Abstract
Exposure of imbibed seeds to low temperature (typically 4°C) is widely used to break seed dormancy and to improve the frequency of germination. However, the mechanism by which temperature accelerates germination is largely unknown. Using DNA microarray and gas chromatography–mass spectrometry analyses, we found that a subset of gibberellin (GA) biosynthesis genes were upregulated in response to low temperature, resulting in an increase in the level of bioactive GAs and transcript abundance of GA-inducible genes in imbibed Arabidopsis thaliana seeds. Using a loss-of-function mutant, the cold-inducible GA biosynthesis gene, AtGA3ox1, was shown to play an essential role in mediating the effect of low temperature. Besides temperature, AtGA3ox1 also is positively regulated by active phytochrome and negatively regulated by GA activity. We show that both red light and GA deficiency act in addition to low temperature to elevate the level of AtGA3ox1 transcript, indicating that multiple signals are integrated by the AtGA3ox1 gene to control seed germination. When induced by low temperature, AtGA3ox1 mRNA was detectable by in situ RNA hybridization in an additional set of cell types relative to that in red light–induced seeds. Our results illustrate that the GA biosynthesis and response pathways are activated during seed imbibition at low temperature and suggest that the cellular distribution of bioactive GAs may be altered under different light and temperature conditions.
INTRODUCTION
Gibberellins (GAs) are phytohormones that are essential for many processes of plant development, such as seed germination, stem elongation, leaf expansion, flowering, and seed development (Davies, 1995). GAs are tetracyclic diterpenoids that are synthesized from geranylgeranyl diphosphate produced mainly through the methylerythritol phosphate pathway (Kasahara et al., 2002). Geranylgeranyl diphosphate is converted to biologically active GAs by terpene cyclases, P450 monooxygenases, and 2-oxoglutarate–dependent dioxygenases (Hedden and Kamiya, 1997). As summarized in Figure 1, most of the genes encoding GA biosynthesis and catabolism enzymes now have been identified (Olszewski et al., 2002). Previous studies have indicated that GA-mediated developmental processes are regulated in part by changing the cellular concentration of bioactive GAs. Therefore, it has been thought that GA concentrations must be carefully modulated, perhaps by integrating various endogenous and external signals (Hedden and Phillips, 2000; Yamaguchi and Kamiya, 2000).
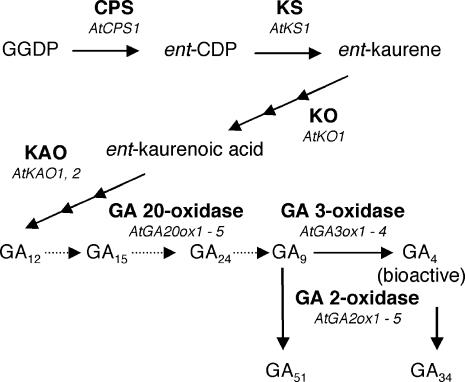
Figure 1.
The Major GA Biosynthesis Pathway in A. thaliana.
Dotted lines indicate the steps that are catalyzed by GA 20-oxidase. CPS, ent-copalyl diphosphate synthase; ent-CDP, ent-copalyl diphosphate; GGDP, geranylgeranyl diphosphate; KAO, ent-kaurenoic acid oxidase; KO, ent-kaurene oxidase; KS, ent-kaurene synthase.
It has been well documented that GA promotes seed germination in many plant species. In Arabidopsis thaliana, severe GA deficient–mutants, such as ga1-3 and ga2-1, are defective in seed germination (Koornneef and van der Veen, 1980). In addition, chemical inhibitors of GA biosynthesis enzymes, such as uniconazole and paclobutrazol, inhibit germination (Nambara et al., 1991; Jacobsen and Olszewski, 1993). These observations indicate that de novo GA biosynthesis is necessary for seed germination after imbibition in A. thaliana (Hedden and Kamiya, 1997). In fact, the level of bioactive GAs recently has been shown to increase just before radicle protrusion in germinating A. thaliana seeds (Ogawa et al., 2003).
Light is a critical environmental determinant for seed germination in some small-seeded plants, such as Lactuca sativa (lettuce), Lycopersicon esculentum (tomato), and A. thaliana (Shinomura, 1997). The effect of light on seed germination is primarily mediated by the red (R) and far-red (FR) light photoreceptor phytochrome (Borthwick et al., 1952; Butler et al., 1959). The best-characterized phytochrome control of seed germination is called the low-fluence response, in which R light promotes and FR light reversibly inhibits germination (Shinomura et al., 1995, 1996). There is some evidence that GA biosynthesis is regulated by phytochrome in germinating L. sativa and A. thaliana seeds, in which genes encoding GA 3-oxidases (Figure 1) are regulated by R and FR light in a photoreversible manner (Toyomasu et al., 1998; Yamaguchi et al., 1998). Therefore, it has been suggested that the control of bioactive GA levels by phytochrome is responsible, at least in part, for light-dependent seed germination in these species.
Temperature is another crucial external cue that controls seed germination (Bewley and Black, 1982). In many plant species, including A. thaliana, exposure of seeds to low temperatures (typically 2 to 5°C) immediately after imbibition promotes germination (Shropshire et al., 1961; Cone and Spruit, 1983). Although such cold treatment, often called stratification, is widely used to improve the frequency and synchronization of germination, the molecular mechanism underlying this thermoregulation remains unclear. Because of its positive role in seed germination, the effect of cold treatment on GA content previously has been examined in Pyrus malus (apple), Corylus avellana (hazel), and A. thaliana seeds (Ross and Bradbeer, 1971; Sińska et al., 1973; Williams et al., 1974; Derkx et al., 1994). These studies indicate that bioactive GAs were more abundant in cold-treated seeds than in non-cold-treated samples. However, it is not clear from these initial studies whether low temperature acted as a signal to modulate the GA metabolism pathway because cold treatment also involved a much longer total imbibition period (e.g., 7 d at 2°C and then 8 h at 24°C) compared with the control sample (e.g., 8 h at 24°C) (Derkx et al., 1994). To understand the molecular basis of how cold treatment promotes seed germination, it is therefore important to distinguish the effect of temperature from that of imbibition, which results in a gradual increase in water content.
Besides the concentration of bioactive GAs, tissue sensitivity to GA is another parameter to be considered to elucidate how GA controls developmental processes. In A. thaliana seeds, both light and temperature conditions can alter the amount of GA required for seeds to germinate (Derkx and Karssen, 1993a; Yang et al., 1995; Koornneef et al., 2002). For example, when GA-deficient ga1 mutant seeds were preincubated at 2°C for 7 d in the dark, a lower concentration of exogenous GA4 was sufficient for the mutant seeds to germinate in comparison with the case without cold treatment (Derkx and Karssen, 1993a). This observation indicates that the sensitivity to bioactive GAs may be enhanced by cold treatment (low temperature or long imbibition) in A. thaliana seeds, although the molecular nature that determines GA sensitivity is still unknown.
To address how cold treatment stimulates seed germination, we performed large-scale expression analysis during imbibition of after-ripened A. thaliana dry seeds at 4°C. This analysis indicates that a number of GA-related genes are differentially expressed between dry and cold-treated seeds. To clarify whether low temperature acts as a signal to modulate GA biosynthesis and/or response pathways, another experimental system has been developed, in which the effects of different temperatures during imbibition are directly compared. Our gas chromatography–mass spectrometry (GC-MS) and quantitative reverse transcription (QRT) PCR analyses show that GA biosynthesis is activated in response to low temperature in dark-imbibed seeds at least in part through changes in the transcript abundance of particular GA biosynthesis genes. Using a loss-of-function mutant of the cold-inducible AtGA3ox1 gene, we show that AtGA3ox1 is required for cold-promoted synthesis of bioactive GAs and seed germination. Furthermore, our data show that the low temperature signal increases the number of cell types in which the AtGA3ox1 transcript is detectable by in situ RNA hybridization analysis, suggesting a complex regulatory mechanism in which the spatial distribution of bioactive GAs is determined under varying environmental conditions.
