Abstract
Stem cell maintenance is essential for growth and development of plants and animals. Similar to animal studies, transcription factors play a critical role in plant stem cell maintenance, however the regulatory logic is not well understood. Shoot apical meristems (SAMs) harbor a pool of pluoripotent stem cells and they provide cells for the development of all above-ground organs. Molecular genetic studies spanning more than a decade have revealed cell-cell communication logic underlying stem cell homeostasis. WUSCHEL (WUS), a homeodomain transcription factor expressed in cells of the organizing center specifies stem cells in overlying cells of the central zone (CZ) and also activates a negative regulator-CLAVATA3 (CLV3). CLV3, a small secreted peptide, binds to CLAVATA1 (CLV1) and also possibly to CLV1-related receptors to activate signaling which restricts WUS transcription. Though the CLV-WUS feedback network explains the cell-cell communication logic of stem cell maintenance, how WUS communicates with adjacent cells had remained elusive. In October 15 2011 issue of Genes and Development, we report that WUS protein synthesized in cells of organizing center migrates into adjacent cells via cell-cell movement and activates CLV3 transcription by directly binding to promoter elements.
Keywords: shoot apical meristem, CLAVATA3, CLAVATA1, central zone and peripheral zone
Shoot apical meristems (SAMs) of Arabidopsis are multilayered structures with three clonally distinct layers of cells.1 The cells in the outermost L1 layer and the sub-epidermal L2 layer divide in anticlinal orientation (perpendicular to the SAM surface), while the underlying L3/corpus forms a multi-layered structure where cells divide in random planes. WUS is expressed in few cells of the L3 layer in a region dubbed as organizing center (OC)/rib-meristem (RM) functions as niche and provides cues to overlying cells of the central zone (CZ) to specify them as stem cells.2 WUS restricts its own transcription by activating CLAVATA3 in cells of the CZ.3,4 Molecular genetic analysis have revealed that maintenance of WUS levels in cells of the OC is critical for stem cell homeostasis.3 Despite several efforts, forward genetic screens have not yielded new molecules that can explain the mode and molecular mechanisms of WUS-mediated cell-cell communication.
Clues about the nature of WUS-mediated stem cell promoting signal originated from analysis of CLV3 function in live imaging experiments.5 This study showed that transient downregulation of CLV3 leads to a sequential and cell by cell radial expansion of CLV3 promoter activity suggesting that WUS-mediated CLV3 activating signal may be a short range diffusible molecule.5 In plants, transcription factor migration across cells forms a short-range non-cell autonomous signaling in patterning developmental fields.6 We tested the possibility of WUS migration across cells as a possible cell-cell communication mechanism. Visualization of WUS protein localization with the help of chimeric GFP:WUS protein revealed a WUS protein gradient that extends from the OC into adjacent cells showing migration of WUS protein.7 We not only detected WUS protein in cells of the CZ but also in cells of the PZ showing both apical and lateral migration of WUS.
Loss of function clv1 alleles and plants experiencing ectopic overexpression of WUS have been shown to over accumulate CLV3 transcripts, however, the molecular basis of this regulation is not understood.3,8 In light of WUS protein migration into cells of the CZ, we tested the possibility of WUS activating transcription of CLV3 by directly binding to its promoter elements. A series of in vivo and in vitro biochemical experiments revealed that WUS binds to CLV3 promoter at multiple positions all of which contained TAAT sequence core and these elements were sufficient to activate transcription of reporter gene in heterologous system.7 Finally, a computational model was developed to explain the significance of WUS protein migration in the context of known regulatory network of stem cell homeostasis.9,10 In silico models with minimal set of interactions were able to explain WUS-mediated modulation of CLV3 expression domain.7
Though this study has solved a long-standing puzzle of cell-cell communication in SAMs, however, it raises several intriguing questions. (1) WUS-mediated activation of CLV3 is a mechanism designed to restrict WUS levels but it does not explain stem cell specification. Again forward genetic approach has not provided many clues. It remains to be seen whether stem cell specification is a systems property that arises as a result of collective behavior of several genes. (2) WUS can bind to CLV3 promoter, then why does it not activate CLV3 in the OC where it is synthesized and in cells of the peripheral zone upon misexpression11? Several possibilities could be envisaged to explain this observation. WUS has been shown to function both as an activator and a repressor of transcription.12,13 In this context, localized expression of co-activators and co-repressors may explain specific activation of CLV3 transcription in cells of the CZ. Cell type specific gene expression map may help in identifying potential localized co-activators and co-repressors that function with WUS.14 Alternately, WUS may be transcriptionally inactive in the OC where it is synthesized whereas acquires transcriptional activity during cell-cell trafficking. (3) WUS protein gradient is important for stem cell homeostasis. What regulates this gradient? Can simple diffusion from the source alone explain the gradient or additional signals within or outside the WUS expression domain/OC regulate gradient? Finding answers to these questions requires new line of work which may involve identifying new molecules that function in the WUS pathway, molecules of protein movement pathways, and also explaining functions of already identified molecules at a higher spatio-temporal resolution.15,16 From descriptive realm, high-resolution analysis with better fluorescent probes is required to understand cell connectivity patterns (symplastic domains) and how they are regulated.17
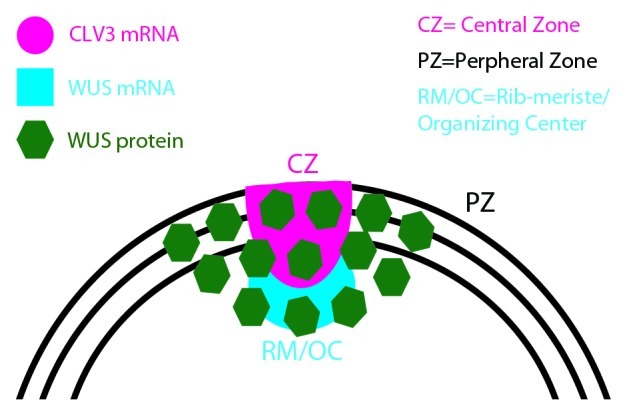
Figure 1. A schematic of shoot apical meristem (SAM) showing distinct functional domains; The CZ/stem cell domain, the PZ/differentiation zone, the RM/OC/cells of the niche. mRNA expression patterns of CLAVATA3 and WUSCHEL indicated along with the spatial pattern of WUSCHEL protein localization.
