Abstract
Effectiveness of DNA cross-linking drugs in the treatment of bladder cancer suggests that bladder cancer cells may have harbored an insufficient cellular response to DNA cross-link damage, which will sensitize cells to DNA cross-linking agents. Cell sensitivity benefits from deficient DNA damage responses, which, on the other hand, can cause cancer. Many changed cellular signaling pathways are known to be involved in bladder tumorigenesis; however, DNA cross-link damage response pathway [Fanconi anemia (FA) pathway], whose alterations appear to be a plausible cause of the development of bladder cancer, remains an under-investigated area in bladder cancer research. In this study, we found FAVL (variant of FA protein L—FANCL) was elevated substantially in bladder cancer tissues examined. Ectopic expression of FAVL in bladder cancer cells as well as normal human cells confer an impaired FA pathway and hypersensitivity to Mitomycin C, similar to those found in FA cells, indicating that FAVL elevation may possess the same tumor promotion potential as an impaired FA pathway harbored in FA cells. Indeed, a higher level of FAVL expression can promote the growth of bladder cancer cells in vitro and in vivo, which, at least partly, results from FAVL perturbation of FANCL expression, an essential factor for the activation of the FA pathway. Moreover, a higher level of FAVL expression was found to be associated with chromosomal instability and the invasiveness of bladder cancer cells. Collectively, FAVL elevation can increase the tumorigenic potential of bladder cancer cells, including the invasive potential that confers the development of advanced bladder cancer. These results enhance our understanding the pathogenesis of human bladder cancer, holding a promise to develop additional effective tools to fight human bladder cancer.
Keywords: FA tumor suppressor pathway, FAVL, bladder cancer, cell invasiveness, chromosomal instability
Introduction
Bladder cancer is the fourth most common cancer and the ninth leading cause of cancer death in the United States.1 Most of bladder cancers are transitional cell carcinomas (TCC). Other types include squamous cell carcinoma and adenocarcinoma. In most patients with bladder cancer, death results from metastatic disease and high recurrence. In bladder cancer treatment, surgery, chemotherapy, radiation therapy, immunotherapy and vaccine therapy are all critical elements. Among chemotherapies, cisplatin or mitomycin C (MMC) DNA cross-linking agent is one of most commonly used intravesical drugs for treating non-invasive bladder cancer. About 50–68% of patients with superficial bladder cancer (non-invasive) have a very good response to intravesical therapy. This type of chemotherapeutic agents is also considered in the treatment of advanced bladder cancer (invasive) that has extended beyond the bladder wall. Therefore, DNA cross-link agents appear to be very effective in killing bladder cancer cells, suggesting the cellular machinery in processing DNA cross-link damage is somehow insufficient in bladder cancer cells, eventually dying of irreparable DNA damage. Bladder cancer development/progression attributed to many defective cellular signaling pathways, including HRAS,2 p53 and RB,2-4 MMP-9,5 IL-8,6 VEGF,7 EGFR8,9 and many others.10-12 The cellular response specifically to DNA cross-link damage is the Fanconi Anemia (FA) /FA-BRCA pathway.13 However, it is largely unclear whether the FA/FA-BRCA pathway is insufficient in bladder cancer cells, and the insufficiency then contributes to the development of bladder cancer.
FA is a rare autosomal recessive or X-linked disease characterized by severe bone marrow failure, developmental abnormalities and an extremely high incidence of both hematological and non-hematological malignancies.14-19 FA cells display chromosome abnormalities and hypersensitivity to DNA cross-linking agents,14,20,21 which can be recapitulated in many types of non-FA cells by disrupting the function of FA genes in our studies22-25 and those of others.26,27 To date, there are 14 or 15 classified FA groups FANC-A, B, C, D1, D2, E, F, G, I, J, L, M, N, O and P,14,15,18-21,28-37 each of which can be accounted for by mutations in a given gene unique to that group. The sensitivity of FA cells from all complementation groups to DNA cross-linking agents and their similar clinical phenotypes suggest that all FA proteins function in a common DNA damage response pathway. Eight of the known FA proteins (FANCA, FANCB, FANCC, FANCG, FANCE, FANCF, FANCL and FANCM)15,30,32,38 and other known39,40 and unknown proteins function together by forming a multi-protein complex to serve as an E3 ubiquitin ligase to monoubiquitinate FANCD2 at lysine 561 (K561)26,41-48 and its paralog FANCI at K523.49 This pathway is activated during DNA replication as well as following DNA damage triggered especially by DNA cross-linking agents, such as mitomycin C (MMC), diepoxybutane (DEB) or cisplatin,50,51 and the monoubiquitinated FANCD2 and FANCI along with FANCD1/BRCA2,42 FANCN/PALB2,30,32,38,52 FANCJ/BRIP153,54 and others work in concert to repair DNA damage. FANCD2 thus is the central player of the pathway,28 and monoubiquitinated FANCD2 can be used as a measure for activation of this signaling pathway. Of the eight FA complex proteins, FANCL was identified as a catalytic subunit of the complex E3 ubiquitin ligase, because it contains a C-terminal PHD finger domain that exhibits E3 ubiquitin ligase activity in vitro.26
In FA patients, cancer incidence is about 5-fold higher than that of the general population and up to several hundred-fold higher for particular malignancies.55 This cancer-prone phenomenon indicated that FA genes play crucial roles in tumor suppression. Thus, the signaling pathway constituted by FA proteins has been termed the FA tumor suppressor pathway. This pathway is also called the FA-BRCA tumor suppressor pathway, because three FA proteins, FANCD1, FANCJ and FANCN, are BRCA or BCRA-related proteins, which are BRCA2, BRIP1 (BRCA1 interaction protein 1) and PALB2 (partner and localizer of BRCA2), respectively. Gene therapy was found to improve the clinical symptoms of FA in the hematopoietic system;56 however, patients still eventually develop cancer57 and die, with a mean age of 16 y.58 The FA tumor suppressor pathway has long been suggested,59 but the implications of this disordered signaling pathway directly in non-FA human cancer just began to be appreciated in our recent work.22 Despite many studies aimed at investigating various aspects of this cancer-prone genetic disease using a variety of methods, including those of genetics, biochemistry and the clinical sciences,21,26,46,60-65 much remains to be learned. For example, how the FA pathway influences specific types of human cancer development is unknown. In this study, we report for the first time that FAVL impairment of the FA pathway has a substantial effect on the development of human bladder cancer, providing novel insights into the development of additional effective tools to fight bladder cancer.