RESULTS
Some GA Biosynthesis Genes Are Responsive to Low Temperature
We analyzed differential transcript accumulation between after-ripened dry seeds (stored at room temperature) and seeds imbibed at 4°C in the dark for 48 h using an oligonucleotide-based microarray consisting of ∼8200 genes. This type of cold treatment (stratification) is known to be effective to promote and synchronize seed germination. Throughout this study, we refer to seed imbibition at 4°C in the dark as cold treatment. Thus, cold treatment in this study involves both imbibition and exposure to 4°C. In this study, both dry and cold-treated seeds yielded approximately equal amounts of total RNA and labeled target RNAs. Therefore, our comparison of relative mRNA levels in this microarray analysis roughly represents relative amounts per seed basis.
Our microarray analysis indicated that transcript levels of 161 and 152 genes were upregulated and downregulated, respectively, when dry seeds were subjected to cold treatment. Genes were classified as being differentially expressed if the signal values deviated either positively or negatively fourfold or more between the two samples in duplicated experiments using independent seed batches. Identities of such genes are shown in the supplemental data online. We found that several GA biosynthesis genes, such as AtGA20ox3 and AtGA3ox1, were upregulated upon cold treatment of dry seeds as shown in Table 1. In addition, it was noted that cold treatment–responsive genes include several genes that also were regulated by GA during seed germination (Ogawa et al., 2003). Among 161 genes that were upregulated upon cold treatment, 38 genes (24%) were classified as being GA inducible based on their responsiveness to 50 μM GA4 in imbibed ga1-3 mutant seeds (Ogawa et al., 2003; see supplemental data online). Similarly, 38 genes (25%) that were downregulated upon cold treatment correspond to GA-repressible genes identified in the same study (Ogawa et al., 2003; see supplemental data online). Taken together, these results suggest that GA biosynthesis might be substantially upregulated during cold treatment, resulting in an upregulation of at least part of the GA response pathway.
Table 1.
Expression of GA Biosynthesis Genes in Dry Seeds versus Seeds after Cold Treatment
| Signal Valuea
|
||||||
|---|---|---|---|---|---|---|
| Enzyme | Gene | Probe Set | AGI Gene Codeb | Dry Seeds | Seeds After Cold Treatment | Differential Expressionc |
| ent-copalyl diphosphate synthase | AtCPS1 | 17498_s_at | At4g02780 | NDd | ND | |
| ent-kaurene synthase | AtKS1 | 15301_s_at | At1g79460 | ND | ND | |
| ent-kaurene oxidase | AtKO1 | 17543_s_at | At5g25900 | 35.1 | 36.4 | |
| ent-kaurenoic acid oxidase | AtKAO1 | 20493_at | At1g05160 | 15.8 | 2.4 | |
| AtKAO2 | 13021_at | At2g32440 | 8.2 | 7.1 | ||
| GA 20-oxidase | AtGA20ox1 | 17299_s_at | At4g25420 | ND | 3.4 | Yes |
| AtGA20ox2 | 17739_at | At5g51810 | ND | 1.7 | ||
| AtGA20ox3 | 12368_at | At5g07200 | ND | 13.7 | Yes | |
| GA 3-oxidase | AtGA3ox1 | 17549_at | At1g15550 | ND | 63.4 | Yes |
| AtGA3ox2 | 17492_at | At1g80340 | ND | 2.3 | ||
| AtGA3ox3 | 19250_at | At4g21690 | ND | ND | ||
| GA 2-oxidase | AtGA2ox1 | 16292_at | At1g78440 | ND | ND | |
| AtGA2ox2 | 13789_at | At1g30040 | ND | 2.1 | Yes | |
| AtGA2ox3 | 19480_s_at | At2g34500 | ND | ND | ||
Signal values from a single replicate are shown.
Arabidopsis Genome Initiative (2000).
Genes are classified as being differentially expressed when signal values deviate fourfold or more in duplicated experiments.
The signal is not detectable (ND) by microarray analysis.
Because we compared after-ripened dry seeds with seeds imbibed in the dark at 4°C by microarray analysis, the genes identified by this analysis would have responded to a difference in the state of imbibition (water content), light, and/or temperature. Therefore, to examine whether GA-related genes responded to low temperature, another set of experiments was designed. It has been shown previously that two genes encoding GA 3-oxidases, AtGA3ox1 and AtGA3ox2 (Figure 1), are regulated by light through phytochrome (Yamaguchi et al., 1998). To eliminate the effect of phytochrome to study temperature regulation of GA biosynthesis, wild-type seeds were imbibed in the dark, irradiated with an FR light pulse, and then incubated in the dark for 48 h at 22 or 4°C (Figure 2A). The FR pulse treatment is necessary to inactivate phytochrome that is stored in dry seeds in the active form (Hayes and Klein, 1974). Under the light condition given in Figure 2A, wild-type seeds do not germinate at both temperatures (up to 96 h). Also, both samples undergo the same period of seed imbibition. Therefore, we reasoned that any difference observed between these two samples would reflect the effect of temperature.
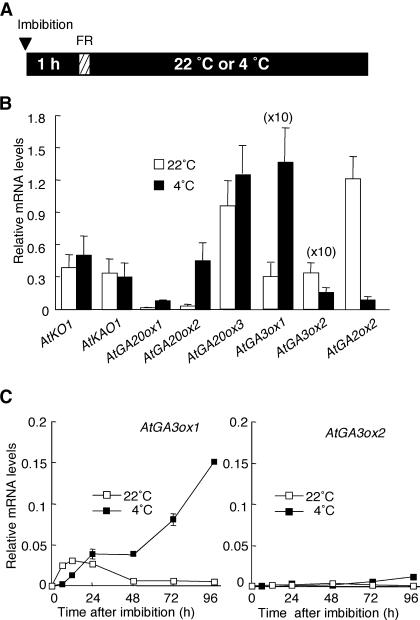
Figure 2.
Expression of GA Biosynthesis Genes at 4 and 22°C in Dark-Imbibed Seeds.
(A) Diagram showing light and temperature treatments. Wild-type seeds were imbibed in the dark and then irradiated with an FR light pulse (hatched box). The seeds then were incubated in the dark at 22 or 4°C until harvested (see below). The triangle indicates the start time of imbibition.
(B) Transcript accumulation of GA biosynthesis genes as determined by QRT-PCR. Wild-type seeds were imbibed as shown in (A) and harvested 48 h after FR light pulse. Values are means with standard errors from three measurements. Values for AtGA3ox1 and AtGA3ox2 are multiplied by 10 for clarity.
(C) Time-course changes in transcript accumulation of AtGA3ox1 and AtGA3ox2 genes as determined by QRT-PCR. Wild-type seeds were imbibed as shown in (A). Start of imbibition was set as 0 h. Experiments were repeated twice with similar results. Data from one of the replicates are shown.
Figure 2B illustrates the result of QRT-PCR analysis of GA metabolism genes in wild-type seeds incubated at 22 or 4°C. In this experiment, we focused on GA metabolism genes that show a higher transcript level in cold-treated seeds than in dry seeds by microarray analysis (Table 1). Among transcripts that encode GA 20-oxidases, those of AtGA20ox1 and AtGA20ox2 accumulated at much higher levels at 4°C than at 22°C (Figure 2B). No significant difference was observed in the level of AtGA20ox3 mRNA at the two temperatures. AtGA3ox1 and AtGA3ox2 are the major genes encoding GA 3-oxidases that are expressed in light-imbibed A. thaliana seeds (Yamaguchi et al., 2001). Although transcript abundance of AtGA3ox1 was highly elevated at 4°C, that of AtGA3ox2 remained low at both temperatures (Figure 2B). These results indicate that particular members of the AtGA20ox and AtGA3ox gene families are upregulated by low temperature. Transcript levels of earlier GA biosynthesis genes, AtKO and AtKAO1, were not significantly higher at 4°C relative to those at 22°C (Figure 2B). Whereas GA 20-oxidase and GA 3-oxidase are involved in the biosynthesis of bioactive GAs, GA 2-oxidases are responsible for deactivating GAs via 2β-hydroxylation (Figure 1). Our QRT-PCR analysis indicated that in contrast with AtGA3ox1, the level of AtGA2ox2 transcript was lower at 4°C than at 22°C.
