Abstract
Abscisic acid (ABA) regulates many aspects of plant growth and development, yet many ABA response mutants present only subtle phenotypic defects, especially in the absence of stress. By contrast, the ABA-insensitive8 (abi8) mutant, isolated on the basis of ABA-resistant germination, also displays severely stunted growth, defective stomatal regulation, altered ABA-responsive gene expression, delayed flowering, and male sterility. The stunted growth of the mutant is not rescued by gibberellin, brassinosteroid, or indoleacetic acid application and is not attributable to excessive ethylene response, but supplementing the medium with Glc improves viability and root growth. In addition to exhibiting Glc-dependent growth, reflecting decreased expression of sugar-mobilizing enzymes, abi8 mutants are resistant to Glc levels that induce developmental arrest of wild-type seedlings. Studies of genetic interactions demonstrate that ABA hypersensitivity conferred by the ABA-hypersensitive1 mutation or overexpression of ABI3 or ABI5 does not suppress the dwarfing and Glc dependence caused by abi8 but partially suppresses ABA-resistant germination. By contrast, the ABA-resistant germination of abi8 is epistatic to the hypersensitivity caused by ethylene-insensitive2 (ein2) and ein3 mutations, yet ABI8 appears to act in a distinct Glc response pathway from these EIN loci. ABI8 encodes a protein with no domains of known function but belongs to a small plant-specific protein family. Database searches indicate that it is allelic to two dwarf mutants, elongation defective1 and kobito1, previously shown to disrupt cell elongation, cellulose synthesis, vascular differentiation, and root meristem maintenance. The cell wall defects appear to be a secondary effect of the mutations because Glc treatment restores root growth and vascular differentiation but not cell elongation. Although the ABI8 transcript accumulates in all tested plant organs in both wild-type and ABA response mutants, an ABI8-β-glucuronidase fusion protein is localized primarily to the elongation zone of roots, suggesting substantial post-transcriptional regulation of ABI8 accumulation. This localization pattern is sufficient to complement the mutation, indicating that ABI8 acts either at very low concentrations or over long distances within the plant body.
INTRODUCTION
Abscisic acid (ABA) regulates many important events during both vegetative and reproductive growth of plants. These range from relatively slow effects, such as promotion of seed storage reserve synthesis, acquisition of desiccation tolerance and dormancy, and induction of stress tolerance, to rapid effects, such as stomatal closure (reviewed in Leung and Giraudat, 1998; Finkelstein and Rock, 2002). Many lines of evidence indicate that there are multiple mechanisms for both ABA perception and signaling. Genetic studies, especially in Arabidopsis thaliana, have identified a large number of loci involved in ABA response, and digenic analyses indicate that these loci are likely to be acting in multiple overlapping response pathways (reviewed in Finkelstein et al., 2002). To date, >50 loci affecting ABA response have been cloned and found to encode proteins that affect processes including transcription, protein phosphorylation or farnesylation, RNA processing, and phosphoinositide metabolism (reviewed in Finkelstein et al., 2002; Himmelbach et al., 2003; Kuhn and Schroeder, 2003). In addition, many likely signaling intermediates correlated with ABA response (e.g., ABA-activated or -induced kinases or phospholipases and DNA binding proteins that specifically bind ABA-responsive elements) have been identified by molecular and biochemical studies (reviewed in Finkelstein et al., 2002; Himmelbach et al., 2003), but the relationships among most of these proteins are unclear. Mechanistic analyses of ABA response are further complicated by stage- and tissue-specific differences, such that a given locus may appear to function as both a positive and a negative regulator, depending on the response. For example, enhanced response to ABA3 (era3) mutants are hypersensitive to ABA inhibition of germination but resistant to inhibition of root growth by ABA (Ghassemian et al., 2000).
Although ABA response mutants were first identified in direct screens for loss of ABA response or ABA hypersensitivity, many additional mutations affecting ABA response have been identified in screens for defects in other signaling pathways. Consequently, there now is substantial evidence for cross talk between signaling pathways regulating response to ABA and assorted stresses (e.g., drought, salinity, and cold) (Ishitani et al., 1997), gibberellins (Lovegrove and Hooley, 2000), brassinosteroids (Steber and McCourt, 2001), jasmonic acid (Berger et al., 1996), auxin and ethylene (Wilson et al., 1990; Beaudoin et al., 2000; Ghassemian et al., 2000), sugars (Arenas-Huertero et al., 2000; Finkelstein and Lynch, 2000b; Huijser et al., 2000; Laby et al., 2000), and even meristem function (Ziegelhoffer et al., 2000). The fact that mutations in only some of the hormone response genes appear to affect multiple signaling pathways suggests that interactions among these pathways are relatively specific. Possible mechanisms of cross talk, particularly among ABA, sugar, and ethylene signaling, are discussed in many recent reviews (McCourt, 1999; Sheen et al., 1999; Gibson, 2000; Coruzzi and Zhou, 2001; Gazzarrini and McCourt, 2001; Eckardt, 2002; Finkelstein and Gibson, 2002).
