Abstract
The disease-resistant Arabidopsis thaliana aberrant growth and death2 (agd2-1) mutant has elevated levels of the defense signal salicylic acid (SA), altered leaf morphology, and mild dwarfism. AGD2 and its close homolog ALD1 (for AGD2-LIKE DEFENSE RESPONSE PROTEIN1) encode aminotransferases that act on an overlapping set of amino acids in vitro. However, kinetic parameters indicate that AGD2 and ALD1 may drive the aminotransferase reaction in opposite directions. ALD1-deficient mutants have the opposite phenotypes from agd2-1, showing reduced SA production and increased disease susceptibility. Furthermore, ALD1 transcript levels are elevated in agd2-1 and are induced in the wild type by bacterial pathogen infection. ALD1 is responsible for some of the elevated SA content and a majority of the disease resistance and dwarfism of agd2-1. A complete knockout of AGD2 renders embryos inviable. We suggest that AGD2 synthesizes an important amino acid–derived molecule that promotes development and suppresses defenses, whereas ALD1 generates a related amino acid–derived molecule important for activating defense signaling.
INTRODUCTION
Extracellular bacterial pathogens such as Pseudomonas syringae infect a wide variety of plants (Gardan et al., 1999), causing water soaking, cell death, and chlorophyll loss (chlorosis) (Agrios, 1997). Diseases caused by P. syringae require bacterial effector proteins that are delivered via a type III secretion apparatus to plants and promote disease by a largely unknown mechanism (Greenberg and Vinatzer, 2003). In some plant genotypes, effectors encoded by avirulence genes can be specifically recognized, and a resistance response is mounted to limit the infection in a so-called gene-for-gene interaction (Greenberg and Vinatzer, 2003). However, it is now widely recognized that even without the gene-for-gene interaction, plants mount an active defense response that partially limits virulent P. syringae (Glazebrook and Ausubel, 1994; Glazebrook et al., 1996; Rogers and Ausubel, 1997). This general resistance can vary in effectiveness depending on the host-pathogen combination. It is not clear what triggers general resistance, but it may be the perception of damage by the plant, the recognition of pathogen-associated molecular patterns, such as flagella (Asai et al., 2002), and/or the weak recognition of effectors (Chang et al., 2002).
Many lines of evidence point to the small phenolic compound salicylic acid (SA) in playing a prominent role as a signal molecule in general resistance. First, SA levels rise during P. syringae infection (Rassmussen et al., 1991). Second, exogenous application of SA can induce resistance to P. syringae (Ryals et al., 1996). Third, numerous mutants compromised for SA accumulation or constitutively producing SA show enhanced susceptibility or resistance, respectively, to P. syringae. In particular, Arabidopsis thaliana mutants, such as eds16/sid2 (for enhanced disease susceptibility16/salicylic acid induction-deficient2) and eds5/sid1, that have very low levels of SA during pathogen infection, probably because of a defect in SA biosynthesis, are disease susceptible (Nawrath and Métraux, 1999; Wildermuth et al., 2001; Nawrath et al., 2002). Plants engineered to produce more SA also are more resistant to viral and fungal infections, but resistance to bacterial pathogens has not been specifically tested (Verberne et al., 2000).
Not surprisingly, several mutants with heightened susceptibility to P. syringae show altered but not abolished accumulation of SA upon infection. These include pad4 (for phytoalexin deficient4; Zhou et al., 1998), eds1 (Feys et al., 2001), eds3 (Glazebrook et al., 1996, 2003), eds8 (Glazebrook et al., 1996, 2003), pad1 (Glazebrook et al., 1997), pad2 (Glazebrook et al., 1997, 2003), eds4 (Gupta et al., 2000), and acd6-T (for accelerated cell death6-T; Lu et al., 2003). These mutants affect SA accumulation to varying extents, with some only compromising the early production of SA. ndr1 (for nonrace-specific disease resistance1) mutants also are compromised for SA production after infection with some avirulent P. syringae isolates (Shapiro and Zhang, 2001). PAD4 encodes a possible lipase and may generate a lipid signal (Jirage et al., 1999). ACD6 is a probable transmembrane protein with an ankyrin repeat region that may act by making protein–protein contacts (Lu et al., 2003). Finally, NDR1 is thought to mediate SA induction through the production of reactive oxygen (Shapiro and Zhang, 2001). However, the basis for the reduction in SA accumulation in most mutants is not well understood. In some cases, genes that regulate SA production may regulate other signals as well, as has been hypothesized for PAD4 (Glazebrook et al., 2003).
Constitutive disease-resistant mutants also have provided a way to identify potential regulators of SA and other defense signals. One such mutant we previously characterized, called agd2-1 (for aberrant growth and death2), showed elevated SA and resistance to P. syringae (Rate and Greenberg, 2001). This mutant also had some spontaneous cell death and enlarged cells, and the overall size of the plants was smaller than that of the wild type. The morphology of agd2-1 was greatly altered by depleting SA with the nahG transgene, whose product converts SA to catechol (Gaffney et al., 1993). agd2-1-nahG plants had tumor-like growths and cells with highly endoreduplicated DNA, a phenotype that could be suppressed by application of an SA agonist (Rate and Greenberg, 2001; Vanacker et al., 2001). This led us to suggest that some genes that control defenses also play a role in development (Vanacker et al., 2001). Indeed, we found that the nonexpressor of PR1-1 mutant blocked for SA signaling also had a developmental defect, showing a decrease in cell number and an increase in the ploidy of mesophyll cells when compared with the wild type (Vanacker et al., 2001). Other examples of mutants with developmental and defense defects also have been reported recently (i.e., see Holt et al., 2002; Jin et al., 2002; Barth and Conklin, 2003).
In addition to defense components sometimes affecting cell growth and/or death, they also interact with other pathways. For example, many abiotic stresses, such as ozone, activate SA synthesis and SA-dependent defenses (Sharma et al., 1996). Amino acid starvation or treatment with the abiotic elicitor α-aminobutyric acid can induce SA-dependent and SA-independent defense markers and the phytoalexin camalexin, possibly through a common pathway (Zhao et al., 1998). However, no clear mechanism for how these stimuli are sensed and transduced has yet emerged. Amino acid imbalances also can alter development. Transgenic plants that accumulate 10-fold more Lys than the wild type have striking developmental defects (Frankard et al., 1992). These transgenic Nicotiana sylvestris plants have decreased chlorophyll content, lack apical dominance, and show altered leaf size and morphology. Whether these Lys-overexpressing plants also alter the defense response has not been investigated. In plants, amino acids are precursors to hormones or secondary metabolites and serve to transport nitrogen from source to sink organs. As such, the control of amino acid synthesis in plants affects many aspects of growth and development.
In this report, we describe the cloning and characterization of AGD2, which encodes a novel aminotransferase. A. thaliana has one homolog of AGD2, called ALD1 (for AGD2-LIKE DEFENSE RESPONSE PROTEIN1), sharing 62% identity (77% similarity) with AGD2. The recombinant AGD2 and ALD1 proteins have overlapping substrate specificity on several amino acids but act in different directions in the aminotransferase reaction in vitro. AGD2 expression is suppressed by dark treatment and during senescence, whereas ALD1 is induced by bacterial pathogen infection and senescence. ald1 loss-of-function mutants had the opposite phenotypes from agd2-1, showing increased P. syringae susceptibility and reduced SA accumulation. Additionally, most of the agd2-1 phenotypes were at least partially attributed to ALD1 function. Finally, we show that complete loss of AGD2 function results in inviable embryos, suggesting a developmental role for AGD2.
RESULTS
Cloning of AGD2 and Identification of the Homologous ALD1 Gene
We fine-mapped AGD2 to a 65-kb interval encompassed on BAC clone T16L1 between the C5-2 and C7-2N markers. AGD2 was encoded by the At4g33680 gene according to a number of criteria. First, complementation test results using 15- to 20-kb overlapping T16L1 subclones uniquely identified the 7-kb At4g33680 region to be capable of rescuing agd2-1. Second, defined genomic and cDNA clones corresponding only to At4g33680 complemented agd2-1 (Figure 1A; data not shown). Third, the same single nucleotide change was detected in both cDNA and genomic DNA from the At4g33680 gene in agd2-1. The agd2-1 allele contained a C-to-A point mutation resulting in a Pro-to-Ser change at amino acid 398 (Figure 1B).
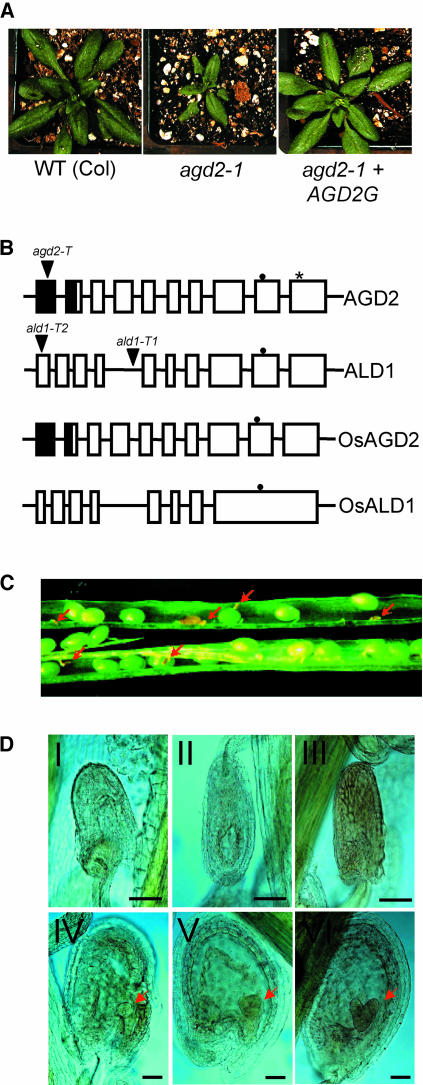
Figure 1.
Features of AGD2, ALD1, and agd2-T.
(A) Complementation of agd2-1. agd2-1 plants were transformed with the genomic clone of AGD2 (AGD2G, at right). The plants shown at the left and in the middle are untransformed wild-type and agd2-1 control plants. agd2-1 plants transformed with the genes adjacent to AGD2 were not complemented (data not shown).
(B) Molecular structures of AGD2, ALD1, OsAGD2, and OsALD1. T-DNA insertion sites are indicated by triangles. The asterisk indicates the Pro-to-Ser change caused by the agd2-1 allele. Closed and open boxes represent exons, and closed boxes represent possible chloroplast transit peptides. The closed circles indicate the pyridoxal-5′-phosphate attachment sites.
(C) Immature seeds inside a heterozygous agd2-T/AGD2 silique. Aborted dead seeds are indicated by red arrows.