To study the effect of low temperature on the GA biosynthesis pathway in more detail, we determined time-course changes in the level of AtGA3ox1 and AtGA3ox2 transcripts during incubation of wild-type seeds at 4 or 22°C as shown in Figure 2A. Figure 2C shows the level of AtGA3ox1 transcript increased gradually during incubation at 4°C. However, AtGA3ox1 expression at 22°C was elevated only slightly during early time points and remained at low levels after 48 h. In contrast with the induction of AtGA3ox1 at 4°C, the level of AtGA3ox2 mRNA remained low at both temperatures. These results confirm that AtGA3ox1 transcript abundance is upregulated by low temperature, whereas this response was not observed for the AtGA3ox2 gene.
Levels of Bioactive GAs Increase at 4°C
Our QRT-PCR studies revealed that the transcript levels of AtGA20ox1, AtGA20ox2, and AtGA3ox1 are significantly higher at 4°C than at 22°C. By contrast, AtGA2ox2, which is involved in GA deactivation, was downregulated at 4°C relative to that at 22°C (Figure 2B). These results suggest that bioactive GAs accumulate at higher levels at 4°C than at 22°C. However, the modified transcript abundance of GA metabolism genes alone does not allow us to assess the content of bioactive GAs at the two temperatures, given that these genes also are regulated by GA activity through feedback inhibition or feedforward induction (Chiang et al., 1995; Phillips et al., 1995; Yamaguchi et al., 1998; Thomas et al., 1999). In addition, it was not clear whether the increase or decrease in the transcript levels caused a change in the corresponding enzyme activity in seeds. To demonstrate that cold treatment leads to an increase in seed bioactive GA levels, quantitative analysis of precursor, active, and deactivated GAs was conducted using GC-MS (Figure 3). Wild-type seeds were imbibed at 4 or 22°C under the light conditions given in Figure 2A and harvested 96 h after the start of imbibition for GA analysis. Figure 3 illustrates that the levels of bioactive GAs, GA4 (the major bioactive GA in A. thaliana), and GA1 (the amount of which is usually ∼10% of that of GA4) were significantly higher at 4°C than at 22°C in wild-type seeds. With the exception of GA29, the level of which was lower at 4°C than at 22°C, there was no significant difference in the level of GA precursors and catabolites between 4 and 22°C. These results suggest that multiple biosynthetic steps are modulated to increase the amount of bioactive GAs at 4°C; therefore, a substantial increase or decrease in the level of particular precursor and/or deactivated forms was not detectable. This speculation would be consistent with the observed upregulation or downregulation of GA biosynthesis/catabolism genes at different metabolic steps in the GA pathway (Figure 2B). It also is possible that most of the biosynthetic steps are not rate limiting and are sensitive to the relative pools of precursor and product.
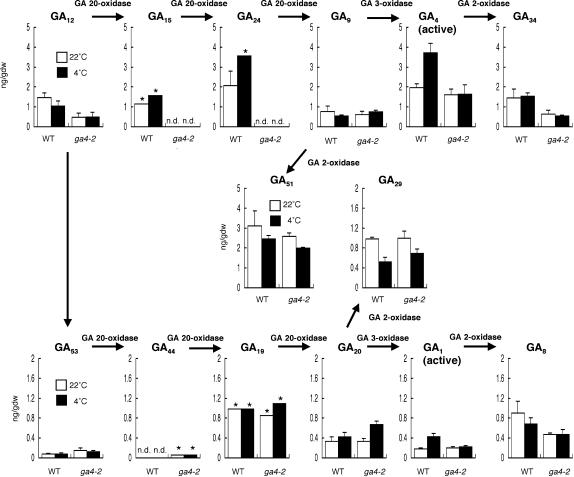
Figure 3.
Endogenous GA Levels at 4 and 22°C in Dark-Imbibed Seeds.
As illustrated in Figure 2A, wild-type seeds were imbibed in the dark, irradiated with an FR light pulse, and then incubated in the dark at 22 or 4°C for 96 h until harvested. Data are means with standard errors from three measurements using independent seed batches unless otherwise indicated. The asterisks indicate means from two measurements (with similar results). Note that the y axis scale for 13-nonhydroxylated GAs (top; GA12,15,24,9,4,34,51) and that for 13-hydroxylated GAs (bottom; GA53,44,19,20,1,8,29) are different for clarity. gdw, grams dry weight; n.d., detectable but could not be quantified because of comigration of impurities.
Because AtGA3ox1 is upregulated during imbibition at 4°C (Figures 2B and 2C), we examined whether this gene is necessary for the increase in bioactive GA levels at 4°C using the loss-of-function mutant ga4-2. The ga4-2 mutant has a T-DNA insertion in the AtGA3ox1 gene and does not contain detectable AtGA3ox1 mRNA (Chiang et al., 1995). Our GC-MS analysis showed that in contrast with wild-type seeds, the levels of bioactive GAs in the ga4-2 mutant were comparable at 4 and 22°C (Figure 3). This result demonstrates that AtGA3ox1 is required for the increase in the amounts of bioactive GAs in response to low temperature. This finding also is consistent with the fact that among four genes encoding (putative) GA 3-oxidases, only AtGA3ox1 was upregulated at 4°C (Figure 2B, data not shown for AtGA3ox3 and AtGA3ox4).
GA-Inducible Genes Are Upregulated at 4°C
To assess if the increase in bioactive GA levels at 4°C caused activation of the GA response pathway, we determined transcript abundance of GA-inducible genes that were identified previously by DNA microarray analysis (Ogawa et al., 2003). To verify GA responsiveness of such genes, we incubated seeds of the GA-deficient ga1-3 mutant in the absence or presence of varying concentrations of GA4. As shown in Figure 4A, five genes (At-EXP1, At-EXP2, AtCP1, At-XTH5, and At-XTH31) responded to exogenous GA4 in a similar dose-response manner. To analyze the response of these GA-inducible genes to low temperature, their transcript levels were determined in wild-type seeds imbibed at 4 or 22°C under conditions depicted in Figure 2A. QRT-PCR analysis demonstrated that GA-inducible At-EXP1, AtCP1, At-EXP2, and At-XTH5 genes were upregulated at 4°C, which is consistent with the increase in active GA levels under the same condition (Figure 4B). However, transcript abundance of the GA-upregulated At-XTH31 gene was lower at 4°C than at 22°C, in contrast with other GA-inducible genes (Figure 4B). These results indicate that imbibition at 4°C results in upregulation of a subset of GA-inducible genes but not of all of them.
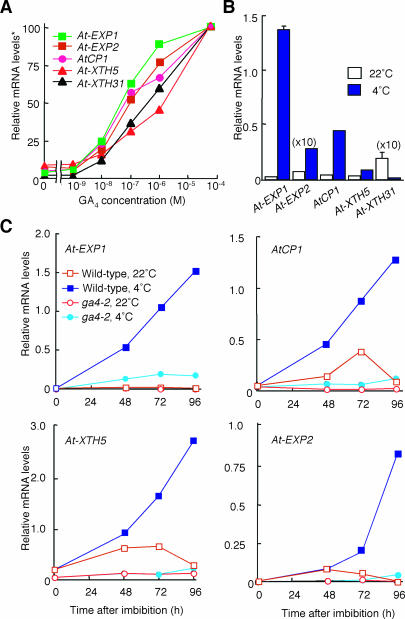
Figure 4.
Expression of GA-Inducible Genes at 4 and 22°C in Dark-Imbibed Seeds.
(A) GA responsiveness of genes used in this study. The ga1-3 mutant seeds were imbibed for 2 d at 4°C in the dark and then incubated at 22°C for 1 d under continuous white light as described by Ogawa et al. (2003). The seeds then were exposed to varying concentrations of GA4 for 6 h. At-EXP1 and At-EXP2 encode expansins (http://www.bio.psu.edu/expansins/arabidopsis.htm). AtCP1 is a gene for putative Cys proteinase. At-XTH5 and At-XTH31 code xyloglucan endotransglycosylase/hydrolase (Rose et al., 2002). Asterisk, the transcript level at 50 μM GA4 was set as 100.
