Abstract
The development of an arbuscular mycorrhizal (AM) symbiosis is a non-synchronous process with typical mycorrhizal root containing different symbiotic stages at one time. Methods providing cell type-specific resolution are therefore required to separate these stages and analyze each particular structure independently from each other.
We established an experimental system for analyzing specific proteomic changes in arbuscule-containing cells of Glomus intraradices colonized Medicago truncatula roots. The combination of laser capture microdissection (LCM) and liquid chromatography-tandem mass chromatography (LC-MS/MS) allowed the identification of proteins with specific or increased expression in arbuscule-containing cells. Consistent with previous transcriptome data, the proteome of arbuscule-containing cells showed an increased number of proteins involved in lipid metabolism, most likely related to the synthesis of the periarbuscular membrane. In addition, transcriptome data of non-colonized cells of mycorrhizal roots suggest mobilization of carbon resources and their symplastic transport toward arbuscule-containing cells for the synthesis of periarbuscular membranes. This highlights the periarbuscular membrane as important carbon sink in the mycorrhizal symbiosis.
Keywords: Medicago truncatula, arbuscular mycorrhizal symbiosis (AM symbiosis), laser capture microdissection (LCM), liquid chromatography-tandem mass spectrometry (LC/MS/MS), proteomics
Introduction
The arbuscular mycorrhizal symbiosis is a mutualistic association between vascular plants and fungi of the phylum Glomeromycota. The key element of this endosymbiosis is a bi-directional exchange of nutrients and carbohydrates. AM fungi are able to take up mineral nutrients from the soil via the extraradicular hyphal network, which grows through the soil. Phosphate as well as ammonium and nitrate and possibly also other nutrients are transported in the fungal hyphae into the roots. The fungus forms intracellular structures, called arbuscules in the root cortical cells, where the nutrient transfer from fungus to plant takes place. Arbuscule-containing cells are completely reprogramed and highly specialized for symbiotic nutrient transfer.1-4 A novel membrane type, the periarbuscular membrane surrounds the fungal structures and harbors a number of specific transporter proteins.5,6 These plant transporter proteins mediate the uptake of the nutrients from the periarbuscular space into the plant cell cytoplasm.5,7,8 There is currently no evidence for the origin of the precursors for the synthesis of the periarbuscular membrane and the secretion pathway of periarbuscular membrane-specific proteins is unknown.
A recent cell type-specific transcriptome analysis of arbuscule-containing and non-arbuscule-containing cortical cells of mycorrhizal roots showed that the proportion of regulated genes related to lipid metabolism is strongly increased in arbuscule-containing cells,1 which is in line with the de novo development of the periarbuscular membrane in arbuscule containing cells.
The Proteome of Arbuscule-Containing Cells Shows an Elevated Number of Proteins Involved in Lipid Metabolism
To investigate proteome changes in arbuscule-containing cells, we combined LCM with LC-MS/MS to determine the specific protein composition in these symbiotic structures. Using LCM, arbuscule-containing cortical cells (arb) and cortical cells of non-mycorrhizal Medicago truncatula roots (cor), were isolated. Roots sections for LCM were prepared as described previously.1 For each replicate, proteins of 12,000 LCM-isolated cells (arb and cor) were digested with trypsin. These tryptic peptide mixtures were analyzed by LC/MS/MS using nanoflow HPLC (Proxeon Biosystems) and a linear ion trap instrument (LTQ-Orbitrap, Thermo Scientific) as mass analyzer. All data are available at our website: mycproteom.mpimp-golm.mpg.de. Proteins were identified by tandem mass spectrometry (MS/MS) by information-dependent acquisition of fragmentation spectra of multiple-charged peptides. Full scans were obtained at a resolution of full width at half maximum (FWHM) of 60,000. CID spectra were acquired in the LTQ. Spectra were searched against a M. Truncatula protein database (ftp://ftpmips.helmholtz-muenchen.de/plants/medicago/MT_3_0/Mt3.0_proteins_20090702_NAMED.fasta) using the MASCOT algorithm (version 2.2.0; Matrix Science, UK, www.matrixscience.com). The protein abundance was determined by the exponentially modified protein abundance index (emPAI), which was calculated from the number of observed spectra for each protein divided by the number of possible observable peptides.9,10 The emPAI values for each protein in a given cell type were divided by the total sum of emPAI values for all proteins in this cell type to calculate the relative abundance of each protein in the sample (mol %).
