Abstract
Correct classification of cancer patients into subtypes is a prerequisite for acute diagnosis and effective treatment. Currently this classification relies mainly on histological assessment, but gene expression analysis by microarrays has shown great promise. Here we show that high accuracy, quantitative proteomics can robustly segregate cancer subtypes directly at the level of expressed proteins. We investigated two histologically indistinguishable subtypes of diffuse large B-cell lymphoma (DLBCL): activated B-cell-like (ABC) and germinal-center B-cell-like (GCB) subtypes, by first developing a general lymphoma stable isotope labeling with amino acids in cell culture (SILAC) mix from heavy stable isotope-labeled cell lines. This super-SILAC mix was combined with cell lysates from five ABC-DLBCL and five GCB-DLBCL cell lines. Shotgun proteomic analysis on a linear ion trap Orbitrap mass spectrometer with high mass accuracy at the MS and MS/MS levels yielded a proteome of more than 7,500 identified proteins. High accuracy of quantification allowed robust separation of subtypes by principal component analysis. The main contributors to the classification included proteins known to be differentially expressed between the subtypes such as the transcription factors IRF4 and SPI1/PU.1, cell surface markers CD44 and CD27, as well as novel candidates. We extracted a signature of 55 proteins that segregated subtypes and contained proteins connected to functional differences between the ABC and GCB-DLBCL subtypes, including many NF-κB-regulated genes. Shortening the analysis time to single-shot analysis combined with use of the new linear quadrupole Orbitrap analyzer (Q Exactive) also clearly differentiated between the subtypes. These results show that high resolution shotgun proteomics combined with super-SILAC-based quantification is a promising new technology for tumor characterization and classification.
Clinical heterogeneity in terms of patient survival rates and response to therapy is a major challenge in cancer treatment. This difficulty partly stems from grouping together molecularly distinct tumor entities as one clinical type and treating them in the same manner. Transcript-based profiling technology enables the segregation of subtypes based on their gene expression signatures (1, 2). However, it is often difficult to interpret such signatures with respect to the biology of the disease (1). In addition, gene expression signatures do not provide information if or to what extent the detected transcript is translated into proteins, and it ignores the effects of post-translational modifications. An in-depth, high accuracy quantitative proteomics approach capable of revealing common and distinct functional features between tumor entities may provide valuable insights into cancer subtypes of potential clinical relevance.
MS-based proteomics has recently evolved into an important tool in mining deregulated signaling pathways in cancer because of its ability to move one step closer toward the cancer phenotype and because of substantial progress in technology and methodology (3, 4). These advances in MS now allow the identification of thousands of proteins in a single experiment as a result of enhanced sensitivity, accuracy, and speed of analysis (5–7). In addition, a variety of quantitative proteomic approaches can monitor expression changes of thousands of proteins and post-translational modifications in a reproducible manner (8, 9). Stable isotope labeling with amino acids in cell culture (SILAC)1 is a particularly accurate method of quantitative proteomics (10, 11), but until recently it was limited to cell lines or animals that could be metabolically labeled with heavy amino acids. This limitation of SILAC in studying patient tumor samples has been overcome through the use of a mix of multiple SILAC-labeled cell lines as an internal standard, a technique called super-SILAC (12). This mix achieved superior quantification accuracy compared with a single SILAC-labeled cell line (13). In particular, a narrow ratio distribution was obtained with 90% of proteins contained within an easily quantifiable 4-fold range between the tumor and the SILAC mix. We reasoned that this ability to quantify several thousand proteins with high accuracy might enable confident proteomic classification of tumors in different subtypes.
The subclassification of diffuse large B-cell lymphoma (DLBCL), the most common lymphoma in adults, by gene expression profiling was a major breakthrough because it resulted in the identification of two histologically indistinguishable subtypes that differ in their outcomes after multiagent chemotherapy (14). The germinal-center B-cell-like (GCB) subgroup has a gene expression signature characteristic of normal germinal center B-cells, whereas the activated B-cell-like (ABC) subgroup, being the one with worse prognosis, has a gene expression signature characteristic of B-cells activated through their B-cell receptor. One of the key pathways that are differentially activated between DLBCL subgroups is signaling to NF-κB family transcription factors, which are constitutively active in the ABC subgroup (15). In B-cells, NF-κB controls the expression of genes necessary for both proliferation and survival in response to stimulation, including antigen recognition by the B-cell receptor (BCR). The IRF4 transcription factor, an NF-κB target, plays multiple roles in B lymphocyte development and function and is critical for plasma cell differentiation. Its high expression in ABC-DLBCL reflects constitutive NF-κB activity and plasmacytic differentiation. Recently, mutations leading to “chronically active” BCR signaling have been described as a mechanism providing aberrant cellular survival signals in ABC-DLBCL (16). In these cases, the constitutive NF-κB activation in ABC-DLBCL depends on the multiprotein CARD11-BCL10-MALT1 (CBM) complex (17–19). Such findings may open the door for new therapeutic modalities that target components of BCR signaling upstream of NF-κB. Furthermore, improvements in DNA sequencing technologies have paved the way to the discovery of novel aspects of DLBCL pathology, such as impairments in chromatin methylation and evasion of T cell immune surveillance (20). This shows that the deployment of novel methodologies continuously enhances our understanding of the complex biology of lymphomas.
Despite the success of gene expression profiling in differentiating between tumor subtypes, the extracted transcriptional signatures do not always suffice to identify biological drivers of tumor pathogenesis. Furthermore, their adoption in the clinic, where protein-based assays are more commonly used, has been slow. A long standing aim of the proteomics community is to directly study human cancer at the protein rather than the transcript level (3). Here, we use high resolution shotgun proteomics combined with a super-SILAC quantitative approach in an attempt to segregate DLBCL subtypes. If applicable, the super-SILAC technology should be particularly accurate, robust, and reproducible because it provides an entire reference proteome consisting of thousands of heavy labeled proteins for comparison of a large number of tumor samples. We evaluate the super-SILAC spike-in approach for distinguishing cell lines derived from ABC- and GCB-DLBCL patients. Choosing such closely related disease entities sets a high bar for our quantitative proteomics technology. Furthermore, the fact that specific biological differences between ABC and GCB are already known allows us to evaluate proteomics results in light of those differences.
EXPERIMENTAL PROCEDURES
Cell Culture Sample Preparation
ABC-DLBCL cell lines (HBL1, OciLy3, RIVA, TMD8, and U2932) and GCB-DLBCL cell lines (BJAB, DB, HT, SUDHL-4, and SUDHL-6) were grown in RPMI medium (Invitrogen) supplemented with 20% fetal bovine serum. Cell lysis was performed using a buffer consisting of 4% SDS, 0.1 m DTT, and 0.1 m Tris-HCl pH 7.5 followed by incubation at 95 °C for 5 min. The lysates were sonicated using a Branson type sonicator and then centrifuged at 16,100 × g for 10 min.
Cell lines selected for inclusion in the super-SILAC mix were grown in RPMI medium containing 13C615N2-lysine (Lys8) and 13C615N4-arginine (Arg10) (Cambridge Isotope Laboratories) instead of the natural amino acids and supplemented with 20% dialyzed fetal bovine serum. The cells were cultured for at least six passages until they were fully labeled as assessed by quantitative mass spectrometry. Less than 1% of tryptic peptides contained unlabeled arginine or lysine in the nine labeled cell lines (Ramos, Mutu, BL-41, U2932, OciLy3, BJAB, L1236, L428, and DB) and less than 0.3% of identified peptides showed evidence of Arg to Pro conversion. Equal amounts of the heavy lysates were mixed to generate the super-SILAC mix.