Results
FAVL expression is elevated in bladder cancer tissues
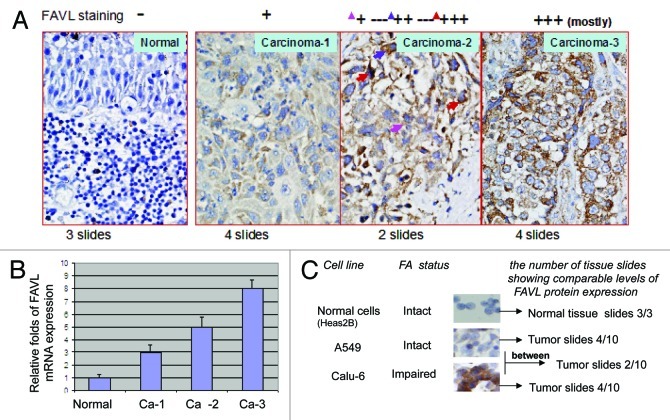
Effectiveness of DNA cross-link drugs in the treatment of bladder cancer suggests that bladder cancer cells may have harbored an insufficient cross-link DNA damage response that allows cells to be sensitive to DNA cross-link agents. Cell sensitivity benefits from deficient DNA damage responses, which, on the other hand, can cause cancer. Many altered cellular signaling pathways are known to be involved in bladder tumorigenesis; however, roles of DNA cross-link damage response pathway known as the FA pathway has never been studied in the development of bladder cancer. To this end, we decided to investigate the roles of FAVL in the development of bladder cancer, which is a tumor promotion factor we found previously,22,23 by disrupting the FA signaling pathway. Using immunohistochemistry (IHC) assay, we found positive FAVL staining on bladder carcinoma tissue slides showing diffuse cytoplasmic staining with various staining intensities (Fig. 1A). Compared with the three normal tissue samples examined, FAVL expression was elevated to some extend in 10 tumor samples examined. Specifically, four samples display a low density of FAVL staining; four samples show a higher density of FAVL staining; and two samples carry mixed densities with various levels of FAVL staining intensity at different spots as indicated in Figure 1A. We also confirmed elevated FAVL mRNA level in these bladder cancer tissues by quantitative RT-PCR, which is consistent with the levels of FAVL expression detected by IHC (Fig. 1B). Moreover, we compared the immunostaining intensity of FAVL expression in bladder cancer tissues to those in cell lines under the same IHC conditions, and these cells are showed an impaired or an intact status of the FA pathway.22 We found more than half of the bladder carcinoma cases tested have a similar FAVL staining intensity, as shown in cells that harbor a defective FA pathway (Fig. 1C). This suggests that nearly half of bladder carcinomas might have harbored an impaired FA pathway that may have contributed to the development of bladder cancer.
Figure 1. FAVL expression is elevated in bladder cancer tissue samples. (A) Immunohistochemical analysis of FAVL expression in bladder cancer tissues. The elevated level of FAVL expression is presented by the various degrees of brown color. Four normal tissues are negative for FAVL staining, as shown in panel 1. Two of 10 carcinoma tissue samples show a light brown color; 4 of 10 show a darker brown, and the remaining 2 appear to show a heterogeneous staining with both light and dark brown spots. (B) Quantitative RT-PCR analysis of FAVL expression in a panel of bladder cancer tissues. The VIC-FAVL splice junction (26mer, synthesized from ABI) was used as the Taqman probe, and 6FAM-actin (purchase from ABI) served as an internal control. The relative level of FAVL mRNA expression is higher in bladder cancer tissues compared with normal control. The intensity of brown color correlates with the FAVL mRNA expression detected by qRT-PCR (the error bar indicates the standard deviation derived from triplicate). (C) Nearly a half of bladder carcinomas detected express a higher level of FAVL, which is a comparable level shown in cells carrying an impaired FA status triggered by FAVL.
FAVL elevation enhances the growth potential of bladder cancer cells in vitro and in vivo
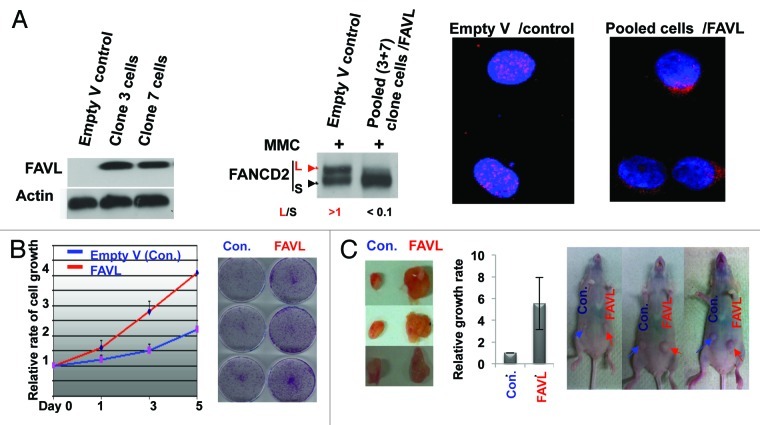
FAVL was found substantially elevated in human bladder cancer tissues examined (Fig. 1). Whether FAVL elevation contributes to the development of bladder cancer appears to be an imminent question. We thus systematically examined how the growth rate of bladder cancer cells is affected by elevated FAVL. HTB-4 bladder cancer cell line, expressing FAVL at the lowest level among several TCC cell lines tested and carrying an intact FA pathway (data not shown), appears to be a suitable cell system to study the effect of elevated FAVL on the growth of bladder cancer cells. Using HTB-4 cells, we generated stable cell pairs that express FAVL at a higher or a normal level and, thus, harbor an intact or impaired status of the FA pathway, respectively (Fig. 2A). The status of the FA pathway in stable cells was further confirmed by the FANCD2 focus study (Fig. 2B), another measure for the status of the FA signaling pathway. Subsequently, we conducted cell proliferation assay and found FAVL clearly promoted cell growth (Fig. 2C). Whether elevated FAVL possesses a strong potential to enhance the growth rate of bladder cancer cells in vivo was also assessed. We found that the xenograft tumors generated from HTB-4 cells carrying a higher level of FAVL expression grow much faster compared with the corresponding controls, suggesting FAVL elevation can also increase in vivo growth potential (Fig. 2D). Together, these results reveal that FAVL is an unrecognized cause of bladder cancer development, which may contribute heavily to bladder cancer development and/or progression.
