Abstract
The Hippo signaling pathway regulates cellular proliferation and survival, thus exerting profound effects on normal cell fate and tumorigenesis. We previously showed that the pivotal effector of this pathway, YAP, is amplified in tumors and promotes epithelial-to-mesenchymal transition (EMT) and malignant transformation. Here, we report that overexpression of TAZ, a paralog of YAP, in human mammary epithelial cells promotes EMT and, in particular, some invasive structures in 3D cultures. TAZ also leads to cell migration and anchorage-independent growth in soft agar. Furthermore, we identified amphiregulin (AREG), an epidermal growth factor receptor (EGFR) ligand, as a target of TAZ. We show that AREG functions in a non-cell-autonomous manner to mediate EGF-independent growth and malignant behavior of mammary epithelial cells. In addition, ablation of TEAD binding completely abolishes the TAZ-induced phenotype. Last, analysis of breast cancer patient samples reveals a positive correlation between TAZ and AREG in vivo. In summary, TAZ-dependent secretion of AREG indicates that activation of the EGFR signaling is an important non-cell-autonomous effector of the Hippo pathway, and TAZ as well as its targets may play significant roles in breast tumorigenesis and metastasis.
Keywords: amphiregulin (AREG), epithelial-to-mesenchymal transition (EMT), Hippo pathway, TAZ, tumor metastasis
Introduction
The Hippo signaling pathway regulates cellular proliferation and survival, thus exerting profound effects on normal cell fate and tumorigenesis.1-3 The key downstream effectors of the Hippo pathway, YAP (Yes-associated protein) and TAZ (transcriptional co-activator with PDZ-binding motif), are tightly regulated by a number of upstream molecules, such as Mst1/2, Lats1/2 and RASSF family proteins. Inactivation of YAP/TAZ by the Hippo pathway via cytoplasmic sequestration from 14-3-3 binding is well known.4,5 In addition, a novel Hippo pathway-independent restriction of YAP/TAZ by angiomotin and their ubiquitin-mediated degradation have been recently reported,6-8 adding another layer to the complexity of YAP/TAZ regulation. We previously identified by array comparative genomic hybridization (aCGH) that YAP, the pivotal effector of the Hippo pathway, is amplified in mouse and human breast cancers.9 Overexpression of YAP induces epithelial-to-mesenchymal transition (EMT), which has been recently shown to play multiple roles in tumor metastasis.10,11 Furthermore, we have shown that overexpression of YAP led to suppression of apoptosis, growth factor-independent proliferation and anchorage-independent growth in soft agar.9 Comprehensive survey of the most common solid cancer types revealed widespread and frequent YAP overexpression in lung, ovarian, pancreatic, colorectal, hepatocellular and prostate carcinomas.1
TAZ, also known as WWTR1 for WW domain containing transcription regulator 1, was first identified in a cDNA screen for 14-3-3-binding proteins.12 The TAZ gene was mapped to chromosome 3q24.13 Human TAZ shares 45% amino acid identity with YAP and also contains the WW domain, coiled-coil region and PDZ-binding motif. Therefore, TAZ is regarded as a paralog of YAP, and both can function as transcriptional co-activators.12 Conceivably, some overlapping functions have been reported for TAZ and YAP. For example, both TAZ and YAP interact with and activate the TEAD/TEF-1 family transcription factors.14-18 In addition, a recent study reported that TAZ and YAP are nuclear relays of mechanical signals exerted by extracellular matrix (ECM) rigidity and cell shape, and thus serve as sensors and mediators of mechanical cues instructed by the cellular microenvironment.19
However, there also exist structural and characteristic differences between TAZ and YAP, which are likely responsible for some distinct functions performed by either protein. For example, YAP contains an SH3-binding motif and a proline-rich region at its N terminus, both contributing to its binding to Yes, whereas TAZ lacks these domains as well as the ability to bind Yes. It is known that TAZ interacts with a wide variety of DNA-binding transcription factors, such as RUNX-2,20,21 TTF-1,22 polyomavirus T antigens,23 TBX524 and Pax3.25 Interestingly, TAZ, but not YAP, has been reported as a critical transcriptional modulator of mesenchymal stem cell differentiation.21 It plays a molecular rheostat role by fine-tuning the balance between osteoblast and adipocyte development through co-activation of the Runx2-dependent gene transcription and repression of the PPARγ-dependent gene transcription.21 A recent study also found that TAZ binds heteromeric Smad2/3–4 to maintain self-renewal of human embryonic stem cells.26 In addition, TAZ has been recently reported to be involved in the cancer stem cell (CSC) property of breast cancer and glioma.27,28 Of note, although TAZ is proposed to play an important role in cell transformation, tumorigenesis and metastasis, the elusive underlying mechanisms are yet to be unraveled.
In the present study, we further investigated the transforming role of TAZ in cell-based three-dimensional (3D) cultures. More importantly, we identified amphiregulin (AREG), an epidermal growth factor receptor (EGFR) ligand, as a target of TAZ. We specifically showed that AREG may function in a non-cell-autonomous manner to mediate EGF-independent growth and malignant behavior of mammary epithelial cells. Last, we observed a striking correlation between TAZ and AREG in breast cancer patients. In summary, TAZ-dependent secretion of AREG and the resultant activation of EGFR signaling may be an important non-cell-autonomous effector of the Hippo pathway, with implications on the regulation of both physiological and malignant cell behaviors.
