Abstract
Among mammalian sirtuins, SIRT7 is the only enzyme residing in nucleoli where ribosomal DNA is transcribed. Recent reports established that SIRT7 associates with RNA Pol I machinery and is required for rDNA transcription. Although defined by its homology to the yeast histone deacetylase Sir2, current knowledge suggests that SIRT7 itself has little to no deacetylase activity. Because only two SIRT7 interactions have been thus far described: RNA Pol I and upstream binding factor, identification of proteins and complexes associating with SIRT7 is critical to understanding its functions. Here, we present the first characterization of SIRT7 interaction networks. We have systematically investigated protein interactions of three EGFP-tagged SIRT7 constructs: wild type, a point mutation affecting rDNA transcription, and a deletion mutant lacking the predicted coiled-coil domain. A combinatorial proteomics and bioinformatics approach was used to integrate gene ontology classifications, functional protein networks, and normalized abundances of proteins co-isolated with SIRT7. The resulting refined proteomic data set confirmed SIRT7 interactions with RNA Pol I and upstream binding factor and highlighted association with factors involved in RNA Pol I- and II-dependent transcriptional processes and several nucleolus-localized chromatin remodeling complexes. Particularly enriched were members of the B-WICH complex, such as Mybbp1a, WSTF, and SNF2h. Prominent interactions were validated by a selected reaction monitoring-like approach using metabolic labeling with stable isotopes, confocal microscopy, reciprocal immunoaffinity precipitation, and co-isolation with endogenous SIRT7. To extend the current knowledge of mechanisms involved in SIRT7-dependent regulation of rDNA transcription, we showed that small interfering RNA-mediated SIRT7 knockdown leads to reduced levels of RNA Pol I protein, but not messenger RNA, which was confirmed in diverse cell types. The down-regulation of RNA Pol I protein levels placed in the context of SIRT7 interaction networks led us to propose that SIRT7 plays a crucial role in connecting the function of chromatin remodeling complexes to RNA Pol I machinery during transcription.
The correlation between histone acetylation status and transcriptional regulation is well established in that hyperacetylation is commonly associated with transcriptional activation, whereas hypoacetylation is associated with transcriptional repression (1–3). In eukaryotes, Sir2-like proteins form a family of enzymes known as sirtuins (4). Distinct from class I and II histone deacetylases (HDACs), which facilitate hydrolysis of acetyl-lysine as a bimolecular reaction (5), sirtuins consume an equimolar amount of nicotinamide adenine dinucleotide (NAD+) for each hydrolysis reaction and generate nicotinamide, O-acetyl ADP-ribose, and deacetylated substrate as end products (4). Based on homology of the sirtuin core domain, there are seven sirtuins (SIRT1–7) in human cells with functions not limited to histone deacetylation, because many additional cellular proteins have been identified as substrates (4, 6). In addition, the localizations of human sirtuins are distributed across multiple organelles including nucleolus, nucleus, cytoplasm, and mitochondria (7).
Systematic monitoring of the cellular localizations of all human sirtuins has demonstrated that sirtuin 7 (SIRT7) is the only enzyme that localizes to nucleoli, similar to that of the founding family member Sir2 in yeast (7). Studies of the yeast histone deacetylase Sir2 have shown its involvement in several biological processes, the majority of these functions being related to transcriptional silencing (8). However, unlike yeast Sir2, there have been no reports to indicate involvement of SIRT7 in transcriptional silencing; instead, several lines of evidence have shown that SIRT7 plays a positive role in activating rDNA transcription (9, 10). It has been proposed that SIRT7 physically interacts with RNA polymerase I (denoted as Pol I) machinery and that its deacetylation activity is crucial for maintaining the elongation phase of Pol I (9). However, several important issues remain unresolved in this model because in vivo substrates have not been identified to date, and the evidence for deacetylation activity based on in vitro assays is conflicting (7, 9, 11). In addition, there appears to be some uncertainty as to whether SIRT7 directly associates with Pol I, because one study showed that instead of interacting with subunits within the Pol I machinery, SIRT7 preferentially interacts with the nucleolar upstream-binding factor (UBF) (10). Moreover, the disruption of SIRT7 nucleolar localization followed by inhibition of rDNA transcription is not a generally observed effect across different cell lines (9, 10). Therefore, the mechanisms by which SIRT7 regulates rDNA transcription remain unclear.
Based on phylogenetic analysis of sirtuin core domains, sirtuins are further divided into five separate classes (6). Although yeast Sir2 and human SIRT7 share the same nucleolar localization, they belong to different phylogenetic classes, I and IV, respectively (6). This may explain why SIRT1, which belongs to the same class (class I) as Sir2, has a well established deacetylase activity and numerous substrates (4), whereas SIRT6 and SIRT7, which are the only two members of class IV, have 1000-fold less or undetectable deacetylase activities (12). Because it has been reported that SIRT7 has no ADP-ribosylation activity, it is necessary to identify its interacting proteins to understand the role of this enzyme in rDNA transcription and other cellular processes.
Here, we have carried out, to our knowledge, the first proteomic study aimed at establishing the interacting proteins of SIRT7. We have systematically investigated the protein networks of EGFP-tagged SIRT7 from three different constructs including a full-length wild type form and two mutant forms containing either a point mutation affecting rDNA transcription or a deletion of exon 2 within the predicted coiled-coil domain. Candidate interactions that showed significant enrichment in SIRT7 isolations relative to EGFP control isolations were subjected to gene ontology classification and functional network analysis, revealing a subset of nucleolar-enriched proteins, chromatin remodeling and modification factors, and transcriptional regulators. By correlating normalized immunoisolated protein abundance with these prominent functional categories, we refined the proteomic data set into targets of significant biological interest. These included Pol I- and II-dependent transcriptional processes as well as several nucleolar chromatin remodeling complexes, in particular the B-WICH complex. The interactions were then validated using a combinatorial approach integrating confocal microscopy, reciprocal immunoaffinity precipitation, metabolic labeling with stable isotopes, and co-isolation with endogenous SIRT7. We further demonstrated that knockdown of SIRT7 leads to down-regulation of Pol I machinery at the protein level. This down-regulation was SIRT7-specific, being partially rescued in a siRNA-resistant SIRT7 cell line. This observation indicates that a loss of SIRT7 functions may lead to arrest or halting of the Pol I complexes and subsequent Pol I degradation. We propose a model in which SIRT7 regulates RNA Pol I transcription through interaction with chromatin remodeling complexes.
EXPERIMENTAL PROCEDURES
Materials
Antibodies used were in-house developed rabbit polyclonal anti-GFP (13), rabbit polyclonal anti-SIRT7 (a generous gift from Dr. Izumi Horikawa of NCI, National Institutes of Health) (7), mouse monoclonal anti-GFP (Roche Applied Science), mouse monoclonal anti-UBF (F-9; Santa Cruz), mouse monoclonal anti-RPA194 (C-1; Santa Cruz), rabbit polyclonal anti-hSNF2H (H-300; Santa Cruz), rabbit polyclonal anti-RIF1 (Bethyl Laboratories Inc.), rabbit polyclonal anti-ATRX (Bethyl Laboratories Inc.), rabbit polyclonal anti-Mybbp1a (Bethyl Laboratories Inc.), and rabbit polyclonal anti-c-Myc (Cell Signaling). All of the oligonucleotide primers designed for this study were ordered from Integrated DNA Technologies and are listed in supplemental Table S1. Human SIRT7 siRNA and a universal siRNA negative control were purchased from Sigma-Aldrich. Protein A/G Plus-agarose was purchased from Santa Cruz. All other reagents were purchased from Sigma-Aldrich unless otherwise specified.
Generation of EGFP-tagged SIRT7, SIRT7 Mutants, and siRNA-resistant SIRT7 Cell Lines
HEK293 (Human embryonic kidney 293), HeLa (cervical carcinoma cell), A549 (human lung adenocarcinoma epithelial cell line), U2OS (human osteosarcoma cell line), and ZR-75–1 (human breast carcinoma cell line) were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum in 37 °C with 5% CO2. HL60 (human promyelocytic leukemia cells) were cultured in RPMI supplemented with 10% fetal bovine serum. The plasmids containing SIRT7 cDNAs with full-length (plasmid 20268), S111A point mutant (plasmid 20269), and exon 2 deletion (plasmid 13818) were purchased from Addgene. SIRT7 cDNAs were amplified by PCR using SIRT7-specific primers (supplemental Table S1), purified, and digested with EcoRI and XhoI restriction enzymes. The digestion products were ligated to the 5′ end of EGFP cDNA (pEGFP-N1; Clontech) that we cloned into a pLXSN retroviral vector (Clontech), thus generating pLXSN-SIRT7-EGFP-FLAG retroviral plasmids. RNA interference-resistant clones were generated by site-directed mutagenesis using primers listed in supplemental Table S1. HEK293 cell lines stably expressing EGFP-FLAG or SIRT7-EGFP-FLAG full-length wild type or mutants were generated using the PhoenixTM retrovirus expression system (Orbigen, San Diego, CA). Briefly, the different pLXSN-SIRT7-EGFP-FLAG plasmids were transfected into Phoenix cells using FuGENE (Roche Applied Science), the cells were grown to 90% confluency, and the resulting retrovirus was collected and used to transduce HEK293 cells. The cells stably expressing SIRT7-EGFP-FLAG and derivatives were selected with 400 μg/liter G418 (EMD, Gibbstown, NJ) for 2 weeks and sorted by fluorescence-activated cell sorting (Vantage S.E. with TurboSort II; Becton Dickinson, Franklin Lakes, NJ). The expression of all EGFP fusion proteins was further confirmed by confocal microscopy and immunoblotting.
siRNA-resistant clones were generated based on site-directed mutagenesis procedures. In brief, the primer set (supplemental Table S1) containing several silent mutations at third bases of codons (wobble position) was used in PCR amplification using pLXSN-SIRT7-EGFP-Flag plasmid as template. The reaction mixture was digested with DpnI restriction enzyme to remove the wild type strain. Aliquots of the reaction containing siRNA-resistant clones were transformed into DH5α strain and selected by ampicillin. The cell lines containing siRNA-resistant clones were generated according to the protocol mentioned above.
Isolation of SIRT7-containing Protein Complexes
SIRT7 and control immunoaffinity purifications on magnetic beads were performed via EGFP, as previously described (13, 14). HEK293 cell lines stably expressing EGFP alone, SIRT7WT-EGFP-FLAG (WT), SIRT7S111A-EGFP-FLAG (S111A), and SIRT7dE2-EGFP-FLAG (where dE2 indicates deletion of exon 2; see Fig. 1A) were washed with 10 ml of ice-cold D-PBS (Invitrogen) per 15-cm plate, then harvested by scraping with a plastic spatula (Fisher Scientific) on ice, pelleted at 200 × g at 4 °C for 5 min, and washed with ice-cold D-PBS (Invitrogen). The washed cell pellet was gently mixed with 100 μl/1 g of cells of 20 mm HEPES-NaOH, pH 7.5, containing 1.2% polyvinylpyrrolidone (w/v) and 1:100 (v/v) protease inhibitor mixture (Sigma), then frozen in liquid nitrogen, and subjected to cryogenic lysis using a Retsch MM 301 Mixer Mill (10 steps × 2 min at 30 Hz) (Retsch, Newtown, PA). All of the subsequent steps were performed at 4 °C unless otherwise noted. The cell powder (0.8 g) was suspended in 10 ml of ice-cold optimized lysis buffer (20 mm HEPES-KOH, pH 7.4, containing 0.1 m potassium acetate, 2 mm MgCl2, 0.1% Tween 20, 1 μm ZnCl2, 1 μm CaCl2, 0.5% Triton X-100, 250 mm NaCl, 4 μg/ml DNase, 1/100 (v/v) protease, and phosphatase inhibitor cocktails). The optimization of the lysis buffer for efficient solubilization and isolation of SIRT7-EGFP was performed as reported (15) and is illustrated in supplemental Figs. S3–S5 and described in the supplemental text. The resulting cell suspension was subjected to homogenization using a Polytron (2 × 15 s) (Kinematica), and the insoluble material was removed by centrifugation at 8,000 × g for 10 min. The cell lysates (supernatant) were incubated for 45 min with 7 mg of magnetic beads (M270 Epoxy Dynabeads; Invitrogen) conjugated with rabbit anti-GFP antibody, as described (16). The magnetic beads were then washed six times with lysis buffer (without protease and phosphatase inhibitors) and once with distilled H2O. The washed beads were incubated with 40 μl of 1× LDS sample buffer (Invitrogen) for 10 min at 70 °C, followed by shaking for 10 min at room temperature. Eluted proteins were reduced with DTT (50 mm for 10 min at 70 °C) and alkylated with iodoacetamide (100 mm, 30 min at room temperature). The samples were either stored at −20 °C or immediately processed for proteomic analysis, as described below.