Despite recent major strides toward elucidating ABA signaling, our view of the relevant pathways still is fragmented. We have yet to identify sufficient signaling elements to constitute any complete pathway or network. Biochemical and genetic characterization of additional ABA response loci can help fill these gaps. Toward this end, we have been studying a severe ABA response mutant, ABA-insensitive8 (abi8), isolated by screening for reduced sensitivity to ABA inhibition of germination. In addition to ABA resistance, abi8 mutants have pleiotropic growth defects resulting in a severely dwarfed phenotype or death. Consistent with this, molecular identification of the ABI8 locus revealed that abi8 is allelic to two dwarf mutants, elongation defective1 (eld1) and kobito1 (kob1) (Cheng et al., 2000; Pagant et al., 2002), whose hormone signaling defects were not recognized. This locus encodes a protein whose biochemical function is unknown and therefore is likely to represent a component of a novel signaling mechanism.
RESULTS
Mutant Isolation
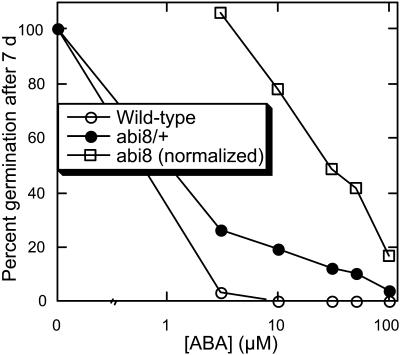
The abi8 mutant was isolated on the basis of resistance to inhibition of germination by 3 μM ABA (Figure 1). The mutation is fully recessive, such that heterozygotes have no apparent defects, but most homozygotes die as seedlings. The few survivors are severely stunted (1 to 2 cm tall at maturity), grow very slowly, flower many weeks later than wild-type plants grown under identical environmental conditions, and are male sterile (data not shown). Consequently, the mutant was backcrossed and maintained as a heterozygote (all physiological studies were performed with individuals identified in segregating populations). All of the characteristics described below cosegregated in lines that had been backcrossed three times. To determine the extent of resistance to ABA inhibition of germination, seeds of the wild type and abi8/+ were incubated on media supplemented with up to 100 μM ABA. The normalized abi8 data points indicate the percentage of germination of the abi8 segregants produced by an abi8/+ parent. Although 3 μM ABA inhibits germination of wild-type seeds, some abi8 seeds germinate even in the presence of 100 μM ABA (Figure 2).
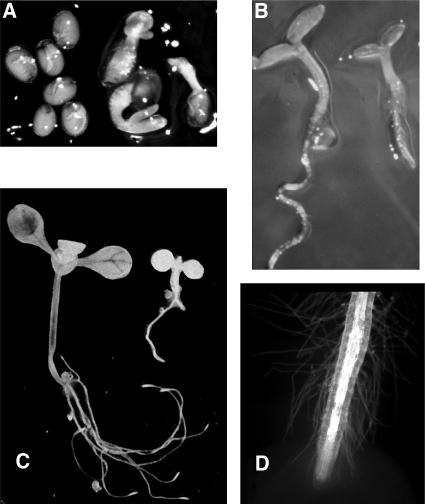
Figure 1.
Morphology of abi8 Mutant Seedlings.
Comparison of wild-type and mutant growth. The wild type is at left, and abi8 is at right in (A), (B), and (C).
(A) Seeds incubated 4 d poststratification on 3 μM ABA.
(B) 3-d-old seedlings grown on minimal medium.
(C) 11-d-old seedlings grown on GM.
(D) Tip of abi8 root.
Figure 2.
ABA Sensitivity of Germination Inhibition for Wild-Type versus abi8 Mutant Seeds.
Seeds were chilled for 3 d before incubation in continuous light at 22°C for 7 d. The normalized abi8 data points indicate the percentage of germination of the abi8 segregants produced by an abi8/+ parent.