Results
TAZ induces malignant cell behaviors
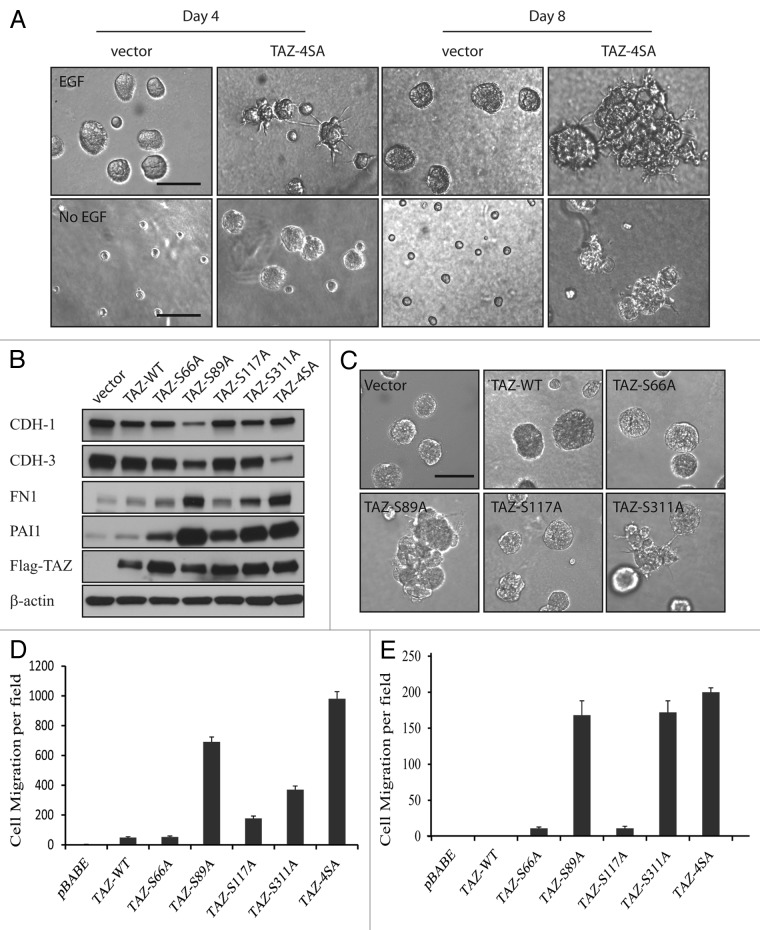
YAP/TAZ is known to be inhibited by the upstream Hippo pathway component Lats1/2 via phosphorylation of the serine residue in HXRXXS motifs.4 Therefore, we first tested the effect of aberrant TAZ activation due to serine to alanine mutations at four HXRXXS motifs (TAZ4SA) on the behavior of human non-transformed mammary epithelial MCF10A cells. Overexpression of TAZ4SA induced morphological alterations of the MCF10A cells from cobblestone-like epithelial phenotype to fibroblast-like mesenchymal phenotype, typically known as the epithelial-to-mesenchymal transition (EMT) (Fig. S1A). To confirm that the observed morphological alterations were indeed an EMT process, we analyzed the expression of epithelial and mesenchymal markers in the vector control and TAZ4SA-overexpressing cells. As expected, there was a dramatic decrease of the epithelial markers [e.g., E-cadherin (CDH1) and P-cadherin (CDH3)] and an increase of the mesenchymal markers [e.g., fibronectin-1, plasminogen activator inhibitor-1 (PAI1) and vimentin] as revealed by immunoblot (Fig. S1B). 3D epithelial culture systems are a valuable tool for modeling cancer genes and pathways in a structurally appropriate context.29 We thus performed 3D cultures of the vector control and TAZ4SA-transduced MCF10A cells. To our great interest, we observed enlarged multi-acini formation accompanied by a unique phenotype of striking invasive projections in the TAZ4SA cells in as early as four days (Fig. 1A, top). To further test whether TAZ can induce growth factor-independent proliferation of MCF10A cells, we performed 3D cultures in the absence of epithelial growth factor (EGF). Interestingly, we found that TAZ4SA but not vector control cells were capable of forming acini and projections under EGF deprivation (Fig. 1A, bottom). To dissect the effects of TAZ activation from S-A mutation at the various HXRXXS motifs, we created individual mutations, i.e., S66A, S89A, S117A and S311A, at each motif and examined their effects on MCF10A cells. As a result, we observed that overexpression of wild-type (wt) and each individual S-A mutant TAZ induced EMT (Fig. S2), and immunoblot confirmed the changes of the epithelial and mesenchymal markers (Fig. 1B). However, in 3D cultures, wt-TAZ, TAZS66A and TAZS117A promoted the acinar formation similar to control cells, whereas TAZS89A produced enlarged multi-acini, and TAZS311A induced projection formation without obviously enlarged acinar formation (Fig. 1C). To further examine the ability of various TAZ mutants to induce cell migration, we performed the Transwell cell migration assay. All of the TAZ mutations as well as wt-TAZ were capable of significantly increasing the migratory ability of MCF10A cells, but at different efficiency, with the strongest effect observed for TAZS89A and TAZ4SA (Fig. 1D). Discrepancies were also observed in the soft agar colony formation. Although significant difference in the anchorage-independent growth was observed for TAZS66A- or TAZS117A-transduced cells compared with the vector control (p < 0.05), TAZS89A, TAZS311A and TAZ4SA much more strikingly increased the cell growth in soft agar (p < 0.001, Fig. 1E). Our results indicated that modification of the serine site in each individual HXRXXS motif plays a distinct role in the TAZ-induced cell migration and cellular transformation.
Figure 1. TAZ induces malignant cell behavior. (A) Top: 3D cultures of vector control and TAZ4SA-transduced MCF10A cells in the presence of EGF on day 4 and day 8. (Scale bar, 100 µm); bottom: 3D cultures of vector control and TAZ4SA-transduced MCF10A cells in the absence of EGF on day 4 and day 8. (Scale bar, 100 µm) (B) Expression of wild-type (wt) and mutant TAZ results in the loss of epithelial markers and gain of mesenchymal markers. Immunoblot reveals an increase in E-cadherin (CDH1) and P-cadherin (CDH3) as well as a decrease in fibronectin-1 (FN1) and plasminogen activator inhibitor-1 (PAI1). Flag antibody detects the ectopic expression of TAZ. β-Actin was used as a loading control. (C) Overexpressing wt and mutant TAZ as well as control vector in presence of EGF in MCF10A 3D culture . (Scale bar, 100 µm) (D) TAZ mutations promote cell migration. Control and mutant TAZ-transduced MCF10A cells were plated onto 8-µm Transwell filters and allowed to migrate for 24 h. Data are the mean number of migrated cells per × 20 field of four fields from each of the triplicate wells. Error bars equal ± SD of three independent experiments. (E) Effect of TAZ on anchorage-independent growth in soft agar. Vector control and mutant TAZ-transduced MCF10A cells were plated in soft-agar assays and allowed to grow for 21 d. Data are mean number of colonies per six-well plate culture of 5 × 104 cells. Error bars equal ± SD of three independent experiments.
