cells were harvested, resuspended in TBT + 18 mg/liter PMSF + 0.4 mg/liter pepstatin A, and lysed by passing through a Microfluidizer. 6 liters of wild-type DF5α yeast were grown in YPD to log phase at 30 °C. The cells were harvested and mechanically lysed using a planetary ball mill as described (93). Yeast powder was resuspended in ∼4 volumes of TBT + 18 mg/liter PMSF + 0.4 mg/liter pepstatin A. Yeast and bacterial lysates were clarified with a 90-min spin at 145,000 × gav in a Ti50.2 rotor (Beckman) at 4 °C. To ensure that no nonspecific interactions to the Dynabeads TALON would occur, this clarified lysate was incubated with 3 ml of fresh TALON-Sepharose three times. The lysate was then stored at 4 °C until ready for use. Immediately before each use, the lysate was passed through a 0.22-μm filter, and an additional 18 mg/liter PMSF + 0.4 mg/liter pepstatin A was added.
Bead Binding Assay to Measure Kap/FG Nup Affinity
Dynabeads TALON (Dynal Biotech; Invitrogen) or Dynabeads His tag isolation and pulldown (Invitrogen) were equilibrated in TALON wash buffer (50 mm NaH2PO4, 300 mm NaCl, 0.1% Tween 20). 0.5 mg of beads was used per data point, i.e. 6.5 mg of beads for a 13-point curve. The beads were then resuspended in TALON wash buffer + 0.8 m urea + 25% saturated ammonium sulfate + 18 mg/liter PMSF + 0.4 mg/liter pepstatin A, and protein was added as appropriate. For our standard assays, for each data point 15–50 pmol of FG nup and 25 μg of GFP-His6 were combined and added to 0.5 mg of beads. The large amount of GFP ensured that no empty TALON surface remained. Based on Coomassie Blue staining intensity of the bead complete eluate (analyzed by SDS-PAGE), we estimate that ∼1 μg of FG nup was bound for each data point. The density of the beads was ∼2.5 × 108 beads/mg, and each bead had a surface area of 3.8 μm2 (Invitrogen; Dynabead product information). Therefore under these conditions, each FG nup was separated from its neighbor by ∼6.6 nm. For dilute conditions, three times as many beads and three times as much GFP were used, such that each FG nup was separated from its neighbor by ∼9 nm. For crowded assays, (supplemental Fig. 12) the beads were separated into two aliquots. The appropriate amount of FG nup was incubated with Dynabeads such that there was 1 μl of Dynabeads/μg protein. The remaining beads were saturated with GFP. The two aliquots were washed separately and then mixed together, so that the FG nups and GFP each saturated a subset of those beads. Under these conditions, each FG nup was separated from its neighbor by ∼2 nm. In all cases, the beads were incubated with the protein for >10 min at room temperature to bind the His-tagged proteins to the beads. The beads were then washed three times with TALON wash buffer + 18 mg/liter PMSF + 0.4 mg/liter pepstatin A, combined if necessary (for “crowded”), and divided equally among tubes of Kap solutions of various concentrations. Kap solutions were made in TBT + 0.3% PVP (a blocking agent (94)) + 18 mg/liter PMSF + 0.4 mg/liter pepstatin A (plus competitor proteins when appropriate), and the volumes were adjusted so that at all concentrations there was always greater than 3-fold molar excess of Kap over nup. To ensure adequate mixing, the volumes were never below 0.3 ml for Dynabeads TALON and never below 3 ml for Dynabeads His tag isolation and pulldown. The beads were incubated with Kap solutions overnight on a rotating wheel at 4 °C. After incubation, the beads from each data point were washed twice with 1 ml of TBT + 18 mg/liter PMSF + 0.4 mg/liter pepstatin A and resuspended in 20 μl of SDS-PAGE sample buffer. The beads were heated for 10 min to 95 °C, and 15 μl of the supernatant was loaded on a Novex 4–20% TG gel. The gels were stained with Coomassie Blue R-250, the images were digitized, and background-subtracted band intensities were quantified using ImageJ (95). Kd values were calculated by fitting the resulting curves to Equation 1 in SigmaPlot (Systat Software).
The distribution of FG nup and GFP on the beads was confirmed by fluorescence microscopy. The FG domain of Nsp1, Nsp1FG-Cys-His6, was expressed and purified as described previously (48), dialyzed into PBS, and labeled with Alexa 633 (Invitrogen) according to the manufacturer's instructions. The beads were prepared as described above for visual examination. The images were collected with a Hamamatsu C4742-95 cooled CCD camera attached to a Zeiss Axioplan 2 microscope, using a 63× objective lens (NA 1.4). GFP and Alexa 633 were imaged using FITC and CY5 excitation/dichroic/emission filter sets, respectively, by Chroma. The images were acquired, colorized, and combined using OpenLab microscopy software (Improvision). The linear contrast was optimized using Photoshop (Adobe), without altering the gamma values.
Overlay Assay
As FG nups are natively unfolded (24), they can be run on a denaturing SDS-PAGE gel, transferred to nitrocellulose, and still be recognized by Kaps (46, 47, 88, 96). We took advantage of this ability to probe the interactions between transport factors and FG nups. For these assays, enriched Kaps were dialyzed into PBS buffer and labeled with Alexa Fluor 488 or 633 (Invitrogen) according to the manufacturer's instructions. FG nups were diluted to 0.05 mg/ml in SDS-PAGE sample buffer and heated for 10 min at 95 °C. 10 μl (0.5 μg) was loaded per lane in 10-well 4–20% Novex TG gels (Invitrogen). The proteins were transferred to nitrocellulose overnight at 35 V using standard procedures. The membranes were stained with Ponceau S to mark the location of the FG nup bands, dried, and cut into strips with two gel lanes per strip. Strips were blocked with 2 ml of TBT + 0.3% PVP for >1 h at room temperature. The strips were then incubated overnight in the dark on a rocker at 4 °C with solutions of fluorescent Kaps (48). Kap solutions were made in TBT + 0.3% PVP, and volumes were adjusted so that there was always greater than 3-fold molar excess of Kap over FG nup. To ensure proper coverage and agitation of the membrane strip, the volume of the Kap-containing solution was never less than 900 μl. The membranes were washed with TBT + 0.3% PVP three times quickly and then three times for 5 min each. The strips were then scanned at on a Typhoon 9400 variable imager (Amersham Biosciences, Rockefeller University Proteomics Resource Center) at the appropriate settings for Alexa 488 or 633. Band intensities were quantified in ImageJ as above.
Additional Validation Experiments
Control experiments indicated that Kap does not bind to beads coated with only GFP; therefore the binding observed was specifically between Kaps and FG nups (supplemental Fig. 3). We ensured that there was no bias from the gel loading order (supplemental Fig. 4). It has been shown that the FG repeat domains of the FG nups are necessary and sufficient for interaction with Kaps (19, 37, 80–82). Our findings agreed with this; full-length Nup100 and the isolated FG domain interact similarly with Kap95 (supplemental Fig. 5). To further ensure that the urea did not lead to nonspecific binding of Kaps to the normally structured anchor domains of the FG nups (92), we measured Kap binding to the isolated non-FG domain of Nup100. Although Kaps bound to full-length Nup100, they showed no significant binding to the non-FG portions of Nup100 when it was purified under either native or denaturing conditions (supplemental Fig. 5). Therefore, the binding observed in these assays appears to be specifically between Kaps and just the natively unfolded FG domains of FG nups.
We tested different incubation periods in our assay and found that the assays were not sensitive to incubation time. Binding between Kap/FG nup starts rapidly and saturates after several hours. As expected, although the total amount of Kap binding increases with time, the apparent Kd did not change (supplemental Fig. 6). However, to ensure that the binding reactions had reached equilibrium and to maximize the signal, Kaps and FG nups were allowed to bind overnight. Controls using fluorescently tagged FG nups indicated that there was minimal mixing of proteins from bead to bead during the incubation with Kap (supplemental Fig. 7).
RESULTS
To address the apparent discrepancy between in vitro and in vivo measurements of Kap/FG nup interactions (Fig. 1), we set out to systematically measure the apparent affinities between three representative Kaps and ten yeast FG nups. To ensure there was no bias because of the experimental design, we used two different well established assays to measure these affinities: a bead binding assay (Fig. 2) (37, 68, 82) and a far Western assay (88). A detailed description of both assays and their controls can be found under “Experimental Procedures.”
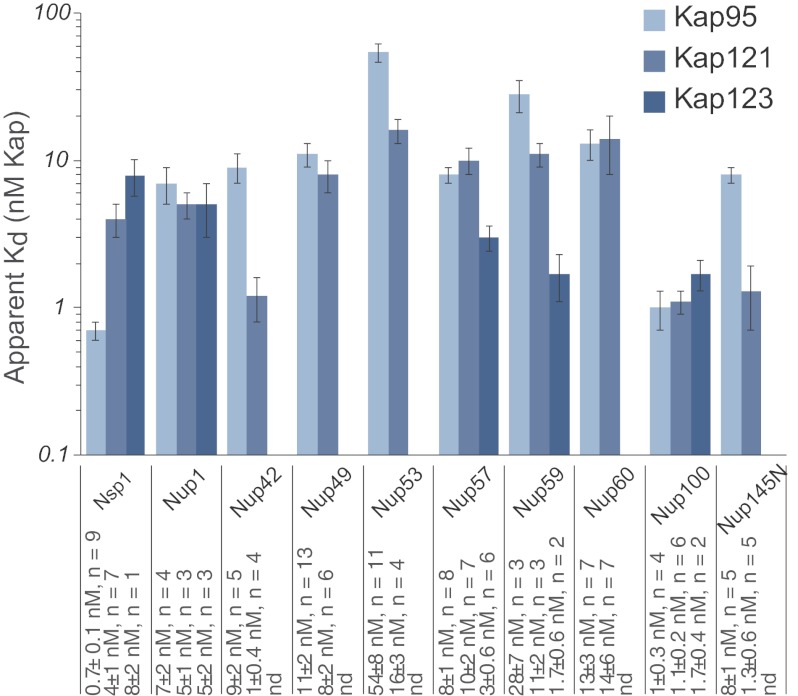
The apparent affinities between Kap95, Kap121, and Kap123 with the 10 FG nups are presented in Figs. 3 and 4 and supplemental Table 1 and Figs. 8 and 9 and are consistent in range with previous findings (36, 37, 68, 69). These measurements were robust to the presence or absence of reducing agent and a range of salt (0–150 mm) or detergent (0.01–0.1%) concentrations (supplemental Table 1). Importantly, all of the experiments were done in the presence of excess of blocking agents (PVP and Tween 20) (94), and we correspondingly saw no nonspecific binding to beads with no FG nup. Moreover, these blocking agents work to prevent nonspecific sticking and would therefore affect the nonspecific background binding but would not impact the apparent affinity. Addition of another standard blocking agent (BSA) also did not change either the apparent affinity (see below) or the total amount of Kap bound per unit nup at saturation (data not shown).