(B) Transcript levels of GA-upregulated genes at 4 and 22°C. Wild-type seeds were imbibed at 4 or 22°C under the light conditions depicted in Figure 2A. The transcript abundance was determined 48 h after the start of imbibition. Values are means with standard errors from three replicates.
(C) Effect of the ga4-2 mutation on transcript levels of GA-upregulated genes at 4 and 22°C. Wild-type and ga4-2 mutant seeds were imbibed at 4 or 22°C under the light conditions depicted in Figure 2A. The transcript abundance was determined 48, 72, and 96 h after the start of imbibition. Experiments were repeated twice with similar results. Data from one of the replicates are shown.
It has been demonstrated previously that cold treatment increases sensitivity to exogenous GA in imbibed A. thaliana seeds (Derkx and Karssen, 1993b; Debeaujon and Koornneef, 2000). Therefore, the elevated transcript abundance of GA-upregulated genes at 4°C relative to that at 22°C (Figure 4B) could be in part because of an increase in the sensitivity to GA. To evaluate the influence of the twofold increase in active GA levels at 4°C (Figure 3), we examined transcript abundance of GA-inducible genes in the ga4-2 mutant, in which bioactive GA levels remained constant at 4 and 22°C. As illustrated in Figure 4C, the levels of GA-inducible At-EXP1, AtCP1, At-EXP2, and At-XTH5 transcripts in wild-type seeds steadily increased during the incubation at 4°C, but this increase was not observed at 22°C. The induction of these GA-inducible genes by low temperature was significantly reduced in ga4-2 mutant seeds, indicating that a cold-induced increase in GA4 content at 4°C is at least partially responsible for activating the GA response pathway in wild-type seeds. Slightly higher transcript levels of At-EXP1 and AtCP1 at 4°C than at 22°C in the ga4-2 mutant (Figure 4C) may be attributable to an elevated sensitivity of seeds to GA at 4°C.
AtGA3ox1 Is Required for Cold-Stimulated Seed Germination
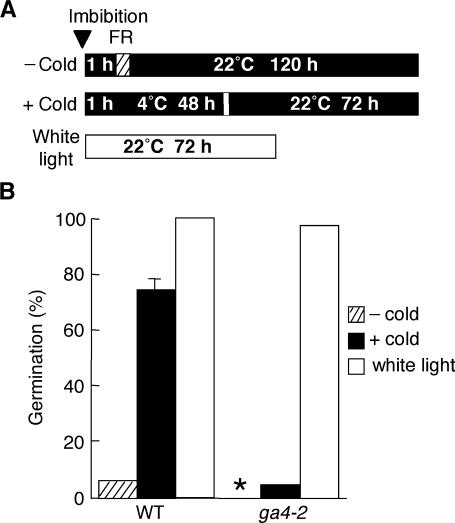
The data shown in Figures 3 and 4C demonstrate that a functional AtGA3ox1 gene is required for increasing the level of bioactive GAs and for upregulating several GA-inducible genes at 4°C. To evaluate the physiological importance of AtGA3ox1 for cold-stimulated seed germination, we examined germination ability of the loss-of-function AtGA3ox1 mutant ga4-2. Under continuous white light at 22°C, the germination of ga4-2 seeds was comparable to wild-type seeds (Figure 5B), presumably as a result of the activity of AtGA3ox2 because the AtGA3ox2 transcript is relatively abundant in light-imbibed seeds (Yamaguchi et al., 1998). When an FR light pulse was given to wild-type seeds followed by incubation in the dark at 22°C, they failed to germinate. However, under the same light conditions, 74% of wild-type seeds were able to germinate when preincubated at 4°C before transfer to 22°C (Figure 5). This observation illustrates that the inhibition of germination by inactivating phytochrome is partially removed by exposure to 4°C. Importantly, this cold-promoted germination was not observed for the ga4-2 seeds (Figure 5B). These data demonstrate that AtGA3ox1 is required for stimulating seed germination by the exposure to 4°C under these experimental conditions.
Figure 5.
Effects of Cold Treatment on Germination of Wild-Type and ga4-2 Mutant Seeds.
(A) Diagram showing temperature and light conditions. Wild-type and ga4-2 mutant seeds were imbibed in the dark, irradiated with an FR light pulse (hatched box), and then incubated in the dark at 4°C for 48 h before being placed at 22°C for 72 h in the dark to allow germination (+ cold). As a control, the incubation at 4°C for 48 h was omitted (− cold). Seeds also were imbibed at 22°C under continuous white light. The triangle indicates the start time of imbibition.
(B) Germination rates under conditions described in (A). Asterisk, no germination (0%).
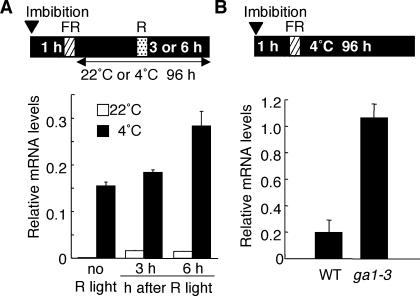
Regulation of AtGA3ox1 by Multiple Signals
This study and our previous studies have shown that the AtGA3ox1 gene is regulated by at least three factors: light (phytochrome), temperature, and GA activity (feedback inhibition). Low temperature, R light, and GA deficiency elevate the level of AtGA3ox1 transcript. To study how these multiple signals determine the abundance of AtGA3ox1 mRNA, we attempted to show whether the transcript level of AtGA3ox1 produced in response to cold treatment could be further increased by R light and/or GA deficiency. In the experiment shown in Figure 6A, an R light pulse was given to wild-type seeds during the incubation in the dark at 4°C. QRT-PCR analysis showed that the R light irradiation further elevated transcript accumulation of AtGA3ox1. Figure 6B shows that the GA-deficient ga1-3 seeds accumulated the AtGA3ox1 transcript at a much higher level than wild-type seeds, indicating that the cold-elevated AtGA3ox1 mRNA level is further increased by the ga1-3 mutation that causes GA deficiency.
Figure 6.
Effects of R light and GA deficiency on AtGA3ox1 Transcript Accumulation at 4°C.
Schematic diagram is shown at the top of each panel for light and temperature conditions. Graphs below the diagram show relative AtGA3ox1 mRNA levels. Values are means with standard errors from three replicates.
(A) Effect of an R light pulse during cold treatment. Wild-type seeds were imbibed in the dark, irradiated with an FR light pulse (hatched box), and then incubated in the dark at 4°C for 96 h. During the dark incubation at 4°C, the seeds were treated with a brief R light pulse (dotted box) 3 or 6 h before being harvested.
(B) Effect of the ga1-3 mutation at 4°C. Wild-type and ga1-3 mutant seeds were imbibed in the dark, irradiated with an FR light pulse, and then incubated in the dark at 4°C for 96 h.