Identification of AREG as a non-cell-autonomous effector of TAZ
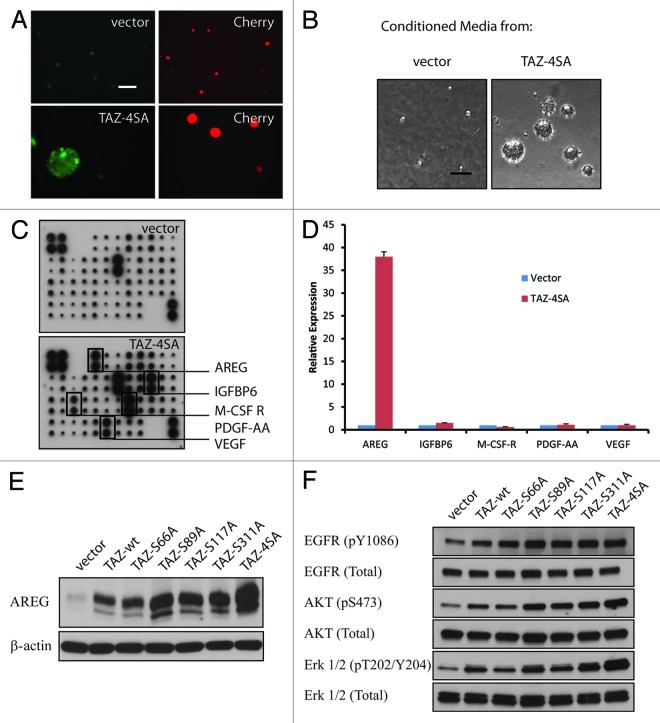
Normal cells require mitogenic growth signals to proliferate, whereas tumor cells often generate their own proliferative signals through the secretion of growth factors or the activation of growth factor receptors.30 To investigate the mechanism underlying TAZ-induced, EGF-independent cell proliferation, we performed a mixing experiment using CherryRed-tagged cells co-cultured with cells transduced by GFP-labeled vector control or TAZ4SA (Fig. 2A). As expected, in the absence of EGF, only MCF10A cells overexpressing GFP-TAZ4SA, but not the CherryRed vector cells, formed acini in 3D cultures. However, to our great interest, acinar formation of CherryRed cells was also observed in the co-culture with TAZ4SA cells. This finding strongly suggested a paracrine or non-cell-autonomous effect from TAZ4SA, i.e., some growth factor(s) might have been secreted into the media and promote the CherryRed vector cell proliferation. To confirm that TAZ induced secretion of growth factor(s), we performed the conditioned media experiments. Media from either the vector control or TAZ4SA-transduced MCF10A in 3D cultures was collected and applied onto the parental MCF10A cells. Conditioned media from TAZ4SA-transduced, but not vector, cells induced acinar formation (Fig. 2B), indicating that there must be some secreted growth factors exclusively induced by TAZ4SA. Next, to identify the putative factor(s) secreted by TAZ4SA-expressing MCF10A cells, we applied conditioned media onto a cytokine antibody array representing 41 growth factors and cytokines.31 TAZ4SA media revealed five distinctively enriched proteins: amphiregulin (AREG), insulin-like growth factor binding protein-6 (IGFBP-6), macrophage colony-stimulating factor-receptor (M-CSF-R), platelet-derived growth factor-AA (PDGF-AA) and vascular endothelial growth factor (VEGF) (Fig. 2C). To distinguish the specific TAZ-induced factor(s) from by-products of MCF10A proliferation, we analyzed mRNA level under EGF deprivation by real-time qRT-PCR. It was found that only AREG mRNA was significantly induced in the TAZ4SA cells (p < 0.001, Fig. 2D). AREG thus constituted the primary candidate for a TAZ-induced secreted factor involved in cellular proliferation. We further confirmed by immunoblot that AREG was also induced by wt-TAZ and other TAZ mutants, particularly S89A (Fig. 2E). Finally, since AREG is an EGFR ligand,32,33 we asked whether the EGFR signaling pathway was indeed activated by TAZ. Immunoblot revealed an increase of phospho-EGFR as well as the activation of down stream PI3K/AKT and MAPK pathways by wt- and mutant TAZ (Fig. 2F). In summary, we identified AREG as a secreted growth factor induced by TAZ, and it aberrantly activated the EGFR signaling pathway.
Figure 2. Identification of AREG as a non-cell-autonomous effector of TAZ. (A) Non-cell-autonomous effect of TAZ. CherryRed-tagged vector and GFP-tagged TAZ4SA-transduced MCF10A cells were co-cultured in 3D 8 d without EGF. Representative light and fluorescence images are shown. (Scale bar, 100 µm) (B) TAZ4SA conditioned media induces EGF-independent growth of parental MCF10A cells in 3D cultures. Representative phase contrast images are shown. (Scale bar, 100 µm) (C) Secreted growth factor screen. Human growth factor antibody array analysis was performed using conditioned media (day 12) from vector- (top) or TAZ4SA-transduced (bottom) MCF10A cells. The membrane was printed with antibodies for 41 growth factors and receptors, with four positive and four negative controls in the upper left corner. Five proteins were exclusively enriched in TAZ4SA conditioned medium (rectangles). (D) Induced AREG mRNA in TAZ4SA-transduced cells in the absence of EGF, as detected by qRT-PCR. GAPDH was used as an internal control. Data represent mean ± SD of three independent experiments. (E) Induction of AREG by wild-type and mutant TAZ in the absence of EGF, as revealed by immunoblot. β-Actin was used as a loading control. (F) Activation of the EGFR signaling induced by overexpression of wild-type and mutant TAZ.