Defects in Sugar Mobilization and Sensing
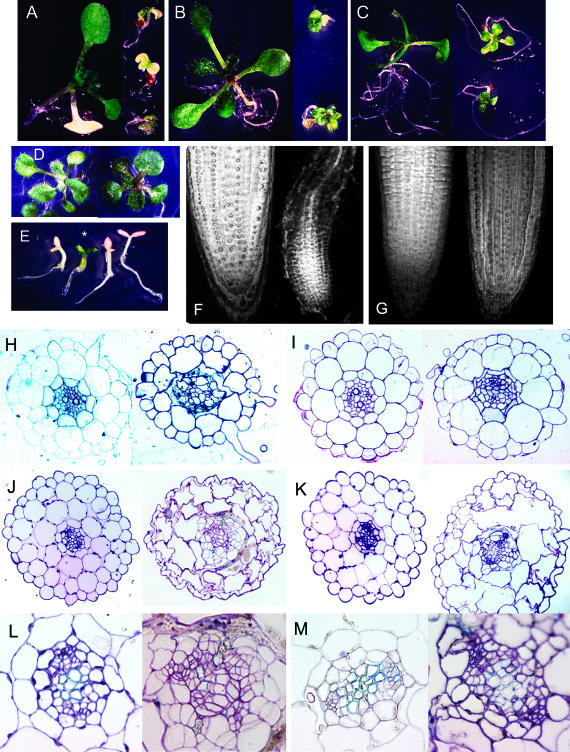
A striking feature of the mutant is that on a medium containing only essential mineral salts, radicle emergence is followed by swelling, rather than elongation, of the root and hypocotyl (Figure 1). Scanning electron microscopic comparisons of epidermal cell length showed that abi8 hypocotyl cells were approximately sevenfold shorter than those of the wild type (data not shown). On this minimal medium, the root remains short, and lateral roots are rarely initiated. Even though abundant root hairs are produced, root surface area is greatly reduced. This stunting of root growth is exacerbated by even transient exposure to ABA, indicating that the mutant is not completely insensitive to ABA but responds inappropriately to it. Attempts to rescue the mutant by gibberellin, brassinosteroid, or indoleacetic acid application to stimulate cell elongation (and/or root growth, in the case of indoleacetic acid) were unsuccessful, indicating that the stunting did not reflect deficiencies in these hormones (data not shown). However, the growth defect is partially rescued by supplementing the medium with Glc but not sucrose (Figures 3A to 3C). Whereas root growth is much more vigorous, resulting in ∼100% survival of seedlings, shoot growth is severely reduced on medium supplemented with only 1% Glc. Addition of 4% Glc mildly inhibits wild-type growth but further promotes abi8 growth, such that both genotypes grow similarly under these conditions (Figure 3D). Anatomical comparisons showed that the mutation severely disrupts cell division patterns and organization of the stele and leads to loss of the root apical meristem in plants grown on minimal media (Figures 3F and 3H). By contrast, 1% Glc treatment led to restoration of the endodermal layer, properly oriented divisions in the vascular tissue, and maintenance of the root meristem (Figures 3G and 3I) but did not substantially increase elongation of individual cells (data not shown). Examination of hypocotyl anatomy in Glc-treated plants showed that despite the improved vascular development, mutant cells were ∼2.5-fold shorter and much wider than those of the wild type and had some incomplete walls (Figures 3J to 3M), reminiscent of mutants with defects in cell walls.
Figure 3.
Growth of abi8 Mutants Is Both Glc Dependent and Glc Resistant.
Comparison of wild-type and mutant growth. In all panels, the wild type is at left, abi8 is at right, and each pair is shown at the same scale.
(A) to (D) Growth of 2- to 3-week-old plants on minimal medium (A), GM + 1% Suc (B), GM + 1% Glc (C), and minimal + 4% Glc (D).
(E) One-week-old progeny of abi8/+ cultured on minimal medium + 6% Glc. abi8 segregant is designated by an asterisk.
(F) to (I) Root anatomy. Confocal longitudinal views of root tips grown on minimal medium (F) or minimal + 1% Glc (G). Toluidine blue–stained cross-sections of a differentiated zone of roots grown on minimal medium (H) or minimal + 1% Glc (I).
(J) and (K) Hypocotyl anatomy after 11 d of growth on minimal medium (J) or GM + 1% Glc (K).
(L) and (M) High-magnification (100×) views of sections shown in (J) and (K), respectively.
Upon discovering that stunting was more severe after preselection on ABA, we began identifying abi8 segregants based on their stunted growth when grown on hormone-free media supplemented with 1% Glc either continuously or after 3 to 5 d on minimal nutrient salts. Individuals cultured for several weeks on Glc-supplemented medium developed extensive roots, enabling them to routinely survive transfer to soil, but the shoots still grew very slowly and produced male sterile flowers. Although it was not possible to develop a source of homozygous seed, we were able to generate sufficient tissue for physiological and biochemical analyses of vegetative organs of mutant plants grown on either solid or liquid media supplemented with Glc.
