Abstract
The expansins comprise a family of proteins that appear to be involved in the disruption of the noncovalent bonds between cellulose microfibrils and cross-linking glycans, thereby promoting wall creep. To understand better the expansion process in Petunia hybrida (petunia) flowers, we isolated a cDNA corresponding to the PhEXP1 α-expansin gene of P. hybrida. Evaluation of the tissue specificity and temporal expression pattern demonstrated that PhEXP1 is preferentially expressed in petal limbs during development. To determine the function of PhEXP1, we used a transgenic antisense approach, which was found to cause a decrease in petal limb size, a reduction in the epidermal cell area, and alterations in cell wall morphology and composition. The diminished cell wall thickness accompanied by a reduction in crystalline cellulose indicates that the activity of PhEXP1 is associated with cellulose metabolism. Our results suggest that expansins play a role in the assembly of the cell wall by affecting either cellulose synthesis or deposition.
INTRODUCTION
The final morphology of plant organs is achieved through a tight regulation of cell division and cell expansion (Meyerowitz, 1997). The direction and magnitude of primary cell wall extension appear to be primary determinants of the expansion pattern and, thus, of the final shape and size that cells assume (Kotilainen et al., 1999, Martin et al., 2001; Smith, 2003).
The involvement of phytohormones such as auxin, gibberellins, cytokinins, ethylene, and brassinolides in polar cell elongation processes has been clearly demonstrated (Shibaoka and Nagai, 1994; Szekeres et al., 1996; Creelman and Mullet, 1997; Kende and Zeevaart, 1997; Kieber, 1997). Recently, genetic proof has been obtained that shows that directed cell expansion is dependant on cellulose synthesis and deposition (Fagard et al., 2000; Schindelman et al., 2001; Pagant et al., 2002). Proteins in the cell wall are believed to play important roles in regulation of cell wall extensibility, which is a key parameter in determining cell expansion. Among the cell wall proteins studied to date, expansins are unique in their ability to induce immediate cell wall extension in vitro and cell expansion in vivo (Brummell et al., 1999; Cosgrove, 1999; Cho and Cosgrove, 2000; Pien et al., 2001; Choi et al., 2003). Expansins are classified as primary wall loosening agents, referring to their capacity to induce stress relaxation of the cell wall in a pH dependent manner, possibly by disrupting hydrogen bonds that link cellulose and hemicellulose wall components. This results in the slippage between these polymers leading to secondary water absorption by the cell and, therefore, in expansion of the cell wall (Cosgrove, 1997b). The purification and sequencing of the Cucumis sativus (cucumber) expansin protein enabled the isolation of the first expansin gene (Shcherban et al., 1995). Subsequently, homologous genes were identified in gymnosperms and in both monocots and dicots among the angiosperms. Examples include Lycopersicon esculentum (tomato) leaves (Keller and Cosgrove, 1995), Avena sativa (oat) coleoptiles (Cosgrove and Li, 1993), Zea mays (maize) roots (Wu et al., 1996), Nicotiana tabacum (tobacco) cell cultures (Link and Cosgrove, 1998), and various fruits (Civello et al., 1999; Rose et al., 2000). Expansin genes appear to be highly conserved throughout plant evolution (Cosgrove, 2000a), whereas closely related expansin-like sequences also have been found in Dictyostelium discoideum, suggesting that these cell wall proteins have a very deep evolutionary origin (Li et al., 2002).
Two families of expansin genes have been recognized: namely, the α-expansins, which were discovered first, and the β-expansins. These two expansin families have only ∼20% amino acid identity but share several conserved motifs. Members of both families have similar wall-loosening activities (Cosgrove, 1997b, 2000a). Expansins occur as multigene families in Arabidopsis thaliana, Oryza sativa (rice), C. sativus, L. esculentum, Z. mays, and other species in which they have been examined in detail (Cosgrove, 1999; Wu et al., 2001b). The A. thaliana and O. sativa α-expansin family is larger than the β-expansin family (Cosgrove, 2000b; Choi et al., 2003). The large number of expansin genes—38 in A. thaliana (Li et al., 2002), at least 30 in Z. mays (Wu et al., 2001a), and 80 in O. sativa (Li et al., 2003)—suggests multiple developmental or tissue-specific roles for these proteins. Although a substantial body of work has been performed in the identification and characterization of the expansin genes, only limited data are available that define their precise biological roles during plant development.
Several reports have examined the presence and activity of expansin proteins or the expression of expansin genes in growing tissues (such as C. sativus hypocotyls, deepwater O. sativa internodes, Gossypium hirsutum [cotton] fibers, xylem cells in Zinnia elegans [zinnia] and Populus trichocarpa [poplar], and adventitious roots in Pinus taeda [pine]), shoot meristems, and developing fruits as mediators of plant cell growth (Shcherban et al., 1995; Cho and Kende, 1997; Fleming et al., 1997; Orford and Timmis, 1998; Reinhardt et al., 1998; Sterky et al., 1998; Hutchison et al., 1999; Im et al., 2000). Expansins also are expressed in nongrowing tissues, such as ripening fruits (Rose et al., 1997; Brummell et al., 1999; Civello et al., 1999), and have been associated with developmental events, such as ripening, abscission (Cosgrove, 1997a), and endosperm weakening (Chen and Bradford, 2000). The role of expansin in the growth and modification of the cell wall also has been evaluated by manipulating the expression of expansin genes in transgenic plants. In particular, altered expression of LeEXP1 points to its involvement in fruit softening (Brummell et al., 1999) and modification of AtEXP3 expression highlighted its role in the control of leaf growth and pedicel abscission (Cho and Cosgrove, 2000). Transient local microinduction of CsEXP1 in N. tabacum meristem induced morphogenesis, leading to the formation of phenotypically normal leaves, whereas CsEXP1 induction at early stages of leaf development altered local lamina growth and produced leaves with altered morphology (Pien et al., 2001). Transgenic O. sativa plants in which the expansin gene OsEXP4 was downregulated by an antisense/sense approach were shorter on average than control plants, whereas transgenic plants with inducible OsEXP4 expression showed a close correlation between OsEXP4 protein levels and seedling growth (Choi et al., 2003).
In a previous study, we defined a developmental map of Petunia hybrida (petunia) petals by analyzing the expression of in situ cell cycle markers and by reconstructing the pattern of cell expansion through measurement of cell size (Reale et al., 2002). During flower development, floral organs typically assume their final shape long after the major mitotic activity has ceased, indicating that cell expansion plays an important role in determining organ shape (Pyke et al., 1991; Tsuge et al., 1996). To integrate expansin expression in this map, we have cloned the P. hybrida α-expansin gene PhEXP1 and analyzed its tissue specificity and temporal expression pattern. Our results show that PhEXP1 is preferentially expressed in petal limbs during development. Moreover, to analyze the in vivo function of PhEXP1, P. hybrida was transformed with an antisense construct of PhEXP1 cDNA under the control of the constitutive 35S promoter. This allowed for the demonstration that downregulation of PhEXP1 influences the amount of crystalline cellulose of plant cell walls and affects cell expansion, area, and shape, leading to phenotypic changes of petal limbs.
RESULTS
Isolation of a P. hybrida α-Expansin cDNA
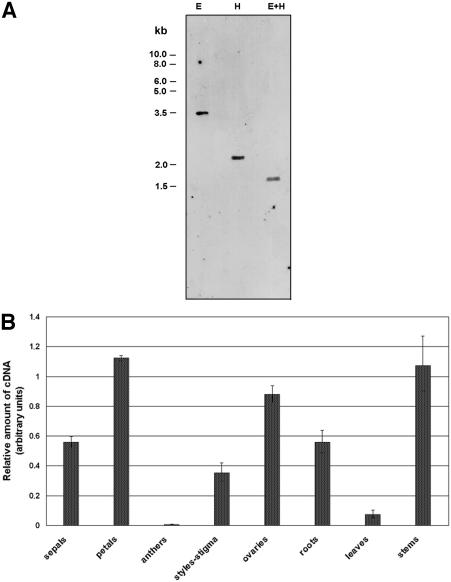
Based on the sequences corresponding to conserved regions of α-expansin proteins, two degenerate oligonucleotides were designed and used for PCR of an ovary-specific cDNA library of P. hybrida. A single fragment of ∼500 bp was amplified and cloned. DNA sequence analysis indicated that this fragment coded for a peptide sharing high amino acid identity with known α-expansins. To isolate a full-length cDNA, the 500-bp fragment was used as a probe to screen the ovary-specific library. Of the 4 × 105 clones screened, six were selected on the basis of their hybridization signals, and after a second round of purification, only the clone that gave rise to the strongest hybridization signal was selected for further analysis. The resulting cDNA was 1250 bp in length and contained a 780-bp open reading frame expected to encode a protein of 260 amino acids with a molecular mass of 25 kD. Alignment with expansin sequences revealed that the predicted amino acid sequence exhibits all of the anticipated characteristics of α-expansins. Therefore, this P. hybrida cDNA was termed PhEXP1. A cDNA probe was generated by amplification of the 3′ untranslated region of PhEXP1 and used for DNA gel blot analysis. At high stringency, a single hybridization signal was observed, indicating that PhEXP1 is present in the P. hybrida genome as a single copy gene and that the probe is gene specific (Figure 1A).