AREG mediates the effect of TAZ on growth factor-independent growth
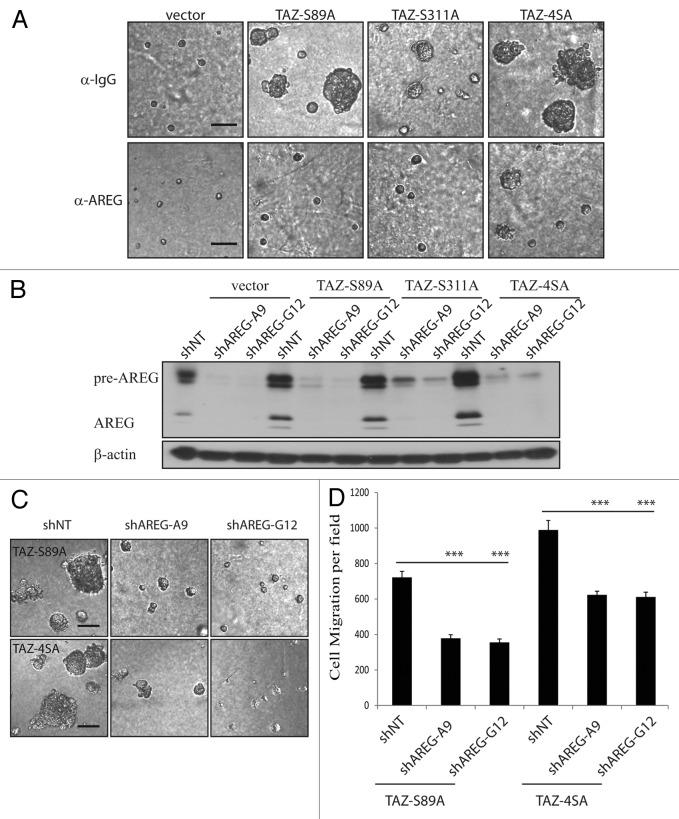
To further validate AREG as the secreted factor responsible for TAZ-induced EGF-independent MCF10A cell proliferation and phenotype change, we first tested whether neutralizing antibody against AREG could suppress TAZ-mediated 3D acinar formation. Anti-AREG neutralizing antibody (1 μg ml−1) suppressed TAZ-induced acinar formation by ~80%, whereas control IgG antibody had no effect (Fig. 3A). We then investigated the effect of AREG knockdown with two independent lentiviral shRNA constructs in MCF10A cells. Efficient knockdown of AREG was confirmed by immunoblot (Fig. 3B). As expected, in the AREG-knockdown cells, significantly reduced acinar formation was observed compared with control cells in the EGF-deprivation condition (Fig. 3C). Last, knockdown of AREG also partially reduced the TAZS89A- and TAZ4SA-induced cell migration (Fig. 3D). Taken together, these results support the functional importance of AREG in TAZ-induced EGF-independent growth and malignant phenotype observed in 3D cultures.
Figure 3. AREG mediates the effect of TAZ. (A) AREG-neutralizing antibody blocks TAZS89A and TAZ4SA-induced EGF-independent 3D growth. Vector control and TAZ-expressing cells were cultured in 3D assay in the absence of EGF for 12 d, together with a neutralizing antibody against AREG. Normal goat IgG is used as a control. Representative images are shown. (Scale bar, 100 µm) (B) Efficient knockdown of AREG by shRNA in vector control or TAZS89A and TAZ4SA-transduced MCF10A cells, as revealed by immnoblot. β-Actin was used as a loading control. (C) Knockdown of AREG abolishes the 3D phenotype induced by TAZS89A and TAZ4SA in absence of EGF (Scale bar, 100 µm). (D) Knockdown of AREG reduces the TAZS89A- and TAZ4SA-induced cell migration.
Induction of AREG by TAZ is mediated through TEAD
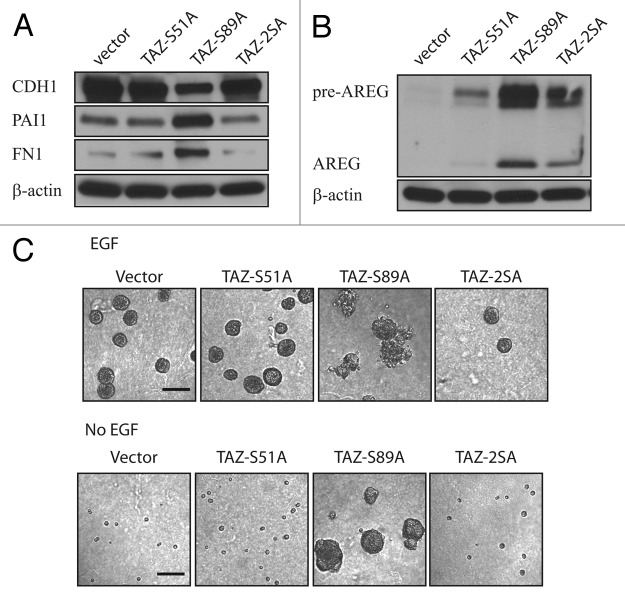
The TEAD transcription factor family has been suggested as a major mediator of the Hippo pathway function through interacting with YAP and TAZ.17,18 To test whether TAZ induced AREG through TEAD, we made a single S-A mutation at the 51st amino acid of TAZ, which disrupts the TEAD binding, as well as double mutations of S51A and S89A on TAZ.18 We found that disruption of the TAZ-TEAD binding abrogated TAZS89A-induced EMT (Fig. 4A). Additionally, induction of AREG by TAZ was also significantly reduced by the S51A mutation (Fig. 4B), manifesting again an important role of TEAD in the transforming capability of TAZ. Last, we performed 3D culture experiments in the presence and absence of EGF. Very strikingly, the S51A mutation completely abolished the TAZS89A-induced enlarged multi-acini formation both in the presence and absence of EGF (Fig. 4C, top and bottom), demonstrating once again an indispensable role of TEAD in the oncogenic function of TAZ.