Protein Digestion and Fractionation
The super-SILAC mix (100 μg) was combined with an equal amount of the unlabeled cells and further processed by the filter-aided sample preparation (FASP) method (21). In short, the sample was loaded on Microcon filters with a 30-kDa cutoff (Millipore, Billerica, MA), which allows the replacement of SDS with a urea containing buffer. The proteins were then alkylated with iodoacetamide followed by overnight trypsin digestion at 37 °C in 50 mm ammonium bicarbonate. Peptides were collected from the filter after centrifugation and elution with water (2×).
Using strong anion exchange chromatography, 40 μg of the peptide mixture from each replicate was fractionated (22). In summary, the strong anion exchange (SAX) was performed in tip columns prepared from 200-μl micropipet tips stacked with six layers of a 3M Empore anion exchange disk (1214-5012; Varian, Palo Alto, CA). We used Britton & Robinson universal buffer composed of 20 mm acetic acid, 20 mm phosphoric acid, and 20 mm boric acid and titrated with NaOH to the desired pH for column equilibration and fraction elution. After loading the peptides at pH 11 and collecting it, five additional fractions were collected consecutively with buffers of pH 8, 6, 5, 4, and 3. The eluted fractions were desalted on reversed phase C18 Empore disc StageTips (23). Peptide elution was performed twice with 20 μl of buffer B containing 80% ACN in 0.5% acetic acid. Organic solvents were removed by a SpeedVac concentrator to prepare the samples for analysis by LC-MS/MS.
Liquid Chromatography and MS for Fractionation Experiments
Eluted peptides were separated on an in-house-made 15-cm column with a 75-μm inner diameter packed with ReproSil-Pur C18-AQ 3 μm resin (Dr. Maisch GmbH, Ammerbuch-Entringen, Germany) using an Easy nanoflow HPLC system (Proxeon Biosystems, now Thermo Fisher Scientific). The HPLC was coupled via a nanoelectrospray ion source (Proxeon Biosystems) to an LTQ-Orbitrap Velos mass spectrometer (Thermo Fisher Scientific) (24). Approximately 2 μg of peptides were loaded in buffer A (0.5% (v/v) acetic acid) with a flow rate of 500 nl min−1 and eluted with a 200-min linear gradient at a flow rate of 200 nl min−1. Four different gradients were applied for optimal separation based on average peptide hydrophobicity. A gradient of 2–25% buffer B to separate the pH 11 fraction; 7–25% buffer B for the pH 8 fraction; 7–30% buffer B for the pH 6 and 5 fractions; and 7–37% buffer B for the pH 4 and 3 fractions. After each gradient, the column was washed, reaching 90% buffer B followed by re-equilibration with buffer A.
The mass spectra were acquired with an automatic switch between a full scan and up to 10 data-dependent MS/MS scans. Target value for the full scan MS spectra were 1,000,000 and resolution was 30,000 at m/z 400. Up to the 10 most intense ions (minimum signal threshold of 5,000) were sequentially isolated and accumulated to a target value of 40,000 with a maximum injection time of 150 ms and were fragmented by higher energy collisional dissociation (25). For a subset of measurements, MS/MS target values were set to 50,000. The spectra of the fragmented ions were acquired in the Orbitrap analyzer with resolution of 7,500 at m/z 400.
Liquid Chromatography and MS for Single-shot Experiments
The peptides were separated on an in-house-made 50-cm column with a 75-μm inner diameter packed with 1.8 μm C18 resin (Dr. Maisch GmbH, Ammerbuch-Entringen, Germany). The Thermo EASY-nLC 1000 system with a binary buffer system consisting of 0.5% formic acid (buffer A) and 80% acetonitrile in 0.5% formic acid (buffer B) was used for reverse phase chromatography. Peptides (∼4 μg) were eluted with a 220-min linear gradient of buffer B up to 30% at a flow rate of 250 nl min−1. The column temperature was kept at 40 °C by an in-house designed oven with a Peltier element (26). The LC was coupled to a Q Exactive mass spectrometer (27) (Thermo Fisher Scientific) via the nanoelectrospray source (Proxeon Biosystems, now Thermo Fisher Scientific). Mass spectra were acquired on the Q Exactive in a data-dependent mode with an automatic switch between a full scan and up to 10 data-dependent MS/MS scans. Target value for the full scan MS spectra was 3,000,000 with a maximum injection time of 20 ms and a resolution of 70,000 at m/z 400. The 10 most intense ions with charge two or more from the survey scan were selected with an isolation window of 1.6 Th and fragmented by higher energy collisional dissociation (25) with normalized collision energies of 25. The ion target value for MS/MS was set to 1,000,000 with a maximum injection time of 60 ms and a resolution of 17,500 at m/z 400. These settings lead to constant injection times of 60 ms, fully in parallel with transient acquisition of the previous scan, ensuring fast cycle times. Repeat sequencing of peptides was kept to a minimum by dynamic exclusion of the sequenced peptides for 25 s.
Data Analysis
The acquired raw files were analyzed by MaxQuant (28) (version 1.2.0.34). Andromeda, a probabilistic search engine incorporated into the MaxQuant framework (29), was used to search the peak lists against the IPI human database version 3.68 which contains 87,083 entries. Common contaminants were added to this database. The search included cysteine carbamidomethylation as a fixed modification and N-terminal acetylation and methionine oxidation as variable modifications. The second peptide identification option in Andromeda was enabled (29). For statistical evaluation of the data obtained, the posterior error probability and false discovery rate were used. The false discovery rate was determined by searching a reverse database. A false discovery rate of 0.01 for proteins and peptides was required. Enzyme specificity was set to trypsin allowing N-terminal cleavage to proline. Two miscleavages were allowed, and a minimum of six amino acids per identified peptide were required. Peptide identification was based on a search with an initial mass deviation of the precursor ion of up to 6 ppm, and the allowed fragment mass deviation was set to 20 ppm. The mass accuracy of the precursor ions was improved by retention time-dependent mass recalibration (28). To match identifications across different replicates and adjacent fractions, the “match between runs” option in MaxQuant was enabled within a time window of 2 min. Quantification of SILAC pairs was performed by MaxQuant with standard settings using a minimum ratio count of 2. Bioinformatics analysis was done with Perseus tools available in the MaxQuant environment.
When needed for the analysis, the missing values were replaced using data imputation. The idea of our algorithm for imputation of missing values is that they should simulate signals of low abundant proteins. To accomplish this, we first determine the mean and standard deviation of all valid values in the matrix. Then we draw numbers for the missing entries from a suitable probability distribution in an independently, identically distributed way. For that purpose, we use a normal distribution with a mean and standard deviation adjusted in such a way as to simulate signals of low abundant proteins. This is necessary because the missing values are biased toward the detection limit of the LC-MS/MS measurement. Optimal values for the down shift parameter were adjusted in a way that the distribution of imputed values adjusts smoothly to the lower end of the distribution of measured values. We iteratively adjusted the values to avoid too large or too small down shifts. The former would result in a separation of imputed and measured values (a bi-modal total distribution), whereas the latter would introduce too much noise into the system and would potentially destroy protein signatures. The two values for downshifting and width adjustment are determined once but then apply to all the cell lines. These optimal values were different for the label-free and SILAC reference cases. For label-free data, we employed a width of 0.3 and a downshift of 1.8; in the super-SILAC data, the width was 0.3, and the downshift was 0.5, each in units of the standard deviation of the distribution of present values.