From both cell types, arb and cor cells, we could identify altogether 401 M. truncatula proteins with similarities to annotated M. truncatula (Medicago genome version 3.0) (www.medicagohapmap.org) proteins. Additional 99 proteins showed similarities to fungal proteins, and are therefore assumed to be of fungal origin. Of the 401 plant proteins, 53 were exclusively detected in cortical cells of non-mycorrhizal roots, whereas 188 proteins were exclusively found in arbuscule-containing cells.
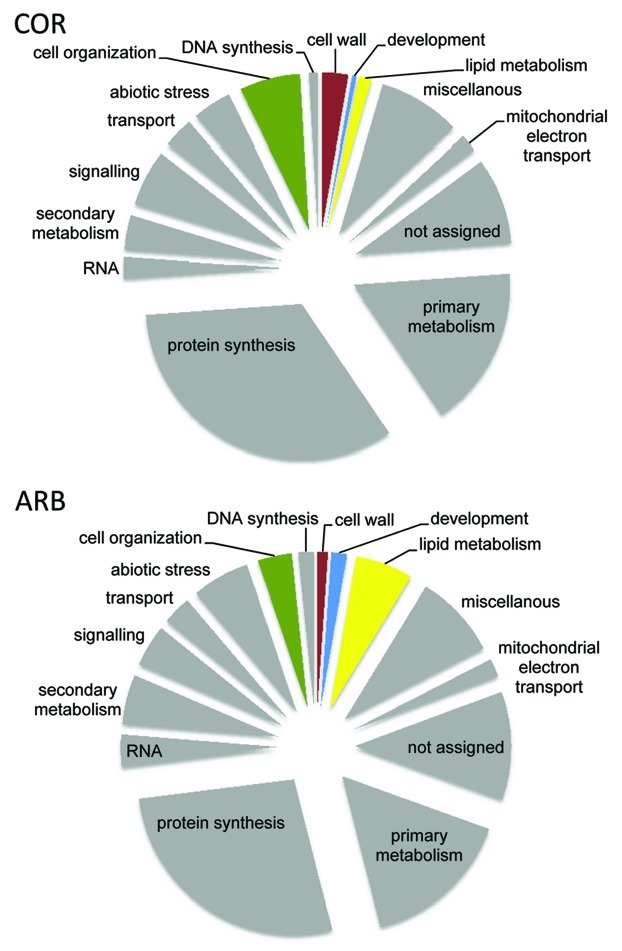
The 401 plant proteins were placed into functional categories using MapMan (Fig. 1). When comparing the relative number of identified proteins of each category between both cell types, we observed an elevated number of proteins involved in lipid metabolism and development in arbuscule-containing cells. This is consistent with the cellular changes during arbuscule development in particular with regard to the periarbuscular membrane synthesis. For example, a number of accumulating proteins involved in the de novo fatty acid synthesis such as a ACC-carboxylase, a biotin carboxylase and a β-ketoacyl synthase were identified (Table 1), which were not detected in LCM-harvested cortical cells of non-mycorrhizal roots. This clearly points to either a specific or a strongly increased expression of these proteins in arbuscule-containing cells. Additionally, several lipases and lipoxygenases participating in lipid breakdown and jasmonate biosynthesis were identified, which is consistent with the increase in jasmonate biosynthesis in arbuscule-containing cells11 and elevated levels of this phytohormone in mycorrhizal roots.12 Of the 16 proteins which were exclusively detected in arb cells, four (medtr4g161110, medtr3g103330, medtr3g033970 and medtr1g125800) were also strongly (log fold change cutoff + 2) transcriptionally induced in arb cells.1
Figure 1. Distribution of M. truncatula proteins identified in LCM-harvested arbuscule-containing cell of mycorrhizal roots (arb) and cortical cells of non-mycorrhizal roots (cor). Functional categories were assigned using MapMan.15
Table 1. Identified proteins related to lipid metabolism in cortical cells of non-mycorrhizal roots and arbuscule-containing cells.
