Abstract
Protein kinase D (PKD) is a cytosolic serine/threonine kinase implicated in regulation of several cellular processes such as response to oxidative stress, directed cell migration, invasion, differentiation, and fission of the vesicles at the trans-Golgi network. Its variety of functions must be mediated by numerous substrates; however, only a couple of PKD substrates have been identified so far. Here we perform stable isotope labeling of amino acids in cell culture-based quantitative phosphoproteomic analysis to detect phosphorylation events dependent on PKD1 activity in human cells. We compare relative phosphorylation levels between constitutively active and kinase dead PKD1 strains of HEK293 cells, both treated with nocodazole, a microtubule-depolymerizing reagent that disrupts the Golgi complex and activates PKD1. We identify 124 phosphorylation sites that are significantly down-regulated upon decrease of PKD1 activity and show that the PKD target motif is significantly enriched among down-regulated phosphorylation events, pointing to the presence of direct PKD1 substrates. We further perform PKD1 target motif analysis, showing that a proline residue at position +1 relative to the phosphorylation site serves as an inhibitory cue for PKD1 activity. Among PKD1-dependent phosphorylation events, we detect predominantly proteins with localization at Golgi membranes and function in protein sorting, among them several sorting nexins and members of the insulin-like growth factor 2 receptor pathway. This study presents the first global detection of PKD1-dependent phosphorylation events and provides a wealth of information for functional follow-up of PKD1 activity upon disruption of the Golgi network in human cells.
The protein kinase D (PKD)1 family comprises three closely related members: PKD1, PKD2, and PKD3, which belong to the calcium- and calmodulin-dependent kinase family of serine/threonine kinases (1). Dependent on stimulus and cell type, active PKD localizes to organelles such as the Golgi complex, mitochondria, nucleus, plasma membrane, and the F-actin cytoskeleton to control various cellular processes including survival responses to oxidative stress (2), directed cell migration (3–5), invasion (6, 7), differentiation (8–10), and fission of the cell surface-directed transport carriers at the trans-Golgi network (TGN) (11, 12). In most cases, involvement of PKD in these processes has been demonstrated by expression of a kinase dead PKD protein (PKDkd), which acts in a dominant-negative fashion toward the endogenous PKD proteins and thereby presents a functional knock-out. For example, expression of PKD1kd induces the formation of tubule-like structures, thus blocking secretion of basolateral cargo at the Golgi complex (11). Conversely, expression of a constitutively active PKD (PKD1ca) induces extensive fragmentation of Golgi membranes (13). Likewise, PKD1kd enhances directed migration of breast cancer cells, whereas PKD1ca suppresses migration of these cells (3, 4).
These multiple functions of PKD are obviously mediated through numerous substrates. During the past years the knowledge on these substrates has increased dramatically. For instance, it was shown that PKD controls directed cell migration by direct phosphorylation of the cofilin phosphatase slingshot 1 (4, 5, 14), the kinase PAK4 (15), cortactin (16), and the tumor suppressor RIN1 (17), thereby affecting dynamic F-actin remodeling at the leading edge. At the TGN, PKD directly phosphorylates the lipid kinase PI4KIIIβ (18) and the lipid transfer proteins CERT (19) and OSBP (20), thus mediating the fission of vesicles destined for the cell surface. However, knockdown of CERT did not suppress soluble cargo secretion as effectively as a kinase dead, dominant-negative PKD1 variant (19), demonstrating that yet unidentified PKD substrates contribute to proper Golgi function. The microtubule-depolymerizing reagent nocodazole, which disrupts the Golgi complex to generate Golgi mini-stacks, activates PKD, and this nocodazole-dependent fragmentation of the Golgi can be blocked by expression of a kinase dead PKD1 protein (21). The PKD signaling pathways involved in nocodazole-dependent Golgi dispersal, however, remain to be investigated.
Mass spectrometry-based proteomics is increasingly used in global detection of kinase substrates in eukaryotic cells. Modern, gel-free biochemical approaches for phosphopeptide enrichment (22) are used in combination with specific inactivation of kinases to perform quantitative phosphoproteomic readouts of kinase activity. Specific inhibition of analog-sensitive kinases (23, 24) and subsequent SILAC-based quantitative phosphoproteomics has recently been used to identify CDK1- and Aurora-dependent phosphorylation events in budding and fission yeast, respectively (25, 26). Likewise, chemical inhibition of endogenous kinases has been used to identify phosphorylation events downstream of the mTORC1, Polo-like, and Aurora kinases in human cells (27–29). In a groundbreaking study, a combination of the above approaches was used to systematically inactivate all protein kinases and phosphatases in budding yeast and derive the first comprehensive kinase/phosphatase substrate network of an eukaryotic microorganism (30).
Here we perform a global, SILAC-based quantitative phosphoproteomic study to identify phosphorylation events dependent on PKD activity. We compare two nocodazole-treated HEK293 strains, expressing constitutively active or dominant-negative (kinase dead) forms of PKD1. We demonstrate that 124 phosphorylation events are down-regulated upon the loss of PKD activity under conditions of nocodazole-dependent Golgi complex fragmentation. We perform functional enrichment analysis and show that proteins with localization and function at the membrane are significantly enriched among detected PKD targets. We further classify the PKD-dependent phosphorylation events based on their level of confidence, perform analysis of PKD1 target sequence motifs, and show that proline residues at position +1 relative to a PKD1-dependent phosphorylation site act as an inhibitory cue for PKD activity.
EXPERIMENTAL PROCEDURES
Cell Culture
Flp-In T-Rex-293 cells (Invitrogen) were grown in Dulbecco's modified Eagle's medium containing 10% FCS, zeocin at 100 μg/ml, and blasticidin at 15 μg/ml. These cells stably express the Tet repressor and contain a single FRT site and were used to generate the Flp-In-PKD1 lines. The cells were cotransfected with pcDNA5/FRT/TO-EGFP-PKD1ΔPH (PKD1 constitutively active or PKD1ca) (31) and PKD1K612W (PKD1 kinase dead or PKD1kd) (31), respectively, and the Flp recombinase expression plasmid pOG44 at a ratio of 1:10 and then selected with hygromycin at 100 μg/ml. Induction of protein expression with doxycycline was at 10 ng/ml.
SILAC Experiments
All of the cell lines were maintained in custom-made SILAC Dulbecco's modified Eagle's medium for 14 days. PKD1 constitutively active cells (FlpIN T-Rex293 PKD1ΔPH-GFP or PKD1ca) were labeled with SILAC heavy label (Lys8–Arg10); Parental FlpIN T-Rex293 cells were labeled with SILAC medium-heavy label (Lys4–Arg6); and PKD1 kinase dead cells (PKD1kd-GFP or PKD1kd) were labeled with the SILAC light label (Lys0–Arg0). Incorporation levels of the SILAC labels were in all cases higher than 95%. For phosphoproteome analysis, cells were plated on 15-cm dishes (7.5*106 cells/dish) followed by induction of protein expression 24 h later for additional 24 h. Prior to harvesting, the cells were left untreated or were stimulated with nocodazole (5 ng/μl) for 30 min as described previously (21). Three different SILAC experiments were performed. First, nocodazole-stimulated PKD1kd and nocodazole-stimulated PKD1ca cells were compared with each other, [PKDca/PKDkd]Noco+. Second, unstimulated PKD1kd-expressing cells were compared with unstimulated parental cells, [parental/PKDkd]Noco−. Finally, nocodazole-stimulated PKD1kd-expressing cells were compared with nocodazole-stimulated parental cells, [parental/PKDkd]Noco+.