Acknowledgments
Work in Reddy lab is supported by National Science Foundation grant (IOS-1147250).
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/19793
References
- 1.Steeves TA, Sussex IM. Patterns in Plant Development: Shoot Apical Meristem Mutants of Arabidopsis thaliana. New York: Cambridge University Press 1989. [Google Scholar]
- 2.Mayer KF, Schoof H, Haecker A, Lenhard M, Jürgens G, Laux T. Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell. 1998;95:805–15. doi: 10.1016/S0092-8674(00)81703-1. [DOI] [PubMed] [Google Scholar]
- 3.Schoof H, Lenhard M, Haecker A, Mayer KF, Jürgens G, Laux T. The stem cell population of Arabidopsis shoot meristems in maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell. 2000;100:635–44. doi: 10.1016/S0092-8674(00)80700-X. [DOI] [PubMed] [Google Scholar]
- 4.Brand U, Fletcher JC, Hobe M, Meyerowitz EM, Simon R. Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science. 2000;289:617–9. doi: 10.1126/science.289.5479.617. [DOI] [PubMed] [Google Scholar]
- 5.Reddy GV, Meyerowitz EM. Stem-cell homeostasis and growth dynamics can be uncoupled in the Arabidopsis shoot apex. Science. 2005;310:663–7. doi: 10.1126/science.1116261. [DOI] [PubMed] [Google Scholar]
- 6.Gallagher KL, Paquette AJ, Nakajima K, Benfey PN. Mechanisms regulating SHORT-ROOT intercellular movement. Curr Biol. 2004;14:1847–51. doi: 10.1016/j.cub.2004.09.081. [DOI] [PubMed] [Google Scholar]
- 7.Yadav RK, Perales M, Gruel J, Girke T, Jönsson H, Reddy GV. WUSCHEL protein movement mediates stem cell homeostasis in the Arabidopsis shoot apex. Genes Dev. 2011;25:2025–30. doi: 10.1101/gad.17258511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fletcher JC, Brand U, Running MP, Simon R, Meyerowitz EM. Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science. 1999;283:1911–4. doi: 10.1126/science.283.5409.1911. [DOI] [PubMed] [Google Scholar]
- 9.Gordon SP, Chickarmane VS, Ohno C, Meyerowitz EM. Multiple feedback loops through cytokinin signaling control stem cell number within the Arabidopsis shoot meristem. Proc Natl Acad Sci U S A. 2009;106:16529–34. doi: 10.1073/pnas.0908122106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jönsson H, Heisler M, Reddy GV, Agrawal V, Gor V, Shapiro BE, et al. Modeling the organization of the WUSCHEL expression domain in the shoot apical meristem. Bioinformatics. 2005;21(Suppl 1):i232–40. doi: 10.1093/bioinformatics/bti1036. [DOI] [PubMed] [Google Scholar]
- 11.Yadav RK, Tavakkoli M, Reddy GV. WUSCHEL mediates stem cell homeostasis by regulating stem cell number and patterns of cell division and differentiation of stem cell progenitors. Development. 2010;137:3581–9. doi: 10.1242/dev.054973. [DOI] [PubMed] [Google Scholar]
- 12.Ikeda M, Mitsuda N, Ohme-Takagi M. Arabidopsis WUSCHEL is a bifunctional transcription factor that acts as a repressor in stem cell regulation and as an activator in floral patterning. Plant Cell. 2009;21:3493–505. doi: 10.1105/tpc.109.069997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Perales M, Reddy GV. Stem cell maintenance in Arabidopsis shoot apex. Curr Opin Plant Biol. 2012;15:10–6. doi: 10.1016/j.pbi.2011.10.008. [DOI] [PubMed] [Google Scholar]
- 14.Yadav RK, Girke T, Pasala S, Xie M, Reddy GV. Gene expression map of the Arabidopsis shoot apical meristem stem cell niche. Proc Natl Acad Sci U S A. 2009;106:4941–6. doi: 10.1073/pnas.0900843106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu XM, Wang J, Xuan Z, Goldshmidt A, Borrill PG, Hariharan N, et al. Chaperonins facilitate KNOTTED1 cell-to-cell trafficking and stem cell function. Science. 2011;333:1141–4. doi: 10.1126/science.1205727. [DOI] [PubMed] [Google Scholar]
- 16.Koizumi K, Wu S, MacRae-Crerar A, Gallagher KL. An essential protein that interacts with endosomes and promotes movement of the SHORT-ROOT transcription factor. Curr Biol. 2011;21:1559–64. doi: 10.1016/j.cub.2011.08.013. [DOI] [PubMed] [Google Scholar]
- 17.Bayer E, Thomas C, Maule A. Symplastic domains in the Arabidopsis shoot apical meristem correlate with PDLP1 expression patterns. Plant Signal Behav. 2008;3:853–5. doi: 10.4161/psb.3.10.6020. [DOI] [PMC free article] [PubMed] [Google Scholar]