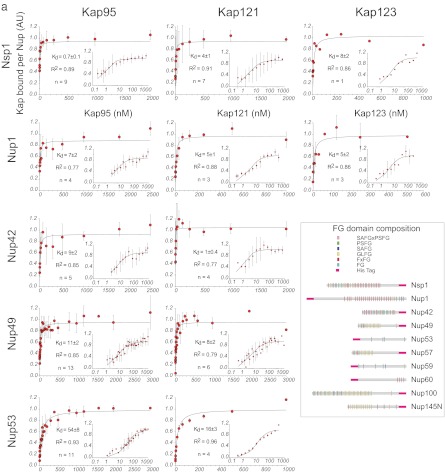
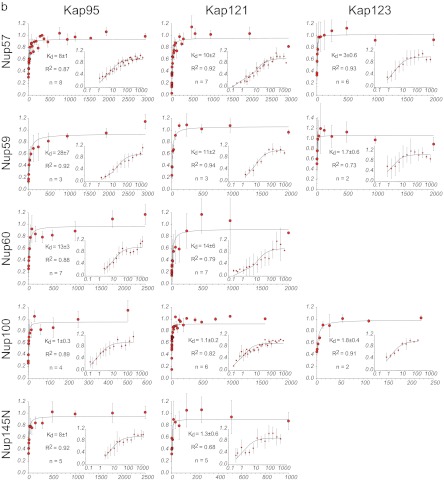
Fig. 3.
Plots of the Kap/FG nup affinities as measured by Dynabead binding assay in vitro. The assays were performed as shown in Fig. 2. The larger plots show the data on a linear scale. The insets show the same data on a log/linear scale. Schematics of the FG nup amino acid constructs are shown in the inset (adapted from Ref. 82); the His tag was always placed on the ordered domain, to mimic the site of attachment to the NPC in vivo.
Fig. 4.
Summary of the Kap/FG nup affinities as measured by bead binding assay. Kd values and the number of repeats are indicated below the plot. The error bars are the error of the fit as calculated by Sigma Plot using Equation 1. For clarity, the data are shown on a log/linear scale.
Interestingly, all of the Kap/FG nup interactions in our study appear to saturate at roughly a 1:1 stoichiometry, because the gel bands of Kaps and FG nups saturate at approximately the same Coomassie Blue intensity (Fig. 2), although without further quantitation this can only be considered very approximate. Nevertheless, excessive stoichiometries of many Kaps binding per FG nup seem unlikely.
Kap/FG Nup Affinities as Measured in Vitro Are All Very Tight
Importantly, the measured affinities in our hands are still several orders of magnitude tighter than the ∼μm to ∼mm Kd values expected from in vivo transport kinetics (13, 23, 63, 64). This difference could be due to (i) experimental design, (ii) important differences between the arrangement of FG nups in our experiment compared with in the NPC, (iii) specific, previously unidentified cofactor(s) missing in the in vitro set-up, or (iv) the fact that the proteins are not being studied in their native environment. The problem is very unlikely to be in the specific experimental design used here, because tight Kap/FG nup affinities have been observed by numerous classic and careful studies using a wide range of in vitro techniques (microtiter binding (36), surface plasmon resonance (69), experiments using Sepharose beads (37, 38, 68), overlay assays, and experiments using Dynabeads (this work)). Hence, the effect must be due either to the arrangement of the FG nups or effects from cofactors or the environment. We set out to identify the influence of both of these factors on Kap/FG nup binding by altering our experimental conditions to better mimic the native environment that these proteins experience in a living cell.
The Presence of Other Macromolecules Has a Major Effect on Kap/FG Nup Binding Affinities
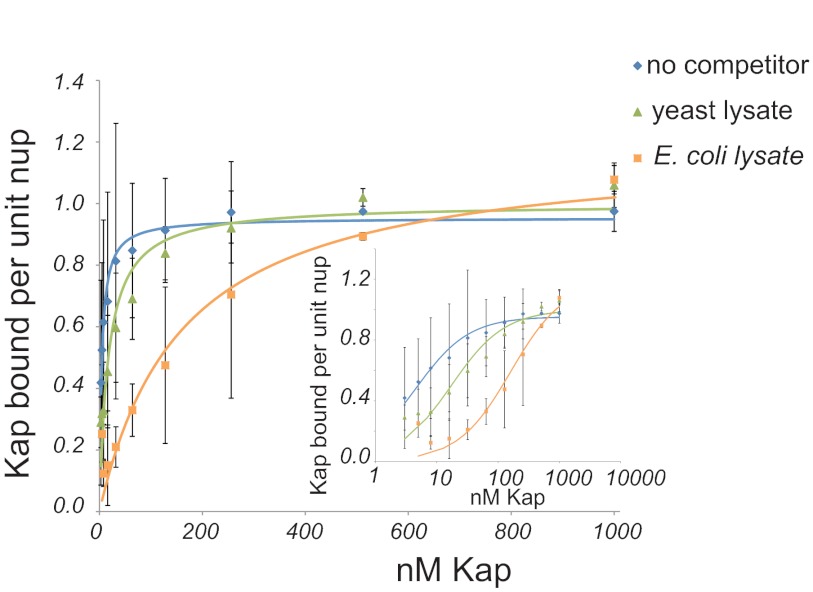
Nucleocytoplasmic transport in vivo occurs in the presence of many specific transport cofactors and a cellular milieu of concentrated macromolecules. To examine the effect of cytosolic proteins present during normal transport, we measured the apparent affinities of recombinant Kap123 and Nup100 in the presence or absence of varying concentrations of clarified ΔKap123 yeast lysate, which contains all of the soluble factors seen by Kap123 in vivo, but the only Kap123 available was exogenously added. We found that a significant reduction in the apparent affinity of Kap123 for Nup100 was produced by as little as 0.1 mg/ml yeast lysate, which is 1000× more dilute than in living cells (97) (Fig. 5). Thus, yeast cytosol acts as a competitor that weakens the apparent affinities between Kaps and FG nups in vivo and has a significant effect even in the presence of a ∼30-fold excess of blocking agent.
Fig. 5.
The presence of competitor significantly influences apparent affinities between Kap123 and Nup100. Kd curves of Kap123/Nup100 as measured without competitor (blue diamonds) or in the presence of 0.1 mg/ml yeast (green triangles) and bacterial (orange squares) lysate. The inset shows the same data presented on a log/linear scale.