(D) Microscopy analysis of aborted and normal seeds in agd2-T/AGD2 heterozygotes. Embryos are indicated by red arrows. Top panel (I to III), dying and dead seeds; bottom panel (IV to VI), normal seeds. IV, Globular stage; V, triangular stage; VI, heart stage. Bars = 80 μm.
AGD2 had 9 introns (Figure 1B) and encoded a 461–amino acid protein. Using the simple modular architecture research tool (http://smart.embl-heidelberg.de/), we found that residues 148 to 460 had modest similarity (E = 1.5e-8) to the aminotransferase class I and class II protein families (Pfam 9.0) and contained a pyridoxal-5′-phosphate attachment site (Mehta et al., 1989). The PSORT and TargetP algorithms (http://psort.ims.u-tokyo.ac.jp/ and http://www.cbs.dtu.dk/services/TargetP/, respectively) both predicted AGD2 to be chloroplast localized (Figure 1B).
A. thaliana has one close homolog of AGD2 on chromosome 2 (At2g13810), which we named ALD1. ALD1 had 62% identity and 77% similarity with AGD2 at the amino acid level. ALD1 had 10 exons (Figure 1B) and encoded a 456–amino acid protein. It was predicted to localize to the cytoplasm by the PSORT algorithm and to the chloroplasts by the TargetP algorithm. Because of this ambiguity, we have tentatively indicated that ALD1 lacks a chloroplast transit signal (Figure 1B).
The Oryza sativa (rice) genome had two genes: one called OsJNBa00664E16.9 and one that we annotated from genomic region AC105731 (tentatively named OsALD1), whose products had high similarities to both AGD2 and ALD1 (Table 1). We isolated full-length O. sativa cDNAs for OsJNBa00664E16.9 (cOsAGD2, GenBank accession number AY338235) and OsADL1 (cOsALD1, GenBank accession number AY338236) and found that they had gene structures similar to AGD2 and ALD1, respectively (Figure 1B). Additionally, the product of cOsAGD2 was predicted to have a subcellular location similar to AGD2, whereas the product of cOsALD1 was predicted to be cytoplasmic. Because of the high similarities of the predicted O. sativa proteins to both AGD2 and ALD1 (Table 1), localization assignments for these proteins may change when more data is available.
Table 1.
Percentage Identities and Similarities (in Parentheses) among AGD2, ALD1, OsAGD2, and OsALD1 Proteins
| AGD2 | ALD1 | OsAGD2 | |
|---|---|---|---|
| ALD1 | 62 (77) | ||
| OsAGD2 | 76 (95) | 58 (86) | |
| OsALD1 | 61 (82) | 58 (86) | 62 (85) |
Loss of AGD2 Causes Embryo Lethality
Analysis of the AGD2P398S mutant recombinant enzyme activity corresponding to the product encoded by agd2-1 (see below; Figure 6) suggested that the agd2-1 allele was not null. To ascertain the phenotype of AGD2 null mutant plants, we isolated a heterozygous mutant (agd2-T/AGD2) with a kanamycin-resistant (kanR) T-DNA insertion in the first exon of AGD2 (Figure 1B). The progeny of heterozygous agd2-T/AGD2 plants yielded 20 wild-type, 44 heterozygous, and no homozygous mutant plants, suggesting that AGD2 is an essential gene. Furthermore, of 352 seeds from the heterozygous plant that were germinated on kanamycin plates, 116 kanamycin-sensitive (kanS) and 236 kanR seeds were observed. This is a good fit to the 1:2:0 hypothesis that the homozygous embryos were not viable (χ2 = 0.012, P > 0.9). In accordance with the lack of homozygous seeds, the seed yield per silique in the wild type (54.5 ± 0.9 sd, n = 4) and heterozygous agd2-T (40.3 ± 2.2 sd, n = 4) was significantly different (P = 0.0009). The seeds in wild-type plants all looked normal and were of uniform size. By contrast, some seeds of the heterozygous plants were very tiny and dry and looked aborted (Figure 1C). This phenotype was detected in all of the >100 heterozygous plants assayed.
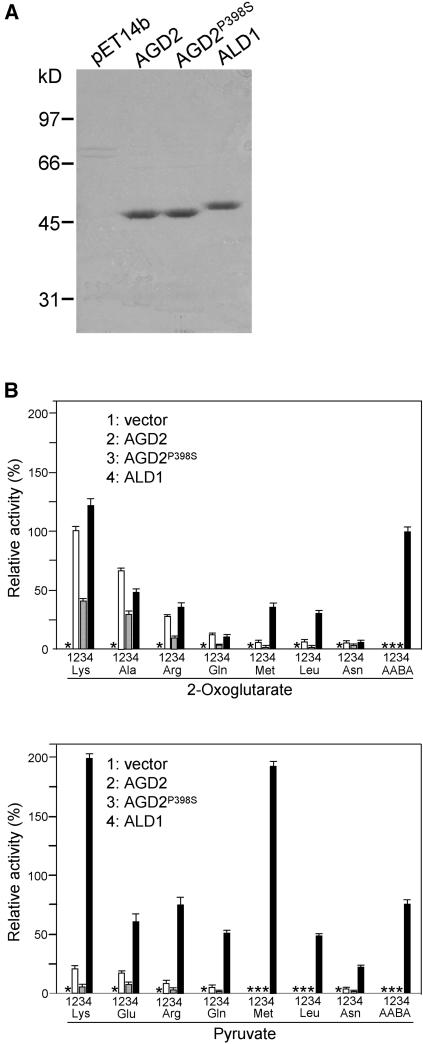
Figure 6.
Purification and Relative Activities of AGD2, AGD2P398S, and ALD1.
(A) Electrophoretic patterns of purified recombinant proteins through column elution using Ni2+-NTA.
(B) Relative aminotransferase activities of AGD2, AGD2P398S, and ALD1. Substrates were at a 50-mM concentration. Aminotransferase activity was determined by measuring concentration of the reaction product Glu (using 2-oxoglutarate as the cosubstrate) or Ala (using pyruvate as the cosubstrate). The activity of AGD2 with Lys and 2-oxoglutarate was arbitrarily set at 100%. Asterisks indicate no detectable activities. No detectable activity was seen with all of the other standard amino acids tested. Bars indicate standard deviations (n = 3).
To determine when in development AGD2 might act, we examined at which stage the homozygous embryos died. Before the globular stage, all of the developing embryos in agd2-T/AGD2 heterozygotes were normal. However, when most of the embryos reached the globular stage in agd2-T/AGD2 heterozygotes, some seeds already had died or were severely malformed (Figure 1D). The aborted seed size was much smaller than that of normal seeds, and the embryo proper could no longer be seen. It seems likely that after fertilization, homozygous embryos had normal development at the beginning and that later the embryos died. To investigate the ratio of lethal-to-healthy embryos at the heart stage, we examined the contents of individual silique chambers. The heterozygous plant had 20.8 ± 0.66 sd (n = 5) normal and 7.0 ± 0.32 sd (n = 5) dead seeds per silique chamber, whereas the control wild-type plants had 27.8 ± 0.37 sd (n = 5) apparently living seeds. These ratios of live-to-dead seeds were significantly different in the heterozygous mutant and the wild type (P < 0.0001). Furthermore, the embryo lethality of the agd2-T mutant was complemented by the AGD2 genomic clone. This was evidenced by the fact that numerous viable T2 plants (12) homozygous for agd2-T only were recovered when the AGD2 transgene was present. In reciprocal crosses, we found no indication that agd2-T haploid pollen or ovules had any transmission defects (data not shown). Therefore, agd2-T affected the embryo and resulting diploid seeds but not haploid pollen or ovules. These results also were consistent with the 1:2 segregation of kanS:kanR of seeds from agd2-T/AGD2 heterozygotes.
AGD2 Localizes to Chloroplasts
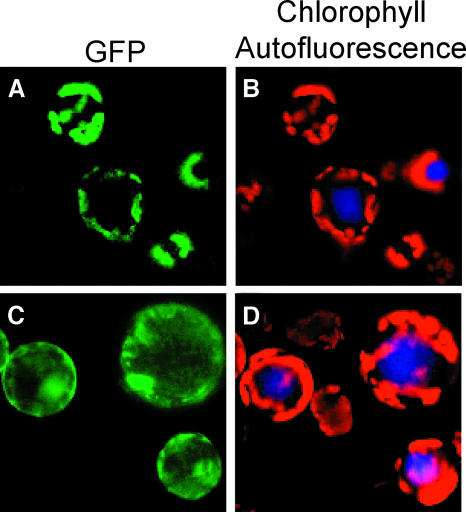
To investigate where in the cell AGD2 might function, we examined the subcellular location of the full-length protein fused to the green fluorescent protein (GFP). We drove expression of the AGD2 cDNA fused to gfp from the 35S promoter of Cauliflower mosaic virus (CaMV). This construct complemented the agd2-1 mutant (data not shown). The GFP fluorescence patterns in protoplasts isolated from A. thaliana expressing AGD2-GFP colocalized with the red autofluorescence patterns of the chloroplasts (Figures 2A and 2B). When GFP lacking the AGD2 moiety was expressed, the GFP fluorescence was not specifically colocalized with chloroplast autofluorescence but was distributed throughout the cells (Figures 2C and 2D). The chloroplast localization of AGD2-GFP is consistent with its possible role in amino acid synthesis because many amino acids are known to be synthesized in chloroplasts (Bryan, 1990).
Figure 2.
Subcellular Localization of AGD2.
The pGFP and pAGD2-GFP under control of the CaMV 35S promoter were transformed into A. thaliana. GFP fluorescence of the protoplasts was observed using a fluorescence microscope.
(A) and (B) pAGD2-GFP.
(C) and (D) pGFP.