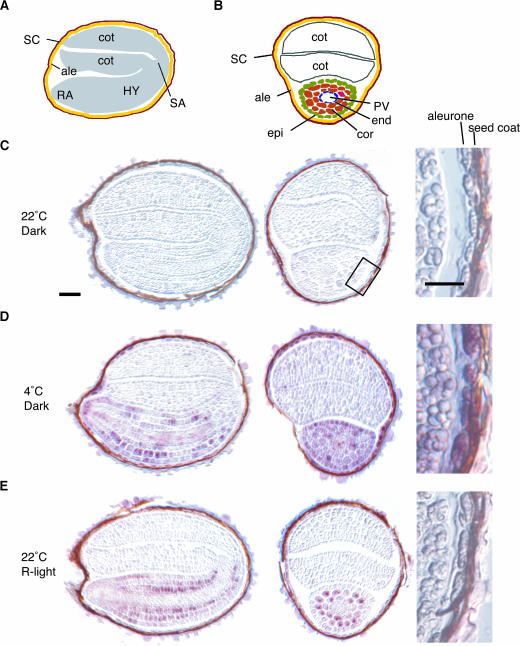
Cellular Localization of AtGA3ox1 mRNA at 4°C
In situ RNA hybridization was employed to determine cell-type specificity of AtGA3ox1 expression in seeds after the induction by low temperature. AtGA3ox1 mRNA was clearly observed in the entire embryonic axes and in the aleurone layer of cold-treated seeds (Figure 7D), whereas no hybridization signal was detectable in seeds imbibed at 22°C (Figure 7C). It has been shown previously that AtGA3ox1 mRNA accumulates mainly in the cortex and endodermis of embryonic axes, but not in the provasculature and aleurone layer, when wild-type seeds were imbibed under continuous white light at 22°C without cold treatment (Yamaguchi et al., 2001). These results suggest that the spatial expression pattern of AtGA3ox1 in imbibed seeds is modified under different light and temperature conditions. To verify this hypothesis, it is necessary to ensure that the observed difference in cell-type specificity is not an artifact because of an overall change in transcript abundance. As shown in Figure 6A, when AtGA3ox1 expression was induced by R light after a long incubation in the dark (90 to 93 h after the FR light pulse), the AtGA3ox1 transcript level was much lower than that induced by low temperature. However, when an R pulse is given after a shorter incubation in the dark (24 h after the FR light pulse; Figure 7E), as previously reported (Yamaguchi et al., 1998), the level of AtGA3ox1 mRNA was found to be similar to that induced by low temperature (Figure 7D), as determined by QRT-PCR analysis (data not shown). We therefore compared cell-type specificity of AtGA3ox1 mRNA in these two samples (Figures 7D and 7E). In seed treated with R light without cold treatment (Figure 7E), AtGA3ox1 mRNA was mainly detected in the endodermis, cortex, and some epidermal cells of the embryonic axes, but only weak or no signal was detected in the provasculature and aleurone layer. To further examine the signal(s) that possibly modifies the spatial expression pattern of AtGA3ox1, the effect of GA deficiency was tested using ga1-3 mutant seeds imbibed in the light (data not shown). When upregulated as a result of GA deficiency, the AtGA3ox1 transcript still accumulated mainly in the cortex and endodermis of embryonic axes in a similar manner to R light–treated wild-type seeds. Taken together, these results indicate that cold treatment induces the accumulation of AtGA3ox1 transcript in an additional set of cell types relative to that in light-induced seeds.
Figure 7.
In Situ Hybridization Analysis of AtGA3ox1 mRNA.
(A) Schematic drawing of a longitudinal section. ale, aleurone; cot, cotyledon; HY, hypocotyl; RA, radicle; SA, shoot apical meristem; SC, seed coat.
(B) Schematic drawing of a transverse section. ale, aleurone; cor, cortex; cot, cotyldon; end, endodermis; epi, epidermis; PV, provasculature; SC, seed coat.
(C), (D), and (E) Longitudinal (left) and transverse (center) sections of imbibed seeds were hybridized with digoxigenin-labeled antisense AtGA3ox1 RNA probe. Magnified views (right) highlight the aleurone layer (the area surrounded by a box in (C) was magnified). Wild-type seeds were incubated at 22°C in (C) or 4°C in (D) for 96 h after FR light pulse as depicted in Figure 2A. All incubations were performed at 22°C in (E) as described by Yamaguchi et al. (1998). Wild-type seeds were incubated at 22°C for 24 h after FR light pulse and then harvested 4 h after the R light pulse. Bars = 50 μm.
DISCUSSION
Using DNA microarray analysis, we have identified a group of genes, whose transcript level is significantly altered when after-ripened dry seeds are imbibed in the dark at 4°C for 2 d. We found that this group contains several GA biosynthesis genes (Table 1). In addition, ∼25% of cold treatment–regulated transcripts found in this study previously have been shown to be GA regulated based on their response to exogenous GA4 in ga1-3 mutant seeds (Ogawa et al., 2003; see supplemental data online). As GA-inducible and GA-repressible genes represent only 2.8 and 1.5%, respectively, of the total genes examined by previous microarray analysis (Ogawa et al., 2003), GA-regulated genes make up a high proportion of the cold-responsive gene group identified in this study. These observations suggest that GA plays a substantial role in stimulating seed germination during cold treatment. We also have shown, using the ga4-2 mutant, that an increase in bioactive GA levels in response to low temperature is required for cold-induced seed germination in the dark (Figures 3 and 5). Although an essential role of GA in A. thaliana seed germination has long been established, as shown by the nongerminating phenotype of severe GA-deficient mutants (Koornneef and van der Veen, 1980), our data provide evidence that cold treatment leads to an increase in bioactive GA levels, which in turn promotes seed germination.
Our QRT-PCR analyses have shown that the effect of 4°C on imbibed A. thaliana seeds is targeted to particular members of gene families that encode GA 20-oxidase, GA 3-oxidase, and GA 2-oxidase genes (Table 1, Figure 2B). Therefore, in addition to cold-induced upregulation of de novo synthesis of bioactive GAs, cold-induced downregulation of GA catabolism also might play a role in increasing the levels of active GAs. Certainly, imbibition at 4°C seems to regulate GA biosynthesis and catabolism in a coordinated manner. In our previous study, we predicted that de novo GA biosynthesis, rather than catabolism by GA 2β-hydroxylation, was mainly responsible for determining bioactive GA levels in light-imbibed seeds (Ogawa et al., 2003) because expression of all (putative) AtGA2ox genes stayed at low levels throughout germination. Moreover, the time-course curve for GA34 (a deactivated form of GA4) nearly paralleled that of GA4, suggesting that the amount of GA34 is dependent on the level of GA4 (Ogawa et al., 2003). In this study, we found that the AtGA2ox2 transcript accumulated at a higher level in the dark at 22°C than at 4°C (Figure 2B). This result suggests that repression of GA 2-oxidation also might contribute to elevating bioactive GA levels at 4°C. However, the effect of AtGA2ox2 downregulation on bioactive GA levels is not yet clear because the amounts of GA8 and GA34 did not change significantly at 4 and 22°C. Loss-of-function AtGA2ox mutants would be useful to examine the possible regulatory role of 2β-hydroxylation in cold-stimulated seed germination.
The effect of low temperature on GA biosynthesis has been studied previously in Thlaspi arvense (pennycress), which requires cold for flowering (Hazebroek and Metzger, 1990; Hazebroek et al., 1993). In the shoot tip of this species, in vivo and in vitro metabolism studies suggested that the conversion of ent-kaurenoic acid (KA) into ent-7α-hydroxy KA is upregulated by low temperature. The conversion of KA into GA12 through ent-7α-hydroxy KA is catalyzed by a single multifunctional P450 monooxygenase (Figure 1; Helliwell et al., 2001). In this study, we did not see cold-induced transcript accumulation of AtKAO1 and AtKAO2 genes in A. thaliana seeds (Table 1, Figure 2B). These observations suggest that the low temperature signal may be targeted to distinct steps in the GA biosynthesis pathway in different plant species and/or at different developmental stages. To assess this hypothesis, it would be necessary to examine whether the temperature regulation of KA metabolism involves a change in transcript accumulation of KA oxidase gene(s) in T. arvense.
Recently, large-scale expression studies have been performed during the exposure of A. thaliana seedlings to low temperatures (in most cases, 4°C) using DNA microarrays (Fowler and Thomashow, 2002; Kreps et al., 2002; Seki et al., 2002; Provart et al., 2003). Unlike our results from imbibed seeds, no GA biosynthesis gene was classified as being cold responsive in A. thaliana seedlings upon exposure to 4°C (Fowler and Thomashow, 2002; Kreps et al., 2002; Seki et al., 2002). Moreover, only a few genes were determined to be cold responsive commonly in both seeds and seedlings based on this study and published microarray data. These observations indicate that the response to low temperature is drastically modified at different developmental stages. However, when light-grown A. thaliana seedlings at 22°C were exposed to a moderately low temperature (12°C), the transcript level of AtGA3ox1 increased and that of AtGA2ox2 decreased (Provart et al., 2003). These results suggest that GA biosynthesis might be regulated by temperature in A. thaliana seedlings as well. However, it is not clear if the alteration in transcript abundance of GA biosynthesis genes results in an increase in the amount of bioactive GAs in seedlings because such a change in mRNA accumulation also would occur through feedback and feedforward mechanisms when active GA levels are lowered (Chiang et al., 1995; Phillips et al., 1995; Yamaguchi et al., 1998; Thomas et al., 1999). In addition, physiological roles of GA during the temperature shift from 22 to 12°C in seedling development also should be clarified.