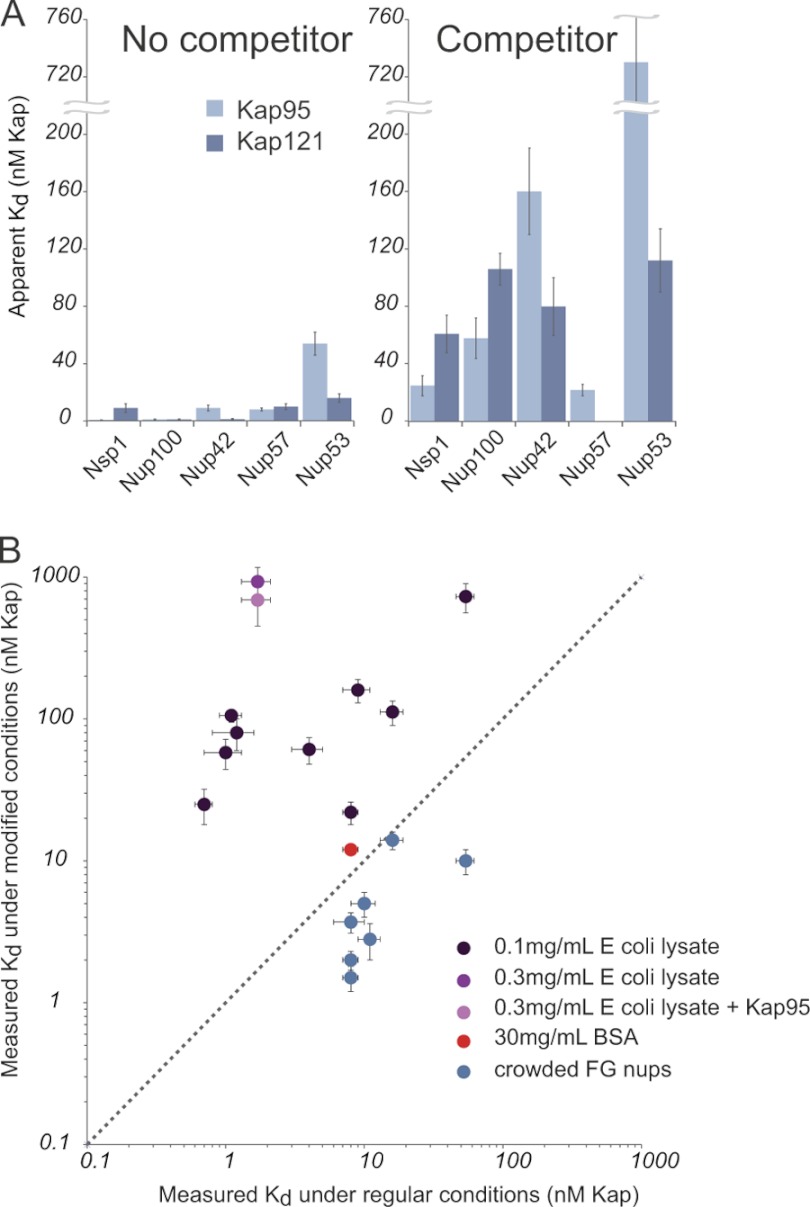
We then investigated whether the effect on apparent Kap/FG nup affinity is due to competition from other transport factors in particular or from yeast cytosol more generally. To best represent the bulk cellular proteins without introducing additional Kaps, FG nups, or NLS (Nuclear Localization Signal) cargo, we used lysate from E. coli as the competitor. As prokaryotes, E. coli contain many analogs of the components of eukaryotic cytoplasm but do not contain any proteins that function in nucleocytoplasmic transport. We found that the addition of bacterial lysate also caused a dramatic reduction in apparent Kap/FG nup affinities. We saw this effect for all three Kaps, and all examples of FG nups tested; these include nups classified as predominantly containing FXFG, GLFG, FG, and SAFGXPSFG repeats (98) (showing the effect was not specific to a certain subset of FG nups) and both symmetric and asymmetric nups (showing that those nups normally exposed only to the cytoplasm or nucleoplasm are not immune to the effect). Again, our experiments routinely used lysate concentrations that were 1000-fold lower than found in vivo (97). Nevertheless, even this small amount of competitor protein reduced all of the apparent affinities between Kaps and FG nups by between 1 and 2 orders of magnitude (supplemental Table 3 and Fig. 6). The higher the concentration of lysate added, the greater the effect on apparent Kap/FG nup affinity (Fig. 6). Increasing concentrations of lysate also decreased the total amount of Kap bound per unit nup at saturation (data not shown). Therefore, we suggest that components in yeast and bacterial lysates have hydrophobic patches on their surface (or other transiently adherent moieties) and are thus able to interact specifically with either the Kaps and/or the FG nups. This distinguishes the effect of competitor protein (lysate) from a blocking agent (PVP/Tween 20 or BSA; see above), because a blocking agent would not change the measured affinities between Kaps and FG nups, whereas a competitor would. Indeed, these assays were done in the presence of an excess of blocking agent (PVP/Tween 20) relative to lysate, showing that the only the latter acts as a competitor. We further confirmed that the decrease in apparent affinity was not due to degradation of the FG nups by proteases from the lysate (supplemental Fig. 8), was independent of assay design (supplemental Fig. 9), and is not a specific competitor (as the effect occurs with both prokaryotic and eukaryotic lysates), and the effect was likely due to protein rather than small molecules or nucleic acids (supplemental Fig. 10).
Fig. 6.
Lysate, but not crowding or BSA, significantly decreases the apparent affinities between all Kaps and FG nups measured. A, Kap/FG nup affinities as measured with and without indicated competitor. The pale blue bars are Kap95, and the dark blue bars are Kap121. B, plot of the measured affinity using the bead binding assay in unmodified (x axis) and modified (y axis) conditions: 0.1 mg/ml E. coli lysate, 0.3 mg/ml E. coli lysate, and 0.3 mg/ml E. coli lysate + Kap95 (dark, medium, and pale purple data points, respectively), 30 mg/ml BSA (red), and in the crowded condition (blue) (supplemental Fig. 12). The dotted line shows where the affinities would be if they were equal in the two conditions. Increasing FG nup density on the beads slightly increases the measured affinities, while adding small quantities of lysate significantly reduced the measured affinities. Adding up to 30 mg/ml BSA did not significantly change the affinity between Kap121 and Nup49, whereas merely 0.1 mg/ml of lysate did.
Heterogeneous extracts like E. coli lysates have been used as blocking agents for protein-protein interactions (99–101). The addition of these lysates typically reduces nonspecific, i.e. off target, interactions, but not the specific interaction affinities. This is the basis of blocking agents in all biological assays. For example, we found that E. coli lysate had no effect on the binding of protein A to rabbit IgG in our assays (supplemental Fig. 9). The overwhelming consensus that in vitro assays can be used to elucidate in vivo behavior necessitates that most macromolecular interactions are relatively insensitive to the normal cellular milieu; were all interactions so sensitive, it is hard to imagine how the integrity of most cellular processes could be maintained. The tremendous sensitivity of specific Kap-FG nup interaction to such additives as seen here is thus very surprising; however, it is consistent with the recent finding that interactions with the cytosol stabilize the disordered state of a natively unfolded protein (102), because the FG repeats are themselves such disordered protein regions. The sensitivity of Kap/nup interactions to competitors is also consistent with the documented tendency of Kaps to bind cytosolic proteins nonspecifically (72).
The Apparent Affinity between Kaps and FG Nups Does Not Strongly Depend on Either FG Repeat Composition or the Location of the FG Nup within a NPC
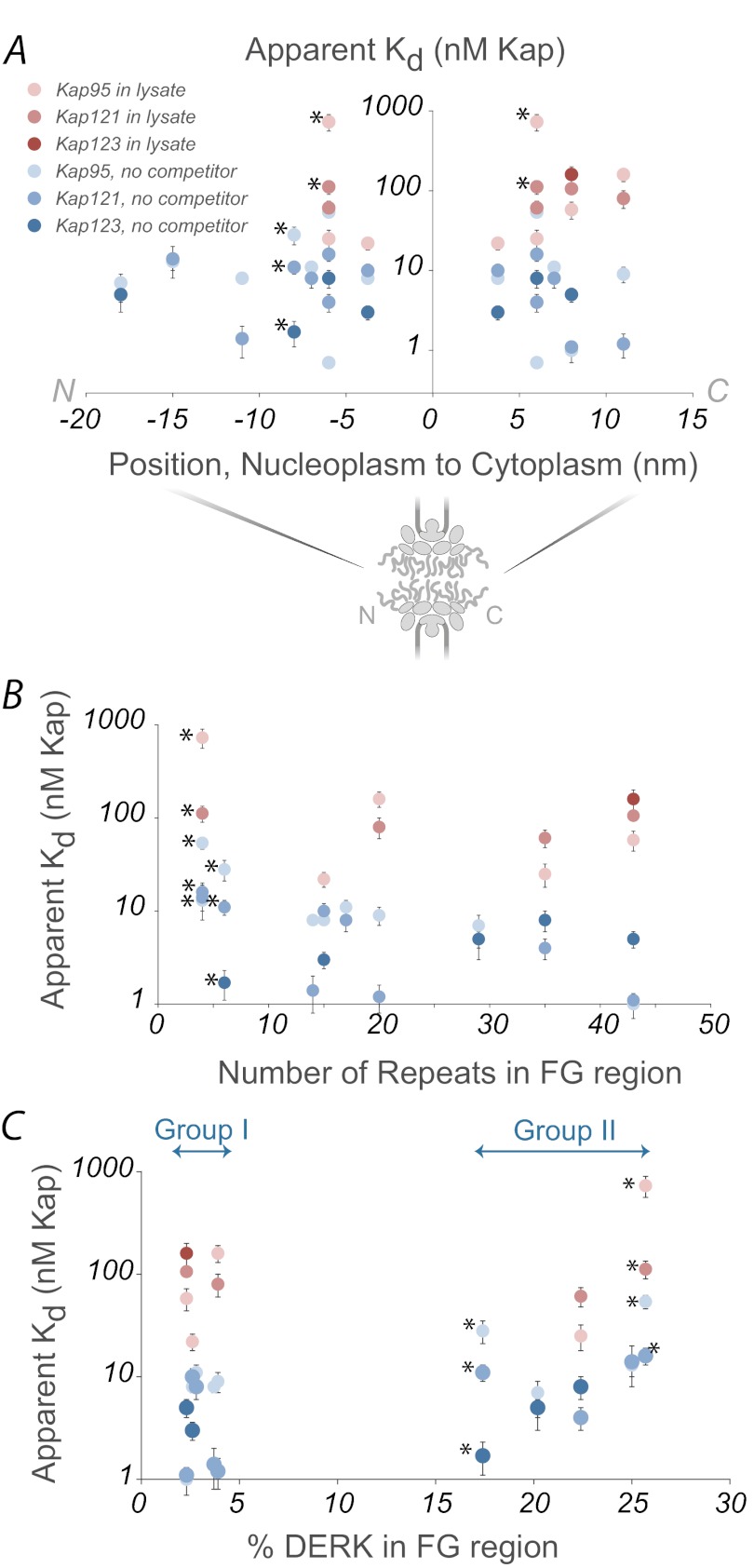
Given the strong influence of lysate on the apparent affinities between Kaps and FG nups, we looked to see what else may impact Kap/FG nup apparent affinities (Fig. 7). There may be a slight correlation between the number of FG repeats and affinity (although we did not explore this relationship further). We found no correlation between apparent affinity and location within the NPC (36, 68), again arguing against an affinity gradient as a general mechanism for transport; although it is known that very high affinity docking sites at the nucleoplasmic or cytoplasmic extremes of the NPC are important in terminating import or export, respectively, of particular cargoes (36, 68), they were not explored here. In vivo Kap concentration or FG nup amino acid composition either in the absence or presence of lysate as a competitor also did not correlate with apparent affinity and location within the NPC (Fig. 7). This is consistent with the finding that empty Kap95 seems to shuttle back and forth through the NPC with no directional bias in vivo (103). Directionality of cargo transport therefore is likely solely because of the binding and unbinding of cargo caused by the presence or absence of RanGTP on either side of the nuclear envelope (13, 104–106) and not because of a gradient of affinity between Kaps and FG nups.
