Abstract
The heat stress (HS) response in eukaryotes is mainly regulated by heat shock factors (HSFs). Genetic disruption of the master HSF gene leads to dramatically reduced HS response and thermotolerance in several model organisms. However, it is not clear whether organisms devoid of the master regulator can still acclimate to heat. Previously, we showed that Arabidopsis HsfA1a, HsfA1b, and HsfA1d act as master regulators in the HS response. In this study, we examined the heat acclimation capacity of the Arabidopsis quadruple and triple T-DNA knockout mutants of HsfA1a, HsfA1b, HsfA1d, and HsfA1e. Our data showed that in the absence of the master regulators, a minimal but significant level of acquired thermotolerance could be attained in the Arabidopsis mutants after acclimation. The optimum acclimation temperature for the HsfA1 quadruple mutant was lower than that for the wild type plants, suggesting that plant cells have two HS-sensing mechanisms that can be distinguished genetically. The acquired thermotolerance of the quadruple mutant was likely due to the induction of a small number of HsfA1-independent HS response genes regulated by other transcription factors. Here, we discuss the possible candidates and propose a working model of the transcription network of the HS response by including the HsfA1-dependent and -independent pathways.
Keywords: Arabidopis, heat shock factor, bZIP28, ER stress, acquired thermotolerance
In response to elevated temperature, living cells immediately switch on the transcription of a multitude of genes encoding protective proteins or enzymes that enable it to minimize injury or sustain a subsequent harsher heat challenge. This universal phenomenon in all organisms studied to date is known as the heat stress or heat shock response (HSR).1 In eukaryotic cells, the HSR is mediated by structurally conserved transcription regulators, named heat shock factors (HSFs), which form active trimeric conformations that bind to the conserved cis-elements in the promoters of many HSR genes.2,3 Genetic disruption or knockdown of the master regulator gene(s) has been shown to dramatically diminish the HSR and thermotolerance in chlamydomonas,4 drosophila,5 mammalian cells,6,7 and plants.8-10 However, whether organisms lacking the master regulator can still acclimate to heat is not clear. This question is of great interest in plants as the sessile organisms frequently face the challenge of HS in the natural environment, and the existence of an additional HSR pathway independent of the master HSFs could be advantageous for plants adapting to HS.
In higher plants, homologs of HSFs form a multigene family, whose members can be classified into 3 major classes (class A, B, and C) and several subclasses.11 Recently, genetic evidence has revealed diverse functions for some of these HSF homologous genes.8-10,12-21 The members of the HsfA1 subclass have been shown to be the master regulators of the HSR. Transcriptional knockdown of tomato HsfA1a expression dramatically reduced basal and acquired thermotolerance of the transgenic plants, suggesting that HsfA1a plays a major role in the HSR of tomato.9 In Arabidopsis, there are four members belonging to the HsfA1 subclass, HsfA1a, HsfA1b, HsfA1d, and HsfA1e. Disruption of Arabidopsis HsfA1a or both HsfA1a and HsfA1b by T-DNA insertions does not cause significant defect in thermotolerance, as is the case in tomato.22 Thus, it was thought that the four HsfA1 genes are likely redundant in function. To evaluate this possibility, we generated a quadruple T-DNA knockout (QK) and four triple KO mutants, named aTK, bTK, dTK, and eTK, where the prefixed letters represent the remaining intact HsfA1.8 These mutants are derived from single mutants of Columbia (Col) or Wassilewskija (Ws) ecotype backgrounds. We showed that the QK and eTK mutants are extremely sensitive to various HS treatments from seed to adult plant stages compared with either the Col or Ws wild types. Moreover, the HSR was dramatically compromised in these mutants, while in the aTK, bTK, and dTK mutants the thermotolerance and HSR were not or were only partially affected. These results suggest that HsfA1a, HsfA1b, and HsfA1d share the role of master regulator of the HSR in Arabidopsis.8 In this study, we were interested to know whether acquired thermotolerance can be attained in the absence of the master transcription regulators.