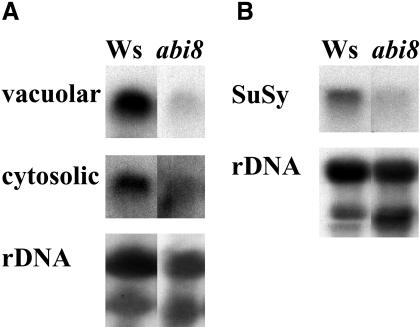
The differential response to Glc versus sucrose suggested a defect in transport, uptake, or metabolism of sucrose presumably supplied either exogenously or as a photosynthate. Sucrose is the most commonly translocated form of sugar in plants and may be cleaved by invertases present in the cell wall, cytosol, or vacuole or by sucrose synthase. Consistent with the poor growth of abi8 in the presence of sucrose, total invertase activity, assayed by histochemical staining as described by Finkelstein and Lynch (2000b), was reduced slightly in abi8 mutant roots (data not shown). Studies of antisense expression of assorted sugar-mobilizing enzymes have shown that cell wall invertases are the most critical isoforms for establishing sink strength and resulting growth of taproots, but vacuolar invertases and sucrose synthase also are important to root growth, contributing to the osmotic potential driving cell expansion (reviewed in Sturm and Tang, 1999). Using class-specific but not gene-specific probes, we found that both vacuolar and cytosolic invertase expression was reduced in mutant roots (Figure 4A). Expression of another sucrose-cleaving enzyme, sucrose synthase, also was reduced in mutant roots (Figure 4B). Cell wall invertase transcripts were undetectable in either genotype (data not shown), suggesting that they are less critical for promoting growth of seedling roots than they are for mature taproots.
Figure 4.
Expression of Genes Encoding Sucrose-Cleaving Enzymes in Wild-Type versus abi8 Roots.
Seedlings were grown on minimal media for 4 d to permit identification of abi8 segregants based on their stunted growth and then transferred to minimal media with 1% Glc for 10 d before harvesting roots for RNA extraction. Blots were probed with clones corresponding to a vacuolar invertase (At-β-fruct3) and a cytosolic invertase (A) and a sucrose synthase (SuSy, ATSUS1) (B). Each lane contains 10 or 3 μg total RNA ([A] and [B], respectively); an rDNA probe was used to assay uniformity of loading.
To determine whether the sugar metabolism defects of the abi8 mutant also are reflected in altered accumulation of soluble or insoluble sugars, we assayed levels of Glc, sucrose, fructose, and starch in young seedlings germinated on minimal medium. Under these conditions, the segregating progeny of abi8 heterozygotes are readily distinguishable by 4 to 5 d postgermination and can be assayed without the complication of the contributions from the exogenous Glc that is required to maintain abi8 seedling growth. Despite the decreased levels of invertase and sucrose synthase, there was only a small increase in starch accumulation and very little change in soluble sugars in the mutant (data not shown).
In addition to their roles in plant nutrition and in affecting the osmotic potential of growing cells, sugars are well known as signaling molecules in plants (reviewed in Rolland et al., 2002). Evidence for interactions between ABA and sugar signaling mechanisms has been provided by the isolation of new mutant alleles in ABA response or biosynthetic loci in screens for sugar-resistant seedling growth or gene expression (Arenas-Huertero et al., 2000; Huijser et al., 2000; Laby et al., 2000; Rook et al., 2001), induction of hypersensitivity to Glc by overexpression of ABI3, ABI4, or ABI5 (Finkelstein et al., 2002), Glc-induced expression of all three of these transcription factors and ABA biosynthesis (Arenas-Huertero et al., 2000; Brocard et al., 2002; Cheng et al., 2002), and suppression of ABA-inhibited germination by low levels of Glc (Finkelstein and Lynch, 2000b). To determine whether abi8 also affects sugar signaling, seeds of abi8/+ individuals were sown on medium containing 6% Glc. After 1 week, the wild-type segregants have arrested and turned pink, but the abi8 mutants remain green and display their usual stunted growth (Figure 3E). Controls with isoosmotic sorbitol indicated that the arrest of the wild type was not attributable to an osmotic effect, and abi8 segregants were not significantly resistant to osmotic inhibition of growth (data not shown). This suggests that ABI8 may be another member of the subset of ABA response loci and biosynthetic genes that affect sugar signaling.