Fig. 7.
Lysate concentration, but not FG nup position, number of FG repeats, or amino acid composition, correlates with measured affinity. Kap/FG nup affinities as measured in vitro show little apparent correlation with location in the NPC (A), number of FG repeats (B), or FG nup amino acid composition (C) in the presence (shades of red) or absence (shades of blue) of lysate. The palest points are Kap95, medium points are Kap121, and the darkest points are Kap123. (*) identifies points corresponding to Nup53 and Nup59, which contain few FG repeats and are not always considered to be true FG nups.
Neither Additional Kaps nor FG Nup Packing Density Has a Significant Effect on Apparent Affinity
In addition to competitor proteins in the nucleoplasm and cytoplasm, Kaps and FG nups in living cells are also exposed to other Kaps and other FG nups. Each yeast Kap is present at roughly 1–5 μm in the cytoplasm (72). We probed the influence of other transport factors within the cellular milieu by using E. coli lysate containing an overexpressed, recombinant, untagged Kap95 as a competitor. This lysate showed a similar response in modulating the interaction between Kap123/Nup100 to lysate without transport factor, indicating that lysate itself is potent competitor (Fig. 6). FG nups in vivo are also highly concentrated, on the order of many μm, with ∼130 FG domains occupying the limited space within the NPC (21, 37, 98). We therefore examined the effect of altering distribution of FG nup on the surface of the beads, to see whether this could also be contributing to the apparent discrepancy between in vivo and in vitro apparent affinity measurements with Kaps. Under the normal conditions of our binding assays, each FG nup molecule is spaced from its neighboring FG nup molecules by 6.6 nm (see methods). The FG domains, being natively unfolded, are somewhat less dense than ordered proteins. Their Stokes radii have been determined by size exclusion chromatography to be between 3 and 6 nm. Thus, in our standard condition, there may already be some overlap between the FG Nups on the surface (28). We further concentrated the nups on a small subset of the beads while saturating the remaining beads with GFP, still maintaining the same total amount of nup and GFP as in the regular condition, such that each nup is spaced from its neighbors by ∼2 nm. This is close enough to form cohesive interactions between the FG repeats (80). We finally diluted the same amount of nup among three times more beads fully pacivated with three times more GFP (Fig. 6 and supplemental Fig. 11), each FG nup in this condition being spaced from other FG nups by ∼9 nm. It should be emphasized that the amount of FG nup (as well as the ratio of GFP to beads) present in each experiment was the same in all three conditions; only the distribution of the FG nup differed. We found that for the FG nups tested, the apparent affinity for Kap95 and Kap121 were modestly but significantly tighter in the crowded condition and that there was no significant difference between the regular and dilute conditions (Fig. 6). This modest increase in avidity might be expected for an increase in the local concentration of binding sites but seems inconsistent with models in which packing of individual FG nups induces a phase change into an alternative state that binds with a dramatically reduced apparent affinity to transport factors. Although we acknowledge that we cannot achieve the exact level of crowding in the living NPC, changes in packing density or interactions between FG repeats appear to be unable to explain the apparent discrepancy between in vitro apparent affinities and the rapid in vivo transport kinetics.
Interpretation of Our Findings with Simple Mathematical Models
Our data on Kap/FG nup binding have revealed unexpected behaviors that we wish to understand to explain key features of nucleocytoplasmic transport. To do this, we constructed models of the interactions between Kaps, FG nups, and competitors that, although simplified, nevertheless capture key features of these interactions.
Kaps and FG Nups Bind through Multivalent Interactions
Because we know that each Kap molecule can bind multiple FG repeats (29, 34, 66, 67, 86, 107), the tight Kap/FG nup binding affinities observed in vitro appear to be the result of combined monovalent interactions between individual FG repeats and their multiple cognate Kap binding sites. Multivalency results in multiple weaker monovalent interactions combining to produce a stronger collective interaction (108, 109). These monovalent interactions are potentially in competition with interactions made by proteins found in the cellular milieu. This is because the monovalent site on FG nups is a flexible hydrophobic ligand capable of interacting not only with specific binding pockets on transport factors but also with exposed hydrophobic patches found on many other proteins; similarly, FG repeats could possibly the binding pockets on a Kap can be potentially occupied by hydrophobic motifs other than the FG repeats (32, 67, 110, 111).
We chose to consider in detail the Kap95/Nup100 interaction, which had an apparent Kd in vitro of 1 nm (Fig. 4). We modeled this interaction by a series of differential rate equations (supplemental material) in which we considered the stepwise binding of each FG repeat in Nup100 into a Kap95 binding pocket (Fig. 8). There are four experimentally verified and six additional proposed FG repeat binding sites on Kap95 (67, 86), and so we considered cases of between one and ten binding pockets. We incorporated the observed competition caused by lysate in our model as a single competitor making only monovalent interactions with each repeated hydrophobic Phe ligand, although this is also functionally equivalent in our calculations to competitors binding instead to single hydrophobic pockets on Kaps.
Fig. 8.
Schematic of the model. A diagram showing an example (number of Kap/FG nup binding sites, N = 3) of the rate equations used in the mathematical model of Kap/FG nup binding. See text and supplemental materials for a detailed description.
We assume that each binding site has the same off-rate, koffAB (s−1). For simplicity we assume that the first binding event occurs with a first order rate constant, konAB (m−1 s−1) of the equivalent monovalent Kap/FG interaction (Fig. 8). Based on the work of Whitesides and co-workers (108), we express the zero order rate of formation of higher valency interactions as konAB [Beff], where [Beff] can be thought of as the effective local concentrations of FGs in the vicinity of each subsequent cognate Kap binding pocket. In this model, the presence of other proteins can have two independent effects on Beff. First, the competitor can directly bind to the repeats, reducing the concentration of available Fs to bind a Kap, i.e. reducing Beff. Second, because FG nups should be as exquisitely sensitive to their environment as other natively unfolded proteins studied so far (for review, see Ref. 62), proteins in the cellular milieu likely change the structure and behavior of the protein (102) and thus change Beff. In the model, the individual monovalent Kap/FG nup binding sites are indistinguishable, and the maximum valency is N, which for simplicity is taken to be the same value for both the Kap and the FG nup (Fig. 8). For this model, there are therefore only two parameters: the single-site dissociation constant (Kdsingle = koffAB/konAB), and Beff as described above. In the limit of high Beff (i.e. strongly tending toward the fully bound state), the apparent multivalent dissociation constant is approximately Kdapp α Beff(Kdsingle/Beff)N. To compare different valencies, we assumed that Beff was independent of N. We approximated Beff as 150 mm based on the observation that 43 repeats of the FG region of Nup100 occupy a ∼5-nm radius sphere (28). For each value of N, we calculated Kdsingle so as to reproduce the value of Kdapp = 1 nm measured for this interaction. Although we cannot independently determine Kdsingle and Beff exactly, it is important to note that this model proved relatively insensitive to the exact values chosen.
The Strength of the Monovalent Competitors Depends on the Degree of Valency
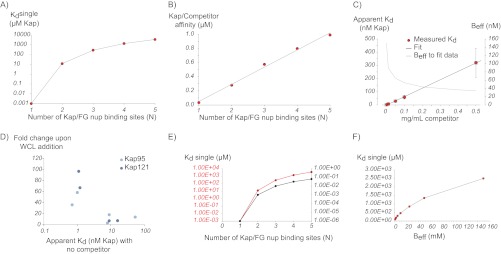
We began by modeling the effect of competing monovalent interactors on Kap/FG nup binding for different values of N (Fig. 9). As expected, in the absence of competitor, as the number of Kap/FG nup binding sites increased, the affinity of each individual FG binding (Kdsingle) site necessary to reproduce the measured apparent affinity decreased (Fig. 9), such that at N = 5, Kdsingle = 3.5 mm. Notably, this value of Kdsingle is of the same order of magnitude as Kd estimates for nonspecific protein-protein interactions (72, 112); thus, such nonspecific interactions could significantly modulate Kap/FG nup binding.
Fig. 9.
Plots illustrating results from the simple model. A, calculated values of Kdsingle required to reproduce the experimental data for different numbers of Kap/FG nup binding sites (N) for Beff = 150 mm. B, calculated Kap/competitor affinities needed to reproduce the experimental affinities for different values of N, showing that competitor has a stronger effect for higher valency interactions. C, experimental Kap/FG nup affinities depend strongly on concentration of competitor protein (red dots), which is fit with our model (black line) by allowing Beff to depend on competitor concentration (gray line). D, experimental Kap/FG nup affinities are inversely correlated with the fold change upon the addition of competitor as predicted by the model. Light blue circles are Kap95 and, dark blue circles are Kap121. E, calculated Kdsingle needed to reproduce the experimental affinities for different values of N and Beff (red line, Beff = 150 mm; black line, Beff = 1 mm), showing that our model is quantitatively but not qualitatively dependent on the choice of this parameter. F, calculated values of Kdsingle required to reproduce the experimental data for N = 3 at different values for Beff.