First, to determine whether there are differential temperature optimums to induce acquired thermotolerance in the HsfA1 mutants, 7-d-old seedlings were first acclimated at different temperatures from 31 to 39°C for 1 h, allowed to recover at 22°C for 2 h, then challenged by severe HS at 44°C for 150 min. Viability of the seedlings was then assessed after 7 d of recovery from treatment. The results showed that 37°C was the most effective temperature in inducing acquired thermotolerance in the wild type, aTK, bTK, and dTK plants (Fig. 1A and B). The acclimation treatments at different temperatures could not confer even slight tolerance to the severe HS treatment in the QK and eTK mutants (data not shown), which was a more severe result than that seen in the Hsp101 KO line. Hsp101 encodes a molecular chaperone that has an important role in thermotolerance.23,24 These results are consistent with our previous findings: HsfA1a is slightly more effective than HsfA1d, while HSFA1b is the least effective at conferring acquired thermotolerance and the QK and eTK are unable to attain acquired thermotolerance under the assay conditions.8 However, we suspected that the challenge treatment at 44°C for 150 min might have been too harsh to reveal the heat acclimation effect in the QK and eTK, which could both be too weak. Thus, we reduced the severity of our conditions to 43°C for 25 min for the HS challenge. This condition is lethal to non-acclimated QK and eTK seedlings, but not to the wild type or even the Hsp101 KO plants. Figure 1C and D show that acclimation at 33°C to 37°C could significantly enhance thermotolerance in eTK and QK, and the optimum acclimation temperature was 35°C. Despite the fact that the survival rate of the QK mutant was as high as that of the wild type if acclimated first at 35°C, the mutant plants showed retarded growth after the HS challenge (Fig. 1C), indicating a delicate nature of the acquired thermotolerance without the participation of HsfA1s. The viability of the eTK was slightly lower than that of the QK at different acclimation temperatures (Fig. 1C and D), which is consistent with our previous findings that HsfA1e is not required for thermotolerance.8
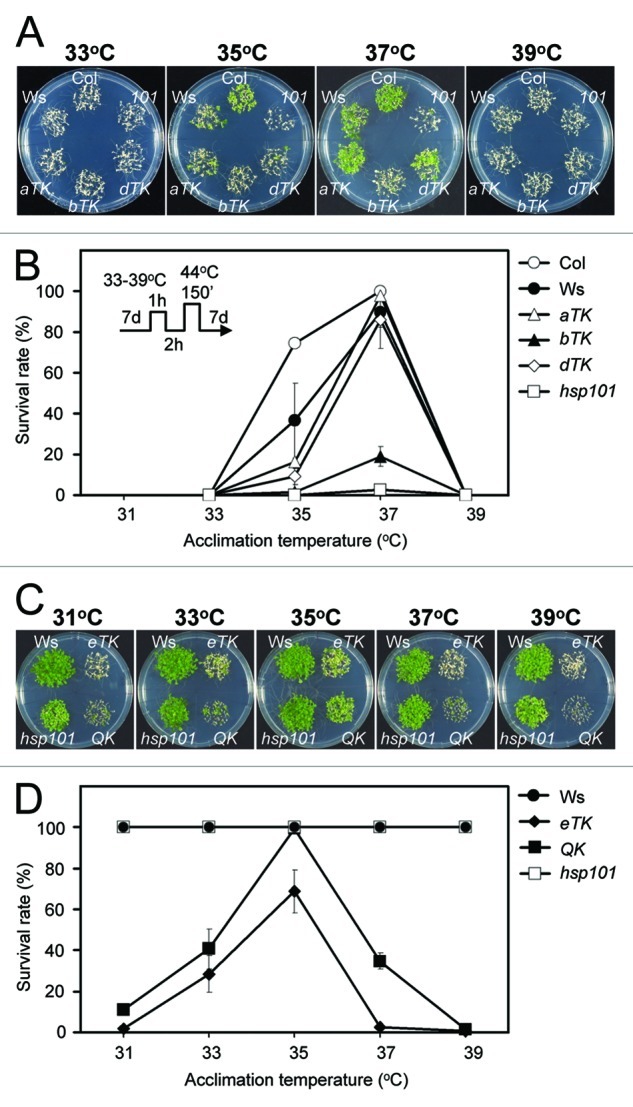
Figure 1. Arabidopsis seedlings attained low levels of acquired thermotolerance in the absence of the HsfA1s. Seedlings of the wild type (Col and Ws), aTK, bTK, dTK, eTK, QK, and Hsp101 KO mutant (hsp101 or 101) lines were subjected to HS treatments with HS regimes schematically shown inside the panel B. The 7-d-old seedlings were first acclimated at 31–39°C as indicated, allowed to recover at 22°C for 2 h, and then challenged at 44°C for 150 min (A and B) or 43°C for 25 min (C and D). The plants were allowed to grow for 7 d after the HS treatments before being photographed. The effect of acclimation temperatures on acquired thermotolerance was measured by growth performance (A and C) and survival rate (B and D). The survival rates are presented as mean values of three replicates ± SD (n ≥ 50 each).