RESULTS AND DISCUSSION
Development of a Lymphoma Super-SILAC Mix
To accurately quantify proteome differences between lymphoma subtypes, we set out to generate a super-SILAC mix that would be optimally suited as an internal standard for a broad range of B-cell lymphomas. We considered commonly used cell lines derived from patients with different types of the disease. First we selected two lines, L428 and L1236, to represent Hodgkin's lymphoma. Of the non-Hodgkin's lymphomas, we selected three cell lines of patients with Burkitt's lymphoma, which is characterized by a c-Myc t(8;14) translocation. For DLBCL, we started out with the five ABC type cell lines and five GCB type cell lines that we wished to segregate by proteomics. From these, we chose two ABC type cell lines (Oci-Ly3 and U2932), as well as two GCB type cell lines (BJAB and DB).
Next, we wished to select an optimal subset of these nine representative cell lines (green in Fig. 1A). Instead of empirically testing different combinations, we reasoned that an in-depth proteome of each of the nine cell lines should be sufficient to mathematically determine the best combination. For this purpose, we performed a six-fraction FASP-SAX-based analysis, with 4-h gradients on an LTQ-Orbitrap Velos and higher energy collisional dissociation-based fragmentation (Fig. 1A and “Experimental Procedures”). This involved a single day of measurement time for each of the nine proteomes.
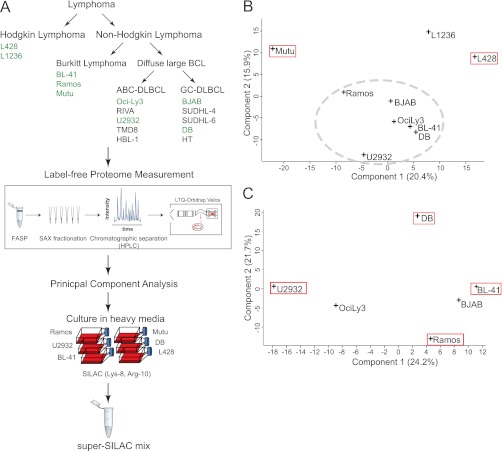
Fig. 1.
Rational construction of lymphoma super-SILAC mix. A, label-free proteomics of nine B-cell lymphoma cell lines was performed after FASP-SAX processing and analyzed using high resolution precursor and fragment measurements on an Orbitrap Velos. They included two Hodgkin lymphoma cell lines (L428 and L1236) and seven non-Hodgkin lymphoma cell lines (Ramos, Mutu, BL-41, OciLy3, U2932, BJAB, and DB). B, PCA of nine B-cell lymphoma cell lines based on their protein expression profiles. The red boxes indicate cell lines selected for the super-SILAC mix. The gray dashed ellipse groups non-Hodgkin lymphoma cell lines to be further analyzed by a second PCA. C, PCA of the six non-Hodgkin lymphoma cell lines encircled in B. The red boxes indicate cell lines selected for the super-SILAC mix.
To compare the label-free proteomes of the cell lines to each other, we performed principal component analysis (PCA). PCA transforms large data sets into points in a data space of orthogonal components, such that the first component captures most of the variability. Because PCA analysis requires a complete data set (in this case label-free protein intensities for all identified proteins in all samples), we employed “data imputation” as described in “Experimental Procedures.” We aimed to create a mixture of cells that capture the largest diversity. Therefore, we searched for those that were most distant from one another. Mutu(−), one of the Burkitt-derived cell lines, was the furthest outlier (Fig. 1B). L1236 and L428, the only Hodgkin's lymphoma cell lines, were also outliers. We therefore selected Mutu(−) and one of the two Hodgkin's lymphoma cell lines (L428). We then performed a second round of PCA on the remaining seven non-Hodgkin cell lines and selected the four outermost in the resulting PCA space (U2932, DB, BL-41, and Ramos) (Fig. 1C).
To produce the super-SILAC mix from the selected six cell lines, we grew them in heavy SILAC media and mixed them in equal proportions. For a first evaluation, we spiked the mix into lysate of an unlabeled ABC-DLBCL cell line (HBL-1) that was not part of the original selection. The histogram of fold changes between cell line proteins and super-SILAC proteins was narrow, with 96% of the values within a 4-fold range. To check the overall selection procedure, we also performed this experiment with a mix of all nine initially selected cell lines. The width of the distribution was essentially unchanged, indicating that the six-cell line mix already adequately represented the proteome. Finally, a three-cell line mix of only the largest outliers of the PCA analysis (Mutu, L428, and U2932; Fig. 1B) also performed surprisingly well, attesting to the usefulness of our selection procedure (supplemental Fig. 1).
In-depth Proteome Coverage Using the Lymphoma Super-SILAC Mix
We spiked the super-SILAC mix into five unlabeled ABC and five GCB cell lines and analyzed them as described above for the label-free experiment, except that each proteome was measured in triplicate (Fig. 2A). Joint analysis of the resulting 180 LC MS/MS files (30 days measuring time) in MaxQuant identified a total of 7,756 different protein groups, by far the largest B-cell lymphoma proteome reported to date. Of these proteins, 6,263 were quantified in at least two replicates of the same cell line (supplemental Tables I and II).
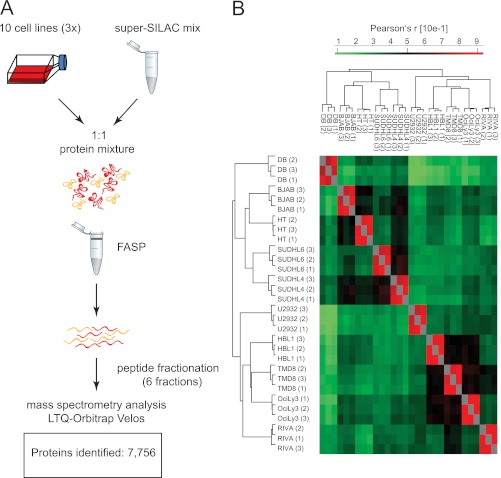
Fig. 2.
Proteomic workflow and overall results. A, the super-SILAC mix developed on the basis of label-free proteome comparison was used as an internal standard for 10 different DLBCL cell lines. The samples were processed by FASP-SAX followed by triplicate 1-day proteome analyses. B, heat map of Pearson correlation coefficients showing reproducibility between replicates.
At this depth of proteome coverage, we identified and quantified a large number of known members of the B-cell receptor-initiated signal transduction pathway (supplemental Fig. 2). Likewise, many transcription factors relevant to B-cell biology were measured. Altogether, we identified 285 proteins annotated by Gene Ontology to have sequence-specific DNA-binding transcription factor activity (supplemental Table II). This list included many transcription factors playing important roles in B-cells, such as basic leucine zipper transcription factor ATF-like (BATF), B-cell lymphoma 3 protein (BCL3), B-cell lymphoma 6 protein (BCL6), immunoglobulin transcription factors 1 and 2 (ITF1 and ITF2), Ets domain-containing PU.1, and the B-lineage specifying transcription factor PAX-5.
Next, we quantified all 30 proteome measurements against each other based on the ratios to the super-SILAC mix and calculated their Pearson correlation coefficients (r). Unsupervised clustering of the rows and columns of the matrix of the 30 × 30 coefficients co-clustered the triplicates in each case (Fig. 2B). Good reproducibility is further indicated by the high average Pearson coefficients of the triplicates (r = 0.87).
