Abstract
Despite the potential benefits of selective redox-modulating strategies for cancer therapy, an efficacious methodology for testing therapies remains elusive because of the difficulty in measuring intracellular redox potentials over time. In this report, we have incorporated a new FRET-based biosensor to follow in real time redox-sensitive processes in cells transformed to be tumorigenic and cultured in a microfluidic channel. A microfluidic network was used to control micro-scale flow near the cells and at the same time deliver drugs exogenously. Subsequently, the response of a redox homeostasis circuit was tested, namely reduced glutathione (GSH)/oxidized glutathione(GSSG), to diamide, a thiol oxidant, and two drugs used for cancer therapies: BSO (l-buthionine-[SR]-sulfoximine) and BCNU (carmustine). The main outcome from these experiments is a comparison of the temporal depletion and recovery of GSH in single living cells in real-time. These data demonstrate that mammalian cells are capable of restoring a reduced intracellular redox environment in minutes after an acute oxidative insult is removed. This recovery is significantly delayed by (i) the inhibition of GSH biosynthesis by BSO; (ii) the inactivation of glutathione reductase by BCNU; and (iii) in tumorigenic cells relative to an isogenic non-tumorigenic control cell line.
Introduction
As a cancerous tumor expands, it quickly outgrows the diffusion-limited blood supply leading to a hypoxic microenvironment and oxidative stress. Oxidative stress, in particular, has been hypothesized to influence tumors since the decreased susceptibility of tumor cells to stimuli that induce apoptosis has been linked to their inherently reduced reduction/oxidation (redox) potential.1–6 Thus, redox homeostasis has become a target for cancer chemotherapy. Generally, cells depend vitally on the balance struck between redox reactions, which can be disrupted by factors such as increases in the level of reactive oxygen species (ROS) that develop during inflammation or exposure to environmental chemicals that are metabolized to antioxidant-depleting electrophiles. Consequently, cells have evolved sophisticated regulatory mechanisms to buffer the reduction potential and mitigate oxidative stress. A key question that remains is whether tumor cells have lost the ability to mount the apparent changes in intracellular redox potential that accompany normal cell growth or the ability to sense these changes.
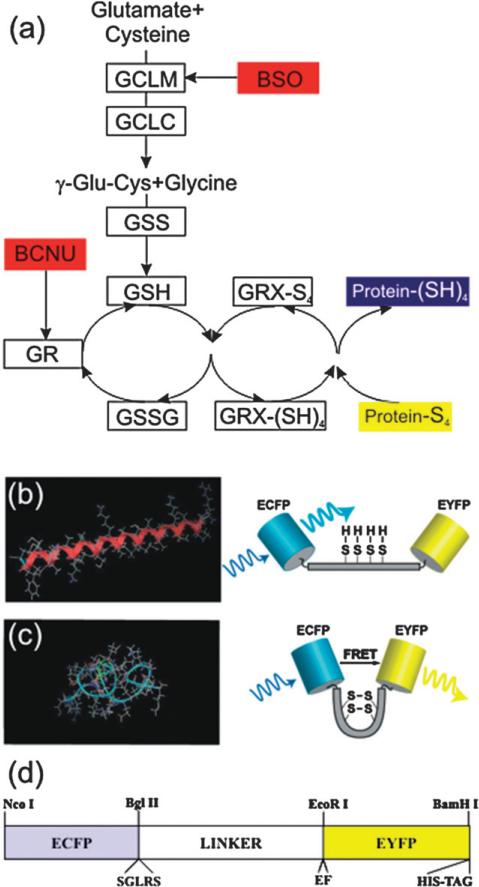
Although redox homeostasis most likely affects cell fate more than any other biochemical phenomenon, the molecular mechanisms mediating redox sensitivity and regulation are poorly defined and difficult to study.7,8 Being the most abundant redox buffer in eukaryotic cells, the glutathione (GSH)/oxidized glutathione (GSSG) homeostasis circuit plays a crucial role in the response to oxidative stress. As illustrated in Fig. 1(a), GSH biosynthesis is catalyzed by two ATP-dependent enzymes: γ-glutamyl-cysteine ligase (GCL), which is comprised of two independent subunits GCLC and GCLM; and glutathione synthetase (GSS).9–13 GSH protects cells against oxidative stress through termination of oxidants with concomitant oxidation of GSH to GSSG. GSH reacts spontaneously with electrophilic substances such as free radicals and ROS producing glutathione disulfide (GSSG). When GSH becomes oxidized to form GSSG, it can then effectively be recycled by the antioxidant flavoenzyme glutathione reductase (GR). GR catalyzes the reduction of oxidized glutathione by NADPH and elevated levels of GSH are often detected in drug-resistant tumors.13
Fig. 1.
Glutaredoxin pathway of reduction of protein disulfides and a FRET biosensor to probe it. (a) The glutaredoxin system acts via the GSH/GSSG redox buffer. Its high electron-donating capacity (high negative redox potential) combined with high intracellular concentration (millimolar levels) generate great reducing power. GRX-(SH)4 and GRX-S4 indicate the reduced and oxidized forms, respectively, of glutaredoxin. In the Figure the enzymes are indicated by boxes and their inhibitors are in red. (b,c) A schematic of the redox sensitive switching mechanism for a single molecule. Under oxidative conditions, intermolecular disulfide bonds can form shifting the free energy minimum from the (b) α-helix, to a (c) ‘clamped-coil’ state (similar to a helix–coil transition). The coiled state brings the donor (ECFP) and acceptor (EYFP) in closer proximity than in the extended helix state, where they exchange excitation energy more efficiently (i.e. a high FRET state). The extent of energy transfer is easily quantified from the increased emission of the acceptor. (d) Diagram of the CY-RL7 construct, indicating the location of the redox linker inserted between two GFP variants, ECFP and EYFP. The position of the restriction sites for NcoI, BlgII and BamHI, EcoRI used for cloning are also indicated in the map along with the location of the hexa histidine-tag, identified by the trademarked name His-tag, which is used for protein purification.