Figure 1.
PhEXP1 Is a Single Copy Gene in the P. hybrida Genome and Is Expressed at Various Levels in Different P. hybrida Organs.
(A) DNA gel blot analysis of genomic DNA. Ten micrograms of DNA was digested with EcoRI (E), HindIII (H), and EcoRI and HindIII (E+H), separated on a 0.8% agarose gel, and subjected to DNA gel blot analysis. A purified PCR product from the 3′ untranslated region of PhEXP1 was labeled with fluorescein-11-dUTP by random priming and used as a probe.
(B) Analysis of the expression pattern in different organs of PhEXP1 by semiquantitative real-time RT-PCR. Total RNA was extracted from wild-type plants grown under standard greenhouse conditions. Analyses were performed with specific primers homologous to the 3′ untranslated region of PhEXP1. Error bars represent standard deviation.
PhEXP1 Is Preferentially Expressed in Petal Limbs
The developmental expression pattern of PhEXP1 was studied in different organs using semiquantitative, real-time reverse transcriptase (RT) PCR. High transcript levels of PhEXP1 were detected in petals, elongating stems, and ovaries, and intermediate levels were observed in sepals, roots, and stigmas and styles. In leaves, PhEXP1 mRNA levels were very low, and in anthers, the transcript levels were barely detectable (Figure 1B). A developmental map of P. hybrida petals previously has been defined that describes the patterns of cell division and expansion throughout 14 different developmental stages (Reale et al., 2002). During the first three stages of petal development, growth is sustained by cell divisions that are uniformly distributed in the apical and basal portion of the petal. Starting from stage 4 (4 mm of petal length), the expansion of petal cells begins in the basal portion and then gradually extends to the apical portion (Reale et al., 2002).
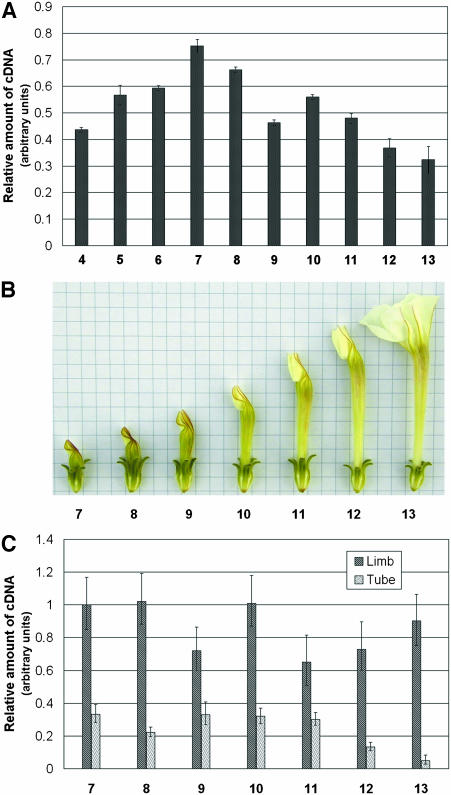
To position PhEXP1 in this map, we first analyzed PhEXP1 expression at the same developmental stages, starting from stage 4. PhEXP1 transcripts were detectable throughout the entire period of petal development. Expression levels increased during development until stage 7 (15 to 19.5 mm of petal length), decreased until stage 9, reaching a second peak at stage 10, and subsequently decreased again to a minimum level at stage 13 (Figure 2A). Notably, the accumulation of PhEXP1 reaches a peak at stages 7 and 10. These stages mark the onset of cell expansion in mesophyll of petal limbs (stage 7) and the expansion of the epidermal cells involved in flower unfolding (stage 10). PhEXP1 expression also was analyzed separately in the tube and limb petal subportions from stages 7 to 13 of petal development (Figure 2B). At these stages, the tube and limb are morphologically distinct and, therefore, they can be dissected manually. Throughout development, PhEXP1 expression levels always were significantly higher (t test, P < 0.01) in the limb than in the tube, and PhEXP1 transcript levels were ∼3.5 times higher in the limb with respect to the tube (Figure 2C).
Figure 2.
Analysis of the Temporal Expression Pattern of PhEXP1 by Semiquantitative Real-Time RT-PCR.
Total RNA was extracted from wild-type plants grown under standard greenhouse conditions. Analyses were performed with specific primers homologous to the 3′ untranslated region of PhEXP1.
(A) Temporal expression of PhEXP1 during petal development. RNA was extracted from petals sampled at stages 4 to 13 of P. hybrida flower development as described by Reale et al. (2002).
(B) P. hybrida var Mitchell flowers at stages 7 to 13 of development.
(C) Temporal expression of PhEXP1 during limb and tube development. RNA was extracted from petals sampled at stages 7 to 13 of P. hybrida flower development.
Error bars represent standard deviation.
PhEXP1 Antisense Plants Show Phenotypic Changes in Petal Limbs
Real-time RT-PCR studies suggested that PhEXP1 is preferentially expressed in petal limbs. Based on this observation, it was hypothesized that PhEXP1 may play a role in regulating cell expansion in petal limbs. To test this hypothesis, we cloned PhEXP1 cDNA downstream of a 35S promoter of Cauliflower mosaic virus (CaMV) in the antisense orientation.
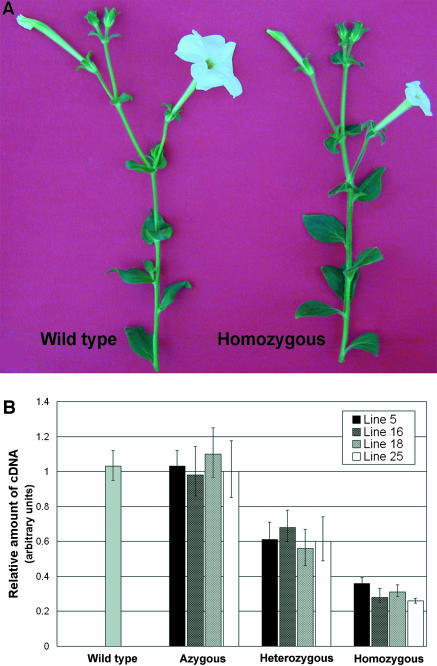
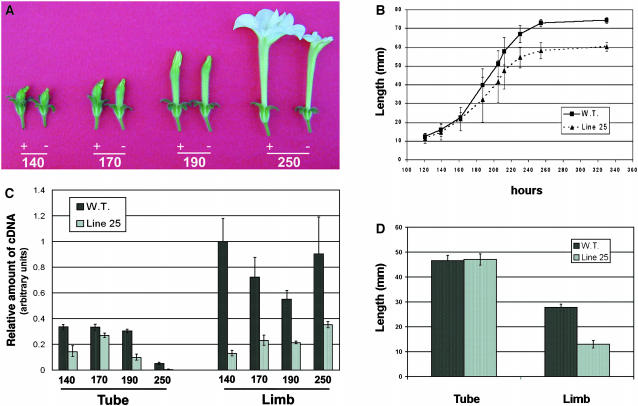
The resulting construct (PhEXP1 antisense) was introduced into P. hybrida (var Mitchell) using Agrobacterium tumefaciens–mediated gene transfer (Horsch et al., 1985). Seeds resulting from self-pollination of the transformants (T0) were scored for kanamycin resistance. Four T1 progenies giving a 3:1 (resistant:susceptible) segregation of the kanamycin marker were identified, and 10 plants for each progeny were phenotypically analyzed under standard greenhouse conditions. In the four T1 families analyzed, plants with the same morphological alterations affecting petal size were observed. In particular, in families 18 and 25, alterations affected 2 out of 10 plants, whereas in families 5 and 16, alterations affected 3 out of 10 plants. Segregation analysis in T2 and subsequent generations indicated that the mutant phenotype was found only in individuals that had become homozygous for the transgene. Plant size, habit, growth rate, phyllotaxis, length of internodes, and the number, shape, and size of leaves of homozygous plants were indistinguishable from those of the wild type (Figure 3A). No morphological alterations were observed in any other floral organ. The pistil exhibited normal style, stigma, and ovary without any visible morphological alterations; likewise, the anthers and stamen filaments were normal, pollen was viable, and megagametogenesis proceeded normally in the ovary, and homozygous plants were fertile and formed viable seeds (data not shown). At anthesis, sepals appeared normal in the homozygous plants, whereas the size of the petal was highly reduced in comparison to the wild type (Figure 3A).
Figure 3.
Phenotypical Alteration of the Homozygous Transgenic Plant and PhEXP1 Expression in Segregating T1 Families.
(A) Homozygous transgenic plant compared with the wild type.
(B) Analysis of the PhEXP1 expression in wild-type, azygous, heterozygous, and homozygous T1 plants by semiquantitative real-time RT-PCR. Total RNA was extracted from petal pools collected at 140, 180, and 250 h AFBA. Error bars represent standard deviation.