To model direct monovalent competition, one could either incorporate the effect of competition from the cytosol into Beff (see above) or explicitly and separately include it. Although both are equivalent, we initially chose the latter as a more straightforward representation. To this end, we allowed each FG repeat binding site to interact with either a Kap binding pocket (Kdsingle) or a monovalent competitor protein (KdBC). The limiting case of N = 1 is a simple competitor model, where Kap and competitor binding to the nup are mutually exclusive. In our model, we assumed that the lysate behaved as one averaged protein and used the empirically determined average molecular mass of 50 kDa to determine the competitor concentration (thus 0.1 mg/ml = 2 μm). We first reproduced the Kap95/Nup100 binding affinity as measured in the presence of 0.1 mg/ml lysate, using different values of N in the model. We found that as the number of Kap/FG nup binding sites increased, the affinity between monovalent competitor and FG nup necessary to reproduce this experimental data dramatically decreased (Fig. 9). For example, for the limiting case of N = 1, the monovalent competitor must bind with an unreasonably high affinity of KdBC = 35 nm (i.e. in the same apparent affinity range as specific interactions). However, when N = 5, well in the range of the estimated number of binding sites between Kaps and FG nups (67, 86), a protein interacting monovalently need only have the weak and physiologically reasonable binding affinity of KdBC = 1 μm to successfully compete with Kap/FG nup interactions.
If Kdsingle is similar for all of the Kap/FG nup interactions, then the strongest Kap/FG nup interactions would therefore have the highest valency and so would be most sensitive to competitor. We can therefore test our model, by determining whether there is a direct relationship between the strength of Kap/FG nup interaction and the sensitivity to competitor. Indeed, such a relationship was observed (Fig. 9).
Modeling Environmentally Induced Conformational Changes
The simple competition model predicts that the relationship between apparent affinity and competitor concentration should depend on the valency of the interaction. If there are N binding sites for competitor, then Kdapp α [C]N. In other words, we should see an exponential relationship between competitor concentration and the measured Kd. We therefore experimentally determined this relationship for Kap95/Nup100 binding. However, our data show that this relationship is approximately linear (Fig. 9) over a 50-fold range in competitor concentration, supporting the idea that direct competitive binding is not the only effect of competition (see above); the presence of competitor proteins in the cellular milieu thus may also lead to conformational changes in the FG nup (and/or Kap), changing Beff and thus altering the strength of the Kap/FG nup interaction. Even though we do not know the exact nature of these environment-induced conformational changes, we can represent them in our model by allowing Beff to depend on the concentration of competitor. Indeed, we can reproduce our experimental data (Fig. 9) by fitting Beff for each competitor concentration given a fixed Kdsingle in the absence of an explicit competitor. We find that increasing the concentration of competitor decreases Beff. Recall that in the limit of high Beff, Kdapp α Beff1−N, and so, as in the case of the simple competition model, the higher the valency of the interaction (the higher N), the more sensitive the interaction is to changes in competitor, as we saw for the model with fixed Beff above.
DISCUSSION
Competition and the Kinetics of Nuclear Transport
Several models have been proposed to explain how the disordered FG repeat regions cooperate to mediate nucleocytoplasmic transport. It has variously been suggested that FG domains reduce access to all or part of the central channel by occluding it as a polymer brush (9, 22, 25, 26, 113–116), as a series of physiochemically and functionally distinct quasi-structured regions (27, 28, 82, 83), as a hydrophobic gel/“hydrogel” (2, 80, 81, 117), as a reversibly collapsible mesh (114, 115), and as a layer of collapsed domains around the sides of the channel (118). Our current work provides insight into details of transport that are not dependent on nor included in any of these models and can provide a conceptual framework for understanding the kinetics of transport through the NPC. Importantly, this framework only depends on well established features regarding the nature of Kap/FG nup binding that are common to all proposed molecular mechanisms (9, 13, 82, 117–119) and on the presence of competitors during transport.
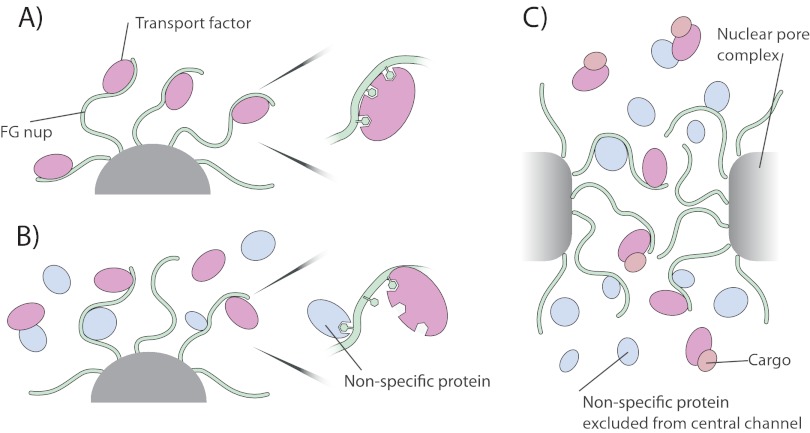
Several lines of evidence support the idea that the transport pathway of NPCs in living cells are crowded with proteins, at concentrations comparable with or exceeding those in the surrounding cellular milieu (Fig. 10) (76). First, the FG repeats are known to extend tens of nanometers into the nucleoplasm and cytoplasm, mingling with macromolecules in both the nucleoplasm and cytoplasm (22, 120) Second, the nonspecific flux of macromolecules across the NPC has been well documented, and multiple types of proteins up to ∼40 kDa can diffuse with some alacrity through the NPC (76) providing a substantial pool of potential competitors within the central channel. Third, although cargos are being specifically transported, they themselves make no specific interactions with FG nups and thus themselves can act as competitors occupying space within the central channel. Fourth, the FG repeats are themselves highly concentrated in the NPC, potentially transiently interacting with each other, and competing with each other for space and binding (27, 28, 121). Although we cannot replicate exactly the geometry or environment of the NPCs in our in vitro assay, we have examined the influence of specific and nonspecific competitor proteins to try and mimic the in vivo conditions as closely as possible. The increased crowding within the cylindrical channel of the NPC will likely increase the influence of competition, as predicted by Zilman (121).
Fig. 10.
Potential implications of our results for nucleocytoplasmic transport. A, in the absence of monovalent competitors, Kaps bind tightly to the FG Nups, preventing rapid exchange. B, however, monovalent competitor reduces the valency and so avidity of Kaps for FG Nups, allowing a rapid and dynamic exchange of the two (a zoomed example of competitor interacting with FG repeats is shown; competitor interaction with Kap is also possible). C, within the nuclear pore complex, Kaps and FG nups are in contact with other FG nups, transport factors, cargo, and small proteins that can rapidly diffuse through the central channel. All of these factors contribute to rapid and effective transport of Kap/cargo complexes through the NPC and serve to block the passage of non-Kap-bound macromolecules.
Notably, we have previously shown another example—also involving Kaps—where competition from the cellular milieu modulates a specific interaction. In this case, competition of cytosolic proteins for Kaps and cargo is a key step that limits the efficiency of NLS-mediated nuclear import (72). By contrast, in the case of Kaps and FG nups, competition serves to facilitate rapid passage of the Kap across the NPC. However, both Kap/NLS and Kap/FG nup interactions share the features of multivalent binding through relatively weak monovalent interactions with high off-rates (122, 123).
We have previously discussed how competition might influence transport, that because the multivalency of Kap/FG nup binding provides a stronger interaction than those of monovalent competitors, competition for binding sites and space in the central channel of the NPC will facilitate Kap-mediated transport at the expense of the passage of the competitors (124–126). Many groups have also emphasized the importance of the multivalency of Kap/FG nup binding to nuclear transport (2, 17, 24, 42, 67, 84, 86, 114, 125). Here we expand upon these ideas, introducing evidence that competition actually modulates the effective affinity of a transport factor for the NPC. The hypothesis, supported by our data and modeling, that such competition effects profoundly influence the transport process, potentially resolves several contradictions between observed behaviors of nucleocytoplasmic transport in vivo and observed affinities in vitro, and with respect to Kap/FG nup interactions may add significant insights into the likely behavior of transport factors as they cross the NPC. First, if one only considers the in vitro measured nanomolar Kap/FG nup affinities and the estimated micromolar concentration of transport factors in vivo (72), the NPC in living cells would be fully saturated at all times with transport factors, blocking efficient transport. However, by invoking competition, our data and model suggest that instead the other cellular proteins reduce Kap/FG nup effective affinities and so prevent such NPC saturation. Second, nanomolar Kap/FG nup affinities would also mean very slow off-rates, of the order of hours, precluding rapid exchange of Kaps across the NPC. However, in our model, competition dramatically increases the off-rates—and so transport rates—of Kaps at the NPC. Third, as with NLS/Kap interactions (72), the primary influence on Kap/FG nup affinity is interactions with the cellular cytosol rather than direct competition from other transport factors, such that Kaps need not significantly compete with each other in the NPC for FG nups. Fourth, the local concentration of available FG repeats from a given FG nup (Beff) could be lowered enough such that a Kap could not distinguish between a nearby repeat on the same FG nup and a repeat on neighboring FG nups. This would enable Kaps to easily and quickly jump from FG nup to FG nup through the NPC.
Conclusions
To understand the in vivo binding kinetics of nucleocytoplasmic transport, much work has been done using classic, tried and tested approaches that have worked so well in the past: binding assays between pairs of interactors in defined, proven biologically compatible buffers. However, we show here that in the case of the FG nups, and likely for many natively unfolded proteins, such assays can be misleading with respect to their actual behavior in the living cell. NPCs exist in the crowded environment of a living cell and themselves present a crowded environment to macromolecules in transit; based on the results presented here, these multiple weak competitors in the cellular milieu likely act collectively and significantly impact the binding behavior of Kaps and FG nups. Therefore, to truly understand how these proteins function in vivo, future studies must examine these proteins in as close to their native context as possible.
Acknowledgments
We acknowledge all members of the Rout laboratory for discussions and support, in particular Josef Franke for the plasmid containing the non-FG domain of Nup100. We are also grateful to Brian Chait, Tijana Jovanovic-Talisman, Georges Belfort, Reiner Peters, Sandy Simon, Tom Muir, Fred Cross, Seth Darst, and Anton Zilman for helpful discussions.