Despite the potential benefits of a selective GSH-modulating strategy for cancer therapy, an efficacious methodology remains elusive because of the difficulty in measuring intracellular redox potentials over time. Various methods have been employed to quantify the redox environment in cells, including the use of fluorescent sensors based on GFP proteins,8,14–16 peptide scaffolds,17,18 or small molecules;19 each having strengths and weaknesses. For example, dyes force redox measurements to be made in a static mode, thus providing only limited evidence about redox dynamics.20 For such applications, genetically encoded fluorophores are more suitable.8,21
We report here on the development of a novel genetic construct that enables real-time assessment of intracellular redox, and its use in tracking the redox response of tumorigenic cells to glutathione perturbations introduced through a microscale flow. This genetic construct senses intracellular redox using resonance energy transfer (FRET), a powerful technique for monitoring interactions, associations, and structural modifications of biological molecules. In our case, oxidation induces a change in the molecular conformational that alters the distance between the FRET donor–acceptor pair, which in turn causes a detectable change in FRET efficiency.22 Using this FRET sensor, protocols were first established to test the redox response of Chinese hamster ovary (CHO) cells cultured in a microfluidic channel. In contrast to a canonical culture vessel, the microfluidic device affords us stringent control over the spatial and temporal microenvironment of the cell. Then the redox response of two isogenic porcine cell lines was examined: one being nontransformed control fibroblasts, which exhibit contact-inhibition (161-C) versus a transformed fibroblast cell line that harbors six oncogenes (161-T), hTERT, p53DD, cyclin D1, CDK4R24C, c-MycT58A and H-RasG12V, known to perturb pathways commonly corrupted in human cancer.23 This cell line is tumorigenic when implanted into the host animal, and it was used supposing that tumorigenesis in pigs is similar to that in humans, even at a molecular level.23
The main outcome from these experiments is a comparison of the real-time kinetics of GSH depletion and recovery in single normal and cancer cells subjected to oxidant challenges. Specifically, the responses of 161-C and 161-T cells to a pulsatile challenge by diamide ((CH3)2NCON=NCON(CH3)2), an oxidant probe for thiols, were examined after treatment with two drugs used in cancer therapy: BSO (l-buthionine-[SR]-sulfoximine) and BCNU (1,3-bis[2-chloroethyl]-2-nitrozourea, commonly known as Carmustine). These data demonstrate that mammalian cells are capable of restoring a reduced intracellular redox environment in minutes after an acute oxidative insult is removed. But the recovery is significantly delayed by both (i) the inhibition of GSH biosynthesis by BSO; and (ii) the inactivation of glutathione reductase by BCNU.
Results and discussion
Fluorescence microscopy is ideally suited to elucidate the spatio-temporal distribution and functional states of constituent intracellular molecules.24,25 In this work it was used to both demonstrate the utility of a FRET-based biosensor, CY-RL7 (cyan-yellow FRET pair with a newly designed redox linker 7 (RL7)) and illuminate the intracellular redox environment driven specifically by the 2GSH/GSSG couple in live cells in real time. A schematic of the redox-sensitive switching mechanism for a single molecule is shown in Fig. 1(b,c). An oxidation event induces a conformational change in the sensor altering the distance between the FRET donor–acceptor pair, which in turn causes a detectable change in FRET efficiency. Under oxidative conditions intermolecular disulfide bonds form, shifting the free energy minimum from the α-helix, shown in Fig. 1(b) to a ‘clamped-coil’ state represented in Fig. 1(c), which forces the donor (ECFP) and acceptor (EYFP) into closer proximity. The efficacy of redox biosensors with related structures was previously demonstrated in vitro by analyzing purified proteins by steady-state fluorescence spectroscopy.22 The lack of linkers capable of both separating donor from acceptor and conferring redox sensitivity is a major drawback in engineering FRET-based redox biosensors, but that issue was resolved in our previous study. In this most recent design, developed using the cloning strategy represented schematically in Fig. 1(d), the linker is comprised of eight –EAAAK–sequences with 4 cysteine residues flanked by the two GFP variants, ECFP and EYFP.
As a test of its suitability for use in vivo, FRET measurements using CY-RL7 in living CHO cells were performed. The live cells were first cultured in a microfluidic channel like that shown in Fig. 2(a,b). The microchannel enables stringent control of the cell's microenvironment and allows us to broadcast drugs exogenously.26–28 To establish sensitivity of the FRET sensor to changes in intracellular redox potential, the CHO cells were subsequently subjected in a pulsatile manner to oxidative stress using 1 mM diamide in DMEM.
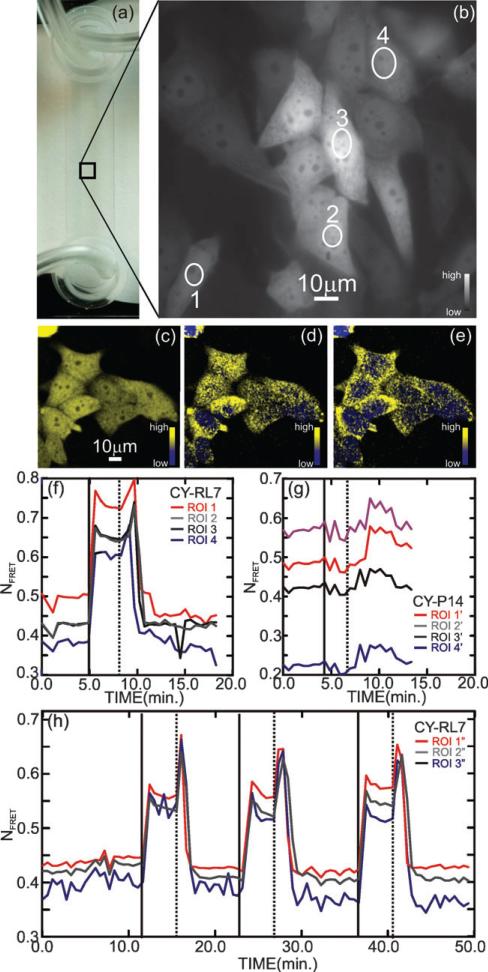
Fig. 2.
Validation of the CY-RL7 redox sensor in CHO cells. (a) Microfluidic channel showing the tubing used for fluid exchange (left) and CHO cell FRET channel image with the regions of interest (ROI) enumerated (right). (b) Fluorescence micrograph showing CHO cells imaged in the microfluidic channel in a low flow (48 μL min–1) at t = 0, just prior to a diamide pulse (see the corresponding Fig. (f) as well). (c,d,e) Confocal images of CHO cells show that, on exposure to a 1 mM diamide pulse, the spatial distribution and intensity of the FRET signal changes, indicating a change in the redox environment in the cytoplasm. The three confocal images compare (c) the integrated two dimensional projection of the three dimensional confocal image constructed from the change in YFP channel with (d,e) showing slices taken from (c) along the optic axis at z = 0.5 μm and z = 4.5 μm respectively. (f,g) Time lapse of the fluorescence of a few ROIs demonstrates the response of the CY-RL7 and CY-P14 FRET constructs respectively expressed in CHO cells after treatment with 1 mM exogenous oxidant diamide (solid vertical line at t = 5 min) followed by removal of oxidant (dashed vertical line, t = 8 min). The ROIs associated with CY-RL7 shown in (f) are indicated in (b), while the ROIs associated with CY-P14 are taken from other cells not shown. (h) Three sequential cycles, comprised of an oxidative insult with 1 mM diamide (solid vertical lines) followed by removal of oxidant by brief washing (dashed vertical lines) 4 min later, were applied to CHO cells (not shown), illustrating the reversible nature of the CY-RL7 response. The data are representative of 12 independent experiments using a minimum of 4 ROIs.
A cell contains millimolar concentrations of GSH (~10 mM) that is maintained in this reduced form by a cytosolic NADPH-dependent reaction catalyzed by GR. Thus, protein disulfide bonds that drive the FRET sensor rarely form in the cytosol because of the high concentrations of GSH. However, upon exposure of the cells to diamide, concentrations of intracellular GSH are rapidly depleted.29,30 The diamide penetrates cell membranes within seconds and reacts within the cell at a high rate (seconds to minutes) at physiological pH, enabling the study of cell functions altered by a temporary perturbation of the thiol status.