Defects in Stomatal Regulation
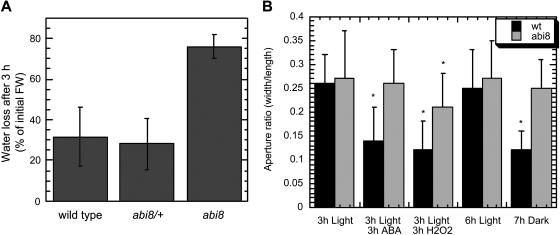
Stomatal closure is a well-known response to ABA, but only a subset of ABA-resistant mutants have defects in this response. To determine whether abi8 mutants are defective in stomatal regulation, water loss in excised plants was measured after a 3-h incubation. Comparison of wild-type, abi8, and abi8/+ heterozygotes shows that homozygous mutants lose at least twice as much water as the wild-type or heterozygous plants (Figure 5A). Because wilting also can result from defective vascular tissue (Postlethwait and Nelson, 1957; Rock and Ng, 1999), we tested stomatal response directly by measuring stomatal apertures of excised rosette leaves from 3- to 4-week-old plants that had been grown on agar medium supplemented with 1% Glc. Leaves were floated in opening solution for 3 h in light, followed by an additional 3 h in light with or without ABA or H2O2. Photoregulation was measured by comparing apertures of stomata examined either after 2 to 7 h of dark incubation of leaves or immediately after overnight incubation of intact plants in darkness (Figure 5B; data not shown). Although there was substantial variability in both wild-type and mutant responses, differences between wild-type and abi8 apertures were significant for all except the light-incubated controls (Figure 5B). Whereas no closure of abi8 stomata was observed in response to either ABA or darkness, partial closure was induced by H2O2, indicating that the mutant stomata still are capable of changing their apertures. However, the failure to promote full closure under any conditions prevented us from testing whether the mutation also disrupts ABA inhibition of stomatal opening.
Figure 5.
Stomatal Regulation in Wild-Type versus abi8 Mutant Plants.
(A) Water loss in excised plants during 3-h incubation. Comparison of wild-type, abi8, and abi8/+ heterozygotes (genotypes determined by analysis of abi8 segregation in progeny).
(B) Stomatal apertures of rosette leaves from 3- to 4-week old plants floated in opening solution for 3 h in light, followed by an additional 3 h in light ±10 μM ABA or 100 μM H2O2 or measured after 7-h incubation in darkness.
Asterisks designate samples with significantly different (P < 0.004) apertures from light-incubated controls. Differences between wild-type and abi8 apertures were significant for all except the light treatments.
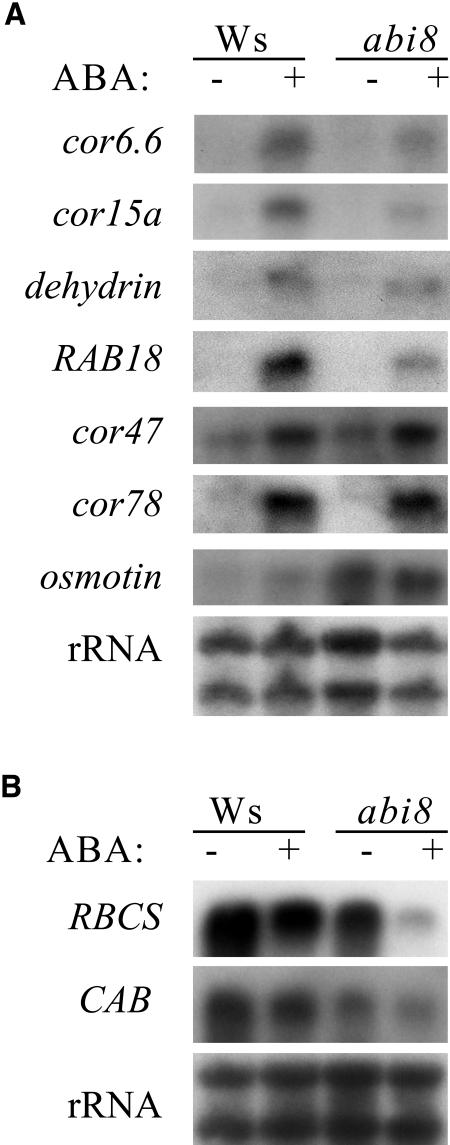
ABI8-Dependent ABA-Regulated Gene Expression
To determine whether ABI8 is required for ABA-dependent gene expression, we compared ABA inducibility of a variety of ABA- and stress-induced transcripts in 2-week-old wild-type and abi8 plants grown on agar medium supplemented with 1% Glc. Although some of these show reduced ABA response in the abi8 mutant (cor6.6, cor15a, dehydrin, and RAB18), others show similar ABA response in wild-type and mutant segregants (cor47 and cor78) or even constitutive expression (osmotin) in the mutant (Figure 6A). We also have compared reporter activity of Dc3:GUS and RAB18:GUS transgenes in seeds and found that GUS activity does not vary significantly among wild-type and mutant segregants (data not shown), indicating that ABI8 function is not required for embryonic expression of these genes. Several light-regulated genes (e.g., ribulose biphophate carboxylase small subunit [RBCS] and chlorophyll a/b binding protein [CAB]) have been shown to be ABA repressed in wild-type plants (Weatherwax et al., 1996). These genes were underexpressed in abi8 mutants grown under control conditions and further repressed by ABA treatment (Figure 6B). These results support the conclusion that the abi8 mutation alters, but does not eliminate, response to ABA.