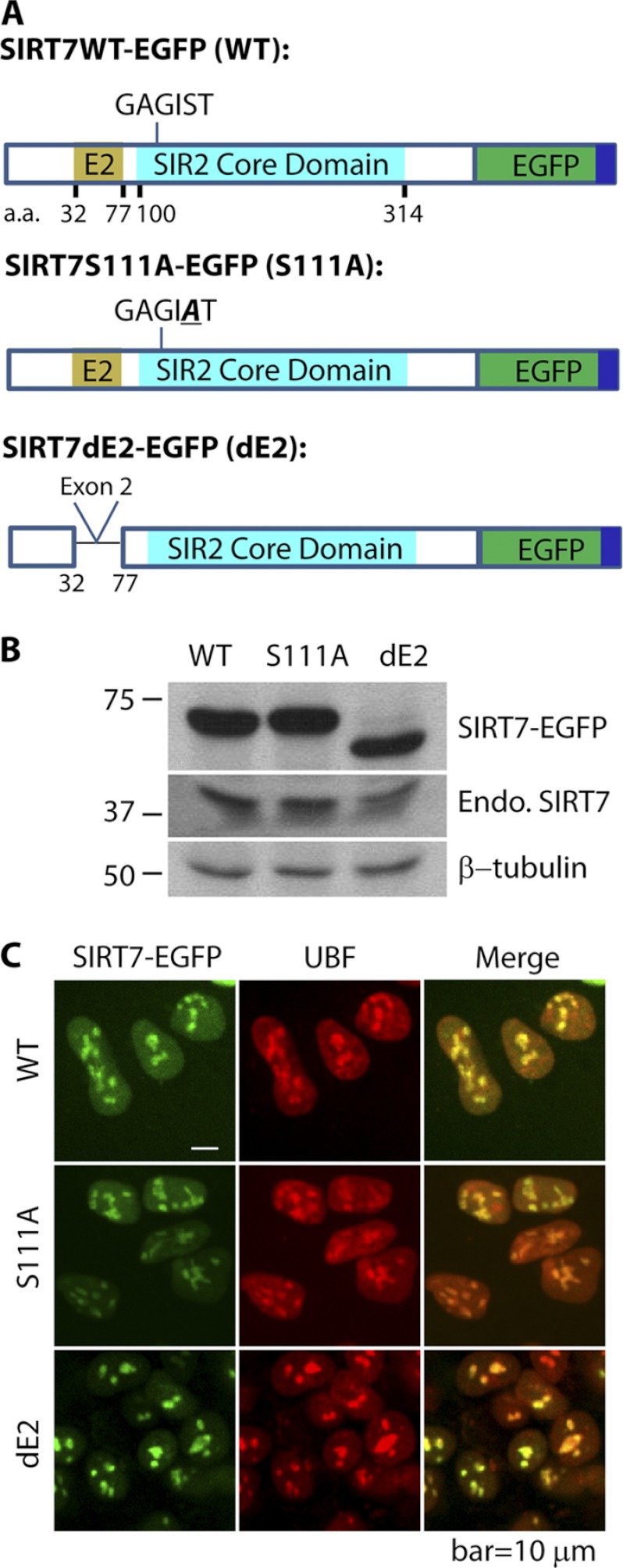
Fig. 1.
Characterization of cell lines expressing EGFP-tagged SIRT7 derivatives. A, diagram of three SIRT7 derivatives tagged with EGFP (dark green) and FLAG (dark blue)—wild type SIRT7 (WT), SIRT7 S111A mutant, containing a Ser to Ala mutation in the conserved GAGIST motif within the core domain (turquoise), and SIRT7 dE2 with deleted exon 2 (E2, light brown). B, immunoblotting with anti-GFP and anti-SIRT7 antibodies confirms the expression and approximate molecular masses of EGFP-tagged SIRT7 variants. C, the three EGFP-tagged SIRT7 variants have predominant nucleolar localizations and co-localize with nucleolar protein UBF, as confirmed by immunofluorescence using anti-UBF and anti-GFP antibodies in HEK293 cells. A 60× oil immersion lens was used. Endo. SIRT7, endogenous SIRT7. Bar, 10 μm.
Validation of Interactions via Reciprocal Isolations
Reciprocal immunoaffinity purifications were performed using 0.1 g of cell pellet from SIRT7WT-EGFP-FLAG (WT) expressing cells for each reaction and the lysis buffer (20 mm HEPES-KOH, pH 7.4, containing 0.1 m potassium acetate, 1 mm MgCl2, 0.1% Tween 20, 0.5% Triton X-100, 150 mm NaCl, 4 μg/ml DNase, 1/100 (v/v) protease inhibitor mixture (Sigma)). The proteins were immunoisolated with 4 μg of antibody to either control IgG or identified SIRT7 protein interactions (e.g. UBF, RPA194, and Mybbp1a) for 1 h, followed by 2 h of incubation with 30 μl of protein A/G Plus-agarose beads (Santa Cruz). The agarose beads were subsequently washed twice with lysis buffer and twice with D-PBS, resuspended in 50 μl of 6× Laemmli sample buffer and heated to 95 °C for 10 min. The identities of the co-isolated proteins were analyzed by Western blot analysis.
Confocal Microscopy
HEK293 cell lines stably expressing different SIRT7-EGFP-FLAG fusion proteins were cultured on glass-bottomed dishes, pretreated with poly-d-lysine (Sigma). After 48 h, the cells were fixed with 2% paraformaldehyde, washed with D-PBS, incubated with 1 μg/ml 4′,6′-diamino-2-phenylindole in D-PBS (PBST) for 15 min, and visualized by confocal microscopy on a PerkinElmer Life Sciences RS3 spinning disk using a 60× oil immersion lens. For co-localization analysis, the cells were cultured and fixed as above, permeabilized with 0.1% Triton X-100 in PBST for 15 min, and blocked in 2% (w/v) bovine serum albumin, 0.2% (v/v) Tween 20 in PBST at room temperature for 60 min. Incubation with a primary antibody was performed at room temperature for 60 min in blocking buffer. The cells were then washed with PBST and incubated with secondary antibodies conjugated to Alexa 546 or 633 (Invitrogen). Finally, the cells were washed and incubated with 1 μg/ml TO-PRO-3 iodide (Invitrogen) in PBST for 15 min, then washed again, and mounted on glass slides with a drop of Aqua Poly/Mount media (Polysciences) to proceed with confocal imaging.
Proteomic and Mass Spectrometry Analyses of SIRT7 Complexes
Primary eluates from SIRT7 immunoisolations were partially resolved (∼2 cm) on 4–12% Bis-Tris NuPAGE gels and stained with SimplyBlue Coomassie stain (Invitrogen). Each lane corresponding to a single immunoisolation was excised, cut into 1-mm slices, and pooled into six equal fractions. The gel pieces were destained in 50 mm ammonium bicarbonate, 50% acetonitrile (ACN), followed by two rounds of dehydration, and rehydration in 100% ACN and 50 mm ammonium bicarbonate, respectively. Washed gel pieces were dehydrated a final time in 100% ACN and then incubated with 12.5 ng/μl of sequencing grade trypsin (Promega, Madison, WI) overnight at 37 °C. The peptides were extracted in 0.5% formic acid for 4 h at room temperature, followed by a second extraction in 0.5% formic acid, 50% ACN for 2 h. The peptides were concentrated by vacuum centrifugation to ∼10 μl, and half of the sample was analyzed by nLC-MS/MS on a Dionex Ultimate 3000 RSLC coupled directly to an LTQ-Orbitrap Velos ETD mass spectrometer (ThermoFisher Scientific). The peptides were washed onto the trap column (Magic C18 AQ, 3 μm, 100 μm × 2.5 cm; Michrom Bioresources, Inc.) in 0.5% TFA, 1% ACN, 98.5% water and desalted for 5 min at 5 μl/min and then separated by reverse phase chromatography (Acclaim PepMap RSLC, 1.8 μm, 75 μm × 25 cm) at a flow rate of 250 nl/min using a 90-min discontinuous gradient of ACN as follows: 4% to 20% B over 50 min, 20% to 40% B over 40 min (Mobile phase A: 0.1% formic acid, 0.1% acetic acid in water, Mobile phase B: 0.1% formic acid, 0.1% acetic acid in 97% ACN). The mass spectrometer was operated in data-dependent acquisition mode with FT preview scan disabled and predictive AGC and dynamic exclusion enabled (repeat count, 1; exclusion duration, 70 s). A single acquisition cycle comprised a single full scan mass spectrum (m/z = 350–1700) in the Orbitrap (r = 30,000 at m/z = 400), followed by CID fragmentation on the top 20 most intense precursor ions (minimum signal = 1E3) in the dual pressure linear ion trap. The following instrument parameters were used: FT MS and IT MSn (tandem MS) target values of 1E6 and 5E3, respectively; FT MS and IT MSn maximum injection time of 300 and 100 ms, respectively. CID fragmentation was performed at an isolation width of 2.0 Th, normalized collision energy of 30, and an activation time of 10 ms.
Data Processing and Functional Protein Analyses
MS/MS spectra from raw files corresponding to single biological samples (one gel lanes, n = 6 fractions) were extracted by Proteome Discoverer (version 1.3; ThermoFisher Scientific) and submitted to SEQUEST (version 1.20) for database searching against the UniProt SwissProt sequence database (downloaded November 2010) containing the subset of human, herpesvirus, and common contaminant sequences (21,570 entries). Spectra were searched against indexed peptide databases, generated from the forward and reverse protein sequence entries, using the following settings: full enzyme specificity, maximum of two missed cleavages, parent and fragment ion mass tolerances of 10 ppm and 0.5 Da, respectively, static modification of carbamidomethylcysteine (+57 Da), variable modifications of methionine oxidation (+16 Da), phosphoserine, threonine, and tyrosine (+80 Da), and acetyl-lysine (+42 Da). Peptide spectrum matches were loaded into Scaffold (ver. 3.2; Proteome Software, Inc.) for post-search validation by sequential searching using PeptideProphet and ProteinProphet (17, 18) followed by X!Tandem (GPM 2010.12.1.1) re-search of the peptide spectrum matches using the subset database parameter and additional variable modification of deamidation of asparagine and glutamine (+1 Da) and pyro-Glu formation of peptide N-terminal glutamate (+17 Da). The high mass accuracy search option was enabled. Peptide spectrum matches across all conditions (see Fig. 2B) were loaded into a single Scaffold session, and the following confidence filters were selected to reduce peptide and protein global false discovery rate to <1%: 99% protein confidence, 95% peptide confidence, minimum of two unique peptides/protein in at least one biological samples. Proteins passing these filters were exported with their respective “unweighted spectrum counts” to Excel for further processing.
