Abstract
Ubiquitination plays a key role in protein degradation and signal transduction. Ubiquitin is a small protein modifier that is adducted to lysine residues by the combined function of E1, E2, and E3 enzymes and is removed by deubiquitinating enzymes. Characterization of ubiquitination sites is important for understanding the role of this modification in cellular processes and disease. However, until recently, large-scale characterization of endogenous ubiquitination sites has been hampered by the lack of efficient enrichment techniques. The introduction of antibodies that specifically recognize peptides with lysine residues that harbor a di-glycine remnant (K-ε-GG) following tryptic digestion has dramatically improved the ability to enrich and identify ubiquitination sites from cellular lysates. We used this enrichment technique to study the effects of proteasome inhibition by MG-132 and deubiquitinase inhibition by PR-619 on ubiquitination sites in human Jurkat cells by quantitative high performance mass spectrometry. Minimal fractionation of digested lysates prior to immunoaffinity enrichment increased the yield of K-ε-GG peptides three- to fourfold resulting in detection of up to ∼3300 distinct K-GG peptides in SILAC triple encoded experiments starting from 5 mg of protein per label state. In total, we identify 5533 distinct K-ε-GG peptides of which 4907 were quantified in this study, demonstrating that the strategy presented is a practical approach to perturbational studies in cell systems. We found that proteasome inhibition by MG-132 and deubiquitinase inhibition by PR-619 induces significant changes to the ubiquitin landscape, but that not all ubiquitination sites regulated by MG-132 and PR-619 are likely substrates for the ubiquitin-proteasome system. Additionally, we find that the proteasome and deubiquitinase inhibitors studied induced only minor changes in protein expression levels regardless of the extent of regulation induced at the ubiquitin site level. We attribute this finding to the low stoichiometry of the majority ubiquitination sites identified in this study.
Ubiquitination is a post-translational modification that plays a central role in regulating protein half-life through degradation via the 26S proteasome (1). Aside from this archetypal role, ubiquitination is essential for regulating myriad other processes including protein endocytosis, lysosomal targeting, and chromatin remodeling (2). Similar to phosphorylation, ubiquitination is reversible, ATP-dependent, and enzymatically driven (3). In contrast, ubiquitin itself is a conserved 76 amino acid protein that is adducted to its substrates in a highly modular method. Attachment of ubiquitin to the epsilon-amino group of lysine residues is facilitated through the sequential action of three discrete enzymes, the ubiquitin activating enzyme (E1), the ubiquitin-conjugating enzyme (E2), and the ubiquitin-ligating enzyme (E3). In the first step of the ubiquitin cascade, an active site cysteine residue of the E1 enzyme forms a thioester linkage with the C-terminal carboxyl group of ubiquitin (3). The ubiquitin molecule is then transferred to a cysteine residue on an E2 enzyme and is finally linked to its substrate protein through interactions made with the E2 conjugating enzyme and the E3 ligase. Ubiquitin is bound to substrate through an amide bond between the C terminus of ubiquitin and the epsilon amino group of a Lysine in the substrate (Fig. 1). Approximately 10 E1, 40 E2, and >600 E3s are encoded in the human genome resulting in increased complexity and specificity following each step of the ubiquitin cascade (3, 4).
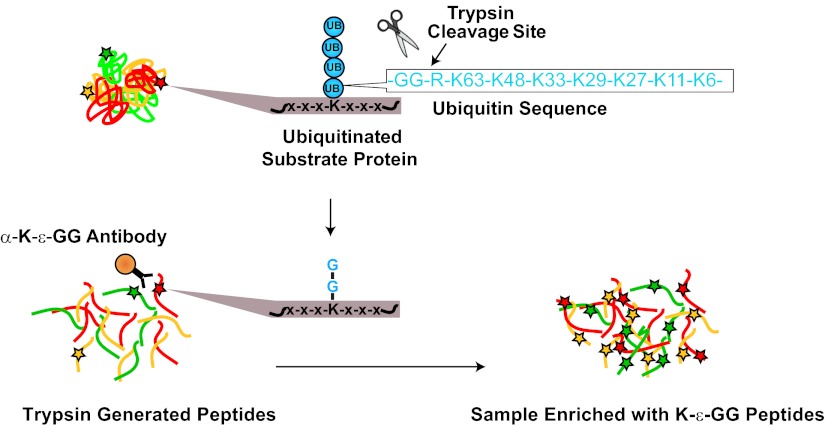
Fig. 1.
Schematic of the enrichment of K-ε-GG peptides with an anti- K-ε-GG antibody. Following tryptic digestion two glycine residues from the C terminus of ubiquitin remain linked to the epsilon amino group of a modified lysine residue. An anti-K-ε-GG antibody is used to efficiently enrich these peptides away from non- K-ε-GG peptides.
Removal of ubiquitin molecules is catalyzed by deubiquitinating enzymes (DUBs)1 (4). Approximately 80–90 DUBs are encoded in the human genome and are subdivided into five classes: ubiquitin-specific proteases (USPs), ubiquitin C-terminal hydrolases (UCHs), ovarian tumor proteases (OTUs), Joesephines, and JAMM/MPN+ metalloenzymes (5). USPs, UCHs, OTUs, and Josephines are all cysteine proteases (3, 5). Aside from functioning to remove ubiquitin from substrate molecules, DUBs can also function to trim polyubiquitin chains or generate free ubiquitin from ubiquitin precursors (5). Additionally, three well conserved DUBs associate with the proteasome and regulate substrate degradation via chain trimming or through removal of the ubiquitin chain en bloc (6, 7). As a result, proteasomal DUBs facilitate the recycling of cellular ubiquitin molecules upon substrate degradation (5).
The cellular signaling events facilitated by ubiquitination are highly versatile because of the diversity in ubiquitin chain topology. For example, substrate lysine residues can be either monoubiquitinated or polyubiquitinated with a variety of chain lengths. In addition, all seven lysine residues of ubiquitin as well as the N terminus can form polyubiquitin linkages with lysine residues on additional ubiquitin molecules in either a homotypic or heterotypic manner (2). Distinct ubiquitin binding domains recognize particular ubiquitin chain products and function to trigger a diverse array of signaling events (2). Polyubiquitination often functions as a signal for degradation through the ubiquitin-proteasome system (UPS) to maintain cellular homeostasis. The 26S proteasome is a large, multisubunit protein complex that works to unfold and proteolytically degrade ubiquitinated proteins (6).
Until recently, proteomic techniques for characterizing specific sites of ubiquitination in a large-scale manner have been deficient relative to well established methods for characterizing thousands of phosphorylation sites (7, 8). The analysis of ubiquitinated proteins is typically faced with the following challenges: (1) ubiquitin itself is a protein, (2) ubiquitination is substoichiometric, (3) ubiquitination frequently leads to protein degradation, and (4) the topology of ubiquitin chains is heterogeneous. Previously, proteomic methods have utilized affinity-tagged ubiquitin, ubiquitin specific antibodies, or reagents consisting of ubiquitin binding domains to enrich substiochiometrically modified proteins (5–7). Although these methods have enabled the characterization of up to several hundred sites of ubiquitination, they are inherently hampered by the increase in complexity that results from the presence of a large number of nonubiquitinated peptides derived from unmodified regions of the enriched proteins after digestion (9).
Digestion of ubiquitinated, NEDD8ylated, or ISG15ylated proteins with trypsin results in cleavage of all but the two C-terminal glycine residues of ubiquitin, NEDD8, or ISG15 that remain linked to the epsilon amino group of any modified lysine residues (K-ε-GG) in digested substrate proteins (Fig. 1) (10). The modified lysine residues are not cleaved by trypsin, and therefore the K-ε-GG modifications are internal to the resulting tryptic peptides of the substrate proteins. Antibodies have recently been developed that specifically recognize and immunoprecipitate peptides containing the K-ε-GG remnant (10–12). Here we used a commercially available anti-K-ε-GG antibody to enrich K-ε-GG peptides from human Jurkat cells (Fig. 1) to study the effects of proteasome inhibition and DUB inhibition on endogenous ubiquitination sites. MG-132 was used to inhibit the chymotryptic-like activity of the proteasome. For DUB inhibition, we used the nonselective, reversible DUB inhibitor PR-619 (2,6-diaminopyradine-3,5-bis(thiocynate)) that targets four of the five DUB families (13). In total, we successfully identified 5533 distinct K-ε-GG peptides from 2039 proteins and quantified 4907 K-ε-GG peptides from 1883 proteins. Technical improvements involving minimal fractionation of protein digests prior to liquid chromatography-tandem MS (LC-MS/MS) allowed us to obtain this level of coverage and quantification in SILAC triple encoded samples using a few milligram of protein lysate from each label state.