Once in the cell, the diamide may directly oxidize the biosensor as it does cell extracts.36 Alternatively, the diamide insult, which is governed by the following reactions:
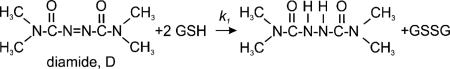 |
(1) |
| (2) |
| (3) |
| (4) |
affects the glutathione homeostasis circuitry, shifting the cell to an environment conducive to the oxidation of the biosensor. Overall, the diamide reacts with glutathione more rapidly than other thiol substrates according to reaction (1); it is characterized by a rate constant of k1 = 18 mM–1 min–1.29 On the other hand, the diamide reacts very slowly with non-GSH thiols and even more slowly with non-thiols. Moreover, the diamide is more likely to react with a small thiol like GSH than with a protein thiol due to steric hindrance.29 Thus, it was reasoned that the diamide challenge predominately perturbs the glutathione circuitry and, since glutathione is the major nonprotein thiol in most cells, it is possible to gain information about the role of GSH in redox homeostasis this way.
In most living organisms GR maintains a high cytosolic ratio (~100:1) of reduced GSH over GSSG by catalyzing reaction (2). However, following a 1 mM diamide challenge, the GSH is exhausted, which drives reaction (2) toward the production of more GSH. In particular, the net reaction described by reaction (4) may shift the FRET sensor to a ‘clamped-coil’ state and a more efficient exchange of the excitation energy. Thus, while the FRET response seems to follow the concentration of GSH, it is difficult to distinguish between the direct oxidation of the probe and an alteration in the redox potential of the glutathione couple.
Fig. 2 shows the FRET response after exposure to 1 mM diamide. The microfluidic flow is switched from a perfusion comprised of only DMEM to a flow with DMEM plus 1 mM diamide (solid vertical line) for 3 min. Fig. 2(c)–(e) are confocal microscopic images that illustrate the dramatic change in FRET after exposure to diamide. The (false-colored) yellow is related to the increase in FRET response. Fig. 2(c) is a false-color, two-dimensional projection integrated over the three-dimensional confocal image of the cells, showing the FRET channel observed two minutes after exposure to 1 mM diamide, while Fig. 2(d,e) show two slices of the image in (c) taken at z = 0.5 μm and z = 4.5 μm along the optic axis from the same CHO cells under the same conditions. From these images it appears that the response is compartmentalized. The change in fluorescence is spatially dependent on a scale well beyond the lateral and axial resolution, which is estimated to be 0.61*λ/NA = 285 nm and 2λ/(NA)2 = 778 nm, respectively. It is well known that redox potential is compartmentalized in eukaryotes.31,32 Within organelles and intracellular compartments, a cell maintains specific redox potentials in correspondence with its functionality.33 Consistent with this assertion, the sensor response is observed to vary spatially throughout the cell indicating that the redox potential is changing at various locations through the cell.
Figs. 2(f,g) indicate that sensitivity of our FRET redox biosensor in the ROIs is exclusively due to the nature of the redox linker. These Figures compare the time lapse response of the FRET constructs with two different linkers: (f) RL7 comprised of l-helical sequence with four embedded cysteine residues and (g) P14 comprised of 14 proline residues, expressed in CHO cells, to sequential treatment with diamide. Positive responses to diamide were not observed for CY-P14, the negative control construct. On the other hand, the initial state of the CY-RL7 biosensor (before oxidative challenge) is characterized by a low FRET signal, which demonstrates that CY-RL7 is fully reduced in accordance with the physiological state of the cell. The FRET response was normalized against ECFP and EYFP expression levels and to remove the stochastic variation from cell to cell (as described in the Methods section); typically ΔNFRET ≈ 0.15, but the low FRET signal varies from one ROI to the next, which may be attributed (at least in part) to the spatial variation of the FRET within the cell. Finally, the CY-RL7 construct shows a better response to GSH/GSSG ratios representing a significant improvement over previously reported constructs.22 Various attributes of the sensor response are described in more detail elsewhere.34
Subsequently, the time development of FRET was followed in regions of interest (ROIs) in the cell. As illustrated in the supplemental Fig. S1,† the cell recovery time, measured by a return to the FRET level prior to diamide exposure, depends on the strength of the diamide challenge. However, the change in FRET depends only weakly on concentration, as illustrated in Fig. S2.† So, to minimize the recovery time, while maximizing the change in FRET, the cells were washed, switching from a diamide in DMEM flow back to the DMEM-only flow (dashed line), with a minimized wash duration. The diamide treatment does not seem to cause any irreversible damage. After the oxidative insult is removed the original thiol status is apparently recovered as evident from Fig. 2(f). To unequivocally demonstrate the reversible character of the sensor, three consecutive cycles of oxidative challenge were applied to the same cells as illustrated by Fig. 2(h). Each treatment cycle includes perfusion with DMEM medium containing 1 mM diamide (solid vertical lines), time lapse image acquisition at oxidized state in presence of diamide followed by perfusion in DMEM-only at 48 μL min–1 (dashed vertical lines). A rapid cell recovery is evident from the decrease in FRET to the initial level observed for all three cycles.
Biosensor oxidation occurs quickly (in < 12 s) with exposure to diamide, which is evidenced by the rapid increase of the FRET signal shown in Fig. 2(f) near t = 5 min. The diamide eventually overwhelms the GSH buffer and so disulfide bonds form in CY-RL7 forcing the coiled state. In steady-state, the rate of the oxidation of GSH has to be equal to the rate of the chemical reaction regenerating GSH. After the diamide is washed away, reaction (2) eventually re-establishes the reducing environment of the cell after a delay corresponding to the time required to exhaust the remaining diamide. Thus, the FRET signal tracks the recovery in the GSH concentration according to reactions (2) and (3) as demonstrated in Fig. S1.†
To further illuminate the kinetics of redox changes characterized by the GSSG/2GSH couple, the effect of GSH deficiency on the redox restoration was examined by the imaging of live CHO cells pretreated with BSO or BCNU followed by an oxidative insult with diamide. First, to establish a control, untreated CHO cells were exposed to 1 mM diamide in DMEM (Fig. 3(b), solid vertical line). After a 4–5 min oxidative stress, diamide was washed out with DMEM. The control shown in Fig. 3(a,b) indicates that cell oxidation occurs quickly with exposure to diamide, as shown by the rapid increase of the FRET signal near t = 4 min. Following the diamide insult, the cells recover quickly, within 2–3 min. Subsequently, the CHO cells were exposed to another form of oxidative stress: 0.5 mM BSO in DMEM for 24 h. BSO is a non-toxic and selective inhibitor of the enzyme GCL in GSH biosynthesis and thereby generates oxidative stress by decreasing the GSH pool.35 This assertion is supported by our prior observation of a > 10-fold decrease in GSH by reaction with monochlorobomane (mBCl), a common technique to measure GSH in cultured cells.22,36 After 24 h treatment with BSO, the image-acquisition of cells was started. The cell's appearance, depicted in Fig. 3(c), confirms that pretreatment with BSO for 24 h does not produce noticeable changes in the redox sensor response. This can be explained by the low FRET state of the biosensor during the first four minutes before oxidative insult with diamide. These data are supported by similar (static) results on roGFP2 in mammalian cells.37 This BSO pretreatment was followed by a second oxidant insult using 1 mM diamide in DMEM for 4–5 min with a subsequent wash in DMEM. The time dependence of the FRET signal in several ROIs shown in Fig. 3(d) consistently demonstrates a rapid increase in FRET immediately following the diamide insult, but a delayed return to the initial reduced state. Although the delay in restoration of redox homeostasis varied from cell to cell, ranging from 10 to 25 min, it is in stark contrast to the 2 min recovery time exhibited by cells oxidized only by diamide illustrated in Fig. 3(b).