Footnotes
* This work was supported by National Institutes of Health Grants U54 RR022220, R01 GM62427, and R01 GM071329 (to M. R.) and F32 GM087854 (to L. H.), a Revson Senior Fellowship (to L. H.), and a Howard Hughes Medical Institute predoctoral fellowship (to J. T.-N.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- NPC
- nuclear pore complex
- Kap
- karyopherin
- FG
- phenylalanine-glycine
- IDP
- intrinsically disordered protein
- GFP
- green fluorescent protein
- Nup
- nucleoporin
- NLS
- nuclear localization signal
- PVP
- polyvinylpyrrolidone.
REFERENCES
- 1. Paine P. L. (1975) Nucleocytoplasmic movement of fluorescent tracers microinjected into living salivary gland cells. J. Cell Biol. 66, 652–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Frey S., Richter R. P., Görlich D. (2006) FG-rich repeats of nuclear pore proteins form a three-dimensional meshwork with hydrogel-like properties. Science 314, 815–817 [DOI] [PubMed] [Google Scholar]
- 3. Dworetzky S. I., Feldherr C. M. (1988) Translocation of RNA-coated gold particles through the nuclear pores of oocytes. J. Cell Biol. 106, 575–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Adam S. A., Gerace L. (1991) Cytosolic proteins that specifically bind nuclear localization signals are receptors for nuclear import. Cell 66, 837–847 [DOI] [PubMed] [Google Scholar]
- 5. Adam E. J., Adam S. A. (1994) Identification of cytosolic factors required for nuclear location sequence-mediated binding to the nuclear envelope. J. Cell Biol. 125, 547–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Görlich D., Prehn S., Laskey R. A., Hartmann E. (1994) Isolation of a protein that is essential for the first step of nuclear protein import. Cell 79, 767–778 [DOI] [PubMed] [Google Scholar]
- 7. Görlich D., Vogel F., Mills A. D., Hartmann E., Laskey R. A. (1995) Distinct functions for the two importin subunits in nuclear protein import. Nature 377, 246–248 [DOI] [PubMed] [Google Scholar]
- 8. Leslie D. M., Grill B., Rout M. P., Wozniak R. W., Aitchison J. D. (2002) Kap121p-mediated nuclear import is required for mating and cellular differentiation in yeast. Mol. Cell. Biol. 22, 2544–2555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rout M. P., Aitchison J. D., Magnasco M. O., Chait B. T. (2003) Virtual gating and nuclear transport: The hole picture. Trends Cell Biol. 13, 622–628 [DOI] [PubMed] [Google Scholar]
- 10. Mosammaparast N., Pemberton L. F. (2004) Karyopherins: From nuclear-transport mediators to nuclear-function regulators. Trends Cell Biol. 14, 547–556 [DOI] [PubMed] [Google Scholar]
- 11. McLane L. M., Pulliam K. F., Devine S. E., Corbett A. H. (2008) The Ty1 integrase protein can exploit the classical nuclear protein import machinery for entry into the nucleus. Nucleic Acids Res. 36, 4317–4326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Panté N., Kann M. (2002) Nuclear pore complex is able to transport macromolecules with diameters of about 39 nm. Mol. Biol. Cell 13, 425–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Macara I. G. (2001) Transport into and out of the nucleus. Microbiol. Mol. Biol. Rev. 65, 570–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Köhler A., Hurt E. (2007) Exporting RNA from the nucleus to the cytoplasm. Nat. Rev. Mol. Cell Biol. 8, 761–773 [DOI] [PubMed] [Google Scholar]
- 15. Stewart M. (2007) Molecular mechanism of the nuclear protein import cycle. Nat. Rev. Mol. Cell Biol. 8, 195–208 [DOI] [PubMed] [Google Scholar]
- 16. Terry L. J., Wente S. R. (2009) Flexible gates: Dynamic topologies and functions for FG nucleoporins in nucleocytoplasmic transport. Eukaryot. Cell 8, 1814–1827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Peters R. (2009) Translocation through the nuclear pore: Kaps pave the way. Bioessays 31, 466–477 [DOI] [PubMed] [Google Scholar]
- 18. Rout M. P., Wente S. R. (1994) Pores for thought: Nuclear pore complex proteins. Trends Cell Biol. 4, 357–365 [DOI] [PubMed] [Google Scholar]
- 19. Allen N. P., Huang L., Burlingame A., Rexach M. (2001) Proteomic analysis of nucleoporin interacting proteins. J. Biol. Chem. 276, 29268–29274 [DOI] [PubMed] [Google Scholar]
- 20. Tran E. J., Wente S. R. (2006) Dynamic nuclear pore complexes: Life on the edge. Cell 125, 1041–1053 [DOI] [PubMed] [Google Scholar]
- 21. Alber F., Dokudovskaya S., Veenhoff L. M., Zhang W., Kipper J., Devos D., Suprapto A., Karni-Schmidt O., Williams R., Chait B. T., Sali A., Rout M. P. (2007) The molecular architecture of the nuclear pore complex. Nature 450, 695–701 [DOI] [PubMed] [Google Scholar]
- 22. Rout M. P., Aitchison J. D., Suprapto A., Hjertaas K., Zhao Y., Chait B. T. (2000) The yeast nuclear pore complex: Composition, architecture, and transport mechanism. J. Cell Biol. 148, 635–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ribbeck K., Görlich D. (2001) Kinetic analysis of translocation through nuclear pore complexes. EMBO J. 20, 1320–1330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Denning D. P., Patel S. S., Uversky V., Fink A. L., Rexach M. (2003) Disorder in the nuclear pore complex: The FG repeat regions of nucleoporins are natively unfolded. Proc. Natl. Acad. Sci. U.S.A. 100, 2450–2455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lim R. Y., Huang N. P., Köser J., Deng J., Lau K. H., Schwarz-Herion K., Fahrenkrog B., Aebi U. (2006) Flexible phenylalanine-glycine nucleoporins as entropic barriers to nucleocytoplasmic transport. Proc. Natl. Acad. Sci. U.S.A. 103, 9512–9517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Miao L., Schulten K. (2009) Transport-related structures and processes of the nuclear pore complex studied through molecular dynamics. Structure 17, 449–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Krishnan V. V., Lau E. Y., Yamada J., Denning D. P., Patel S. S., Colvin M. E., Rexach M. F. (2008) Intramolecular cohesion of coils mediated by phenylalanine-glycine motifs in the natively unfolded domain of a nucleoporin. PLoS Comput. Biol. 4, e1000145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yamada J., Phillips J. L., Patel S., Goldfien G., Calestagne-Morelli A., Huang H., Reza R., Acheson J., Krishnan V. V., Newsam S., Gopinathan A., Lau E. Y., Colvin M. E., Uversky V. N., Rexach M. F. (2010) A bimodal distribution of two distinct categories of intrinsically-disordered structures with separate functions in FG nucleoporins. Mol. Cell. Proteomics 9, 2205–2224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Otsuka S., Iwasaka S., Yoneda Y., Takeyasu K., Yoshimura S. H. (2008) Individual binding pockets of importin-β for FG-nucleoporins have different binding properties and different sensitivities to RanGTP. Proc. Natl. Acad. Sci. U.S.A. 105, 16101–16106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bailer S. M., Siniossoglou S., Podtelejnikov A., Hellwig A., Mann M., Hurt E. (1998) Nup116p and nup100p are interchangeable through a conserved motif which constitutes a docking site for the mRNA transport factor gle2p. EMBO J. 17, 1107–1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bayliss R., Kent H. M., Corbett A. H., Stewart M. (2000) Crystallization and initial x-ray diffraction characterization of complexes of FxFG nucleoporin repeats with nuclear transport factors. J. Struct. Biol. 131, 240–247 [DOI] [PubMed] [Google Scholar]
- 32. Bayliss R., Leung S. W., Baker R. P., Quimby B. B., Corbett A. H., Stewart M. (2002) Structural basis for the interaction between NTF2 and nucleoporin FxFG repeats. EMBO J. 21, 2843–2853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bayliss R., Littlewood T., Stewart M. (2000) Structural basis for the interaction between FxFG nucleoporin repeats and importin-β in nuclear trafficking. Cell 102, 99–108 [DOI] [PubMed] [Google Scholar]
- 34. Bayliss R., Littlewood T., Strawn L. A., Wente S. R., Stewart M. (2002) GLFG and FxFG nucleoporins bind to overlapping sites on importin-β. J. Biol. Chem. 277, 50597–50606 [DOI] [PubMed] [Google Scholar]
- 35. Bayliss R., Ribbeck K., Akin D., Kent H. M., Feldherr C. M., Görlich D., Stewart M. (1999) Interaction between NTF2 and xFxFG-containing nucleoporins is required to mediate nuclear import of RanGDP. J. Mol. Biol. 293, 579–593 [DOI] [PubMed] [Google Scholar]
- 36. Ben-Efraim I., Gerace L. (2001) Gradient of increasing affinity of importin β for nucleoporins along the pathway of nuclear import. J. Cell Biol. 152, 411–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gilchrist D., Mykytka B., Rexach M. (2002) Accelerating the rate of disassembly of karyopherin.cargo complexes. J. Biol. Chem. 277, 18161–18172 [DOI] [PubMed] [Google Scholar]
- 38. Gilchrist D., Rexach M. (2003) Molecular basis for the rapid dissociation of nuclear localization signals from karyopherin α in the nucleoplasm. J. Biol. Chem. 278, 51937–51949 [DOI] [PubMed] [Google Scholar]
- 39. Grant R. P., Neuhaus D., Stewart M. (2003) Structural basis for the interaction between the Tap/NXF1 UBA domain and FG nucleoporins at 1A resolution. J. Mol. Biol. 326, 849–858 [DOI] [PubMed] [Google Scholar]