EXPERIMENTAL PROCEDURES
Cell Culture
For stable isotope labeling by amino acids in cell culture (SILAC) experiments (14), Jurkat E6–1 cells (ATCC) were cultured in Roswell Park Memorial Institute 1640 media deficient in l-arginine and l-lysine (custom preparation from Caisson Laboratories, North Logan, UT). Media was supplemented with 10% dialyzed fetal bovine serum (SAFC-Sigma), penicillin, streptomycin, and glutamine. Cells were grown in media containing either l-arginine (Arg0) and l-lysine (Lys0), l-arginine-13C6 (Arg6) and l-lysine-4,4,5,5-d4 (L) (Lys4), or l-arginine-13C6, 15N4 (Arg10) and l-lysine-13C6,15N2 (Lys8) (Sigma Isotech) for ∼7 doublings. Five hours prior to harvest, cells were treated with either 0.5% v/v dimethyl sulfoxide, 5 μm MG-132 (Calbiochem, San Diego, CA), or 5 μm PR-169 (Lifesensors). The described cell culture procedure was used to prepare two biological replicates of each SILAC triple encoded experiment. SILAC amino acid incorporation was monitored by analyzing peptide samples prior to mixing of SILAC encoded states.
Cell Lysis, Protein Digestion
Cell pellets were lysed at 4 °C in 8 m urea, 50 mm Tris pH 7.5, 150 mm NaCl, and 1 mm EDTA containing 2 μg/ml aprotinin, 10 μg/ml leupeptin, 1 mm phenylmethylsulfonyl fluoride, 50 μm PR-619, and 5 mm chloroacetamide. A bicinchoninic acid (BCA) protein assay (Pierce, Waltham, MA) was used to estimate protein concentration. SILAC-labeled samples were combined using 5 mg of protein per SILAC state for each experimental replicate. Approximately 5 mg of protein were obtained from 1 T175 flask. Samples were treated with 5 mm dithiothreitol for 45 min at room temperature. Carbamidomethylation was completed by treatment with 10 mm iodoacetamide for 45 min at room temperature. The protein mixture was diluted to 2 m urea with 50 mm Tris/HCl pH 7.5 and digested with sequencing grade trypsin (Promega, Madison, WI) overnight at room temperature. Peptide mixtures were acidified with trifluoroacetic acid (TFA) and desalted using a 500 mg tC18 Sep-Pak SPE cartridge (Waters, Milford, MA). Desalted peptide mixtures were frozen and subsequently lyophilized using a FreeZone Triad Freeze Dry System (Labconco, Kansas City, MO).
Strong Cation Exchange (SCX) Chromatography
Lyophilized peptides (5 mg per SILAC state) were resuspended in SCX buffer A (7 mm KH2PO4 pH 2.65, 30% acetonitrile) and separated on a Polysulfoethyl A column (250 × 9.4 mm, 5 μm, 200 Å) (PolyLC) using an Akta Purifier 10 system (GE Healthcare). A 160 min SCX gradient was used for separation at a flow rate of 3 ml/min. The gradient consisted of a 20 min equilibration phase with 100% buffer A, a linear increase to 30% buffer B (7 mm KH2PO4, pH2.65, 350 mm KCl, 30% MeCN) within 30 min, a second linear increase to 75% buffer B in 80 min, 100% B for 10 min, and a final equilibration with Buffer A for 20 min. Upon injection of the sample, 60 fractions were collected with a P950 fraction collector throughout the run. Pooling of SCX fractions was guided by the UV 214 nm trace. For proteome analysis, 5% of each SCX fraction was aliquoted, and fractions were combined into 24 proteome samples. For experiment 1/replicate 2 only 23 proteome SCX fractions were analyzed. For K-ε-GG analysis 95% of each SCX fraction was used, and the SCX fractions were combined into four K-ε-GG samples. All fractions were lyophilized, desalted, and again lyophilized.
K-ε-GG Peptide Enrichment
Lyophilized peptides were reconstituted in 50 mm MOPS pH 7.2, 10 mm sodium phosphate, and 50 mm NaCl (IP buffer). K-ε-GG ubiquitin remnant motif antibody bead conjugates (80μl 1:1 slurry as supplied by vendor) (Cell Signaling Technologies, Danvers, MA) were washed three times with ice cold IP buffer and equally separated into four tubes. Peptides mixtures were added to K-ε-GG antibody beads and incubated overnight at 4 °C with gentle rotation. Antibody beads were washed three times with ice cold IP buffer followed by two washes with ice cold H2O. For peptide elution, 50 μl of 0.15% trifluoroacetic acid (TFA) were added to the antibody beads two times and supernatants were collected. All peptides were desalted using C18 StageTips (15). Prior to sample loading, StageTips were washed with 50 μl of 60% acetonitrile/0.1% TFA, followed by two washes with 0.1% TFA. Peptide mixtures were then loaded directly onto StageTips, which were subsequently washed two times with 100 μl of 0.1% TFA. Peptides were eluted from StageTips with 100 μl of 60% acetonitrile/0.1% TFA and dried to completion.
LC-MS/MS
Peptide mixtures were reconstituted in 1% formic acid/3% acetonitrile and analyzed on an LTQ Orbitrap Velos mass spectrometer (Thermo Fisher Scientific) coupled online to an Agilent 1200 HPLC system. Samples were injected directly on a capillary column (360 μm o.d. × 75 μm i.d.) packed in-house with 12 cm of 3 μm ReproSil-Pur C18-AQ resin (Dr. Maisch GmbH). The capillary column was equipped with an integrated 10 μm tip. The HPLC mobile phase A was 0.1% formic acid and the mobile phase B was 90% acetonitrile/0.1% formic acid. Peptides were eluted from the column at 200 nL/min using a reversed phase HPLC gradient with a 70 min linear gradient (0.29%B/min) from 10% solvent A (0.1% formic acid in water) to 30% solvent B (0.1% formic acid/90% acetonitrile). The total run time including sample loading and column reconditioning was 140 min.
The LTQ Orbitrap Velos was operated in the data-dependent mode where an MS1 scan was acquired in the Orbitrap analyzer at a resolution of 60,000 and a target value of 1,000,000 ions. The MS1 scan was followed by collision-induced dissociation MS/MS scans on the top 16 most abundant ions. All MS/MS scans were acquired in the ion trap with a target value of 10,000 ions, normalized collision energy of 30, and an isolation width of 2.5 amu. Dynamic exclusion was enabled with an exclusion duration of 20 s.
MS Data Analysis
Peptides and proteins were identified and quantified using the MaxQuant software package, version 1.2.2.5, and the terminology and settings described below are specific to this software, (16, 17). MS/MS spectra were searched against the human International Protein Index (IPI) database (version 3.68) which contained 87,062 entries and 248 frequently observed laboratory contaminants as provided by the MaxQuant software package. Precursor mass tolerance was set to 20ppm for the first search where initial mass recalibration was completed. For the main search, a 6ppm precursor mass tolerance was used. Product ions were searched with a mass tolerance 0.5 Da. The maximum precursor ion charge state used for searching was 7. Carbamidomethylation of cysteines was searched as a fixed modification and oxidation of methionines, acetylation of protein N termini, deamidation of asparagine, and presence of GlyGly on the sidechains of lysine residues were searched as variable modifications. Enzyme specificity was set to trypsin and a maximum of two missed cleavages was allowed for searching. The target-decoy-based false discovery rate (FDR) for peptide and protein identification was set to 1% for peptides and proteins (18). The filter labeled amino acids and the peptide requantification function was enabled for the analysis. Unmodified, oxidized methionine, deamidated asparagine, and N-terminally acetylated peptides were utilized for protein quantification.