In agreement with the elevation of bioactive GA levels at 4°C relative to those at 22°C (Figure 3), our QRT-PCR analyses have shown that several GA-inducible genes are upregulated at the lower temperature (Figure 4). However, unlike other GA-inducible transcripts, the level of At-XTH31 mRNA was not higher at 4°C than at 22°C (Figure 4B) despite its dose-dependent response to exogenous GA4 in light-imbibed ga1-3 seeds at 22°C (Figure 4A). This result suggests that the capacity of individual GA-regulated genes to respond to GA in seeds may be modified under different environmental conditions. This hypothesis should be further verified by analyzing the effect of exogenous GA4 on the abundance of a larger number of GA-responsive transcripts at the two temperatures. If the distribution of bioactive GAs is changed under different environmental conditions, the failure of AtXTH31 to be upregulated by low temperature also may be explained by the fact that it is only expressed in cells in which bioactive GAs do not accumulate at a sufficient level to activate it. Although previous physiological experiments have suggested that the tissue sensitivity to GA is modulated by light and temperature signals, the molecular basis for GA sensitivity is not yet clear (Derkx and Karssen, 1993a; Yang et al., 1995; Debeaujon and Koornneef, 2000). Our finding that only a fraction of GA-inducible genes responded to an increase in endogenous GA levels at 4°C suggests that a difference in GA sensitivity may be visualized by grouping GA-regulated genes on a large scale based on their response to GA at varying environmental parameters. Our QRT-PCR analysis showed that a GA catabolism gene, AtGA2ox2, is expressed at a lower level at 4°C than at 22°C (Figure 2B). If GA 2-oxidation (deactivation) activity was reduced at 4°C, this might be partly responsible for the increase in tissue sensitivity to exogenous GA at low temperature.
In situ RNA hybridization analysis suggests that exposure of seeds to 4°C results in an increase in the number of cell types that accumulate AtGA3ox1 mRNA compared with R light–treated seeds (Figure 7). We predict that the signal responsible for this spatial modification might be low temperature because the induction of AtGA3ox1 by R light and GA deficiency did not cause detectable transcript accumulation in the provasculature and aleurone layer. However, the role of low temperature in modifying the spatial expression pattern needs to be carefully discussed because total incubation time for R light induction of AtGA3ox1 mRNA (Figure 7E) was shorter than that for the induction by low temperature (Figure 7D) in our experiments. We employed these experimental conditions because the two samples contained similar overall AtGA3ox1 mRNA levels (as determined by QRT-PCR; data not shown), which allowed us to assess any difference in the spatial expression patterns directly. As shown in Figure 6A, when an R light pulse was given at 22°C after a long incubation in the dark (∼90 h), the level of AtGA3ox1 mRNA after an R light pulse was much lower than that induced by low temperature. Therefore, it is possible that a long imbibition (and a resulting change in the developmental state) also may play a role in the upregulation of AtGA3ox1 in the provasculature and aleurone layer. As reported earlier, AtGA3ox1 mRNA accumulates predominantly in the cortex and endodermis of embryonic axes when seeds are imbibed under continuous white light at 22°C (without cold treatment). Under this condition, the cell-type-specific expression pattern of AtGA3ox1 was consistent at several time points during germination (from imbibition to radicle protrusion) (Yamaguchi et al., 2001; S. Yamaguchi, unpublished data). This observation suggests that the cell-type specificity of AtGA3ox1 mRNA accumulation does not drastically change at developmental stages during germination and supports the idea that the observed spatial change would be caused principally by a difference in environmental parameters.
We previously have found that the GA-inducible AtCP1 transcript accumulates mainly in the provasculature, epidermis, and aleurone in light-imbibed wild-type seeds at 22°C, whereas the major GA biosynthesis genes are predominantly expressed in the endodermis and cortex of embryonic axes. Based on the distinct cellular locations between GA biosynthesis and GA-responsive transcripts, we predicted that GA itself or its secondary signal moves across different cell layers (Ogawa et al., 2003). In this study, exposure of imbibed seeds to 4°C for a long period substantially elevated the accumulation of AtGA3ox1 mRNA in the provasculature and aleurone layer (Figure 7D). These observations suggest that cold treatment might play a unique role in stimulating seed germination by increasing bioactive GA levels in the provasculature and aleurone, which, without cold treatment, may rely largely on GA or GA signal from other cell types. To predict the physiological significance of this hypothetical mechanism, it would be helpful to determine if cellular localization of any GA-regulated transcript is modified between the R light–treated and cold-treated seeds. Such studies also will be valuable to assess if the distribution of bioactive GAs is in fact modulated according to the modification of cell types that accumulate the AtGA3ox1 mRNA.
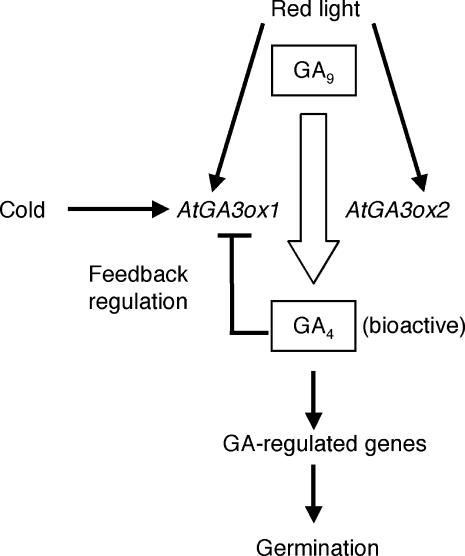
In conclusion, this study has demonstrated that GA biosynthesis is activated by low temperature during seed imbibition at least in part through modulating transcript abundance of particular GA biosynthesis genes. Combined with our previous studies, regulation of the final biosynthesis steps to produce bioactive GAs can be summarized as shown in Figure 8. It has long been known that light (phytochrome) and temperature are critical determinants for A. thaliana seed germination. Our work has indicated that light and temperature signals are targeted to the transcript abundance of AtGA3ox1 (Figures 6 and 8). Integration of these environmental signals into the GA biosynthesis pathway emphasizes the crucial role of this hormone in controlling seed germination. We speculate that the light and temperature regulation of GA biosynthesis might be important for seeds to ensure that germination occurs at the most advantageous time in nature. Besides its regulation by such external cues, AtGA3ox1 is subject to feedback regulation by GA activity during seed germination (Yamaguchi et al., 1998). These findings now allow us to further investigate the mechanisms by which bioactive GA levels are determined by multiple factors, for example, through identifying cis-regulatory elements for individual signals in the AtGA3ox1 gene. Such studies will be beneficial for understanding the potential interactions among light, temperature, and GA signals at other developmental stages as well. Finally, in addition to GA-related genes that have been studied in detail here, our microarray analysis identified a number of transcripts whose abundance is modulated upon imbibition of dry seeds at 4°C in the dark (see supplemental data online). Future work will be valuable that aims to elucidate how such genes contribute to stimulating seed germination.
Figure 8.
Summary of the Regulation of AtGA3ox1 and AtGA3ox2 during Seed Germination.
Thin arrows indicate positive regulation. Feedback inhibition is shown by the T bar. Conversion of GA9 into bioactive GA4 is indicated by the thick arrow. Regulation by phytochrome and by GA activity has been reported previously (Chiang et al., 1995; Yamaguchi et al., 1998).
