Abstract
Comment on: Rodriguez GP, et al. Proc Natl Acad Sci USA 2012; 109:6153-8.
DNA mismatch repair (MMR) is important for preventing mutations due to errors occurring during replication. Mismatches that are not removed by the proofreading function of the replicative DNA polymerases are recognized by a MutS protein in bacteria or usually in eukaryotes by a MutSα or MutSβ heterodimer.1 After recognition, downstream events involving MutL in bacteria and usually MutLα in eukaryotes result in the removal of the newly synthesized DNA strand and resynthesis of the region, avoiding a change in DNA sequence.1 However, there are many situations in which MMR is active outside of the context of genome replication, and an important question is what determines which strand of DNA is removed in those cases once a mismatch is detected. Using a reversion assay that detects point mutations in the TRP5 gene of yeast, we recently demonstrated that mispairs that escape detection at the replication fork can be recognized later by MMR and repaired in a manner independent of the replicating strand.2 In that case, MMR was found to be acting in a mutagenic manner and could restore growth to cells that were in a nondividing state.
When a mismatch is detected, how does MMR determine which strand to remove? A variety of experiments have demonstrated that eukaryotic MMR is directed to remove the DNA strand containing a nick, which could occur in the newly synthesized DNA strand through a variety of processes.1,3 Evidence suggests that an important source of those nicks is activation of the latent endonuclease activity of MutLα; MutLα interacts with PCNA in a manner that could discriminate the template and primer stands of replication and thus give proper strand discrimination to MMR.4 Strand discrimination also likely involves MutSα and MutSβ, as both complexes have been shown to have robust interactions with PCNA.1 However, two MMR pathways involving MutSα have recently been observed: one with MutSα as an integral component of replication factories and the other in which MutSα presumably scans the genome for mismatches and is independent of PCNA interaction.5 This latter pathway of MutSα, untethered to normal replication, is of interest here.
One major function of MMR outside of replication is its anti-recombination role, by which it prevents aberrant recombination between non-identical sequences.6 MMR also functions in meiotic recombination and is responsible for the formation of gene conversion gradients, which can best be understood as arising from directed MMR near the site of initiating double-strand breaks, and randomly directed MMR further from the site of such strand discrimination signals.7 Although in meiotic recombination it appears that MMR acts without strand discrimination in some cases, the result is not an increase in mutation, as the end result is a choice between two pre-existing alleles.
There are at least three situations in which MMR acting outside of replication appears to play a pro-mutagenic role. One is in the expansion of triplet repeat sequences, which play a role in a number of neurodegenerative diseases.3,4 Although MMR would be expected to prevent repeat slippage during replication, there is biochemical evidence that slippage loops formed in triplet repeat sequences in a non-replicating state could load a PCNA-MutLα complex that would nick DNA in a random manner, leading to large expansions.4 There is also evidence from various mouse model systems that suggest a role for MMR in promoting repeat expansions.3,8
The second situation is somatic hypermutation. It had been found in 1998 that somatic hypermutation was, contrary to expectations, dependent on the presence of active MMR,9 but there was no mechanistic way to explain that dependence. A combination of patch replication by a relatively inaccurate translesion DNA polymerase and MMR acting randomly in terms of DNA strand, offers the best rationale for this process.2,3
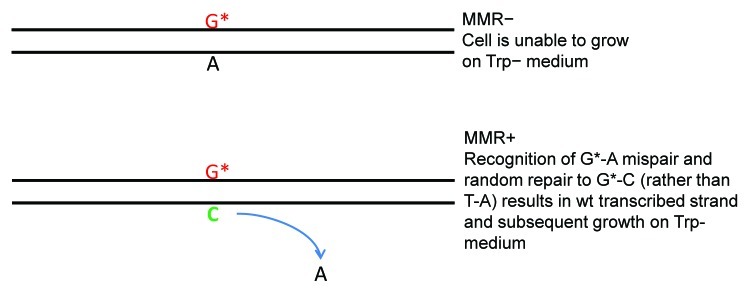
The third situation, which also has the broadest sweep, is that of cancer. If several different pathways must be altered by mutation in order for a tumor to form, how could that happen in cells that either divide very slowly, or are in a nondividing state? Two years after the initial discovery of the linkage between MMR defects and cancer, MacPhee hypothesized that MMR acting in a “randomly templated” mode could be responsible for the formation of mutations in nondividing cells, which could reenter a growth phase as a result of those mutations;10 that corresponds to the observation we recently made in yeast, illustrated in Figure 1.2 This is precisely the type of activity envisioned by MacPhee. What is not yet known is the signal giving strand discrimination to MMR; it could possibly be a random loading of MutLα,4 or it could be the presence of a random nick in the DNA strand close enough to the mismatch to be used for strand discrimination.
Figure 1. Mutagenic MMR in nondividing cells. Cells were electroporated with 8-oxoGTP and then plated on Trp medium. The 8-oxoGTP was incorporated throughout the genome, including in some cells at the position indicated above which must revert via a TA→GC transversion in order for the cell to become Trp+. Once plated, the Trp- cells were unable to undergo even one round of replication, although they remained viable for over two weeks. In the absence of MMR, very few revertants appeared. However, in the presence of MMR, many revertants arose within the 3 d expected for cells that would have begun growth immediately after plating, but an even larger number of revertant colonies arose later on the plates up to a period of a week later. Thus only cells with active MMR were able to regain growth from a nondividing state due to the mutation created by MMR activity.
The importance of MMR in preventing mutations during replication is unquestioned. However, the multiple activities of MMR outside of replication tend to be less appreciated. When MMR recognizes mismatches in DNA outside of the context of replication, the issue of what gives strand discrimination to MMR becomes critical. If the signal is randomly generated, MMR activity can be mutagenic, as we have found.2
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/21259
References
- 1.Iyer RR, et al. Chem Rev. 2006;106:302–23. doi: 10.1021/cr0404794. [DOI] [PubMed] [Google Scholar]
- 2.Rodriguez GP, et al. Proc Natl Acad Sci USA. 2012;109:6153–8. doi: 10.1073/pnas.1115361109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peña-Diaz J, et al. Trends Biochem Sci. 2012;37:206–14. doi: 10.1016/j.tibs.2012.03.001. [DOI] [PubMed] [Google Scholar]
- 4.Pluciennik A, et al. Proc Natl Acad Sci USA. 2010;107:16066–71. doi: 10.1073/pnas.1010662107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hombauer H, et al. Cell. 2011;147:1040–53. doi: 10.1016/j.cell.2011.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.George CM, et al. Crit Rev Biochem Mol Biol. 2012;47:297–313. doi: 10.3109/10409238.2012.675644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Argueso JL, et al. Mol Cell Biol. 2003;23:873–86. doi: 10.1128/MCB.23.3.873-886.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McMurray CT. DNA Repair. 2008;7:1121–34. doi: 10.1016/j.dnarep.2008.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cascalho M, et al. Science. 1998;279:1207–10. doi: 10.1126/science.279.5354.1207. [DOI] [PubMed] [Google Scholar]
- 10.MacPhee DG. Mismatch repair, somatic mutations, and the origins of cancer. Cancer Res. 1995;55:5489–92. [PubMed] [Google Scholar]