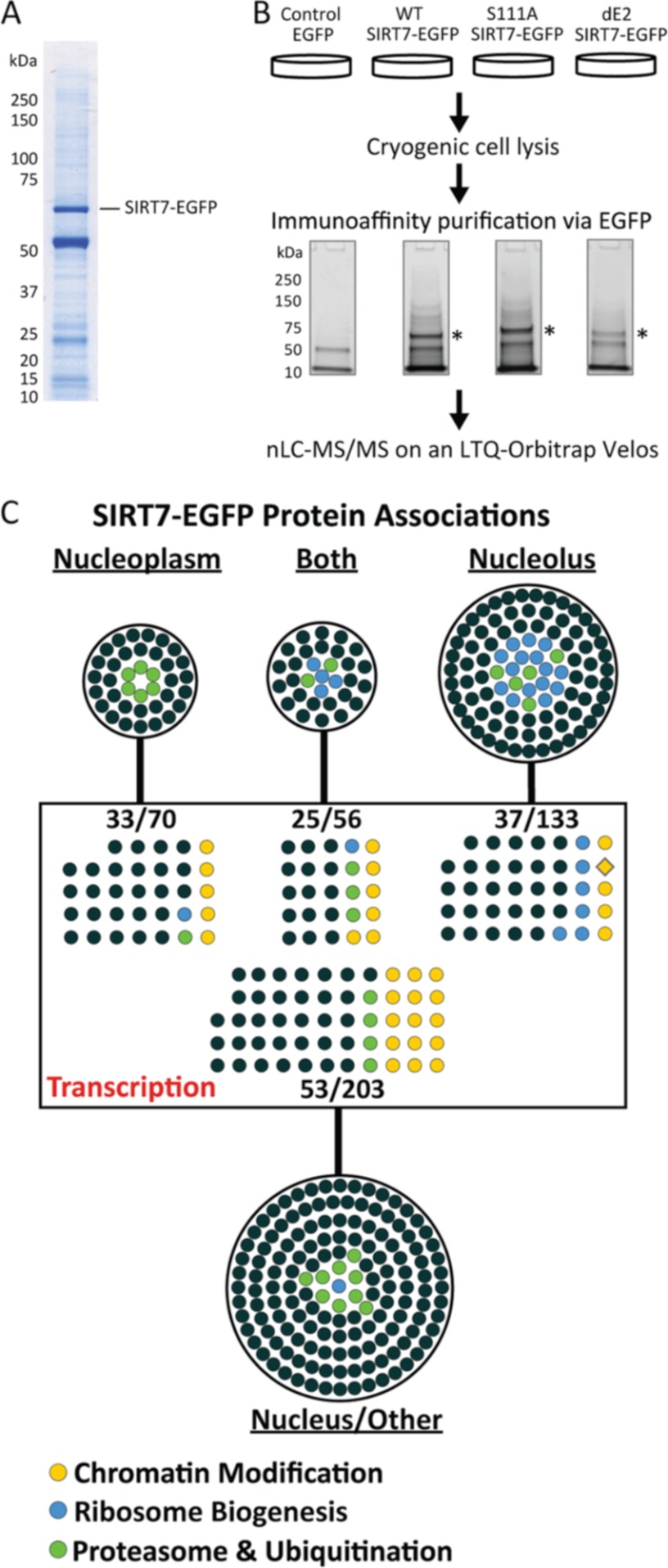
Fig. 2.
Immunoisolation and gene ontology analysis of SIRT7 protein complexes. A, representative one-dimensional SDS-PAGE Coomassie-stained gel demonstrating the immunoisolation of SIRT7WT-EGFP, corresponding to the most prominent stained band (∼70 kDa). B, experimental design for parallel immunoaffinity purifications of EGFP, SIRT7WT-EGFP, SIRT7S111A-EGFP, and SIRT7dE2-EGFP. Immunoisolates were resolved ∼2 cm by one-dimensional SDS-PAGE. Under these conditions, the molecular mass difference between SIRT7WT- and SIRT7dE2-EGFP cannot be visualized. In-gel trypsin digestion was performed on the whole gel lane. The peptides were pooled into six fractions per condition and subsequently analyzed by nLC-tandem mass spectrometry on an LTQ-Orbitrap-Velos. *, SIRT7-EGFP. C, Cytoscape network visualization of putative SIRT7-EGFP protein interactions classified by gene ontology. A total of 462 proteins were visualized, representing two biological replicates and passing defined specificity (3-fold spectral count enriched versus EGFP) and functional (nuclear localization) filters. The proteins were grouped by annotated subcellular localization (UniProtKB Gene Ontology Annotation) into nucleus/unclassified (203 proteins), nucleoplasm (70 proteins), nucleolus (133 proteins), or both (56 proteins), as well as by biological processes containing the keyword “transcription.” Specific ontological functions are illustrated by colored circles.
To determine co-isolated proteins that were enriched in SIRT7 derivatives versus the EGFP control, a spectral counting approach was performed as follows: 1) a spectral count of 1 was added to all total spectral counts to facilitate calculation of fold enrichment; 2) spectral counts from biological replicates were averaged; 3) proteins with <8 average spectral counts in at least one condition were excluded; 4) spectral counts for co-isolated proteins from the S111A and dE2 conditions were normalized by the factor: (SIRT7WT spectral counts/SIRT7mutant spectral counts); and finally 5) proteins with ≥3-fold enrichment in SIRT7-EGFP isolation (versus EGFP) were retained for further gene ontology analysis (supplemental Table S2). Proteins identified across the SIRT7 immunoisolations were imported into Cytoscape, with nodes representing protein identifications and edges connecting to the respective immunoisolation (WT, S111A, or dE2) in which it was identified. Full gene ontologies (OBO v1.2) and human gene annotations were downloaded from online and respective ontologies were imported into Cytoscape (19) as node attributes. Proteins classified by a “cytoplasmic only” cellular component were excluded (supplemental Table S4). The remaining “nuclear” or “unassigned” proteins (supplemental Table S5) were classified into functional subgroups according to biological processes (supplemental Table S7). All STRING network analyses were performed using protein accessions as input and with the following parameters: “Experimental” and “Databases” evidences @ medium (0.4) confidence level. For visualization, STRING networks were exported to PSI-XML and imported into Cytoscape.
Relative protein abundance between co-isolated proteins was calculated by the NSAF method (20) from proteins in nucleus-enriched functional group and expressed as a percent of the total abundance (supplemental Table S5). Briefly, normalized spectral counts were calculated by averaging each protein's spectral counts between biological replicates and then dividing by the protein's length. Then, normalized spectral counts for each protein were divided by the sum of all normalized spectral counts, and expressed as a percent. To account for effects of cellular abundance on immunoisolation protein abundance, individual NSAF values were normalized by the estimated proteome abundance of the protein obtained from the PAX-DB, by the following equation: log2 (104 × NSAF/PAX). NSAF/PAX values were imported into Cytoscape as node attributes, and the relative abundance was expressed by node size and color, relative to proteins within the network.
Targeted SRM-like I-DIRT Analyses
An overview of the I-DIRT workflow is illustrated in Fig. 4A. Briefly, WT HEK293 cells and HEK293 cells expressing SIRT7WT-EGFP, described above, were cultured for ≤6 passages in heavy isotope (13C6-lysine and 13C6-arginine, Cambridge Isotopes) and light isotope (12C6 and 12C6-arginine) medium, respectively, containing 10% dialyzed FBS (Hyclone). The cells were harvested and lysed by cryogenic disruption. For light/heavy normalization, an aliquot of the mixed cell lysates was reserved and resolved by SDS-PAGE. To correct for the relative loading of the light (L) and heavy (H) samples, two fractions of tryptic peptides from the in-gel digest of input material were analyzed by data-dependent nLC-MS/MS and Proteome Discoverer/SEQUEST, as described above, except for the inclusion of the precursor ion quantification module. This module calculates L/H peptide ratios from L and H precursor areas derived from their respective extracted ion chromatograms. A histogram of L/H protein ratios (n = 594) was constructed and fit to a Gaussian curve. The median curve fit value was used to normalize L/H ratios calculated from targeted, data-independent experiments described below.
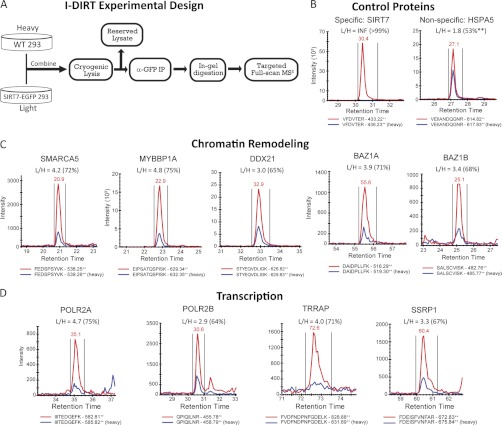
Fig. 4.
Validation of SIRT7-associated chromatin remodeling and transcription proteins by I-DIRT. A, experimental design for performing I-DIRT, a mass spectrometry-based method to assess the relative specificity of interacting proteins from immunoaffinity isolations. HEK293 cells expressing SIRT7WT-EGFP were grown in medium containing natural abundance amino acids for at least six passages, whereas in parallel, wild type HEK293 cells were grown in medium containing 13C6-arginine and 13C6-lysine. The cells were combined and lysed, and SIRT7-EGFP was isolated. Immunoisolates were resolved by SDS-PAGE and digested in-gel with trypsin. Targeted acquisition of full scan MS2 spectra was performed for light/heavy isopeptide pairs selected from unique peptides that were previously observed in label-free studies. B–D, proteins were selected for validation from control (B), chromatin modeling (C), and transcription groups (D). XICs of light (red traces) and heavy (blue traces) isopeptides from full scan MS/MS spectra were reconstructed and integrated using Skyline. Light/heavy (L/H) peptide ratios were calculated directly from the XICs. The percentages in parentheses reflect the relative interaction specificity, % = (L/H)/(1 + L/H), which was normalized by the average ratio (i.e. loading correction of L and H samples) calculated from a Gaussian curve fit to the distribution of input ratios (supplemental Fig. S8A). A detailed example of XIC construction and additional confirmatory peptides are included in supplemental Figs. S8 and S9).
To assess specificity of SIRT7-EGFP interacting proteins, a targeted data-independent experiment was performed. SIRT7-EGFP and interacting proteins were co-isolated by immunoaffinity purification from the mixed cell lysates, resolved by SDS-PAGE, and subjected to in-gel digestion using trypsin. For the data-independent experiments, the Skyline software platform (21) was used to 1) generate spectral libraries from large scale, non-I-DIRT SIRT7 immunoisolation experiments, 2) select the top two unique peptides per protein for targeted SRM-like analysis, 3) generate data-independent LTQ full scan MS2 methods, and 4) perform post-acquisition reconstruction and manual validation of precursor-product ion XICs. A transition report containing all peptides sequences and corresponding proteins targeted for full scan MS2 analyses is provided in supplemental Table S9. Peptides from the I-DIRT SIRT7WT immunoisolation were resolved by nLC as described above and then analyzed in the ion trap of an LTQ-Orbitrap Velos operated in data-independent mode. One complete cycle consisted of a maximum of 16 IT MS2 spectra plus one FT MS spectrum (r = 7500 at m/z 400), using the following instrument parameters: max injection time, 100 ms; MSn (tandem MS) target value, 5E3; isolation width 2.5; normalized collision energy, 35). Peptide identification was confirmed by manual assignment of fragment ions, and at least two fragment ions were manually selected for construction of precursor product ion XICs. L/H ratios were normalized by the median curve fit value calculated from the input material and then converted to a specificity value expressed as [L/H/(1 + L/H)] × 100.
siRNA Knockdown and Real Time Quantitative PCR
siRNA (5′-GGGAGUACGUGCGGGUGUUdTdT; 5′-AACACCCGCACGUACUCCCdTdT) targeting to SIRT7 mRNA was purchased from Sigma. Transfection of siRNA was performed using Lipofectamine RNAiMAX (Invitrogen). Final concentration of siRNA used in each experiment was 20 nm. After 2 days of incubation, the cell lysates were prepared, and knockdown of SIRT7 was confirmed by immunoblotting using anti-GFP and anti-SIRT7 antibodies.
The mRNA levels of different genes upon siRNA or scrambled siRNA treatment were monitored by real time quantitative PCR. In brief, mRNA was purified using TRIzol (Invitrogen) reagent and converted to cDNA according to the reverse transcription protocol (Retroscript, Ambion). Primer sets representing the genes of interest (supplemental Table S1) were mixed with cDNA and the DNA polymerase mixture containing SYBR green I Dye (Power SYBR green PCR master mix; ABI) for real time quantitative PCR (ABI 7900HT). GAPDH was used as internal control for normalization of expression levels.
RESULTS
Characterization of Cell Lines Expressing EGFP-tagged SIRT7 Derivatives
To systematically identify SIRT7-associated proteins, we generated HEK293 cell lines stably expressing EGFP-tagged SIRT7 fusion proteins and EGFP alone and performed immunoaffinity purifications of SIRT7-associated proteins via the EGFP tag. Fig. 1A illustrates SIRT7 domains in WT and two mutants: S111A and dE2 (deletion of exon 2). Overexpression of the SIRT7 Ser-111 to Ala mutant (S111A), which was shown to lack deacetylase activity in other Sir2 homologs (22), has been reported to reduce rDNA transcription in human HEK293T and U2OS cells (9). The S111A mutation is located within a motif (GAGIST; Fig. 1A) conserved in all human SIRTs; its corresponding site in a yeast Sir2 homolog (i.e. HST2) was shown to localize at the C-site of the Rossmann fold that is proposed to be involved in nicotinamide cleavage (22, 23). The second SIRT7 mutant utilized in our study lacks exon 2 (dE2; Fig. 1A), corresponding to a sequence predicted to be a coiled-coil domain (supplemental Fig. S1). Because coiled-coil domains are commonly found in transcription factors to form homodimer (24) and heterodimers (25), exon 2 could contribute to a subset of SIRT7 interactions.