Segregation between DLBCL Subtypes
To investigate whether our proteomics measurements can segregate ABC from GCB proteomes and to determine an optimal data analysis strategy, we started by performing unsupervised hierarchical clustering of all proteome measurements. We required that proteins were present in at least 50% of the 30 measurements and filled any missing values by “data imputation” (“Experimental Procedures”). Again, replicate measurements were always clustered together. Intriguingly, the two major branches of the dendrogram precisely grouped all the ABC and all the GCB subtypes together and apart from each other. This indicates that these subtypes have quite different protein expression patterns at a global level that are capable of defining them as distinct entities.
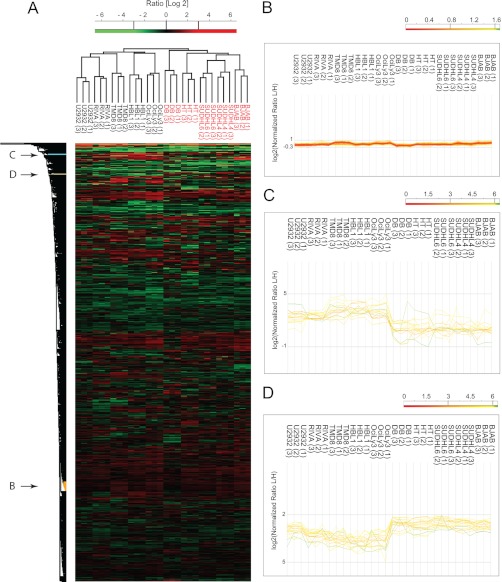
The cluster indicated with arrow B in Fig. 3A, consists of 107 proteins, 70 of which are annotated as ribosomal, 12 of which are components of the 20 S proteasome, and 14 of which are components of the 26 S proteasome (CORUM annotation) (15). As shown in Fig. 3B, their expression varies little across the cell lines; thereby they serve as “loading controls” and validate correct normalization and imputation of the proteome samples by MaxQuant. This ensures that the variation of protein expression values between ABC and GCB can directly be attributed to biological differences between these cell types rather than experimental artifacts. Fig. 3C shows the differences in expression of two clusters in the upper part of Fig. 3A (indicated with arrows C and D) with large differential expression patterns between the two main branches of the dendrogram. The first cluster consists of 16 proteins that are up-regulated in the ABC subtype relative to GC. This cluster includes proteins such as CD44, FOXP1, IL4I1, VAV2, and BID (supplemental Table IV). The second cluster consists of 19 proteins that are up-regulated in the GCB subtype and includes proteins such as CD81, KIND3, WIP, INPP5B, PAG, and BRDG1 (supplemental Table V).
Fig. 3.
Unsupervised hierarchical clustering. A, unsupervised clustering of protein expression profiles of 10 DLBCL cell lines after filtering for 50% valid values and imputation of missing values. B, expression patterns for a cluster enriched for ribosomal and proteasomal proteins. C, expression patterns for a cluster of proteins with higher expression levels in ABC relative to GCB. D, expression patterns for a cluster of proteins with higher expression levels in GCB relative to ABC.
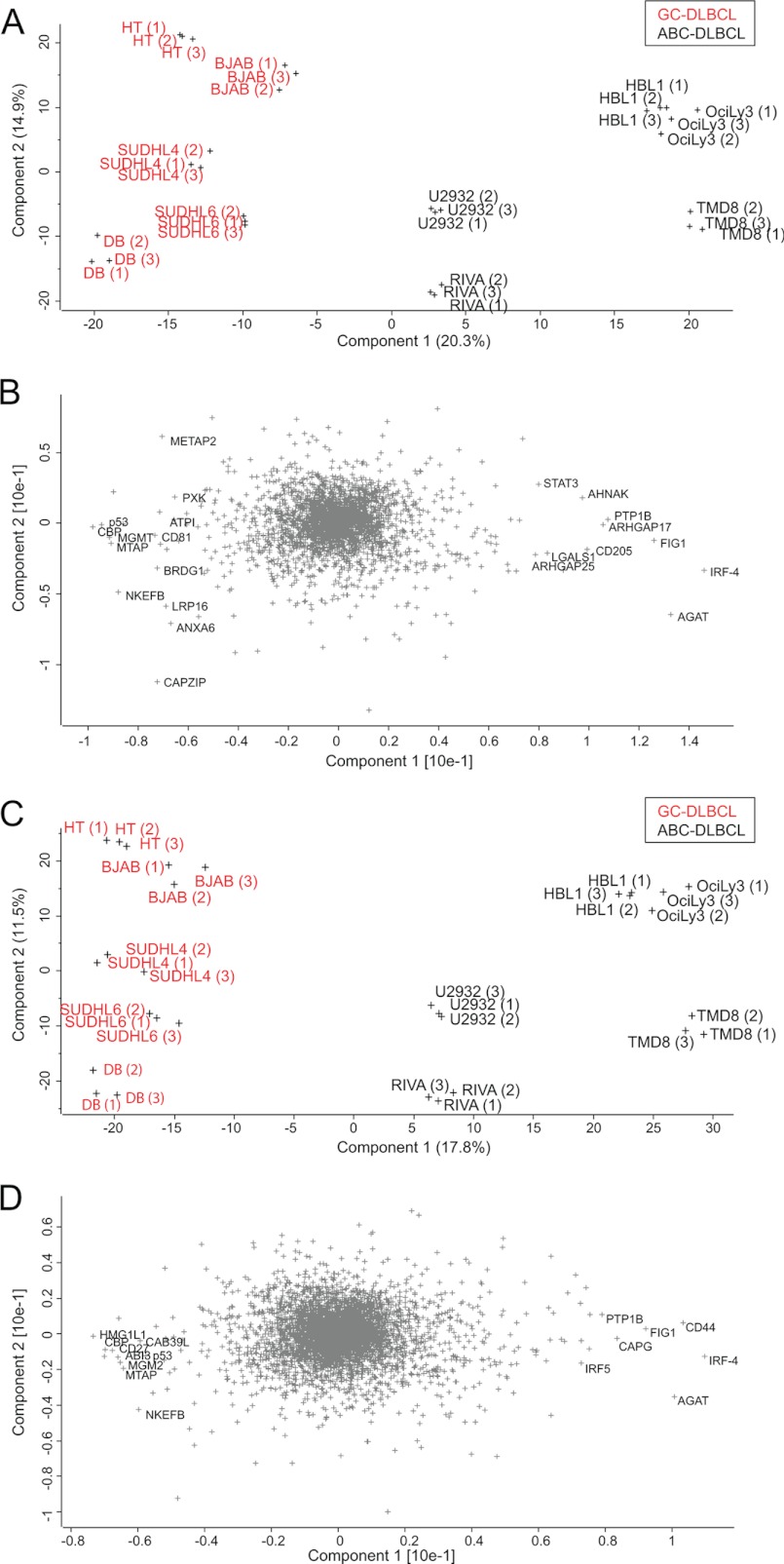
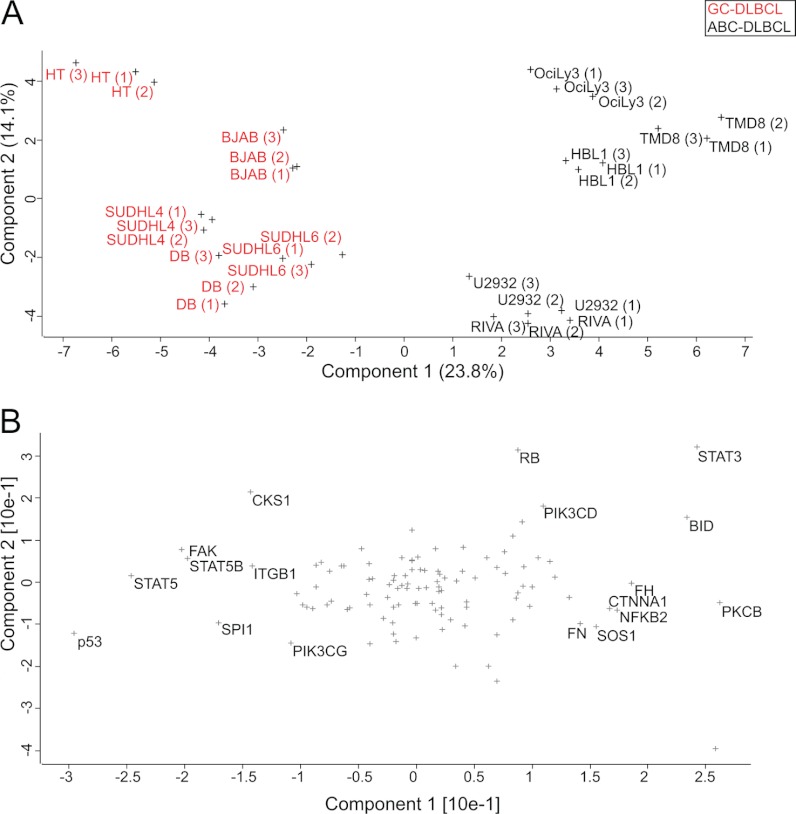
Principal component analysis was performed to project the SILAC-based proteome measurements into a two-dimensional data space. We first applied PCA for the subgroup of proteins that were quantified in each of the 30 proteome measurements (100% valid values; 3,007 protein groups). Component 1 of the PCA, which accounts for 20.5% of total variability (horizontal axis in the two-dimensional plot of Fig. 4A), clearly separates GCB (group on the left side) from ABC (group on the right side). Furthermore, Fig. 4A shows that the distance between the replicates is much smaller than the separation between the groups, supporting the robustness of the segregation.