The cells were lysed in lysis buffer (6 m urea, 2 m thiourea, 10 mm Tris, pH 8.0, 1% octylglucoside, 1- Complete protease inhibitor mixture; missing paren Roche Applied Science) and 1× PhosSTOP phosphatase inhibitor mixture (Roche Applied Science)) for 30 min at room temperature and then sonified for 1 min on ice (Bandelin SONOPULS HD 200, Program MS73D). The proteins were precipitated using ice-cold acetone/methanol for 30 min on ice. The proteins were pelleted by centrifugation (2000 × g, 20 min, 4 °C) and washed three times with 80% ice-cold acetone. Dried proteins were resolved in digestion buffer (6 m urea, 2 m thiourea, 10 mm Tris, pH 8.0).
In Vitro Phosphorylation Assay
HEK293T cells were maintained in Dulbecco's modified Eagle's medium SILAC heavy or SILAC light medium for 14 days (incorporation > 95%). The cells were plated on 15-cm dishes (15 × 106 cells/dish), and 24 h later the cells were harvested in phosphorylation buffer (50 mm Tris, pH 7.5, 10 mm MgCl2, 1× complete protease inhibitor) and lysed by sonification for 7 min in ice-cold water (Bio Rupter, Diagenode, 30-s cycles mean intensity). The lysates were clarified by centrifugation (16,000 × g, 10 min, room temperature) and incubated 20 min at room temperature to allow protein dephosphorylation by endogenous phosphatases to occur. Subsequently, 1 mg of lysate (2.5-ml volume) was incubated with 700 μl of purified active PKD1 (32) (SILAC heavy) or PBS (SILAC light). Furthermore, DTT (2 mm), ATP (5 mm), and PhosSTOP were added to the reaction mix and incubated 30 min at 37 °C. Acetone/methanol protein precipitation was performed as described above. The dried protein pellets were resolved in water.
SDS-PAGE and Western Blot Analysis
SDS-PAGE and Western blot analysis have been described elsewhere (17). The proteins were visualized using the ECL detection system (Pierce) according to the manufacturer's specifications. Infrared dye-labeled secondary antibodies were visualized using the Odyssey infrared imaging system (LICOR Biosciences). The antibody specific for PKD autophosphorylation at serine 910 has been described elsewhere (21). The PKD pMOTIF antibody was from Cell Signaling. The GFP-specific mouse monoclonal antibody was obtained from Roche Applied Science. The tubulin α-specific monoclonal antibody was from Neomarkers. Secondary antibodies used were goat anti-rabbit IgG HRP-coupled (Dianova, Germany) and goat anti-mouse infrared dye-labeled secondary antibodies (LICOR Biosciences). The antibodies specific for phosphorylated serine 294 in PI4KIIIβ and phosphorylated serine 910 in human PKD1 were described elsewhere (18).
In Vitro FRET Using G-PKDrep-live
FRET measurements were performed with an Infinite 200 reader (TECAN, Mainz, Germany). Parental and PKD1kd-GFP-expressing HEK293FlpIN cells were transiently transfected with G-PKDrep-live (33) using TransIT293 (Mirus, Houston, TX) according to the manufacturer's instructions. Expression of PKD1kd-EGFP was induced 24 later by doxocyline (10 ng/ml). 48 h later, the cells were stimulated with nocodazole (5 μg/ml) for 30 min. The cells were solubilized in lysis buffer (50 mm Tris-HCl, pH 7.4, 100 mm NaCl, 1 mm EDTA, 0.2% Triton X-100, Complete protease inhibitor and PhosSTOP (Roche Applied Science)), and the lysates were clarified by centrifugation at 16,000 × g for 15 min. 100 μl of lysate were measured in a black flat-bottomed 96-well plate. mCFP was excited at 430 nm, and the emission of mCFP and cpVENUS was detected at 460 nm (CFP) and 550 nm (YFP-C), respectively. The YFP-C/CFP ratio was calculated for each sample. The background of nontransfected cell lysates was subtracted for each cell type.
Phosphorylation of G-PKDrep
Parental and HEK293FlpIn cells expressing PKD1kd-GFP were transiently transfected with a plasmid encoding G-PKDrep (21) using TransIT293 (Mirus) according to the manufacturer's instructions. Expression of PKD1kd-EGFP was induced 24 h later by doxocycline (10 ng/ml). 48 h later, the cells were stimulated with nocodazole (5 μg/ml) for 30 min. The cells were solubilized in lysis buffer (20 mm Tris, pH 7.4, 150 mm NaCl, 1% Triton X-100, or Nonidet P-40, 1 mm EGTA, 1 mm EDTA, Complete protease inhibitor and PhosSTOP), and SDS-PAGE and Western blotting were performed as described above. Quantitative analysis was performed with the Odyssey software (Licor-Biosciences, Germany). Phosphorylation of G-PKDrep was normalized with the level of expression. The densitometry of the control sample was arbitrarily set to 1.0.
Immunofluorescence
HEK293FlpIN cells expressing PKD1kd-EGFP were grown on coverslips, washed with PBS, fixed in 4% para-formaldehyde at room temperature for 15 min, washed, permeabilized with 0.1% Triton X-100 (5 min at room temperature) and blocked with blocking buffer (5% fetal calf serum in PBS) for 30 min. The cells were incubated with the primary antibodies (giantin-specific polyclonal rabbit, Abcam; and p230-specific monoclonal mouse antibody, BD Biosciences) in blocking buffer for 2 h, washed, incubated with secondary antibodies (Alexa 546-coupled anti-mouse and anti-rabbit IgG; Invitrogen) in blocking buffer for 1 h, washed, and mounted in ProLong Gold (Invitrogen) supplemented with 1 μg/ml 4′,6′-diamino-2-phenylindole. The cells were analyzed by confocal laser scanning microscopy (LSM 710; Zeiss). EGFP was excited with the 488-nm line of the argon laser, and fluorescence was detected at 490–550 nm. Alexa 546 was excited at 561 nm using a diode-pumped solid state laser, and emission was detected at 566–680 nm. 4′,6′-diamino-2-phenylindole was excited with the 405-nm line of the argon laser, and fluorescence was detected at 415–485 nm.
Protein Digestion and Isoelectric Focusing
Protein extracts derived from the “light” and “heavy” labeled cell cultures were mixed in a 1:1 ratio according to measured protein amounts. Ten milligrams of the mixture was digested in solution with trypsin as described previously (34). For proteome analysis, 2% of the total tryptic peptides were fractionated by isoelectric focusing on an OffGel 3100 Fractionator (Agilent) according to the manufacturer's instructions. Focusing was done with 24 cm (24 well) Immobiline DryStrips pH 3–10 (Bio-Rad) at a maximum current of 50 μA for 50 kVh. In SILAC experiments [parental/PKD1kd]Noco− and [parental/PKD1kd]Noco+ 100 μg of protein extracts were separated on a one-dimensional gel and in-gel digested by trypsin as described previously (34). Peptide fractions were collected and desalted separately using C18 StageTips (35).