SILAC peptide ratios were obtained from the MaxQuant Evidence table. All peptides identified as reverse or contaminant hits were removed from the data set. Within a given experiment, all non-K-ε-GG peptide SILAC ratios were used to normalize K-ε-GG SILAC ratios. Arg and Lys containing peptides were normalized independently to correct for incomplete amino acid incorporation. After normalization, K-ε-GG SILAC ratios were log2 transformed and a median value was calculated for all nondistinct forms of a given K-ε-GG peptide in a given experiment. Peptides that differed only by charge state, methionine oxidation, asparagine deamidation, or N-terminal acetylation, were not considered distinct. To aid in peptide grouping, K-ε-GG site localization information was simplified such that an original localization score of ≥0.75 was deemed a confidently localized site and denoted with a score of 1, an original localization score of <0.75 and ≥0.5 was deemed 50% localized and denoted with a score of 0.5; and an original localization score of <0.5 was deemed unlocalized and therefore no localization score was given. Peptide identifications in which GlyGly sites were localized to a C-terminal Lys of a peptide were considered false positives (as trypsin would not cut adjacent to a K-ε-GG site as has been previously demonstrated (19)) and were manually removed from the data set. In the case of peptides that contain only two Lys residues and also end with LysLys, the GlyGly modification was deemed to be localized to the second to the last C-terminal Lys residue.
Protein level information was obtained from the MaxQuant Protein Groups table. Leading protein groups identified as reverse hits or contaminants were removed from the data set. All reported protein groups were identified by two or more distinct peptides and quantified with three or more ratio counts. A protein SILAC ratio was derived by calculating a median value over normalized, log2 transformed peptide SILAC ratios assigned to a given protein group. Arginine and lysine containing peptides were normalized separately to account for incomplete labeling. K-ε-GG peptides were not considered in protein level quantification.
Using the two replicate measurements for each treatment condition, differentially (up or down) regulated peptides and proteins were identified using the moderated t-statistic (20). This statistic is similar to the ordinary t-statistic, with the exception that the standard errors are calculated using an empirical Bayes method utilizing information across all peptides, thereby making inference about each individual peptide more robust. The p values arising from moderated t-statistics are nominal p values (like those obtained from an ordinary t test), and are corrected for multiple testing by controlling the false discovery rate (FDR), as proposed by Benjamini and Hochberg (21). Peptides and proteins with an FDR adjusted p value of less than 0.1 were deemed to be reproducibly regulated. A corrected p value threshold of 0.1 was chosen for this study because the standard p value cutoffs of 0.05 or 0.01 result in significant loss of sensitivity because of the small number of replicates used and the large number of regulated peptides.
Hyperlinks embedded in supplemental Table S1 link to a Spectrum Mill viewer that shows an annotated MS/MS spectrum for K-ε-GG peptides. URLs >256 characters will not have an associated hyperlink and need to be pasted into the internet browser to access the viewer. We recommend using Internet Explorer on Windows for best results. Raw data are available at https://proteomecommons.org and they are mirrored at ftp://ftp.broadinstitute.org/distribution/proteomics/public_datasets/Udeshi_MCP_2012/.
Gene-Ontology Enrichment
The DAVID Bioinformatics resource was used to identify enriched gene ontology (GO) terms in our data set (22, 23). Enrichment analysis was completed using the functional annotation tool within DAVID. The background gene population used in all analyses was the entire human gene set. GO terms assigned a Benjamini-Hochberg adjusted p value of less than 0.05 by DAVID were deemed to be enriched over the background gene set.
K-ε-GG Sequence Window Analysis
To study the representation of amino acids proximal to K-ε-GG sites, sequence windows of ± 10 amino acids were created for ∼5000 confidently localized K-ε-GG sites from our data set. In addition, same-sized sequence windows were extracted for ∼5000 lysine residues chosen at random from the human IPI database (version 3.66) to serve as a background. A log-odds ratio was calculated for the frequency of an amino acid at every position within the sequence window between K-ε-GG sites and the background data set. To visualize these results, a heat map was generated based on the log-odds ratio using GENE-E (24).
RESULTS
Prefractionation Significantly Increases the Number of Identified K-ε-GG Peptides in SILAC Triple Encoded Samples
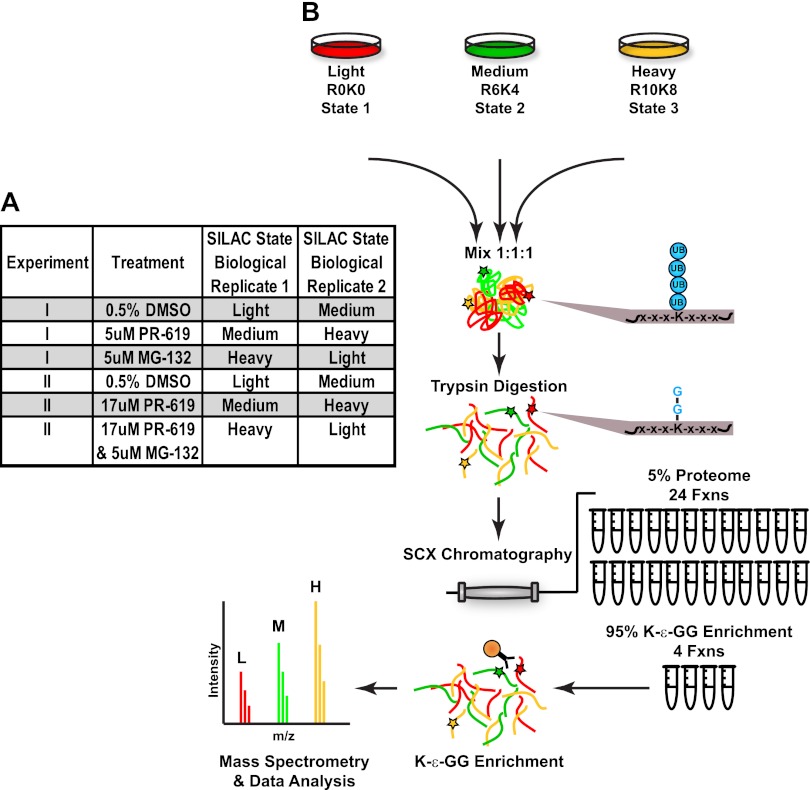
A major focus of our work was to establish practical methods for quantifying global changes in both the ubiquitinome and proteome in cells undergoing various perturbation conditions. In the present study we set out to quantify changes in the abundance of K-ε-GG peptides after cellular treatment with the proteasome inhibitor MG-132 or the broad specificity DUB inhibitor PR-619 relative to vehicle-treated (Fig. 2A). We employed triple-encoding SILAC, a well established and powerful method for quantifying changes in the abundance of peptides and proteins across multiple cellular states (14). In our large-scale experiment, we investigated the usefulness of pre-fractionating samples prior to K-ε-GG enrichment by comparing the number of K-ε-GG peptides identified with limited SCX fractionation relative to direct enrichment. For the comparison, three of our SILAC triple encoded samples (Fig. 2A) were divided such that 50% of the sample was directly enriched for K-ε-GG peptides whereas the other 50% was first separated into four SCX fractions (Fig. 2B). Pooling of fractions for the K-ε-GG IP was guided by results obtained from preliminary work to maximize the yield of K-ε-GG peptides per IP (supplemental Fig. S1A). Each SCX fraction was separately enriched for K-ε-GG peptides (total amount of antibody used in all comparisons was equivalent) and analyzed by LC-MS/MS.
Fig. 2.
Outline of the SILAC experimental design and schematic of the workflow. Two SILAC triple-encoded experiments were completed in biological duplicates with label switching as outlined in (A). Our workflow is depicted in (B). SILAC labeled Jurkat cells were lysed, proteins reduced, alkylated, and digested with trypsin and subsequently fractionated off-line by SCX chromatography. A small percentage of each SCX fraction was combined to create 24 total pools for the proteome level analysis. The remaining SCX samples were combined to create four total pools for K-ε-GG enrichment. Peptide mixtures were enriched for K-ε-GG peptides and analyzed by LC-MS/MS.