Figure 2. FAVL promotes the bladder cancer cell growth in vitro and in vivo. (A) Two sub-lines, derived from bladder cancer cells, stably express a higher level of FAVL (left panel). Pooled stable cells were verified to carry an impaired FA pathway, indicated by the compromised level of monoubiquitinated FANCD2 (middle panel) and decreased focus formation (right panel). (B and C) Cells expressing FAVL at a higher level grow faster compared with control cells (empty-V transfected) in vitro (B) and in vivo (C).
FAVL can promote the invasive potential of bladder cancer cells
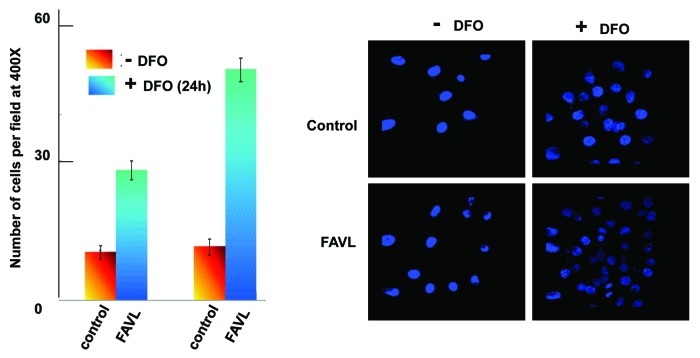
Cancer results from accumulated genetic alterations that, over time, cause genomic instability. As such, tumor formation proceeds through relatively phase-specific transformations. These phases generally are hyperplasia, tumor in situ and invasion/metastasis. The progression of bladder cancer through these phases mainly depends on the invasiveness of cancer cells, which is a major cause of bladder cancer death. To further understand the role of FAVL in bladder tumorigenesis, we extended the experiments done on growth potentials by revealing whether FAVL is capable of promoting tumor cell invasion. As shown in Figure 3, bladder cancer cells expressing FAVL at a higher level show a substantially increased capability to penetrate the other side of mesh, which represents the ability of cancer cells to invade and metastasize to other places. Interestingly, when cells growth under normoxia condition, we did not observe a clear difference in bladder cells expressing different levels of FAVL. However, when cells were pre-cultured in hypoxic condition, the invasiveness triggered by FAVL elevation was clearly detected. These data suggest that FAVL elevation remains to be an important tumor promotion factor responsible for the formation of advanced tumors, which, in general, would carry hypoxia.
Figure 3. Invasiveness is promoted in cells expressing a higher level of FAVL. A total of 5000 HTB-4 cells containing empty vector as a control or expressing a higher level of FAVL protein were plated respectively in the chamber in triplicate as described in the procedure provided by manufacture. The chamber member was then taken out of the chamber 2 h after seeding, fixed and stained with DAPI. The number of cells per microscopic field was generated by averaging 10 fields randomly selected. A set of representative images were shown.
FAVL impairs the FA pathway in bladder cancer cells by affecting the availability of FANCL, a catalytic subunit of the FA complex E3 for the activation of the FA pathway
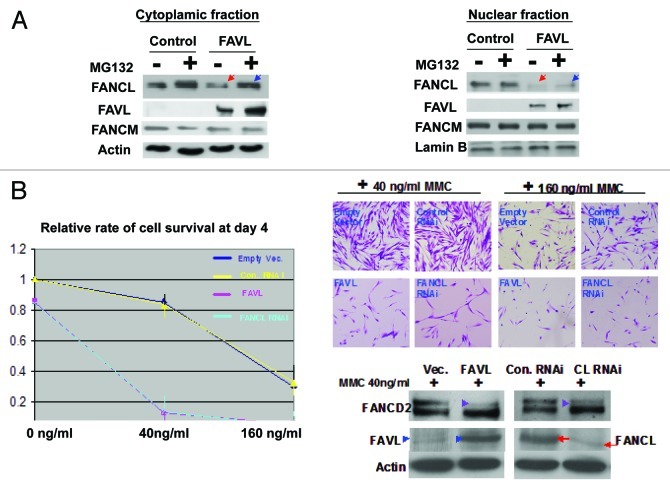
FAVL disrupts the FA pathway through sequestration of FANCL in the cytoplasm and promoting its degradation in osteosarcoma cells (U2OS).22 To known whether FAVL impairment of the FA pathway in bladder cancer cells in a manner similar to the one we found in U2OS cells, we examined FAVL and FANCL protein levels in cytoplasmic and nuclear fractions prepared from bladder stable cell pairs expressing normal or an elevated level of FAVL. We found that in HTB-4 cells carrying a high level of FAVL, FANCL expression level in the nuclear fraction was much lower when compared to the corresponding expression level in HTB-4 cells harboring a relatively low level of FAVL expression (Fig. 4A). When cells are treated with a proteasome inhibitor, MG132, the level of FANCL in the cytoplasm is relatively increased (Fig. 4A). Indeed, FAVL impairs the FA pathway in bladder cancer cells is in a manner analogous to the one we revealed in U2OS cells by affecting FANCL availability for the FA complex E3.22 Therefore, FANCL availability interrupted by FAVL elevation appears to be a common mechanism underlying the tumor promotion activity initiated by FAVL. To further validate the relationship between FANCL and FAVL, we asked whether downregulation of FANCL and overexpression of FAVL show similar phenotypes in “normal” human cells (WI-38). We found that WI-38 cells either silencing FANCL or overexpressing FAVL show a similar cell survival rate (Fig. 4B), suggesting that FAVL elevation and FANCL downregulation function in a common signaling pathway. Therefore, downregulated FANCL is crucial for the tumor promotion activity triggered by FAVL.