To verify whether the alteration observed in petal size correlated with the downregulation of PhEXP1, plants of the four selected families azygous, heterozygous, and homozygous for the transgene were analyzed by semiquantitative real-time RT-PCR and compared with the wild type (Figure 3B). Similar results were obtained in the four families analyzed. PhEXP1 transcript levels were lowered on average by 75% in homozygous plants (t test, P < 0.01) and by 40% in heterozygous plants (t test, P < 0.01) in comparison to the wild type (Figure 3B). No significant difference was observed between azygous and wild-type plants (t test, P < 0.01). Taken together, these results showed that the reduction of PhEXP1 expression depended on the dose of the transgene, suggesting that the flower phenotype appeared when a threshold level of transcript reduction was overcome. Therefore, homozygous transgenic plants were considered to be PhEXP1 antisense silenced plants.
Because there were no variations in the flower phenotype among homozygous antisense plants, we chose homozygous plants belonging to one plant line (line 25) for further analysis.
Expansin Downregulation Is Specific of PhEXP1
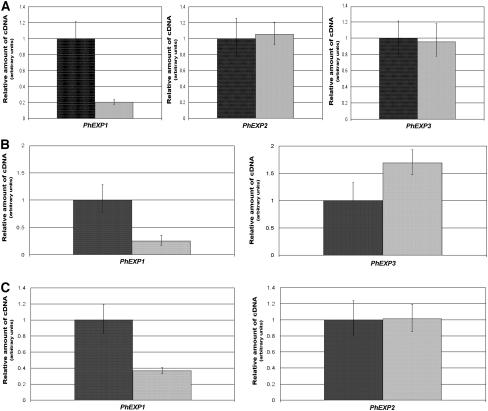
Because expansin genes are highly conserved, the question arose of whether other expansin genes also were downregulated in phenotypically altered antisense plants. Therefore, we analyzed the expression of PhEXP1 and two other α-expansin–encoding genes, namely PhEXP2 and PhEXP3, in antisense and wild-type plants. PhEXP2 and PhEXP3 are members of the P. hybrida expansin gene family that have been isolated and characterized. PhEXP2 is expressed in petals and sepals, whereas PhEXP3 expression was detected in petals and ovaries (S. Zenoni and M. Pezzotti, unpublished data). The transcript levels of these genes were analyzed by semiquantitative, real-time RT-PCR. Expression analysis was performed in petals for PhEXP1, PhEXP2, and PhEXP3, in ovaries for PhEXP1 and PhEXP3, and in sepals for PhEXP1 and PhEXP2 in line 25 and wild-type plants (Figure 4). Pools of petals, ovaries, and sepals were collected and analyzed at three different stages of development: 140, 180, and 250 h after flower bud appearance (AFBA). In the antisense line, PhEXP1 expression levels were significantly lowered by 75% in petals and ovaries (t test, P < 0.01) (Figures 4A and 4B) and by 65% in sepals (t test, P < 0.01) (Figure 4C) in comparison to the wild type. The expression levels of the other expansin genes, PhEXP2 and PhEXP3 in petals (Figure 4A) and PhEXP2 in sepals (Figure 4C), were unaffected by the presence of the transgene. In antisense ovaries, PhEXP3 expression levels significantly increased (t test, P < 0.01) compared with wild-type ovaries (Figure 4B). Thus, it is reasonable to conclude that the mutant phenotype is specifically attributable to partial gene silencing of the endogenous PhEXP1 gene (Van Houdt et al., 2000; Kim et al., 2001; Kapoor et al., 2002).
Figure 4.
Specific Transcript Level Reduction of PhEXP1.
Total RNA was extracted from petals, ovaries, and sepals. Each sample represents pools of material collected at 140, 180, and 250 h AFBA. Semiquantitative real-time RT-PCR analyses were performed with specific primers homologous to the 3′ untranslated region of PhEXP1, PhEXP2, and PhEXP3.
(A) Semiquantitative real-time RT-PCR analysis of PhEXP1, PhEXP2, and PhEXP3 transcript levels in wild-type (dark gray) and line 25 (light gray) petals.
(B) Semiquantitative real-time RT-PCR analysis of PhEXP1 and PhEXP3 transcript levels in wild-type (dark gray) and line 25 (light gray) ovaries.
(C) Semiquantitative real-time RT-PCR analysis of PhEXP1 and PhEXP2 transcript levels in wild-type (dark gray) and line 25 (light gray) sepals.
Error bars represent standard deviation.
PhEXP1 Downregulation Is Responsible for Reduction of Petal Limb Size
To determine if the rate of petal growth also was affected in the antisense line, corolla length of wild-type and line 25 flowers was measured during flower development on a temporal basis (hours AFBA). No differences were detected until 187 h AFBA, after which the petal of the antisense line showed a decreased growth rate that resulted, at anthesis, in a significant reduction (t test, P < 0.01) of 14 mm in length compared with the wild type (Figures 5A and 5B). To determine the petal subportion to which this difference was attributable, tube and limb length of the wild type and line 25 were measured at anthesis (Figure 5D). The length of the petal limb, but not that of the tube, was significantly reduced (t test, P < 0.01) in the antisense line in comparison to the wild type.
Figure 5.
Morphological Alteration during Petal Development in Line 25 Is Correlated with a Reduction in PhEXP1 Transcript Levels.
(A) Wild-type (+) and line 25 (−) flowers at 140, 170, 190, and 250 h AFBA.
(B) Petal length comparison at 120 to 340 h AFBA of wild-type and line 25 plants.
(C) Analysis of PhEXP1 transcript levels by semiquantitative real-time RT-PCR of wild-type and line 25 petal tube and limb at 140, 170, 190, and 250 h AFBA.
(D) Tube and limb length at anthesis of the wild type and line 25.
Error bars represent standard deviation.
To analyze how the reduction in expression of PhEXP1 occurred in tube and limb during petal development, PhEXP1 transcript accumulation was studied by real-time RT-PCR (Figure 5C). Tube and limb RNAs of the antisense line 25 were analyzed separately and compared with the wild type throughout development. PhEXP1 transcript levels were significantly reduced in both tube (t test, P < 0.05) and limb (t test, P < 0.01) of the antisense line in comparison to the wild type (Figure 5C). In general, the reduction in PhEXP1 expression was more pronounced in the limb than in the tube throughout petal development. Taken together, these results indicate a clear correlation between the decreased size of petal limb observed in the antisense line 25 plants and the reduction in PhEXP1 expression in the same flower portion.
Petal Size Reduction Is Correlated with a Decrease in Epidermal Cell Size
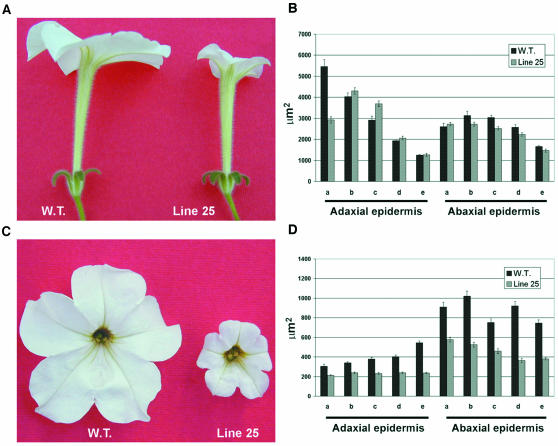
To verify whether the reduced size of the petal limbs in the antisense plants correlates with differences in the size of epidermal cells, petals of antisense line 25 and wild-type flowers were microscopically compared at anthesis (Figures 6A and 6C). Tube and limb tissues were divided into five subsamples, starting from the basal part of the organ. Length, width, and surface area of cells on both the adaxial and the abaxial epidermises were measured for each tube and limb subsample (Figures 6B and 6D). No significant differences (t test, P < 0.01) were observed in the cell surface area of the tube subsamples in either epidermis between antisense and wild-type plants (Figure 6B). In the five limb subsamples, cell surface areas of the adaxial and abaxial epidermises of antisense plants were significantly smaller (t test, P < 0.01) than the corresponding cell areas of wild-type plants (Figure 6D). Total limb surface also was calculated, revealing that wild-type limbs were 3.7 times larger than those of line 25.
Figure 6.
Morphological Alteration of Line 25 Petals Is Correlated with Reduction of the Epidermal Cell Surface in the Limb Portion of Line 25.
(A) and (C) Comparison between wild-type and line 25 petal tubes (A) or petal limbs (C) at anthesis.
(B) and (D) Cell surface of the adaxial and abaxial epidermises of wild-type and line 25 petal tubes (B) and petal limbs (D). Both tube and limb epidermises were divided into five subportions (a, b, c, d, and e), starting from the basal part of the organ. Error bars represent standard error; n = 80.
To determine whether limb size differences also were attributable to alteration of cell number, we calculated the total cell number constituting the abaxial limb epidermis of antisense line 25 and wild-type plants. The total cell number averaged 1,041,847 in wild-type plants and 1,009,192 in antisense line 25. Thus, there were no significant alterations in cell number (t test, P < 0.01).