Our data demonstrate that in Arabidopsis the ability to acquire enhanced thermotolerance by acclimation is dramatically reduced in the absence of HsfA1a/b/d, but is not completely abolished. This is consistent with the transcriptional profiles that have previously shown that the heat-induction of a relatively small number of HS-upregulated genes is independent of the master regulators.8 Moreover, some highly heat-induced genes remain inducible by heat treatment in the QK mutant, although to a lesser degree.8 These genes may contribute to the acquired thermotolerance of the QK and eTK mutants. So far, it is not clear what transcription factors are responsible for the heat-induction of these genes. Other HSF genes, such as HsfA4a, HsfA4c, or HsfA7a, whose transcripts are relatively abundant under normal condition,11 are possible candidates. These transcription factors might play a minor role in acquired thermotolerance. It was reported that disruption of HsfA7a due to T-DNA insertion causes a defect in acquired thermotolerance.25 However, the mechanism of how this may occur has not been reported. In our work, we did not observe obvious and consistent thermotolerance defects in the same HsfA7a KO line (SALK_080138) under various HS conditions (ref. 13 and unpublished data). The role and mechanism of this HSF in acquired thermotolerance remain to be shown. Moreover, we could not exclude the possible participation of HsfA2, Dreb2A, and HsfB1/2b, which have been implicated in acquired thermotolerance.13,14,18,26 Of note, HsfA2, HsfA7a, Dreb2A, and HsfB1/2b are HsfA1-dependent HSR genes.8,10
Alternatively, there might be other types of transcription factors regulating the expression of the HsfA1-independent HSR genes, such as BiP1 (At5g28540), BiP2 (At5g42020), and UTR3 (At1g14360).8 BiP1 and BiP2 are molecular chaperones localized in the endoplasmic reticulum (ER), while UTR3 is a UDP-galactose transporter localized both at the ER and Golgi.27 Recently, these genes were shown to be regulated by an ER membrane-tethered basic domain/leucine zipper (bZIP) transcription factor, bZIP28, during ER and heat stresses.28,29 Upon exposure to stress conditions, the transcription factor domain of bZIP28 is released from the ER membrane by proteolysis and redistributed to the nucleus.28,29 The Arabidopsis T-DNA KO mutant of bZIP28 (At3g10800) exhibits increased sensitivity to HS and is unable to trigger the heat-inducible expression of BiP1/BiP2 and UTR3.28 In contrast, the expression of Hsp17.4-CIII (At1g54050), a target gene of HsfA1, is not affected in the bZIP28 mutant.28 Although it remains to be seen to what extent bZIP28 is involved in acquired thermotolerance, this membrane-tethered transcription factor may act as a sensor of HS in a pathway parallel to that mediated by HsfA1. Interestingly, bZIP28 itself is heat-inducible.28 Our microarray data showed that the heat-induced but not the basal expression of bZIP28 was substantially reduced in the absence of the HsfA1s (Fig. 2), suggesting that at least one of the HsfA1s positively regulates bZIP28 under HS conditions. The basal expression of bZIP28 apparently was sufficient to upregulate its target genes. Actually, the expression of BiP1/BiP2 and UTR3 were even higher in the QK mutant than in the wild type under HS conditions (Fig. 2), suggesting that HsfA1s act as negative regulators of these genes under HS conditions. More experiments are needed to confirm these relationships.