Figure 4. Induction of AREG by TAZ is mediated through TEAD. (A) S51A abolishes TAZS89A-induced EMT of MCF10A cells. Epithelial and mesenchymal markers were detected by immunoblot. β-Actin was used as a loading control. (B) Induction of AREG by TAZS89A is inhibited by S51A as revealed by immnoblot. β-Actin was used as a loading control. (C) S51A abolishes the 3D phenotype induced by TAZS89A both in the presence (top) and absence (bottom) of EGF. Representative phase contrast images are shown. (Scale bar, 100 µm)
TAZ and AREG are positively correlated in breast cancer patient samples
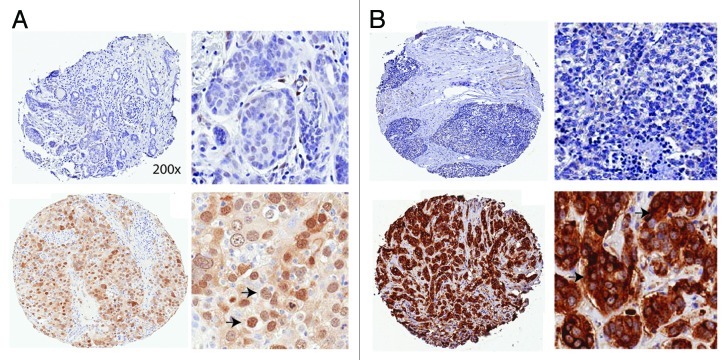
To investigate the regulation of TAZ and AREG in vivo, we analyzed tissue microarrays (TMAs) of breast cancer patients by immunohistrochemistry staining. We observed staining of TAZ in both cytoplasm and nucleus, with stronger nuclear staining concurrent with higher TAZ expression level (data not shown). We thus focused on the nuclear staining of TAZ. For the AREG variable, cytoplasmic intensity score was used. Further, these variables were stratified into low (TAZ multiplication index = 0, Fig. 5A, top) and high (TAZ multiplication index > 0, Fig. 5A, bottom). For the AREG variable, stratification was based on intensity score, i.e., < = 1 (low, Fig. 5B, top) and intensity score > 1 (high, Fig. 5B, bottom). Following these criteria, we had 269 total observations, with 107 high-TAZ and 129 low-TAZ (with 33 TMA not available) as well as 123 high-AREG and 117 low-AREG scores. We then asked the question whether there existed any correlation between TAZ and AREG in clinical samples. Intriguingly, there was a significant positive correlation between TAZ and AREG in the breast cancer patient samples (p = 0.0147). This recapitulation of the correlation between TAZ and AREG in patient samples strongly corroborated our previous in vitro findings and may have implications in practical application.
Figure 5. Positive correlation between TAZ and AREG in breast cancer patients. (A) Example of IHC staining of low (top) and high (bottom) TAZ expression in breast cancer TMAs. Arrows: positive TAZ staining. Low (left) and high (right) resolutions are presented. (B) Example of IHC staining of low (top) and high (bottom) AREG expression in breast cancer TMAs. Arrows: positive AREG staining. Low (left) and high (right) resolutions are presented.
Discussion
In the present study, we report that TAZ induces EMT and increases cell migration and cellular transformation, which is consistent with the previous reports.4,34 Importantly, we have identified amphiregulin (AREG), an EGFR ligand, as a target of TAZ. In addition, we specifically showed that AREG may function in a non-cell-autonomous manner to mediate EGF-independent growth and malignant behavior of mammary epithelial cells. It is of interest to note that we have previously discovered AREG as a non-cell-autonomous component of the Hippo pathway induced by YAP.31 Due to their structural similarities, overlapping targets between YAP and TAZ are expected and have been reported, e.g., connective tissue growth factor (CTGF) is a direct target of both YAP and TAZ.18,35,36 Indeed, compared with the effect of YAP,31 AREG is induced at an even higher level by TAZ (Fig. 2D). Furthermore, the effect of TAZ on EGF-independent growth of MCF10A cells manifested as early as four days and exhibited unique formation of striking invasive projections (Fig. 1A). Conceivably, although targeting a same molecule, YAP and TAZ may still function at different strength and elicit distinct outcomes.
The malignant cell behaviors we observed with ectopic TAZ expression are consistent with previous reports.4,34 In particular, moderate effects resulting from TAZS66A and TAZS117A (Fig. 1C–E) may suggest these HXRXXS motifs as less important than the sites of 89 and 311. This is understandable, because TAZS89A is reported to be significantly resistant to the inhibitory phosphorylation by Lats2.4 Although TAZS311A is only partially resistant to the phosphorylation by Lats2, it also prevents the ubiquitination and proteasomal degradation of the protein,6,37 and therefore exerted a strong effect. In particular, TAZS311A seems to be mostly responsible for the striking invasive projection structure in 3D cultures (Fig. 1C), suggesting that various HXRXXS motifs may be involved in the regulation of different aspects of the TAZ function.
It has been reported that TAZ- and TAZ4SA increased cell proliferation through alteration of cell cycle.4 However, such an effect was not observed by us in 2D cultures by the MTT assay (data not shown) or by some other groups.27,28 Still, we observed TAZ-induced enlarged multi-acini in 3D cultures, especially in the absence of EGF and as early as 4 days, which might result from stimulated proliferation. The invasive structures observed in 3D cultures as well as the increased anchorage-independent growth in soft agar indicated, for some, transforming capacity of TAZ. Although we showed in this study another overlapping effect of YAP and TAZ, future work may emphasize on their specific functions. For example, the YAP-null mice die at embryonic day 8.5,38 whereas TAZ-null mice are viable,39-41 clearly pointing to some separable functions of YAP and TAZ. Since YAP and TAZ do not bind to DNA directly, but rather interact with transcription factors, their differential functions may depend on the utilization of distinct DNA-binding partners or might be guided by YAP and TAZ themselves.1 Indeed, TAZ, but not YAP, has been recently reported to play a critical role in conferring cancer stem cell (CSC)-related traits on breast cancer28 and glioma cells.27 Interestingly, it has been reported that amphiregulin mediated self-renewal in immortalized mouse mammary epithelial cells with stem cell characteristics.42 It will be of great interest to further test whether AREG contributes to the TAZ-induced breast cancer stem cell properties.
The in vivo role of TAZ in tumorigenesis and metastasis has been reported. For example, the Hong group analyzed 126 breast cancer samples by immunohistochemistry.34 They found that 27 (21.4%) samples overexpressed TAZ, and most (21 samples) of the TAZ-expressing cancers were of invasive (infiltrating) ductal carcinoma (IDC). In the present study, we analyzed 269 breast cancer patient samples by immunohistochemistry and had a comparable TAZ overexpression rate of 39%. More importantly, we identified a striking positive correlation between TAZ and AREG in those patients. To our knowledge, this is the first report of a TAZ-correlated target in patient samples. Of particular note, AREG can be secreted into the patient blood system, and its detection could be noninvasive and quite simple. In conclusion, we identified here AREG as an important non-cell-autonomous target of TAZ in mediating its oncogenic roles both in vitro and in vivo. Our work may shed light on more effective treatment of carcinomas, especially their metastasis, and provide a quick and easy biomarker for cancer diagnosis and prognosis.