Phosphopeptide Enrichment
Phosphopeptide enrichment and phosphoproteome analysis was done as described previously (36) with minor modifications: ∼95% of the peptides were separated by strong cation exchange (SCX) chromatography with a gradient of 0 to 35% SCX solvent B resulting in eleven fractions that were subjected to phosphopeptide enrichment by TiO2 beads. Fractions containing a lot of peptides were subjected to TiO2 enrichment multiple times. Elution from the beads was performed three times with 100 μl of 40% ammonia hydroxide solution in 60% acetonitrile (pH > 10.5). Enrichment of phosphopeptides from the SCX flow-through was done in three cycles.
Mass Spectrometry
LC-MS/MS analyses were performed on an EasyLC nano-HPLC (Proxeon Biosystems) coupled to an LTQ Orbitrap XL or an LTQ Orbitrap Elite mass spectrometer (Thermo Scientific) as described previously (26). The peptide mixtures were injected onto the column in HPLC solvent A (0.5% acetic acid) at a flow rate of 500 nl/min and subsequently eluted with a 106-min gradient of 5–33% HPLC solvent B (80% ACN in 0.5% acetic acid). During peptide elution the flow rate was kept constant at 200 nl/min.
For proteome analysis, the 10 (Orbitrap XL) or 20 (Orbitrap Elite) most intense precursor ions were sequentially fragmented in each scan cycle. The phosphoproteome analysis was exclusively done on the Orbitrap XL, with the five most intense precursor ions fragmented by multistage activation of neutral loss ions at −98, −49, and −32.6 Th relative to the precursor ion (37). In all measurements, sequenced precursor masses were excluded from further selection for 90 s. The target values for the LTQ were 5000 charges (MS/MS) and 106 charges (MS); the maximum allowed fill times were 1000 ms. The lock mass option was used for real time recalibration of MS spectra (38). The experiments [PKD1ca/PKD1kd]Noco+ and [parental/PKDkd]Noco− were performed in two biological replicates, whereas the experiment [parental/PKDkd]Noco+ was performed in three biological replicates. For the measurement of the in vitro phosphorylation assay, an inclusion list containing 51 selected phosphopeptide masses detected as down-regulated (PKD1-dependent) in the monolayer cell culture experiment were used.
MS Data Processing
The MS data of all SILAC experiments were processed using default parameters of the MaxQuant software (v1.0.14.3) (39). Peak lists were derived using the Quant module of the software and subsequently submitted to Mascot (Matrix Science, UK) database search engine (v2.2.0) to query a custom made target decoy database (40) consisting of the IPI human proteome database (v3.84, 90,166 protein entries), 262 commonly observed contaminants, and two GFP-tagged versions of PKD1.
In a database search, full tryptic specificity was required, and up to two missed cleavages were allowed. Carbamidomethylation of cysteine was set as fixed modification; protein N-terminal acetylation, oxidation of methionine, and phosphorylation of serine, threonine, and tyrosine were set as variable modifications. Initial precursor mass tolerance was set to 7 ppm and 0.5 Da at the fragment ion level.
Identified peptides were parsed using the Identify module of MaxQuant and further processed for statistical validation of identified and quantified peptides, phosphorylation sites (p-sites) and protein groups. For protein group quantitation a minimum of two quantified peptides were required. False discovery rates were set to 1% at peptide, phosphorylation site, and protein group level. Downstream analysis of all resulting text files was performed using R (v 2.13.0).
Normalization of Detected Phosphorylation Sites
To assign a p-site to a specific residue, we used a minimal reported localization probability of 0.75, and we refer to these as localized phosphorylation sites. Quantified proteins were used to normalize the raw p-site ratios (supplemental Table 1). For each quantified p-site for which the corresponding protein was also quantified, we divided the p-site ratio by the protein ratio. Given the distributions of measured intensities and ratios of all quantified peptides, we calculated a p value for each heavy to light ratio indicating its significance as described in Ref. 39 (“Significance B”). Derived p values were further corrected for multiple hypothesis testing using the method of Benjamini and Hochberg (41). All p-sites having a Benjamini-Hochberg corrected p value below 0.05 were regarded as significantly regulated.
Functional Enrichment Analysis
To test whether specific Gene Ontology and Kyoto Encyclopedia of Genes and Genomes terms were significantly enriched among proteins carrying down or up-regulated p-sites, respectively, we used Fisher's exact test and tested against the entire detected phosphoproteome. All terms with Benjamini-Hochberg corrected p values below 0.05 were regarded as significantly overrepresented.
Consensus Motif Analysis
The frequencies of the 20 amino acids at ±6 flanking positions of down-regulated p-sites were compared with the frequencies of all detected localized p-sites at the corresponding positions. The hypergeometric distribution was used as a null model, and Fisher's exact test was applied to derive p values. p values below 1e-6 were regarded to be statistically significant. To address the problem of multiple testing, we used the Bonferroni correction to control the family wise error rate of the 260 performed statistical tests. Corrected p values below 0.01 (uncorrected p values below 3.85e-5) were considered to be statistically significant.
RESULTS
In this study we aimed to perform a global detection of PKD-dependent phosphorylation events in HEK293 cells. We first SILAC-labeled constitutively active PKD1 strain (PKD1ca, heavy label) and kinase dead PKD1 strain (PKD1kd, light label). We verified that the endogenous PKD1 activity was decreased in the PKD1kd compared with the PKD1ca-expressing cells and localized to the Golgi apparatus. Prior to harvesting, we treated both strains with nocodazole, a microtubule-depolymerizing reagent that disrupts the Golgi complex and activates PKD (21). To detect potential bias caused by the use of the PKDca strain and nocodazole, we performed two additional SILAC experiments, comparing PKD1kd and parental strain with and without nocodazole treatment. Upon cell lysis, equal amounts of protein extracts were mixed and digested with trypsin. A small portion of the resulting digest was prepared for proteome analysis by LC-MS. A larger portion of the digest was subjected to two stages of phosphopeptide enrichment, using SCX and TiO2 chromatographies. All of the samples were measured on an LTQ-Orbitrap XL and LTQ-Orbitrap Elite mass spectrometers (Thermo) coupled to an Easy-LC nano-HPLC (Proxeon/Thermo). The LC-MS data were processed by MaxQuant software. All of the experiments were performed biological duplicates or triplicates. The phosphoproteomic workflow is depicted in Fig. 1.
Fig. 1.
The phosphoproteomic workflow. Heavy and light SILAC-labeled cells (expressing PKD1ca and PKD1kd, respectively) were treated with nocodazole for 30 min. Upon lysis, protein extracts were mixed and digested with trypsin. A part of the resulting digest was used for proteome analysis, whereas the other part was subjected to two stages of phosphopeptide enrichment, by SCX and TiO2 chromatographies. All of the samples were measured by LC-MS/MS; measured phosphopeptide ratios were normalized by the corresponding protein ratios.