Figure 4. Downregulated FANCL is a mechanism underlying the function of elevated FAVL. (A) FAVL interferes with the nuclear expression level of FANCL protein Both HTB-4 control and FAVL-overexpressed cells were first treated with or without 5 μM of MG132 for 12 h. Subsequently, cytoplasmic and nuclear fractions were prepared for western blotting analysis on FANCL, FANCM, FAVL and loading controls (actin for cytoplasmic fraction; lamin for nuclear fraction). Expression level of FANCL protein was decreased in both cytoplasmic and nuclear fractions when cells carrying a higher level of FAVL (Red arrowhead), but is relatively increased when treated with MG132 (blue arrowhead compared with the red one accordingly). The level of FANCM protein remains to be similar in cells expressing different levels of FAVL, or with or without MG132 treatment. (B) Silencing FANCL and overexpression of FAVL function in a common signaling pathway. WI-38 cells were transfected with the plasmid containing FAVL cDNA or RNAi oligos targeting FANCL as well as corresponding empty vector or non-specific RNAi oligo control, respectively. Transfected cells were treated with or without MMC in quadruplicate for three consecutive days. Subsequently, we counted these cells and took corresponding images. The rate of cell survival is similar between FAVL cDNA and FANCL RNAi transfected cells (left panel, plotted from the total number of survived cells; the corresponding images shown in right top panel). The amount of FAVL plasmid or FANCL RNAi oligos were enough to compromise FANCD2 activation (right bottom panel, marked with the purple arrowheads). The protein levels of FAVL marked with blue arrowheads) or FANCL (marked with red arrowheads) were increased or decreased, respectively, in WI-38 cells harboring FAVL cDNA or FANCL RNAi oligos.
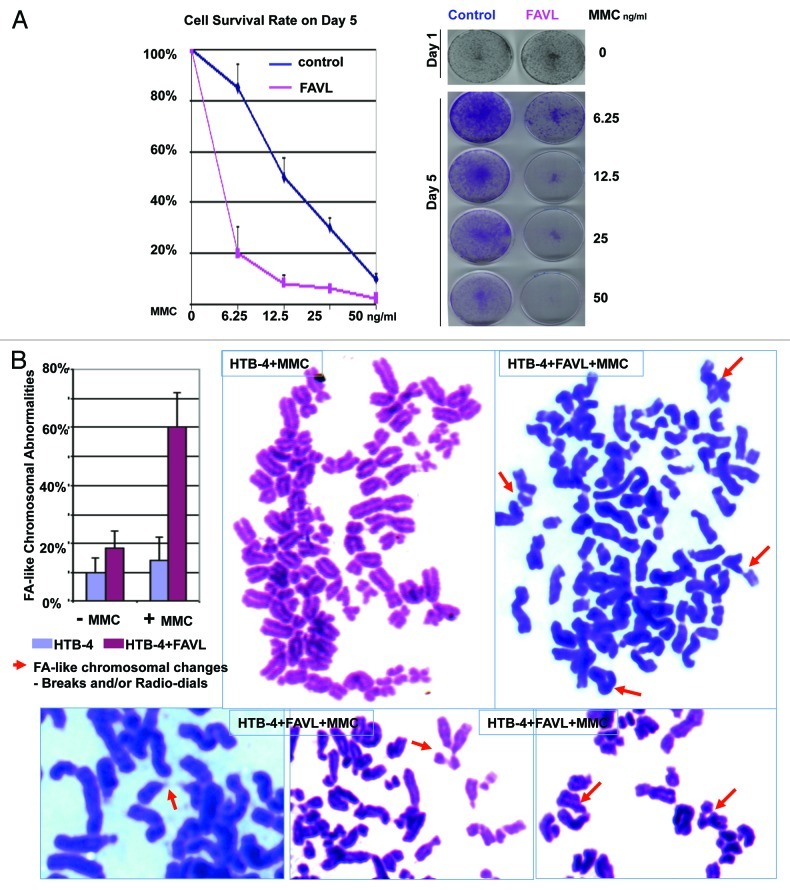
FAVL disruption of the FA pathway promotes bladder cancer cell chromosome abnormalities and drug sensitivity
We have observed that FAVL elevation in a significant portion of bladder carcinoma cases detected (Fig. 1), which may lead to a deficient FA pathway, as suggested in Figure 3. Therefore, we questioned whether chromosomal abnormalities, a hallmark of cancer, would be promoted by FAVL impairment of the FA pathway harbored in bladder cancer cells. Human HTB-4 bladder cancer cells expressing different levels of FAVL were used to prepare conventional chromosomal spreads. We found that HTB-4 cells expressing FAVL at a higher level showed a dramatically increase in the percentage of FA-chromosomal abnormalities compared with the corresponding control cells (Fig. 5A). Therefore, as a defective FA pathway promotes chromosomal abnormalities in FA cells, FAVL impairment of the FA/FA-BRCA pathway in bladder cancer cells can also promote chromosomal instability, conferring a hallmark of bladder carcinomas. We also asked whether an impaired FA pathway triggered by FAVL elevation renders the hypersensitivity of bladder cancer cells to DNA cross-linking agents. Cell survival assay using HTB-4 derivative cells expressing a low or high level of FAVL was performed. As shown in Figure 5B, cells carrying a high level of FAVL are hypersensitive to MMC compared with the control cells, indicating FAVL can sensitize cells to MMC, which might be a critical factor conferring the sensitivity of bladder cancer cells to the platinum chemotherapy known in clinic, providing a novel molecular basis for a fact that platinum-related drugs are the most commonly used in the treatment of human bladder cancer.
Figure 5. FAVL elevation is associated with Chromosomal Instability and cisplatin sensitivity. (A) Both control and FAVL-overexperssion cells were treated with a series of MMC for 5 d. Cell viability was plotted upon the readings from MTT assay on cells treated for 5 d with difference concentrations of MMC, with non-treated cells as 100%. The images shown on the right are a set of cells fixed and stained with crystal blue. Cells carrying FACL are more sensitive to cell death triggered by MMC. (B) The same set of HTB-4 derivative cells were split and subsequently treated with 100 ng/ml MMC for 24 h. Both control and FAVL-overexpressing cells were prepared for chromosomal spread study. The percentage of chromosomal abnormalities was scored only upon breaks and tri/tetra radio-dials, which are FA-like chromosomal abnormalities. FAVL elevation enhances the percentage of FA-like chromosomal changes. Images shown are the whole or partial chromosome spreads prepared from treated cells.