To validate the successful expression of EGFP-tagged SIRT7, the molecular mass of the fusion protein from each cell line was analyzed by Western blotting (Fig. 1B). Antibodies against EGFP and SIRT7 confirmed the presence of the fusion proteins (SIRT7-EGFP) and endogenous SIRT7 (Endo. SIRT7), respectively. The mass difference between WT and the dE2 mutant was apparent, corresponding to truncation of 45 amino acids (∼5 kDa) (Fig. 1B). Earlier studies (7, 9, 10) and our results (supplemental Fig. S2) have shown that endogenous SIRT7 localizes at the nucleolus, evidenced by its co-localization with UBF. To assess the localization patterns of the EGFP-tagged SIRT7 (overexpressed by ∼13-fold compared with endogenous SIRT7; supplemental Fig. S2), immunofluorescence studies were performed. Using confocal microscopy, we observed co-localization of all SIRT7-EGFP constructs with endogenous UBF (Fig. 1C). These results suggest that the putative deacetylase activity and coiled-coil domain are not required for the localization of SIRT7 to nucleoli. Collectively, our results indicate that the SIRT7-EGFP fusion proteins have the appropriate masses and localization to the nucleoli and are therefore suitable for exploring SIRT7 interaction networks.
Directed Proteomics of SIRT7 Protein Interactions Reflects Nucleolar Localization and Roles in Transcription
To perform immunoprecipitation of EGFP-tagged SIRT7 fusion proteins, we first optimized the extraction conditions by examining SIRT7WT-EGFP levels in different lysis buffer conditions (supplemental Fig. S3). Optimal isolation conditions were chosen (supplemental Fig. S3, buffer 3) based on the yield of SIRT7 and other co-isolated proteins (supplemental Fig. S4, condition A) among the different combinations of salt (NaCl) and detergent (Triton X-100) concentrations. Under the optimal conditions, SIRT7WT-EGFP was efficiently isolated and eluted from magnetic beads (>90% efficiency; supplemental Fig. S5, lane E1 versus E2 and Beads) as assessed by Western blotting (supplemental Fig. S5, lane E1 versus Pellet and FT). A representative Coomassie-stained gel of a SIRT7WT-EGFP immunoisolate under the optimized conditions is shown in Fig. 2A.
Using the optimized lysis buffer condition, EGFP control, SIRT7WT-EGFP, SIRT7S11A-EGFP, and SIRT7dE2-EGFP were isolated by immunoaffinity capture from HEK293 stable cell lines. Immunoisolates were partially resolved by SDS-PAGE, digested in-gel with trypsin (six fractions per lane), and analyzed by nLC-tandem MS (MS/MS) on an LTQ-Orbitrap Velos mass spectrometer (Fig. 2B and “Experimental Procedures”). Two independent biological replicates were performed for each condition (Fig. 2B). MS/MS spectra corresponding to LC runs (n = 6) from each single condition and biological replicate were analyzed by SEQUEST database searches (Proteome Discoverer) and loaded into Scaffold to facilitate comparison across biological conditions and replicates. After filtering of peptides and protein identifications across all conditions (≥2 unique peptides, <1% peptide, and protein false discovery rate; “Experimental Procedures”), spectral counting analysis was employed to assess relative enrichment of each interaction for SIRT7-EGFP versus the EGFP control. To retain high confidence interacting partners, multiple steps of filtering were performed. First, spectral counts were averaged between biological replicates. Then proteins with fewer than eight spectral counts in at least one condition were excluded, as were nonspecific proteins, which were defined as proteins showing less than 3-fold spectral count enrichment in at least one SIRT7-EGFP condition (versus EGFP). Additionally, given the nucleolar localization of endogenous and EGFP-tagged SIRT7 (Fig. 1C and supplemental Fig. S2), proteins with a cytoplasm cellular component gene ontology term, but lacking nuclear, nucleoplasm, or nucleolus gene ontology terms were excluded (supplemental Table S4 for excluded cytoplasmic proteins). Although the gene ontology annotation is still incomplete and therefore some of the excluded proteins could associate with SIRT7 (e.g. ribosomal proteins), because this is the first study on SIRT7 interactions, we selected to be more stringent and focus on the proteins with annotated nucleolar and nuclear localizations.
In total, 462 proteins passed these criteria (supplemental Table S5), 97% (448) of which were identified in all three SIRT7 proteins isolated. There were no proteins detected as unique to a single SIRT7 isolation; however, one protein, ATM, was not detected in SIRT7S111A, whereas 13 proteins were not detected in the SIRT7dE2 sample (supplemental Table S6). Because coiled-coil domains are efficient in intermolecular interactions, we performed a coiled-coil domain analysis on these 13 proteins and found that 7 contained putative coiled-coil domain (supplemental Fig. S6). As with most immunoaffinity purification strategies, the 462 identified proteins represent both direct and indirect associations. In fact, the majority of co-isolated proteins are likely to represent indirect associations. The fact that a high number of proteins was isolated with SIRT7 can be interpreted from different angles. The lysine deacetylases that are best studied, such as SIRT1 and HDAC1, are known to be part of multiple large protein complexes and have numerous interactions. Therefore, these putative SIRT7 interactions may also represent different complexes and functions of SIRT7. Nevertheless, several cautionary notes have to be considered. The proteins that are real interactions of SIRT7 can act as new interacting sites for nonspecific associations with high abundance proteins or proteins that would encounter each other only after cell lysis. This type of nonspecific association would not be controlled for by parallel immunoaffinity purification of EGFP alone. Additionally, the high sensitivity of current mass spectrometric instrumentation allows the identification of traces of co-isolated proteins. Therefore, their enrichment in the SIRT7 isolation and specificity must be evaluated by alternative approaches such as I-DIRT, co-localization, and functional analyses.
First, to pursue an unbiased assessment of the function of putative SIRT7 interactions, proteins were classified according to their subcellular localization (within the nucleus) and biological functions (Fig. 2C and supplemental Table S7). Among the proteins having a specific annotated subnuclear localization (nucleoplasm, nucleolus, or both), ∼74% (189 of 257) had nucleolar localization, consistent with the cellular localization of SIRT7. The most prominent category for the biological function ontology contained 150 proteins assigned to transcription (Fig. 2C, in the rectangle), which included broad terms such as DNA-dependent transcriptional regulation, as well as more specific classifications related to RNA polymerase I and II transcription, of which there were 4 and 56 proteins, respectively. Consistent with recent reports implicating SIRT7 in regulation of rDNA transcription, 28 proteins were ascribed to ribosomal biogenesis (Fig. 2C, blue circles). Interestingly, 32 and 30 putative interacting proteins were classified into chromatin modification/remodeling and proteasome/ubiquitination processes (supplemental Table S7), respectively. Given the lack of knowledge regarding the molecular mechanisms of SIRT7 function within the nucleus, we speculated that the specific protein functions represented within chromatin modification and protein degradation categories could provide new insights into the potential roles of SIRT7 in coordinating the regulation of gene and protein expression.
Functional Network Analysis Predicts a Prominent Role for Chromatin Modification and Remodeling Complexes in SIRT7-mediated Functions
To test this hypothesis, we analyzed the 32 proteins that belonged to the chromatin modification group by STRING, a knowledge base of protein associations that statistically evaluates multiple experimental and predictive evidence lines to cluster proteins with similar or related functionality (26). As shown in Fig. 3A, 26 of these proteins form a highly interconnected network representing diverse chromatin-related protein functions, including the chromatin remodeling factors BPTF and RSF1, and several core components of the SWI/SNF complex (ARID1A, SMARCA4, SMARCC1, and SMARCC2)—a chromatin remodeling complex that preferentially associates with regulatory elements connected to transcription and chromosome organization (27). In addition, chromatin modification enzymes involved in deacetylation (HDAC2) and methylation (DNMT1, KDM1A, and PRMT5) were also integrated into this network. Of interest was the notable absence of a functional connection between SIRT7 and the chromatin modification network (Fig. 3A and supplemental Fig. S7A), reflecting the fact that SIRT7 association to chromatin remodeling complexes has not been previously described. Because these proteins represent a multitude of genetic regulatory mechanisms, we sought to further refine the scope of factors that could be of prime importance in regulating SIRT7 function and also to explore alternative processes through which SIRT7 could be functionally linked to chromatin modification.
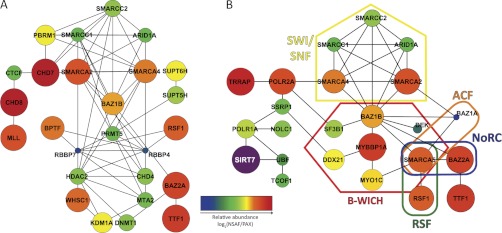
Fig. 3.
Protein network analysis links SIRT7 to chromatin modification and remodeling complexes. Protein networks were assembled by STRING and visualized using Cytoscape. Genes without known functional connections were excluded and are presented in supplemental Fig. S7. Node color and size represent relative protein abundance (NSAF) within SIRT7 immunoisolates that were normalized by overall human proteome abundance (PAX). Both NSAF and PAX abundance values are calculated from mass spectrometric spectral counting data (see “Experimental Procedures”). A, protein network assembled by STRING from genes with chromatin modification or chromatin remodeling biological process ontology terms (Fig. 2C, yellow circles, and supplemental Table S6). 26 of 33 genes (yellow colored nodes) were connected into a functional network. B, functional network representing 22 proteins that link SIRT7 to chromatin remodeling complexes via transcriptional regulation. STRING analysis was conducted on the subset of chromatin remodeling complex members from A, supplemented with previously known SIRT7 interactions and members of chromatin remodeling complexes that were also in the nuclear classification of SIRT7 immunoisolates (Fig. 2A and supplemental Table S7).
To address the former, we first calculated the average relative abundance of nuclear-enriched proteins in the SIRT7 isolations (supplemental Table S5) using NSAF values (20). We hypothesized that proteins representing strong associations would have greater relative abundance in the SIRT7 immunoisolation and therefore could be core factors essential for the regulation of its function. The total spectral count abundance was calculated as the sum of normalized spectral counts for all nuclear-enriched proteins (n = 462) (see “Experimental Procedures”). Additionally, to correct for the effect of total cellular abundance on immunoisolation abundance, individual NSAF values were normalized to the respective abundance of each protein in the PAX database (28), which contains estimated protein abundances calculated from a collection of large scale spectral counting proteome analyses in the PeptideAtlas database (29) (supplemental Table S5). To compare the protein abundance relationships within a network, log2-transformed abundance values were represented in the Cytoscape networks (Fig. 3) by node color and size, with larger and warmer-colored nodes representing the most abundant proteins within an individual network. This quantitative analysis revealed that a subset of chromatin remodeling complex members were more abundant than others (Fig. 3A, orange-to-red nodes), including SMARCA2, SMARCA4, RSF1, BAZ1B, BAZ2A, and TTF1. In contrast, proteins with roles in direct histone modification processes, such as HDAC2, RBBP7, RBBP4, CHD4, MTA2, and PRMT5, were less abundant (Fig. 3A, blue-to-green nodes). Interestingly, a dichotomy in abundance existed between SWI/SNF complex members (SMARCA2/4 versus SMARCC1/2 and ARID1A). Because these proteins can exist within multiple distinct complexes, it suggests that additional chromatin remodeling complexes containing these abundant interactions (Fig. 3A, orange-to-red nodes) could participate in the cellular activities of SIRT7.