PKD1 Activity and Localization in the PDK1kd Strain
The major prerequisite for global detection of PKD1-dependent phosphorylation events is a loss of endogenous PKD1 activity in the PKD1kd relative to the PKD1ca HEK293 strain. To test this, we performed a Western blot analysis of the PKD1kd and PKD1ca cell lysates using an antibody specific for PKD1 and PKD2 autophosphorylation at serine 910 and 876, respectively (21). The analysis showed an increase of PKD phosphorylation upon nocodazole stimulation in the parental HEK293FlpIN cells. In contrast, a clear reduction of nocodazole-induced endogenous PKD phosphorylation was observed in the PKD1kd strain, demonstrating the loss of endogenous PKD activity (supplemental Fig. 1A). In addition, we analyzed endogenous PKD activity using the PKD-specific reporter constructs G-PKDrep (21) and G-PKDrep-live, respectively (33). This was done as follows: parental and PKD1kd-GFP-expressing HEK293FlpIN cells were transiently transfected with plasmids encoding G-PKDrep and G-PKDrep-live, respectively. Doxycycline was added 24 h later to induce expression of PKD1kd-GFP, and the cells were incubated for additional 24 h. Afterward, the cells were stimulated with nocodazole for 30 min. Phosphorylation of G-PKDrep and G-PKD-rep-live was quantified using Western blot and FRET analysis, respectively, as described elsewhere (21, 33). The results are shown in Fig. 2. In both experiments, PKD-mediated phosphorylation of the reporter constructs was increased upon nocodazole stimulation, and this was decreased upon expression of PKD1kd (Fig. 2). Of note, a dominant-negative effect of PKD1kd was not visible in unstimulated cells. These results demonstrated that (i) PKD1kd acts in a dominant-negative manner toward endogenous PKD and (ii) this dominant-negative function requires stimulation of cells with nocodazole. Furthermore, we analyzed the localization of PKD1kd-GFP in HEK293FlpIN cells. As shown in supplemental Fig. 1B, the GFP-tagged PKD1kd protein colocalized with the Golgi complex specific marker proteins giantin (42) and p230 (43). This is in line with previous results (31) and points to a Golgi-specific block of endogenous PKD function by PKD1kd. Of note, nocodazole activates PKD specifically at the Golgi compartment (21).
Fig. 2.
Expression of PKD1kd-GFP inhibits endogenous PKD activity upon nocodazole stimulation. A, parental and PKD1kd-GFP-expressing HEK293FlpIN cells were transiently transfected with G-PKDrep, left untreated, or stimulated with nocodazole for the indicated time. The cells were lysed, and reporter phosphorylation was assessed by immunoblotting with the pS294 specific antibody. Expression of the G-PKDrep fusion protein was verified with a GFP-specific antibody. Representative immunoblots (left panel) and quantitative data of G-PKDrep phosphorylation normalized with the level of G-PKDrep (right panel) are shown. B, FRET response of lysates obtained from parental and PKD1kd-GFP-expressing HEK293FlpIN cells transfected with G-PKDrep-live and treated as indicated. Statistical analysis was performed using the Kruskal-Wallis test for nonparametric distribution and Dunn's multiple comparison test (n ≥ 4; *, p < 0. 05).
Quantitative Phosphoproteome of the Nocodazole-stimulated PKD1ca and PKD1kd Cells
The combined proteome and phosphoproteome analysis of the nocodazole-stimulated PKD1ca and PKD1kd cells, [PKDca/PKDkd]Noco+, resulted in identification of 180,416 MS/MS spectra corresponding to 39,541 nonredundant peptide sequences and 5,952 protein groups. The estimated false discovery rate was 0.43% at the peptide level and 2.05% at the protein group level. We detected a total of 6,889 phosphorylation events, of which 5,402 were localized to a specific amino acid residue with a median localization probability 99.8%; 4,843 phosphorylation events were localized on Ser, 505 events were localized on Thr, and 54 events were localized on Tyr. All of the detected protein groups and phosphorylation events are presented in supplemental Table 1.
Of 5,402 detected phosphorylation sites, 124 were significantly down-regulated, and 49 were up-regulated in at least one replicate upon the loss of PKD activity (Fig. 3A and Table I). This number excludes phosphorylation events that showed inconsistent or opposite regulation in biological replicates. The overall correlation between the phosphorylation levels measured in the two biological replicates was high (supplemental Fig. 3A). Importantly, PKD target motifs were clearly enriched among down-regulated phosphopeptides, demonstrating that many of them are likely direct PKD1 substrates (supplemental Table 2). To exclude the quantitation bias caused by possible changes in protein expression, we also quantified the changes at the proteome level, and we normalized phosphopeptide ratios by protein ratios. Comparison of normalized and measured phosphopeptide and protein ratios showed relatively small expression changes at the protein level (supplemental Fig. 2A).
Fig. 3.
Distribution of ratios of all detected phosphorylation events measured in the three SILAC experiments. A, results of the [PKDca/PKDkd]Noco+ analysis. The PKD target motif was overrepresented among phosphorylation sites down-regulated upon the loss of PKD1 activity, pointing to the presence of direct PKD1 substrates. B, results of the [parental/PKDkd]Noco− analysis. C, results of the [parental/PKDkd]Noco+ analysis. Blue triangles represent the phosphorylation sites matching the known PKD target motif (L/V/I)X(R/K)XX(S/T).
Table I. Overview of detected phosphorylation events in the three SILAC experiments performed in this study.
Detailed results are presented in supplemental Table I. Overlap of all localized sites is presented in supplemental Fig. 6.
| Phosphorylation events | Localized phosphosites | Down-regulated phosphosites | Up-regulated phosphosites | |
|---|---|---|---|---|
| [PKDca/PKDkd]Noco+ | 6889 | 5402 | 124 | 49 |
| [parental/PKDkd]Noco− | 9382 | 7450 | 22 | 37 |
| [parental/PKDkd]Noco+ | 9180 | 7253 | 50 | 17 |
Quantitative Phosphoproteome of the Parental and PKD1kd Cells
To increase the PKD1 activity, our main experiment involved the PKD1ca strain and nocodazole stimulation, both of which could potentially result in detection of nonendogenous PKD substrates. To identify phosphorylation events influenced by nocodazole, we performed two additional SILAC experiments, comparing parental and PKD1kd strains with and without nocodazole stimulation. In spite of more than 9,000 phosphorylation sites detected in each experiment and high reproducibility between the biological replicates (supplemental Fig. 3, B and C), the number of down-regulated phosphorylation events was significantly smaller than in the first experiment, in which PKD1ca strain was used (Fig. 3, B and C, and Table I). The difference was especially pronounced in the unstimulated cells, [parental/PKDkd]Noco−, showing that the levels and the activity of the endogenous PKD were too small to measure the PKDkd effect in our assay. This result was in full agreement with the biological data using PKD-specific reporter constructs (see above and Fig. 2). Comparison of the parental and PKD1kd strains under stimulation with nocodazole, [parental/PKDkd]Noco+ led only to a moderate increase of down-regulated phosphorylation events. Interestingly, in addition to a couple of known PKD1 substrates (PKD1 itself and c-Jun), most of the down-regulated sites did not conform to the canonical PKD1 target motif or expected cellular function (supplemental Figs. 4 and 5), most likely because the activity of the endogenous kinase was still too low to detect significant reduction of PKD-dependent phosphorylation events. Therefore, we suggest that most of the down-regulated phosphorylation events in this experiment were likely dependent on nocodazole under reduced PKD1 activity. Importantly, the overlap of the significantly regulated phosphorylation events in the three experiments was minimal (supplemental Fig. 6), demonstrating that the PKD activation through the use of the PKD1ca strain and nocodazole stimulation was necessary to detect the phosphorylation events dependent on the PKD1 activity. Protein groups and phosphorylation events detected in all SILAC experiments are presented in supplemental Table 1.