Epidermal Cells of Antisense Petals Show Characteristic Morphology
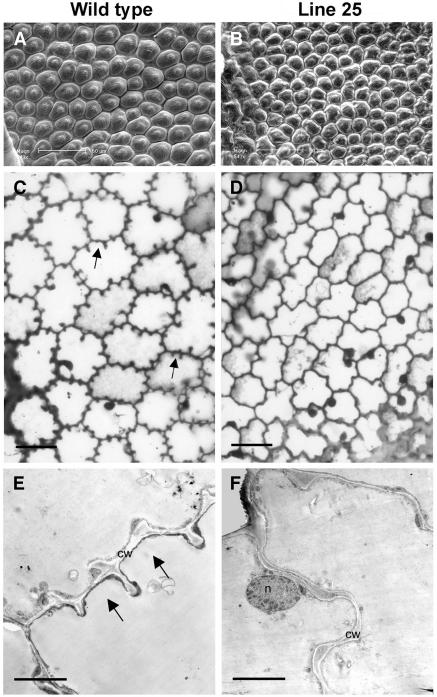
The external morphology of cells in the abaxial epidermis limb of wild-type and antisense plants at anthesis was analyzed by scanning electron microscopy. Cells of the abaxial epidermis of wild-type petals usually have a pentagonal base, and the outer walls are strongly convex with a conical tip in the center (Figure 7A). The corresponding cells of the antisense line 25 were smaller with a less-pronounced conical tip (Figure 7B).
Figure 7.
Changes in Cell Shape in Line 25.
(A) and (B) Scanning electron micrographs of abaxial epidermal limb cells at anthesis. Bars = 50 μm.
(C) and (D) Tangential semithin sections of abaxial epidermis of petal limb at anthesis. Arrows indicate the characteristic cell wall lobes. Bars = 25 μm.
(E) and (F) Transmission electron micrographs of tangential ultrathin sections of the abaxial epidermis of petal limb at anthesis. cw, cell wall; n, nucleus. Bars = 5 μm.
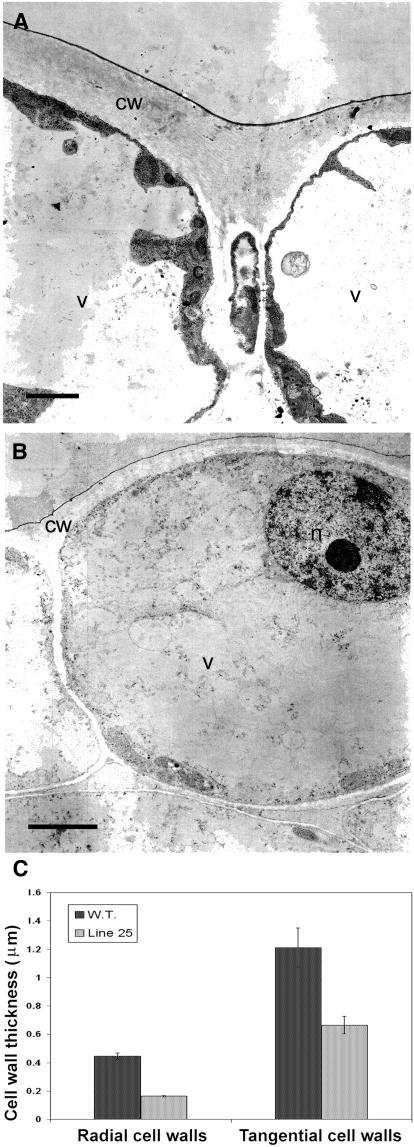
To further study the differences between the antisense line 25 and wild-type plants, tangential sections of limb abaxial epidermis were analyzed by light microscopy. Differences in cell areas were evident, and antisense line 25 cells were smaller than wild-type cells (Figures 7C and 7D). Wild-type cells usually had a nearly isodiametric shape that was uniformly lobed (Figure 7C), whereas antisense cells were more irregularly shaped, and lobes were almost completely absent (Figure 7D). Transmission electron microscopy observation of ultrathin tangential sections of limb abaxial epidermis confirmed and highlighted cell wall morphology differences between the wild type (Figure 7E) and the antisense line (Figure 7F). Tangential and radial cell walls appeared thinner in the antisense line compared with the wild type (Figures 8A and 8B). Wild-type radial and tangential cell walls were 2.8 and 1.8 times thicker than the corresponding cell walls of the antisense line, respectively (Figure 8C).
Figure 8.
Changes in Cell Wall Thickness in Line 25.
(A) and (B) Transmission electron micrographs of transversal sections of abaxial epidermal limb cells at anthesis of the wild type (A) and line 25 (B). c, cytoplasm; cw, cell wall; n, nucleus; v, vacuoles. Bars = 2 μm.
(C) Thickness of radial cell walls and tangential cell walls in the wild type and line 25. Error bars represent standard errors; n = 20 different cells from 20 different micrographs. Each value is the average of three measurements in one cell.
Downregulation of PhEXP1 Reduces the Amount of Crystalline Cellulose in Epidermal Cell Walls
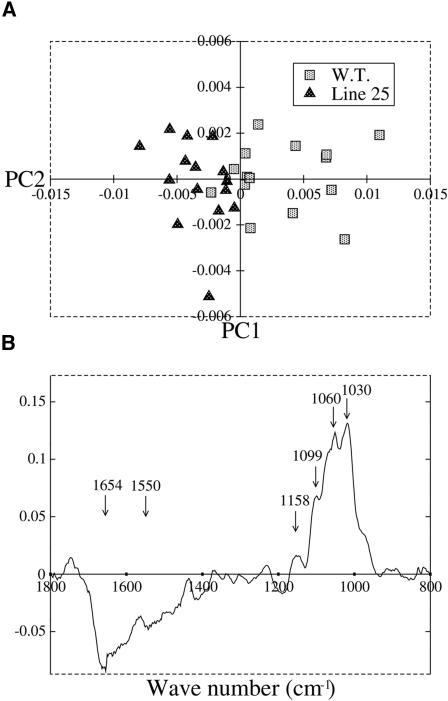
Limb abaxial epidermal cells also were analyzed by Fourier transform infrared (FTIR) spectroscopy. This technique has been successfully used to rapidly analyze the composition of plant cell walls (Chen et al., 1998; Fagard et al., 2000; Carpita et al., 2001a, 2001b). To reveal the major spectral differences between the wild type and the antisense line, a principal component analysis (PCA) was applied to all spectra (Chen et al., 1998; Kemsley, 1998).
PCA demonstrated that wild-type spectra can be separated from antisense line 25 spectra using a combination of two principal component (PC) scores (Figure 9A). PC1 score explained 68.32% of the variance. The loading for PC1 (Figure 9B) showed characteristics of purified cellulose in the fingerprint region (peaks at 1038, 1064, 1100, and 1162 cm−1) and of protein (peaks at 1650 and 1550 cm−1). Peaks at 1650 and 1550 cm−1 correlated negatively with the cellulose fingerprint peaks. The PC scores of the spectra of the line 25 petal abaxial epidermis samples were negative relative to the mean (Figure 9A). These results indicate that the petal epidermis cell walls of the antisense line are relatively richer in protein and poorer in cellulose than the cells of the wild type. Thus, the reduction in thickness of the antisense line cell walls may be attributed to a deficiency in crystalline cellulose.
Figure 9.
Line 25 Petal Limb Cell Walls Are Cellulose Deficient.
(A) FTIR analysis of petal limb cell walls. PCA was performed using 15 FTIR spectra from the wild type and line 25. PC1 explained 68.3% of the variance between spectra from wild-type plants and spectra from line 25 plants.
(B) The PC1 loading showed positive peaks characteristic of cellulose in the fingerprinting region (1158, 1099, 1060, and 1030 cm−1), indicating that line 25 cell walls were deficient in cellulose relative to the wild type. Two other major peaks (1654 and 1550 cm−1) appeared negatively in this PC loading. These peaks corresponded to amide groups of proteins, suggesting an enrichment of proteins in line 25 walls relative to the wild type.
To confirm the observed changes chemically, cell walls were isolated from wild-type and line 25 petal limbs for crystalline cellulose quantification, which was determined as cell wall material resistant to acid hydrolysis. Measurements of four independent replicates showed that there was a significant difference (t test, P < 0.01) between wild-type limbs, which had a mean value of 95 ± 6 μg cellulose (mg cell wall)−1, and line 25 limbs, which had a mean value of 63 ± 2 μg cellulose (mg cell wall)−1 (Table 1). In conclusion, chemical analysis of the cell wall of line 25 limbs confirmed the reduction in crystalline cellulose observed by FTIR microspectroscopy, suggesting that PhEXP1 directly or indirectly plays a role in cellulose metabolism.
Table 1.
Crystalline Cellulose Analysisa
| Sample | Average OD (490 nm) | Glucose Quantity (μg) | Cell Wall Starting Material (mg) | Glucose Quantity (μg)/Cell Wall Starting Material (mg) |
|---|---|---|---|---|
| Wild type A | 1.257 | 162 | 1.8 | 90 |
| Wild type B | 1.434 | 185 | 1.8 | 103 |
| Wild type C | 1.223 | 158 | 1.7 | 93 |
| Wild type D | 1.301 | 168 | 1.8 | 93 |
| Line 25 A | 0.880 | 112 | 1.8 | 62 |
| Line 25 B | 1.041 | 133 | 2.0 | 66 |
| Line 25 C | 0.941 | 120 | 1.9 | 63 |
| Line 25 D | 0.912 | 116 | 1.9 | 61 |
Cellulose quantity was determined as described by Schindelman et al. (2001).