Discussion
Among all types of cancer, cisplatin is a relatively most effective chemotherapeutic drug in the treatment of bladder cancer, suggesting cellular signaling pathways responsible for repairing cross-link damage harbored in bladder cancer cells are most likely defective, leading cells to be sensitive to the treatment. Indeed, the FA-BRCA signaling pathway, responsible especially for repairing cross-link DNA damage, was found to be impaired in more than 35% of bladder carcinoma cases, resulting from an elevated level of FAVL expression, which is comparable to the one in cells found to have an impaired FA pathway (Fig. 1). Similar to FA cells that are sensitive to cross-linking agents, bladder cancer cells displayed hypersensitivity to cross-linking agents when ectopically expressing FAVL (Fig. 5). More importantly, elevated FAVL promotes not only the growth of bladder cancer cells in vitro and in vivo, but also their invasiveness (Figs. 2 and 3), which confers the development of advanced bladder cancer. Therefore, FAVL appears to be a tumor promotion factor that may function through all phases of bladder tumor development. For bladder cancer, tumor development undergoes specific phase-transitions, such as, Ta, non-invasive papillary carcinoma; Tis, carcinoma in situ, “flat tumor”; T1, tumor invades subepithelial connective tissue; T2, tumor invades muscularis propria; T2a, tumor invades inner half muscle; T2b, tumor invades outer half muscle; T3, tumor invades perivesical tissue and T4, tumor invades other organs and pelvic wall. How FAVL affects the development of particular phases needs to be further defined in the future.
Research advances in the field of molecular biology have provided a new understanding of the genetic background of human bladder cancer. For example, it is known that bladder tumorigenesis results from numerous defective cellular signaling pathways, including HRAS,2 p53 and RB,2,3 MMP-9,5 IL-8,6 VEGF7 and EGFR8 pathways. However, bladder cancer remains to be one of leading causes of cancer death, indicating studies on improving our understanding of pathogenesis of human bladder cancer are much needed. We here for the first time found that FAVL impairment of the FA pathway contributes to bladder tumorigenesis. Knowledge learned herein, like genetic abnormalities and biologic aberrations known to have roles in tumorigenesis,2,3 will be the basis for investigations on the development of specific methods for bladder cancer prevention, diagnosis and/or treatment.
Increasing studies provide evidence linking the compromised FA-BRCA signaling cascade to sporadic human cancers. In these cancers, the FA-BRCA pathway is impaired by epigenetic silencing and somatic or inherited mutations of one or several FA genes, supporting the long proposed concept that the FA-BRCA pathway is a tumor suppressor pathway. We previously found a novel alternative splice variant of FANCL, named FAVL, which was increased in nearly 50% of cancer tissue samples detected. More importantly, the study on FAVL demonstrated, for the first time, the essential role of an intact FA-BRCA signaling in suppressing the development of non-FA human cancer (11). Here, we aimed to address how the FA-BRCA tumor suppressor pathway affects specific types of human cancer. The results presented in this study are among the first to indicate that the FA-BRCA tumor suppressor pathway may emerge to be an important guardian pathway responsible for protecting human cells, particularly bladder cells, from going awry and becoming neoplasm.
Materials and Methods
Cell lines, antibodies, chemicals and RNAi oligos
All cell lines, with the exception of WI-38 from Coriell, were purchased from ATCC. The anti-FANCD2 antibody was purchased from NOVUS (cat#N100–182). The anti-Flag (cat# F3165) and anti-β-actin (cat# 5316) antibodies as well as MMC and cisplatin were from Sigma. FAVL siRNA- ctgaccatggattttactatg and the nonspecific control siRNA -LacZ-siRNA-gtgaccagcgaatacctgt were synthesized from Dharmacon.
Immunohistochemistry, immunoblotting and quantitative RT-PCR
All methods were described as our previous report.22,66 IHC was performed by using FAVL antibody with 1:50 dilation ratio for primary incubation, and followed by using ImmPRESS Reagent Kit (Vector cat#MP-7401). For immunoblotting, primary antibodies were used at a dilution ratio of 1:1000 for FANCD2 and FANCL or 1:5000 for anti-Actin. VIC-FAVL and 6FAM-Actin internal controls were purchased from ABI. Total RNA was extracted using TRIzol Reagent (Invitrogen). The cDNA for PCR template was generated by using First-strand cDNA Synthesis Kit (Invitrigen) as recommended in the manufactures’ protocols.
Transfection
WI-38 cells or HTB4 cells were cultured in DMEM containing 10% FBS. Plasmids and SiRNA transient transfections were performed using reagents Lipfectamine2000 and Oligofectamine, respectively. The procedures of transfection were described as manufacture’s protocols.
Protein fraction preparation
Both cytoplasmic and nuclear fractions were prepared essentially as described in the protocols provided by the manufacturer (PIERCE). The NR-PER Kit (cat#78833) (Nuclear and Cytoplasmic Extraction Reagents) was used.
Cell proliferation
Cells with or without overexpressed FAVL were plated at day 0 with an equal number, and total cell numbers were recounted on day 1, 3 and 5. The cell growth curve was generated by plotting cell numbers counted with SEM (n = 3) from three independent experiments, averaged from triplicate samples for each individual experiment.
Xenograft formation
Nude mice (4–6 weeks old) purchased from NCI Frederick were injected with 200 μl Geltrex flurry (Invitrogen) (prepared at a 1:1 ratio with 1 × PBS) containing 6 million cells constitutively expressing FAVL or containing empty vector control.
Animals
Five- to six-week old male nude mice were obtained from NCI Frederick. The mice were euthanized at the end of experiments using an approved Institutional Animal Care and Use Committee protocol, which followed recommendations of the Panel on Euthanasia of the American Veterinary Medical Association. Xenograft tumors were excised out for weighting mass and pictures.
Immunofluorescence (IF)
IF was performed according to standard procedures. Briefly, cells were first fixed by 1% paraformaldehyde and permeabilized with 0.1% Triton X-100, and followed by blocking with 3% goat serum in 1X PBS and incubating with primary antibody (FANCD2 antibody 1:1,000 dilution in 1X PBS containing 0.05% goat serum).
Biospecimen
All biospecimens used in this study were under an approved IRBe protocol.