PKD1 Target Motif Analysis
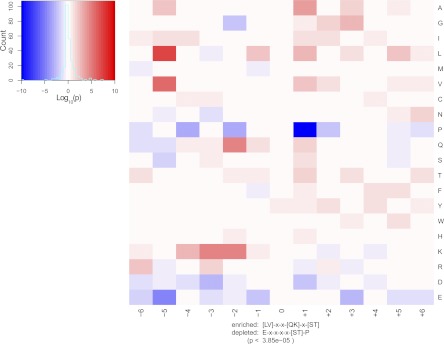
Because the subset of down-regulated sites from the main SILAC experiment [PKDca/PKDkd]Noco+ likely contained direct PKD1 substrates, we further focused on this part of the data set. To identify the PKD1 target motif, we tested the frequencies of 20 common amino acids in positions −6 to +6 relative to the PKD1-dependent phosphorylation site. We performed this analysis on all 124 significantly down-regulated sites; the results showed a significant overrepresentation (p < 3.85E-5) of the amino acids leucine or valine at position −5, as well as lysine at position −3 and lysine or glutamine at position −2 relative to the phosphorylation site. Combined, this was in strong agreement with the previously established PKD target motif (L/V/I)X(R/K)XX(S/T) (44, 45). It should be noted that the amino acid frequencies were tested independently and that the overrepresented residues do not always appear in a strict motif. However, there appears to be a strong requirement for the positively charged region at positions −2 through −4, as well as for a hydrophobic residue (leucine or valine) at position −5 relative to the phosphorylation site. Importantly, we also observed a significant underrepresentation of glutamate at position −5, as well as proline residues at position +1 relative to the phosphorylation site, suggesting that PKD1 disfavors substrates with these residues in close proximity to the phosphorylation site (Fig. 4).
Fig. 4.
PKD1 target motif analysis. Frequencies of the 20 common amino acids were tested on positions −6 to +6 relative to the phosphorylation site. The 124 phosphorylation sites down-regulated upon the loss of PKD1 activity in the [PKDca/PKDkd]Noco+ analysis were used as the test data set. Significantly overrepresented amino acid residues were in agreement with the previously published PKD target motif. Importantly, several residues were significantly underrepresented, especially proline at position +1 relative to the phosphorylation site.
Functional Enrichment Analysis of PKD1-dependent Phosphoproteins
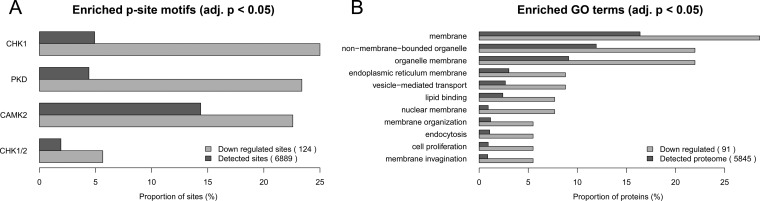
To gain insight into the PKD1-mediated signaling under conditions of nocodazole-dependent Golgi dispersal, we performed a bioinformatic functional enrichment analysis of phosphoproteins that were regulated and therefore dependent on PKD1 activity. In agreement with the known effects of PKD1 under nocodazole treatment, functional enrichment analysis of down-regulated PKD-dependent phosphorylation events pointed to a significant overrepresentation of proteins involved in regulation of membrane organization (Fig. 5). Among them we detected lamin; Hip1-related protein; cortactin; sorting nexin-1, -2, and -17; and insulin-like growth factor 2 receptor. Also in agreement with expected activity of PKD1, we observed a significant overrepresentation of the Golgi localization among proteins carrying down-regulated phosphorylation events. Among them we detected the vesicle-associated membrane protein-associated protein B/C, sorting nexin-1 and -17, oxysterol-binding protein-related protein 10, endoplasmic reticulum-Golgi intermediate compartment protein 2, epidermal growth factor receptor substrate 8, and transmembrane protein 43.
Fig. 5.
Functional enrichment analysis of PKD1-dependent phosphoproteins. In agreement with nocodazole-dependent fragmentation of the TGN and expected PKD1 activation in this process, all GO terms overrepresented in the proteins with PKD1-dependent phosphorylation sites pointed to membrane localization and functions related to membrane organization. A, enriched p-site motifs. B, enriched Gene Ontology/Kyoto Encyclopedia of Genes and Genomes/Pfam terms.
Among down-regulated phosphorylation events, we expected to detect at least some known direct PKD1 targets. Indeed, in addition to serine 910, the known autocatalytic site on PKD1, eight additional phosphorylation sites were down-regulated on PKD1 itself; among them, serines 225 and 548 were significantly regulated and have not been described before. We also detected down-regulation of phosphorylation at Ser298 on cortactin, which was previously shown to be directly phosphorylated by PKD1 and involved in actin polymerization and cell motility (16, 46). Of note, cortactin is also critically involved in actin-supported vesicle fission at the TGN (47). Most of other phosphorylation events previously linked to PKD1 activity (e.g. on slingshot, PAK4, etc.) were detected but not regulated in our data set, most likely because of specific experimental conditions such as nocodazole treatment. Indeed, nocodazole activates PKD1 specifically at the Golgi apparatus (21), thereby channeling PKD activity toward a locally restricted response. However, this response might be independent of vesicle fission events and thus requires different substrates. This hypothesis is confirmed by the fact that nocodazole treatment does not alter the phosphorylation of CERT (48). Other down-regulated phosphorylation events, especially those that fit to the predicted target motif of the kinase, present novel, potentially direct substrates of PKD1. Of note, among these potential direct PKD substrates were also serine/threonine-protein kinases such as SIK3, cdc28, and mitogen-activated protein kinases 1 and 3. Furthermore, we also detected phosphatases such as protein phosphatase 1 regulatory subunit 12A and a phosphatase inhibitor (phosphatase and actin regulator 4). It is thus possible that some of the detected down-regulated phosphorylation events are secondary effects caused by the changes in activity of kinases and phosphatases.
As expected, the small number of up-regulated phosphorylation events (49 events on 29 proteins) did not lead to significant enrichment of any functional category; however, we note that up-regulated phosphorylation events were detected on several kinases, such as Bcr and Cdc2-related kinase. These kinases with up-regulated phosphorylation sites are not direct targets of PKD1 but are likely positioned further downstream in signaling networks shared by PKD1. The complete results of the functional enrichment analysis are presented in supplemental Table 3.