DISCUSSION
PhEXP1 Is Preferentially Expressed in Petal Limbs and Exhibits a Temporal Expression Pattern That Correlates with the Expansion Phases of Petal Development
Analysis of the tissue specificity of PhEXP1 indicated that it is expressed at various levels in different P. hybrida organs, although a very high level of expression was observed in petals. During petal development, PhEXP1 transcript levels were modulated in two distinct induction phases that were coincident with the expansion of mesophyll cells of the petal limb (stage 7) and with expansion of the epidermal cells of the limb (stage 10) involved in flower unfolding (Reale et al., 2002). These results suggest a correlation between PhEXP1 transcript levels and petal cell enlargement during development. The levels of expression of PhEXP1 detected in petal tubes were significantly lower than those in the limb. Mesophyll and epidermal tube cells contribute to the determination of petal shape and size by anisotropic growth, whereas those of the limb grow isotropically (Reale et al., 2002). These growth patterns account for the observation that epidermal cells are highly elongated in the tube and are nearly isodiametric in the limb. Because PhEXP1 is more expressed in the limb than in the tube of developing petals, we hypothesize a possible involvement for it in isotropic growth.
Specific Silencing of PhEXP1 Leads to a Reduction in the Size of Petal Limbs
A transgenic approach was used to study the role of PhEXP1 in plant development and growth. A portion of PhEXP1 cDNA was cloned in a reverse orientation under control of the 35S CaMV promoter to decrease the endogenous transcript content. Antisense expression of PhEXP1 resulted in a novel phenotype, characterized by a severe reduction of the petal size, which correlated with a significant, specific reduction of PhEXP1 transcript levels. No visible alterations affected other plant organs, in spite of the fact that some of them also showed a significant reduction in PhEXP1 transcript levels. Expression analysis demonstrated a specific diminution of PhEXP1 transcript levels in petals, ovaries, and sepals, whereas the levels of PhEXP2 and PhEXP3 transcripts were not significantly lowered by an antisense approach in these organs. This suggests that PhEXP1 plays a key role in the growth and development of the petal, whereas in other organs, its function may be less important. A decrease in the expression of PhEXP1 in ovaries and sepals may have subtle effects that might be compensated for by other members of the expansin family. In addition, comparative analysis of PhEXP1 expression in the limb and tube during development of the wild type and line 25 revealed that PhEXP1 transcript levels were reduced in both petal subportions, although drastic reduction was observed only in the limb. Because the tube was not morphologically affected by the antisense approach in line 25 and PhEXP1 transcript levels in the tube are lower than they are in limbs in wild-type plants, it can be hypothesized that, at least in the tube, PhEXP1 does not play a fundamental role in cell expansion. Alternatively, it could be functionally replaced by other expansin genes. Therefore, we propose that PhEXP1 can play diverse roles in different plant organs and distinct organ subportions (tube and limb) that are characterized by distinct patterns of cell growth (anisotropic and isotropic).
It has been suggested that growth properties in organs with an extended lamina, such as petals, are determined primarily by gene activity in the epidermal layer, which sets the size of the outer skin and is coordinated with the growth of the internal cells through signaling (Vincent et al., 2003). According to this view, PhEXP1 could have the role of modulator of the epidermal cell expansion levels, which determine the final shape of petals.
Our data indicate that the antisense expression of PhEXP1 affects cell expansion but not cell division. Measurements of total cell number and surface areas in the abaxial epidermis of petal limb in the wild type and in line 25 revealed that although the total cell number remained unaltered, the cell surface was significantly reduced. Therefore, as previously observed (Cho and Cosgrove, 2000), altered expression of an expansin gene does not interfere with the cell cycle, and the reduction in epidermal cell area accounted for the size differences in the petal limb between antisense and wild-type plants.
PhEXP1 Antisense Expression Affects Cell Wall Morphology and Composition
A reduction in the epidermis cell area was coupled with modifications of cell morphology. Moreover, FTIR and chemical analyses showed a reduced deposition of crystalline cellulose in the epidermal cells of the antisense line.
A. thaliana cell wall biosynthesis genes have been isolated and characterized in depth (reviewed in Martin et al., 2001; Reiter, 2002), including the functional analysis of cellulose synthase genes by forward genetics. Mutants at the PROCUSTE1 (PRC1) locus showed decreased cell elongation that correlated with cellulose deficiency (Fagard et al., 2000). Cells of the prc1 mutant have broken fragments of walls that are believed to form because the walls are thinner than those of wild-type cells. Moreover, a mutant allele of the cellulose synthase gene AtCesA7 (Zhong et al., 2003) resulted in reduced cellulose content, decreased primary wall thickness, and reduced leaf epidermal cell size. Thus, mutations in the catalytic subunits of cellulose synthase, which plays a key role in the synthesis of cellulose microfibrils in the primary cell wall, inhibit growth by limiting the supply of cellulose for wall extension. The cell wall alterations in PhEXP1 antisense plants are very similar to the above-mentioned A. thaliana cellulose synthase mutant phenotypes, which relate expansin downregulation with reduction of cellulose biosynthesis affecting cell wall assembly and morphology. Additional proteins also have been studied for their involvement in wall assembly and cell expansion. A. thaliana KORRIGAN (KOR) and KOBITO1 (KOB1) encode a plasma membrane–bound endo-1,4-β-d-glucanase and a novel protein possessing a putative N-terminal membrane anchor, respectively (Nicol et al., 1998; Pagant et al., 2002). Mutations in KOR altered the cellulose–xyloglucan network and reduced the number of the cellulose microfibrils affecting the cell wall composition and assembly, resulting in a dwarf phenotype caused by reduced cell expansion. kob1 mutants were cellulose-deficient, with microfibrils randomized and occluded by a layer of pectic material causing a reduction of cell elongation. Therefore, to obtain cell expansion, it is fundamental to have a cell wall that is correctly assembled through coordinated activity of cellulose synthase genes as well as the activity of gene products that prepare wall regions for deposition of microfibrils, thus playing a role in the coordination between cellulose synthesis and cell expansion.
The main role of expansins as cell wall loosening agents has been largely demonstrated (Cosgrove, 2000b). Our results confirm the role of the expansins in cell wall extensibility, but also indicate that downregulation of expansin altered cell wall morphology by reducing the cellulose content. It is likely that the downregulation of the expansin gene interferes with the correct wall region preparation for microfibril deposition, eventually leading to disorganized cellulose deposition and consequent degradation. Furthermore, in line 25, cell lobes were absent and cell wall thickness was significantly reduced in comparison to the wild type. It has been reported that the emergence of lobes is consistently preceded by the reorganization of the cortical microtubules into a series of bands that are associated with the formation of local cellulosic wall thickenings. Lobes subsequently emerge in the thinner regions of the wall, which are more extensible, as the cell expands under the force of the turgor pressure (Smith, 2003). The PhEXP1 antisense line 25 phenotype suggests that the absence of lobes in the epidermal cells could be explained by reduced formation of local cellulosic wall thickenings and/or reduction of relaxation of the cell wall.
Expansin mechanism of action is still poorly understood, but a working model has been proposed (Cosgrove, 1999). In this model, the protein binds tightly with a binding domain to the cell wall, apparently to noncrystalline surface regions of cellulose, and restricts mobility. The putative catalytic domain is likely to function by disrupting the noncovalent bonding between wall polysaccharides, either at the surface of the cellulose microfibrils or more distantly in the matrix between microfibrils. It has been reported that expansin LeEXP1 is involved in fruit cell wall metabolism by potential relaxation of the cell wall directly and a possible control of the access of enzymes that contribute to cell wall polymer modification or depolymerization (Brummell et al., 1999). According to this view, expansins allow the action of enzymes that achieve the precise disassembly of particular cell wall components, and according to our data, they also permit the action of a battery of wall synthesizing and modifying enzymes that collectively regulate cell wall assembly and, therefore, cell size and shape.
Because the walls of plant cells are complex composites of cellulose, cross-linking glycans, proteins, and pectic substances, the quantitative and qualitative modification of one of the complex components can have some effect on the others, as observed in cellulose biosynthesis mutants (Fagard et al., 2000; Pagant et al., 2002).
FTIR spectroscopy is a remarkably efficient means of screening for cell wall mutants, and PC loadings derived are more or less specific to the carbohydrate fingerprinting region of the spectrum. Unfortunately, specific changes at any frequency to a specific sugar or polysaccharide cannot be assigned (Chen et al., 1998). FTIR data on petal epidermal peels oriented our effort toward quantitative analysis of crystalline cellulose, deficiency of which was chemically demonstrated in the antisense line. However, relative changes of wall pectin and hemicellulose also could be responsible for the phenotype observed.