Materials and Methods
Cell culture, transfection and transduction
MCF10A cells were cultured as described.43 Transfection was performed using X-tremeGENE 9 DNA Transfection Reagent following the manufacturer’s protocol (Roche). Packaging of retrovirus and lentivirus, cell transduction and drug selection were performed following standard protocols. For knockdown experiments, shRNA hairpins targeting human AREG were obtained from the RNAi Consortium (The Broad Institute). The target sequences are listed (in the 5'-3' direction): sh Non-taret-Ctrl: CCGGCAACAAGATGAAGAGCACCAACTCGAGTTGGTGCTC TTCATCTTGTTGTTTTT; shAREG/A9: CCGGCACTGCCAAGTCATAGCCATAC TCGAGTATGGCTATGACTTGGCAGTGTTTTTG and shAREG/G12: CCGGGAACG AAAGAAACTTCGACAACTCGAGTTGTCGAAGTTTCTTTCGTTCTTTTTG.
Plasmid construction and mutagenesis
The human TAZ ORF was obtained from Dr. Yue Xiong (University of North Carolina at Chapel Hill School of Medicine, Chapel Hill). PCR-based mutagenesis was performed with complementary primers containing the desired mutation site (synthesized by IDT) using Expand High Fidelity PCR System (Roche) and cloned into pBABE-puro vector as a Flag-tagged BamHI-EcoRI fragment. Mutant constructs were confirmed by DNA sequencing. pBABE-GFP and pBABE-GFP- TAZ4SA were constructed, respectively, by inserting the GFP cDNA or N-terminal-tagged-GFP-TAZ4SA sequence into the pBABE vector.
Antibodies and immunoblot analysis
TAZ, total Akt, pS473-Akt, total Erk1/2 (P44/42 MAPK) and pT201/Y204- Erk1/2 (P44/42 MAPK) antibodies were purchased from Cell Signaling Technology; β-actin antibody from Upstate; amphiregulin antibody from R&D Systems; fibronectin and Flag (M2) antibodies from Sigma-Aldrich; E-cadherin (CDH1), P-cadherin (CDH3), Vimentin, PAI1 and total EGFR antibodies from BD Biosciences; pY1086-EGFR from Invitrogen. For protein extraction, cells were washed with phosphate-buffered saline and collected with IP buffer: 20 mM TRIS-HCl (pH 8.0), 150 mM NaCl, 20% glycerol, 0.5% NP-40, plus 1× Complete™ EDTA-free Protease Inhibitor Cocktail (Roche) or 1x HaltTM EDTA-free Protease and Phosphotase Inhibitor (Thermo Scientific). Cell lysate was cleared by centrifugation at 14,000 rpm for 20 min at 4°C. Lysate was loaded onto 4–15% MINI-PROTEAN TGX gel (Bio-Rad) with 4X SDS sample buffer. For immunoblot, proteins were transferred onto Immobilon-P membrane (Millipore), detected by various antibodies and visualized with ECL Plus western blotting detection reagents (GE Healthcare).
Cytokine antibody array
Media collected from the three-dimensional culture of TAZ4SA MCF10A cells was applied onto RayBio Human Cytokine Antibody Array (RayBiotech) and assayed following the manufacturer’s protocol.
Real-time RT-PCR
For RNA preparation and qRT-PCR, RNA was extracted using the Trizol reagent (Invitrogen). cDNA synthesis was performed using the First-Strand cDNA Synthesis Kit (GE Healthcare) and quantitative real-time RT-PCR was performed using Power SYBR Green PCR Master Mix (Invitrogen). Sequences of the qPCR primer pairs (in the 5′-3′ direction) are as follows: GAPDH-F: GGTGAAGGTCGGAGTCAACGG; GAPDH-R: GAGGTCAATGAAGGGGTCATTG; TAZ-F: AGTACCCTGAGCCAGCAGAA; TAZ-R: GATTCTCTGAAGCCGCAGTT; AREG-F: GGGAGTGAGATTTCCCCTGT; AREG-R: AGCCAGGTATTTGTGGTTCG; PDGF-AA-F: CCATGCCACTAAGCATGTGC; PDGF-AA-R: GGAATCTCGTAAATGACCG; IGFBP-6-F: CAGAGACCAACAGAGGAATC; IGFBP-6-R: CTCGGTAGACCTCAGTCTGG; VEGF-F: CTACCTCCACCATGCCAAGT; VEGF-R: GCAGTAGCTGCGCTGATAGA; M-CSF-R-F: GGAAGGTGATGTCCATCAGC; and M-CSF-R-R: CCAGCTCTGCAGGCACCAG. All measurements were performed in triplicate and standardized to the levels of GAPDH.
Three-dimensional morphogenesis assay
Cells were cultured in Growth Factor Reduced BD Matrigel™ Matrix Basement Membrane (BD Biosciences) in Nunc Lab-Tek II Chamber Slide (Thermo Scientific) as described.43 In the mixing culture experiment, CherryRed-labeled MCF10A cells were co-cultured with GFP-labeled vector control or TAZ4SA MCF10A cells at a ratio of 1:1. In the experiment using conditioned media, media from vector control or TAZ4SA cells was collected and applied onto parental MCF10A cells. In neutralizing antibody experiment, normal goat IgG (Santa Cruz) and AREG antibody were applied into culture media at 1 µg ml−1. Cell lines were assayed in three independent experiments.
Transwell migration assay
Transwell cell migration assay was performed as previously described.9
Soft agar assay
5 × 104 cells were added to 1.5 ml of growth medium with 0.4% agar and layered onto 2 ml of 0.5% agar beds in six-well plates. Cells were fed with 1 ml of medium with 0.4% agar every seven days for three weeks, after which colonies were stained with 0.02% iodonitrotetrazolium chloride (Sigma-Aldrich) and photographed. Colonies larger than 50 μm in diameter were counted as positive for growth. Assays were conducted in triplicate in three independent experiments.
Immunohistochemistry of clinical TMA
Tissue microarrays (TMA) of breast cancer patients at RPCI was analyzed with informed consent. Anti-human TAZ antibody from Cell Signaling and anti-human AREG antibody from Proteintech were used for the immunohistochemistry. Cell nuclei were counterstained with Hematoxylin. Each sample on TMAs was read and scored at three microscopic observations by a pathologist at two independent readings. TAZ nuclear index was obtained through multiplication of scores. Correlation between TAZ and AREG was analyzed by Chi-square test.