We found that limited fractionation into four SCX fractions prior to enrichment increased the number of identified K-ε-GG peptides by three- to fourfold over the direct enrichment approach in three separate evaluations (supplemental Table S1). Starting with ca. 5 mg of protein per SILAC state, prefractionation enabled the identification of up to ∼3300 distinct K-ε-GG peptides from a single SILAC triple encoded sample (supplemental Table S1). On average, non-K-ε-GG peptides constituted ∼30–50% of all distinct peptides identified in each IP. K-ε-GG peptides were found to follow the expected trend of having increased precursor charge state with increasing SCX eluting time (supplemental Fig. S1B). Comparing sequenced peptide precursors between K-ε-GG and non-K-ε-GG peptides revealed that tryptic K-ε-GG peptides have higher precursor charge states, with the majority of K-ε-GG peptides having a charge state of >2 (supplemental Fig. S1C). This finding correlates with the fact that K-ε-GG peptides tend to contain more amino acids than non-K-ε-GG peptides because of the inability of trypsin to cleave at the modified lysine residue and the additional di-glycine remnant (supplemental Fig. S1D). Ultimately, we conclude that reducing complexity, even minimally, significantly increases the number of identified K-ε-GG peptides in a given sample without greatly increasing the amount of time necessary to complete a quantitative biological comparison. Additionally, employing SCX separation prior to enrichment combines the fractionation for whole proteome analysis and for K-ε-GG analysis into a single step thereby facilitating acquisition of both global proteome and global ubiquitination data on the same sample.
Production of Iodoacetamide-induced Artifacts is not Significant in Complex Mixtures
It was previously reported that in gel digestion protocols using high concentrations of iodoacetamide (55 mm) can induce lysine adducts that are identical in atomic composition to the di-glycine remnant (25). Although acetamide adducts may occur after 55 mm iodoacetamide treatment of a gel slice, we reasoned that the degree of iodoacetamide-induced artifacts would be nominal in the context of complex mixtures under our alkylation conditions, which treat with 10 mm iodoacetamide in Tris HCl buffer. Additionally, we would not expect the anti-K-ε-GG antibody to enrich acetamide adducts. To determine if iodoacetamide induces significant levels of di-acetamide artifacts in complex mixtures under our working conditions we treated 2 mg of protein from total cell lysate with either 10 mm (our working concentration) or 55 mm (high concentration) with D4 iodoacetamide following reduction with 5 mm dithiothreitol. Each sample was prepared as described in supplemental methods. Tryptic peptides were analyzed before and after enrichment with the anti-K-ε-GG antibody. These experiments revealed that the number of peptide spectral matches (PSMs) identified with two heavy acetamide adducts on lysine residues in K-ε-GG pre-enrichment samples and K-ε-GG enriched samples was less than 0.1% for our working concentration of 10 mm iodoacetamide (supplemental Table S2). Additionally, increasing the iodoacetamide concentration to 55 mm did not result in a significant increase in the number of heavy di-acetamide adducts on lysines. Taken together, we conclude that treatment of total cell lysates with 10 mm iodoacetamide under our reaction conditions does not contribute a significant level of the lysine adducted artifact which mimics the di-glycine remnant.
Proteasome Inhibition Induces Large Effects in the Ubiquitin Landscape
We investigated the effects of proteasome inhibition on protein ubiquitination by treating Jurkat cells with the small molecule inhibitor MG-132 (Fig. 2). This compound is a potent inhibitor of the chymotryptic-like activity of the 20S proteasome and is commonly used to study ubiquitinated proteins (1, 26). Initial experiments by Western blotting confirmed that MG-132 does increase the levels of ubiquitination with increasing treatment concentrations in Jurkat cells (supplemental Fig. S2).
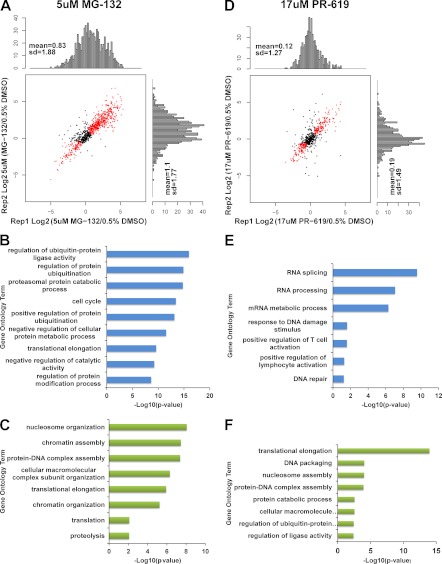
For the SILAC experiment, cells were treated for four hours with 5 μm MG-132 which significantly altered the abundance of a large number of K-ε-GG peptides (Fig. 3A). We found that ∼50% of K-ε-GG peptides were up-regulated >2-fold and as much as 30-fold by MG-132 (supplemental Table S1). GO Biological Process (GOBP) term enrichment for up-regulated K-ε-GG modified proteins revealed that ubiquitination machinery is highly enriched in this population (Fig. 3B). Sites of increased ubiquitination following MG-132 treatment were observed on proteins from each of the three enzyme classes that make up the ubiquitin cascade. Specifically, up-regulated sites were identified on the ubiquitin-activating enzyme UBA1, several ubiquitin-conjugating enzymes, including UBE2D3, UBE2T, and UBE2N, and several E3 ligases, including RNF168, RNF216, Rad18, TRIM41, and E6-AP (supplemental Table S1). Ubiquitination sites were also significantly increased on both α- and β-type subunits of the 20S proteasome, consistent with previous findings (27).
Fig. 3.
The effects of MG-132 and PR-619 on K-ε-GG peptides. Scatter plots and histograms of the SILAC ratios of biological replicate 1 versus 2 for (A) K-ε-GG peptides after 5 μm MG-132 treatment. Ratios deemed to be reproducibly regulated by the moderated t-statistic (Benjamini-Hochberg p value <0.1) are indicated in red. Enriched GOBP terms were derived using the DAVID bioinformatics resource. A selection of significantly enriched terms (Benjamini-Hochberg p value <0.05) were plotted for K-ε-GG modified proteins that were (B) up-regulated and (C) down-regulated by MG-132. Scatter plots and histograms of SILAC ratios of biological replicate 1 versus 2 for (D) K-ε-GG peptides after 17 μm PR-619 treatment. A selection of significantly enriched GOBP terms (p < 0.05) were plotted for K-ε-GG peptides ratios that were (E) up-regulated or (F) down-regulated by 17 μm PR-619 treatment.
In addition to the large degree of up-regulation observed with MG-132 treatment, ∼15% of K-ε-GG peptides were reproducibly down-regulated (Fig. 3A and supplemental Table S1). GOBP term enrichment for down-regulated K-ε-GG proteins (Fig. 3C) indicated that chromatin related proteins including histones H2A, H2A.x, macro-H2A, H2B, and H1 are highly enriched in this subpopulation (supplemental Table S1). This result is supported by previous work that found reduced levels of histone ubiquitination on histones H2A and H2B upon proteasomal inhibition and proteotoxic stress (28, 29). Additionally, this down-regulated subpopulation was enriched in translational machinery including the 40S ribosomal protein S20, the 60S ribosomal proteins L9, L11, L30, L19, and L24, and the elongation factor 1-alpha.
DUB Inhibition by PR-619 Increases Protein Ubiquitination Levels but is Less Effective than Proteasome Inhibition by MG-132
Next we evaluated the general effects of the DUB inhibitor PR-619 in Jurkat cells. Western blotting was initially used to assess the global effects of PR-619 on ubiquitinated proteins. This initial experiment confirmed that PR-619 functions to increase ubiquitination levels in Jurkat cells with increasing treatment concentrations (supplemental Fig. S2). For the SILAC-based study, we treated Jurkat cells with two different concentrations of PR-619, 5 μm (low) or 17 μm (high), for 4 h (Fig. 2A). Final treatment concentrations were chosen based on the reported IC50 values (5uM–20 μm range) of this compound for inhibiting DUBs or ubiquitin-like isopeptidases as well as, information from the vendor (Lifesensors Inc.) indicating that treatment with >30 μm PR-619 may induce cytotoxic effects. In addition to treatment with either low- or high-concentration PR-619, Jurkat cells were also treated with a combination of 17 μm PR-619 and 5 μm MG-132 to increase the abundance of those peptides that may be rapidly degraded by the proteasome following DUB inhibition (Fig. 2A).