Quantitative Functional Network Analysis Highlights a Central Role for the B-WICH Remodeling Complex in SIRT7 Function via RNA Polymerase Signaling Pathways
We next explored the possibility that our initial unbiased ontological and network analysis did not reveal the full range of associated remodeling complexes. Using prior literature knowledge and protein abundance (NSAF/PAX) as a reference, we interrogated the nuclear-enriched proteins from SIRT7 immunoisolations, searching for potential chromatin remodeling complex members. Surprisingly, this analysis uncovered the presence of nearly all components (seven of eight) of the B-WICH chromatin remodeling complex, which is known to associate with the Pol I machinery and to facilitate rDNA transcription (30). Although BAZ1B was previously assigned to the chromatin remodeling network (Fig. 3A), the six additional members (MYBBP1A, SMARCA5, MYO1C, DDX21, SF3B1, and DEK) were absent because of either their incomplete annotation in gene ontology (MYBBP1A, DDX21, MYO1C, and SF3B1) or their initial exclusion by our stringent criteria of 3-fold enrichment in the SIRT7 immunoisolations versus EGFP (SMARCA5 and DEK). Importantly, the majority of these members, in particular MYBBP1A, MYO1C, and SMARCA5, were of high abundance, in a similar range as BAZ1B. Inclusion of these factors in the STRING analysis revealed several additional remodeling complexes (Fig. 3B). These included 1) the RSF complex, which facilitates nucleosome deposition (31), 2) the nucleolar remodeling complex (NoRC), which affects heterochromatin formation and nucleosome position through its interaction with promoter-associated RNA (23), and 3) the (ATP-dependent chromatin assembly and remodeling factor) (ACF) complex involved in regulation of nucleosome spacing (32).
To predict putative cellular processes through which SIRT7 may connect to these chromatin remodeling complexes, the previously reported SIRT7 interactions that were recapitulated in our study, Pol I (POLR1A) and UBF, were included in the STRING network analysis (Fig. 3B). Because Pol II (POLR2A) had a high abundance similar to the B-WICH members, and Pol II-dependent transcription reflects a top biological process annotation, POLR2A and several proximal transcription proteins enriched in our data set were included for analysis (supplemental Table S8). In contrast to the chromatin remodeling network that lacked connectivity to SIRT7 (Fig. 3A), the addition of key transcriptional factors resulted in several potential links between SIRT7 and chromatin remodeling chromatin (Fig. 3B). Overall, these data support the hypothesis that SIRT7 function is either directly linked to chromatin remodeling or cooperate with chromatin remodeling complexes to regulate Pol I- and/or Pol II-dependent transcription.
Targeted Validation of SIRT7 Interaction with Transcription and Chromatin Remodeling Factors by Quantitative Mass Spectrometry
In the above experiments, putative specificity of SIRT7 interactions was assessed by enrichment compared with equivalent EGFP control isolations, which controls for nonspecific associations to the tag and resin. As a complementary approach for the assessment of interaction specificity, we designed a targeted SRM-like method that employs the I-DIRT technique (33). I-DIRT uses metabolic labeling with stable isotopes and mass spectrometry as a readout for interaction specificity, controlling for proteins that bind nonspecifically to SIRT7 protein complexes during protein solubilization and/or affinity purification. As illustrated in Fig. 4A, samples were prepared for I-DIRT by post-harvest mixing of wild type HEK293 cells cultured in medium containing heavy (13C) isotope-containing amino acids and HEK293 cells expressing SIRT7WT-EGFP in medium containing natural abundance amino acids. The mixed cells were subjected to cryogenic lysis and suspended in optimized lysis buffer, and an aliquot of input lysates was set aside for loading normalization. SIRT7WT-EGFP was immunoisolated as above (Figs. 2B and 4A), followed by quantitative mass spectrometry analysis using a hypothesis-driven SRM-like approach. Specific interactions were differentiated from random interactions by quantifying the relative ratio between light (L) and heavy (H) isopeptide pairs using targeted acquisition of full scan MS2 spectra in the ion trap of an LTQ-Orbitrap Velos, followed by reconstruction of precursor-product ion XICs using Skyline (21) (supplemental Fig. S8). Scan speed of the ion trap limited acquisition of full scan MS2 spectra to eight isopeptide pairs per experiment plus one full scan FT-MS spectra at a resolution of 7,500. Selection of optimal peptides for quantification was based on experimental spectra libraries created from the previous SIRT7 immunoisolations (Fig. 2). As expected, the relative quantification of the tryptic peptide, VFDVTER, derived from enriched SIRT7WT-EGFP, resulted in robust detection of the light isopeptide, whereas the heavy peptide (from WT HEK293 cells) was not detected (Fig. 4B, left panel). In contrast, the light/heavy (L/H) ratio for 78-kDa glucose-regulated protein (HSPA5) was 1.8 (Fig. 4B, right panel). After correction for the difference in light and heavy components of the input material (supplemental Fig. S8), the normalized HSPA5 ratio was 1.1, corresponding to 53%, and therefore was considered nonspecific.
Given the limitations of metabolically labeled sample quantity and the number of peptides that can be concurrently monitored, we focused the SRM-like I-DIRT analysis on components of the B-WICH complex, which our functional analysis suggested could participate in SIRT7-mediated regulation of Pol I transcription (Fig. 3B). Quantification of peptides from Mybbp1a, SNF2h (gene name SMARCA5), RH-II/GU (gene name DDX21), and WSTF (gene name BAZ1B) resulted in an average specificity of 70%, with individual values ranging from 65% (DDX21) to 75% (Mybbp1a) (Fig. 4C), consistently higher than those measured for the nonspecific protein, HSPA5 (Fig. 4B). Because the interaction between SIRT7 and Pol II has not been previously reported, two subunits of Pol II, POLR2A and POLR2B, as well as TRRAP and SSRP1, which were functionally connected to POLR2A (Fig. 3B), were also assessed for their specificity. Although POLR2A was of similar specificity (75%) to Mybbp1a, POLR2B had the lowest measured specificity of 64%. Quantification of additional unique peptides was performed for the majority of the selected proteins (supplemental Fig. S9), resulting in similar specificity values as in Fig. 4. Complementing our previous label-free analyses, these data support the inclusion of the B-WICH complex and RNA polymerase II-related proteins as SIRT7-interacting proteins.
SIRT7 Interacts with Pol I, UBF and B-WICH Components in the Nucleoli
Numerous proteins identified by our proteomic strategy that co-isolate with SIRT7 have been shown to be part of protein complexes that play important roles in regulating rDNA transcription. WSTF (gene name BAZ1B), SNF2h (gene name SMARCA5), Mybbp1a, SAP155 (gene name SF3B1), RH-II/GU (gene name DDX21), and myosin I (gene name MYO1C) are part of the B-WICH complex (34). B-WICH is an ATP-dependent chromatin remodeling complex that has been shown to associate with the Pol I machinery, facilitating rDNA transcription (30). Following our targeted quantitative mass spectrometry approach, we further confirmed these interactions with SIRT7 using immunofluorescence experiments to examine whether Pol I and Mybbp1a, the most abundant B-WICH component quantified in our SIRT7 isolations (Fig. 3B), co-localize in the nucleoli. Our results demonstrated that EGFP-tagged SIRT7 co-localized with both Pol I, as shown by its subunit RPA-194 (Fig. 5A), and Mybbp1a (Fig. 5B). Moreover, SIRT7 co-localization with Pol I and Mybbp1a was not dependent on the putative deacetylation activity or coiled-coil domain of SIRT7, because both S111A and dE2 mutants still displayed strong co-localization within nucleoli (Fig. 5, A and B). These data are consistent with a previous study showing that SNF2h, which was the second most prominent B-WICH component in the SIRT7 isolation (Fig. 3B), also co-localized with Pol I in the nucleoli (30). SNF2h corresponds to the human imitation switch (ISWI) enzyme that exhibits ATP-dependent chromatin remodeling function in the B-WICH complex (34).
Fig. 5.
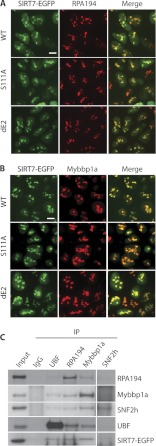
Validation of SIRT7 association with Pol I, UBF, and B-WICH components via co-localization and reciprocal isolation. A, co-localization of Pol I (RPA194) with three SIRT7-EGFP fusion proteins. Cellular localization was assessed in HEK293 cells stably expressing the EGFP-tagged SIRT7 variants by immunofluorescence microscopy using anti-RPA194 (red) and anti-GFP (for SIRT7-EGFP, green) antibodies. Bars, 10 μm. B, co-localization of Mybbp1a with SIRT7-EGFP fusion proteins. Immunofluorescence microscopy was performed as in A using anti-Mybbp1a antibodies. C, reciprocal immunoprecipitations (IP) using antibodies against endogenous UBF, RPA194, Mybbp1a, and SNF2h. Equal amount of IgG was conjugated to protein A/G-agarose beads as negative control. Precipitates were analyzed by SDS-PAGE followed by immunoblotting using individual antibodies.
Using targeted affinity purification followed by Western blotting approaches, earlier studies have shown that SIRT7 tagged with either GST or TAP associates with UBF and Pol I in HeLa and U2OS cells, respectively (9, 10). To further validate the association of SIRT7 with components of the B-WICH complex, UBF and Pol I in HEK293 cells, we performed reciprocal immunoaffinity purifications using antibodies against the endogenous proteins. Our results confirmed that SIRT7 associates with UBF and Pol I in HEK293 cells (Fig. 5C). Importantly, the affinity purification of endogenous Mybbp1a led to the co-isolation of EGFP-tagged SIRT7 (Fig. 5C). SNF2h also co-isolated SIRT7-EGFP, although at a lower level, possibly because of the presence of SNF2h in multiple protein complexes (e.g. B-WICH, ACF, and NoRC; Fig. 3B) or the suboptimal affinity of the anti-SNF2h antibody for immunoaffinity purifications (as indicated by the weak band of bait SNF2h). In addition, we found Mybbp1a to associate with Pol I, UBF, and SNF2h, consistent with the previous report that the B-WICH complex associates with the Pol I machinery (30). None of the targeted proteins were co-isolated in the nonspecific IgG control (Fig. 5C). Altogether, our localization and reciprocal isolation studies demonstrated that UBF, Pol I (RPA194), and Mybbp1a associate with each other and with SIRT7, suggesting that B-WICH and SIRT7 work cooperatively to regulate rDNA transcription.
Endogenous SIRT7 Associates with Pol I, UBF and the B-WICH Complex
Because we were able to co-isolate B-WICH components, together with Pol I and UBF, with EGFP-tagged SIRT7, and confirm these interactions by reciprocal isolations, we next assessed these associations with endogenous SIRT7. Although a previous study has performed an affinity purification of Pol I and showed co-isolation of endogenous SIRT7 by Western blotting (9), to our knowledge, isolations of endogenous SIRT7 and determination of its association with endogenous Pol I and UBF have not been reported. Similarly, the novel association of components of the B-WICH complex has not been demonstrated with either tagged or endogenous SIRT7. We performed small scale immunoaffinity purifications of endogenous SIRT7 and used targeted nLC-MS/MS analyses focused on these proteins. Our results validated that endogenous SIRT7 associates with Pol I, UBF, and importantly, components of the B-WICH complex—Mybbp1a, SAP155 (SF3B1), SNF2h (SMARCA5), and RH-II/GU (DDX21), which were confirmed by the presence of at least two unique peptides per protein (supplemental Fig. S10). These results further support the association of SIRT7 with B-WICH components, Pol I, and UBF, and the hypothesis that B-WICH and SIRT7 may function together to modulate Pol I transcription.
Knockdown of SIRT7 Leads to a Decrease in RNA Pol I Protein, but Not mRNA, Levels
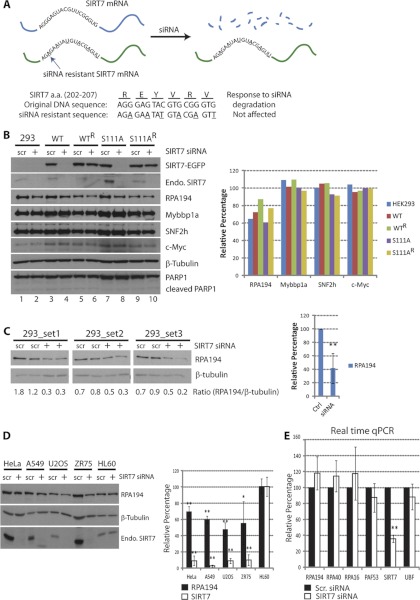
Given that SIRT7 knockdown is known to inhibit rDNA transcription (9, 10) and the co-localization and isolation reciprocity of SIRT7, Pol I, and B-WICH components, we hypothesized that the levels of these interacting proteins would be affected by SIRT7 knockdown. We first assessed the efficiency of SIRT7-EGFP and endogenous SIRT7 knockdown in HEK293 cells. As shown in Fig. 6B, endogenous SIRT7, SIRT7WT- and SIRT7S111A-EGFP protein levels were significantly reduced upon siRNA-mediated knockdown (lanes 2, 4, and 8) but not when a scrambled siRNA was used (lanes 1, 3, and 7). The cells treated with siRNA under these conditions grew normally, and no apoptosis was detected, as indicated by the absence of PARP1 cleavage (Fig. 6B). To control for the possibility of off-target effects and test specific dependence on SIRT7, we generated cell lines resistant to the siRNA-mediated knockdown of SIRT7. As depicted in Fig. 6A, by changing the SIRT7 DNA sequence only at wobble bases, the transcribed mRNA would not be recognized by the SIRT7 siRNA, while still encoding the correct amino acid sequence of SIRT7. Indeed, stable cell lines expressing the siRNA knockdown-resistant SIRT7 constructs (WTR and S111AR) prevented knockdown of SIRT7-EGFP following siRNA treatment (Fig. 6B, lanes 6 and 10), compared with the knockdown-sensitive cell lines (Fig. 6B, lanes 4 and 8). Additionally, the protein levels of endogenous SIRT7, encoded by the siRNA-recognizable sequence, were reduced in all cell lines (Fig. 6B, Endo. SIRT7 in lanes 2, 4, 6, 8, and 10).