Figure 6.
ABA-Regulated Gene Expression in Wild-Type versus abi8 Mutant Plants.
RNA gel blot analyses of ABA effects on expression of ABA-inducible (A) and light-inducible/ABA-repressible (B) genes in 2-week-old plants. Blots were hybridized to indicated probes; loading uniformity was assayed by hybridization to rRNA.
Genetic Interactions with Other Hormone-Signaling Components
To determine whether ABI8 interacts genetically with some of the known ABA response loci, we constructed lines combining the abi8 mutation and a variety of mutations or transgenes conferring ABA hypersensitivity or ABI-regulated GUS expression. These include ABA-hypersensitive1 (abh1) (Hugouvieux et al., 2001) and hypersensitivity conferred by overexpression of ABI transcription factors (Parcy et al., 1994; Brocard et al., 2002). Comparison of germination sensitivities to ABA and seedling morphology in these lines have shown that overexpression of ABI3 or ABI5 does not restore normal seedling growth in abi8 mutant segregants but results in intermediate ABA sensitivity at germination (Table 1). Similarly, seedlings of abi8 abh1 double mutants display abi8-like dwarfing but are only weakly resistant to ABA inhibition of germination. The intermediate ABA sensitivities of the abi8 35S:ABI and abi8 abh1 seeds suggest action in parallel pathways. Comparisons of ABI-regulated GUS expression show that ABI3:GUS, ABI4:GUS, and ABI5:GUS expression all appear normal in abi8 mutant seedlings (data not shown), which also is consistent with action of ABI8 in a separate pathway from the ABI transcription factors.
Table 1.
Genetic Interactions between abi8 and Other ABA Response Loci
| Genotype | Germination (%) 0.3 μM ABA | (n) | Germination (%) 1 μM ABA | (n) | Dwarf (%) | (n) |
|---|---|---|---|---|---|---|
| Ws | 73.3 | (359) | 19.1 | (393) | 0 | |
| abi8/+ | 96.4 | (501) | 49.2 | (528) | 22 | (463) |
| abi8 | 100 | (125) | 100 | (132) | ||
| 35S:ABI3 | 45.2 | (126) | 16.9 | (136) | 0 | |
| abi8/+ 35S:ABI3 | 68 | (409) | 21.4 | (252) | 20.4 | (216) |
| abi8 35S:ABI3 | 92.7 | (82) | 80.7 | (88) | ||
| 35S:ABI5 | 2 | (197) | 0 | (181) | 0 | |
| abi8/+ 35S:ABI5 | 26.7 | (533) | 15.6 | (449) | 24.1 | (429) |
| abi8 35S:ABI5 | 54.1 | (133) | 62.5 | (112) | ||
| abh1 | 31.4 | (118) | 1.5 | (134) | 0 | |
| abi8/+ abh1 | 15.4 | (280) | 4.3 | (232) | 23.2 | (302) |
| abi8 abh1 | 57 | (70) | 17.3 | (58) |
ABA sensitivity was assayed by measuring germination (scored as emergence of entire seedling from seed coat) after 4 d of incubation on 0.3 μM ABA or 8 d of incubation on 1 μM ABA. Among germinating progeny of abi8/+ 35S:ABI or abi8/+ plants, abi8 segregants were identified by their swollen radicles. The normalized percentage of germination was calculated relative to the anticipated fraction of double homozygotes. Dwarfing as a result of the abi8 mutation was assayed in progeny of abi8/+ plants grown on hormone-free medium; dwarfing was not observed in lines lacking the abi8 mutation. The results are expressed as a percentage of the total number of seeds plated (n).