Statistical analysis
Statistical analysis of data was performed using the SPSS statistics software package (SPSS, IL). All results are expressed as mean ± SD. *, p < 0.05; **, p < 0.001; ***, p < 0.0001.
Supplementary Material
Acknowledgments
We thank Drs. Yue Xiong (University of North Carolina) and Quanying Lei (Fudan University) for pBABE-wt-TAZ and -TAZ4SA. We thank Ms. Noreen Ersing for her expertise in immunohistochemistry. This work was supported by the Roswell Park Cancer Institute and National Cancer Institute (NCI) grant #P30 CA016056 (to J.Z). This research was supported, in part, by the American Cancer Society Institutional Research Grant 02–197–04 (to J.Z.).
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/21386
References
- 1.Pan D. The hippo signaling pathway in development and cancer. Dev Cell. 2010;19:491–505. doi: 10.1016/j.devcel.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhao B, Tumaneng K, Guan KL. The Hippo pathway in organ size control, tissue regeneration and stem cell self-renewal. Nat Cell Biol. 2011;13:877–83. doi: 10.1038/ncb2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mauviel A, Nallet-Staub F, Varelas X. Integrating developmental signals: a Hippo in the (path)way. Oncogene. 2012;31:1743–56. doi: 10.1038/onc.2011.363. [DOI] [PubMed] [Google Scholar]
- 4.Lei QY, Zhang H, Zhao B, Zha ZY, Bai F, Pei XH, et al. TAZ promotes cell proliferation and epithelial-mesenchymal transition and is inhibited by the hippo pathway. Mol Cell Biol. 2008;28:2426–36. doi: 10.1128/MCB.01874-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hao Y, Chun A, Cheung K, Rashidi B, Yang X. Tumor suppressor LATS1 is a negative regulator of oncogene YAP. J Biol Chem. 2008;283:5496–509. doi: 10.1074/jbc.M709037200. [DOI] [PubMed] [Google Scholar]
- 6.Zhao B, Li L, Tumaneng K, Wang CY, Guan KL. A coordinated phosphorylation by Lats and CK1 regulates YAP stability through SCF(beta-TRCP) Genes Dev. 2010;24:72–85. doi: 10.1101/gad.1843810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chan SW, Lim CJ, Chong YF, Pobbati AV, Huang C, Hong W. Hippo pathway-independent restriction of TAZ and YAP by angiomotin. J Biol Chem. 2011;286:7018–26. doi: 10.1074/jbc.C110.212621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao B, Li L, Lu Q, Wang LH, Liu CY, Lei Q, et al. Angiomotin is a novel Hippo pathway component that inhibits YAP oncoprotein. Genes Dev. 2011;25:51–63. doi: 10.1101/gad.2000111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Overholtzer M, Zhang J, Smolen GA, Muir B, Li W, Sgroi DC, et al. Transforming properties of YAP, a candidate oncogene on the chromosome 11q22 amplicon. Proc Natl Acad Sci USA. 2006;103:12405–10. doi: 10.1073/pnas.0605579103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gasparotto D, Polesel J, Marzotto A, Colladel R, Piccinin S, Modena P, et al. Overexpression of TWIST2 correlates with poor prognosis in head and neck squamous cell carcinomas. Oncotarget. 2011;2:1165–75. doi: 10.18632/oncotarget.390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zlobec I, Lugli A. Epithelial mesenchymal transition and tumor budding in aggressive colorectal cancer: tumor budding as oncotarget. Oncotarget. 2010;1:651–61. doi: 10.18632/oncotarget.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kanai F, Marignani PA, Sarbassova D, Yagi R, Hall RA, Donowitz M, et al. TAZ: a novel transcriptional co-activator regulated by interactions with 14-3-3 and PDZ domain proteins. EMBO J. 2000;19:6778–91. doi: 10.1093/emboj/19.24.6778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Walter MA, Spillett DJ, Thomas P, Weissenbach J, Goodfellow PN. A method for constructing radiation hybrid maps of whole genomes. Nat Genet. 1994;7:22–8. doi: 10.1038/ng0594-22. [DOI] [PubMed] [Google Scholar]
- 14.Mahoney WM, Jr., Hong JH, Yaffe MB, Farrance IK. The transcriptional co-activator TAZ interacts differentially with transcriptional enhancer factor-1 (TEF-1) family members. Biochem J. 2005;388:217–25. doi: 10.1042/BJ20041434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vassilev A, Kaneko KJ, Shu H, Zhao Y, DePamphilis ML. TEAD/TEF transcription factors utilize the activation domain of YAP65, a Src/Yes-associated protein localized in the cytoplasm. Genes Dev. 2001;15:1229–41. doi: 10.1101/gad.888601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kitagawa M. A Sveinsson’s chorioretinal atrophy-associated missense mutation in mouse Tead1 affects its interaction with the co-factors YAP and TAZ. Biochem Biophys Res Commun. 2007;361:1022–6. doi: 10.1016/j.bbrc.2007.07.129. [DOI] [PubMed] [Google Scholar]
- 17.Zhao B, Ye X, Yu J, Li L, Li W, Li S, et al. TEAD mediates YAP-dependent gene induction and growth control. Genes Dev. 2008;22:1962–71. doi: 10.1101/gad.1664408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang H, Liu CY, Zha ZY, Zhao B, Yao J, Zhao S, et al. TEAD transcription factors mediate the function of TAZ in cell growth and epithelial-mesenchymal transition. J Biol Chem. 2009;284:13355–62. doi: 10.1074/jbc.M900843200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dupont S, Morsut L, Aragona M, Enzo E, Giulitti S, Cordenonsi M, et al. Role of YAP/TAZ in mechanotransduction. Nature. 2011;474:179–83. doi: 10.1038/nature10137. [DOI] [PubMed] [Google Scholar]
- 20.Cui CB, Cooper LF, Yang X, Karsenty G, Aukhil I. Transcriptional coactivation of bone-specific transcription factor Cbfa1 by TAZ. Mol Cell Biol. 2003;23:1004–13. doi: 10.1128/MCB.23.3.1004-1013.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hong JH, Hwang ES, McManus MT, Amsterdam A, Tian Y, Kalmukova R, et al. TAZ, a transcriptional modulator of mesenchymal stem cell differentiation. Science. 2005;309:1074–8. doi: 10.1126/science.1110955. [DOI] [PubMed] [Google Scholar]