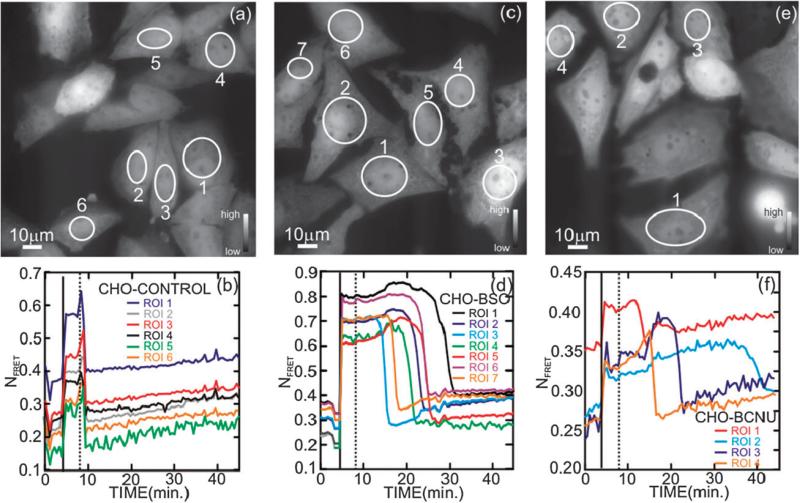
Fig. 3.
Effect of diamide with and without BSO and BCNU pretreatment on CHO cell recovery. Fluorescence micrographs showing CHO cells stably transfected with CY-RL7, imaged in the microfluidic channel under low flow conditions (48 μL min–1) at t = 0, just prior to a diamide pulse (a) without pretreatment, (c) after pretreatment with 0.5 mM BSO for 24 h and (e) after 2 h of BCNU pretreatment. (b), (d), and (f) show the corresponding ratiometric analyses of the CY-RL7 fluorescence for the ROIs indicated in the respective Figures. The imaging commenced under low flow conditions (48 μL min–1) at t = 0, just prior to exposure to a diamide pulse. Generally, the FRET changes dramatically after exposure to 1 mM diamide in DMEM (solid vertical line) at t = 4 min. The FRET signal recovers rapidly (within 2 min) once the diamide is washed out at t = 8 min (dashed vertical line) without BSO/BCNU pretreatment, (see (b)), but recovers only after a delay with a pretreatment (d,f). These data are representative of 3 independent experiments using a minimum of 4 ROIs.
Finally, the effect of GSH deficiency was also studied by pre-exposure of CHO cells to 0.5 mM BCNU in DMEM for 2 h. After in situ decomposition, the alkylating cytostatic drug, BCNU, produces oxidative stress by inactivating GR, producing an accumulation of GSSG.38,39 According to reaction (2), GR catalyzes the reduction of oxidized glutathione by NADPH, which sensitizes a cell to oxidative stress. Fig. 3(e,f) show that BCNU has a similar effect to BSO. Just like BSO, the pretreatment with BCNU does not produce noticeable changes in the redox sensor response. Subsequently, the pretreatment with BCNU was followed by 1 mM diamide in DMEM for 4–5 min. Again, a rapid response to the onset of the diamide challenge and a rapid, but delayed return of the FRET sensor to the initial reduced state was observed. Although the delay in restoration of redox homeostasis ranged from 10 to 30 min depending on the cell, the recovery time is much longer than that observed in cells oxidized only by diamide. Apparently, in the absence of diamide in the medium, the recovery time in CHO cells is delayed by thiol regeneration, which requires consumption of the remaining reagent. This assertion is supported by the shortening of the recovery time with decreasing diamide concentration shown in Fig. S1.† Therefore, regeneration is slow to occur in cells lacking GR due to inhibition by BCNU, or in cells in which biosynthesis of GSH is blocked.
Taken altogether, these data demonstrate unequivocally, not only the efficacy of the new CY-RL7 biosensor for tracking redox, but also that mammalian cells are capable of restoring a reduced intracellular redox environment on a time scale as short as ~2 min after an acute oxidative insult is removed. Neither BSO nor BCNU was observed to oxidize the biosensor. On the other hand, diamide, which is a de-facto thiol oxidant, oxidizes the biosensor when it permeates the cell membrane. However, thiol regeneration kinetics after the oxidative insult is removed must drive the biosensor reduction since the response time changes with BSO or BCNU treatments, which deplete the intracellular GSH pool. Thus, it is inferred that the biosensor tracks the initial steps in cytosolic regeneration of GSH depleted by the pretreatment with BSO or BCNU and the subsequent diamide insult. To our knowledge, these are the first data on the time-response of depletion of GSH in real-time after an oxidative insult. In this regard, the data highlight the importance of considering this rapid cellular response when conducting studies on the role of GSH in redox homeostasis.
Elevated and prolonged oxidative stress has been associated with several pathophysiologies, including cancer. But until recently the limitations with respect to tools for measuring the intracellular redox status has crippled research at the molecular level. The new redox-sensitive FRET-based CY-RL7 biosensor remedies some of these limitations and provides an opportunity to examine perturbations of intracellular redox homeostasis. The efficacy of therapies targeting redox homeostasis was tested in an experimental setting directly related to cancer biology using the CY-RL7 redox sensor. As illustrated in Fig. 4, the same challenge and recovery experiments described above were performed with 161-C and 161-T cells transiently transfected with CY-RL7, evaluating the effect of oxidative stress by BSO or BCNU followed by a second oxidant insult to the same cells using 1 mM diamide in DMEM for 4–5 min. Only a limited number of cells fluoresce within a field of view because of the inefficiency of the transfection. However, the results reported are typical of those obtained from multiple experiments following the same protocol.