Acknowledgments
This study is supported by NIH grant (CAR01CA136532 to PF). We thank Dr. Weidong Wang (NIH-NIA) for providing FANCL antibody and FANCL cDNA, and the Fanconi Anemia Research Foundation (FARF) for FANCA, FANCM and FANCF antibodies. We also thank Department of Laboratory Medicine and Pathology, Mayo Clinic and The University of Hawaii Cancer Center, University of Hawaii for their support.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/21400
References
- 1.Jemal A, et al. Cancer genetics. CA: a Cnacer Journal of Clinicians. 2007;57:43–66. doi: 10.3322/canjclin.57.1.43. [DOI] [Google Scholar]
- 2.Mitra AP, Birkhahn M, Cote RJ. p53 and retinoblastoma pathways in bladder cancer. World J Urol. 2007;25:563–71. doi: 10.1007/s00345-007-0197-0. [DOI] [PubMed] [Google Scholar]
- 3.George B, Datar RH, Wu L, Cai J, Patten N, Beil SJ, et al. p53 gene and protein status: the role of p53 alterations in predicting outcome in patients with bladder cancer. J Clin Oncol. 2007;25:5352–8. doi: 10.1200/JCO.2006.10.4125. [DOI] [PubMed] [Google Scholar]
- 4.Jee HJ, Kim AJ, Song N, Kim HJ, Kim M, Koh H, et al. Nek6 overexpression antagonizes p53-induced senescence in human cancer cells. Cell Cycle. 2010;9:4703–10. doi: 10.4161/cc.9.23.14059. [DOI] [PubMed] [Google Scholar]
- 5.Deryugina EI, Quigley JP. Matrix metalloproteinases and tumor metastasis. Cancer Metastasis Rev. 2006;25:9–34. doi: 10.1007/s10555-006-7886-9. [DOI] [PubMed] [Google Scholar]
- 6.Izawa JI, Slaton JW, Kedar D, Karashima T, Perrotte P, Czerniak B, et al. Differential expression of progression-related genes in the evolution of superficial to invasive transitional cell carcinoma of the bladder. Oncol Rep. 2001;8:9–15. doi: 10.3892/or.8.1.9. [DOI] [PubMed] [Google Scholar]
- 7.Crew JP, O’Brien T, Bradburn M, Fuggle S, Bicknell R, Cranston D, et al. Vascular endothelial growth factor is a predictor of relapse and stage progression in superficial bladder cancer. Cancer Res. 1997;57:5281–5. [PubMed] [Google Scholar]
- 8.Chow NH, Liu HS, Yang HB, Chan SH, Su IJ. Expression patterns of erbB receptor family in normal urothelium and transitional cell carcinoma. An immunohistochemical study. Virchows Arch. 1997;430:461–6. doi: 10.1007/s004280050056. [DOI] [PubMed] [Google Scholar]
- 9.Sayan AE, Stanford R, Vickery R, Grigorenko E, Diesch J, Kulbicki K, et al. Fra-1 controls motility of bladder cancer cells via transcriptional upregulation of the receptor tyrosine kinase AXL. Oncogene. 2012;31:1493–503. doi: 10.1038/onc.2011.336. [DOI] [PubMed] [Google Scholar]
- 10.Sugano G, Bernard-Pierrot I, Laé M, Battail C, Allory Y, Stransky N, et al. Milk fat globule--epidermal growth factor--factor VIII (MFGE8)/lactadherin promotes bladder tumor development. Oncogene. 2011;30:642–53. doi: 10.1038/onc.2010.446. [DOI] [PubMed] [Google Scholar]
- 11.Ahmad I, Morton JP, Singh LB, Radulescu SM, Ridgway RA, Patel S, et al. β-Catenin activation synergizes with PTEN loss to cause bladder cancer formation. Oncogene. 2011;30:178–89. doi: 10.1038/onc.2010.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Askham JM, Platt F, Chambers PA, Snowden H, Taylor CF, Knowles MA. AKT1 mutations in bladder cancer: identification of a novel oncogenic mutation that can co-operate with E17K. Oncogene. 2010;29:150–5. doi: 10.1038/onc.2009.315. [DOI] [PubMed] [Google Scholar]
- 13.Sengerová B, Wang AT, McHugh PJ. Orchestrating the nucleases involved in DNA interstrand cross-link (ICL) repair. Cell Cycle. 2011;10:3999–4008. doi: 10.4161/cc.10.23.18385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bagby GC., Jr. Genetic basis of Fanconi anemia. Curr Opin Hematol. 2003;10:68–76. doi: 10.1097/00062752-200301000-00011. [DOI] [PubMed] [Google Scholar]
- 15.Taniguchi T, D’Andrea AD. Molecular pathogenesis of Fanconi anemia: recent progress. Blood. 2006;107:4223–33. doi: 10.1182/blood-2005-10-4240. [DOI] [PubMed] [Google Scholar]
- 16.Huang TT, D’Andrea AD. Regulation of DNA repair by ubiquitylation. Nat Rev Mol Cell Biol. 2006;7:323–34. doi: 10.1038/nrm1908. [DOI] [PubMed] [Google Scholar]
- 17.Fei P, Yin J, Wang W. New advances in the DNA damage response network of Fanconi anemia and BRCA proteins. FAAP95 replaces BRCA2 as the true FANCB protein. Cell Cycle. 2005;4:80–6. doi: 10.4161/cc.4.1.1358. [DOI] [PubMed] [Google Scholar]
- 18.Huang TT, Nijman SM, Mirchandani KD, Galardy PJ, Cohn MA, Haas W, et al. Regulation of monoubiquitinated PCNA by DUB autocleavage. Nat Cell Biol. 2006;8:339–47. doi: 10.1038/ncb1378. [DOI] [PubMed] [Google Scholar]
- 19.Wang W. Emergence of a DNA-damage response network consisting of Fanconi anaemia and BRCA proteins. Nat Rev Genet. 2007;8:735–48. doi: 10.1038/nrg2159. [DOI] [PubMed] [Google Scholar]
- 20.D’Andrea AD. The Fanconi road to cancer. Genes Dev. 2003;17:1933–6. doi: 10.1101/gad.1128303. [DOI] [PubMed] [Google Scholar]
- 21.D’Andrea AD, Grompe M. The Fanconi anaemia/BRCA pathway. Nat Rev Cancer. 2003;3:23–34. doi: 10.1038/nrc970. [DOI] [PubMed] [Google Scholar]