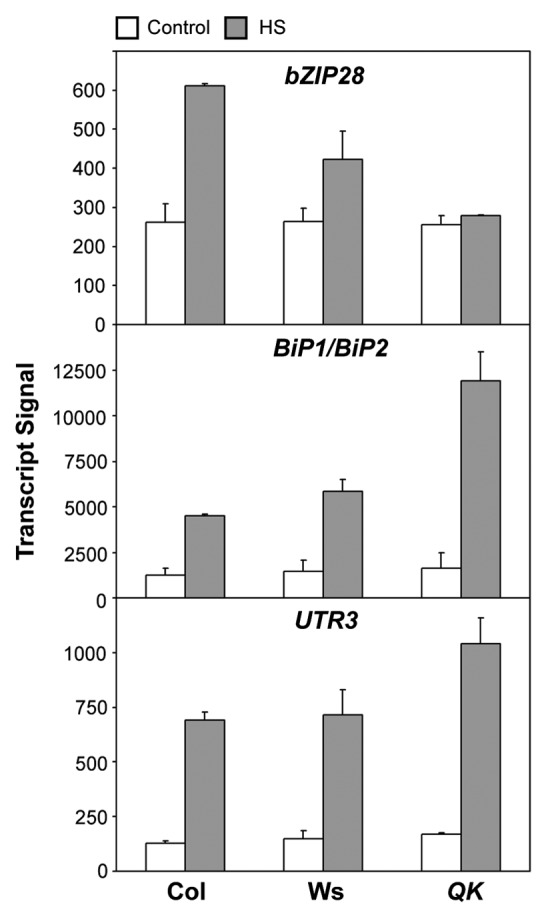
Figure 2. The expression levels of bZIP28, BiP1/BiP2, and UTR3 in the wild type and QK mutant. The data were derived from the microarray results published previously.8 Statistical significance of these data passed the scrutiny of the false discovery rate (FDR) at the stringent level of 0.05.
Taken together, our results suggest that acquired thermotolerance can be conferred in part by the transcription pathways independent of the HsfA1s in Arabidopsis. A simplified working model is proposed to summarize the current knowledge and the points discussed above (Fig. 3). In this model, the HSR is triggered by post-translational modifications of the transcription factors, HsfA1, bZIP28, and possibly others, which then regulate the transcription of HSR genes. It has been well-documented that HSF activity is associated with post-translational modifications in eukaryotes.30-35 In Arabidopsis, phosphorylation36 and sumoylation37,38 of HsfA1a and HsfA2, respectively, have been reported. Activation of bZIP28 by proteolysis has been nicely demonstrated.28,29 The pathways mediated by HsfA1 and bZIP28 can be independently induced at an early stage of the HSR, but may get cross-wired due to the effect of HsfA1 on the expression of bZIP28 and its downstream genes. So far, we do not know whether this is a direct or indirect effect and how this effect could physiologically influence the HSR and thermotolerance in the long run. Further studies are anticipated to address these questions.
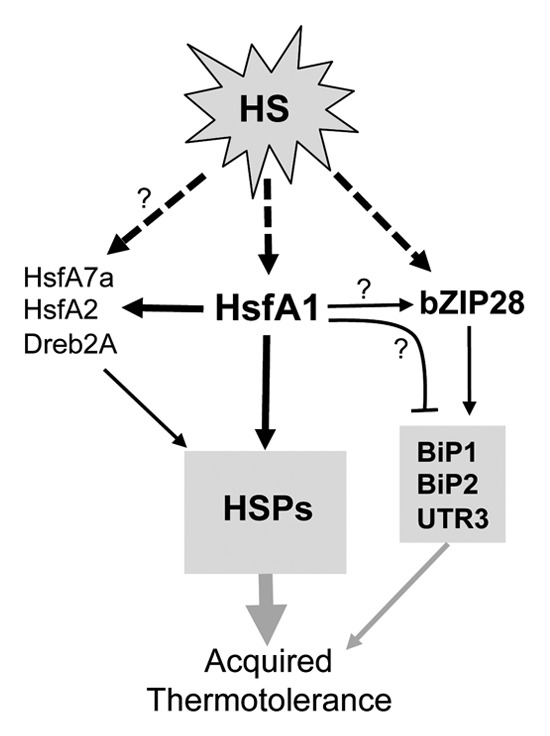
Figure 3. Simplified working model of the transcription network of the HSR in Arabidopsis. HsfA1 represents HsfA1a/b/d. The arrows with dashed lines indicate activation of the transcription factors by posttranslational modifications. The black arrows and a bar-head with solid lines indicate positive and negative transcription regulation, respectively. The question marks denote links to be confirmed. The proteins shown in gray boxes are a sample list of target genes of the transcription factors. The gray arrows of different thicknesses indicate speculative degrees of acquired thermotolerance conferred by different HSR pathways.