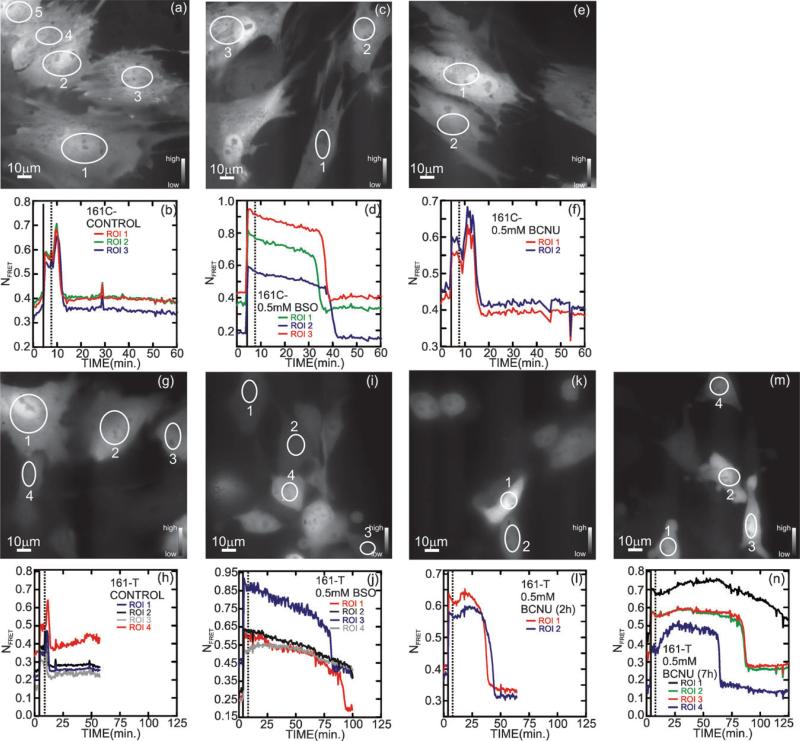
Fig. 4.
Effect of diamide with and without BSO and BCNU pretreatment on recovery of 161-C control (nontransformed) and transformed 161-T cells. Fluorescence micrographs showing 161-C cells transiently transfected with CY-RL7, imaged in the microfluidic channel under low flow conditions (48 μL min–1) at t = 0, just prior to a diamide pulse (a) without pretreatment, (c) after pretreatment with 0.5 mM BSO for 24 h and (e) after 2 h of BCNU pretreatment. (b), (d) and (f) show the corresponding ratiometric analyses of ROIs after cell exposure to 1 mM diamide in DMEM (solid vertical line) followed by brief washing (dashed vertical line) under pretreatment conditions indicated in (a), (c) and (e), respectively. The imaging commenced under low flow conditions (48 μL min–1) at t = 0, just prior to exposure to a diamide pulse. Generally, the FRET changes dramatically in the cytoplasm after exposure to 1 mM diamide in DMEM (solid vertical line) at t = 4 min. The FRET signal recovers rapidly (within 2 min) once the diamide is washed out at t = 8 min (dashed vertical line) without BSO/BCNU pretreatment, but recovers only after a delay with a pretreatment. These data are representative of 3 independent experiments using a minimum of 4 ROIs. Fluorescence micrographs showing 161-T cells transiently transfected with CY-RL7, imaged in the microfluidic channel under low flow conditions (48 μL min–1) at t = 0, just prior to a diamide pulse (g) without pretreatment, (i) after pretreatment with 0.5 mM BSO for 24 h and (k) after 2 h of BCNU pretreatment. Panels (h), (j) and (l) show the corresponding ratiometric analyses of ROIs after cell exposure to 1 mM diamide in DMEM (solid vertical line) followed by brief washing (dashed vertical line) under pretreatment conditions indicated in (g), (i) and (k), respectively. The imaging commenced under low flow conditions (48 μL min–1) at t = 0, just prior to exposure to a diamide pulse. Generally, the FRET changes dramatically in the cytoplasm after exposure to 1 mM diamide in DMEM (solid vertical line) at t = 4 min. The FRET signal recovers rapidly (within 2 min) once the diamide is washed out at t = 8 min (dashed vertical line) with a pretreatment, but recovers only after a much longer delay with a pretreatment. To emphasize the affect of the duration of the pretreatment on the delay, panels (m) and (n) present data from 7 h of pretreatment with BCNU. These data are representative of 3 independent experiments using a minimum of 4 ROIs.
The 161 cell line provides a valuable opportunity to interrogate the contributions of redox signalling in well defined cancer signalling pathways. The two cells lines tested, derived from embryonic pig cells, consist of a nontransformed control (161-C), which is subject to contact-inhibited cell growth, and a companion tumorigenic cell line (161-T) that harbors six oncogenes, which have been rigorously defined in regards to their tumorigenic properties.23 The response and recovery to diamide challenge with and without pretreatments with 0.5 mM BSO for 24 h and 0.5 mM BCNU for 2 h is quantitatively similar to that observed with CHO cells described above. Fig. 4(b) shows a ratiometric analysis of the CY-RL7 fluorescence for three ROIs indicated in Fig. 4(a). The FRET parameter changes dramatically upon exposure to 1 mM diamide in DMEM (solid vertical line) at t = 4 min and recovers rapidly, within 2 min once the diamide is washed out at t = 8 min (dashed vertical line). On the other hand, after a pretreatment with 0.5 mM BSO for 24 h, there is a rapid but delayed return of the FRET sensor to the initial reduced state following the 1 mM diamide challenge; the total recovery time is extended to 35–42 min, which is about twice as long as the recovery time measured in CHO cells under the same conditions as shown in Fig. 4(c,d). Likewise, as illustrated in Fig. 4(e,f) after treatment with 0.5 mM BCNU for 2 h, the recovery time after a 1 mM diamide challenge and wash is extended to 15–18 min, which is within the range of responses found for CHO cells. In each case, the recovery time is much longer than the 2 min delay observed in cells oxidized only by diamide.
As illustrated in Fig. 4(g–n), the same challenge and recovery experiments were performed with the isogeneic 161-T cancer cell line transiently transfected with CY-RL7. While the recovery seems to be qualitatively similar to the results obtained with 161-C cells, i.e. there is a rapid, but delayed return of the FRET sensor to the initial reduced state, the 161-T cells either did not or took a substantially longer time to recover from the diamide oxidative challenge. Fig. 4(h) shows a ratiometric analysis of the CY-RL7 fluorescence for four ROIs indicated in Fig. 4(g). The NFRET parameter changes dramatically upon exposure to 1 mM diamide in DMEM (solid vertical line) at t = 4 min and recovers rapidly, within 2 min once the diamide is washed out at t = 8 min (dashed vertical line). On the other hand, after a pretreatment with 0.5 mM BSO for 24 h, there is a rapid but delayed return of the FRET sensor to the initial reduced state following the 1 mM diamide challenge; the total recovery time is extended to 80–90 min (Fig. 4(i,j)). Likewise, as illustrated in Fig. 4(k,l) after treatment with 0.5 mM BCNU for 2 h, the recovery time after a 1 mM diamide challenge is extended to 37–50 min. In each case, the recovery time is much longer (>2×) than the delay observed in 161-C control cells and longer still than the 2 min delay encountered in 161-T oxidized only by diamide.