- 22.Zhang J, Zhao D, Park HK, Wang H, Dyer RB, Liu W, et al. FAVL elevation in human tumors disrupts Fanconi anemia pathway signaling and promotes genomic instability and tumor growth. J Clin Invest. 2010;120:1524–34. doi: 10.1172/JCI40908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang J, Wang X, Lin CJ, Couch FJ, Fei P. Altered expression of FANCL confers mitomycin C sensitivity in Calu-6 lung cancer cells. Cancer Biol Ther. 2006;5:1632–6. doi: 10.4161/cbt.5.12.3351. [DOI] [PubMed] [Google Scholar]
- 24.Zhang J, Zhao D, Wang H, Lin CJ, Fei P. FANCD2 monoubiquitination provides a link between the HHR6 and FA-BRCA pathways. Cell Cycle. 2008;7:407–13. doi: 10.4161/cc.7.3.5156. [DOI] [PubMed] [Google Scholar]
- 25.Park HK, Wang H, Zhang J, Datta S, Fei P. Convergence of Rad6/Rad18 and Fanconi anemia tumor suppressor pathways upon DNA damage. PLoS ONE . 2010;5:e13313. doi: 10.1371/journal.pone.0013313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meetei AR, de Winter JP, Medhurst AL, Wallisch M, Waisfisz Q, van de Vrugt HJ, et al. A novel ubiquitin ligase is deficient in Fanconi anemia. Nat Genet. 2003;35:165–70. doi: 10.1038/ng1241. [DOI] [PubMed] [Google Scholar]
- 27.Taniguchi T, Tischkowitz M, Ameziane N, Hodgson SV, Mathew CG, Joenje H, et al. Disruption of the Fanconi anemia-BRCA pathway in cisplatin-sensitive ovarian tumors. Nat Med. 2003;9:568–74. doi: 10.1038/nm852. [DOI] [PubMed] [Google Scholar]
- 28.Hahn SA, Greenhalf B, Ellis I, Sina-Frey M, Rieder H, Korte B, et al. BRCA2 germline mutations in familial pancreatic carcinoma. J Natl Cancer Inst. 2003;95:214–21. doi: 10.1093/jnci/95.3.214. [DOI] [PubMed] [Google Scholar]
- 29.Meetei AR, Medhurst AL, Ling C, Xue Y, Singh TR, Bier P, et al. A human ortholog of archaeal DNA repair protein Hef is defective in Fanconi anemia complementation group M. Nat Genet. 2005;37:958–63. doi: 10.1038/ng1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reid SSD, Hanenberg H, Barker K, Hanks S, Kalb R, Neveling K, et al. Biallelic mutations in PALB2 cause Fanconi anemia subtype FA-N and predispose to childhood cancer. Nat Genet 2007; 39:162-4. doi: 10.1038/ng1947. [DOI] [PubMed] [Google Scholar]
- 31.Roest HP, Baarends WM, de Wit J, van Klaveren JW, Wassenaar E, Hoogerbrugge JW, et al. The ubiquitin-conjugating DNA repair enzyme HR6A is a maternal factor essential for early embryonic development in mice. Mol Cell Biol. 2004;24:5485–95. doi: 10.1128/MCB.24.12.5485-5495.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xia BDJ, Ameziane N, de Vries Y, Rooimans MA, Sheng Q, Pals G, et al. Fanconi anemia is associated with a defect in the BRCA2 partner PALB2. Nat Genet 2007; 39:159-61. doi: 10.1038/ng1942. [DOI] [PubMed] [Google Scholar]
- 33.Kim Y, Lach FP, Desetty R, Hanenberg H, Auerbach AD, Smogorzewska A. Mutations of the SLX4 gene in Fanconi anemia. Nat Genet. 2011;43:142–6. doi: 10.1038/ng.750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Levy-Lahad E. Fanconi anemia and breast cancer susceptibility meet again. Nat Genet. 2010;42:368–9. doi: 10.1038/ng0510-368. [DOI] [PubMed] [Google Scholar]
- 35.Meindl A, Hellebrand H, Wiek C, Erven V, Wappenschmidt B, Niederacher D, et al. Germline mutations in breast and ovarian cancer pedigrees establish RAD51C as a human cancer susceptibility gene. Nat Genet. 2010;42:410–4. doi: 10.1038/ng.569. [DOI] [PubMed] [Google Scholar]
- 36.Stoepker C, Hain K, Schuster B, Hilhorst-Hofstee Y, Rooimans MA, Steltenpool J, et al. SLX4, a coordinator of structure-specific endonucleases, is mutated in a new Fanconi anemia subtype. Nat Genet. 2011;43:138–41. doi: 10.1038/ng.751. [DOI] [PubMed] [Google Scholar]
- 37.Vaz F, Hanenberg H, Schuster B, Barker K, Wiek C, Erven V, et al. Mutation of the RAD51C gene in a Fanconi anemia-like disorder. Nat Genet. 2010;42:406–9. doi: 10.1038/ng.570. [DOI] [PubMed] [Google Scholar]
- 38.Rahman NSS, Thompson D, Kelly P, Renwick A, Elliott A, Reid S, et al. Breast Cancer Susceptibility Collaboration (UK). PALB2, which encodes a BRCA2-interacting protein, is a breast cancer susceptibility gene. Nat Genet 2007; 39:165-7. doi: 10.1038/ng1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ling C, Ishiai M, Ali AM, Medhurst AL, Neveling K, Kalb R, et al. FAAP100 is essential for activation of the Fanconi anemia-associated DNA damage response pathway. EMBO J. 2007;26:2104–14. doi: 10.1038/sj.emboj.7601666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ciccia A, Ling C, Coulthard R, Yan Z, Xue Y, Meetei AR, et al. Identification of FAAP24, a Fanconi anemia core complex protein that interacts with FANCM. Mol Cell. 2007;25:331–43. doi: 10.1016/j.molcel.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 41.Morgan NV, Tipping AJ, Joenje H, Mathew CG. High frequency of large intragenic deletions in the Fanconi anemia group A gene. Am J Hum Genet. 1999;65:1330–41. doi: 10.1086/302627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Howlett NG, Taniguchi T, Olson S, Cox B, Waisfisz Q, De Die-Smulders C, et al. Biallelic inactivation of BRCA2 in Fanconi anemia. Science. 2002;297:606–9. doi: 10.1126/science.1073834. [DOI] [PubMed] [Google Scholar]