High-concentration PR-619 treatment resulted in a greater percentage of regulated K-ε-GG peptides relative to low-concentration treatment (supplemental Fig. S3). High- and low-concentration PR-619 treatment altered the abundance of ∼20 and 40% of quantified K-ε-GG peptides, respectively (Fig. 3D and supplemental Fig. S4). Analysis of GOBP terms associated with proteins found to have up-regulated K-ε-GG sites after high-concentration PR-619 treatment revealed an enrichment for proteins involved in RNA processing functions including HNRNP M, HNRPN K, HNRNP H, as well as the ATP-dependent RNA helicase eIF4A-1 and 46, and the DNA-directed RNA polymerase II subunit RPB2 (Fig. 3E and supplemental Table S1). Also in accordance with the findings of Altun et al. (13) we find that DUB inhibition by PR-619 increases the level of ubiquitination on sites of several proteasome subunits including PROS26 and PSMA5 but does not significantly change the level of ubiquitination on the known NEDD8ylated substrate CUL1 at K410 (13). Interestingly, similar to MG-132 treatment, high concentration PR-619 caused down-regulation of K-ε-GG sites on histone proteins as well as ribosomal proteins (Fig. 3F and supplemental Table S1).
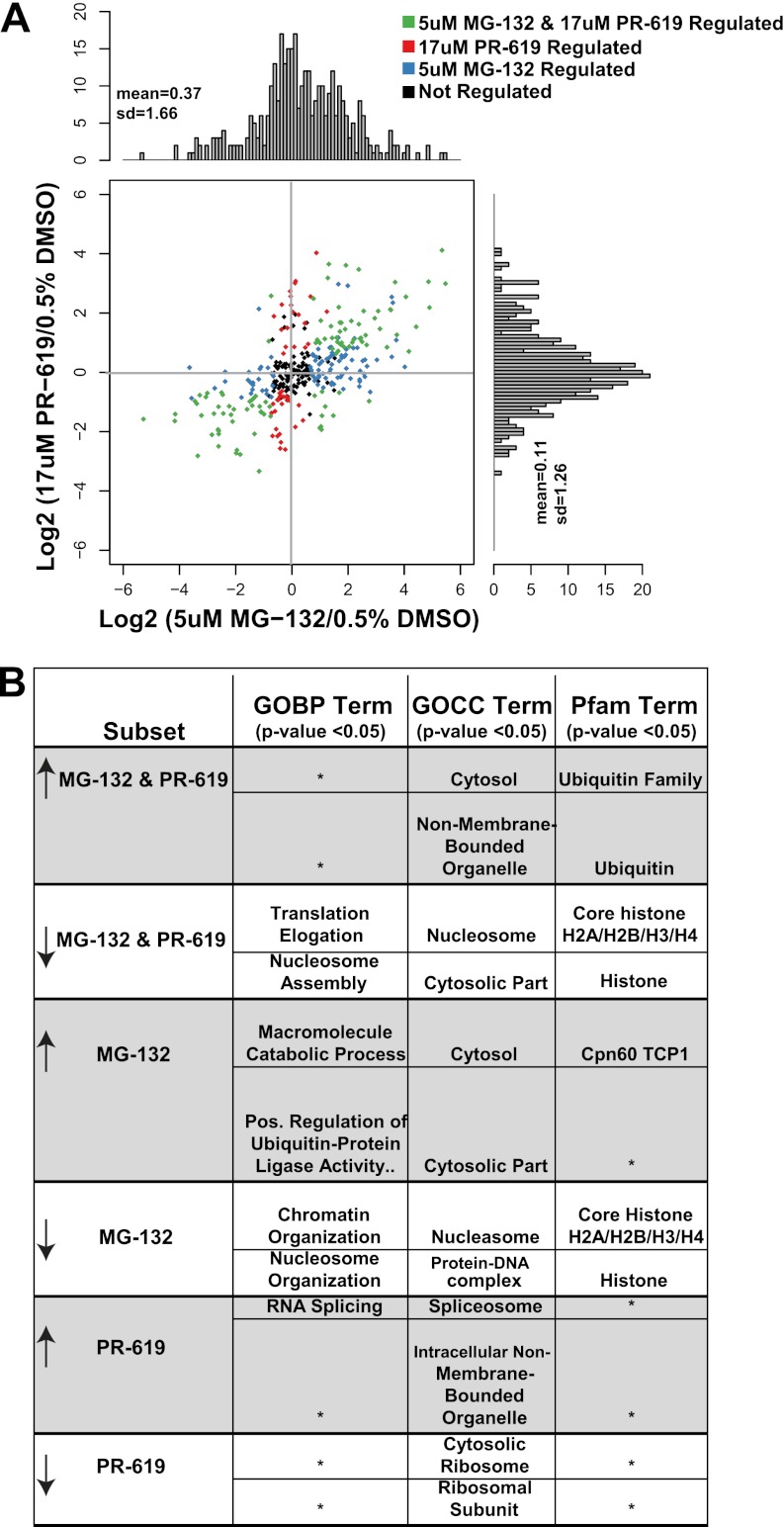
K-ε-GG peptides quantified across all SILAC experiments were used to directly compare the effects of 5 μm MG-132 versus 17 μm PR-619 (Fig. 4A). Approximately 30% of K-ε-GG peptides compared were reproducibly regulated by both PR-619 and MG-132 (Fig. 4A). However, MG-132 significantly increased levels of nearly twice as many K-ε-GG sites than PR-619. As expected from the individual analysis of each treatment condition, the subset of K-ε-GG sites down-regulated by both MG-132 and PR-619 were highly enriched in chromatin related proteins and translational machinery as indicated by GO cellular component, GOBP, and Pfam (a comprehensive database of over 13,000 conserved protein families, http://pfam.sanger.ac.uk/) term enrichment (Fig. 4B). As expected, both compounds increased the levels of K-ε-GG sites on ubiquitin itself and also resulted in increased K-ε-GG levels on the ubiquitin-like proteins NEDD8 and SUMO-2 (Fig. 4B and supplemental Table S1). Approximately 30% of K-ε-GG peptides were significantly regulated by only MG-132 treatment. MG-132 up-regulated K-ε-GG sites were found on proteasome subunits alpha type-1 and -6 as well as on the regulatory subunits 4 and 7 (supplemental Table S1) (30). The subset of proteins with K-ε-GG sites that increased only after PR-619 treatment was enriched with tubulins (supplemental Table S1) (31).
Fig. 4.
Comparison of effects induced by MG-132 and PR-619 on the same K-ε-GG peptide. Scatter plot and histograms of SILAC ratios for (A) K-ε-GG peptides after 5 μm MG-132 versus 17 μm PR-619 treatment. To generate this plot, the average log2 SILAC ratio between two biological replicates was calculated for each treatment condition. Regulation status was determined using the moderated t-statistic (Benjamini-Hochberg p value <0.1). Points colored in black were not deemed reproducibly regulated in either of the treatment conditions. Enriched GOBP, GOCC and Pfam terms were derived using the DAVID bioinformatics resource. The two most significantly enriched terms (Benjamini-Hochberg p value <0.05) are tabulated (B) for K-ε-GG modified proteins within each of the colored subsets. Asterisks indicate that no significantly enriched ontology term was identified.
DUB inhibition by PR-619 could result in rapid degradation of proteins in the absence of proteasome inhibition. Therefore, to study this, treatment with a high concentration of PR-619 was carried out in the presence and absence of MG-132. This comparison revealed that nearly 40% of K-ε-GG peptides were regulated under both conditions and that the level of regulation for the majority of these K-ε-GG sites was similar (supplemental Fig. S5 and supplemental Table S1). Approximately 15% of sites were only up-regulated following the combination of PR-619 and MG-132 treatment. This subset of K-ε-GG-peptides was compared with the set obtained following treatment with MG-132 to determine which K-ε-GG sites were distinctly regulated by the combined treatment of PR-619 and MG-132. In total, 13 K-ε-GG-modified proteins observed in the MG-132 treatment data contained K-ε-GG sites that were only reproducibly up-regulated by the combined treatment of PR-619 and MG-132. This list included modification of K21 of the proteasomal ubiquitin receptor ADRM1 and K54 of NEDD8 (supplemental Table S1). Proteins harboring K-ε-GG sites that were up-regulated with a combination of PR-619 and MG-132 treatment are potential candidates for degradation by the UPS following DUB inhibition. Overall we find that many K-ε-GG sites are regulated to a similar extent by PR-619 and MG-132. However, among the K-ε-GG sites studied, we find that inhibition of the proteasome by MG-132 results in almost twice the number of up-regulated K-ε-GG sites relative to PR-619 treatment alone.