We propose that expansins may have at least two roles. First, they may act in the disruption of the noncovalent bonds between cellulose microfibrils and cross-linking glycans, thereby promoting wall creep. Second, expansins may have a role in the preparation of the cell wall for cellulose deposition by separating wall matrix during expansion by a yet undefined mechanism. Expansins appear to have a key function in the regulation of cell expansion by coordinating cellulose synthesis, deposition, and spatial organization in relation to other polymers and protein constituents of the cell wall.
METHODS
Plant Materials and Growth Conditions
The P. hybrida var Mitchell and transgenic plants were grown under normal greenhouse conditions. To analyze the segregation, T1 seeds were germinated on selective MS medium (Sigma, St. Louis, MO) supplemented with kanamycin (100 mg/L).
Cloning of PhEXP1
A cDNA for PhEXP1 was isolated by screening an ovary cDNA library of P. hybrida constructed in Uni-ZAP XP (Stratagene, La Jolla, CA). Two degenerate oligonucleotides were designed based on the amino acid sequences conserved in plant expansins: a forward primer EXP1 (5′-GGIGCI(A/C)G(C/T)GGITA(C/T)GGIAA-3′) and a reverse primer EXP2 (5′-TGCCA(A/G)TT(C/T)TGICCCCA(A/G)TT-3′). PCR was performed using the cDNA library as a template (1 μL of 1 × 106 plaque forming units/μL) in a volume of 50 μL containing 200 μM deoxynucleotide triphosphate, 0.4 μM of each primer, 50 mM KCl, 10 mM Tris-HCl, pH 8.3, 1.5 mM MgCl2, and 0.01% (w/v) gelatin. After PCR, the amplified product (∼500 bp) was gel purified and subcloned into the Topo TA cloning vector (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. A single clone was sequenced by the dideoxy method of Sanger (Sanger et al., 1977). Approximately 3.5 × 106 plaque forming units from a P. hybrida ovary library were screened under high stringency conditions using the 32P-radiolabeled 500-bp fragment as a probe according to standard procedures (Sambrook et al., 1989). In vivo excision of the plaque-purified cDNA clones in pBluescript KSII+ was performed as described by the manufacturer (Stratagene). The cDNA sequence was determined by the Sanger method (Sanger et al., 1977).
DNA Isolation and Gel Blot Analysis
Genomic DNA was isolated from P. hybrida mature leaves as described by Souer et al. (1995). Genomic DNA (10 μg) was digested with EcoRI, HindIII, and EcoRI and HindIII overnight, separated on a 0.8% agarose gel, and blotted onto a Hybond-N+ membrane (Amersham Biosciences, Piscataway, NJ). Blots were hybridized to a probe labeled with Gene Images random-prime labeling module (Amersham Biosciences). The PhEXP1 specific probe was amplified by PCR with primers corresponding to the 3′ untranslated region of PhEXP1 (5′- GTACTACTAGCTCATAATTTC-3′ and 5′-GCAAATTGTAACTAGTAGAAAC-3′). Hybridization was visualized using the CDP-Star detection module, and Hyperfilm ECL (Amersham Biosciences) was used for autoradiography.
RNA Isolation and Analysis
Total cellular RNA from various P. hybrida organs was extracted with Trizol reagent (Invitrogen) and treated with DNase I (Invitrogen). cDNA was obtained from 0.8 μg of total RNA using the SuperScript first-strand synthesis system (Invitrogen) with the oligo(dT)12-18 as the primer.
To analyze organ-specific expression patterns of the expansin gene, total RNA was extracted from petals, sepals, anthers, ovaries, styles and stigma, stems, leaves, and roots. Each sample represents pools of material collected at three different developmental stages. For the temporal expression patterns of PhEXP1 in petals, total RNA was extracted from flowers at stages 4 to 13 of P. hybrida flower development as described by Reale et al. (2002). For the analysis of PhEXP1 transcript expression levels in wild-type, azygous, heterozygous, and homozygous plants, total RNA was extracted from petals of material collected at three different developmental stages calculated in hours (140, 180, and 250) AFBA. To verify the specificity of PhEXP1 antisense RNA, total RNA was extracted from petals, ovaries, and sepals representing pools of material collected at three different developmental stages (140, 180, and 250 h AFBA). For the analysis of PhEXP1 transcript expression levels in homozygous transgenic plants during petal development, total RNA was extracted separately from both limb and tube portions of petals collected at 140, 170, 190, and 250 h AFBA.
Semiquantitative Real-Time RT-PCR
Semiquantitative real-time RT-PCR was performed using the Gene Amp 5700 sequence detection system (Applera, Foster City, CA) with SYBR Green PCR Master Mix reagent (Applera). Specific primers for PhEXP1, PhEXP2, and PhEXP3 were designed in the 3′ untranslated region: EXP1 1A (5′-AGCTCCTACACTTCTCTTCCA-3′) and EXP1 2A (5′-GGAAAAGTAAGGTTCTTATTATGAGGT-3′); EXP2 1A (5′-TGTTTTATGATTTCTTCCCTCTTAAA-3′) and EXP2 2A (5′-GCATATTACTATTCCTATTCCGAATGT-3′); and EXP3 1A (5′-GATTTTGAGGGTATTGCTATTAGATTGT-3′) and EXP3 2A (5′-CCACCTCTGCTTACTCAGGC-3′).
All of the real-time RT-PCR experiments were performed with two independent sets of RNA samples. For the experiments reported in Figure 2A, data derived from three independent sets of RNA samples. For each sample, three replicates were performed in a final volume of 50 μL containing 1 μL of cDNA, 0.2 μm of each primer, and 25 μL of 2× SYBR Green PCR Master Mix according to the manufacturer's instructions. All PCRs were performed using the Gene Amp 5700 sequence detection system for 10 min at 94°C and then 40 cycles consisting of 30 s at 94°C, 20 s at 52°C, and 30 s at 72°C. All quantifications were normalized to actin cDNA fragments amplified in the same conditions by primers ACT1 (5′-ATCCCAGTTGCTGACAATAC-3′) and ACT2 (5′-GGCCCGCCATACTGGTGTGAT-3′). Each PCR product was cloned and sequenced to confirm the specificity of the real-time RT-PCR approach. Moreover, each real-time assay was tested by a dissociation protocol to ensure that each amplicon was a single product.
The data were organized according to the comparative method described in User Bulletin 2 (Applera). Data were submitted to statistical analysis (t test).
Construction of the Binary Vector and Plant Transformation
The binary vector pBIN19 containing fbp2 cDNA (Angenent et al., 1994) was digested with BamHI and XhoI to excise the MADS box gene. A fragment of 780 bp containing PhEXP1 cDNA was amplified by PCR using two oligonucleotide primers: 5′-CCCTCGAGGGGAATTCCAGGTGTTTATAGTGG-3′ and 5′-CGGGATCCCGTTAAATTCTAAAGCTTCTTTGCCC-3′. The underlined regions denote the XhoI and BamHI sites, respectively. After digestion with both enzymes, the cDNA region was ligated in pBIN19.
The chimeric construct was transformed via A. tumefaciens (LBA4404) into P. hybrida var Mitchell using the leaf disk transformation method (Horsch et al., 1985). Regeneration of transformants was performed as described by van Tunen et al. (1989). After regeneration on selective medium with kanamycin, transformed P. hybrida lines were checked for the presence of the transgene by PCR performed using the following primers: 35S (5′-GCCGACAGTGGTCCCAAAGATG-3′) and ANTI (5′-GCCCGCAGCATTCTTAGAACAATA-3′). The T2 and T1 plants were obtained by self-pollination of the T0 PhEXP1 antisense primary transformants.
Epidermis Cell Area and Total Cell Number Measurement
The petals of wild-type and antisense line 25 plants were collected at anthesis and divided into limbs and tubes. Both limb and tube were subdivided, starting from the basal part of the organ, into five portions termed a, b, c, d, and e. Petal epidermises were peeled, and both adaxial and abaxial epidermises were analyzed by light microscopy. To measure cell length, width, and area, Leica QWin image analysis software (Leica Microsystems, Heerbrugg, Switzerland) was used.
To measure the total surface of petal limbs, 10 limbs from wild-type and antisense plants were collected at anthesis and digitally photographed with a Nikon Coolpix 950 (Nikon, Tokyo, Japan). Total limb surface was calculated using Image-Pro Plus image analysis software (Media Cybernetics, Carlsbad, CA). To measure the total cell number, two portions of each petal limb were analyzed by light microscopy. The area and number of abaxial epidermal cells were calculated using Image-Pro Plus image analysis software. The average cell number per surface unit of wild-type and antisense line 25 plants was multiplied by the total area. Data regarding total cell number was elaborated by statistical analysis (t test).