- 22.Park KS, Whitsett JA, Di Palma T, Hong JH, Yaffe MB, Zannini M. TAZ interacts with TTF-1 and regulates expression of surfactant protein-C. J Biol Chem. 2004;279:17384–90. doi: 10.1074/jbc.M312569200. [DOI] [PubMed] [Google Scholar]
- 23.Tian Y, Li D, Dahl J, You J, Benjamin T. Identification of TAZ as a binding partner of the polyomavirus T antigens. J Virol. 2004;78:12657–64. doi: 10.1128/JVI.78.22.12657-12664.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murakami M, Nakagawa M, Olson EN, Nakagawa O. A WW domain protein TAZ is a critical coactivator for TBX5, a transcription factor implicated in Holt-Oram syndrome. Proc Natl Acad Sci USA. 2005;102:18034–9. doi: 10.1073/pnas.0509109102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murakami M, Tominaga J, Makita R, Uchijima Y, Kurihara Y, Nakagawa O, et al. Transcriptional activity of Pax3 is co-activated by TAZ. Biochem Biophys Res Commun. 2006;339:533–9. doi: 10.1016/j.bbrc.2005.10.214. [DOI] [PubMed] [Google Scholar]
- 26.Varelas X, Sakuma R, Samavarchi-Tehrani P, Peerani R, Rao BM, Dembowy J, et al. TAZ controls Smad nucleocytoplasmic shuttling and regulates human embryonic stem-cell self-renewal. Nat Cell Biol. 2008;10:837–48. doi: 10.1038/ncb1748. [DOI] [PubMed] [Google Scholar]
- 27.Bhat KP, Salazar KL, Balasubramaniyan V, Wani K, Heathcock L, Hollingsworth F, et al. The transcriptional coactivator TAZ regulates mesenchymal differentiation in malignant glioma. Genes Dev. 2011;25:2594–609. doi: 10.1101/gad.176800.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cordenonsi M, Zanconato F, Azzolin L, Forcato M, Rosato A, Frasson C, et al. The Hippo transducer TAZ confers cancer stem cell-related traits on breast cancer cells. Cell. 2011;147:759–72. doi: 10.1016/j.cell.2011.09.048. [DOI] [PubMed] [Google Scholar]
- 29.Debnath J, Brugge JS. Modelling glandular epithelial cancers in three-dimensional cultures. Nat Rev Cancer. 2005;5:675–88. doi: 10.1038/nrc1695. [DOI] [PubMed] [Google Scholar]
- 30.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/S0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 31.Zhang J, Ji JY, Yu M, Overholtzer M, Smolen GA, Wang R, et al. YAP-dependent induction of amphiregulin identifies a non-cell-autonomous component of the Hippo pathway. Nat Cell Biol. 2009;11:1444–50. doi: 10.1038/ncb1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Busser B, Sancey L, Brambilla E, Coll JL, Hurbin A. The multiple roles of amphiregulin in human cancer. Biochim Biophys Acta. 2011;1816:119–31. doi: 10.1016/j.bbcan.2011.05.003. [DOI] [PubMed] [Google Scholar]
- 33.Zhang Z, Stiegler AL, Boggon TJ, Kobayashi S, Halmos B. EGFR-mutated lung cancer: a paradigm of molecular oncology. Oncotarget. 2010;1:497–514. doi: 10.18632/oncotarget.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chan SW, Lim CJ, Guo K, Ng CP, Lee I, Hunziker W, et al. A role for TAZ in migration, invasion, and tumorigenesis of breast cancer cells. Cancer Res. 2008;68:2592–8. doi: 10.1158/0008-5472.CAN-07-2696. [DOI] [PubMed] [Google Scholar]
- 35.Zhao B, Wei X, Li W, Udan RS, Yang Q, Kim J, et al. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 2007;21:2747–61. doi: 10.1101/gad.1602907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang J, Smolen GA, Haber DA. Negative regulation of YAP by LATS1 underscores evolutionary conservation of the Drosophila Hippo pathway. Cancer Res. 2008;68:2789–94. doi: 10.1158/0008-5472.CAN-07-6205. [DOI] [PubMed] [Google Scholar]
- 37.Liu CY, Zha ZY, Zhou X, Zhang H, Huang W, Zhao D, et al. The hippo tumor pathway promotes TAZ degradation by phosphorylating a phosphodegron and recruiting the SCFbeta-TrCP E3 ligase. J Biol Chem. 2010;285:37159–69. doi: 10.1074/jbc.M110.152942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morin-Kensicki EM, Boone BN, Howell M, Stonebraker JR, Teed J, Alb JG, et al. Defects in yolk sac vasculogenesis, chorioallantoic fusion, and embryonic axis elongation in mice with targeted disruption of Yap65. Mol Cell Biol. 2006;26:77–87. doi: 10.1128/MCB.26.1.77-87.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hossain Z, Ali SM, Ko HL, Xu J, Ng CP, Guo K, et al. Glomerulocystic kidney disease in mice with a targeted inactivation of Wwtr1. Proc Natl Acad Sci USA. 2007;104:1631–6. doi: 10.1073/pnas.0605266104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tian Y, Kolb R, Hong JH, Carroll J, Li D, You J, et al. TAZ promotes PC2 degradation through a SCFbeta-Trcp E3 ligase complex. Mol Cell Biol. 2007;27:6383–95. doi: 10.1128/MCB.00254-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Makita R, Uchijima Y, Nishiyama K, Amano T, Chen Q, Takeuchi T, et al. Multiple renal cysts, urinary concentration defects, and pulmonary emphysematous changes in mice lacking TAZ. Am J Physiol Renal Physiol. 2008;294:F542–53. doi: 10.1152/ajprenal.00201.2007. [DOI] [PubMed] [Google Scholar]
- 42.Booth BW, Boulanger CA, Anderson LH, Jimenez-Rojo L, Brisken C, Smith GH. Amphiregulin mediates self-renewal in an immortal mammary epithelial cell line with stem cell characteristics. Exp Cell Res. 2010;316:422–32. doi: 10.1016/j.yexcr.2009.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Debnath J, Muthuswamy SK, Brugge JS. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods. 2003;30:256–68. doi: 10.1016/S1046-2023(03)00032-X. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.