Enrichment of K-ε-GG Peptides Facilitates the Analysis of Perturbation Induced Effects on Polyubiquitin Linkages
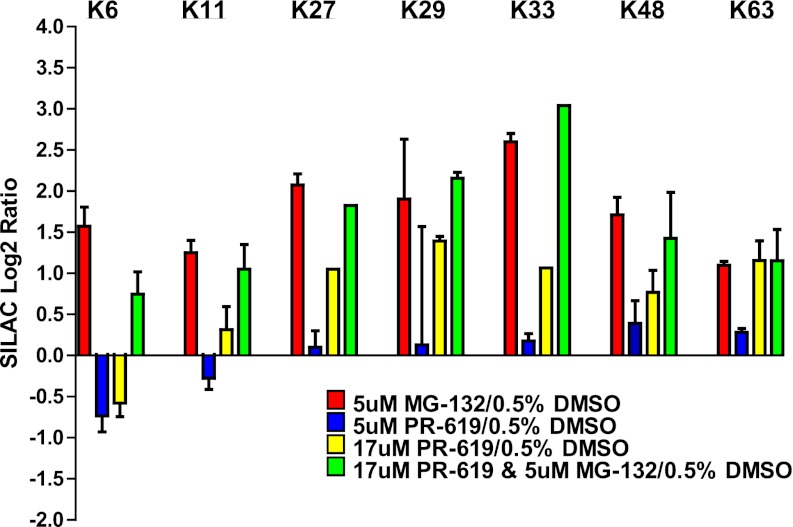
The described methodology can also be used to measure changes in global polyubiquitin chain topology following cellular perturbation. SILAC ratios were plotted for GlyGly-modified lysine residues of ubiquitin for all cellular treatment conditions (Fig. 5). Similar to previous findings, we observe that proteasome inhibition increases the abundance of all polyubiquitin linkages, but has the mildest effect on K63, a site that is known to have nonproteolytic functions (11, 12, 32, 33). PR-619 induced milder effects on polyubiquitin chain abundance relative to MG-132 for nearly all linkages. This may suggest that the inhibitory effect of PR-619 on the proteasome is low compared with MG-132, as is indicated by the vendor, or that the majority of polyubiquitin chains affected by PR-619 are rapidly degraded by the proteasome. Interestingly, PR-619 treatment reduces levels of K6 polyubiquitin linkages Comparing results from PR-619 treatment with and without the presence of MG-132 indicates that the combined treatment of PR-619 and MG-132 is not essential for significantly increasing the levels of polyubiquitin chains (Fig. 5). Overall, we find that MG-132 is more efficient at increasing the levels of polyubiquitin linkages relative to PR-619.
Fig. 5.
Effects of treatment on polyubiquitin linkages. SILAC log2 ratios are plotted for GlyGly modified lysine residues of ubiquitin. Ratios for all nondistinct K-ε-GG modified lysine residues of ubiquitin were used for this analysis.
Representation of Amino Acids Proximal to K-ε-GG Sites
Large scale ubiquitin site mapping efforts facilitates the study of amino acid representation proximal to K-ε-GG sites, which can aid in understanding the specificity of lysine ubiquitination. To search for over- or underrepresented amino acids proximal to K-ε-GG sites, we compared the frequency of all amino acids within a ± 10 amino acid sequence window (extracted from the protein sequence) between confidently localized K-ε-GG sites from our data set to a background of randomly chosen lysine residues from the human IPI database. The log-odds ratio was calculated for the frequency of an amino acid at every position in the sequence window and results were visualized as a heat map (supplemental Fig. S6). Similar to recent findings by Kim et al., the most pronounced effects observed were underrepresentation of the basic amino acids lysine and histidine especially N-terminal to the K-ε-GG site and extending out to the −4 position (12). In addition we find that proline residues are depleted at the ± 1 position. We also observed some degree of over-representation of glycine, serine, threonine, tyrosine, glutamine, and asparagine residues either N- or C-terminal to the K-ε-GG site. No strong sequence motif was identified in our data set using motif enrichment (data not shown) (34). We note that these observations could be reflective of sequence biases of the anti-K-ε-GG antibody.
Steady-State Protein Levels are Largely Unaffected Following Proteasome and DUB Inhibition
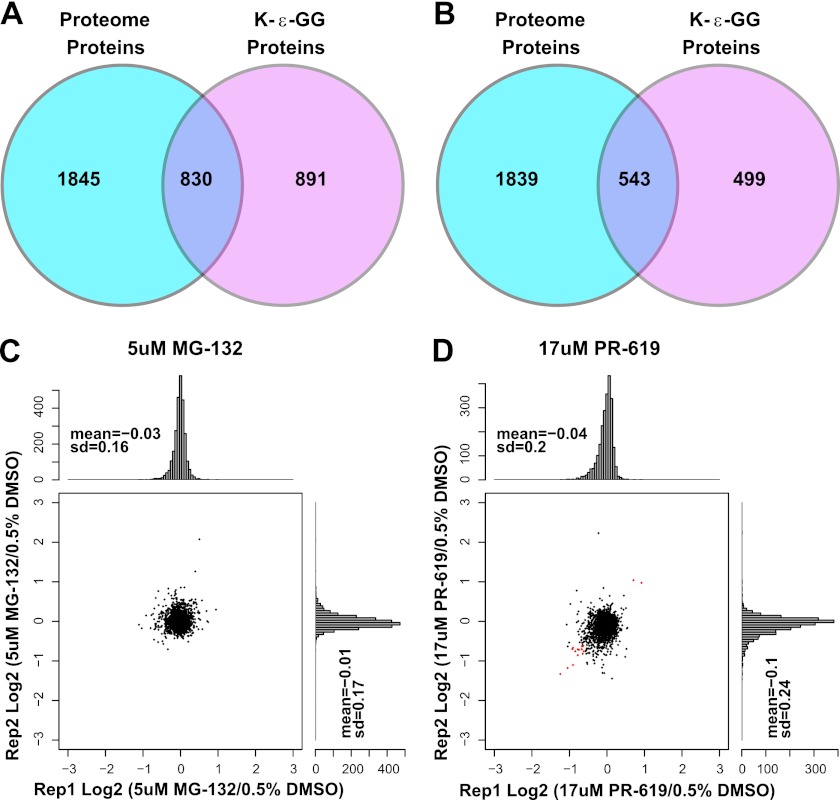
In addition to the enrichment of K-ε-GG peptides, we conducted whole-proteome analyses (without Ub enrichment) on the same samples to study the effects of MG-132 and PR-619 on protein steady-state levels. We reproducibly quantified 2675 and 2382 proteins quantified by at least three distinct peptides ratios in the SILAC experiments I and II, respectively (Fig. 2 and supplemental Table S1). As a result, we obtained proteome level information for approximately half of K-ε-GG modified proteins identified in both SILAC triple encoded samples (Figs. 6A and 6B). At this depth of coverage, we obtained whole proteome data for many important regulatory enzymes modulating protein ubiquitination including >10 E2s, >20 E3s, and ∼15 DUBs. We were initially surprised to find that nearly all protein levels were unaffected following MG-132 or PR-619 treatment, regardless of the extent of regulation at the K-ε-GG level, as the canonical function for ubiquitination is degradation by the UPS (Figs. 6C and 6D). Interestingly, the protein level of ubiquitin itself was found to be unchanged under all treatment conditions. These results correlate with data from a recent large-scale proteomic effort which also studied the effects of proteasome inhibition on whole protein levels (12). The authors suggest that minimal observed changes to protein levels following proteasome inhibition may be linked to a low stoichiometry of the ubiquitinated form of a given protein (12). For K-ε-GG sites quantified after MG-132 treatment, we compared the SILAC ratios for the K-ε-GG peptide, the unmodified peptide containing the modified lysine residue, and the measured protein SILAC ratio to study K-ε-GG site stoichiometry (supplemental Figs. S8A, S8B, and S8C) (35). We note that comparison of the exact peptide in the K-ε-GG and non-K-ε-GG state is not possible for the majority of cases as ubiquitination of a lysine residue results in a missed cleavage by trypsin. The comparison of the non-K-ε-GG peptide ratio to the protein ratio indicated that only a small percentage of non-K-ε-GG peptide SILAC ratios differed significantly from the measured protein ratio for regulated K-ε-GG sites (supplemental Fig. S8B). Further comparison of the non-K-ε-GG and K-ε-GG SILAC ratios after MG-132 treatment also revealed that the majority of regulated K-ε-GG sites did not show a reciprocal change in the non-K-ε-GG SILAC ratio (supplemental Fig. S8C). Taken together, our data supports the hypothesis that the majority of K-ε-GG sites measured in this study have low stiochiometry, which would result in minimal observed changes to the protein level.