Fig. 4.
Principal component analysis. A, the proteomes of 10 DLBCL cell lines measured in triplicate segregated into ABC-DLBCL and GCB-DLBCL subtypes after filtering for 100% valid values (3,007 proteins). B, loadings of A reveal proteins that strongly drive the segregation in PCA component 1. C, the same analysis as in A but after filtering for 50% valid values (4,991 proteins) and filling the missing values by data imputation results in even stronger separation. D, loadings of C uncover additional known and unknown markers that segregate the ABC and GCB subtypes.
The proteins that are most responsible for separating the proteomes in the PCA can be seen in the “loadings.” The loadings of component 1, which capture the differences between the two groups, include the transcription factor IRF4, mentioned above as one of the main drivers of the functional differences between GCB and ABC lymphomas (Fig. 4B). In fact, high expression of IRF4 in ABC-DLBCL is tied to the constitutive activity of NF-κB that is required for survival of this subtype of lymphoma cells (15). This transcription factor, which was quantified in 30 of 30 proteomes, is the strongest differentiator in this unbiased large scale analysis. PTP1B was another one of the strongest loadings of component 1. PTP1B is a key tyrosine phosphatase implicated in the regulation of JAK/STAT signaling. The preferential expression of PTP1B in ABC-DLBCL is already known, and its overexpression has been suggested to contribute to the enhanced STAT6 dephosphorylation that is observed in these tumors upon IL-4 stimulation (30, 31).
The above analysis required quantification of the proteins in every proteome measurement, which could exclude many interesting proteins, such as those exclusively expressed in only one subtype. We therefore employed imputation of missing values to make a larger subset of the proteome amenable to PCA analysis. We first filtered for at least 50% valid values (4,991 proteins) and imputed the missing values. Incorporation of the information from these additional proteins led to an even stronger separation of the subtypes (Fig. 4C). The GCB cell lines appear to cluster more tightly together, whereas the ABC cell lines U2932 and RIVA are somewhat separated from the other ABC cell lines. The loadings in Fig. 4D reveal additional known markers such as the cell surface markers CD44 for ABC-DLBCL (quantified exclusively in ABC) and CD27 for GCB-DLBCL (Fig. 4D). The above analysis demonstrates that requiring less than 100% valid values and imputing missing values is a valid and robust strategy for segregation of subtype groups, as well as for finding individual differentiators by proteomics.
Category-based Analysis of Subtype Differences
The above analyses were global and unbiased in that they considered the entire proteome. To determine whether specific groups of proteins by themselves could differentiate the subtypes, we extracted the proteins belonging to specific KEGG categories from the quantified proteome. We then performed PCA analysis on this subset as described above. Interestingly, the category “pathways in cancer” (108 quantified proteins) was able to clearly separate the groups, albeit to a lesser degree than the full proteome (Fig. 5A). The strongest loadings preferential for GCB in this category were p53, STAT5, STAT5B, and SPI1/PU.1 (Fig. 5B). SPI1 has a major role in maintaining germinal center B-cells through repressing the expression of plasma cell transcriptional regulators and thus blocking plasma cell differentiation (32). The strongest loadings preferential for ABC included the anti-apoptotic protein BCL2, overexpression of which is a known mechanism by which NF-κB driven tumors evade apoptosis. Surprisingly, the pro-apoptotic protein BID was also in this group. The loadings preferential for ABC include STAT3. It has been shown that NF-κB signaling in ABC induces the expression of IL-6 and IL-10, which act through JAK kinases and STAT3 as autocrine signals (33). The constitutive activity of STAT3 promotes proliferation and cell survival in the ABC subtype (34). This explains the synergistic effect of blocking JAK signaling and NF-κB signaling in killing ABC cells (33). PKCB is another interesting driver of the ABC subtype because its overexpression is a strong marker for refractory or fatal DLBCL and a recognized drug target (35). Our finding that it is preferentially expressed in the aggressive ABC subtype compared with the GCB subtype may therefore be of clinical interest. The observation that a small group of proteins can separate the subtypes prompted us to search for such groups in the entire quantified data set.
Fig. 5.
Category-based analysis of subtype differences. A, PCA of 10 lymphoma cell lines after filtering for proteins annotated by KEGG to be involved in cancer (KEGG category: pathways in cancer). B, loadings of PCA in A.
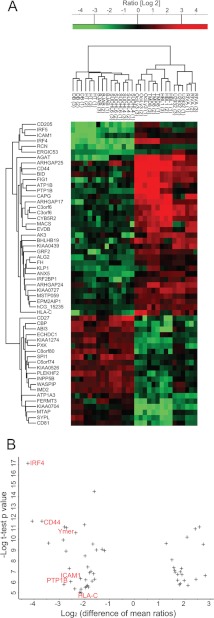
t Test Signature
To identify in a supervised manner a set of proteins that significantly distinguishes the ABC from the GCB subtype, we performed a t test between the cell lines using a permutation-based false discovery rate (0.05). This analysis resulted in a set of 55 proteins (Fig. 6A) that strongly segregated the subtypes as seen after PCA analysis (supplemental Fig. 3). Interestingly, cell lines of the GCB subtype collapse into a single cluster, indicating that the proteins most strongly differentiating ABC and GCB subtypes do not distinguish different GCB cell lines from each other (variation between cell lines is equal to the variation between replicates). In contrast, replicates of ABC cell lines remained distinguishable, which indicates a larger degree of heterogeneity in ABC-type cell lines as we observed previously.