In Vitro Validation of PKD1-dependent Phosphorylation Events
To test whether some of the detected down-regulated phosphorylation sites are direct PKD1 targets, we performed a comprehensive in vitro assay of PKD1 activity on the whole and SILAC heavy and light labeled HEK293 protein extracts. To achieve partial protein dephosphorylation through endogenous phosphatase activity, prior to treatment with PKD1 the lysates were left at room temperature without phosphatase inhibitors. Equal amounts of protein extracts were then incubated either with purified active PKD1 (SILAC heavy extract) or with the corresponding volume of PBS (SILAC light extract) in ATP-containing phosphorylation buffer (supplemental Fig. 7), mixed, and digested with trypsin. After phosphopeptide enrichment and LC-MS measurement, we detected 7,205 localized phosphorylation sites, of which 777 were at least 2-fold up-regulated upon incubation with PKD1. Among them we detected 70 phosphorylation sites that were significantly down-regulated upon PKD1 inhibition in the monolayer cell culture experiment. The median ratio of these 70 phosphorylation sites was 1.84, pointing out that the majority of them were also phosphorylated by PKD1 in vitro (supplemental Table 4). Importantly, in this experiment we confirmed PKD1 dependence of phosphorylation sites on many proteins with functions related to Golgi membrane, such as sorting nexins, vesicle-associated membrane protein-associated protein B/C, Vti1B, and epidermal growth factor receptor substrate 8.
In a second in vitro validation approach, we overexpressed a potential PKD1 target, protein MgcRacGAP, in HEK293 cells. To verify PKD1-mediated direct phosphorylation of serine 203, a Class 1 site from our cell culture experiment, immunoprecipitated MgcRacGAP was incubated with or without purified PKD1 in ATP-containing phosphorylation buffer. After the reaction, the MgcRacGAP protein was tryptically digested, and the intensity of the phosphorylated serine 203 peptide was analyzed by mass spectrometry for both samples (supplemental Fig. 8). Indeed, the MgcRacGAP peptide phosphorylated at serine 203 was detected in the sample incubated with PKD1 but not in the control sample, verifying direct phosphorylation by PKD1.
DISCUSSION
Although the mechanisms of regulation of PKD and its importance in diverse cellular processes have been intensively characterized in the past, the identification of specific substrates that transmit the PKD signal is still an important issue to address. Here we have made use of a SILAC-based screen to identify phosphoproteins and phosphosites directly and indirectly controlled by PKD activity. We used HEK293 cells that inducibly and stably expressed a kinase dead or a constitutively active PKD1 protein, and we stimulated cells with nocodazole, which has previously been shown to activate PKD at the Golgi compartment (21). Moreover, PKD1 is critical for nocodazole-mediated fission of Golgi membranes into so-called Golgi ministacks. It is important to note that stimulation of cells is essential to identify PKD substrates and downstream signaling pathways, because expression of PKD1kd-GFP will only result in a dominant-negative phenotype under conditions of Golgi dispersal, whereas expression of PKD1ca bypasses external signaling. Previous studies have shown that PKD controls fission of vesicles destined for the plasma membrane by direct phosphorylation of the lipid kinase PI4KIIIβ (18), and the lipid transfer proteins CERT (19) and OSBP (20). However, we could only observe a significant change in OSBP phosphorylation in our cellular system. This indicates that other proteins mediate Golgi complex fission downstream of nocodazole-induced PKD activation. In line with this is a previous study showing that phosphorylation of CERT does not alter upon nocodazole treatment (48). Of note, phosphorylation of the PKD substrate cortactin at serine 298 decreased significantly in PKD1kd-expressing cells upon nocodazole treatment. Cortactin has been previously described to localize to dynamic F-actin-containing structures at the plasma membrane such as lamellipodia. Recently, it was demonstrated that PKD-mediated phosphorylation of cortactin at serine 298 controls F-actin binding and thus nucleation complex turnover (16). In addition, cortactin is also critically involved in actin-supported vesicle fission at the trans-Golgi network (47). It is thus intriguing to speculate that PKD-mediated phosphorylation of cortactin could mediate nocodazole-induced Golgi dispersal. In addition to cortactin and OSBP, several other proteins such as c-Jun, MgcRacGAP, sorting nexin 2, VAP-B/C, Vti1B, epidermal growth factor receptor substrate 8, and insulin-like growth factor 2 receptor (CI-MPR) have been identified as potential PKD substrates in nocodazole-induced Golgi fragmentation. In line with our results, the transcription factor c-Jun has been identified as a direct PKD substrate in previous studies (49, 50).
The main function of the insulin-like growth factor 2 receptor network is trafficking of lysosomal enzymes from the TGN to the endosomes and their subsequent transfer to lysosomes. Nocodazole inhibits the initial rate of mannose 6-phosphate receptor transport from endosomes to the TGN in vitro (51), indicating the importance of microtubules in this process. Interestingly, a couple of components of the insulin-like growth factor 2 receptor network such as Vti1B were also significantly regulated and could thus be PKD1 targets. Because PKD is not recruited to endosomes (52), it is most likely that the kinase interacts with these proteins at the TGN under conditions of nocodazole treatment. It will be a future challenge to clarify the interplay of PKD and these proteins in Golgi structure and maintenance.
Data Availability
The mass spectrometry data associated with this manuscript may be downloaded from the ProteomeCommons.org Tranche network using the following hash code oNBq7tFA2/2IHQUNY7nrGTBtAeiUV3VUnKTDSrMnwaOIArgw xHLQLzAS3tQIfkxyBTtleFn34alcVRBS6acUbvOex/AAAAAAA ESnXA==. The hash code may be used to prove exactly what files were published as part of the data set of this manuscript, and the hash code may also be used to check that the data has not changed since publication. At the time of acceptance, the new MS data were still being transferred to Tranche. Until the upload is complete, all annotated spectra may be downloaded at: ftp://proteom-centrum.de/franz/ with the password “nipusa12.”
Footnotes
* This work was supported by Baden-Württemberg Stiftung (Juniorprofessoren-Program), the German Research Foundation, and PRIME-XS (to B. M.) and by German Cancer Aid, German Research Foundation, and the Heidelberger Akademie der Wissenschaften (WIN-Kolleg) (to A. H.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- PKD
- protein kinase D
- PKD1kd
- PKD1 kinase dead
- PKD1ca
- PKD1 constitutively active
- TGN
- trans-Golgi network
- SILAC
- stable isotope labeling by amino acids in cell culture
- GFP
- green fluorescent protein
- EGFP
- enhanced GFP
- CFP
- cyan fluorescent protein
- YFP
- yellow fluorescent protein
- SCX
- strong cation exchange
- p-site
- phosphorylation site; International Protein Index (IPI).