Fig. 6.
Results of Proteome Analysis. Overlap of proteins and K-ε-GG modified peptides identified in SILAC (A) Experiment I and (B) Experiment II. Scatter plots and histograms are shown for SILAC ratios of biological replicate 1 versus 2 for Proteome-level protein ratios after (C) 5 μm MG-132 treatment and (D) 17 μm PR-619 treatment.
DISCUSSION
A major focus of this work was to develop practical methods for routine, robust, and relatively fast global proteomic studies of ubiquitination. We designed our process to enable ubiquitination as well as whole proteome analysis on the same sample (versus independent samples) to both conserve on sample and to reduce the amounts of costly labeling reagents and instrument time used. The workflow we developed employing limited prefractionation of peptides by SCX is compatible with K-ε-GG enrichment and affords a three- to fourfold increase in the number of identified K-ε-GG peptides in a given SILAC triple-encoded experiment. Using ca. 5 mg of input protein per SILAC label state, we successfully identified 5533 distinct K-ε-GG peptides of which 4907 were quantified. These amounts of input protein per state can be readily derived from a few (e.g. 4–5) dishes of cells, and acquisition of K-ε-GG data requires only 1.5 days of on-instrument time. Thus the strategy presented here represents a practical and readily implementable approach to the analysis of global ubiquitination in a wide range of cellular perturbational studies. Other groups have recently shown that using the anti K-ε-GG antibody, large numbers of K-ε-GG peptides (5000–7000) can be identified in a given sample by limiting SILAC-based experiments to only two states (12, 36) or by label-free experiments (11) all in combination with greatly increased amounts of input protein (up to 35 mg). Comparison of our findings with the SILAC based experiments performed by Wagner et al. (11) shows similar numbers of K-ε-GG site identifications across two biological replicates for both studies (supplemental Fig. S7 and supplemental Table S2). Most importantly, this comparison reveals a high degree of similarity in the biological processes found to be regulated by MG-132 treatment in studies conducted in two independent laboratories. We believe that further improvements in overall yield of quantified K-ε-GG peptides and proteins from limiting amounts of biological samples may be possible through optimization of both the antibody enrichment strategy as well as the specific choice of pre-fractionation method used. Such improvements will facilitate application of quantitative ubiquitination analysis to primary cells and other instances where protein amounts can be limiting.
In this study we show that both proteasome inhibition by the commonly used proteasome inhibitor MG-132 and DUB inhibition by the broad-specificity DUB inhibitor PR-619 induce extremely large effects on protein ubiquitination levels. As expected, proteasome inhibition significantly increased the levels of nearly all polyubiquitin linkages and resulted in up-regulation of over half of all K-ε-GG sites measured (supplemental Table S1). DUB inhibition also increased levels of ubiquitination on a large number of proteins, including those involved in pre-mRNA processing. We found that polyubiquitin linkages were generally less affected by DUB inhibition relative to proteasome inhibition, suggesting that cellular treatment with proteasome inhibitors is a more effective approach for increasing the levels of polyubiquitinated proteins that may be candidates for degradation by the UPS. Overall we found that proteasome inhibition by MG-132 is more effective in increasing the levels of ubiquitination sites relative to PR-619.
Cellular ubiquitin is distributed in a variety of forms including mono- and polyubiquitin linkages as well as a free molecule pool (37). Previous work has shown that proteasome inhibitors cause deubiquitination of histone proteins and has linked this phenomenon to redistribution of free ubiquitin into ubiquitin conjugates, which occurs in response to the proteotoxic stress induced by these compounds (28, 37–39). From our work, we find that both proteasome inhibition as well as broad DUB inhibition results in decreased levels of histone ubiquitination, suggesting that both compounds induce proteotoxic stress on the system through disruption of ubiquitin equilibrium. Knowing this, we suggest that careful experiment design should be considered when using inhibitors that globally affect ubiquitin processing for studying in vivo perturbation induced changes to protein ubiquitination.
Whole proteome-level analysis revealed that the steady-state levels of proteins were largely unaffected by proteasome or DUB inhibition. Although initially unexpected, our proteome level results are consistent with recent large-scale proteomic efforts that also examined the effects of proteasome inhibition on protein steady-state levels (12). The apparent lack of regulation observed for protein levels may suggest that many ubiquitinated proteins targeted for degradation by the UPS exist at low stoichiometry relative to their nonubiquitinated form (12). In this case, only a small percentage of proteins that are ubiquitinated would change in abundance through proteosomal clearance, whereas the levels of the bulk of proteins would appear to remain largely unchanged by global proteomic methods. Additionally, it has been suggested that up to 30% of newly synthesized proteins are degraded by the proteasome as a quality control mechanism for removing aberrant protein products of translation or mis-folded proteins (40). In our study we observed regulation of K-ε-GG sites on a number of proteins involved with translational elongation and processing, which may cause a degree of dysregulation in the synthesis of new proteins, thereby reducing the observable changes in protein steady-state levels. Last, it has been shown that degradation of ubiquitinated proteins by the proteasome can have a dependence on polyubiquitin chain length, which may influence the degree of observed protein changes observed in our study, especially in the case of DUB inhibition (41–43).
Ultimately, we find that the introduction of antibodies recognizing ubiquitin remnant peptides has been a huge breakthrough for studying protein ubiquitin. We show that this technique, coupled with quantitative mass spectrometry, is extremely powerful for quantifying perturbation induced changes in the ubiquitin landscape. We are certain that these techniques will be extremely useful for understanding how components of the ubiquitin system are is involved in human disease and will hopefully help to accelerate the future development of drugs that target the ubiquitin system (3).
Footnotes
* Support for this work was provided in part by the Broad Institute of MIT and Harvard and by grants from the National Cancer Institute (U24CA160034) and National Heart Lung and Blood Institute (HHSN268201000033C and R01HL096738) to SAC. We would like to acknowledge James Strickler of LifeSensors Inc. for helpful discussions.
 This article contains supplemental Methods, Figs. S1 to S8, and Tables S1 and S2.
This article contains supplemental Methods, Figs. S1 to S8, and Tables S1 and S2.
1 The abbreviations used are:
- DUB
- deubiquitinase
- SILAC
- stable isotope labeling by amino acids in cell culture
- CID
- collision-induced dissociation
- SCX
- strong cation exchange
- UPS
- ubiquitin-proteasome system
- GO
- Gene Ontology.