Materials and Methods
The plant materials, growth conditions, and thermotolerance assay were described previously.8 The expression data shown in Figure 2 were obtained from the microarray data published previously, which can be accessed in the Gene Expression Omnibus at the National Center for Biotechnology Information (NCBI; accession number GSE26266).8
Acknowledgments
This work was funded by the National Science Council (97–2311-B-001–007-MY3), Taiwan, R.O.C. We thank Dr. AndreAna Peña for English editing. We also thank the anonymous reviewer for the helpful suggestions.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/19803
References
- 1.Lindquist S. The heat-shock response. Annu Rev Biochem. 1986;55:1151–91. doi: 10.1146/annurev.bi.55.070186.005443. [DOI] [PubMed] [Google Scholar]
- 2.Morimoto RI. Regulation of the heat shock transcriptional response: cross talk between a family of heat shock factors, molecular chaperones, and negative regulators. Genes Dev. 1998;12:3788–96. doi: 10.1101/gad.12.24.3788. [DOI] [PubMed] [Google Scholar]
- 3.Wu C. Heat shock transcription factors: structure and regulation. Annu Rev Cell Dev Biol. 1995;11:441–69. doi: 10.1146/annurev.cb.11.110195.002301. [DOI] [PubMed] [Google Scholar]
- 4.Schulz-Raffelt M, Lodha M, Schroda M. Heat shock factor 1 is a key regulator of the stress response in Chlamydomonas. Plant J. 2007;52:286–95. doi: 10.1111/j.1365-313X.2007.03228.x. [DOI] [PubMed] [Google Scholar]
- 5.Jedlicka P, Mortin MA, Wu C. Multiple functions of Drosophila heat shock transcription factor in vivo. EMBO J. 1997;16:2452–62. doi: 10.1093/emboj/16.9.2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McMillan DR, Xiao X, Shao L, Graves K, Benjamin IJ. Targeted disruption of heat shock transcription factor 1 abolishes thermotolerance and protection against heat-inducible apoptosis. J Biol Chem. 1998;273:7523–8. doi: 10.1074/jbc.273.13.7523. [DOI] [PubMed] [Google Scholar]
- 7.Zhang Y, Huang L, Zhang J, Moskophidis D, Mivechi NF. Targeted disruption of hsf1 leads to lack of thermotolerance and defines tissue-specific regulation for stress-inducible Hsp molecular chaperones. J Cell Biochem. 2002;86:376–93. doi: 10.1002/jcb.10232. [DOI] [PubMed] [Google Scholar]
- 8.Liu HC, Liao HT, Charng YY. The role of class A1 heat shock factors (HSFA1s) in response to heat and other stresses in Arabidopsis. Plant Cell Environ. 2011;34:738–51. doi: 10.1111/j.1365-3040.2011.02278.x. [DOI] [PubMed] [Google Scholar]
- 9.Mishra SK, Tripp J, Winkelhaus S, Tschiersch B, Theres K, Nover L, et al. In the complex family of heat stress transcription factors, HsfA1 has a unique role as master regulator of thermotolerance in tomato. Genes Dev. 2002;16:1555–67. doi: 10.1101/gad.228802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yoshida T, Ohama N, Nakajima J, Kidokoro S, Mizoi J, Nakashima K, et al. Arabidopsis HsfA1 transcription factors function as the main positive regulators in heat shock-responsive gene expression. Mol Genet Genomics. 2011;286:321–32. doi: 10.1007/s00438-011-0647-7. [DOI] [PubMed] [Google Scholar]
- 11.von Koskull-Döring P, Scharf K-D, Nover L. The diversity of plant heat stress transcription factors. Trends Plant Sci. 2007;12:452–7. doi: 10.1016/j.tplants.2007.08.014. [DOI] [PubMed] [Google Scholar]