Light Microscopy, Transmission Electron Microscopy, and Scanning Electron Microscopy
Corollas were dissected from flowers at anthesis and divided into small portions at different distances from the margin. Samples were fixed in glutaraldehyde (5% [v/v]) overnight at room temperature and postfixed in OsO4 (1% [w/v]) for 4 h (both in 0.075 M cacodylate buffer, pH 7.2). After dehydration with a graded ethanol series and propylene oxide, samples were embedded in Epon resin (2-dodecenylsuccinic anhydride and methylnadic anhydrid mixture) (Loreto et al., 2001). Semithin (1 to 2 μm thick) and ultrathin (70 to 90 nm thick) sections were cut with an OmU2 ultramicrotome (Reichert, Heidelberg, Germany) equipped with a glass blade or a diamond blade, respectively. The semithin sections were stained with toluidine blue and mounted in Eukitt for light microscopy observation. Photomicrographs were taken using a Leica DMR HC photomicroscope (Leica Microsystems). The ultrathin sections were mounted on uncoated copper grids (200 mesh) and were contrasted by adding uranyl acetate and an aqueous solution of lead nitrate before observation with a TEM 400 T transmission electron microscope (Philips, Eindhoven, The Netherlands).
For scanning electron microscopy, petal portions dissected from flower buds at anthesis were fixed for 3 h at room temperature in 5% glutaraldehyde in 0.07 M sodium cacodylate buffer, postfixed for 1 h in buffered 1% OsO4, and dehydrated in a graded ethanol series. After critical point drying in liquid CO2, samples were mounted on aluminum stubs, sputter coated with gold, and viewed with the Stereoscan 90B (Cambridge Instruments, Cambridge, UK).
FTIR Spectroscopy
Abaxial epidermal peels of limb petals at anthesis (15 for each condition) were peeled and dried at 37°C for 1 h. An area of epidermis (3 mm2) was selected for spectral collection. Sixty-four interferograms were collected on a MAGNA-IR Nicolet 760 (Nicolet Instrument Technologies, Madison, WI) spectrometer using 4-cm−1 resolution in the spectral range between 700 and 2500 cm−1 for each sample. Because absorbance varies with sample thickness, all data sets were corrected for baseline and normalized for area. PCA was performed with Win-DAS software (E.K. Kemsley, Institute of Food Research, Norwich, UK). This statistical method allows for characterization of samples by their scores on a small number of variables, PCs, which are ordered in terms of decreasing variance. PC scores can be plotted against one another to reveal clustering or structure in the data set. It also is possible to mathematically derive a spectrum related to the PC score (termed a PC loading) to identify the molecular factors responsible for the separation of groups of spectra.
Reference infrared absorption spectra of cellulose and other β-glucans were obtained from the literature (Tsuboi, 1957; Liang and Marchessault, 1959; Séné et al., 1994).
Cellulose Analysis
Petal limbs at anthesis weighing 0.5 to 1.5 g were frozen at −80°C, and quantification of crystalline cellulose was performed as described in Schindelman et al. (2001).
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession numbers AY487167, AY487168, and AY487169.
Acknowledgments
Urs Fischer is gratefully acknowledged for kindly providing gene-specific primers of PhEXP2 and PhEXP3. Flavia Guzzo is thanked for her help during experiments involving total area and total cell number measurement and light microscopy. Luigi Russi is thanked for his help in the statistical analyses. We also are grateful to Fabio Finotti for technical assistance in the greenhouse and to Tom Gerats, Francesca Quattrocchio, and Ronald Koes for critical reading of the manuscript. This work was supported in part by a grant of University of Verona: Alterazione dell'espressione di un gene di Petunia hybrida, petalo specifico, codificante espansina. Caratterizzazione di knock out, antisenso sovraespressi e mutagenizzati con l'elemento endogeno dTPh1.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Mario Pezzotti (mario.pezzotti@univr.it).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.018705.
References
- Angenent, G.C., Franken, J., Busscher, M., Weiss, D., and van Tunen, A.J. (1994). Co-suppression of the petunia homeotic gene fbp2 affects the identity of the generative meristem. Plant J. 5, 33–44. [DOI] [PubMed] [Google Scholar]
- Brummell, D.A., Harpster, M.H., Civello, P.M., Palys, J.M., Bennett, A.B., and Dunsmuir, P. (1999). Modification of expansin protein abundance in tomato fruit alters softening and cell wall polymer metabolism during ripening. Plant Cell 11, 2203–2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpita, N., Tierney, M., and Campbell, M. (2001. a). Molecular biology of the plant cell wall: Searching for the genes that define structure, architecture and dynamics. Plant Mol. Biol. 47, 1–5. [PubMed] [Google Scholar]
- Carpita, N.C., Defernez, M., Findlay, K., Wells, B., Shoue, D.A., Catchpole, G., Wilson, R.H., and McCann, M.C. (2001. b). Cell wall architecture of the elongating maize coleoptile. Plant Physiol. 127, 551–565. [PMC free article] [PubMed] [Google Scholar]
- Chen, F., and Bradford, K.J. (2000). Expression of an expansin is associated with endosperm weakening during tomato seed germination. Plant Physiol. 124, 1265–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, L., Carpita, N.C., Reiter, W.D., Wilson, R.H., Jeffries, C., and McCann, M.C. (1998). A rapid method to screen for cell-wall mutants using discriminant analysis of Fourier transform infrared spectra. Plant J. 16, 385–392. [DOI] [PubMed] [Google Scholar]
- Cho, H.T., and Cosgrove, D.J. (2000). Altered expression of expansin modulates leaf growth and pedicel abscission in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 97, 9783–9788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho, H.T., and Kende, H. (1997). Expression of expansin genes is correlated with growth in deepwater rice. Plant Cell 9, 1661–1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, D., Lee, Y., Cho, H.T., and Kende, H. (2003). Regulation of expansin gene expression affects growth and development in transgenic rice plants. Plant Cell 15, 1386–1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Civello, P.M., Powell, A.L., Sabehat, A., and Bennett, A.B. (1999). An expansin gene expressed in ripening strawberry fruit. Plant Physiol. 121, 1273–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove, D.J. (1997. a). Creeping walls, softening fruit, and penetrating pollen tubes: The growing roles of expansins. Proc. Natl. Acad. Sci. USA 94, 5504–5505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove, D.J. (1997. b). Relaxation in a high-stress environment: The molecular bases of extensible cell walls and cell enlargement. Plant Cell 9, 1031–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove, D.J. (1999). Enzymes and other agents that enhance cell wall extensibility. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 391–417. [DOI] [PubMed] [Google Scholar]
- Cosgrove, D.J. (2000. a). New genes and new biological roles for expansins. Curr. Opin. Plant Biol. 3, 73–78. [DOI] [PubMed] [Google Scholar]
- Cosgrove, D.J. (2000. b). Loosening of plant cell walls by expansins. Nature 407, 321–326. [DOI] [PubMed] [Google Scholar]
- Cosgrove, D.J., and Li, Z.C. (1993). Role of expansin in cell enlargement of oat coleoptiles (analysis of developmental gradients and photocontrol). Plant Physiol. 103, 1321–1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creelman, R.A., and Mullet, J.E. (1997). Oligosaccharins, brassinolides, and jasmonates: Nontraditional regulators of plant growth, development, and gene expression. Plant Cell 9, 1211–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagard, M., Desnos, T., Desprez, T., Goubet, F., Refregier, G., Mouille, G., McCann, M., Rayon, C., Vernhettes, S., and Hofte, H. (2000). PROCUSTE1 encodes a cellulose synthase required for normal cell elongation specifically in roots and dark-grown hypocotyls of Arabidopsis. Plant Cell 12, 2409–2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming, A.J., McQueen-Mason, S., Mandel, T., and Kuhlemeier, C. (1997). Induction of leaf primordia by the cell wall protein expansin. Science 276, 1415–1418. [Google Scholar]
- Horsch, R.B., Rogers, S.G., and Fraley, R.T. (1985). Transgenic plants. Cold Spring Harb. Symp. Quant. Biol. 50, 433–437. [DOI] [PubMed] [Google Scholar]
- Hutchison, K.W., Singer, P.B., McInnis, S., Diaz-Sala, C., and Greenwood, M.S. (1999). Expansins are conserved in conifers and expressed in hypocotyls in response to exogenous auxin. Plant Physiol. 120, 827–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im, K.H., Cosgrove, D.J., and Jones, A.M. (2000). Subcellular localization of expansin mRNA in xylem cells. Plant Physiol. 123, 463–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor, S., Kobayashi, A., and Takatsuji, H. (2002). Silencing of the tapetum-specific zinc finger gene TAZ1 causes premature degeneration of tapetum and pollen abortion in petunia. Plant Cell 14, 2353–2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller, E., and Cosgrove, D.J. (1995). Expansins in growing tomato leaves. Plant J. 8, 795–802. [DOI] [PubMed] [Google Scholar]
- Kemsley, E.K. (1998). Chemometric Methods for Classification Problems: Discriminant Analysis and Modelling of Spectroscopic Data (Chichester, UK: John Wiley & Sons) .