Conclusion
We have demonstrated that mammalian cells are capable of restoring a reduced intracellular redox environment on a time scale of a few minutes after an acute oxidative insult is removed. This is significant because the redox environment exerts a profound influence on normal cellular processes like DNA synthesis, enzyme activation, selective gene expression, cell cycle progression, proliferation, differentiation, and apoptosis. This recovery time is therefore relevant to all these normal cellular processes.40 In addition, oxidative stress is thought to be a strongly influential factor in aging, cancer, and various inflammatory disorders.41 In regards to cancer, there is evidence indicating that the decreased susceptibility of tumors to stimuli that induce apoptosis can be linked to their inherently reduced redox potential.42,43 Although less defined at the molecular level, intracellular redox status also affects cell cycle progression.44,45 For example, several redox-sensitive proteins involved in cell cycle progression have been identified, including p53,46 AP-1,47 NF-κB,48 PKC,49 and low molecular-weight protein tyrosine phosphatases.50 These and possibly other unidentified cell cycle proteins harboring redox-sensitive motifs may constitute a mechanistic link between the intracellular redox environment and cell cycle progression.51–53 Therefore, a means to investigate and characterize macromolecular interactions is crucial for elucidating the mechanisms underlying tumorigenesis.
The tools developed in this work provide a new and unique method to examine the fundamental mechanisms underpinning tumorigenesis. The initial tests of these tools on tumorigenic cells indicate that BSO and BCNU grossly affect thiol regeneration preferentially in cancer cells, which in turn increases the duration of oxidative stress on the cell. Some, but not all cancer cells seem to lose or at least have a compromised ability to sense and mount apparent changes in intracellular redox potential, since the response time seems to be at least 2× longer than observed in normal control cells. But it remains to be seen how the longer response time directly relates to the effectiveness of selective redox-modulating drug therapies.
These tools could ultimately affect the development and dosing recommendations of GR inhibitors as drugs against tumor cells, as well. For example, Fig. 4(m,n) show that by increasing the exposure to BCNU from 2 h to 7 h, there is a concomitant increase in the recovery time: after a 1 mM diamide challenge the recovery time is extended to 63–125 min. However, these results should be interpreted cautiously because the targeted cells could survive even when GR is completely inhibited because the thioredoxin system, working in parallel, seems to be capable of maintaining high GSSG fluxes.54
Experimental
Cell culture
The porcine control (161-C) and tumor (161-T) cell lines were maintained at 37 °C in 5% CO2 in Dulbecco's modfied eagle medium (DMEM)/F-10, supplemented with 15% fetal bovine serum, streptomycin (5000 U L–1), penicillin (5000 U L–1), and fungizone (250 μg L–1) in T-75 cell culture flasks (Costar, Cambridge, MA). Chinese hamster ovary (CHO) fibroblasts were cultured in Dulbecco's modified eagle medium (DMEM) supplemented with 10% FetalPlex and above mentioned antibiotics. Mammal cells transfected with the sensor were cultured in presence of G418 (15 μl ml–1).
Genetic constructs
Redox-sensitive linker (RL) RL7 and polyproline linker P14 were designed using methods similar to those described earlier.22 New constructs CY-RL7 and CY-P14 (Fig. 1(b,c) were built by the insertion of linkers between ECFP/EYFP pair cloned into mammalian vector pECFP-C.
Microchannel
All cells were subsequently seeded in Ibidi μ-Slide VI with tissue culture treated coating (Model #: 80606) at density close to 50% confluency, approximately 24 h prior to observation. The flowchamber has six microchannels with the following dimensions: 0.4 mm × 3.8 mm × 17 mm long and with a 30 μl volume. Cells were observed in DMEM medium without phenol red and subsequently oxidized with 1 mM diamide ((CH3)2NCON=NCON(CH3)2, Sigma Chemical) in DMEM solution. The addition of diamide affected the pH of the solution only marginally. DMEM without diamide showed a pH of 7.95, while DMEM with diamide showed a pH of 8.27. For physiological redox recovery experiments the diamide was washed out after 3–5 min with the initial DMEM medium. A photograph of a microchannel with tubing for fluid exchange is shown in Fig. 2(a). Fluids are always removed from one side by a waste collection tube and refilled from the other side by either DMEM ll-in tube or diamide in DMEM ll-in tube (Fig. 2(a)). The wash passed twice the reservoir volume (2×70 μL) through the 30 μL channel.
FRET microscopy
The time development of FRET in ROIs in the cell was examined. Cells were imaged on a Zeiss Axio-Observer inverted microscope with a Zeiss Achroplan 100× oil immersion objective (1.3NA). The microscope is equipped with an environmental chamber that is temperature controlled to 37 °C during live cell imaging. In addition, a μ-slide (Ibidi, Germany) was placed inside a closed microincubator (BioscienceTools, TC-MI) for more accurate CO2 control. A Pecon CO2 controller controls 5% CO2 content as measured inside the controller and not at the slide location. Images were taken with 1024 × 1024 EMCCD camera (Andor, DU-888) operated at –70 °C.
The inverted microscope has a three-filter cube system for FRET assay.25 A three-filter cube system eliminates the cross-talk between donor and acceptor fluorescence with carefully selected fluorophore pairs. Thus, FRET analysis was performed with three channels: FRET, CFP and YFP. The FRET channel was defined by an excitation filter with a 438/24 nm center wavelength/bandwidth, a dichroic mirror with a 458 nm edge wavelength, and an emission filter 542/27 nm. Similarly, the CFP channel filter parameters were excitation 438/24 nm, dichroic edge at 458 nm, emission 483/32 nm. The YFP had an excitation filter 500/24 nm with a dichroic edge at 520 nm and an emission filter at 542/27 nm. The excitation was provided by a EXFO X-Cite 120 metal halide lamp operated at 12% iris to reduce photobleaching. To minimize photobleaching the exposure times were kept at 20 ms using a camera high gain (typically 50 to 100 on the scale 0–255) with images taken every 15–60 s. Data acquisition and analysis was automated using NI Labview.
Fluorescence images were background-corrected by manual selection of cell-free regions. FRET analysis was based on mean values taken over regions of interest (ROI) within the cells or was carried out pixel by pixel as shown in Fig. 2. To account for slight cell movements that occur during the time lapse acquisition, the ROIs were automatically adjusted to track the same regions. ROIs were chosen randomly for each cell and a net FRET (nF) was calculated according to: nF = IFRET – IYFP × a – ICFP × b where IFRET, IYFP and ICFP are fluorescence intensities in each ROI. Coefficients a and b are the norms of bleed-through to FRET channel from EYFP and ECFP, respectively.25 The coefficient a was determined as a fraction of yellow light excited by 438 nm light relative to 500 nm excitation when cells expressing only EYFP are used. Similarly, b is determined as a fraction, IFRET /ICFP, when cells expressing only ECFP are measured. For our system a = 0.19, b = 0.47.
In these experiments, the expression level of the FRET biosensor varied from cell to cell. To reliably quantify FRET signal in cells independent of the expression levels of redox biosensor, the net FRET was normalized against expression levels of ECFP and EYFP through as described elsewhere.25 This normalization procedure is seen to remove the stochastic variation in FRET from cell to cell. Moreover, while the FRET channel accurately reflects the oxidative state of the cell, the dynamics in the recovery region exhibited a common increase in intensities in all channels. nF, the amplitude of the response to oxidation, increased but an increase of the signal in the recovery region was also observed. The NFRET normalization removes this effect and shows a return to the original redox state found prior to the application of diamide.