| Identifier | Description | Average cor mol% | Average arb mol% |
|---|---|---|---|
| medtr4g161110 |
CAC2 (acetyl co-enzyme A carboxylase biotin carboxylase subunit) |
0.00 |
0.59 |
| medtr2g025430 |
ACP2 (ACYL CARRIER PROTEIN 2) |
0.57 |
0.57 |
| medtr3g103330 |
3-oxoacyl-(acyl-carrier protein) reductase |
0.00 |
0.52 |
| medtr6g015070 |
BCCP2, CAC1-B | BCCP2 (biotin carboxyl carrier protein 2) |
0.27 |
0.38 |
| medtr3g097100 |
biotin/lipoyl attachment domain-containing protein |
0.00 |
0.27 |
| medtr5g090990 |
biotin/lipoyl attachment domain-containing protein |
0.00 |
0.27 |
| medtr7g012310 |
BCCP2 (biotin carboxyl carrier protein 2) |
0.00 |
0.27 |
| medtr4g165430 |
KAS I (3-ketoacyl-acyl carrier proteon synthaseI, fatty-acid synthase |
0.00 |
0.26 |
| medtr3g033970 |
binding / catalytic/ transferase, Acyl transferase |
0.00 |
0.22 |
| medtr1g125800 |
plastidic pyruvate kinase |
0.00 |
0.22 |
| medtr4g101180 |
ENR1 (enoyl-[acyl-carrier-protein] reductase); (NADH)/ oxidoreductase |
0.00 |
0.20 |
| medtr3g115120 |
transketolase family protein |
0.00 |
0.17 |
| medtr2g126370 |
phospholipase D |
0.10 |
0.10 |
| medtr8g071040 |
ACCD (carboxytransferase β subunit of the Acetyl-CoA carboxylase (ACCase) complex in plastids) |
0.00 |
1.44 |
| medtr3g101350 |
lipid-associated family protein |
0.00 |
1.45 |
| medtr8g088830 |
GDSL-motif lipase/hydrolase family protein |
0.00 |
0.42 |
| medtr3g101380 |
lipid-associated family protein |
0.00 |
0.40 |
| medtr8g021380 |
LOX1 (Lipoxygenase 1) |
0.00 |
0.07 |
| medtr8g021550 |
LOX5 (Lipoxygenase 5) |
0.07 |
0.35 |
| medtr8g021590 |
LOX1 (Lipoxygenase 1) |
0.27 |
0.22 |
| medtr8g021650 |
LOX1 (Lipoxygenase 1) |
0.00 |
0.27 |
| medtr8g021730 | LOX1 (Lipoxygenase 1) | 0.17 | 0.16 |
Transcript Data Provide Evidence for Mobilization of Carbohydrate Resources in Non-Colonized Cells of Mycorrhizal Roots
The increased percentage of proteins involved in lipid synthesis in the proteome of arbuscule-containing cells is most likely related to the de novo synthesis of the periarbuscular membrane. The production of such an extensive novel membrane system is likely to require a vast amount of carbohydrate resources, making arbuscule-containing cells particularly strong carbon sinks. However, we did not find evidence for induction of carbon transporters or internal mobilization of carbon resources in arbuscule-containing cells.1 Instead, we found more evidence for a mobilization of carbohydrate resources reflected by the regulation of genes related to primary metabolism and carbohydrate transport in non-colonized cells of mycorrhizal roots as compared with cells of non-mycorrhizal controls1 (Fig. 2). In arbuscule-containing cells, we observed the downregulation of a neutral invertase along with an induction of a sucrose synthase. In contrast, in non-arbuscule-containing cortical cells in the vicinity of arbuscules, amylase transcripts are induced, indicating a mobilization of sugar resources. In addition, genes for a putative sucrose transporter (MtSut4) and for a hexose transporter (MtHext1) showed enhanced RNA accumulation in non-colonized cells of mycorrhizal roots, which may be involved in the export of sucrose from the vacuole and in the uptake of hexose from the apoplast into the cytosol. Thus both would also lead to a mobilization of carbohydrate resources in cells in the vicinity of arbuscules.1 Consistent with the increased expression of amylase and neutral invertase encoding transcripts this suggests that carbohydrate resources are supplied by non-colonized cortical cells of mycorrhizal roots for periarbuscular membrane synthesis in the arbuscule- containing cells.