AGD2 and ALD1 Have Divergent Expression Patterns
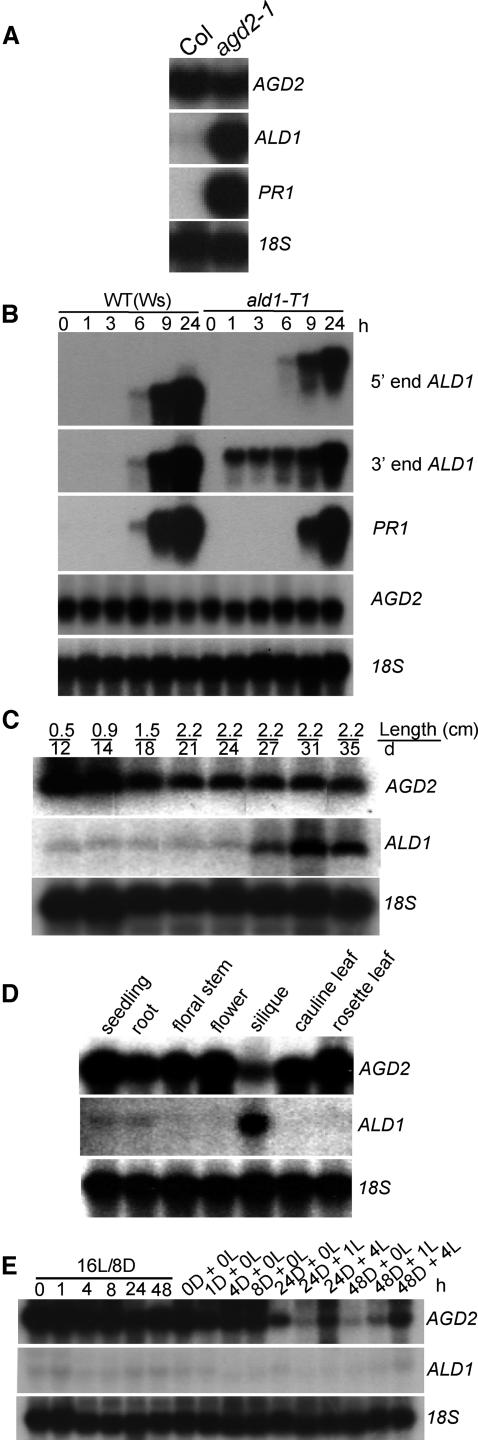
agd2-1 hypomorphic plants are resistant to P. syringae (Rate and Greenberg, 2001) and showed elevated ALD1 expression (Figure 3A). This suggested that AGD2 and/or ALD1 mRNA levels might be modulated during pathogenesis. To test this, we used gene-specific probes to examine their transcript levels after infection of wild-type plants with P. syringae. In response to P. syringae pv maculicola strain DG3 infection, AGD2 mRNA levels were not significantly changed, whereas ALD1 was induced in a pattern similar to that of the known defense-related gene Pathogenesis Related1 (PR1; Figure 3B). Mock inoculation did not alter ALD1 or AGD2 expression (data not shown). Because some pathogen-induced genes also are senescence regulated (Morris et al., 2000), we followed the expression levels of AGD2 and ALD1 in leaf 4 throughout its life span. Visible senescence started on day 21. AGD2 expression was initially high and decreased with time, whereas ALD1 was induced after day 24 (Figure 3C). Thus, the expression of both genes was modulated during senescence.
Figure 3.
RNA Expression of AGD2 and ALD1.
Tissue from the wild type and/or aldT-1 was used for RNA extraction and RNA gel blot analysis.
(A) ALD1 and PR1 expression in the fourth and fifth leaves of 20-d-old wild-type (Col) and agd2-1 plants.
(B) ALD1 and PR1 expression in wild-type (Ws) and ald1-T1 plants. Leaves (fourth and fifth) from 18-d-old plants were infected with P. s. maculicola DG3 (OD600 = 0.01).
(C) Expression of AGD2 and ALD1 during development of the fourth leaf. Leaf length and age of the plant when RNA was extracted are indicated.
(D) Expression of AGD2 and ALD1 in the indicated tissues.
(E) A time course of steady state AGD2 mRNA accumulation. Twenty-day-old plants were kept in a 16-h-light/8-h-dark (16L/8D) cycle or were shifted to continuous dark (D) for the indicated time after 4 h in the light of the normal light cycle (16-h-light/8-h-dark) and then switched back to light (L) for 1 or 4 h. Although the signal was low for the ALD1 transcript, the probe was verified to be high specific activity. It gave a strong signal on the blots probed in parallel (see [A] and [B]).
We also examined the expression levels of AGD2 and ALD1 in various tissues (Figure 3D). AGD2 was strongly expressed in all of the tissues, including seedling, root, stem, flower, and leaves. However, AGD2 expression was relatively lower in siliques, whereas ALD1 was very high in siliques but relatively low in other tissues. The expression of ALD1 is reminiscent of that seen for PR genes that show expression in flowers, fruit, and ripening fruit (Lotan et al., 1989; Salzman et al., 1998). Such expression may confer resistance to pathogens during reproduction.
Amino acid–synthesizing enzymes as well as amino acids (the possible substrates and products of aminotransferases) can vary in abundance depending on whether light is present (Buchanan et al., 2000). Additionally, the expression of some defense genes, such as ACD6, is light dependent (Lu et al., 2003). To test the effect of light on AGD2 and ALD1 expression, plants maintained in a 16-h-light/8-h-dark cycle were moved to constant dark after 4 h of the normal morning light. These are conditions under which ACD6 transcript levels are modulated (Lu et al., 2003). In 16-h-light/8-h-dark conditions, AGD2 expression was unchanged throughout the course of a day (Figure 3E). However, after dark treatment, AGD2 transcript levels were significantly reduced at 24 and 48 h. When the dark-treated plants were re-exposed to light for 4 h, the AGD2 transcript level was restored back to the level seen in the control plants maintained in the normal light/dark cycle conditions (Figure 3E). ALD1 levels were not significantly altered under these conditions. Thus, the expression patterns of AGD2 and ALD1 are spatially, developmentally, and environmentally regulated, with different and sometimes complementary patterns.
ALD1 Is Important for Resistance to the Bacterial Pathogen P. syringae
The pathogen-induced expression of ALD1 suggested it could be important for disease resistance. To test this, we isolated a mutant, ald1-T1, with a T-DNA insertion in the fourth intron of ALD1 (Figure 1B) in ecotype Wassilewskija (Ws). ald1-T1 plants showed two aberrantly large ALD1 transcripts, visualized with 5′ and 3′ end ALD1 probes, because of the presence of the T-DNA insertion (Figure 3B). Reverse transcriptase (RT)-PCR and sequence analysis established that ald1-T1 produced one transcript from the native promoter with the first four ALD1 exons and T-DNA border sequence. The second transcript included a T-DNA sequence at the 5′ end and the six 3′ exons of ALD1 at the 3′ end (data not shown). No full-length transcripts were found, suggesting that little if any functional ALD1 protein was present in the ald-T1 mutant.
The ald1-T1 homozygous plants and seeds were morphologically normal (data not shown). Infection of wild-type and ald1-T1 plants with P. s. maculicola DG3 resulted in chlorotic symptoms in the leaves of ald1-T1 but still green or slight symptom development in wild-type leaves (data not shown). P. s. maculicola DG3 bacteria grew at least 10-fold more in ald1-T1 than in wild-type plants on days 2 and 3 after infection (Figure 4A). As P. s. maculicola DG3 was not very virulent in Ws, we also assayed the growth of P. syringae pv tomato strain DC3000, a virulent isolate. ald1-T1 also was more susceptible than the wild type to P. s. tomato DC3000 (Figure 4A). Ecotype Columbia (Col) plants with RNA interference–mediated downregulation of ALD1 or containing a T-DNA insertion in exon one of ALD1 (ald1-T2; Figure 1B) behaved similarly to ald1-T1 (Figure 5; data not shown). ald1-T1 was recessive and was complemented by a genomic clone of ALD1 (Figure 4B).
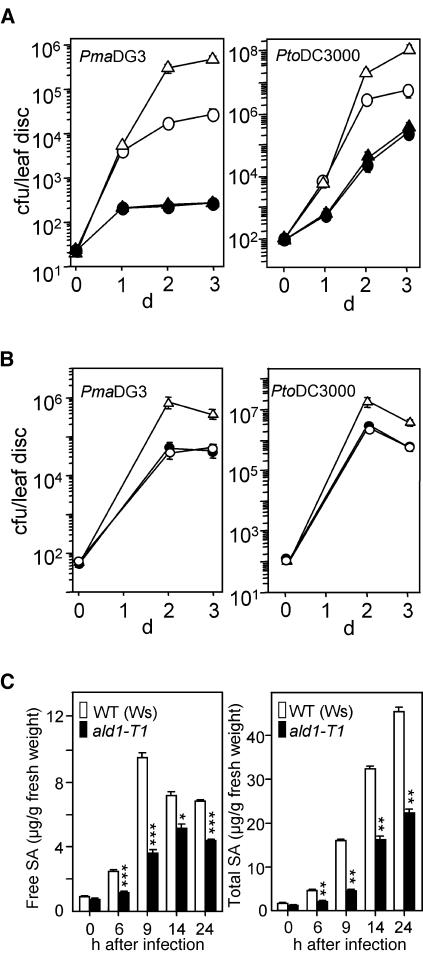
Figure 4.
Increased Disease Susceptibility in the ald1-T1 Mutant.
(A) Disease susceptibility of ald1-T1 plants. Wild-type Ws (open circles) and ald1-T1 (open triangles) plants were infected with P. s. maculicola DG3 (PmaDG3, at left) or with P. s. tomato DC3000 (PtoDC3000, at right) at OD600 = 0.0001. Growth of P. syringae in Ws and ald1-T1 was significantly different on days 2 and 3 (P < 0.01, t test, n = 8). Ws and ald1-T1 plants were pretreated with 100 μM BTH or water for 2 d and then subjected to infection with P. s. maculicola DG3. BTH-treated Ws (closed circles) and ald1-T1 (closed triangles) were not significantly different for bacterial growth (P > 0.5, t test, n = 8). cfu, colony-forming units. Bars indicate standard error. In some cases, the symbols obscure the error bars.
(B) Complementation of the ald1-T1 mutant phenotype. Ws (open circles), ald1-T1 (open triangles), and ald1-T1 transformed with the ALD1 genomic clone (closed circles) were infected with P. s. maculicola DG3 (PmaDG3, at left) or P. s. tomato DC3000 (PtoDC3000, at right) at OD600 = 0.0001. Growth of P. syringae in Ws and ald1-T1 was significantly different on days 2 and 3 (P < 0.01, t test, n = 8), whereas growth in Ws and the transgenic plants was not different (P > 0.7, t test, n = 8).
(C) SA levels in Ws and ald1-T1 plants during pathogen infection. Leaves (fourth and fifth) from 18-d-old plants were infected with P. s. maculicola DG3 (OD600 = 0.01) and were harvested, extracted, and analyzed by HPLC. The free and total SA values ±sd are averages of three sets of samples. The number of asterisks indicates samples that are significantly different from one another at a given confidence level (one asterisk, P < 0.004; two asterisks, P < 0.0008; three asterisks, P < 0.0002).