- 12.Banti V, Mafessoni F, Loreti E, Alpi A, Perata P. The heat-inducible transcription factor HsfA2 enhances anoxia tolerance in Arabidopsis. Plant Physiol. 2010;152:1471–83. doi: 10.1104/pp.109.149815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Charng YY, Liu HC, Liu NY, Chi WT, Wang CN, Chang SH, et al. A heat-inducible transcription factor, HsfA2, is required for extension of acquired thermotolerance in Arabidopsis. Plant Physiol. 2007;143:251–62. doi: 10.1104/pp.106.091322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ikeda M, Mitsuda N, Ohme-Takagi M. Arabidopsis HsfB1 and HsfB2b act as repressors of the expression of heat-inducible Hsfs but positively regulate the acquired thermotolerance. Plant Physiol. 2011;157:1243–54. doi: 10.1104/pp.111.179036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kotak S, Vierling E, Bäumlein H, von Koskull-Döring P. A novel transcriptional cascade regulating expression of heat stress proteins during seed development of Arabidopsis. Plant Cell. 2007;19:182–95. doi: 10.1105/tpc.106.048165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kumar M, Busch W, Birke H, Kemmerling B, Nürnberger T, Schöffl F. Heat shock factors HsfB1 and HsfB2b are involved in the regulation of Pdf1.2 expression and pathogen resistance in Arabidopsis. Mol Plant. 2009;2:152–65. doi: 10.1093/mp/ssn095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pajerowska-Mukhtar KM, Wang W, Tada Y, Oka N, Tucker CL, Fonseca JP, et al. The HSF-like Transcription Factor TBF1 Is a Major Molecular Switch for Plant Growth-to-Defense Transition. Curr Biol. 2012;22:103–12. doi: 10.1016/j.cub.2011.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schramm F, Larkindale J, Kiehlmann E, Ganguli A, Englich G, Vierling E, et al. A cascade of transcription factor DREB2A and heat stress transcription factor HsfA3 regulates the heat stress response of Arabidopsis. Plant J. 2008;53:264–74. doi: 10.1111/j.1365-313X.2007.03334.x. [DOI] [PubMed] [Google Scholar]
- 19.Shim D, Hwang J-U, Lee J, Lee S, Choi Y, An G, et al. Orthologs of the class A4 heat shock transcription factor HsfA4a confer cadmium tolerance in wheat and rice. Plant Cell. 2009;21:4031–43. doi: 10.1105/tpc.109.066902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tejedor-Cano J, Prieto-Dapena P, Almoguera C, Carranco R, Hiratsu K, Ohme-Takagi M, et al. Loss of function of the HSFA9 seed longevity program. Plant Cell Environ. 2010;33:1408–17. doi: 10.1111/j.1365-3040.2010.02159.x. [DOI] [PubMed] [Google Scholar]
- 21.Yamanouchi U, Yano M, Lin H, Ashikari M, Yamada K. A rice spotted leaf gene, Spl7, encodes a heat stress transcription factor protein. Proc Natl Acad Sci U S A. 2002;99:7530–5. doi: 10.1073/pnas.112209199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lohmann C, Eggers-Schumacher G, Wunderlich M, Schöffl F. Two different heat shock transcription factors regulate immediate early expression of stress genes in Arabidopsis. Mol Genet Genomics. 2004;271:11–21. doi: 10.1007/s00438-003-0954-8. [DOI] [PubMed] [Google Scholar]
- 23.Hong SW, Vierling E. Mutants of Arabidopsis thaliana defective in the acquisition of tolerance to high temperature stress. Proc Natl Acad Sci U S A. 2000;97:4392–7. doi: 10.1073/pnas.97.8.4392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Queitsch C, Hong SW, Vierling E, Lindquist S. Heat shock protein 101 plays a crucial role in thermotolerance in Arabidopsis. Plant Cell. 2000;12:479–92. doi: 10.1105/tpc.12.4.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Larkindale J, Vierling E. Core genome responses involved in acclimation to high temperature. Plant Physiol. 2008;146:748–61. doi: 10.1104/pp.107.112060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sakuma Y, Maruyama K, Qin F, Osakabe Y, Shinozaki K, Yamaguchi-Shinozaki K. Dual function of an Arabidopsis transcription factor DREB2A in water-stress-responsive and heat-stress-responsive gene expression. Proc Natl Acad Sci U S A. 2006;103:18822–7. doi: 10.1073/pnas.0605639103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reyes F, León G, Donoso M, Brandizzí F, Weber APM, Orellana A. The nucleotide sugar transporters AtUTr1 and AtUTr3 are required for the incorporation of UDP-glucose into the endoplasmic reticulum, are essential for pollen development and are needed for embryo sac progress in Arabidopsis thaliana. Plant J. 2010;61:423–35. doi: 10.1111/j.1365-313X.2009.04066.x. [DOI] [PubMed] [Google Scholar]