- 43.Lo Ten Foe JR, Rooimans MA, Bosnoyan-Collins L, Alon N, Wijker M, Parker L, et al. Expression cloning of a cDNA for the major Fanconi anaemia gene, FAA. Nat Genet. 1996;14:320–3. doi: 10.1038/ng1196-320. [DOI] [PubMed] [Google Scholar]
- 44.Strathdee CA, Gavish H, Shannon WR, Buchwald M. Cloning of cDNAs for Fanconi’s anaemia by functional complementation. Nature. 1992;356:763–7. doi: 10.1038/356763a0. [DOI] [PubMed] [Google Scholar]
- 45.de Winter JP, Rooimans MA, van Der Weel L, van Berkel CG, Alon N, Bosnoyan-Collins L, et al. The Fanconi anaemia gene FANCF encodes a novel protein with homology to ROM. Nat Genet. 2000;24:15–6. doi: 10.1038/71626. [DOI] [PubMed] [Google Scholar]
- 46.Timmers C, Taniguchi T, Hejna J, Reifsteck C, Lucas L, Bruun D, et al. Positional cloning of a novel Fanconi anemia gene, FANCD2. Mol Cell. 2001;7:241–8. doi: 10.1016/S1097-2765(01)00172-1. [DOI] [PubMed] [Google Scholar]
- 47.Levitus M, Rooimans MA, Steltenpool J, Cool NF, Oostra AB, Mathew CG, et al. Heterogeneity in Fanconi anemia: evidence for 2 new genetic subtypes. Blood. 2004;103:2498–503. doi: 10.1182/blood-2003-08-2915. [DOI] [PubMed] [Google Scholar]
- 48.Taniguchi T, Garcia-Higuera I, Xu B, Andreassen PR, Gregory RC, Kim ST, et al. Convergence of the fanconi anemia and ataxia telangiectasia signaling pathways. Cell. 2002;109:459–72. doi: 10.1016/S0092-8674(02)00747-X. [DOI] [PubMed] [Google Scholar]
- 49.Smogorzewska A, Matsuoka S, Vinciguerra P, McDonald ER, 3rd, Hurov KE, Luo J, et al. Identification of the FANCI protein, a monoubiquitinated FANCD2 paralog required for DNA repair. Cell. 2007;129:289–301. doi: 10.1016/j.cell.2007.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sala-Trepat M, Rouillard D, Escarceller M, Laquerbe A, Moustacchi E, Papadopoulo D. Arrest of S-phase progression is impaired in Fanconi anemia cells. Exp Cell Res. 2000;260:208–15. doi: 10.1006/excr.2000.4994. [DOI] [PubMed] [Google Scholar]
- 51.Grompe M. FANCL, as in ligase. Nat Genet. 2003;35:113–4. doi: 10.1038/ng1003-113. [DOI] [Google Scholar]
- 52.Xia B, Sheng Q, Nakanishi K, Ohashi A, Wu J, Christ N, et al. Control of BRCA2 cellular and clinical functions by a nuclear partner, PALB2. Mol Cell. 2006;22:719–29. doi: 10.1016/j.molcel.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 53.Levitus M, Waisfisz Q, Godthelp BC, de Vries Y, Hussain S, Wiegant WW, et al. The DNA helicase BRIP1 is defective in Fanconi anemia complementation group J. Nat Genet. 2005;37:934–5. doi: 10.1038/ng1625. [DOI] [PubMed] [Google Scholar]
- 54.Litman R, Peng M, Jin Z, Zhang F, Zhang J, Powell S, et al. BACH1 is critical for homologous recombination and appears to be the Fanconi anemia gene product FANCJ. Cancer Cell. 2005;8:255–65. doi: 10.1016/j.ccr.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 55.Alter BP. Cancer in Fanconi anemia, 1927-2001. Cancer. 2003;97:425–40. doi: 10.1002/cncr.11046. [DOI] [PubMed] [Google Scholar]
- 56.Williams DA, Croop J, Kelly P. Gene therapy in the treatment of Fanconi anemia, a progressive bone marrow failure syndrome. Curr Opin Mol Ther. 2005;7:461–6. [PubMed] [Google Scholar]
- 57.Spivak JL. The anaemia of cancer: death by a thousand cuts. Nat Rev Cancer. 2005;5:543–55. doi: 10.1038/nrc1648. [DOI] [PubMed] [Google Scholar]
- 58.Kennedy RD, D’Andrea AD. The Fanconi Anemia/BRCA pathway: new faces in the crowd. Genes Dev. 2005;19:2925–40. doi: 10.1101/gad.1370505. [DOI] [PubMed] [Google Scholar]
- 59.Swift M. Fanconi’s anaemia in the genetics of neoplasia. Nature. 1971;230:370–3. doi: 10.1038/230370a0. [DOI] [PubMed] [Google Scholar]
- 60.Auerbach AD, Wolman SR. Susceptibility of Fanconi’s anaemia fibroblasts to chromosome damage by carcinogens. Nature. 1976;261:494–6. doi: 10.1038/261494a0. [DOI] [PubMed] [Google Scholar]
- 61.Alter BP. Aplastic Anemia, Pediatric Aspects. Oncologist. 1996;1:361–6. [PubMed] [Google Scholar]
- 62.Meetei AR, Yan Z, Wang W. FANCL replaces BRCA1 as the likely ubiquitin ligase responsible for FANCD2 monoubiquitination. Cell Cycle. 2004;3:179–81. doi: 10.4161/cc.3.2.656. [DOI] [PubMed] [Google Scholar]
- 63.Rego MA, Harney JA, Mauro M, Shen M, Howlett NG. Regulation of the activation of the Fanconi anemia pathway by the p21 cyclin-dependent kinase inhibitor. Oncogene. 2012;31:366–75. doi: 10.1038/onc.2011.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wilson JB, Yamamoto K, Marriott AS, Hussain S, Sung P, Hoatlin ME, et al. FANCG promotes formation of a newly identified protein complex containing BRCA2, FANCD2 and XRCC3. Oncogene. 2008;27:3641–52. doi: 10.1038/sj.onc.1211034. [DOI] [PubMed] [Google Scholar]
- 65.Hoskins EE, Gunawardena RW, Habash KB, Wise-Draper TM, Jansen M, Knudsen ES, et al. Coordinate regulation of Fanconi anemia gene expression occurs through the Rb/E2F pathway. Oncogene. 2008;27:4798–808. doi: 10.1038/onc.2008.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Park HK, Panneerselvam J, Dudimah FD, Dong G, Sebastian S, Zhang J, et al. Wip1 contributes to cell homeostasis maintained by the steady-state level of Wtp53. Cell Cycle. 2011;10:2574–82. doi: 10.4161/cc.10.15.15923. [DOI] [PMC free article] [PubMed] [Google Scholar]