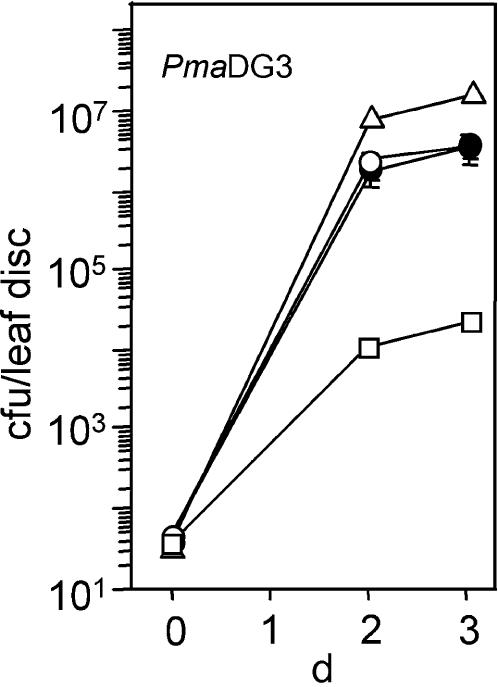
Figure 5.
Bacterial Growth in agd2-1 ald1-T2 Plants.
Wild-type Col (open circles), ald1-T2 (open triangles), agd2-1 (open squares), and agd2-1 ald1-T2 (closed circles) were infected with P. s. maculicola DG3 at OD600 = 0.0001. Growth of P. syringae in Col and agd2-1 ald1-T2 was not significantly different on days 2 and 3 (P > 0.7, t test, n = 8). Bars indicate standard error. In some cases, the symbols obscure the error bars.
ald1-T1 Has Reduced SA Accumulation
Increased susceptibility of plants to P. syringae could result from a defect in SA accumulation. Indeed, the free SA and total SA (including glucoside-conjugated SA) levels in ald1-T1 plants infected with P. s. maculicola DG3 were lower than the levels in infected wild-type plants at all time points tested (Figure 4C). PR1 defense-related gene expression in ald1-T1 plants also was delayed and lower than that seen in the wild type (Figure 3B). Similar results were observed when ald1-T1 and wild-type plants were infected with P. s. tomato DC3000 (data not shown). Treatment of plants with the SA agonist benzo (1,2,3) thiadiazole-7-carbothioic acid (BTH) before infection resulted in ald1-T1 and wild-type plants that were equally disease resistant (Figure 4A). Thus, the defect in SA accumulation was likely responsible, at least in part, for the decreased resistance of ald1-T1.
ALD1 Is Largely Responsible for the Disease Resistance and Small Size of agd2-1
Because agd2-1 had increased resistance to P. syringae and ald1-T1 and ald1-T2 had increased susceptibility to P. syringae, we tested whether ALD1 was required for the elevated disease resistance of agd2-1. Mutant plants with the ald1-T2 allele had no detectable ALD1 transcript (data not shown). ald1-T2 was in the same genetic background as agd2-1, making it suitable for double mutant construction. agd2-1 ald1-T2 plants had similar disease susceptibility to that seen in the wild type but still less than that seen in ald1-T2 alone (Figure 5). Thus, the majority of the disease resistance in agd2-1 was mediated by the ALD1 gene.
We tested whether the suppression of agd2-1 disease resistance in agd2-1 ald1-T2 was correlated with lowered SA levels. Indeed, the agd2-1 ald1-T2 double mutant had a lower basal level of total SA (11.2 ± 0.3 μg/g tissue, n = 3) compared with the agd2-1 single mutant (18.5 ± 0.4 μg/g tissue, n = 3, P = 0.0001). However, the double mutant still had significantly more SA than the wild type (1.3 ± 0.03 μg/g tissue, n = 3, P < 0.0001), even though disease susceptibility was equivalent in wild-type and agd2-1 ald1-T2 plants. This suggests that SA accumulation and ALD1 function can only partially explain the disease resistance of agd2-1.
Because depletion of SA in agd2-1 using a nahG transgene altered the morphology of agd2-1 (Rate and Greenberg, 2001; Vanacker et al., 2001), we also examined the morphology of agd2-1 ald1-T2 plants. agd2-1 ald1-T2 showed tumor-like growths reminiscent of those seen in agd2-1-nahG plants (data not shown), supporting a role for SA in growth control (Vanacker et al., 2001). Strikingly, we also found that the overall size of the agd2-1 ald1-T2 rosettes was nearly restored to that of the wild type (Table 2). These data suggest that at least some agd2-1 phenotypes are mediated by the action of ALD1.
Table 2.
Role of ALD1 in agd2-1 Rosette Size
| Genotype | Diameter (mm) ±sd |
|---|---|
| Col | 73.7 ± 3.4a, n = 12 |
| ald1-T2 | 73.1 ± 3.1a, n = 12 |
| agd2-1 | 31.5 ± 2.7b, n = 12 |
| agd2-1 ald1-T2 | 59.2 ± 3.4c, n = 12 |
The diameters of 35-d-old plants were measured. Superscript letters represent different statistical values using Student's t test (P < 0.0001).
Recombinant AGD2 and ALD1 Have Aminotransferase Activity in Vitro
Protein basic local alignment search tool (BLAST) searches indicated that AGD2 and ALD1 had the closest similarity to an aromatic aminotransferase from Pryococcus hirikoshii with highest activity on the amino acid substrates Tyr, Phe, Glu, Trp, and His (Matsui et al., 2000). However, the similarity was only modest (25% identity between the aromatic aminotransferase from P. hirikoshii and both ALD1 and AGD2). Other much higher BLAST matches were only to putative amino transferases for which activity has not been experimentally validated. To test whether AGD2, AGD2P398S, and ALD1 had amino transferring activity, we overexpressed recombinant AGD2, AGD2P398S, and ALD1 proteins in Escherichia coli and performed aminotransferase activity assays. Optimization of the assays is described in Methods. Six His-tagged truncated versions of the proteins lacking potential organelle targeting signals were overexpressed and then purified using nickel-nitrilotriacetic acid agarose (Ni2+-NTA) beads. The recombinant AGD2, AGD2P398S, and ALD1 proteins were purified to near homogeneity and migrated on SDS-PAGE at the expected molecular masses (Figure 6A).
We first tested the ability of recombinant AGD2, AGD2P398S, and ALD1 proteins to catalyze the transfer of amino groups onto the amino acceptor molecule 2-oxoglutarate in vitro by monitoring the production of glutamate. This amino acceptor was chosen because many characterized aminotransferases are active with this cosubstrate (Mehta et al., 1993). Recombinant AGD2, AGD2P398S, and ALD1 were incubated with each of the standard 20 amino acids (except Glu, which was used to monitor these aminotransferase reactions), β-Ala, α-aminobutyric acid (AABA), β-aminobutyric acid, or γ-aminobutyric acid (GABA) as the amino donor. As a negative control, we used extract from E. coli containing vector alone and purified in the same way as AGD2, AGD2P398S, and ALD1. The highest enzymatic activity was obtained with Lys for both AGD2 and ALD1 (Figure 6B). Both enzymes also exhibited some activity with Ala, Arg, Gln, Met, Leu, and Asn as amino donors. However, they showed no significant activity (the limit of detection was 1% of the activity with Lys) with any other standard amino acids under the conditions tested. It is common for aminotransferases to have activity with several different substrates (Matsui et al., 2000; Nowicki et al., 2001). ALD1 had activity with AABA but not with β-aminobutyric acid or GABA, indicating that ALD1 needs an α-amino group for its activities (Figure 6B; data not shown).
We also tested AGD2, AGD2P398S, and ALD1 for activity with each of the standard amino acids (except Ala, which was used to monitor these aminotransferase reactions), β-Ala, and AABA using pyruvate as another amino acceptor and monitoring the production of Ala. Pyruvate is the second most common amino acceptor for aminotransferases after 2-oxoglutarate (Mehta et al., 1993). AGD2 had lower activity with pyruvate than with 2-oxoglutarate. However, ALD1 had more activity with pyruvate than with 2-oxoglutarate (Figure 6B). As expected, AGD2P398S had reduced activity relative to the wild-type AGD2 protein with all the substrates tested.
Because agd2-1 (Rate and Greenberg, 2001) and ald1-T1 (Figure 4C) mutants have altered SA production, we wondered if ALD2 or AGD2 might act in the SA biosynthetic pathway. One step in this pathway was proposed to convert the oxo-acid prephenate to the amino acid arogenate (Wildermuth et al., 2001). However, we found no detectable activity of AGD2 or ALD1 using prephenate as a cosubstrate with Lys, Ala, or Glu.
In summary, although the BLAST searches with AGD2 and ALD1 revealed modest similarity to an aminotransferase with highest activity with Tyr, Phe, Glu, Trp, and His, the empirical data suggests that AGD2 and ALD1 do not act on these substrates. Rather, AGD2 had its highest activities with Lys, Ala, and Arg, whereas ALD1 had its highest activities with Lys, Ala, Arg, Met, and AABA.
AGD2 and ALD1 Act in Different Directions in the Aminotransferase Reaction in Vitro
To evaluate the kinetic properties of the AGD2 and ALD1 enzymes for three of the best substrates (Lys, Ala, and Arg), the Km values for the individual substrates were determined (Table 3). The Km of AGD2 was 100-fold higher and the kcat/Km value was 100-fold lower for Lys when compared with those of ALD1. The Km values of AGD2 and AGD2P398S were not significantly different, but the kcat value of AGD2P398S was approximately half that of AGD2, causing the mutant protein to have reduced enzyme efficiency (kcat/Km). The Km values for the amino acids Ala and Arg were much lower than that of Lys. They are not good substrates for AGD2 and ALD1, as indicated by the high Km and low kcat/Km values. The Km values of most aminotransferases are within the 0.1 to 5 mM range. For example, the Homo sapiens (human) GABA aminotransferase has a Km of 1.27 mM (Jeon et al., 2000), and a broad substrate specificity aminotransferase from Leishmania mexicana has 0.5 to 2.5 mM Km values with aromatic amino acids and aspartate (Vernal et al., 1998).
Table 3.