METHODS
Plant Materials and Growth Conditions
A. thaliana ecotype Landsberg erecta was used as the wild type in this study. The ga1-3 seeds were originally obtained from Maarten Koornneef (Wageningen University, Wageningen, The Netherlands). The ga4-2 mutant seeds were courtesy of H.M. Goodman (Harvard Medical School, Boston, MA). To propagate seeds for germination experiments, plants were grown on soil under continuous light at 22°C. Harvested mature seeds were stored at room temperature at 30% humidity for at least 2 months to allow after-ripening. For germination, the seeds were washed with 0.02% Triton X solution, rinsed with water, and incubated on wet filter paper (3MM; Whatman, Maidstone, UK). Germination tests were performed using triplicate samples (each containing 50 to 100 seeds). Seeds were scored as germinated when radicle protrusion was visible. For dark incubation, seeds were handled under a dim green safety light. The FR light pulse treatment consisted of 5 min of FR light irradiation (91 μmol·m−2·s−1) supplied from light-emitting diodes (MIL-IF18; Sanyo Biomedical, Osaka, Japan) passed through an FR acrylic filter (Deraglass A900, 2 mm thick; Asahikasei, Tokyo, Japan). The R-light-pulse treatment consisted of 5 min of R light irradiation (120 μmol·m−2·s−1) supplied from light-emitting diodes (MIL-R18; Sanyo Biomedical).
Gibberellin Analysis
One gram of dry seeds was used for each GA measurement. Quantitative analysis of GA was performed using 2H-labeled GAs as internal standards as described previously (Gawronska et al., 1995). GA measurements were performed three times using different seed batches. When data were available, the mean with standard error from triplicate experiments (using different seed batches) was indicated in Figure 3.
Microarray Analysis Using Affymetrix GeneChips Microarray
For microarray analysis, total RNA was extracted from dry or imbibed seeds (starting weight 40 mg of dry seeds) using an RNAqueous RNA isolation kit with plant RNA isolation aid (Ambion, Austin, TX). Double-strand cDNA was synthesized from 5 μg of total RNA using a Super Script Choice cDNA synthesis kit (Invitrogen, Carlsbad, CA) with an oligo(dT)24 primer containing a T7 polymerase promoter site at the 3′ end. Biotin-labeled cRNA was synthesized by T7 RNA polymerase using the double-strand cDNA as a template (Bioarray high yield RNA transcript labeling kit; Enzo Diagnostics, Farmingdale, NY) and purified with the use of the RNeasy RNA purification kit (Qiagen, Valencia, CA). The biotin-labeled cRNA was fragmented and hybridized to a GeneChip microarray (Affymetrix, Santa Clara, CA) for 16 h at 42°C. After hybridization, the arrays were washed and stained with biotinylated antistreptavidin antibody (Vector Laboratories, Burlingame, CA) and a phycoerythrin-streptavidin conjugate (Molecular Probes, Eugene, OR) according to the manufacturer's protocol. Signals were scanned using a confocal microscope scanner (Gene Array Scanner; Hewlett-Packard, Palo Alto, CA) at 570 nm.
Data Analysis
Microarray data were analyzed as described previously (Ogawa et al., 2003). Briefly, signal values for individual genes were obtained using statistical algorithms on Microarray Suite software (version 5.0; Affymetrix). Genes were classified as being responsive to cold treatment if the signal values deviated either positively or negatively fourfold or more relative to those in dry seeds in duplicated experiments using independent seed batches.
QRT-PCR
QRT-PCR using Taq-Man technology (Holland et al., 1991) was employed to determine transcript levels. Total RNA (2 μg) was treated with DNase I (RQ1 RNase-free DNase; Promega, Madison, WI) and used as a template to synthesize first-strand cDNA with random hexamer using a SuperScript first-strand synthesis system according to the manufacturer's instructions (Invitrogen). Quantitative real-time PCR was performed using the first-strand cDNA as a template on a sequence detector system (model 7700; Applied Biosystems, Foster City, CA). By adding transcribed external RNA (encoding enhanced green fluorescent protein originated from pIRES2-EGFP; BD Biosciences, Franklin Lakes, NJ) to the reaction mixture, we confirmed that the efficiency of reverse transcription reactions for different samples was approximately equal. To compare mRNA levels across different genes, the copy number of each target mRNA species was determined by generating standard curves using a series of known concentrations of target sequences. For normalization across samples, 18S rRNA was used as an internal standard. The value at 2 × 10−15 M target RNA in the reaction mixture (25 μL) using 125 ng of total RNA was arbitrarily set as 1.0 throughout this article. For each sample, the mean value from triplicate real-time PCRs was adapted to calculate the transcript abundance. To further confirm the reliability of data in Figures 2B, 4B, and 6, measurements were repeated using three independent plant materials. For time-course analyses (Figures 2C and 4C), experiments were performed twice using different seed batches with similar results. Nucleotide sequences of gene-specific primers and Taq-Man probes are listed in the supplemental data online.
In Situ Hybridization
In situ hybridization experiments were performed using digoxygenin-labeled sense (control) or antisense cRNA probes as described previously (Yamaguchi et al., 2001).
Supplementary Material
Acknowledgments
We thank Eiji Nambara and Damian P. O'Neill (RIKEN) for valuable comments on this manuscript.
On-line version contains Web-only data.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Shinjiro Yamaguchi (shinjiro@postman.riken.go.jp).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.018143.
References
- Arabidopsis Genome Initiative (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815. [DOI] [PubMed] [Google Scholar]
- Bewley, J.D., and Black, M. (1982). Physiology and Biochemistry of Seeds: Viability, Dormancy and Environmental Control, Vol. 2. (Berlin, Germany: Springer-Verlag).
- Borthwick, H.A., Hendricks, S.B., Parker, M.W., Toole, E.H., and Toole, V.K. (1952). A reversible photoreaction controlling seed germination. Proc. Natl. Acad. Sci. USA 38, 662–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler, W.L., Norris, K.H., Siegelman, H.W., and Hendricks, S.B. (1959). Detection, assay, and preliminary purification of the pigment controlling photoresponsive development of plants. Proc. Natl. Acad. Sci. USA 45, 1703–1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang, H.H., Hwang, I., and Goodman, H.M. (1995). Isolation of the Arabidopsis GA4 locus. Plant Cell 7, 195–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cone, J.W., and Spruit, C.J.P. (1983). Imbibition conditions and seed dormancy of Arabidopsis thaliana. Physiol. Plant 59, 416–420. [Google Scholar]
- Davies, P.J. (1995). Plant Hormones: Physiology, Biochemistry and Molecular Biology. (Dordrecht, The Netherlands: Kluwer Academic Publishers).