Fig. 6.
t test signature. A, t test analysis of the proteins from the two groups of cell lines resulted in a signature of 55 proteins that are most significantly different. The panel depicts a heat map of the ratios of these proteins after clustering. B, plot of the difference of mean ratios versus the significance of signature proteins. The proteins on the left are significantly up-regulated in the ABC relative to GCB subtype. The protein names highlighted in red indicate NF-κB regulated genes.
In the signature obtained by unbiased proteomic analysis, there are at least six proteins whose gene expression levels are already described to be different between the subtypes (ABC-like: IRF4, CD44, and PTP1B; GCB-like: CD27, SPI1, and WIP). Because the total number of signature proteins is small, this already validates our proteomic signature and encouraged us to further investigate the new proteins in our signature. These include the recently described GTPase Speckled-like pattern in the germinal center (SLIP-GC), whose expression is limited to germinal center B-cells and to lymphomas derived from the germinal center including diffuse large B-cell lymphomas (36). This finding supports the potential use of SLIP-GC as a potential marker that can be used to differentiate the two subtypes from each other. Another member of the signature set is the surface marker CD81, which has also very recently been reported to be highly expressed in normal germinal B-cells. Further assessment of the role of this cell surface marker in the risk stratification of patients with DLBCL has already been recommended (37). A further novel protein that has a higher expression level in our GCB-DLBCL signature is the signaling adaptor Cbp/PAG. In B-cell non-Hodgkin lymphomas, PAG and Lyn kinase constitute the core of an oncogenic signalosome that results in proliferative and pro-survival signals. The Lyn and PAG signalosome can interact with downstream kinases to mediate these signals in different lymphoma cell lines (38). Our finding that PAG is up-regulated in GCB suggests investigating the modality associated with PAG in this subtype in particular. Ymer, a protein that we previously identified as an effector of EGF signaling (10, 39), is relatively up-regulated in the ABC subtype. Ymer is also known as CCDC50 or C3orf6, and although not studied in the context of DLBCL, this protein has been shown to be required for cell survival in chronic lymphocytic leukemia and mantle cell lymphoma cells (40). It is involved in the control of NF-κB signaling, which is a characteristic of the ABC subtype where it is up-regulated (15). Therefore, in addition to validating differentiators known from gene expression profiling, our proteomic signature reveals a novel set of proteins, some of which have been shown to be involved in lymphomagenesis and might be of clinical relevance.
In an attempt to identify annotated protein categories that are significantly and exclusively up-regulated in one lymphoma subtype relative to the other, we plotted the difference of mean ratios versus the statistical significance of our signature proteins. This revealed that all NF-κB-regulated proteins in the signature are significantly up-regulated in ABC relative to GCB subtype (Fig. 6B). They included IRF4, CD44, Ymer, PTP1B, ICAM, and HLA-C. Thus, our proteomic findings, similar to previous results of gene expression profiling, are consistent with high NF-κB activity in ABC-DLBCL as a hallmark that distinguishes this subtype from GCB-DLBCL.
BCR signaling has been shown to play an important role in lymphomagenesis where malignant B-cells exploit the normal regulatory roles of this pathway for their own purposes (41). We extracted proteins from our data set that are KEGG annotated to be involved in BCR signaling and found that we covered all but eight proteins in this category (59 of 67; supplemental Fig. 2). To better investigate small but reproducible protein changes, we normalized their expression levels by z-scoring across replicates and cell lines. Taking the median values for every subtype revealed four large clusters. The proteins highlighted in red in supplemental Fig. 4A consist of the BCR signaling proteins that exhibit the largest expression differences between the two subtypes and that are higher in the ABC subtype. Interestingly, this cluster included NF-κB1, as well as the two upstream regulatory proteins MALT1 and CARD11 (supplemental Fig. 4B). This is consistent with the role of the multiprotein CARD11-BCL10-MALT1 (CBM) complex in driving the constitutive NF-κB activity in the ABC subtype (17–19). Conversely, the proteins (highlighted in green in supplemental Fig. 4A) are BCR signaling proteins that are higher in GCB (supplemental Fig. 4C).
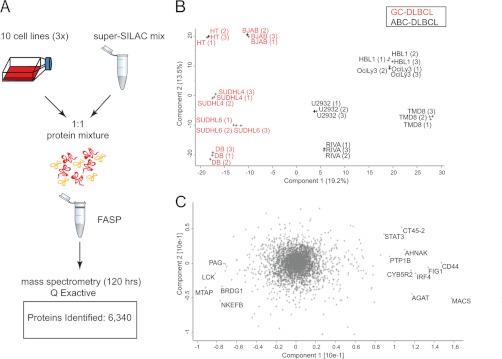
Rapid Lymphoma Classification in Single-shot Runs
Above, we have demonstrated that quantitative proteomics can readily segregate cell lines derived from patients in a robust manner. However, sample amounts and measurement time of our workflow (Fig. 1) would still be an obstacle to clinical application. We therefore wanted to investigate the possibility of making the approach more practical by reducing the measurement time and the amount of sample consumed. Taking advantage of the higher speed and sensitivity of the newly introduced quadrupole Orbitrap mass spectrometer (Q Exactive) (27), we investigated whether we could reach the depth required to segregate the cell lines in a single-shot experiment, that is, without fractionation. The samples were prepared as before, except that FASP-prepared peptides were directly loaded on StageTips and eluted into the autosampler device. Single 4-h gradient runs were performed in triplicates for each of the 10 cell lines, and data files were processed together in MaxQuant. This resulted in the identification of 6,340 proteins and the quantification of 4,611 in at least two replicates of the same cell line (Fig. 7A) (supplemental Tables VI and VII). Filtering for 50% valid values resulted in 3,566 quantified proteins. Upon PCA analysis of the single measurements, we obtained a similar segregation of the two subtypes, and the loadings responsible for the PCA segregation showed a very strong overlap with the previously obtained loadings (Fig. 7B). Interestingly, the data obtained from singlets was sufficient to segregate the two subtypes. This shows that single-shot measurements can reach the depth required for robust separation of lymophoma subtypes, opening up for the analysis of several patient samples per day with sample requirements in the low microgram range.
Fig. 7.
Single-shot proteome measurements to distinguish ABC from GCB. A, unfractionated, FASP-processed peptide mixtures were directly loaded onto a relatively long column (50 cm) after StageTipping. The proteomes were analyzed in triplicates in 4-h runs by an UHPLC (EASY nLC 1000) system coupled to a benchtop quadrupole Orbitrap mass spectrometer (Q Exactive). B, principal component analysis of the single-shot measurements. C, loadings of PCA in B highlighting the proteins that strongly drive the segregation in PCA component 1.
Conclusion and Outlook
Here we have shown that high accuracy, quantitative proteomics based on a super-SILAC approach can robustly segregate closely related cancer subtypes directly at the level of expressed proteins. We developed and used a super-SILAC mix of labeled B-cell lymphoma cell lines as an internal standard to segregate subtypes of DLBCL. We selected the cell lines with the most distinct protein expression profiles to obtain the best coverage of different lymphoma-specific proteins. The mix was spiked into five ABC-DLBCL and five GCB-DLBCL cell lines, which allowed robust, unsupervised segregation of these two histologically indistinguishable lymphomas based on their protein expression profiles. We found that requiring protein quantification values to be present in half of the samples and replicates and imputing the remaining values led to the most robust segregation. The data also revealed a protein expression signature that differentiates the two subtypes. This signature confirmed known markers previously discovered by gene expression studies and highlighted novel ones. Interestingly, our straightforward PCA analysis of the proteome differences revealed proteins such as IRF4, CD44, STAT3, PTP1B, and CD27 as the strongest differentiators between subtypes. The fact that these and a number of other proteins, which all have a strong biological rationale to drive subtype differences, emerge as the top hits in an unbiased analysis, is very intriguing. Furthermore, unbiased and supervised segregation revealed a number of novel proteins, which can now be studied for their involvement in these lymphoma subtypes.