Kinetic Parameters of Recombinant AGD2- and ALD1-Catalyzed Reactions for Amino Acid Degradation Using Either Lys, Ala, or Arg with2-Oxoglutarate
| Substrate | Cosubstrate (mM) | Km (mM) | kcat (s−1) | kcat/Km (mM−1 s−1) | |
|---|---|---|---|---|---|
| AGD2 | Lys | 2-oxoglutarate (100 mM) | 58.8 | 5.33 | 0.091 |
| AGD2P398S | Lys | 2-oxoglutarate (100 mM) | 71.4 | 2.87 | 0.040 |
| ALD1 | Lys | 2-oxoglutarate (50 mM) | 0.53 | 4.42 | 8.34 |
| AGD2 | 2-oxoglutarate | Lys (100 mM) | 1.82 | 2.83 | 1.55 |
| AGD2P398S | 2-oxoglutarate | Lys (100 mM) | 1.92 | 1.83 | 0.95 |
| ALD1 | 2-oxoglutarate | Lys (100 mM) | 2.63 | 3.75 | 1.43 |
| AGD2 | Ala | 2-oxoglutarate (100 mM) | 25.6 | 1.67 | 0.065 |
| AGD2P398S | Ala | 2-oxoglutarate (100 mM) | 23.3 | 0.87 | 0.037 |
| ALD1 | Ala | 2-oxoglutarate (100 mM) | 5.56 | 2.15 | 0.39 |
| AGD2 | Arg | 2-oxoglutarate (100 mM) | 154 | 0.69 | 0.0045 |
| AGD2P398S | Arg | 2-oxoglutarate (100 mM) | 167 | 0.35 | 0.0021 |
| ALD1 | Arg | 2-oxoglutarate (100 mM) | 5.10 | 2.07 | 0.41 |
Km values were obtained from the initial velocity data and Lineweaver-Burk plots. kcat was calculated using equation Vmax = kcat [Enzyme]. Three independent measurements were averaged.
The high Km value of AGD2 with Lys suggests that Lys may not be the in vivo substrate for AGD2. Rather, the function of AGD2 may be to synthesize Lys or some other amino acid. To test this, we ran the aminotransferase reaction in reverse and measured the Km values of AGD2 and ALD1 for the oxo-acid pyruvate as the amino acceptor (6-amino-2-oxohexanoate, the predicted oxo-acid produced by the forward reaction using Lys and2-oxoglutarate, was not commercially available). The Km value of AGD2 was 3.08 mM, 10-fold lower than that of ALD1 (Table 4). Although AGD2 and AGD2P398S showed similar Km values, the kcat/Km value of the wild-type protein was fivefold higher than that of mutant protein. These observations suggest that the reduction of kcat/Km is the cause of the agd2-1 phenotype. These data are consistent with AGD2 being required for the synthesis of an amino acid(s) and ALD1 functioning to convert amino acid(s) to an oxo-acid(s) in vivo.
Table 4.
Kinetic Parameters of Recombinant AGD2- and ALD1-Catalyzed Reactions for Ala Synthesis Using Glutamate and Pyruvate as Cosubstrates
| Substrate | Cosubstrate (mM) | Km (mM) | kcat (s−1) | kcat/Km (mM−1 s−1) | |
|---|---|---|---|---|---|
| AGD2 | Glutamate | Pyruvate (50 mM) | 3.08 | 6.47 | 2.10 |
| AGD2P398S | Glutamate | Pyruvate (50 mM) | 3.12 | 1.48 | 0.47 |
| ALD1 | Glutamate | Pyruvate (50 mM) | 37.0 | 5.80 | 0.16 |
Km values were obtained from the initial velocity data and Lineweaver-Burk plots. Three independent measurements were averaged.
Amino Acid Profiles in agd2-1, AGD2-Overexpressing Plants, ald1-T2, and Wild-Type Plants Are Similar
The aminotransferase activity of AGD2 could indicate a role for AGD2 in the production of Lys and/or other amino acids. To test this, we measured amino acid levels in agd2-1, wild-type (Col), and AGD2-overexpressing transgenic plants. Free amino acid levels were determined in leaves of young (10 d) and older (17 d) plants. There were no large differences in the relative amino acid levels of the wild type versus agd2-1 in the young or older plants, although there were small increases and decreases in some amino acid levels (data not shown). Plants confirmed by RNA gel blot analysis to overexpress AGD2 driven by the CaMV 35S promoter also had similar amino acid levels with wild-type plants (data not shown). Attempts to rescue the agd2-T and agd2-1 plants with major amino acids or mixtures of amino acids using a previously successful approach for rescuing embryo-defective biotin auxotrophs (Patton et al., 1998) were not successful. These data suggest that AGD2 may not be involved in the synthesis of any standard amino acids.
We also tested if ald1-T1 had abnormal amino acid accumulation after P. s. maculicola DG3 infection versus the wild type. Using gas chromatography–mass spectrometry (GC-MS) analysis of methanol extracts, we could see no dramatic differences in the relative levels of the 20 standard amino acids between ald1-T1 and wild-type plants 9 and 24 h after infection. Most other metabolites were not apparently different except free SA, which was 2.5-fold lower in ald1-T1 than the wild type (data not shown), consistent with our HPLC analysis (Figure 4C). These data suggest that standard amino acids may not be the targets of ALD1 action in vivo. We wondered if AABA could be an endogenous ALD1 substrate because it was a good substrate in vitro (Figure 6B). Unfortunately, AABA levels were below the assay detection limit.
DISCUSSION
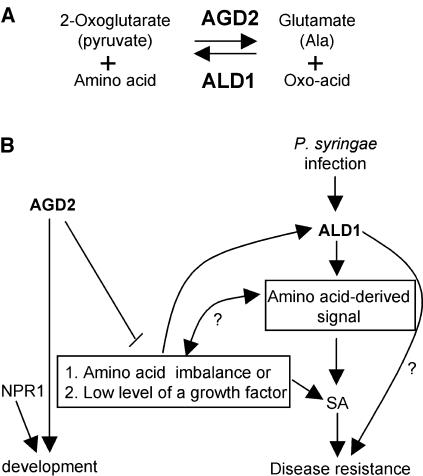
Gene duplications and gene families often encode or are assumed to encode proteins with similar and/or overlapping functions. However, the AGD2 and ALD1 genes of A. thaliana have quite divergent roles in development and defense, despite their high sequence similarity. AGD2 is indispensable for development and also may repress defenses, whereas ALD1 is important for activating defenses and limiting P. syringae growth. The activities of AGD2 and ALD1 in vitro significantly differed from each other. Their enzyme kinetic properties suggest that AGD2 is required for the synthesis of an amino acid(s), whereas ALD1 may utilize the same or a related amino acid(s) to make a defense-regulating molecule(s) (Figure 7A). The two enzymes may act in different subcellular compartments. Transcripts of ALD1 were upregulated by pathogen infection and senescence, whereas those of AGD2 did not accumulate under any of the conditions examined but were downregulated by dark treatment and during senescence.
Figure 7.
Models for the Enzymatic Functions and Roles of AGD2 and ALD1 in Development and Disease Resistance.
(A) Enzymatic reactions that AGD2 and ALD1 may conduct based on the in vitro data. These reactions may be performed in different subcellular compartments.
(B) Integrated model for AGD2 and ALD1 functions in vivo. When plants are infected with P. s. maculicola DG3, ALD1 is needed for SA accumulation and activation of SA signaling responses, such as PR1 expression. ALD1 also is probably required for the generation of an SA-independent defense signal. AGD2 is required for normal development. Reduced activity of AGD2 results in developmental cell death and growth phenotypes and SA accumulation possibly because of the generation of an amino acid–derived signal or an amino acid imbalance. NPR1 also is shown as controlling development as an example of a protein that functions in defense and development (Vanacker et al., 2001).
Specific Physiological Function of AGD2
We established here that many of the agd2-1 phenotypes are partially caused by the presence of the ALD1 gene, possibly through its upregulation. In particular, some of the elevated SA, P. syringae disease resistance, and dwarfism can be suppressed when ALD1 function is removed from agd2-1 plants. However, the fact that in agd2-1 the effects of ALD1 are only partial suggests that there are ALD1-independent AGD2 functions important for the control of SA synthesis, defense activation, and/or development.
agd2-1 hypomorphic plants have some dead cells and some cells with altered size and elevated endoreduplication (Rate and Greenberg, 2001; Vanacker et al., 2001). The loss-of-function agd2-T plants had an early defect in embryogenesis, suggesting that AGD2 has an essential function in plant development. What might this function be? An obvious candidate is the synthesis of one or more amino acid. In particular, our in vitro experiments suggest that Lys would be the preferred product. Furthermore, AGD2 localizes to chloroplasts, the site of synthesis of amino acids (Bryan, 1990). However, several observations lead to questions about how likely this function is for AGD2. First, we saw no significant decrease in the level of Lys (or other amino acids) in agd2-1. Second, we were unable to rescue the agd2-1 or agd2-T mutants with Lys or any of the other major amino acids or mixtures of amino acids. Third, plants are known to make Lys from aspartate via meso-diaminopimelate (Galili, 1995), which is decarboxylated to form Lys using the chloroplast-localized diaminopimelate decarboxylase (Azevedo et al., 1997). A. thaliana has two putative diaminopimelate decarboxylases encoded by the At3g14390 and At5g11880 genes. These Lys biosynthetic enzymes all should be intact in the agd2 mutants. Possibly, Lys production in A. thaliana is primarily formed by decarboxylation of meso-diaminopimelate, but there is a specific requirement for AGD2 during embryogenesis and some stage of leaf development. Our assays may have missed differences in Lys (or other amino acids) accumulation at these specific stages.
An alternative suggestion is that AGD2 is essential for an unknown developmental regulator/signal molecule produced through Lys synthesis or through a nonstandard amino acid. Plant hormones consist of free and conjugated forms. For example, most indole-3-acetic acid (IAA)–based conjugates accumulate as IAA-ester forms to inositol, sugar, or polysaccharides and IAA-amide forms to amino acids or peptides (Cooke et al., 2002; Ljung et al., 2002). The conjugates are generally thought to act as short-term intermediates that can be hydrolyzed to release free forms. Nevertheless, the role of conjugates is still incompletely defined. AGD2 may provide a precursor for a conjugated hormone, such as IAA-Lys, which is present in Agrobacterium tumefaciens and P. savastanoi (Glickmann et al., 1998) and may be present in plants. Such a molecule or one derived from a nonstandard amino acid might be important for embryogenesis.
agd2-1 hypomorphic plants also have increased disease resistance (Rate and Greenberg, 2001), which implicates AGD2 directly or indirectly in repressing defenses. As stated above, this phenotype is partially attributable to ALD1 function. The agd2-T knockout may have a lethal phenotype because of the derepression of defense responses, including programmed cell death. There is precedence for a mutation in a defense component conferring a lethal phenotype. Plants lacking RPM1-interacting protein 4 show lethality (Mackey et al., 2002) that is likely to be because of the direct activation of an R gene pathway that activates cell death (Axtell and Staskawicz, 2003; Mackey et al., 2003). A link between developmental control and defense regulation also has been found for several defense regulatory genes (Vanacker et al., 2001; Holt et al., 2002; Jin et al., 2002; Barth and Conklin, 2003).