REFERENCES
- 1. Fu Y., Rubin C. S. (2011) Protein kinase D: Coupling extracellular stimuli to the regulation of cell physiology. EMBO Rep. 12, 785–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Storz P., Döppler H., Toker A. (2005) Protein kinase D mediates mitochondrion-to-nucleus signaling and detoxification from mitochondrial reactive oxygen species. Mol. Cell. Biol. 25, 8520–8530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Eiseler T., Schmid M. A., Topbas F., Pfizenmaier K., Hausser A. (2007) PKD is recruited to sites of actin remodelling at the leading edge and negatively regulates cell migration. FEBS Lett. 581, 4279–4287 [DOI] [PubMed] [Google Scholar]
- 4. Eiseler T., Döppler H., Yan I. K., Kitatani K., Mizuno K., Storz P. (2009) Protein kinase D1 regulates cofilin-mediated F-actin reorganization and cell motility through slingshot. Nat. Cell Biol. 11, 545–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Peterburs P., Heering J., Link G., Pfizenmaier K., Olayioye M. A., Hausser A. (2009) Protein kinase D regulates cell migration by direct phosphorylation of the cofilin phosphatase slingshot 1 like. Cancer Res. 69, 5634–5638 [DOI] [PubMed] [Google Scholar]
- 6. Eiseler T., Döppler H., Yan I. K., Goodison S., Storz P. (2009) Protein kinase D1 regulates matrix metalloproteinase expression and inhibits breast cancer cell invasion. Breast Cancer Res. 11, R13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhang K., Ye C., Zhou Q., Zheng R., Lv X., Chen Y., Hu Z., Guo H., Zhang Z., Wang Y., Tan R., Liu Y. (2007) PKD1 inhibits cancer cells migration and invasion via Wnt signaling pathway in vitro. Cell Biochem. Funct. 25, 767–774 [DOI] [PubMed] [Google Scholar]
- 8. Sucharov C. C., Langer S., Bristow M., Leinwand L. (2006) Shuttling of HDAC5 in H9C2 cells regulates YY1 function through CaMKIV/PKD and PP2A. Am. J. Physiol. Cell Physiol 291, C1029–C1037 [DOI] [PubMed] [Google Scholar]
- 9. Jensen E. D., Gopalakrishnan R., Westendorf J. J. (2009) Bone morphogenic protein 2 activates protein kinase D to regulate histone deacetylase 7 localization and repression of Runx2. J. Biol. Chem. 284, 2225–2234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kleger A., Loebnitz C., Pusapati G. V., Armacki M., Müller M., Tümpel S., Illing A., Hartmann D., Brunner C., Liebau S., Rudolph K. L., Adler G., Seufferlein T. (2011) Protein kinase D2 is an essential regulator of murine myoblast differentiation. PLoS One 6, e14599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Liljedahl M., Maeda Y., Colanzi A., Ayala I., Van Lint J., Malhotra V. (2001) Protein kinase D regulates the fission of cell surface destined transport carriers from the trans-Golgi network. Cell 104, 409–420 [DOI] [PubMed] [Google Scholar]
- 12. Yeaman C., Ayala M. I., Wright J. R., Bard F., Bossard C., Ang A., Maeda Y., Seufferlein T., Mellman I., Nelson W. J., Malhotra V. (2004) Protein kinase D regulates basolateral membrane protein exit from trans-Golgi network. Nat. Cell Biol. 6, 106–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bossard C., Bresson D., Polishchuk R. S., Malhotra V. (2007) Dimeric PKD regulates membrane fission to form transport carriers at the TGN. J. Cell Biol. 179, 1123–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Barišić S., Nagel A. C., Franz-Wachtel M., Macek B., Preiss A., Link G., Maier D., Hausser A. (2011) Phosphorylation of Ser 402 impedes phosphatase activity of slingshot 1. EMBO Rep. 12, 527–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Spratley S. J., Bastea L. I., Döppler H., Mizuno K., Storz P. (2011) Protein kinase D regulates cofilin activity through p21-activated kinase 4. J. Biol. Chem. 286, 34254–34261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Eiseler T., Hausser A., De Kimpe L., Van Lint J., Pfizenmaier K. (2010) Protein kinase D controls actin polymerization and cell motility through phosphorylation of cortactin. J. Biol. Chem. 285, 18672–18683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ziegler S., Eiseler T., Scholz R. P., Beck A., Link G., Hausser A. (2011) A novel protein kinase D phosphorylation site in the tumor suppressor Rab interactor 1 is critical for coordination of cell migration. Mol. Biol. Cell 22, 570–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hausser A., Storz P., Märtens S., Link G., Toker A., Pfizenmaier K. (2005) Protein kinase D regulates vesicular transport by phosphorylating and activating phosphatidylinositol-4 kinase IIIbeta at the Golgi complex. Nat. Cell Biol. 7, 880–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fugmann T., Hausser A., Schöffler P., Schmid S., Pfizenmaier K., Olayioye M. A. (2007) Regulation of secretory transport by protein kinase D-mediated phosphorylation of the ceramide transfer protein. J. Cell Biol. 178, 15–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nhek S., Ngo M., Yang X., Ng M. M., Field S. J., Asara J. M., Ridgway N. D., Toker A. (2010) Regulation of oxysterol-binding protein Golgi localization through protein kinase D-mediated phosphorylation. Mol. Biol. Cell 21, 2327–2337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fuchs Y. F., Eisler S. A., Link G., Schlicker O., Bunt G., Pfizenmaier K., Hausser A. (2009) A Golgi PKD activity reporter reveals a crucial role of PKD in nocodazole-induced Golgi dispersal. Traffic 10, 858–867 [DOI] [PubMed] [Google Scholar]
- 22. Macek B., Mann M., Olsen J. V. (2009) Global and site-specific quantitative phosphoproteomics: Principles and applications. Annu. Rev. Pharmacol. Toxicol. 49, 199–221 [DOI] [PubMed] [Google Scholar]
- 23. Bishop A. C., Buzko O., Shokat K. M. (2001) Magic bullets for protein kinases. Trends Cell Biol. 11, 167–172 [DOI] [PubMed] [Google Scholar]
- 24. Koch A., Hauf S. (2010) Strategies for the identification of kinase substrates using analog-sensitive kinases. Eur. J. Cell Biol. 89, 184–193 [DOI] [PubMed] [Google Scholar]
- 25. Holt L. J., Tuch B. B., Villén J., Johnson A. D., Gygi S. P., Morgan D. O. (2009) Global analysis of Cdk1 substrate phosphorylation sites provides insights into evolution. Science 325, 1682–1686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Koch A., Krug K., Pengelley S., Macek B., Hauf S. (2011) Mitotic substrates of the kinase aurora with roles in chromatin regulation identified through quantitative phosphoproteomics of fission yeast. Sci Signal. 4, rs6. [DOI] [PubMed] [Google Scholar]