- Debeaujon, I., and Koornneef, M. (2000). Gibberellin requirement for Arabidopsis seed germination is determined both by testa characteristics and embryonic abscisic acid. Plant Physiol. 122, 415–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derkx, M.P.M., and Karssen, C.M. (1993. a). Effects of light and temperature on seed dormancy and gibberellin-stimulated germination in Arabidopsis thaliana: Studies with gibberellin-deficient and gibberellin-insensitive mutants. Physiol. Plant 89, 360–368. [Google Scholar]
- Derkx, M.P.M., and Karssen, C.M. (1993. b). Variability in light, gibberellin and nitrate requirement of Arabidopsis thaliana seeds due to harvest time and conditions of dry storage. J. Plant Physiol. 141, 574–582. [Google Scholar]
- Derkx, M.P.M., Vermeer, E., and Karssen, C.M. (1994). Gibberellins in seeds of Arabidopsis thaliana: Biological activities, identification and effects of light and chilling on endogenous levels. Plant Growth Regul. 15, 223–234. [Google Scholar]
- Fowler, S., and Thomashow, M.F. (2002). Arabidopsis transcriptome profiling indicates that multiple regulatory pathways are activated during cold acclimation in addition to the CBF cold response pathway. Plant Cell 14, 1675–1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gawronska, H., Yang, Y.Y., Furukawa, K., Kendrick, R.E., Takahashi, N., and Kamiya, Y. (1995). Effects of low irradiance stress on gibberellin levels in Pea-seedlings. Plant Cell Physiol. 36, 1361–1367. [Google Scholar]
- Hayes, R.G., and Klein, W.H. (1974). Spectral quality influence of light during development of Arabidopsis thaliana plants in regulating seed germination. Plant Cell Physiol. 15, 643–653. [Google Scholar]
- Hazebroek, J.P., and Metzger, J.D. (1990). Thermoinductive regulation of gibberellin metabolism in Thlaspi arvense L. I. Metabolism of [2H]kaurenoic acid and [14C]gibberellin A12-aldehyde. Plant Physiol. 94, 157–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazebroek, J.P., Metzger, J.D., and Mansager, E.R. (1993). Thermoinductive regulation of gibberellin metabolism in Thlaspi arvense L. II. Cold induction of enzymes in gibberellin biosynthesis. Plant Physiol. 102, 547–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedden, P., and Kamiya, Y. (1997). Gibberellin biosynthesis: Enzymes, genes and their regulation. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48, 431–460. [DOI] [PubMed] [Google Scholar]
- Hedden, P., and Phillips, A.L. (2000). Gibberellin metabolism: New insights revealed by the genes. Trends Plant Sci. 5, 523–530. [DOI] [PubMed] [Google Scholar]
- Helliwell, C.A., Chandler, P.M., Poole, A., Dennis, E.S., and Peacock, W.J. (2001). The CYP88A cytochrome P450, ent-kaurenoic acid oxidase, catalyzes three steps of the gibberellin biosynthesis pathway. Proc. Natl. Acad. Sci. USA 98, 2065–2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland, P.M., Abramson, R.D., Watson, R., and Gelfand, D.H. (1991). Detection of specific polymerase chain reaction product by utilizing the 5′-3′ exonuclease activity of Thermus aquaticus DNA polymerase. Proc. Natl. Acad. Sci. USA 88, 7276–7280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen, S.E., and Olszewski, N.E. (1993). Mutations at the SPINDLY locus of Arabidopsis alter gibberellin signal transduction. Plant Cell 5, 887–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasahara, H., Hanada, A., Kuzuyama, T., Takagi, M., Kamiya, Y., and Yamaguchi, S. (2002). Contribution of the mevalonate and methylerythritol phosphate pathways to the biosynthesis of gibberellins in Arabidopsis. J. Biol. Chem. 277, 45188–45194. [DOI] [PubMed] [Google Scholar]
- Koornneef, M., Bentsink, L., and Hilhorst, H. (2002). Seed dormancy and germination. Curr. Opin. Plant Biol. 5, 33–36. [DOI] [PubMed] [Google Scholar]
- Koornneef, M., and van der Veen, J.H. (1980). Induction and analysis of gibberellin sensitive mutants in Arabidopsis thaliana (L.) Heynh. Theor. Appl. Genet. 58, 257–263. [DOI] [PubMed] [Google Scholar]
- Kreps, J.A., Wu, Y., Chang, H.S., Zhu, T., Wang, X., and Harper, J.F. (2002). Transcriptome changes for Arabidopsis in response to salt, osmotic, and cold stress. Plant Physiol. 130, 2129–2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nambara, E., Akazawa, T., and McCourt, P. (1991). Effects of the gibberellin biosynthetic inhibitor uniconazol on mutants of Arabidopsis. Plant Physiol. 97, 736–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa, M., Hanada, A., Yamauchi, Y., Kuwahara, A., Kamiya, Y., and Yamaguchi, S. (2003). Gibberellin biosynthesis and response during Arabidopsis seed germination. Plant Cell 15, 1591–1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olszewski, N.E., Sun, T.P., and Gubler, F. (2002). Gibberellin signaling: Biosynthesis, catabolism, and response pathways. Plant Cell 14 (suppl.), S61–S80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips, A.L., Ward, D.A., Uknes, S., Appleford, N.E.J., Lange, T., Huttly, A.K., Gaskin, P., Graebe, J.E., and Hedden, P. (1995). Isolation and expression of three gibberellin 20-oxidase cDNA clones from Arabidopsis. Plant Physiol. 108, 1049–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provart, N.J., Gil, P., Chen, W., Han, B., Chang, H.-S., Wang, X., and Zhu, T. (2003). Gene expression phenotypes of Arabidopsis associated with sensitivity to low temperatures. Plant Physiol. 132, 893–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose, J.K., Braam, J., Fry, S.C., and Nishitani, K. (2002). The XTH family of enzymes involved in xyloglucan endotransglucosylation and endohydrolysis: Current perspectives and a new unifying nomenclature. Plant Cell Physiol. 43, 1421–1435. [DOI] [PubMed] [Google Scholar]
- Ross, J.D., and Bradbeer, J.W. (1971). Studies in seed dormancy. V. The content of endogenous gibberellins in seeds of Corylus avellana L. Planta 100, 288–302. [DOI] [PubMed] [Google Scholar]
- Seki, M., et al. (2002). Monitoring the expression profiles of 7000 Arabidopsis genes under drought, cold and high-salinity stresses using a full-length cDNA microarray. Plant J. 31, 279–292. [DOI] [PubMed] [Google Scholar]
- Shinomura, T. (1997). Phytochrome regulation of seed germination. J. Plant Res. 110, 151–161. [DOI] [PubMed] [Google Scholar]
- Shinomura, T., Nagatani, A., Hanzawa, H., Kubota, M., Watanabe, M., and Furuya, M. (1996). Action spectra for phytochrome A- and B-specific photoinduction of seed germination in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 93, 8129–8133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinomura, T., Nagatani, A., Kubota, M., Watanabe, M., and Furuya, M. (1995). Distinct action spectra for phytochrome A-dependent and phytochrome B-dependent seed-germination in Arabidopsis thaliana. J. Cell. Biochem. 21A, 497 (abstr.). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shropshire, J.W., Klein, W.H., and Elstad, V.B. (1961). Action spectra of photomorphogenic induction and photoinactivation of germination in Arabidopsis thaliana. Plant Cell Physiol. 2, 63–69. [Google Scholar]
- Sińska, I., Lewak, S., Gaskin, P., and MacMillan, J. (1973). Reinvestigation of apple-seed gibberellins. Planta 114, 359–364. [DOI] [PubMed] [Google Scholar]
- Thomas, S.G., Phillips, A.L., and Hedden, P. (1999). Molecular cloning and functional expression of gibberellin 2-oxidases, multifunctional enzymes involved in gibberellin deactivation. Proc. Natl. Acad. Sci. USA 96, 4698–4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyomasu, T., Kawaide, H., Mitsuhashi, W., Inoue, Y., and Kamiya, Y. (1998). Phytochrome regulates gibberellin biosynthesis during germination of photoblastic lettuce seeds. Plant Physiol. 118, 1517–1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, P.M., Bradbeer, J.W., Gaskin, P., and MacMillan, J. (1974). Studies in seed dormancy. VIII. The identification and determination of gibberellins A1 and A9 in seed of Corylus avellana L. Planta 117, 101–108. [DOI] [PubMed] [Google Scholar]
- Yamaguchi, S., and Kamiya, Y. (2000). Gibberellin biosynthesis: Its regulation by endogenous and environmental signals. Plant Cell Physiol. 41, 251–257. [DOI] [PubMed] [Google Scholar]
- Yamaguchi, S., Kamiya, Y., and Sun, T. (2001). Distinct cell-specific expression patterns of early and late gibberellin biosynthetic genes during Arabidopsis seed germination. Plant J. 28, 443–453. [DOI] [PubMed] [Google Scholar]
- Yamaguchi, S., Smith, M.W., Brown, R.G., Kamiya, Y., and Sun, T. (1998). Phytochrome regulation and differential expression of gibberellin 3β-hydroxylase genes in germinating Arabidopsis seeds. Plant Cell 10, 2115–2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Y.Y., Nagatani, A., Zhao, Y.J., Kang, B.J., Kendrick, R.E., and Kamiya, Y. (1995). Effects of gibberellins on seed germination of phytochrome-deficient mutants of Arabidopsis thaliana. Plant Cell Physiol. 36, 1205–1211. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.