Specific Physiological Function of ALD1
ALD1 activity is important for P. syringae disease resistance. What might account for the increased disease susceptibility of ald1 mutants? An important clue comes from the observation that SA levels are reduced in ald1 upon pathogen infection and that agd2-1 ald1-T2 plants have less SA than agd2-1 alone. ALD1 possibly synthesizes a signal important for inducing SA synthesis. That the SA agonist BTH can induce disease resistance in ald1 supports this possibility. We have no evidence for ALD1 acting directly in the SA biosynthetic pathway because the enzyme was not active with any proposed SA biosynthetic intermediates.
The control of SA production may not be the sole function of ALD1. The wild-type level of disease susceptibility of agd2-1 ald1-T2 plants that still have ∼8.6-fold elevated SA relative to the wild type suggests that ALD1 also controls the synthesis of another defense signal. Several other genes in A. thaliana, such as PAD1, PAD2, PAD4, EDS1, EDS3, EDS4, EDS8, NDR1, and ACD6, are similar to ALD1 in that mutations in these genes also lead to reduced SA accumulation during infection by at least one strain of P. syringae (Glazebrook et al., 1996, 1997, 2003; Zhou et al., 1998; Gupta et al., 2000; Feys et al., 2001; Shapiro and Zhang, 2001; Lu et al., 2003). However, no demonstrated biochemical activity has been associated with the products of these genes. In the case of pad4, the mutation has been suggested to affect both SA-dependent defenses and possibly additional defenses (Glazebrook et al., 2003). Whether ALD1 acts in the same or separate pathways with PAD4 and/or other SA-regulatory genes remains to be determined.
ALD1 also may have a direct role in synthesizing antimicrobial compounds. Precedent for an aminotransferase having such a function comes from Streptomyces virginae, which uses Lys2-aminotransferase to synthesize a cyclohexadepsipeptide antibiotic (Namwat et al., 2002).
Relationship and Roles of AGD2 and ALD1 in Development and Defense
The opposing defense phenotypes of agd2-1 and ald1 are striking because AGD2 and ALD1 likely drive the aminotransferase reaction on related substrates in opposite directions. A model for how these enzymes function is shown in Figure 7B. AGD2 is shown promoting development based on its requirement for embryogenesis in agd2-T and the abnormal leaf morphology in agd2-1 (Rate and Greenberg, 2001). AGD2 also is shown having a second function repressing defenses. Because agd2-1 showed constitutive defenses before morphological alterations were apparent (Rate, 2000; Rate and Greenberg, 2001), the effect of agd2-1 on defense activation is shown as independent from the effect on development (although formally the developmental phenotype could cause the defense phenotype or vice versa). We speculate that in agd2-1, an amino acid–derived signal (possibly related to the ALD1 substrate), an amino acid imbalance, or the presence of a developmental regulator results in cross talk to the defense pathway. Amino acid starvation is known to activate plant defenses (Zhao et al., 1998). In support of a relationship between the substrates and products of the AGD2 and ALD1 enzymatic reactions, we found that ald1-T2 could in large part suppress the reduced rosette size and other phenotypes of agd2-1.
We have no direct evidence for any large differences in the amino acid compositions of wild-type and agd2-1 plants. However, we most likely missed amino acids that are either conjugated to other molecules (such as hormones), are present only transiently, or are low in abundance. Possibly, any amino acid imbalance caused by agd2-1 is very transient because of the conversion of AGD2 substrates into alternative products. In our model, ALD1 functions to regulate SA synthesis and a second defense signal through an amino acid–derived signal. This is based on the results discussed above showing that agd2-1ald1-T2 plants, which still have residual elevated SA, show pathogen susceptibility equal to that of wild-type plants. The second defense signal postulated to be regulated by ALD1 could be directly produced by ALD1.
In summary, we have shown that two related aminotransferases perform divergent yet critical functions to control plant development and defense. Our study implicates amino and/or oxo-acids as having central roles in these processes.
METHODS
Plant Materials, Treatment, and Pathogen Infection
The agd2-1 A. thaliana mutant in the Col background was described previously (Rate and Greenberg, 2001). T-DNAs in AGD2 and ALD1 (agd2-T and ald1-T1, respectively) in the Ws background were obtained by screening the T-DNA insertion library at the University of Wisconsin (http://www.biotech.wisc.edu/). Another ALD1 T-DNA allele (ald1-T2; SALK_007673) in Col was obtained from the Salk collection (http://signal.salk.edu/cgibin/tdnaexpress/).
Plants were grown and infections done as described previously (Greenberg et al., 2000). P. s. maculicola DG3 derived from P. s. maculicola strain ES4326 was described previously (Guttman and Greenberg, 2001). P. s. tomato DC3000 was obtained from F.M. Ausubel (Massachusetts General Hospital, Boston, MA).
BTH was a gift from Robert Dietrich (Syngenta, Research Triangle Park, NC). BTH treatments were done by spraying 16- to 18-d-old plants until all the leaves were wet. During dark treatment, plants were covered with an aluminum foil–wrapped plastic dome.
Recombination Mapping and Complementation Tests
To map AGD2, agd2-1 (ecotype Col) was crossed to ecotype Landsberg. Homozygous agd2-1 F2 plants were used for recombinant analysis using published cleaved amplified polymorphic sequences (CAPS) and simple sequence length polymorphism markers (Konieczny and Ausubel, 1993; Bell and Ecker, 1994). To identify new CAPS markers, primers were designed using the published sequence (http://www.arabidopsis.org/cereron/). AGD2 was mapped to a 65-kb interval between the CAPS markers C5-2 and C7-2N. The primers used for C5-2 were 5′-GATCGGAGGCATGCATTACAG-3′ and 5′-TCCGAACAATCCAGCATTACC-3′. The primers for C7-2N were 5′-TTCTCCTTCAGCACCTTGAAC-3′ and5′-CTACCCAGAGGTTTAACGACG-3′. Enzymes used for these CAPS markers were ApoI (C5-2) and DraI (C7-2N). The A. thaliana genomic BAC clone T16L1 (The Arabidopsis Information Resource, http://www.arabidopsis.org) covering this region was subcloned into the binary vector pAOV-CaMV. This vector is a modified version of pAOV (Mylne and Botella, 1998) lacking the CaMV 35S promoter, which was removed by digesting pAOV with HindIII and XbaI and filling in the overhangs. Subclones were transformed into agd2-1 as described (Clough and Bent, 1998). A defined AGD2 genomic clone (a 7280-bp SacI fragment from BAC clone T16L1) in pAOV-CaMV and the AGD2 cDNA (AV524925) driven by the CaMV 35S promoter cloned into pAOV were used for complementation of agd2-1. For complementation of ald1-T1 and ald1-T2, a genomic clone of ALD1 was subcloned as a 7970-bp XbaI-ScaI fragment from BAC clone F17L24 into pAOV-CaMV and transformed into ald1-T1 and ald1-T2 plants.
RNA Gel Blot Analysis
Total RNA was isolated as described (Kroczek and Siebert, 1990). The RNA (∼8 μg) was separated by electrophoresis on a 1% agarose gel. Hybridization was performed as described (Sambrook et al., 1989). Gene-specific DNA probes for AGD2 or ALD1 were made using PCR to amplify a portion (3′ end) of each gene from cDNA. The primers used for AGD2 were 5′-CTGCACTTGTTTCAATGGTGC-3′ (sense primer) and 5′-CACTGGTGTCAAAAGTGTCTTC-3′ (antisense primer). The primers for ALD1 were 5′-GAGATACGGTCGGTGAACAACT-3′ (sense primer) and 5′-CAAGACGATGCACATAACACG-3′ (antisense primer). Single-stranded probes were synthesized using the antisense primers.
Purification and Assays of Recombinant AGD2 and ALD1 Proteins
The AGD2 and ALD1 genes were amplified using PCR with the following primers using the cDNA clones of AGD2 (GenBank accession number AV524925), AGD2P398S (obtained using RT-PCR), and ALD1 (GenBank accession number AY057526) as templates. For AGD2 and AGD2P398S, the sequences of primers were 5′-GCAAACCCGGGCAATACTTGCAAATGTGTTGC-3′ (underlined sequence indicates the SmaI site) and 5′-AAACAACCCGGGTCATTTGTAAAGCTGCTTGA-3′ (underlined and boldface sequences indicate the SmaI site and the stop codon, respectively). For ALD1, the sequences of primers were 5′-TGGCTCGAGATTCCCAAGGCTAGTTTGGAC-3′ (underlined sequence indicates the XhoI site) and 5′-TGGCTCGAGATCCTAATTGGTATTAGAAGT-3′ (underlined and boldface sequences indicate the XhoI site and the stop codon, respectively). These recombinant AGD2 (or AGD2P398S) and ALD1 genes used for producing recombinant proteins lacked sequences encoding the first 40 and 20 amino acids, respectively. These N-terminal sequences corresponded to possible plastid-targeting signals. PCR products containing the AGD2, AGD2P398S, and ALD1 genes were digested with SmaI and XhoI and then ligated into the expression vector pET14b (Novagen, Madison, WI) cut with the appropriate restriction enzyme. The recombinant plasmids were transformed into E. coli BL21 (DE3) pLysS. To induce production of the 6x-His-tagged recombinant enzymes, bacterial cultures grown to an A600 of 0.7 to 0.9 in LB medium containing 100 μg/mL ampicillin at 37°C were treated with isopropyl-β-d-thiogalactopyranoside at a final concentration of 1 mM. Incubation was allowed to continue at 20°C for an additional 12 h, and the cells were harvested. Cell pellets were resuspended in one-tenth the culture volume in a solution containing 50 mM Tris-HCl buffer, pH 8.0, and then subjected to two cycles of freezing and thawing. The crude extracts were treated with DNase and MgCl2 to final concentrations of 10 μg/mL and 20 mM, respectively, at room temperature for 30 min. After centrifugation, imidazole and NaCl were added to the supernatant to final concentrations of 5 and 500 mM, respectively. The 6x-His-tagged fusion proteins were purified under native conditions using Ni2+-NTA agarose beads (Qiagen, Valencia, CA). The lysates were incubated for at least 30 min with binding buffer (5 mM imidazole, 50 mM NaCl, and 50 mM Tris-HCl, pH 8.0) containing Ni2+-NTA agarose beads. Elution was performed by a linear gradient of 5 to 200 mM. The recombinant proteins eluted at nearly 80 mM imidazole. Purified enzymes were stored at 4°C for 2 weeks without significant loss of the activity. Protein concentrations were determined as described (Bradford, 1976) using BSA as a standard.