- 27. Hsu P. P., Kang S. A., Rameseder J., Zhang Y., Ottina K. A., Lim D., Peterson T. R., Choi Y., Gray N. S., Yaffe M. B., Marto J. A., Sabatini D. M. (2011) The mTOR-regulated phosphoproteome reveals a mechanism of mTORC1-mediated inhibition of growth factor signaling. Science 332, 1317–1322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yu Y., Yoon S. O., Poulogiannis G., Yang Q., Ma X. M., Villén J., Kubica N., Hoffman G. R., Cantley L. C., Gygi S. P., Blenis J. (2011) Phosphoproteomic analysis identifies Grb10 as an mTORC1 substrate that negatively regulates insulin signaling. Science 332, 1322–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kettenbach A. N., Schweppe D. K., Faherty B. K., Pechenick D., Pletnev A. A., Gerber S. A. (2011) Quantitative phosphoproteomics identifies substrates and functional modules of Aurora and Polo-like kinase activities in mitotic cells. Sci Signal 4, rs5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bodenmiller B., Wanka S., Kraft C., Urban J., Campbell D., Pedrioli P. G., Gerrits B., Picotti P., Lam H., Vitek O., Brusniak M. Y., Roschitzki B., Zhang C., Shokat K. M., Schlapbach R., Colman-Lerner A., Nolan G. P., Nesvizhskii A. I., Peter M., Loewith R., von Mering C., Aebersold R. (2010) Phosphoproteomic analysis reveals interconnected system-wide responses to perturbations of kinases and phosphatases in yeast. Sci Signal. 3, rs4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hausser A., Link G., Bamberg L., Burzlaff A., Lutz S., Pfizenmaier K., Johannes F. J. (2002) Structural requirements for localization and activation of protein kinase C mu (PKC mu) at the Golgi compartment. J. Cell Biol. 156, 65–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Dieterich S., Herget T., Link G., Böttinger H., Pfizenmaier K., Johannes F. J. (1996) In vitro activation and substrates of recombinant, baculovirus expressed human protein kinase C mu. FEBS Lett. 381, 183–187 [DOI] [PubMed] [Google Scholar]
- 33. Eisler S. A., Fuchs Y. F., Pfizenmaier K., Hausser A. (2012) G-PKDrep-live, a genetically encoded FRET reporter to measure PKD activity at the trans-Golgi network. Biotechnol. J. 7, 148–154 [DOI] [PubMed] [Google Scholar]
- 34. Borchert N., Dieterich C., Krug K., Schütz W., Jung S., Nordheim A., Sommer R. J., Macek B. (2010) Proteogenomics of Pristionchus pacificus reveals distinct proteome structure of nematode models. Genome Res. 20, 837–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rappsilber J., Mann M., Ishihama Y. (2007) Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2, 1896–1906 [DOI] [PubMed] [Google Scholar]
- 36. Olsen J. V., Macek B. (2009) High accuracy mass spectrometry in large-scale analysis of protein phosphorylation. Methods Mol. Biol. 492, 131–142 [DOI] [PubMed] [Google Scholar]
- 37. Schroeder M. J., Shabanowitz J., Schwartz J. C., Hunt D. F., Coon J. J. (2004) A neutral loss activation method for improved phosphopeptide sequence analysis by quadrupole ion trap mass spectrometry. Anal. Chem. 76, 3590–3598 [DOI] [PubMed] [Google Scholar]
- 38. Olsen J. V., de Godoy L. M., Li G., Macek B., Mortensen P., Pesch R., Makarov A., Lange O., Horning S., Mann M. (2005) Parts per million mass accuracy on an Orbitrap mass spectrometer via lock mass injection into a C-trap. Mol. Cell. Proteomics 4, 2010–2021 [DOI] [PubMed] [Google Scholar]
- 39. Cox J., Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 40. Elias J. E., Gygi S. P. (2007) Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods 4, 207–214 [DOI] [PubMed] [Google Scholar]
- 41. Benjamini Y., Hochberg Y. (1995) Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. Roy. Stat. Soc. B Met. 57, 289–300 [Google Scholar]
- 42. Linstedt A. D., Hauri H. P. (1993) Giantin, a novel conserved Golgi membrane protein containing a cytoplasmic domain of at least 350 kDa. Mol. Biol. Cell 4, 679–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kooy J., Toh B. H., Pettitt J. M., Erlich R., Gleeson P. A. (1992) Human autoantibodies as reagents to conserved Golgi components. Characterization of a peripheral, 230-kDa compartment-specific Golgi protein. J. Biol. Chem. 267, 20255–20263 [PubMed] [Google Scholar]
- 44. Nishikawa K., Toker A., Johannes F. J., Songyang Z., Cantley L. C. (1997) Determination of the specific substrate sequence motifs of protein kinase C isozymes. J. Biol. Chem. 272, 952–960 [DOI] [PubMed] [Google Scholar]
- 45. Döppler H., Storz P., Li J., Comb M. J., Toker A. (2005) A phosphorylation state-specific antibody recognizes Hsp27, a novel substrate of protein kinase D. J. Biol. Chem. 280, 15013–15019 [DOI] [PubMed] [Google Scholar]
- 46. De Kimpe L., Janssens K., Derua R., Armacki M., Goicoechea S., Otey C., Waelkens E., Vandoninck S., Vandenheede J. R., Seufferlein T., Van Lint J. (2009) Characterization of cortactin as an in vivo protein kinase D substrate: Interdependence of sites and potentiation by Src. Cell Signal. 21, 253–263 [DOI] [PubMed] [Google Scholar]
- 47. Cao H., Weller S., Orth J. D., Chen J., Huang B., Chen J. L., Stamnes M., McNiven M. A. (2005) Actin and Arf1-dependent recruitment of a cortactin-dynamin complex to the Golgi regulates post-Golgi transport. Nat. Cell Biol. 7, 483–492 [DOI] [PubMed] [Google Scholar]
- 48. Chandran S., Machamer C. E. (2008) Acute perturbations in Golgi organization impact de novo sphingomyelin synthesis. Traffic 9, 1894–1904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hurd C., Waldron R. T., Rozengurt E. (2002) Protein kinase D complexes with c-Jun N-terminal kinase via activation loop phosphorylation and phosphorylates the c-Jun N-terminus. Oncogene 21, 2154–2160 [DOI] [PubMed] [Google Scholar]
- 50. Waldron R. T., Whitelegge J. P., Faull K. F., Rozengurt E. (2007) Identification of a novel phosphorylation site in c-Jun directly targeted in vitro by protein kinase D. Biochem. Biophys. Res. Commun. 356, 361–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Itin C., Ulitzur N., Mühlbauer B., Pfeffer S. R. (1999) Mapmodulin, cytoplasmic dynein, and microtubules enhance the transport of mannose 6-phosphate receptors from endosomes to the trans-Golgi network. Mol. Biol. Cell 10, 2191–2197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Maeda Y., Beznoussenko G. V., Van Lint J., Mironov A. A., Malhotra V. (2001) Recruitment of protein kinase D to the trans-Golgi network via the first cysteine-rich domain. EMBO J. 20, 5982–5990 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The mass spectrometry data associated with this manuscript may be downloaded from the ProteomeCommons.org Tranche network using the following hash code oNBq7tFA2/2IHQUNY7nrGTBtAeiUV3VUnKTDSrMnwaOIArgw xHLQLzAS3tQIfkxyBTtleFn34alcVRBS6acUbvOex/AAAAAAA ESnXA==. The hash code may be used to prove exactly what files were published as part of the data set of this manuscript, and the hash code may also be used to check that the data has not changed since publication. At the time of acceptance, the new MS data were still being transferred to Tranche. Until the upload is complete, all annotated spectra may be downloaded at: ftp://proteom-centrum.de/franz/ with the password “nipusa12.”