REFERENCES
- 1. Myung J., Kim K. B., Crews C. M. (2001) The ubiquitin-proteasome pathway and proteasome inhibitors. Med. Res. Rev. 21, 245–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Dikic I., Wakatsuki S., Walters K. J. (2009) Ubiquitin-binding domains [mdash] from structures to functions. Nat. Rev. Mol. Cell Biol. 10, 659–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cohen P., Tcherpakov M. (2010) Will the Ubiquitin System Furnish as Many Drug Targets as Protein Kinases? Cell 143, 686–693 [DOI] [PubMed] [Google Scholar]
- 4. Sacco J. J., Coulson J. M., Clague M. J., Urbé S. (2010) Emerging roles of deubiquitinases in cancer-associated pathways. IUBMB Life 62, 140–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Komander D., Clague M. J., Urbé S. (2009) Breaking the chains: structure and function of the deubiquitinases. Nat. Rev. Mol. Cell Biol. 10, 550–563 [DOI] [PubMed] [Google Scholar]
- 6. Bedford L., Lowe J., Dick L. R., Mayer R. J., Brownell J. E. (2011) Ubiquitin-like protein conjugation and the ubiquitin-proteasome system as drug targets. Nat. Rev. Drug Discov. 10, 29–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Olsen J. V., Blagoev B., Gnad F., Macek B., Kumar C., Mortensen P., Mann M. (2006) Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell 127, 635–648 [DOI] [PubMed] [Google Scholar]
- 8. Huttlin E. L., Jedrychowski M. P., Elias J. E., Goswami T., Rad R., Beausoleil S. A., Villén J., Haas W., Sowa M. E., Gygi S. P. (2010) A tissue-specific atlas of mouse protein phosphorylation and expression. Cell 143, 1174–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Danielsen J. M. R., Sylvestersen K. B., Bekker-Jensen S., Szklarczyk D., Poulsen J. W., Horn H., Jensen L. J., Mailand N., Nielsen M. L. (2010) Mass spectrometric analysis of lysine ubiquitylation reveals promiscuity at site level. Mol. Cell. Proteomics 10, M110.003590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Xu G., Paige J. S., Jaffrey S. R. (2010) Global analysis of lysine ubiquitination by ubiquitin remnant immunoaffinity profiling. Nat. Biotechnol. 28, 868–873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wagner S. A., Beli P., Weinert B. T., Nielsen M. L., Cox J., Mann M., Choudhary C. (2011) A proteome-wide, quantitative survey of in vivo ubiquitylation sites reveals widespread regulatory roles. Mol. Cell. Proteomics. 10, M111.013284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kim W., Bennett E. J., Huttlin E. L., Guo A., Li J., Possemato A., Sowa M. E., Rad R., Rush J., Comb M. J., Harper J. W., Gygi S. P. (2011) Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol. Cell. 44, 325–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Altun M., Kramer, Holger H. B., Willems L. I., McDermott J. L., Leach C. A., Goldenberg S. J., Kumar K. G., Konietzny R., Fischer R., Kogan E., Mackeen M. M., McGouran J., Khoronenkova S. V., Parsons J. L., Dianov G. L., Nicholson B., Kessler B. M. (2011) Activity-based chemical proteomics accelerates inhibitor development for deubiquitylating enzymes. Chem. Biol. 18, 1401–1412 [DOI] [PubMed] [Google Scholar]
- 14. Ong S. E., Blagoev B., Kratchmarova I., Kristensen D. B., Steen H., Pandey A., Mann M. (2002) Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics 1, 376–386 [DOI] [PubMed] [Google Scholar]
- 15. Rappsilber J., Mann M., Ishihama Y. (2007) Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2, 1896–1906 [DOI] [PubMed] [Google Scholar]
- 16. Cox J., Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 17. Cox J., Neuhauser N., Michalski A., Scheltema R. A., Olsen J. V., Mann M. (2011) Andromeda: A Peptide Search Engine Integrated into the MaxQuant Environment. J. Proteome Res. 10, 1794–1805 [DOI] [PubMed] [Google Scholar]
- 18. Elias J. E., Gygi S. P. (2007) Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Meth. 4, 207–214 [DOI] [PubMed] [Google Scholar]
- 19. Seyfried N. T., Xu P., Duong D. M., Cheng D., Hanfelt J., Peng J. (2008) Systematic approach for validating the ubiquitinated proteome. Anal. Chem. 80, 4161–4169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Smyth G. K. (2004) Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3, Article3 [DOI] [PubMed] [Google Scholar]
- 21. Benjamini Y., Hochberg Y. (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. Roy. Statistical Soc. B 57, 289–300 [Google Scholar]
- 22. Huang D. W., Sherman B. T., Lempicki R. A. (2008) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 [DOI] [PubMed] [Google Scholar]
- 23. Huang, da W., Sherman B. T., Lempicki R. A. (2009) Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. GENE-E. http://www.broadinstitute.org/cancer/software/GENE-E/:
- 25. Nielsen M. L., Vermeulen M., Bonaldi T., Cox J., Moroder L., Mann M. (2008) Iodoacetamide-induced artifact mimics ubiquitination in mass spectrometry. Nat. Methods 5, 459–460 [DOI] [PubMed] [Google Scholar]
- 26. Rock K. L., Gramm C., Rothstein L., Clark K., Stein R., Dick L., Hwang D., Goldberg A. L. (1994) Inhibitors of the proteasome block the degradation of most cell proteins and the generation of peptides presented on MHC class I molecules. Cell 78, 761–771 [DOI] [PubMed] [Google Scholar]
- 27. Tanaka K., Tsurumi C. (1997) The 26S proteasome: subunits and functions. Mol. Biol. Reports 24, 3–11 [DOI] [PubMed] [Google Scholar]
- 28. Dantuma N. P., Groothuis T. A. M., Salomons F. A., Neefjes J. (2006) A dynamic ubiquitin equilibrium couples proteasomal activity to chromatin remodeling. The J. Cell Biol. 173, 19–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Xu Q., Farah M., Webster J. M., Wojcikiewicz R. J. H. (2004) Bortezomib rapidly suppresses ubiquitin thiolesterification to ubiquitin-conjugating enzymes and inhibits ubiquitination of histones and type I inositol 1,4,5-trisphosphate receptor. Mol. Cancer Therap. 3, 1263–1269 [PubMed] [Google Scholar]
- 30. Murata S., Yashiroda H., Tanaka K. (2009) Molecular mechanisms of proteasome assembly. Nat. Rev. Mol. Cell Biol. 10, 104–115 [DOI] [PubMed] [Google Scholar]
- 31. Ren Y., Zhao J., Feng J. (2003) Parkin Binds to α/β Tubulin and Increases their Ubiquitination and Degradation. J. Neurosci. 23, 3316–3324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Dammer E. B., Na C. H., Xu P., Seyfried N. T., Duong D. M., Cheng D., Gearing M., Rees H., Lah J. J., Levey A. I., Rush J., Peng J. (2011) Polyubiquitin linkage profiles in three models of proteolytic stress suggest the etiology of Alzheimer disease. J. Biol. Chem. 286, 10457–10465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Deng L., Wang C., Spencer E., Yang L., Braun A., You J., Slaughter C., Pickart C., Chen Z. J. (2000) Activation of the IκB kinase complex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique polyubiquitin chain. Cell 103, 351–361 [DOI] [PubMed] [Google Scholar]
- 34. Schwartz D., Gygi S. P. (2005) An iterative statistical approach to the identification of protein phosphorylation motifs from large-scale data sets. Nat. Biotech. 23, 1391–1398 [DOI] [PubMed] [Google Scholar]
- 35. Olsen J. V., Vermeulen M., Santamaria A., Kumar C., Miller M. L., Jensen L. J., Gnad F., Cox J., Jensen T. S., Nigg E. A., Brunak S., Mann M. (2010) Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci. Signal. 3, ra3- [DOI] [PubMed] [Google Scholar]
- 36. Emanuele M. J., Elia A. E., Xu Q., Thoma C. R., Izhar L., Leng Y., Guo A., Chen Y. N., Rush J., Hsu P. W., Yen H. C., Elledge S. J. (2011) Global identification of modular cullin-RING ligase substrates. Cell 147, 459–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Groothuis T. A., Dantuma N. P., Neefjes J., Salomons F. A. (2006) Ubiquitin crosstalk connecting cellular processes. Cell Div. 1, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Carlson N., Rogers S., Rechsteiner M. (1987) Microinjection of ubiquitin: changes in protein degradation in HeLa cells subjected to heat-shock. J. Cell Biol. 104, 547–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mimnaugh E. G., Chen H. Y., Davie J. R., Celis J. E., Neckers L. (1997) Rapid deubiquitination of nucleosomal histones in human tumor cells caused by proteasome inhibitors and stress response inducers: effects on replication, transcription, translation, and the cellular stress response. Biochemistry 36, 14418–14429 [DOI] [PubMed] [Google Scholar]
- 40. Schubert U., Antón L. C., Gibbs J., Norbury C. C., Yewdell J. W., Bennink J. R. (2000) Rapid degradation of a large fraction of newly synthesized proteins by proteasomes. Nature 404, 770–774 [DOI] [PubMed] [Google Scholar]
- 41. Verhoef L. G., Heinen C., Selivanova A., Halff E. F., Salomons F. A., Dantuma N. P. (2009) Minimal length requirement for proteasomal degradation of ubiquitin-dependent substrates. FASEB J.. 23, 123–133 [DOI] [PubMed] [Google Scholar]
- 42. Zhao M., Zhang N. Y., Zurawel A., Hansen K. C., Liu C.-W. (2009) Degradation of some polyubiquitinated proteins requires an intrinsic proteasomal binding element in the substrates. J. Biol. Chem. 285, 4771–4780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lee B. H., Lee M. J., Park S., Oh D. C., Elsasser S., Chen P. C., Gartner C., Dimova N., Hanna J., Gygi S. P., Wilson S. M., King R. W., Finley D. (2010) Enhancement of proteasome activity by a small-molecule inhibitor of USP14. Nature 467, 179–184 [DOI] [PMC free article] [PubMed] [Google Scholar]