Fig. 6.
Down-regulation of RPA194 upon SIRT7 knockdown. A, diagram depicting the construction and features of the siRNA-resistant SIRT7 construct. The mRNA transcribed from resistant gene remains intact upon siRNA treatment, whereas the mRNA from the original SIRT7 sequence is degraded. Both constructs encode the same amino acids in the 202–207 region; Bottom panel, sequence difference. B, SIRT7 knockdown leads to reduced Pol I protein levels. Left panel, immunoblotting of cell lines treated with scrambled siRNA (scr) and SIRT7 siRNA (+). Right panel, relative changes in protein levels determined by densitometry and calculated as: (Iprotein/Iβ-tubulin)SIRT7 siRNA/(Iprotein/Iβ-tubulin)scrambled siRNA × 100; I = integrated intensity. C, knockdown of endogenous SIRT7 leads to reduced Pol I protein levels. Left panel, parental HEK293 cells were transfected with scrambled or SIRT7 siRNA (n = 3, two technical replicates), and Pol I protein levels were assessed by Western blot using anti-RPA194 antibodies. Right panel, averaged relative changes in Pol I levels. D, down-regulation of RPA194 upon SIRT7 knockdown is a general effect. HeLa, A549, U2OS, ZR75, and HL60 cells were transfected with scrambled (scr) or SIRT7 siRNA for knockdown of endogenous SIRT7. Immunoblotting (left panel) and quantification of changes in protein levels were performed (n = 3) as in B. E, down-regulation at the transcriptional level is not responsible for the reduction of RPA194. mRNA from cells treated with scrambled or SIRT7 siRNA was purified and quantified by real time quantitative PCR (n = 3, four technical replicates). *, p value < 0.05; **, p value < 0.005.
Having confirmed the efficient knockdown of SIRT7 and its “rescue” in siRNA-resistant cell lines, we assessed its effect on the protein levels of Pol I, Mybbp1a, and SNF2h by Western blotting (Fig. 6B, left panel). The relative changes in protein levels caused by SIRT7 knockdown were quantified by densitometry (Fig. 6B, right panel) and expressed as a percentage change relative to the scrambled siRNA condition according to the following equation: (Iprotein/Iβ-tubulin)SIRT7 siRNA/(Iprotein/Iβ-tubulin)scrambled siRNA × 100, where I represents integrated intensity. The protein levels of Mybbp1a and SNF2h, known to be involved in chromatin remodeling complexes at nucleoli, were not affected by SIRT7 knockdown. Additionally, the protein level of a multifunctional transcription factor, c-Myc, which has a positive role in regulating rDNA transcription (35, 36), was examined, and its protein level was found to be unaffected. However, we observed a significant reduction in RPA194 following SIRT7 knockdown in all tested cells. Of note, the overexpression of siRNA-resistant SIRT7 constructs, SIRT7WTR and SIRT7S111AR, partially rescued RPA194 (Fig. 6B, right panel, green and yellow), compared with its reduced levels in wild type HEK293 cells or in the SIRT7WT and SIRT7S111A cell lines (Fig. 6B, right panel, blue, red, and purple). These results indicate that the reduction in RPA194 protein levels is SIRT7-specific but independent of its putative deacetylation activity. To further confirm this result and eliminate potential nonspecific effects of overexpressing SIRT7-EGFP, we performed siRNA knockdown of endogenous SIRT7 in the parental HEK293 cell line. As shown in Fig. 6C, RPA194 protein levels were significantly reduced in replicate experiments (three biological replicates, each with two technical replicates).
To examine whether the SIRT7-dependent down-regulation of RPA194 is a general effect rather than cell type-specific, we determined its protein levels in different cell types upon SIRT7 knockdown. As shown in Fig. 6D, SIRT7 knockdown by siRNA treatment also triggered down-regulation of RP194 protein levels in HeLa, A549, U2OS, and ZR75 cells. This effect was clearly correlated with endogenous SIRT7 levels as for HL60 cells, in which siRNA-mediated knockdown of SIRT7 was ineffective, the protein levels of RPA194 also showed no change (Fig. 6D). Collectively, these results demonstrate that the absence of SIRT7 leads to degradation of Pol I, but not Mybbp1a or SNF2h.
In addition to Pol I, our proteomic analysis identified the association of SIRT7 with transcription networks incorporating Pol II and several transcription factors (e.g. TRRAP) (Figs. 3B and 4C). Therefore, the down-regulation of RPA194 protein levels could be a consequence of a decrease in mRNA transcribed by Pol II. To test this possibility, mRNA from cells treated with either scrambled siRNA or SIRT7 siRNA was purified and quantified by real time quantitative PCR (Fig. 7, n = three biological replicates). In addition to RPA194, we examined several other subunits of the Pol I machinery: RPA40, RPA16, and PAF53. As shown in Fig. 6E, no significant changes in mRNA levels were observed for all the genes tested. Although we have not ruled out the possibility that other Pol I subunits or associated proteins might be down-regulated at the mRNA level, our results demonstrate for the first time that SIRT7 knockdown does not affect the mRNA level of RPA194 but instead modulates its protein level, pointing toward a post-transcriptional or post-translational role in maintaining the stability of Pol I complexes.
Fig. 7.
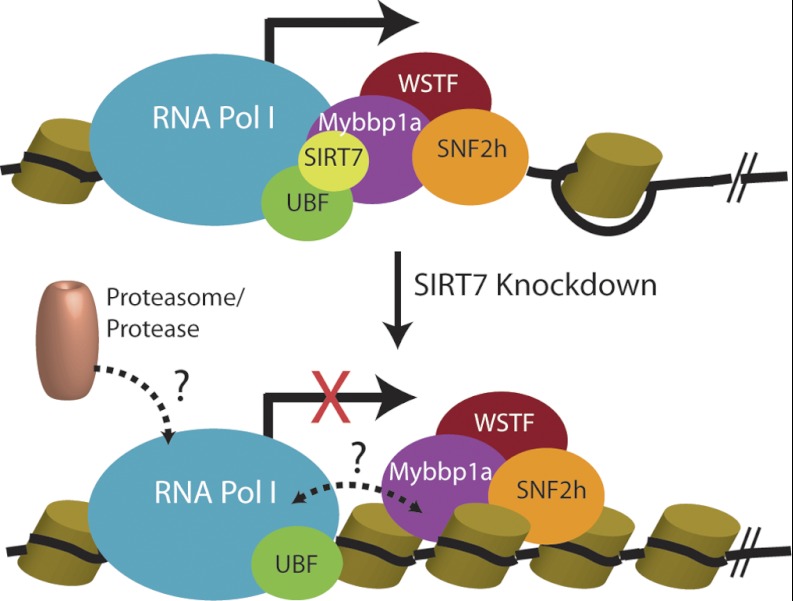
SIRT7 interacting complexes facilitating rDNA transcription. Pol I machinery and UBF interact with SIRT7 and B-WICH complex during transcription (solid arrow). Upon SIRT7 knockdown and inhibition of transcription (red cross), the B-WICH complex may dissociate from Pol I complex (dashed bidirectional arrow), and proteasome or protease may be recruited to the arrested Pol I for degradation (dashed arrow). Cylinders, histones. Black line, DNA.
DISCUSSION
Mammalian SIRT7 is the only sirtuin that localizes at nucleoli where rDNA is continuously transcribed by Pol I (7, 9). Point mutations in the sirtuin-core domain related to deacetylase function have been shown to result in reduction of rDNA transcription (9). However, several studies could not detect in vitro deacetylase activity of SIRT7 from human cells (7, 9), suggesting that SIRT7 may carry out its functions via recruitment of proteins similar to the deacetylation-deficient class IIa HDACs (14). Still, SIRT7 protein interactions remain poorly characterized. Therefore, it is important to identify the interacting proteins of SIRT7 to understand its function in transcription regulation and putative involvement in additional cellular pathways.
Building Regulatory Networks of SIRT7 Protein Interactions
In our current study, we performed the first comprehensive proteomic analysis of SIRT7 interactions. We systematically identified SIRT7-interacting proteins using three different SIRT7 derivatives that encompass the wild type protein, a point mutation in the catalytic domain (S111A), and an in-frame deletion of a putative coiled-coil domain (dE2). We found numerous proteins involved in transcription networks, including Pol I and II (Figs. 2C and 3B), as well as regulators of ribosomal biogenesis (Fig. 2C). These data are consistent with a role for SIRT7 in Pol I-mediated rDNA transcription (9, 10) and the observation that transient overexpression of SIRT7 can up-regulate many Pol II-dependent genes by reporter assays (e.g. c-Myc, E2F) (37). The majority of the co-isolated proteins were common to all SIRT7 derivatives (Fig. 2C), with few putative interactions not being detected in either dE2 or S111A mutants. Interestingly, we found that 7 of the 13 proteins absent in the dE2 isolation may also contain coiled-coil domains (supplemental Fig. S6), such as TTF1 (transcription termination factor 1) and ZBTB11 (zinc finger and BTB domain-containing protein 11). TTF1 is of particular interest given its previously characterized role in silencing of rDNA transcription (38). Although a direct interaction between TTF1 and SIRT7 has not been demonstrated, coiled-coil domains are known to be involved in protein interactions and are commonly found in transcription factors (24, 25), suggesting that the coiled-coil region of SIRT7 may function in this capacity.
Functional network analyses and label-free quantitative mass spectrometry highlighted the prominence of chromatin remodeling complexes in SIRT7 isolations. In this approach, we integrated gene ontology classification with relative protein abundance measurements expressed as NSAF values. Rather than relying solely on the presence or absence of proteins within a particular protein group, NSAF values provide relative protein abundance based on total spectral counts normalized by protein length (20). Critical to this analysis was the additional normalization of NSAF values by relative proteome abundance. Determination of protein abundances within proteomes is a major challenge in systems biology. Although recent advances in targeted proteomic workflows can now cover the full dynamic range of abundances in model organisms such as S. cerevisiae (39), more complex mammalian proteomes are only partially characterized. Recent efforts have begun to integrate large scale proteome data sets, such as those from the PeptideAtlas (29), for semi-quantitative analysis of expressed proteins across organisms (28). To date, this database covers nearly 60% of the human proteome, which was sufficient to obtain estimated protein abundances for all enriched SIRT7 co-isolated proteins for our current study (supplemental Table S5). Normalization of NSAF values by proteome abundance estimates (PAX) and graphical depiction of relative abundances within chromatin modification networks (Fig. 3A) suggested that specific targets, e.g. BAZ1B, BAZ2A, and TTF1, were more highly enriched in SIRT7 immunoisolations with respect to the total proteome abundance. This focused our studies on the nucleolus-localized, B-WICH, and NoRC chromatin remodeling complexes (Fig. 3B). Although network analysis reinforced the existing knowledge of the relationship between chromatin remodeling complexes and gene transcription (compare Fig. 3, A and B), our data now places SIRT7 in a key regulatory role between these two cellular machineries. Moreover, the connection between SIRT7 and chromatin remodeling complexes is predicted to take place via RNA Pol-dependent transcriptional processes. However, because network analyses are largely based on previous experimental or annotated information, we cannot exclude the possibility that SIRT7 could directly modulate or recruit chromatin modification complexes by unknown mechanisms independent of the RNA polymerase machinery. In support of this, SIRT7-EGFP co-localized (Fig. 5B) and was reciprocally co-isolated with the endogenous Mybbp1a (Fig. 5C), suggesting that a significant fraction of B-WICH may associate with SIRT7.