- 40. Iovine M. K., Watkins J. L., Wente S. R. (1995) The GLFG repetitive region of the nucleoporin Nup116p interacts with Kap95p, an essential yeast nuclear import factor. J. Cell Biol. 131, 1699–1713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Isgro T. A., Schulten K. (2007) Cse1p-binding dynamics reveal a binding pattern for FG-repeat nucleoporins on transport receptors. Structure 15, 977–991 [DOI] [PubMed] [Google Scholar]
- 42. Isgro T. A., Schulten K. (2007) Association of nuclear pore FG-repeat domains to NTF2 import and export complexes. J. Mol. Biol. 366, 330–345 [DOI] [PubMed] [Google Scholar]
- 43. Marelli M., Aitchison J. D., Wozniak R. W. (1998) Specific binding of the karyopherin Kap121p to a subunit of the nuclear pore complex containing Nup53p, Nup59p, and Nup170p. J. Cell Biol. 143, 1813–1830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Morrison J., Yang J. C., Stewart M., Neuhaus D. (2003) Solution NMR study of the interaction between NTF2 and nucleoporin FxFG repeats. J. Mol. Biol. 333, 587–603 [DOI] [PubMed] [Google Scholar]
- 45. Strawn L. A., Shen T., Wente S. R. (2001) The GLFG regions of Nup116p and Nup100p serve as binding sites for both Kap95p and Mex67p at the nuclear pore complex. J. Biol. Chem. 276, 6445–6452 [DOI] [PubMed] [Google Scholar]
- 46. Radu A., Blobel G., Moore M. S. (1995) Identification of a protein complex that is required for nuclear-protein import and mediates docking of import substrate to distinct nucleoporins. Proc. Natl. Acad. Sci. U.S.A. 92, 1769–1773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Radu A., Moore M. S., Blobel G. (1995) The peptide repeat domain of nucleoporin Nup98 functions as a docking site in transport across the nuclear-pore complex. Cell 81, 215–222 [DOI] [PubMed] [Google Scholar]
- 48. Jovanovic-Talisman T., Tetenbaum-Novatt J., McKenney A. S., Zilman A., Peters R., Rout M. P., Chait B. T. (2009) Artificial nanopores that mimic the transport selectivity of the nuclear pore complex. Nature 457, 1023–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kowalczyk S. W., Kapinos L., Blosser T. R., Magalhães T., van Nies P., Lim R. Y., Dekker C. (2011) Single-molecule transport across an individual biomimetic nuclear pore complex. Nat. Nanotechnol. 6, 433–438 [DOI] [PubMed] [Google Scholar]
- 50. Lakshmi B. B., Martin C. R. (1997) Enantioseparation using apoenzymes immobilized in a porous polymeric membrane. Nature 388, 758–760 [DOI] [PubMed] [Google Scholar]
- 51. Caspi Y., Zbaida D., Cohen H., Elbaum M. (2008) Synthetic mimic of selective transport through the nuclear pore complex. Nano. Lett. 8, 3728–3734 [DOI] [PubMed] [Google Scholar]
- 52. Uversky V. N. (2010) The mysterious unfoldome: Structureless, underappreciated, yet vital part of any given proteome. J. Biomed. Biotechnol. 2010, 568068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Uversky V. N., Gillespie J. R., Fink A. L. (2000) Why are “natively unfolded” proteins unstructured under physiologic conditions? Proteins 41, 415–427 [DOI] [PubMed] [Google Scholar]
- 54. Bussell R., Jr., Eliezer D. (2001) Residual structure and dynamics in Parkinson's disease-associated mutants of α-synuclein. J. Biol. Chem. 276, 45996–46003 [DOI] [PubMed] [Google Scholar]
- 55. Eliezer D. (2009) Biophysical characterization of intrinsically disordered proteins. Curr. Opin. Struct. Biol. 19, 23–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wright P. E., Dyson H. J. (2009) Linking folding and binding. Curr. Opin. Struct. Biol. 19, 31–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Uversky V. N. (2011) Multitude of binding modes attainable by intrinsically disordered proteins: A portrait gallery of disorder-based complexes. Chem. Soc. Rev. 40, 1623–1634 [DOI] [PubMed] [Google Scholar]
- 58. Metallo S. J. (2010) Intrinsically disordered proteins are potential drug targets. Curr. Opin. Chem. Biol. 14, 481–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Uversky V. N., Dunker A. K. (2010) Understanding protein non-folding. Biochim. Biophys. Acta 1804, 1231–1264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Sigalov A. B. (2010) Protein intrinsic disorder and oligomericity in cell signaling. Mol. Biosyst. 6, 451–461 [DOI] [PubMed] [Google Scholar]
- 61. He B., Wang K., Liu Y., Xue B., Uversky V. N., Dunker A. K. (2009) Predicting intrinsic disorder in proteins: An overview. Cell Res. 19, 929–949 [DOI] [PubMed] [Google Scholar]
- 62. Uversky V. N. (2009) Intrinsically disordered proteins and their environment: Effects of strong denaturants, temperature, pH, counter ions, membranes, binding partners, osmolytes, and macromolecular crowding. Protein J. 28, 305–325 [DOI] [PubMed] [Google Scholar]
- 63. Kubitscheck U., Grünwald D., Hoekstra A., Rohleder D., Kues T., Siebrasse J. P., Peters R. (2005) Nuclear transport of single molecules: Dwell times at the nuclear pore complex. J. Cell Biol. 168, 233–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Yang W., Gelles J., Musser S. M. (2004) Imaging of single-molecule translocation through nuclear pore complexes. Proc. Natl. Acad. Sci. U.S.A. 101, 12887–12892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Grünwald D., Singer R. H. (2010) In vivo imaging of labelled endogenous B-actin mRNA during nucleocytoplasmic transport. Nature 467, 604–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Bayliss R., Corbett A. H., Stewart M. (2000) The molecular mechanism of transport of macromolecules through nuclear pore complexes. Traffic 1, 448–456 [DOI] [PubMed] [Google Scholar]
- 67. Isgro T. A., Schulten K. (2005) Binding dynamics of isolated nucleoporin repeat regions to importin-β. Structure 13, 1869–1879 [DOI] [PubMed] [Google Scholar]
- 68. Pyhtila B., Rexach M. (2003) A gradient of affinity for the karyopherin Kap95p along the yeast nuclear pore complex. J. Biol. Chem. 278, 42699–42709 [DOI] [PubMed] [Google Scholar]
- 69. Lott K., Bhardwaj A., Mitrousis G., Pante N., Cingolani G. (2010) The importin β binding domain modulates the avidity of importin β for the nuclear pore complex. J. Biol. Chem. 285, 13769–13780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Ghaemmaghami S., Huh W. K., Bower K., Howson R. W., Belle A., Dephoure N., O'Shea E. K., Weissman J. S. (2003) Global analysis of protein expression in yeast. Nature 425, 737–741 [DOI] [PubMed] [Google Scholar]
- 71. Paradise A., Levin M. K., Korza G., Carson J. H. (2007) Significant proportions of nuclear transport proteins with reduced intracellular mobilities resolved by fluorescence correlation spectroscopy. J. Mol. Biol. 365, 50–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Timney B. L., Tetenbaum-Novatt J., Agate D. S., Williams R., Zhang W., Chait B. T., Rout M. P. (2006) Simple kinetic relationships and nonspecific competition govern nuclear import rates in vivo. J. Cell Biol. 175, 579–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Peters R. (2009) Functionalization of a nanopore: The nuclear pore complex paradigm. Biochim. Biophys. Acta 1793, 1533–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Kose S., Imamoto N., Tachibana T., Shimamoto T., Yoneda Y. (1997) Ran-unassisted nuclear migration of a 97-kD component of nuclear pore-targeting complex. J. Cell Biol. 139, 841–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Dange T., Grünwald D., Grünwald A., Peters R., Kubitscheck U. (2008) Autonomy and robustness of translocation through the nuclear pore complex: A single-molecule study. J. Cell Biol. 183, 77–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Tu L. C., Musser S. M. (2011) Single molecule studies of nucleocytoplasmic transport. Biochim. Biophys. Acta 1813, 1607–1618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Riddick G., Macara I. G. (2007) The adapter importin-α provides flexible control of nuclear import at the expense of efficiency. Mol. Syst. Biol. 3, 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Riddick G., Macara I. G. (2005) A systems analysis of importin-α-β mediated nuclear protein import. J. Cell Biol. 168, 1027–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Goodri (2006) Binding and Kinetics for Molecular Biologists, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 80. Frey S., Görlich D. (2007) A saturated FG-repeat hydrogel can reproduce the permeability properties of nuclear pore complexes. Cell 130, 512–523 [DOI] [PubMed] [Google Scholar]
- 81. Frey S., Görlich D. (2009) FG/FxFG as well as GLFG repeats form a selective permeability barrier with self-healing properties. EMBO J. 28, 2554–2567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Patel S. S., Belmont B. J., Sante J. M., Rexach M. F. (2007) Natively unfolded nucleoporins gate protein diffusion across the nuclear pore complex. Cell 129, 83–96 [DOI] [PubMed] [Google Scholar]
- 83. Patel S. S., Rexach M. F. (2008) Discovering novel interactions at the nuclear pore complex using bead halo: A rapid method for detecting molecular interactions of high and low affinity at equilibrium. Mol. Cell. Proteomics 7, 121–131 [DOI] [PubMed] [Google Scholar]