To our knowledge, this is the first high accuracy, quantitative proteomics study that unequivocally classified tumor cell lines on par with microarray-based methods. This ability of the super-SILAC proteomic approach to readily segregate between tumor subtypes now opens up the possibility of employing proteomics in many situations that have previously been studied with transcript-based approaches. Toward this goal, we already combined the super-SILAC quantitative approach with single-shot measurements on a benchtop quadrupole Orbitrap instrument. These measurements attained the depth and accuracy required to segregate the two subtypes as exemplified by a number of representative cell lines. Considering the significant reduction in measuring time and in required sample amount, it is conceivable that this workflow could be employed in routine settings to answer practical clinical questions such as tumor classification or drug efficacy.
Acknowledgments
We thank Daniel Krappmann, Berit Jungnickel, Ralf Küppers, Martin Janz, Stephan Mathas, and Georg Lenz for the provision of cell lines. We thank Tamar Geiger and Maria Robles (Charo) for helpful discussions.
The acquired raw data for the fractionated proteome is uploaded to Tranche (https://proteomecommons.org/tranche/) with the hash code: Ib9WXUuYMjCxiCk40HNWo3YM+xvP31WEgYg6RWMjm5295exN5kmoMbyekZXqMvXpG8rvLgDELBXBKcceiScd+R2WqDkAAAAAAACQyQ==.
Footnotes
* This work was supported by European Commission's 7th Framework Program PROSPECTS Grant Agreement HEALTH-F4-2008-201648 and a Deutsche Krebshilfe Onconet2 grant. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
 This article contains supplemental materials.
This article contains supplemental materials.
1 The abbreviations used are:
- SILAC
- stable isotope labeling with amino acids in cell culture
- ABC-DLBCL
- activated B-cell-like diffuse large B-cell lymphoma
- BCR
- B-cell receptor
- DLBCL
- diffuse large B-cell lymphoma
- GCB-DLBCL
- germinal-center B-cell–like DLBCL
- PCA
- principal component analysis
- FASP
- filter-aided sample preparation
- SAX
- strong anion exchange
- STAT
- signal transducers and activators of transcription
- KEGG
- Kyoto Encyclopedia of Genes and Genomes.
REFERENCES
- 1. Quackenbush J. (2006) Microarray Analysis and Tumor Classification. New Engl. J. Med. 354, 2463–2472 [DOI] [PubMed] [Google Scholar]
- 2. McDermott U., Downing J. R., Stratton M. R. (2011) Genomics and the Continuum of Cancer Care. New Engl. J. Med. 364, 340–350 [DOI] [PubMed] [Google Scholar]
- 3. Hanash S., Taguchi A. (2010) The grand challenge to decipher the cancer proteome. Nat. Rev. Cancer 10, 652–660 [DOI] [PubMed] [Google Scholar]
- 4. Choudhary C., Mann M. (2010) Decoding signalling networks by mass spectrometry-based proteomics. Nat. Rev. Mol. Cell Biol. 11, 427–439 [DOI] [PubMed] [Google Scholar]
- 5. Aebersold R., Mann M. (2003) Mass spectrometry-based proteomics. Nature 422, 198–207 [DOI] [PubMed] [Google Scholar]
- 6. Nilsson T., Mann M., Aebersold R., Yates J. R., 3rd, Bairoch A., Bergeron J. J. (2010) Mass spectrometry in high-throughput proteomics: Ready for the big time. Nat. Methods 7, 681–685 [DOI] [PubMed] [Google Scholar]
- 7. Cox J., Mann M. (2011) Quantitative, high-resolution proteomics for data-driven systems biology. Annu. Rev. Biochem. 80, 273–299 [DOI] [PubMed] [Google Scholar]
- 8. Vermeulen M., Selbach M. (2009) Quantitative proteomics: A tool to assess cell differentiation. Curr. Opin. Cell Biol. 21, 761–766 [DOI] [PubMed] [Google Scholar]
- 9. Mallick P., Kuster B. (2010) Proteomics: A pragmatic perspective. Nat. Biotechnol. 28, 695–709 [DOI] [PubMed] [Google Scholar]
- 10. Ong S. E., Blagoev B., Kratchmarova I., Kristensen D. B., Steen H., Pandey A., Mann M. (2002) Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics 1, 376–386 [DOI] [PubMed] [Google Scholar]
- 11. Mann M. (2006) Functional and quantitative proteomics using SILAC. Nat. Rev. Mol. Cell Biol. 7, 952–958 [DOI] [PubMed] [Google Scholar]
- 12. Geiger T., Cox J., Ostasiewicz P., Wisniewski J. R., Mann M. (2010) Super-SILAC mix for quantitative proteomics of human tumor tissue. Nat. Methods 7, 383–385 [DOI] [PubMed] [Google Scholar]
- 13. Geiger T., Wisniewski J. R., Cox J., Zanivan S., Kruger M., Ishihama Y., Mann M. (2011) Use of stable isotope labeling by amino acids in cell culture as a spike-in standard in quantitative proteomics. Nat. Protoc. 6, 147–157 [DOI] [PubMed] [Google Scholar]
- 14. Alizadeh A. A., Eisen M. B., Davis R. E., Ma C., Lossos I. S., Rosenwald A., Boldrick J. C., Sabet H., Tran T., Yu X., Powell J. I., Yang L., Marti G. E., Moore T., Hudson J., Jr., Lu L., Lewis D. B., Tibshirani R., Sherlock G., Chan W. C., Greiner T. C., Weisenburger D. D., Armitage J. O., Warnke R., Levy R., Wilson W., Grever M. R., Byrd J. C., Botstein D., Brown P. O., Staudt L. M. (2000) Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature 403, 503–511 [DOI] [PubMed] [Google Scholar]
- 15. Davis R. E., Brown K. D., Siebenlist U., Staudt L. M. (2001) Constitutive nuclear factor κB activity is required for survival of activated B cell-like diffuse large B cell lymphoma cells. J. Exp. Med. 194, 1861–1874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Davis R. E., Ngo V. N., Lenz G., Tolar P., Young R. M., Romesser P. B., Kohlhammer H., Lamy L., Zhao H., Yang Y., Xu W., Shaffer A. L., Wright G., Xiao W., Powell J., Jiang J. K., Thomas C. J., Rosenwald A., Ott G., Muller-Hermelink H. K., Gascoyne R. D., Connors J. M., Johnson N. A., Rimsza L. M., Campo E., Jaffe E. S., Wilson W. H., Delabie J., Smeland E. B., Fisher R. I., Braziel R. M., Tubbs R. R., Cook J. R., Weisenburger D. D., Chan W. C., Pierce S. K., Staudt L. M. (2010) Chronic active B-cell-receptor signalling in diffuse large B-cell lymphoma. Nature 463, 88–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ngo V. N., Davis R. E., Lamy L., Yu X., Zhao H., Lenz G., Lam L. T., Dave S., Yang L., Powell J., Staudt L. M. (2006) A loss-of-function RNA interference screen for molecular targets in cancer. Nature 441, 106–110 [DOI] [PubMed] [Google Scholar]