Additional validation was performed for several members of chromatin remodeling complexes and transcription-related proteins using targeted SRM-like I-DIRT experiments (Fig. 4A). A major advantage of the I-DIRT approach is that it provides an assessment of specificity complementary to an EGFP control immunoisolation (33). Proteins that bind to SIRT7-EGFP-containing assemblies during protein solubilization or affinity purification will be identified as nonspecific (L/H = 1:1), although this method cannot control for protein interactions that rapidly exchange between the complex and solution, which results in false assignment as nonspecific interactions. In contrast to 78-kDa glucose-regulated protein, which was classified as nonspecific (r = 1.1 or 53%) (Fig. 4B), a consistent enrichment of all targeted B-WICH complex members, as well as Pol I and associated transcription factors, was observed (Fig. 4, C and D, and supplemental Fig. S9). Given limitations imposed by the quantity of metabolically labeled sample and instrument acquisition speed, we could not perform targeted SRM-like experiments for all complex members. Moreover, we initially selected two peptides (four isopeptide pairs) per protein based on empirical observations made in our global proteomic characterization of SIRT7 interactions; however, not all peptides turned out to be suitable candidates for targeted analyses. Although we mitigated this effect by selecting peptides based on previously observed spectra (40), this is a known drawback of SRM- or MRM-like studies, which often require multiple rounds of refinement when selecting target peptides, particularly when global and targeted studies are performed on distinct biological samples. Despite these obstacles, the majority of proteins selected was confirmed and quantified by two unique peptides, each with ≥3 transitions. Altogether, the SRM-like I-DIRT approach, co-localization, and reciprocal immunopurifications confirmed the SIRT7 interaction with Pol I, UBF, and components of the B-WICH complex.
Insights into SIRT7 Roles in Regulating Pol I Transcription from a Proteomic Perspective
It has been shown that the overall transcription of ribosomal RNA genes can be stimulated by many signals (41); however, increased transcription is not due to an increased number of actively transcribed rDNA units but instead is due to changes in the rate of transcription, especially of elongation (42, 43). B-WICH is an ATP-dependent chromatin remodeling complex containing SNF2h, a human ISWI ATPase, and it was shown to associate with Pol I facilitating its transcription (30). The SIRT7 interaction with components of the B-WICH complex supports a hypothesis where SIRT7 regulates the rate of elongation of Pol I through the ATP-dependent remodeling activities of B-WICH.
SIRT7 knockdown is known to inhibit rDNA transcription (9, 10), and our results show for the first time that SIRT7 knockdown also leads to a reduction in the large subunit of Pol I at the protein level but not at the mRNA level. A question to be addressed in future studies is whether this regulation of Pol I protein level occurs at the post-transcriptional (e.g. splicing or exporting mRNA to the cytoplasm) or the post-translational level. One possible mechanism of post-translational level regulation is by proteasome-dependent degradation. Based on our findings, it is tempting to propose a model (Fig. 7) in which the reduced protein levels of RPA194 upon SIRT7 knockdown could be a consequence of the prolonged arrest of Pol I machinery. Under normal conditions, SIRT7 and the B-WICH complex closely associate with the Pol I machinery, facilitating its transcriptional function. Upon down-regulation of SIRT7, the association may be disconnected, and the arrested Pol I machinery may subsequently be ubiquitinated and targeted for proteasomal degradation. Alternatively, the Pol I degradation could involve nonproteasomal proteases (Fig. 7).
However, SIRT7 could also play a more active role in sensing the integrity of the associated components. Because proteasome inhibition leads to reduction of ribosomal RNA (35, 44), it was proposed that the activation of rDNA transcription is dependent on the ubiquitination of transcription factors and their subsequent degradation (44). Furthermore, the 19 S ATPase subunits of the proteasome localize throughout the promoter and coding regions of rDNA, suggesting that the effects of ubiquitination include initiation and elongation (44). In our proteomic analysis of SIRT7 interactions, we also found several ATPase subunits of the proteasome, as well as E3 ubiquitin ligases (Fig. 2C, light green circles, and supplemental Table S7). Therefore, SIRT7 may be involved in the ubiquitination of transcription factors crucial for rDNA transcriptional activation, whereas SIRT7 knockdown would disrupt this ubiquitination-mediated process, resulting in the inhibition of rDNA transcription.
Although this study focused on the most abundant SIRT7 interactions connected to transcription and chromatin remodeling, our proteomic-based studies identified other lower abundance interacting proteins that could provide insights into other functions of SIRT7. For example, ATRX (α-thalassemia/mental retardation syndrome X-linked) (45) was one of the proteins that was reproducibly co-isolated with SIRT7 and not detected in the EGFP controls but that did not pass our criteria of eight average spectral counts. Nevertheless, we observed that ATRX displayed partial co-localization with SIRT7 (supplemental Fig. S11A). ATRX belongs to the family of SNF2 chromatin remodeling factors (46) and was recently shown to be involved in the localization of histone H3.3 to telomeres (47). Regarding its function in the nucleolus, mutations in ATRX were shown to lead to hypomethylation in the rDNA arrays in cells derived from patients with α-thalassemia compared with normal individuals (48). NoRC, which is the major remodeling complex responsible for rDNA silencing, can recruit histone deacetylases and DNA methyltransferases to silencing rDNA units (49, 50). Our results showing that SIRT7 interacts with NoRC (Fig. 3B) and ATRX suggest that SIRT7 may be involved in maintaining the status of inactive rDNA. Silencing rDNA units form heterochromatin regions throughout the cell cycle, independent of the level of rDNA transcription (51). It is likely that SIRT7 associates with different complexes within nucleoli and regulates various aspects of rDNA transcription, activation (e.g. B-WICH) or repression (e.g. NoRC). The connection of SIRT7 to inactive rDNA structure is consistent with the main function of the founding member of the sirtuin family, yeast Sir2, in transcriptional silencing via the formation of epigenetically heritable chromatin structures (52–54). The spreading of silencing complexes leads to position effects in telomeres (55), homothallic mating loci (56), and rDNA (57, 58). Compared with other members involved in the silencing complex in yeast, only Sir2 is essential for the position effect in rDNA (58, 59), suggesting a unique role for Sir2 in rDNA loci. Whether SIRT7 functions in a similar capacity will require further investigation. Another SIRT7 interaction of interest that was identified but not further investigated in this study was RIF1 (Rap1-interacting factor 1 homolog). Human RIF1 can respond to DNA damage, forming foci at DNA strand breakages in an ATM-dependent manner (60). We found that SIRT7 co-localized with RIF1 within nuclear puncta (supplemental Fig. S11B), suggesting that SIRT7 may be involved in DNA damage response. It should be noted that genomic instability was observed in cells depleted of SIRT6, which belongs to the same class IV of sirtuins as SIRT7 (61).
In summary, the goal of our study was to expand the current knowledge of SIRT7 cellular functions. By coupling immunoaffinity purification with targeted proteomics and functional network analysis, we have identified novel SIRT7 interactions within transcriptional and chromatin remodeling functional categories. Specifically, we have identified the nucleoli-localized B-WICH complex as a SIRT7 interaction. This study expands the scope of cellular functions that may be subject to regulation by SIRT7, identifying multiple putative downstream effectors of SIRT7 for future investigation. Furthermore, we showed that knockdown of SIRT7 leads to down-regulation of Pol I at the protein level, which when placed in the perspective of our SIRT7 functional interaction networks led us to propose that SIRT7-dependent regulation of Pol I transcription involves association with chromatin remodeling complexes.
Acknowledgments
We are grateful to Dr. Izumi Horikawa (NCI, National Institutes of Health) for the anti-SIRT7 antibodies. We appreciate the Cristea lab members for critical reading of the manuscript. We thank J. Goodhouse and C. DeCoste (Microscopy and Flow Cytometry Core Facilities, Princeton University).
Footnotes
* This work was supported by National Institute on Drug Abuse Grant DP1DA026192 and Human Frontier Science Program Award RGY0079/2009-C (to I. M. C.) and New Jersey Commission on Cancer Research Postdoctoral Fellowship 10-2409-CCR-EO (to Y. C. T.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- I-DIRT
- isotopic determination of interactions as random or targeted
- NSAF
- normalized spectral abundance factor
- PAX
- proteome abundance estimates
- rDNA
- ribosomal DNA
- SRM
- selected reaction monitoring
- WT
- wild type
- siRNA
- small interfering RNA
- mRNA
- messenger RNA
- ACN
- acetonitrile
- XIC
- extracted ion chromatogram
- NoRC
- nucleolar remodeling complex
- nLC
- nanoLC.
REFERENCES
- 1. Kuo M. H., Brownell J. E., Sobel R. E., Ranalli T. A., Cook R. G., Edmondson D. G., Roth S. Y., Allis C. D. (1996) Transcription-linked acetylation by Gcn5p of histones H3 and H4 at specific lysines. Nature 383, 269–272 [DOI] [PubMed] [Google Scholar]
- 2. de Ruijter A. J., van Gennip A. H., Caron H. N., Kemp S., van Kuilenburg A. B. (2003) Histone deacetylases (HDACs): Characterization of the classical HDAC family. Biochem. J. 370, 737–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Taunton J., Hassig C. A., Schreiber S. L. (1996) A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science 272, 408–411 [DOI] [PubMed] [Google Scholar]
- 4. Sauve A. A., Wolberger C., Schramm V. L., Boeke J. D. (2006) The biochemistry of sirtuins. Annu. Rev. Biochem. 75, 435–465 [DOI] [PubMed] [Google Scholar]
- 5. Finnin M. S., Donigian J. R., Cohen A., Richon V. M., Rifkind R. A., Marks P. A., Breslow R., Pavletich N. P. (1999) Structures of a histone deacetylase homologue bound to the TSA and SAHA inhibitors. Nature 401, 188–193 [DOI] [PubMed] [Google Scholar]
- 6. Frye R. A. (2000) Phylogenetic classification of prokaryotic and eukaryotic Sir2-like proteins. Biochem. Biophys. Res. Commun. 273, 793–798 [DOI] [PubMed] [Google Scholar]
- 7. Michishita E., Park J. Y., Burneskis J. M., Barrett J. C., Horikawa I. (2005) Evolutionarily conserved and nonconserved cellular localizations and functions of human SIRT proteins. Mol. Biol. Cell 16, 4623–4635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Blander G., Guarente L. (2004) The Sir2 family of protein deacetylases. Annu. Rev. Biochem. 73, 417–435 [DOI] [PubMed] [Google Scholar]
- 9. Ford E., Voit R., Liszt G., Magin C., Grummt I., Guarente L. (2006) Mammalian Sir2 homolog SIRT7 is an activator of RNA polymerase I transcription. Genes Dev. 20, 1075–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Grob A., Roussel P., Wright J. E., McStay B., Hernandez-Verdun D., Sirri V. (2009) Involvement of SIRT7 in resumption of rDNA transcription at the exit from mitosis. J. Cell Sci. 122, 489–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Vakhrusheva O., Smolka C., Gajawada P., Kostin S., Boettger T., Kubin T., Braun T., Bober E. (2008) Sirt7 increases stress resistance of cardiomyocytes and prevents apoptosis and inflammatory cardiomyopathy in mice. Circ. Res. 102, 703–710 [DOI] [PubMed] [Google Scholar]