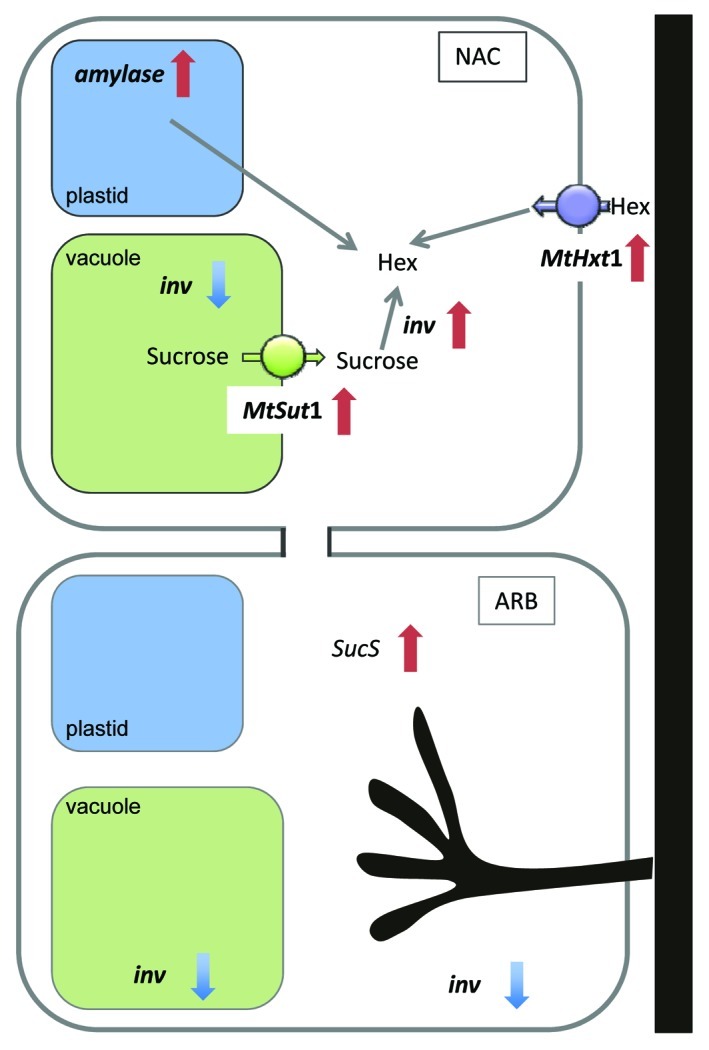
Figure 2. Graphic representation of transcriptomic changes of genes related to major carbohydrate metabolism and carbohydrate transport identified by transcriptome analysis of LMD-isolated arbuscule-containing cell of mycorrhizal roots (arb) and non-arbuscule-containing cells of mycorrhizal roots (nac) as compared with cortical cells (cor) of non-mycorrhizal roots (Gaude et al).1 In nac cells, induced transcript levels of an amylase (medtr3g150290), an invertase (medtr1g122220) (inv), a putative vacuolar sucrose transporter (MtSut1) and a hexose transporter of the plasma membrane (MTHext1) point to a mobilization and import of hexoses (hex) in the cytoplasm. In arb cells, decreased transcript levels of an invertase (medtr1g122200) and of a sucrose synthase (medtr6g090350) (SucS) were observed. A putative vacuolar invertase (medtr8g106250) was downregulated in both cell types of mycorrhizal roots.
Evidences for Symplastic Transport Toward Arbuscule-Containing Cells
Mobilization of carbohydrates in one group of cells and subsequent consumption of these carbohydrates in adjacent cells would require transport of carbon compounds. As increased expression of sugar uptake transporters could not be detected in arbuscule-containing cells as mentioned above, symplastic rather than apoplastic import of carbon must be suggested. The assumption for such a symplastic transport is supported by the occurrence of plasmodesmata with an increased diameter between cells with arbuscules and their neighbors.13 In addition, previous studies of Medicago roots clearly showed that in root nodules, which also represent a strong carbohydrate sink, symplastic rather than apoplastic transport is mediating the exchange of carbon compounds between cells.14 We therefore hypothesize symplastic carbohydrate transfer toward arbuscule-containing cells for periarbuscular membrane synthesis and probably also for delivering carbohydrates to the AM fungus. If mineral nutrients transferred from the fungus to the plant are also transported via the symplastic route from the arbuscule-containing to the adjacent cells and further to the central cylinder remains an open question.