Unexpectedly, a reproducible peak in the FRET signal is observed just after the diamide is flushed from the microfluidic (as illustrated in Fig. 2(f,g)). This peak in the recovery varies in size from experiment to experiment, but is not affected by a change in flow in the microfluidic, and it disappears completely if the diamide concentration is stepped to zero over a 5 min interval, or after cell pretreatment with BSO or BCNU (as illustrated in Fig. 3(g,i)).
Confocal microscopy and image processing
To illustrate explicitly the 3D aspects of the FRET response cells were imaged using a laser scanning confocal microscope (Zeiss LSM 710) with a 40×, 1.2NA water immersion objective and with a pinhole size of set at 31 μm and scaling in x, y, and z were 0.198 × 0.198 × 0.400 μm, respectively. CFP and FRET images were excited with a 458 nm laser and collected using a 465–490 nm and 540–580 nm filters respectively. Eighteen z-stack images were taken every 2 min. Images were deconvolved with Huygens (SVI) and iso-surfaces constructed from confocal image stacks with Imaris software (Bitplane) incorporating MATLAB(Mathworks). We estimate a typical lateral and axial resolution to be 0.61*λ/NA = 285 nm and 2λ/(NA)2 = 778 nm, respectively.
Insight, innovation, integration.
INTEGRATION: This manuscript reports the integration of a tumorigenic cell line with a new FRET redox biosensor and microfluidic for the study of pulsatile perturbations to redox homeostasis in real time. Together these tools provide a new method to examine the fundamental mechanisms underpinning tumorigenesis.
INNOVATION: Despite the potential benefits of selective redox-modulating strategies for cancer therapy, a methodology for testing therapies remains elusive because of the difficulty in measuring intracellular redox potentials over time. To our knowledge, our study provides the first data on the time-response of depletion of (reduced) glutathione, the most abundant redox buffer in eukaryotic cells, in real-time after an oxidative insult.
INSIGHT: The key question we are trying to address with this research is whether tumor cells have lost the ability to mount the apparent changes in intracellular redox potential that accompanies normal cell growth or the ability to sense these changes. Our data indicates that some cancer cells seem to lose or at least have a compromised ability to sense and mount changes in intracellular redox potential.
Acknowledgements
We gratefully acknowledge support from NSF TH2008-01040 ANTC (GT) and from NIHEB004513 (PJAK & HRG).
Footnotes
Electronic supplementary information (ESI) available.
References
- 1.Ballatori N, Krance SM, Notenboom S, Shi SJ, Tieu K, Hammond CL. Biol. Chem. 2009;390(3):191–214. doi: 10.1515/BC.2009.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mena S, Ortega A, Estrela JM. Mutat. Res., Genet. Toxicol. Environ. Mutagen. 2009;674(1–2):36–44. doi: 10.1016/j.mrgentox.2008.09.017. [DOI] [PubMed] [Google Scholar]
- 3.Menon SG, Sarsour EH, Spitz DR, Higashikubo R, Sturm M, Zhang H, Goswami PC. Cancer Res. 2003;63(9):2109–2117. [PubMed] [Google Scholar]
- 4.Hainaut P, Milner J. Cancer Res. 1993;53(19):4469–4473. [PubMed] [Google Scholar]
- 5.Chiarugi P. IUBMB Life. 2001;52(1):55–59. doi: 10.1080/15216540252774775. [DOI] [PubMed] [Google Scholar]
- 6.Conour JE, Graham WV, Gaskins HR. Physiological Genomics. 2004;18(2):196–205. doi: 10.1152/physiolgenomics.00058.2004. [DOI] [PubMed] [Google Scholar]
- 7.Jones DP, Go YM, Anderson CL, Ziegler TR, Kinkade JM, Kirlin WG. FASEB J. 2004;18(9):1246–8. doi: 10.1096/fj.03-0971fje. [DOI] [PubMed] [Google Scholar]
- 8.Bjornberg O, Ostergaard H, Winther JR. Antioxid. Redox Signaling. 2006;8(3–4):354–361. doi: 10.1089/ars.2006.8.354. [DOI] [PubMed] [Google Scholar]
- 9.Meister A, Anderson ME. Annu. Rev. Biochem. 1983;52:711–60. doi: 10.1146/annurev.bi.52.070183.003431. [DOI] [PubMed] [Google Scholar]
- 10.Griffith OW, Mulcahy RT. Adv. Enzymol. Relat. Areas Mol. Biol. 1999;73:209–67. doi: 10.1002/9780470123195.ch7. [DOI] [PubMed] [Google Scholar]
- 11.Ursini F, Maiorino M, Brigeliusflohe R, Aumann K, Roveri A, Schomburg D, Flohe L, Biothiols Pt. B. Methods Enzymol. 1995;252:38–53. doi: 10.1016/0076-6879(95)52007-4. [DOI] [PubMed] [Google Scholar]
- 12.Meister A. J. Biol. Chem. 1988;263(33):17205–8. [PubMed] [Google Scholar]
- 13.Holmgren A, Aslund F. Methods Enzymol. 1995;252:283–92. doi: 10.1016/0076-6879(95)52031-7. [DOI] [PubMed] [Google Scholar]
- 14.Ostergaard H, Henriksen A, Hansen FG, Winther JR. EMBO J. 2001;20(21):5853–62. doi: 10.1093/emboj/20.21.5853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hanson GT, Aggeler R, Oglesbee D, Cannon M, Capaldi RA, Tsien RY, Remington SJ. J. Biol. Chem. 2003;279(13):13044–53. doi: 10.1074/jbc.M312846200. [DOI] [PubMed] [Google Scholar]
- 16.Gutscher M, Pauleau A-L, Marty L, Brach T, Wabnitz GH, Samstag Y, Meyer AJ, Dick TP. Nat. Methods. 2008;5(6):553–559. doi: 10.1038/nmeth.1212. [DOI] [PubMed] [Google Scholar]
- 17.Cline DJ, Thorpe C, Schneider JP. Anal. Biochem. 2004;325(1):144–50. doi: 10.1016/j.ab.2003.10.014. [DOI] [PubMed] [Google Scholar]
- 18.Lee K, Dzubeck V, Latshaw L, Schneider JP. J. Am. Chem. Soc. 2004;126(42):13616–7. doi: 10.1021/ja047300r. [DOI] [PubMed] [Google Scholar]