- 12. Pan P. W., Feldman J. L., Devries M. K., Dong A., Edwards A. M., Denu J. M. Structure and biochemical functions of SIRT6. (2011) Structure and biochemical functions of SIRT6. J. Biol. Chem. 286, 14575–14587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cristea I. M., Williams R., Chait B. T., Rout M. P. (2005) Fluorescent proteins as proteomic probes. Mol. Cell. Proteomics 4, 1933–1941 [DOI] [PubMed] [Google Scholar]
- 14. Greco T. M., Yu F., Guise A. J., Cristea I. M. (2010) Nuclear import of histone deacetylase 5 by requisite nuclear localization signal phosphorylation. Mol. Cell. Proteomics 10, 10.1074/mcp.M110.004317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cristea I. M., Chait B. T. (2011) Affinity purification of protein complexes. Cold Spring Harb. Protoc. 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cristea I. M., Chait B. T. (2011) Conjugation of magnetic beads for immunopurification of protein complexes. Cold Spring Harb. Protoc. 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Keller A., Nesvizhskii A. I., Kolker E., Aebersold R. (2002) Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 74, 5383–5392 [DOI] [PubMed] [Google Scholar]
- 18. Nesvizhskii A. I., Keller A., Kolker E., Aebersold R. (2003) A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 75, 4646–4658 [DOI] [PubMed] [Google Scholar]
- 19. Smoot M. E., Ono K., Ruscheinski J., Wang P. L., Ideker T. (2011) Cytoscape 2.8: New features for data integration and network visualization. Bioinformatics 27, 431–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zybailov B., Mosley A. L., Sardiu M. E., Coleman M. K., Florens L., Washburn M. P. (2006) Statistical analysis of membrane proteome expression changes in Saccharomyces cerevisiae. J. Proteome Res. 5, 2339–2347 [DOI] [PubMed] [Google Scholar]
- 21. MacLean B., Tomazela D. M., Shulman N., Chambers M., Finney G. L., Frewen B., Kern R., Tabb D. L., Liebler D. C., MacCoss M. J. (2010) Skyline: An open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 26, 966–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Min J., Landry J., Sternglanz R., Xu R. M. (2001) Crystal structure of a SIR2 homolog-NAD complex. Cell 105, 269–279 [DOI] [PubMed] [Google Scholar]
- 23. Marmorstein R. (2001) Structure of histone deacetylases: Insights into substrate recognition and catalysis. Structure 9, 1127–1133 [DOI] [PubMed] [Google Scholar]
- 24. Rasmussen R., Benvegnu D., O'Shea E. K., Kim P. S., Alber T. (1991) X-ray scattering indicates that the leucine zipper is a coiled coil. Proc. Natl. Acad. Sci. U.S.A. 88, 561–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Glover J. N., Harrison S. C. (1995) Crystal structure of the heterodimeric bZIP transcription factor c-Fos-c-Jun bound to DNA. Nature 373, 257–261 [DOI] [PubMed] [Google Scholar]
- 26. Szklarczyk D., Franceschini A., Kuhn M., Simonovic M., Roth A., Minguez P., Doerks T., Stark M., Muller J., Bork P., Jensen L. J., von Mering C. (2011) The STRING database in 2011: Functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 39, D561–D568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Euskirchen G. M., Auerbach R. K., Davidov E., Gianoulis T. A., Zhong G., Rozowsky J., Bhardwaj N., Gerstein M. B., Snyder M. (2011) Diverse roles and interactions of the SWI/SNF chromatin remodeling complex revealed using global approaches. PLoS Genet. 7, e1002008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Weiss M., Schrimpf S., Hengartner M. O., Lercher M. J., von Mering C. (2010) Shotgun proteomics data from multiple organisms reveals remarkable quantitative conservation of the eukaryotic core proteome. Proteomics 10, 1297–1306 [DOI] [PubMed] [Google Scholar]
- 29. Deutsch E. W., Lam H., Aebersold R. (2008) PeptideAtlas: A resource for target selection for emerging targeted proteomics workflows. EMBO Rep. 9, 429–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Percipalle P., Fomproix N., Cavellán E., Voit R., Reimer G., Krüger T., Thyberg J., Scheer U., Grummt I., Farrants A. K. (2006) The chromatin remodelling complex WSTF-SNF2h interacts with nuclear myosin 1 and has a role in RNA polymerase I transcription. EMBO Rep. 7, 525–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Loyola A., LeRoy G., Wang Y. H., Reinberg D. (2001) Reconstitution of recombinant chromatin establishes a requirement for histone-tail modifications during chromatin assembly and transcription. Genes Dev. 15, 2837–2851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yang J. G., Madrid T. S., Sevastopoulos E., Narlikar G. J. (2006) The chromatin-remodeling enzyme ACF is an ATP-dependent DNA length sensor that regulates nucleosome spacing. Nat. Struct. Mol. Biol. 13, 1078–1083 [DOI] [PubMed] [Google Scholar]
- 33. Tackett A. J., DeGrasse J. A., Sekedat M. D., Oeffinger M., Rout M. P., Chait B. T. (2005) I-DIRT, a general method for distinguishing between specific and nonspecific protein interactions. J. Proteome Res. 4, 1752–1756 [DOI] [PubMed] [Google Scholar]
- 34. Cavellán E., Asp P., Percipalle P., Farrants A. K. (2006) The WSTF-SNF2h chromatin remodeling complex interacts with several nuclear proteins in transcription. J. Biol. Chem. 281, 16264–16271 [DOI] [PubMed] [Google Scholar]
- 35. Arabi A., Wu S., Ridderstråle K., Bierhoff H., Shiue C., Fatyol K., Fahlén S., Hydbring P., Söderberg O., Grummt I., Larsson L. G., Wright A. P. (2005) c-Myc associates with ribosomal DNA and activates RNA polymerase I transcription. Nat. Cell Biol. 7, 303–310 [DOI] [PubMed] [Google Scholar]
- 36. Grandori C., Gomez-Roman N., Felton-Edkins Z. A., Ngouenet C., Galloway D. A., Eisenman R. N., White R. J. (2005) c-Myc binds to human ribosomal DNA and stimulates transcription of rRNA genes by RNA polymerase I. Nat. Cell Biol. 7, 311–318 [DOI] [PubMed] [Google Scholar]
- 37. Vakhrusheva O., Braeuer D., Liu Z., Braun T., Bober E. (2008) Sirt7-dependent inhibition of cell growth and proliferation might be instrumental to mediate tissue integrity during aging. J. Physiol. Pharmacol. 59, (Suppl. 9) 201–212 [PubMed] [Google Scholar]
- 38. Németh A., Strohner R., Grummt I., Längst G. (2004) The chromatin remodeling complex NoRC and TTF-I cooperate in the regulation of the mammalian rRNA genes in vivo. Nucleic Acids Res. 32, 4091–4099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Picotti P., Bodenmiller B., Mueller L. N., Domon B., Aebersold R. (2009) Full dynamic range proteome analysis of S. cerevisiae by targeted proteomics. Cell 138, 795–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Prakash A., Tomazela D. M., Frewen B., Maclean B., Merrihew G., Peterman S., Maccoss M. J. (2009) Expediting the development of targeted SRM assays: Using data from shotgun proteomics to automate method development. J. Proteome Res. 8, 2733–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Moss T., Langlois F., Gagnon-Kugler T., Stefanovsky V. (2007) A housekeeper with power of attorney: The rRNA genes in ribosome biogenesis. Cell. Mol. Life Sci. 64, 29–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Stefanovsky V., Langlois F., Gagnon-Kugler T., Rothblum L. I., Moss T. (2006) Growth factor signaling regulates elongation of RNA polymerase I transcription in mammals via UBF phosphorylation and r-chromatin remodeling. Mol. Cell 21, 629–639 [DOI] [PubMed] [Google Scholar]
- 43. Stefanovsky V., Moss T. (2006) Regulation of rRNA synthesis in human and mouse cells is not determined by changes in active gene count. Cell Cycle 5, 735–739 [DOI] [PubMed] [Google Scholar]
- 44. Fátyol K., Grummt I. (2008) Proteasomal ATPases are associated with rDNA: The ubiquitin proteasome system plays a direct role in RNA polymerase I transcription. Biochim. Biophys. Acta 1779, 850–859 [DOI] [PubMed] [Google Scholar]
- 45. Gibbons R. J., Wada T., Fisher C. A., Malik N., Mitson M. J., Steensma D. P., Fryer A., Goudie D. R., Krantz I. D., Traeger-Synodinos J. (2008) Mutations in the chromatin-associated protein ATRX. Hum. Mutat. 29, 796–802 [DOI] [PubMed] [Google Scholar]
- 46. Picketts D. J., Higgs D. R., Bachoo S., Blake D. J., Quarrell O. W., Gibbons R. J. (1996) ATRX encodes a novel member of the SNF2 family of proteins: Mutations point to a common mechanism underlying the ATR-X syndrome. Hum. Mol. Genet. 5, 1899–1907 [DOI] [PubMed] [Google Scholar]
- 47. Goldberg A. D., Banaszynski L. A., Noh K. M., Lewis P. W., Elsaesser S. J., Stadler S., Dewell S., Law M., Guo X., Li X., Wen D., Chapgier A., DeKelver R. C., Miller J. C., Lee Y. L., Boydston E. A., Holmes M. C., Gregory P. D., Greally J. M., Rafii S., Yang C., Scambler P. J., Garrick D., Gibbons R. J., Higgs D. R., Cristea I. M., Urnov F. D., Zheng D., Allis C. D. (2010) Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell 140, 678–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Gibbons R. J., McDowell T. L., Raman S., O'Rourke D. M., Garrick D., Ayyub H., Higgs D. R. (2000) Mutations in ATRX, encoding a SWI/SNF-like protein, cause diverse changes in the pattern of DNA methylation. Nat. Genet. 24, 368–371 [DOI] [PubMed] [Google Scholar]
- 49. Santoro R., Li J., Grummt I. (2002) The nucleolar remodeling complex NoRC mediates heterochromatin formation and silencing of ribosomal gene transcription. Nat. Genet. 32, 393–396 [DOI] [PubMed] [Google Scholar]
- 50. Zhou Y., Santoro R., Grummt I. (2002) The chromatin remodeling complex NoRC targets HDAC1 to the ribosomal gene promoter and represses RNA polymerase I transcription. EMBO J. 21, 4632–4640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Conconi A., Widmer R. M., Koller T., Sogo J. M. (1989) Two different chromatin structures coexist in ribosomal RNA genes throughout the cell cycle. Cell 57, 753–761 [DOI] [PubMed] [Google Scholar]
- 52. Hecht A., Laroche T., Strahl-Bolsinger S., Gasser S. M., Grunstein M. (1995) Histone H3 and H4 N-termini interact with SIR3 and SIR4 proteins: A molecular model for the formation of heterochromatin in yeast. Cell 80, 583–592 [DOI] [PubMed] [Google Scholar]
- 53. Moretti P., Freeman K., Coodly L., Shore D. (1994) Evidence that a complex of SIR proteins interacts with the silencer and telomere-binding protein RAP1. Genes Dev. 8, 2257–2269 [DOI] [PubMed] [Google Scholar]
- 54. Strahl-Bolsinger S., Hecht A., Luo K., Grunstein M. (1997) SIR2 and SIR4 interactions differ in core and extended telomeric heterochromatin in yeast. Genes Dev. 11, 83–93 [DOI] [PubMed] [Google Scholar]
- 55. Gottschling D. E., Aparicio O. M., Billington B. L., Zakian V. A. (1990) Position effect at S. cerevisiae telomeres: Reversible repression of Pol II transcription. Cell 63, 751–762 [DOI] [PubMed] [Google Scholar]
- 56. Klar A. J., Strathern J. N., Hicks J. B. (1981) A position-effect control for gene transposition: State of expression of yeast mating-type genes affects their ability to switch. Cell 25, 517–524 [DOI] [PubMed] [Google Scholar]
- 57. Bryk M., Banerjee M., Murphy M., Knudsen K. E., Garfinkel D. J., Curcio M. J. (1997) Transcriptional silencing of Ty1 elements in the RDN1 locus of yeast. Genes Dev. 11, 255–269 [DOI] [PubMed] [Google Scholar]
- 58. Smith J. S., Caputo E., Boeke J. D. (1999) A genetic screen for ribosomal DNA silencing defects identifies multiple DNA replication and chromatin-modulating factors. Mol. Cell. Biol. 19, 3184–3197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Smith J. S., Boeke J. D. (1997) An unusual form of transcriptional silencing in yeast ribosomal DNA. Genes Dev. 11, 241–254 [DOI] [PubMed] [Google Scholar]
- 60. Silverman J., Takai H., Buonomo S. B., Eisenhaber F., de Lange T. (2004) Human Rif1, ortholog of a yeast telomeric protein, is regulated by ATM and 53BP1 and functions in the S-phase checkpoint. Genes Dev. 18, 2108–2119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Mostoslavsky R., Chua K. F., Lombard D. B., Pang W. W., Fischer M. R., Gellon L., Liu P., Mostoslavsky G., Franco S., Murphy M. M., Mills K. D., Patel P., Hsu J. T., Hong A. L., Ford E., Cheng H. L., Kennedy C., Nunez N., Bronson R., Frendewey D., Auerbach W., Valenzuela D., Karow M., Hottiger M. O., Hursting S., Barrett J. C., Guarente L., Mulligan R., Demple B., Yancopoulos G. D., Alt F. W. (2006) Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell 124, 315–329 [DOI] [PubMed] [Google Scholar]