Conclusion
Combined cell-specific proteome and transcriptome analyses suggest that carbohydrates are mobilized in the cortex of mycorrhizal roots and delivered via the symplastic route to arbuscule-containing cells. Here, they are not only transferred to the fungal symbiont, but are also used for the development of the periarbuscular membrane. This means that a notable part of the carbohydrates delivered to the mycorrhizal sink remains in the plant and the carbohydrate loss to the fungus should not be overestimated. The fact that arbuscule-containing cells have a limited life span and resources used for periarbuscular membrane synthesis are recycled during arbuscule degradation could be one of the reasons for the mostly positive outcome of the mycorrhizal symbiosis.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/19650
References
- 1.Gaude N, Bortfeld S, Duensing N, Lohse M, Krajinski F. Arbuscule-containing and non-colonized cortical cells of mycorrhizal roots undergo extensive and specific reprogramming during arbuscular mycorrhizal development. Plant J. 2012;69:510–28. doi: 10.1111/j.1365-313X.2011.04810.x. [DOI] [PubMed] [Google Scholar]
- 2.Guether M, Balestrini R, Hannah M, He J, Udvardi MK, Bonfante P. Genome-wide reprogramming of regulatory networks, transport, cell wall and membrane biogenesis during arbuscular mycorrhizal symbiosis in Lotus japonicus. New Phytol. 2009;182:200–12. doi: 10.1111/j.1469-8137.2008.02725.x. [DOI] [PubMed] [Google Scholar]
- 3.Gomez SK, Javot H, Deewatthanawong P, Torres-Jerez I, Tang Y, Blancaflor EB, et al. Medicago truncatula and Glomus intraradices gene expression in cortical cells harboring arbuscules in the arbuscular mycorrhizal symbiosis. BMC Plant Biol. 2009;9:10. doi: 10.1186/1471-2229-9-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hogekamp C, Arndt D, Pereira PA, Becker JD, Hohnjec N, Küster H. Laser microdissection unravels cell-type-specific transcription in arbuscular mycorrhizal roots, including CAAT-box transcription factor gene expression correlating with fungal contact and spread. Plant Physiol. 2011;157:2023–43. doi: 10.1104/pp.111.186635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Javot H, Penmetsa RV, Terzaghi N, Cook DR, Harrison MJ. A Medicago truncatula phosphate transporter indispensable for the arbuscular mycorrhizal symbiosis. Proc Natl Acad Sci U S A. 2007;104:1720–5. doi: 10.1073/pnas.0608136104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pumplin N, Harrison MJ. Live-cell imaging reveals periarbuscular membrane domains and organelle location in Medicago truncatula roots during arbuscular mycorrhizal symbiosis. Plant Physiol. 2009;151:809–19. doi: 10.1104/pp.109.141879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Benedito VA, Li H, Dai X, Wandrey M, He J, Kaundal R, et al. Genomic inventory and transcriptional analysis of Medicago truncatula transporters. Plant Physiol. 2010;152:1716–30. doi: 10.1104/pp.109.148684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rausch C, Daram P, Brunner S, Jansa J, Laloi M, Leggewie G, et al. A phosphate transporter expressed in arbuscule-containing cells in potato. Nature. 2001;414:462–70. doi: 10.1038/35106601. [DOI] [PubMed] [Google Scholar]
- 9.Ishihama Y, Oda Y, Tabata T, Sato T, Nagasu T, Rappsilber J, et al. Exponentially modified protein abundance index (emPAI) for estimation of absolute protein amount in proteomics by the number of sequenced peptides per protein. Mol Cell Proteomics. 2005;4:1265–72. doi: 10.1074/mcp.M500061-MCP200. [DOI] [PubMed] [Google Scholar]
- 10.Schulze WX, Usadel B. Quantitation in mass-spectrometry-based proteomics. Annu Rev Plant Biol. 2010;61:491–516. doi: 10.1146/annurev-arplant-042809-112132. [DOI] [PubMed] [Google Scholar]
- 11.Hause B, Mrosk C, Isayenkov S, Strack D. Jasmonates in arbuscular mycorrhizal interactions. Phytochemistry. 2007;68:101–10. doi: 10.1016/j.phytochem.2006.09.025. [DOI] [PubMed] [Google Scholar]
- 12.Hause B, Maier W, Miersch O, Kramell R, Strack D. Induction of jasmonate biosynthesis in arbuscular mycorrhizal barley roots. Plant Physiol. 2002;130:1213–20. doi: 10.1104/pp.006007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blee KA, Anderson AJ. Regulation of arbuscule formation by carbon in the plant. Plant J. 1998;16:523–30. doi: 10.1046/j.1365-313x.1998.00315.x. [Review] [DOI] [Google Scholar]
- 14.Complainville A, Brocard L, Roberts I, Dax E, Sever N, Sauer N, et al. Nodule initiation involves the creation of a new symplasmic field in specific root cells of medicago species. Plant Cell. 2003;15:2778–91. doi: 10.1105/tpc.017020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Usadel B, Poree F, Nagel A, Lohse M, Czedik-Eysenberg A, Stitt M. A guide to using MapMan to visualize and compare Omics data in plants: a case study in the crop species, Maize. Plant Cell Environ. 2009;32:1211–29. doi: 10.1111/j.1365-3040.2009.01978.x. [DOI] [PubMed] [Google Scholar]