- 28.Gao H, Brandizzi F, Benning C, Larkin RM. A membrane-tethered transcription factor defines a branch of the heat stress response in Arabidopsis thaliana. Proc Natl Acad Sci U S A. 2008;105:16398–403. doi: 10.1073/pnas.0808463105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu JX, Srivastava R, Che P, Howell SH. An endoplasmic reticulum stress response in Arabidopsis is mediated by proteolytic processing and nuclear relocation of a membrane-associated transcription factor, bZIP28. Plant Cell. 2007;19:4111–9. doi: 10.1105/tpc.106.050021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ahn SG, Thiele DJ. Redox regulation of mammalian heat shock factor 1 is essential for Hsp gene activation and protection from stress. Genes Dev. 2003;17:516–28. doi: 10.1101/gad.1044503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang XZ, Grammatikakis N, Siganou A, Calderwood SK. Regulation of molecular chaperone gene transcription involves the serine phosphorylation, 14-3-3 epsilon binding, and cytoplasmic sequestration of heat shock factor 1. Mol Cell Biol. 2003;23:6013–26. doi: 10.1128/MCB.23.17.6013-6026.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim S-A, Yoon J-H, Lee S-H, Ahn S-G. Polo-like kinase 1 phosphorylates heat shock transcription factor 1 and mediates its nuclear translocation during heat stress. J Biol Chem. 2005;280:12653–7. doi: 10.1074/jbc.M411908200. [DOI] [PubMed] [Google Scholar]
- 33.Reindl A, Schöffl F, Schell J, Koncz C, Bakó L. Phosphorylation by a cyclin-dependent kinase modulates DNA binding of the Arabidopsis heat-shock transcription factor HSF1 in vitro. Plant Physiol. 1997;115:93–100. doi: 10.1104/pp.115.1.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kline MP, Morimoto RI. Repression of the heat shock factor 1 transcriptional activation domain is modulated by constitutive phosphorylation. Mol Cell Biol. 1997;17:2107–15. doi: 10.1128/mcb.17.4.2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Holmberg CI, Hietakangas V, Mikhailov A, Rantanen JO, Kallio M, Meinander A, et al. Phosphorylation of serine 230 promotes inducible transcriptional activity of heat shock factor 1. EMBO J. 2001;20:3800–10. doi: 10.1093/emboj/20.14.3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu H-T, Gao F, Li G-L, Han J-L, Liu D-L, Sun D-Y, et al. The calmodulin-binding protein kinase 3 is part of heat-shock signal transduction in Arabidopsis thaliana. Plant J. 2008;55:760–73. doi: 10.1111/j.1365-313X.2008.03544.x. [DOI] [PubMed] [Google Scholar]
- 37.Cohen-Peer R, Schuster S, Meiri D, Breiman A, Avni A. Sumoylation of Arabidopsis heat shock factor A2 (HsfA2) modifies its activity during acquired thermotholerance. Plant Mol Biol. 2010;74:33–45. doi: 10.1007/s11103-010-9652-1. [DOI] [PubMed] [Google Scholar]
- 38.Miller MJ, Barrett-Wilt GA, Hua Z, Vierstra RD. Proteomic analyses identify a diverse array of nuclear processes affected by small ubiquitin-like modifier conjugation in Arabidopsis. Proc Natl Acad Sci U S A. 2010;107:16512–7. doi: 10.1073/pnas.1004181107. [DOI] [PMC free article] [PubMed] [Google Scholar]