The recombinant AGD2, AGD2P398S, and ALD1 activities were determined by measuring the concentration of the reaction product glutamate or Ala. The assay buffer contained 50 mM Tris-HCl (pH 8.0), 100 mM MgCl2, and 1 μM pyridoxal-5-phosphate. Activities of the recombinant enzymes were found to be highest at 37°C in Tris-HCl, pH 8.0, and 100 mM MgCl2. The optimum temperature for the recombinant enzymes was determined using assay buffer containing 50 mM Tris-HCl (pH 8.0), 100 mM MgCl2, and 1 μM pyridoxal-5-phosphate with 50 mM Lys as the amino donor and 50 mM 2-oxoglutarate as the amino acceptor at 4, 15, 25, 30, 37, 45, 50, and 65°C for 4 h. The pH dependence of the enzymes was determined at 37°C using three different buffer systems. Reactions were performed in 50 mM sodium acetate buffer at pH values ranging from 4.0 to 6.0, in 50 mM sodium phosphate buffer at pH values ranging from 6.0 to 7.0, and in 50 mM Tris-HCl buffer at pH values ranging from 7.0 to 10.0. Effects of various cations on activities also were examined. Cations such as Cu2+, Mn2+, and NH4+ did not significantly activate the activities at 100 mM each, whereas Ca2+, Na+, and K+ showed better activation effects. The highest activities were in 100 mM MgCl2. After the reaction, samples were applied to a thin-layer chromatographic plate (Si250; J.T. Baker, Phillipsburg, NJ) and developed with n-butyl alcohol:acetic acid:water (3:1:1, v/v). The chromatogram, dried at 60°C and cooled to room temperature, was soaked in freshly prepared ninhydrin solution (0.2% in acetone) for 1 min (Bhushan and Martens, 2001). To quantify the signals, the National Institutes of Health Image J program was used (http://rsb.info.nih.gov/ij/).
SA Measurements
Free and total SA (the sum of free and glucosyl SA) was extracted and quantified as described previously (Seskar et al., 1998).
Subcellular Localization of AGD2
The gfp gene was translationally fused at the 3′ end of the cDNA of AGD2 gene in plasmid pAOV (Mylne and Botella, 1998) behind the CaMV 35S promoter to give pAGD2-GFP. The plasmid pAGD2-GFP or pGFP (GFP alone driven by the CaMV 35S promoter) was introduced into A. tumefaciens (GV3101) and then transformed into A. thaliana plants. Protoplasts from leaves of transgenic plants were isolated as described (Asai et al., 2000). GFP localization and chlorophyll autofluorescence were observed using a fluorescence microscope (Axioskop; Carl Zeiss, Jena, Germany) with fluorescein isothiocyanate and UV filters, respectively.
Amino Acid Analysis
Approximately 200 mg of fresh leaves were ground in liquid N2 in the presence of water:methanol:chloroform (2:3.5:8) (Palanivelu et al., 2003). The homogenate was extracted and centrifuged. The supernatant was dried by passing nitrogen gas over it. The residue was resuspended in water and submitted to the Molecular Structure Facility at University of California (Davis, CA) for amino acid analysis by HPLC (Palanivelu et al., 2003).
GC-MS Analysis
Leaves of Ws and ald1-T1 plants were infected with P. s. maculicola DG3 (OD600 = 0.01). The infected leaves (50 mg) at 9 and 24 h after infection were ground in liquid N2 in 1.4 mL of 100% methanol with 50 μL internal standard (2 mg of ribitol/1 mL of water) (Roessner et al., 2000). The mixture was extracted with 1 volume of water and subsequently centrifuged. The supernatant was dried and analyzed by the Michigan State University Mass Spectrometry Facility (East Lansing, MI) using GC-MS. GC-MS conditions and the system for the analysis of metabolites were performed as described (Roessner et al., 2000).
Characterization of O. sativa cDNAs for OsAGD2 and OsALD1
The stop codons of the cDNAs were determined by RT-PCR using primers located at the stop codons of the annotated genomic sequences and/or by homology between the O. sativa and A. thaliana genes. The 5′ ends of the O. sativa cDNAs were determined by 5′ rapid amplification of cDNA ends PCR according to the Invitrogen manual (Carlsbad, CA). GenBank accession numbers of cOsAGD2 and cOsALD1 are AY338235 and AY338236, respectively.
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession numbers AY338235, AY338236, AV524925, and AY057526.
Acknowledgments
We thank Ravishankar Palanivelu for advice and critical reading of the manuscript. We thank Brian Traw and Joy Bergelson for their assistance with SA quantitation. We are grateful to the ABRC at the Ohio State University and Kazusa DNA Research Institute for clones. This work was supported by National Institutes of Health Grant 5R01 GM54292 to J.T.G. and the postdoctoral fellowship program of the Korea Science and Engineering Foundation to J.T.S.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Jean T. Greenberg (jgreenbe@midway.uchicago.edu).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.019372.
References
- Agrios, G.N. (1997) Plant Pathology. (San Diego, CA: Academic Press).
- Asai, T., Stone, J.M., Heard, J.E., Kovtun, Y., Yorgey, P., Sheen, J., and Ausubel, F.M. (2000). Fumonisin B1-induced cell death in Arabidopsis protoplasts requires jasmonate-, ethylene-, and salicylate-dependent signaling pathways. Plant Cell 12, 1823–1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asai, T., Tena, G., Plotnikova, J., Willmann, M.R., Chiu, W.L., Gomez-Gomez, L., Boller, T., Ausubel, F.M., and Sheen, J. (2002). MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415, 977–983. [DOI] [PubMed] [Google Scholar]
- Axtell, M.J., and Staskawicz, B.J. (2003). Initiation of RPS2-specified disease resistance in Arabidopsis is coupled to the AvrRpt2-directed elimination of RIN4. Cell 112, 369–377. [DOI] [PubMed] [Google Scholar]
- Azevedo, R.A., Arruda, P., Turner, W.L., and Lea, P.J. (1997). The biosynthesis and metabolism of the aspartate derived amino acids in higher plants. Phytochemistry 46, 395–419. [DOI] [PubMed] [Google Scholar]
- Barth, C., and Conklin, P. (2003). The lower cell density of leaf parenchyma in the Arabidopsis thaliana mutant lcd1–1 is associated with increased sensitivity to ozone and virulent Pseudomonas syringae. Plant J. 35, 206–218. [DOI] [PubMed] [Google Scholar]
- Bell, C.J., and Ecker, J.R. (1994). Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics 19, 137–144. [DOI] [PubMed] [Google Scholar]
- Bhushan, R., and Martens, J. (2001). Separation of amino acids, their derivatives and enantiomers by impregnated TLC. Biomed. Chromatogr. 15, 155–165. [DOI] [PubMed] [Google Scholar]
- Bradford, M.M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254. [DOI] [PubMed] [Google Scholar]
- Bryan, J.K. (1990) Advances in the biochemistry of amino acid biosynthesis. In The Biochemistry of Plants, J. Miflin, ed (New York, NY: Academic Press), pp. 403–452.
- Buchanan, B.B., Gruissem, W., and Jones, R.L. (2000) Biochemistry and Molecular Biology of Plants. (Rockville, MD: American Society of Plant Physiologists).
- Chang, J.H., Tai, Y.S., Bernal, A.J., Lavelle, D.T., Staskawicz, B.J., and Michelmore, R.W. (2002). Functional analyses of the Pto resistance gene family in tomato and the identification of a minor resistance determinant in a susceptible haplotype. Mol. Plant Microbe Interact. 15, 281–291. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Cooke, T.J., Poli, D., Sztein, A.E., and Cohen, J.D. (2002). Evolutionary patterns in auxin action. Plant Mol. Biol. 49, 319–338. [PubMed] [Google Scholar]
- Feys, B.J., Moisan, L.J., Newman, M.A., and Parker, J.E. (2001). Direct interaction between the Arabidopsis disease resistance signaling proteins, EDS1 and PAD4. EMBO J. 20, 5400–5411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankard, V., Ghislain, M., and Jacobs, M. (1992). Two feedback-insensitive enzymes of the aspartate pathway in Nicotiana sylvestris. Plant Physiol. 99, 1285–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaffney, T., Friedrich, L., Vernooij, B., Negrotto, D., Nye, G., Uknes, S., Ward, E., Kessmann, H., and Ryals, J. (1993). Requirement of salicylic acid for the induction systemic acquired resistance. Science 261, 754–756. [DOI] [PubMed] [Google Scholar]
- Galili, G. (1995). Regulation of lysine and threonine synthesis. Plant Cell 7, 899–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardan, L., Shafik, H., Belouin, S., Broch, R., Grimont, F., and Grimont, P.A. (1999). DNA relatedness among the pathovars of Pseudomonas syringae and description of Pseudomonas tremae sp. nov. and Pseudomonas cannabina sp. nov. (ex Sutic and Dowson 1959). Int. J. Syst. Bacteriol. 49, 469–478. [DOI] [PubMed] [Google Scholar]
- Glazebrook, J., and Ausubel, F.M. (1994). Isolation of phytoalexin-deficient mutants of Arabidopsis thaliana and characterization of their interactions with bacterial pathogens. Proc. Natl. Acad. Sci. USA 91, 8955–8959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazebrook, J., Chen, W., Estes, B., Chang, H.S., Nawrath, C., Métraux, J.P., Zhu, T., and Katagiri, F. (2003). Topology of the network integrating salicylate and jasmonate signal transduction derived from global expression phenotyping. Plant J. 34, 217–228. [DOI] [PubMed] [Google Scholar]
- Glazebrook, J., Rogers, E.E., and Ausubel, F.M. (1996). Isolation of Arabidopsis mutants with enhanced disease susceptibility by direct screening. Genetics 143, 973–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazebrook, J., Zook, M., Mert, F., Kagan, I., Rogers, E.E., Crute, I.R., Holub, E.B., Hammerschmidt, R., and Ausubel, F.M. (1997). Phytoalexin-deficient mutants of Arabidopsis reveal that PAD4 encodes a regulatory factor and that four PAD genes contribute to downy mildew resistance. Genetics 146, 381–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glickmann, E., Gardan, L., Jacquet, S., Hussain, S., Elasri, M., Petit, A., and Dessaux, Y. (1998). Auxin production is a common feature of most pathovars of Pseudomonas syringae. Mol. Plant Microbe Interact. 11, 156–162. [DOI] [PubMed] [Google Scholar]