- 18. Ferch U., Kloo B., Gewies A., Pfänder V., Düwel M., Peschel C., Krappmann D., Ruland J. (2009) Inhibition of MALT1 protease activity is selectively toxic for activated B cell-like diffuse large B cell lymphoma cells. J. Exp. Med. 206, 2313–2320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hailfinger S., Lenz G., Ngo V., Posvitz-Fejfar A., Rebeaud F., Guzzardi M., Penas E. M., Dierlamm J., Chan W. C., Staudt L. M., Thome M. (2009) Essential role of MALT1 protease activity in activated B cell-like diffuse large B-cell lymphoma. Proc. Natl. Acad. Sci. U.S.A. 106, 19946–19951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pasqualucci L., Trifonov V., Fabbri G., Ma J., Rossi D., Chiarenza A., Wells V. A., Grunn A., Messina M., Elliot O., Chan J., Bhagat G., Chadburn A., Gaidano G., Mullighan C. G., Rabadan R., Dalla-Favera R. (2011) Analysis of the coding genome of diffuse large B-cell lymphoma. Nat. Genet. 43, 830–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wiśniewski J. R., Zougman A., Nagaraj N., Mann M. (2009) Universal sample preparation method for proteome analysis. Nat. Methods 6, 359–362 [DOI] [PubMed] [Google Scholar]
- 22. Wiśniewski J. R., Zougman A., Mann M. (2009) Combination of FASP and StageTip-based fractionation allows in-depth analysis of the hippocampal membrane proteome. J. Proteome Res. 8, 5674–5678 [DOI] [PubMed] [Google Scholar]
- 23. Rappsilber J., Ishihama Y., Mann M. (2003) Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal. Chem. 75, 663–670 [DOI] [PubMed] [Google Scholar]
- 24. Olsen J. V., Schwartz J. C., Griep-Raming J., Nielsen M. L., Damoc E., Denisov E., Lange O., Remes P., Taylor D., Splendore M., Wouters E. R., Senko M., Makarov A., Mann M., Horning S. (2009) A dual pressure linear ion trap Orbitrap instrument with very high sequencing speed. Mol. Cell. Proteomics 8, 2759–2769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Olsen J. V., Macek B., Lange O., Makarov A., Horning S., Mann M. (2007) Higher-energy C-trap dissociation for peptide modification analysis. Nat. Methods 4, 709–712 [DOI] [PubMed] [Google Scholar]
- 26. Thakur S. S., Geiger T., Chatterjee B., Bandilla P., Frohlich F., Cox J., Mann M. (2011) Deep and highly sensitive proteome coverage by LC-MS/MS without prefractionation. Mol. Cell. Proteomics 10, M110.003699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Michalski A., Damoc E., Hauschild J. P., Lange O., Wieghaus A., Makarov A., Nagaraj N., Cox J., Mann M., Horning S. (2011) Mass spectrometry-based proteomics using Q exactive, a high-performance benchtop quadrupole Orbitrap mass spectrometer. Mol. Cell. Proteomics 10, M111.011015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cox J., Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 29. Cox J., Neuhauser N., Michalski A., Scheltema R. A., Olsen J. V., Mann M. (2011) Andromeda: A peptide search engine integrated into the MaxQuant environment. J Proteome Res. 10, 1794–1805 [DOI] [PubMed] [Google Scholar]
- 30. Lu X., Malumbres R., Shields B., Jiang X., Sarosiek K. A., Natkunam Y., Tiganis T., Lossos I. S. (2008) PTP1B is a negative regulator of interleukin 4-induced STAT6 signaling. Blood 112, 4098–4108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lu X., Nechushtan H., Ding F., Rosado M. F., Singal R., Alizadeh A. A., Lossos I. S. (2005) Distinct IL-4-induced gene expression, proliferation, and intracellular signaling in germinal center B-cell-like and activated B-cell-like diffuse large-cell lymphomas. Blood 105, 2924–2932 [DOI] [PubMed] [Google Scholar]
- 32. Schmidlin H., Diehl S. A., Nagasawa M., Scheeren F. A., Schotte R., Uittenbogaart C. H., Spits H., Blom B. (2008) Spi-B inhibits human plasma cell differentiation by repressing BLIMP1 and XBP-1 expression. Blood 112, 1804–1812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lam L. T., Wright G., Davis R. E., Lenz G., Farinha P., Dang L., Chan J. W., Rosenwald A., Gascoyne R. D., Staudt L. M. (2008) Cooperative signaling through the signal transducer and activator of transcription 3 and nuclear factor-κB pathways in subtypes of diffuse large B-cell lymphoma. Blood 111, 3701–3713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ding B. B., Yu J. J., Yu R. Y., Mendez L. M., Shaknovich R., Zhang Y., Cattoretti G., Ye B. H. (2008) Constitutively activated STAT3 promotes cell proliferation and survival in the activated B-cell subtype of diffuse large B-cell lymphomas. Blood 111, 1515–1523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Shipp M. A., Ross K. N., Tamayo P., Weng A. P., Kutok J. L., Aguiar R. C., Gaasenbeek M., Angelo M., Reich M., Pinkus G. S., Ray T. S., Koval M. A., Last K. W., Norton A., Lister T. A., Mesirov J., Neuberg D. S., Lander E. S., Aster J. C., Golub T. R. (2002) Diffuse large B-cell lymphoma outcome prediction by gene-expression profiling and supervised machine learning. Nat. Med. 8, 68–74 [DOI] [PubMed] [Google Scholar]
- 36. Richter K., Brar S., Ray M., Pisitkun P., Bolland S., Verkoczy L., Diaz M. (2009) Speckled-like pattern in the germinal center (SLIP-GC), a nuclear GTPase expressed in activation-induced deaminase-expressing lymphomas and germinal center B cells. J. Biol. Chem. 284, 30652–30661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Luo R. F., Zhao S., Tibshirani R., Myklebust J. H., Sanyal M., Fernandez R., Gratzinger D., Marinelli R. J., Lu Z. S., Wong A., Levy R., Levy S., Natkunam Y. (2010) CD81 protein is expressed at high levels in normal germinal center B cells and in subtypes of human lymphomas. Human Pathol. 41, 271–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tauzin S., Ding H., Burdevet D., Borisch B., Hoessli D. C. (2011) Membrane-associated signaling in human B-lymphoma lines. Exp. Cell Res. 317, 151–162 [DOI] [PubMed] [Google Scholar]
- 39. Kratchmarova I., Blagoev B., Haack-Sorensen M., Kassem M., Mann M. (2005) Mechanism of divergent growth factor effects in mesenchymal stem cell differentiation. Science 308, 1472–1477 [DOI] [PubMed] [Google Scholar]
- 40. Farfsing A., Engel F., Seiffert M., Hartmann E., Ott G., Rosenwald A., Stilgenbauer S., Döhner H., Boutros M., Lichter P., Pscherer A. (2009) Gene knockdown studies revealed CCDC50 as a candidate gene in mantle cell lymphoma and chronic lymphocytic leukemia. Leukemia 23, 2018–2026 [DOI] [PubMed] [Google Scholar]
- 41. Lenz G., Staudt L. M. (2010) Aggressive lymphomas. New Engl. J. Med. 362, 1417–1429 [DOI] [PMC free article] [PubMed] [Google Scholar]