Many hormone-signaling mutants have defects in responding to multiple hormones, and several loci have been implicated in mediating cross talk between ABA and ethylene signaling. Because the abundant root hair growth and short swollen hypocotyls of abi8 mutants are reminiscent of ethylene effects on seedlings, we tested whether inhibitors of ethylene synthesis or response (aminoethyoxyvinylglycine and AgNO3, respectively) could suppress the stunted phenotype of the abi8 plants (data not shown). Although these treatments resulted in slightly greater expansion of wild-type seedlings, the abi8 mutants remained stunted, indicating that this phenotype is probably not attributable to defects in ethylene signaling (data not shown). However, because both of these inhibitors affect the initial steps in ethylene signaling (production or receptor binding of the hormone), we also constructed digenic mutants combining abi8 with ethylene-insensitive (ethylene-insenstive2 [ein2] and ein3) mutations affecting later steps in ethylene signaling. EIN2 encodes a membrane protein related to mammalian NRAMP metal transport proteins (Alonso et al., 1999) that appears to mediate activation of EIN3. EIN3 encodes a transcription factor that regulates expression of a class of transcription factors that bind ethylene-responsive elements of ethylene-induced genes (Chao et al., 1997; Solano et al., 1998). Mutations affecting EIN2 have been isolated in screens for altered response to ABA (designated era3) (Beaudoin et al., 2000; Ghassemian et al., 2000), cytokinins (Cary et al., 1995), and auxin transport inhibitors (Fujita and Syono, 1996), suggesting that EIN2 acts at a point of cross talk among hormone signaling pathways. Similarly, ein3 mutants are slightly hypersensitive to ABA for inhibition of germination: only 4% of ein3 seeds germinate after 5 d on media containing 0.3 μM ABA, whereas 77% of wild-type seeds germinate under these conditions. In addition, both ein2 and ein3 mutations confer hypersensitivity to Glc for inhibition of seedling growth (Yanagisawa et al., 2003). Analyses of the progeny of ein abi8/+ plants showed that both homozygous ein2 abi8 and ein3 abi8 segregants exhibited ABA-resistant germination and 1-aminocyclopropane-1-carboxylic acid (ACC)-resistant cotyledon expansion but remained stunted (Table 2) and displayed Glc sensitivity that was intermediate between the parental lines (data not shown). These results support the conclusion that the stunted phenotype of abi8 mutants is not attributable to constitutive ethylene response but show that abi8 acts epistatically to the ein mutations with respect to effects on ABA sensitivity of germination yet acts additively with respect to effects on Glc sensitivity of seedling growth.
Table 2.
Genetic Interactions between abi8 and Ethylene Response Loci
| Seedling Morphology on ACC Triple Response
|
ABA Sensitivity
|
|||||
|---|---|---|---|---|---|---|
| Genotype | − | + | abi8-Like | (n) | Germination | (n) |
| Ws | 0 | 100 | 0 | (100) | 0 | (315) |
| abi8/+ | 0 | 71.4 | 28.6 | (56) | 23 | (513) |
| ein2 | 100 | 0 | 0 | (100) | 0 | (100) |
| abi8/+ ein2 | 75.4 | 0 | 24.7 | (292) | 28 | (500) |
| abi8/+ | 0 | 73.9 | 26.1 | (46) | 22.3 | (417) |
| ein3 | 100 | 0 | 0 | (100) | 0 | (205) |
| abi8/+ ein3 | 73.4 | 0 | 26.6 | (726) | 19.7 | (680) |
Ethylene sensitivity was assayed by plating seeds on 10 μM ACC and scoring seedling morphology after 4 d in the dark at 22°C; the triple response was scored as maintenance of an apical hook, failure to expand cotyledons, and radial swelling. abi8-like seedlings were distinguished from those displaying the triple response by their reduced apical hooks and enhanced radial swelling of the hypocotyl. The ein abi8 double mutants show complete loss of the apical hook. ABA sensitivity was assayed by measuring germination and hypocotyl swelling after 8 or 9 d of incubation on 3 μM ABA for the ein2 and ein3 monogenic and digenic lines, respectively. The results are expressed as a percentage of the total number of seeds plated (n).
ABI8 Encodes a Plant-Specific Protein of Unknown Biochemical Function
Although isolated from a T-DNA insertion line collection, the abi8 mutation did not cosegregate with any of the T-DNA markers. Consequently, we undertook a chromosome walk to identify the affected gene. We fine-mapped ABI8 to within ∼95 kb on chromosome 3 and then tested nearly all of the predicted genes across the interval for complementation by wild-type genomic fragments introduced via floral-dip infiltration (Clough and Bent, 1998) of abi8 LONG HYPOCOTYL2 (HY2)/ABI8 hy2 individuals. Complementation was observed with a 5-kb genomic fragment containing a single complete gene (At3g08550) that encodes a predicted 533–amino acid protein of unknown function. The abi8 mutant allele contains a 5-bp deletion, resulting in a frame shift in the first exon at amino acid 85 and termination shortly thereafter, such that this allele is likely to be a biochemical null (Figure 7A). Basic Local Alignment Search Tool (BLAST) analyses have identified likely orthologs in Nicotiana plumbaginifolia and Oryza sativa (rice) (Figure 7B), and closely related genes are represented in EST collections from diverse plant species. Consistent with this proposed similarity of function, the N. plumbaginifolia gene was identified by virtue of an insertional mutant with a seedling lethal phenotype, sdl-1 (N. Houba, unpublished data). Database searches indicate that abi8 is allelic to two dwarf mutants, eld1 and kob1, previously shown to disrupt cell elongation, cellulose synthesis, vascular differentiation, and root meristem maintenance (Cheng et al., 2000; Pagant et al., 2002). Although highly conserved with other plant proteins, database searches for domains of known function have shown only a suggestion of association with an unidentified membrane via a prokaryotic membrane lipoprotein lipid attachment site and possible targeting to the chloroplast. A variety of potential modifications are suggested by the presence of 4 N-glycosylation sites, 16 phosphorylation sites, 8 myristoylation sites, and an amidation site, but each of these motifs occur frequently in protein sequences, and their function must be verified experimentally.