- 19.Miller EW, Bian SX, Chang CJ. J. Am. Chem. Soc. 2007;129(12):3458–9. doi: 10.1021/ja0668973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fricker M, Runions J, Moore I. Annu. Rev. Plant Biol. 2006;57:79–107. doi: 10.1146/annurev.arplant.57.032905.105239. [DOI] [PubMed] [Google Scholar]
- 21.Chudakov DM, Lukyanov S, Lukyanov KA. Trends Biotechnol. 2005;23(12):605–13. doi: 10.1016/j.tibtech.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 22.Kolossov VL, Spring BQ, Sokolowski A, Conour JE, Clegg RM, Kenis PJA, Gaskins HR. Exp. Biol. Med. 2008;233(2):238–48. doi: 10.3181/0707-RM-192. [DOI] [PubMed] [Google Scholar]
- 23.Adam SJ, Rund LA, Kuzmuk KN, Zachary JF, Schook LB, Counter CM. Oncogene. 2007;26(7):1038–1045. doi: 10.1038/sj.onc.1209892. [DOI] [PubMed] [Google Scholar]
- 24.Miyawaki A, Llopis J, Heim R, McCaffery JM, Adams JA, Ikura M, Tsien RY. Nature. 1997;388(6645):882–7. doi: 10.1038/42264. [DOI] [PubMed] [Google Scholar]
- 25.Xia ZP, Liu YH. Biophys. J. 2001;81(4):2395–2402. doi: 10.1016/S0006-3495(01)75886-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Timp W, Mirsaidov U, Matsudaira P, Timp G. Lab Chip. 2009;9(7):925–934. doi: 10.1039/b810157d. [DOI] [PubMed] [Google Scholar]
- 27.Yu H, Alexander CM, Beebe DJ. Lab Chip. 2007;7:726–730. doi: 10.1039/b618793e. [DOI] [PubMed] [Google Scholar]
- 28.Adler M, Polinkovsky M, Gutierrez E, Groisman A. Lab Chip. 2010;10:388–391. doi: 10.1039/b920401f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kosower NS, Kosower EM. Methods Enzymol. 1995;251:123–133. doi: 10.1016/0076-6879(95)51116-4. [DOI] [PubMed] [Google Scholar]
- 30.Kosower NS, Kosower EM, Wertheim B, Correa WS. Biochem. Biophys. Res. Commun. 1969;37:593–596. doi: 10.1016/0006-291x(69)90850-x. [DOI] [PubMed] [Google Scholar]
- 31.Schafer FQ, Buettner GR. Free Radical Biol. Med. 2001;30(11):1191–212. doi: 10.1016/s0891-5849(01)00480-4. [DOI] [PubMed] [Google Scholar]
- 32.Hansen JM, Go YM, Jones DP. Annu. Rev. Pharmacol. Toxicol. 2006;46:215–34. doi: 10.1146/annurev.pharmtox.46.120604.141122. [DOI] [PubMed] [Google Scholar]
- 33.Hancock JT, Desikan R, Neill SJ, Cross AR. J. Theor. Biol. 2004;226(1):65–68. doi: 10.1016/j.jtbi.2003.08.003. [DOI] [PubMed] [Google Scholar]
- 34.Kolossov VL, Spring BQ, Henry JJ, Sokolowski A, Clegg RM, Kenis PJA, Gaskins HR. doi: 10.1258/ebm.2011.011009. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meister A. J. Biol. Chem. 1994;269(13):9397–9400. [PubMed] [Google Scholar]
- 36.Kamencic H, Lyon A, Paterson PG, Juurlink BHJ. Anal. Biochem. 2000;286(1):35–7. doi: 10.1006/abio.2000.4765. [DOI] [PubMed] [Google Scholar]
- 37.Dooley CT, Dore TM, Hanson GT, Jackson WC, Remington SJ, Tsien RY. J. Biol. Chem. 2004;279(21):22284–93. doi: 10.1074/jbc.M312847200. [DOI] [PubMed] [Google Scholar]
- 38.Cereser C, Boget S, Parvaz P, Revol A. Toxicology. 2001;163(2–3):153–62. doi: 10.1016/s0300-483x(01)00401-2. [DOI] [PubMed] [Google Scholar]
- 39.Muller JG, Bucheler US, Kayser K, Schirmer RH, Werner D, Krauth-Siegel RL. Cell. Mol. Biol. 1993;39:389–396. [PubMed] [Google Scholar]
- 40.Attene-Ramos MS, Kitiphongspattana K, Ishii-Schrade K, Gaskins HR. Am. J. Physiol.: Cell Physiol. 2005;289(5):C1220–C1228. doi: 10.1152/ajpcell.00164.2005. [DOI] [PubMed] [Google Scholar]
- 41.Mates JM, Segura JA, Perez-Gomez C, Rosado L, Olalla L, Blanca M, Sanchez-Jimenez FM. Blood Cells, Mol. Dis. 1999;25(7):103–109. doi: 10.1006/bcmd.1999.0234. [DOI] [PubMed] [Google Scholar]
- 42.Ballatori N, Krance SM, Notenboom S, Shi SJ, Tieu K, Hammond CL. Biol. Chem. 2009;390(3):191–214. doi: 10.1515/BC.2009.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mena S, Ortega A, Estrela JM. Mutat. Res., Genet. Toxicol. Environ. Mutagen. 2009;674(1–2):36–44. doi: 10.1016/j.mrgentox.2008.09.017. [DOI] [PubMed] [Google Scholar]
- 44.Menon SG, Sarsour EH, Spitz DR, Higashikubo R, Sturm M, Zhang H, Goswami PC. Cancer Res. 2003;63(9):2109–2117. [PubMed] [Google Scholar]
- 45.Smith J, Ladi E, Mayer-Proschel M, Noble M. Proc. Natl. Acad. Sci. U. S. A. 2000;97(18):10032–10037. doi: 10.1073/pnas.170209797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hainaut P, Milner J. Cancer Res. 1993;53(19):4469–4473. [PubMed] [Google Scholar]
- 47.Shackelford RE, Kaufmann WK, Paules RS. Free Radical Biol. Med. 2000;28(9):1387–1404. doi: 10.1016/s0891-5849(00)00224-0. [DOI] [PubMed] [Google Scholar]
- 48.Hinz M, Krappmann D, Eichten A, Heder A, Scheidereit C, Strauss M. Mol. Cell. Bio. 1999;19(4):2690–2698. doi: 10.1128/mcb.19.4.2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Black JD. Front. Biosci. 2000;5:D406–D423. doi: 10.2741/black. [DOI] [PubMed] [Google Scholar]
- 50.Chiarugi P. IUBMB Life. 2001;52(1):55–59. doi: 10.1080/15216540252774775. [DOI] [PubMed] [Google Scholar]
- 51.Conour JE, Graham WV, Gaskins HR. Physiol. Genomics. 2004;18(2):196–205. doi: 10.1152/physiolgenomics.00058.2004. [DOI] [PubMed] [Google Scholar]
- 52.Burhans WC, Heintz NH. Free Radical Biol. Med. 2009;47(9):1282–1293. doi: 10.1016/j.freeradbiomed.2009.05.026. [DOI] [PubMed] [Google Scholar]
- 53.Hanahan D, Weinberg RA. Cell. 2000;100(1):57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 54.Kanzok SM, Schirmer RH, Türbachova I, Iozef R, Becker K. J. Biol. Chem. 2000;275(51):40180–40186. doi: 10.1074/jbc.M007633200. [DOI] [PubMed] [Google Scholar]