- 84. Ehrlich P. H. (1979) The effect of multivalency on the specificity of protein and cell interactions. J. Theor. Biol. 81, 123–127 [DOI] [PubMed] [Google Scholar]
- 85. Allen N. P., Patel S. S., Huang L., Chalkley R. J., Burlingame A., Lutzmann M., Hurt E. C., Rexach M. (2002) Deciphering networks of protein interactions at the nuclear pore complex. Mol. Cell. Proteomics 1, 930–946 [DOI] [PubMed] [Google Scholar]
- 86. Liu S. M., Stewart M. (2005) Structural basis for the high-affinity binding of nucleoporin Nup1p to the Saccharomyces cerevisiae importin-β homologue, Kap95p. J. Mol. Biol. 349, 515–525 [DOI] [PubMed] [Google Scholar]
- 87. Lee S. J., Matsuura Y., Liu S. M., Stewart M. (2005) Structural basis for nuclear import complex dissociation by RanGTP. Nature 435, 693–696 [DOI] [PubMed] [Google Scholar]
- 88. Rout M. P., Blobel G., Aitchison J. D. (1997) A distinct nuclear import pathway used by ribosomal proteins. Cell 89, 715–725 [DOI] [PubMed] [Google Scholar]
- 89. Leslie D. M., Zhang W., Timney B. L., Chait B. T., Rout M. P., Wozniak R. W., Aitchison J. D. (2004) Characterization of karyopherin cargoes reveals unique mechanisms of Kap121p-mediated nuclear import. Mol. Cell. Biol. 24, 8487–8503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Kaffman A., Rank N. M., O'Shea E. K. (1998) Phosphorylation regulates association of the transcription factor Pho4 with its import receptor Pse1/Kap121. Genes Dev. 12, 2673–2683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Kapust R. B., Tözsér J., Fox J. D., Anderson D. E., Cherry S., Copeland T. D., Waugh D. S. (2001) Tobacco etch virus protease: Mechanism of autolysis and rational design of stable mutants with wild-type catalytic proficiency. Protein Eng. 14, 993–1000 [DOI] [PubMed] [Google Scholar]
- 92. Devos D., Dokudovskaya S., Williams R., Alber F., Eswar N., Chait B. T., Rout M. P., Sali A. (2006) Simple fold composition and modular architecture of the nuclear pore complex. Proc. Natl. Acad. Sci. U.S.A. 103, 2172–2177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Oeffinger M., Wei K. E., Rogers R., DeGrasse J. A., Chait B. T., Aitchison J. D., Rout M. P. (2007) Comprehensive analysis of diverse ribonucleoprotein complexes. Nature methods 4, 951–956 [DOI] [PubMed] [Google Scholar]
- 94. Haycock J. W. (1993) Polyvinylpyrrolidone as a blocking agent in immunochemical studies. Anal. Biochem. 208, 397–399 [DOI] [PubMed] [Google Scholar]
- 95. Abramoff M., Magelhaes P., Ram S. (2004) Image Processing with ImageJ. Biophotonics Int. 11, 36–42 [Google Scholar]
- 96. Aitchison J. D., Blobel G., Rout M. P. (1996) Kap104p: A karyopherin involved in the nuclear transport of messenger RNA binding proteins. Science 274, 624–627 [DOI] [PubMed] [Google Scholar]
- 97. Zimmerman S. B., Minton A. P. (1993) Macromolecular crowding: Biochemical, biophysical, and physiological consequences. Annu. Rev. Biophys. Biomol. Struct. 22, 27–65 [DOI] [PubMed] [Google Scholar]
- 98. Strawn L. A., Shen T., Shulga N., Goldfarb D. S., Wente S. R. (2004) Minimal nuclear pore complexes define FG repeat domains essential for transport. Nature Cell Biol. 6, 197–206 [DOI] [PubMed] [Google Scholar]
- 99. Schrader N., Stelter P., Flemming D., Kunze R., Hurt E., Vetter I. R. (2008) Structural basis of the nic96 subcomplex organization in the nuclear pore channel. Mol. Cell 29, 46–55 [DOI] [PubMed] [Google Scholar]
- 100. Furuya M., Haramura M., Tanaka A. (2006) Reduction of nonspecific binding proteins to self-assembled monolayer on gold surface. Bioorg. Med. Chem. 14, 537–543 [DOI] [PubMed] [Google Scholar]
- 101. Maehara A., Taguchi S., Nishiyama T., Yamane T., Doi Y. (2002) A repressor protein, PhaR, regulates polyhydroxyalkanoate (PHA) synthesis via its direct interaction with PHA. J. Bacteriol. 184, 3992–4002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Schlesinger A. P., Wang Y., Tadeo X., Millet O., Pielak G. J. (2011) Macromolecular crowding fails to fold a globular protein in cells. J. Am. Chem. Soc. 133, 8082–8085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Cardarelli F., Lanzano L., Gratton E. (2011) Fluorescence correlation spectroscopy of intact nuclear pore complexes. Biophys. J. 101, L27-L29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Izaurralde E., Kutay U., von Kobbe C., Mattaj I. W., Görlich D. (1997) The asymmetric distribution of the constituents of the Ran system is essential for transport into and out of the nucleus. EMBO J. 16, 6535–6547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Floer M., Blobel G., Rexach M. (1997) Disassembly of RanGTP-karyopherin β complex, an intermediate in nuclear protein import. J. Biol. Chem. 272, 19538–19546 [DOI] [PubMed] [Google Scholar]
- 106. Rexach M., Blobel G. (1995) Protein import into nuclei: Association and dissociation reactions involving transport substrate, transport factors, and nucleoporins. Cell 83, 683–692 [DOI] [PubMed] [Google Scholar]
- 107. Bednenko J., Cingolani G., Gerace L. (2003) Nucleocytoplasmic transport: Navigating the channel. Traffic 4, 127–135 [DOI] [PubMed] [Google Scholar]
- 108. Mammen M., Choi S., Whitesides G. (1998) Polyvalent interactions in biological systems: Implications for design and use of multivalent ligands and inhibitors. Angew. Chem. Int. Ed. Engl. 37, 2755–2794 [DOI] [PubMed] [Google Scholar]
- 109. Koshland D. E., Jr., Neet K. E. (1968) The catalytic and regulatory properties of enzymes. Annu. Rev. Biochem. 37, 359–410 [DOI] [PubMed] [Google Scholar]
- 110. Lomize A. L., Pogozheva I. D., Lomize M. A., Mosberg H. I. (2007) The role of hydrophobic interactions in positioning of peripheral proteins in membranes. BMC Struct. Biol. 7, 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Jäkel S., Mingot J. M., Schwarzmaier P., Hartmann E., Görlich D. (2002) Importins fulfil a dual function as nuclear import receptors and cytoplasmic chaperones for exposed basic domains. EMBO J. 21, 377–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Zhou H. X., Rivas G., Minton A. P. (2008) Macromolecular crowding and confinement: Biochemical, biophysical, and potential physiological consequences. Annu. Rev. Biophys. 37, 375–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Lim R. Y., Aebi U., Stoffler D. (2006) From the trap to the basket: Getting to the bottom of the nuclear pore complex. Chromosoma 115, 15–26 [DOI] [PubMed] [Google Scholar]
- 114. Lim R. Y., Fahrenkrog B., Köser J., Schwarz-Herion K., Deng J., Aebi U. (2007) Nanomechanical basis of selective gating by the nuclear pore complex. Science 318, 640–643 [DOI] [PubMed] [Google Scholar]
- 115. Lim R. Y., Köser J., Huang N. P., Schwarz-Herion K., Aebi U. (2007) Nanomechanical interactions of phenylalanine-glycine nucleoporins studied by single molecule force-volume spectroscopy. J. Struct. Biol. 159, 277–289 [DOI] [PubMed] [Google Scholar]
- 116. Lim R. Y., Deng J. (2009) Interaction forces and reversible collapse of a polymer brush-gated nanopore. ACS Nano. 3, 2911–2918 [DOI] [PubMed] [Google Scholar]
- 117. Ribbeck K., Görlich D. (2002) The permeability barrier of nuclear pore complexes appears to operate via hydrophobic exclusion. EMBO J. 21, 2664–2671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Peters R. (2005) Translocation through the nuclear pore complex: Selectivity and speed by reduction-of-dimensionality. Traffic 6, 421–427 [DOI] [PubMed] [Google Scholar]
- 119. Peleg O., Lim R. Y. (2010) Converging on the function of intrinsically disordered nucleoporins in the nuclear pore complex. Biol. Chem. 391, 719–730 [DOI] [PubMed] [Google Scholar]
- 120. Ma J., Yang W. (2010) Three-dimensional distribution of transient interactions in the nuclear pore complex obtained from single-molecule snapshots. Proc. Natl. Acad. Sci. U.S.A. 107, 7305–7310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Zilman A. (2009) Effects of multiple occupancy and interparticle interactions on selective transport through narrow channels: Theory versus experiment. Biophys. J. 96, 1235–1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Conti E., Izaurralde E. (2001) Nucleocytoplasmic transport enters the atomic age. Curr. Opin. Cell Biol. 13, 310–319 [DOI] [PubMed] [Google Scholar]
- 123. Xu D., Farmer A., Chook Y. M. (2010) Recognition of nuclear targeting signals by Karyopherin-Î2 proteins. Curr. Opin. Struct. Biol. 20, 782–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Tetenbaum-Novatt J., Rout M. P. (2010) The mechanism of nucleocytoplasmic transport through the nuclear pore complex. Cold Spring Harbor Symp. Quant. Biol. 75, 567–584 [DOI] [PubMed] [Google Scholar]
- 125. Zilman A., Di Talia S., Chait B. T., Rout M. P., Magnasco M. O. (2007) Efficiency, selectivity, and robustness of nucleocytoplasmic transport. PLoS Comput. Biol. 3, e125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Zilman A., Di Talia S., Jovanovic-Talisman T., Chait B. T., Rout M. P., Magnasco M. O. (2010) Enhancement of transport selectivity through nano-channels by non-specific competition. PLoS Comput. Biol. 6, e1000804. [DOI] [PMC free article] [PubMed] [Google Scholar]