- Kende, H., and Zeevaart, J. (1997). The Five “Classical” Plant Hormones. Plant Cell 7, 1197–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieber, J.J. (1997). The ethylene signal transduction pathway in Arabidopsis. J. Exp. Bot. 48, 211–218. [DOI] [PubMed] [Google Scholar]
- Kim, S.H., Arnold, D., Lloyd, A., and Roux, S.J. (2001). Antisense expression of an Arabidopsis ran binding protein renders transgenic roots hypersensitive to auxin and alters auxin-induced root growth and development by arresting mitotic progress. Plant Cell 13, 2619–2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotilainen, M., Helariutta, Y., Mehto, M., Pollanen, E., Albert, V.A., Elomaa, P., and Teeri, T.H. (1999). GEG participates in the regulation of cell and organ shape during corolla and carpel development in gerbera hybrida. Plant Cell 11, 1093–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, O.Y., Darley, C.P., Ongaro, V., Fleming, A., Schipper, O., Baldauf, S.L., and McQueen-Mason, S.J. (2002). Plant expansins are a complex multigene family with an ancient evolutionary origin. Plant Physiol. 128, 854–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y., Jones, L., and McQueen-Mason, S. (2003). Expansins and cell growth. Curr. Opin. Plant Biol. 6, 603–610. [DOI] [PubMed] [Google Scholar]
- Liang, C.Y., and Marchessault, R.H. (1959). Infrared spectra of crystalline polysaccharides. II. Native celluloses in the region from 640 to 1700 cm−1. J. Polym. Sci. 31, 269–278. [Google Scholar]
- Link, B.M., and Cosgrove, D.J. (1998). Acid-growth response and alpha-expansins in suspension cultures of bright yellow 2 tobacco. Plant Physiol. 118, 907–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loreto, F., Mannozzi, M., Maris, C., Nascetti, P., Ferranti, F., and Pasqualini, S. (2001). Ozone quenching properties of isoprene and its antioxidant role in leaves. Plant Physiol. 126, 993–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, C., Bhatt, K., and Baumann, K. (2001). Shaping in plant cells. Curr. Opin. Plant Biol. 4, 540–549. [DOI] [PubMed] [Google Scholar]
- Meyerowitz, E.M. (1997). Genetic control of cell division patterns in developing plants. Cell 88, 299–308. [DOI] [PubMed] [Google Scholar]
- Nicol, F., His, I., Jauneau, A., Vernhettes, S., Canut, H., and Hofte, H. (1998). A plasma membrane-bound putative endo-1,4-beta-D-glucanase is required for normal wall assembly and cell elongation in Arabidopsis. EMBO J. 17, 5563–5576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orford, S.J., and Timmis, J.N. (1998). Specific expression of an expansin gene during elongation of cotton fibres. Biochim. Biophys. Acta 1398, 342–346. [DOI] [PubMed] [Google Scholar]
- Pagant, S., Bichet, A., Sugimoto, K., Lerouxel, O., Desprez, T., McCann, M., Lerouge, P., Vernhettes, S., and Hofte, H. (2002). KOBITO1 encodes a novel plasma membrane protein necessary for normal synthesis of cellulose during cell expansion in Arabidopsis. Plant Cell 14, 2001–2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pien, S., Wyrzykowska, J., McQueen-Mason, S., Smart, C., and Fleming, A. (2001). Local expression of expansin induces the entire process of leaf development and modifies leaf shape. Proc. Natl. Acad. Sci. USA 98, 11812–11817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyke, K.A., Marrison, J.L., and Leech, R.M. (1991). Temporal and spatial development of the cells of the expanding first leaf of Arabidopsis thaliana (L.) Heynh. J. Exp. Bot. 42, 1407–1416. [Google Scholar]
- Reale, L., Porceddu, A., Lanfaloni, L., Moretti, C., Zenoni, S., Pezzotti, M., Romano, B., and Ferranti, F. (2002). Patterns of cell division and expansion in developing petals of Petunia hybrida. Sex. Plant Reprod. 15, 123–132. [Google Scholar]
- Reinhardt, D., Wittwer, F., Mandel, T., and Kuhlemeier, C. (1998). Localized upregulation of a new expansin gene predicts the site of leaf formation in the tomato meristem. Plant Cell 10, 1427–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter, W.-D. (2002). Biosynthesis and properties of plant cell wall. Curr. Opin. Plant Biol. 5, 536–542. [DOI] [PubMed] [Google Scholar]
- Rose, J.K., Cosgrove, D.J., Albersheim, P., Darvill, A.G., and Bennett, A.B. (2000). Detection of expansin proteins and activity during tomato fruit ontogeny. Plant Physiol. 123, 1583–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose, J.K., Lee, H.H., and Bennett, A.B. (1997). Expression of a divergent expansin gene is fruit-specific and ripening-regulated. Proc. Natl. Acad. Sci. USA 94, 5955–5960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual, 2nd ed. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Sanger, F., Nicklen, S., and Coulson, A.R. (1977). DNA sequencing with chain-terminating inhibitors. Biotechnology 24, 104–108. [PubMed] [Google Scholar]
- Schindelman, G., Morikami, A., Jung, J., Baskin, T.I., Carpita, N.C., Derbyshire, P., McCann, M.C., and Benfey, P.N. (2001). COBRA encodes a putative GPI-anchored protein, which is polarly localized and necessary for oriented cell expansion in Arabidopsis. Genes Dev. 15, 1115–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Séné, C., McCann, M., Wilson, R.H., and Grinter, R. (1994). FT-Raman and FT-infrared spectroscopy: An investigation of five higher plant cell walls and their components. Plant Physiol. 106, 1623–1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shcherban, T.Y., Shi, J., Durachko, D.M., Guiltinan, M.J., McQueen-Mason, S.J., Shieh, M., and Cosgrove, D.J. (1995). Molecular cloning and sequence analysis of expansins–A highly conserved, multigene family of proteins that mediate cell wall extension in plants. Proc. Natl. Acad. Sci. USA 92, 9245–9249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibaoka, H., and Nagai, R. (1994). The plant cytoskeleton. Curr. Opin. Cell Biol. 6, 10–15. [DOI] [PubMed] [Google Scholar]
- Smith, L.G. (2003). Cytoskeletal control of plant cell shape: Getting the fine points. Curr. Opin. Plant Biol. 6, 63–73. [DOI] [PubMed] [Google Scholar]
- Souer, E., Quattrocchio, F., de Vetten, N., Mol, J.N.M., and Koes, R.E. (1995). A general method to isolate genes tagged by a high copy number transposable element. Plant J. 7, 677–685. [DOI] [PubMed] [Google Scholar]
- Sterky, F., et al. (1998). Gene discovery in the wood-forming tissues of poplar: Analysis of 5, 692 expressed sequence tags. Proc. Natl. Acad. Sci. USA 95, 13330–13335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szekeres, M., Nemeth, K., Koncz-Kalman, Z., Mathur, J., Kauschmann, A., Altmann, T., Redei, G.P., Nagy, F., Schell, J., and Koncz, C. (1996). Brassinosteroids rescue the deficiency of CYP90, a cytochrome P450, controlling cell elongation and de-etiolation in Arabidopsis. Cell 85, 171–182. [DOI] [PubMed] [Google Scholar]
- Tsuboi, M. (1957). Infrared spectrum and crystal structure of cellulose. J. Polym. Sci. 25, 159–171. [Google Scholar]
- Tsuge, T., Tsukaya, H., and Uchimiya, H. (1996). Two independent and polarized processes of cell elongation regulate leaf blade expansion in Arabidopsis thaliana (L.) Heynh. Development 122, 1589–1600. [DOI] [PubMed] [Google Scholar]
- Van Houdt, H., Van Montagu, M., and Depicker, A. (2000). Both sense and antisense RNAs are targets for the sense transgene-induced posttranscriptional silencing mechanism. Mol. Gen. Genet. 263, 995–1002. [DOI] [PubMed] [Google Scholar]
- van Tunen, A.J., Mur, L.A., Brouns, G.S., Rienstra, J.D., Koes, R.E., and Mol, J.N.M. (1989). Pollen- and anther-specific chi promoters from petunia: Tandem promoter regulation of the chiA gene. Plant Cell 2, 393–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent, C.A., Carpenter, R., and Coen, E.S. (2003). Interaction between gene activity and cell layers during floral development. Plant J. 33, 765–774. [DOI] [PubMed] [Google Scholar]
- Wu, Y., Meeley, R.B., and Cosgrove, D.J. (2001. a). Analysis and expression of the alpha-expansin and beta-expansin gene families in maize. Plant Physiol. 126, 222–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, Y., Sharp, R.E., Durachko, D.M., and Cosgrove, D.J. (1996). Growth maintenance of the maize primary root at low water potentials involves increases in cell-wall extension properties, expansin activity, and wall susceptibility to expansins. Plant Physiol. 111, 765–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, Y., Thorne, E.T., Sharp, R.E., and Cosgrove, D.J. (2001. b). Modification of expansin transcript levels in the maize primary root at low water potentials. Plant Physiol. 126, 1471–1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, R., Morrison, W.H., 3rd, Freshour, G.D., Hahn, M.G., and Ye, Z.H. (2003). Expression of a mutant form of cellulose synthase AtCesA7 causes dominant negative effect on cellulose biosynthesis. Plant Physiol. 132, 786–795. [DOI] [PMC free article] [PubMed] [Google Scholar]