- Greenberg, J.T., Silverman, F.P., and Liang, H. (2000). Uncoupling salicylic acid-dependent cell death and defense-related responses from disease resistance in the Arabidopsis mutant acd5. Genetics 156, 341–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg, J.T., and Vinatzer, B.A. (2003). Identifying type III effectors of plant pathogens and analysing their interaction with plant cells. Curr. Opin. Microbiol. 6, 20–28. [DOI] [PubMed] [Google Scholar]
- Gupta, V., Willits, M.G., and Glazebrook, J. (2000). Arabidopsis thaliana EDS4 contributes to salicylic acid (SA)-dependent expression of defense responses: Evidence for inhibition of jasmonic acid signaling by SA. Mol. Plant Microbe Interact. 13, 503–511. [DOI] [PubMed] [Google Scholar]
- Guttman, D.S., and Greenberg, J.T. (2001). Functional analysis of type III effectors AvrRpt2 and AvrRpm1 of P. syringae using a single copy genomic integration system. Mol. Plant Microbe Interact. 14, 145–155. [DOI] [PubMed] [Google Scholar]
- Holt, B.F., 3rd, Boyes, D.C., Ellerstrom, M., Siefers, N., Wiig, A., Kauffman, S., Grant, M.R., and Dangl, J. (2002). An evolutionarily conserved mediator of plant disease resistance gene function is required for normal Arabidopsis development. Dev. Cell 2, 807–817. [DOI] [PubMed] [Google Scholar]
- Jeon, S.G., Bahn, J.H., Jang, J.S., Park, J., Kwon, O., Cho, S., and Choi, S.Y. (2000). Human brain GABA transaminase. Eur. J. Biochem. 267, 5601–5607. [DOI] [PubMed] [Google Scholar]
- Jin, H., Axtell, M.J., Dahlbeck, D., Ekwenna, O., Zhang, S., Staskawicz, B., and Baker, B. (2002). NPK1, an MEKK1-like mitogen-activated protein kinase kinase kinase, regulates innate immunity and development in plants. Dev. Cell 3, 291–297. [DOI] [PubMed] [Google Scholar]
- Jirage, D., Tootle, T.L., Reuber, T.L., Frost, L.N., Feys, B.J., Parker, J.E., Ausubel, F.M., and Glazebrook, J. (1999). Arabidopsis thaliana PAD4 encodes a lipase-like gene that is important for salicylic acid signaling. Proc. Natl. Acad. Sci. USA 96, 13583–13588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konieczny, A., and Ausubel, F.M. (1993). A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific markers. Plant J. 4, 403–410. [DOI] [PubMed] [Google Scholar]
- Kroczek, R.A., and Siebert, E. (1990). Optimization of Northern analysis by vaccum-blotting, RNA transfer, visualization and ultraviolet fixation. Anal. Biochem. 184, 90–95. [DOI] [PubMed] [Google Scholar]
- Ljung, K., Hull, A.K., Kowalczyk, M., Marchant, A., Celenza, J., Cohen, J.D., and Sandberg, G. (2002). Biosynthesis, conjugation, catabolism and homeostasis of indole-3-acetic acid in Arabidopsis thaliana. Plant Mol. Biol. 49, 249–272. [PubMed] [Google Scholar]
- Lotan, A.D., Wahleithner, J.A., Lund, M., and Bonnett, H.T. (1989). Pathogenesis-related proteins are developmentally regulated in tobacco flowers. Plant Cell 1, 881–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, H., Rate, R.N., Song, J.T., and Greenberg, J. (2003). ACD6, a novel ankyrin protein, is a regulator and an effector of salicylic acid signaling in the Arabidopsis defense response. Plant Cell 15, 2408–2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackey, D., Belkhadir, Y., Alonso, J.M., Ecker, J.R., and Dangl, J.L. (2003). Arabidopsis RIN4 is a target of the type III virulence effector AvrRpt2 and modulates RPS2-mediated resistance. Cell 112, 379–389. [DOI] [PubMed] [Google Scholar]
- Mackey, D., Holt, B.F., 3rd, Wiig, A., and Dangl, J.L. (2002). RIN4 interacts with Pseudomonas syringae type III effector molecules and is required for RPM1-mediated resistance in Arabidopsis. Cell 108, 743–754. [DOI] [PubMed] [Google Scholar]
- Matsui, I., Matsui, E., Sakai, Y., Kikuchi, H., Kawarabayashi, Y., Ura, H., Kawaguchi, S., Kuramitsu, S., and Harata, K. (2000). The molecular structure of hyperthermostable aromatic aminotransferase with novel substrate specificity from Pyrococcus horikoshii. J. Biol. Chem. 275, 4871–4879. [DOI] [PubMed] [Google Scholar]
- Mehta, P.K., Hale, T.I., and Christen, P. (1989). Evolutionary relationships among aminotransferases. Eur. J. Biochem. 186, 249–253. [DOI] [PubMed] [Google Scholar]
- Mehta, P.K., Hale, T.I., and Christen, P. (1993). Aminotransferases: Demonstration of homology and division into evolutionary subgroups. Eur. J. Biochem. 214, 549–561. [DOI] [PubMed] [Google Scholar]
- Morris, K., MacKerness, S.A., Page, T., John, C.F., Murphy, A.M., Carr, J.P., and Buchanan-Wollaston, V. (2000). Salicylic acid has a role in regulating gene expression during leaf senescence. Plant J. 23, 677–685. [DOI] [PubMed] [Google Scholar]
- Mylne, J., and Botella, J.R. (1998). Binary vectors for sense and antisense expression of Arabidopsis ESTs. Plant Mol. Biol. Rep. 16, 257–262. [Google Scholar]
- Namwat, W., Kinoshita, H., and Nihira, T. (2002). Identification by heterologous expression and gene disruption of VisA as L-lysine2-aminotransferase essential for virginiamycin S biosynthesis in Streptomyces virginiae. J. Bacteriol. 184, 4811–4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrath, C., Heck, S., Parinthawong, N., and Métraux, J.P. (2002). EDS5, an essential component of salicylic acid-dependent signaling for disease resistance in Arabidopsis, is a member of the MATE transporter family. Plant Cell 14, 275–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrath, C., and Métraux, J.P. (1999). Salicylic acid induction-deficient mutants of Arabidopsis express PR-2 and PR-5 and accumulate high levels of camalexin after pathogen inoculation. Plant Cell 11, 1393–1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowicki, C., Hunter, G.R., Montemartini-Kalisz, M., Blankenfeldt, W., Hecht, H., and Kalisz, H.M. (2001). Recombinant tyrosine aminotransferase from Trypanosoma cruzi: Structural characterization and site directed mutagenesis of a broad substrate specificity enzyme. Biochem. Biophys. Acta 1546, 268–281. [DOI] [PubMed] [Google Scholar]
- Palanivelu, R., Brass, L., Edlund, A.F., and Preuss, D. (2003). Pollen tube growth and guidance is regulated by POP2, an Arabidopsis gene that controls GABA levels. Cell 114, 1–20. [DOI] [PubMed] [Google Scholar]
- Patton, D.A., Schetter, A.L., Franzmann, L.H., Nelson, K., Ward, E.R., and Meinke, D.W. (1998). An embryo-defective mutant of Arabidopsis disrupted in the final step of biotin synthesis. Plant Physiol. 116, 935–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rassmussen, J.B., Hammerschmidt, R., and Zook, M.N. (1991). Systemic induction of salicylic acid accumulation in cucumber after inoculation with Pseudomonas syringae pv. syringae. Plant Physiol. 97, 1342–1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rate, D.N. (2000) Novel Genes of the Plant Defense Response Pathway in Arabidopsis thaliana. PhD dissertation (Boulder, CO: University of Colorado at Boulder).
- Rate, D.N., and Greenberg, J.T. (2001). The Arabidopsis aberrant growth and death2 mutant shows resistance to Pseudomonas syringae and reveals a role for NPR1 in suppressing hypersensitive cell death. Plant J. 27, 203–211. [DOI] [PubMed] [Google Scholar]
- Roessner, U., Wagner, C., Kopka, J., Trethewey, R.N., and Willmitzer, L. (2000). Technical advance: Simultaneous analysis of metabolites in potato tuber by gas chromatography-mass spectrometry. Plant J. 23, 131–142. [DOI] [PubMed] [Google Scholar]
- Rogers, E.E., and Ausubel, F.M. (1997). Arabidopsis enhanced disease susceptibility mutants exhibit enhanced susceptibility to several bacterial pathogens and alterations in PR-1 gene expression. Plant Cell 9, 305–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryals, J.A., Neuenschwander, U.H., Willits, M.G., Molina, A., Steiner, H.Y., and Hunt, M.D. (1996). Systemic acquired resistance. Plant Cell 8, 1809–1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzman, R.A., Tikhonova, I., Bordelon, B.P., Hasegawa, P.M., and Bressan, R.A. (1998). Coordinate accumulation of antifungal proteins and hexoses constitutes a developmentally controlled defense response during fruit ripening in grape. Plant Physiol. 117, 465–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual, 2nd ed. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Seskar, M., Shulaev, V., and Raskin, I. (1998). Endogenous methyl salicylate in pathogen-inoculated tobacco plants. Plant Physiol. 116, 387–392. [PMC free article] [Google Scholar]
- Shapiro, A.D., and Zhang, C. (2001). The role of NDR1 in avirulence gene-directed signaling and control of programmed cell death in Arabidopsis. Plant Physiol. 127, 1089–1101. [PMC free article] [PubMed] [Google Scholar]
- Sharma, Y.K., Leon, J., Raskin, I., and Davis, K.R. (1996). Ozone-induced responses in Arabidopsis thaliana: The role of salicylic acid in the accumulation of defense-related transcripts and induced resistance. Proc. Natl. Acad. Sci. USA 93, 5099–5104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanacker, H., Lu, H., Rate, D.N., and Greenberg, J.T. (2001). A role for salicylic acid and NPR1 in regulating cell growth in Arabidopsis. Plant J. 28, 209–216. [DOI] [PubMed] [Google Scholar]
- Verberne, M.C., Verpoorte, R., Bol, J.F., Mercado-Blanco, J., and Linthorst, H.J. (2000). Overproduction of salicylic acid in plants by bacterial transgenes enhances pathogen resistance. Nat. Biotechnol. 18, 779–783. [DOI] [PubMed] [Google Scholar]
- Vernal, J., Cazzalo, J.J., and Nowicki, C. (1998). Isolation and partial characterization of a broad specificity aminotransferase from Leishmania mexicana promastigotes. Mol. Biochem. Parasitol. 96, 83–92. [DOI] [PubMed] [Google Scholar]
- Wildermuth, M.C., Dewdney, J., Wu, G., and Ausubel, F.M. (2001). Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 414, 562–565. [DOI] [PubMed] [Google Scholar]
- Zhao, J., Williams, C.C., and Last, R.L. (1998). Induction of Arabidopsis tryptophan pathway enzymes and camalexin by amino acid starvation, oxidative stress, and an abiotic elicitor. Plant Cell 10, 359–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, N., Tootle, T.L., Tsui, F., Klessig, D.F., and Glazebrook, J. (1998). PAD4 functions upstream from salicylic acid to control defense responses in Arabidopsis. Plant Cell 10, 1021–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]