Figure 7.
ABI8 and Homologous Genes.
(A) Schematic of ABI8 exon/intron stucture and site of abi8 mutation.
(B) Alignments of predicted amino acid sequences with those from homologous genes. Schematic of A. thaliana genes (top); sequence alignment of A. thaliana, O. sativa, and N. plumbaginifolia genes (bottom). Residues conserved across all five sequences are shaded black; residues conserved across three or four sequences are shaded gray. Consensus sequences are shown underneath; conserved groups are as follows from top to bottom: 1 = DN, 3 = ST (hydroxylated), 4 = KR (basic), 5 = FYW (aromatic), and 6 = LIVM (aliphatic or M). Alignment was performed by the Pileup program of the University of Wisconsin Genetics Computer Group software package; shading of conserved residues was accomplished with the GeneDoc program.
A. thaliana contains two predicted genes that encode proteins that are 60 to 70% identical to ABI8 over 400 to 500 amino acids: At3g57200 and At2g41450 (Figure 7B). One of these (At2g41450) encodes an additional 577 amino acids at the N terminus, which includes three conserved domains: a breast cancer C-terminal domain, an FBD domain (found in FBox and breast cancer C-terminal domain–containing plant proteins), and a domain present in N-acetyltransferases. Although there are no published reports of mutant phenotypes resulting from disruption of the A. thaliana homologs of ABI8, multiple insertion lines are available for both of these loci from the SALK SIGnAL collection (http://signal.salk.edu). We have identified lines carrying the desired insertions but see no evidence of a phenotype reminiscent of abi8 (data not shown). In combination with the severity of the abi8 phenotype, this suggests that these loci do not function redundantly.
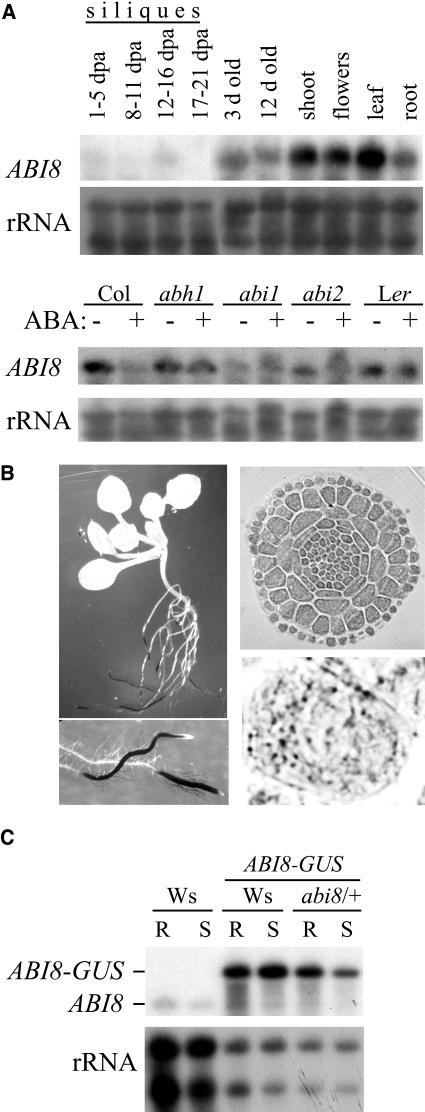
ABI8 Expression and Localization
At the level of mRNA accumulation, ABI8 is expressed in all tissues tested throughout development in wild-type plants, but transcript levels are relatively low in developing siliques (Figure 8A). ABI8 transcript levels show no consistent regulation by ABA exposure, although the gene appears slightly underexpressed in abi1 mutants (Figure 8A). We have assayed tissue and subcellular localization by histochemical staining of transgenic plants containing fusions of an ABI8 genomic fragment, including the entire 5′ region and coding sequence, tagged at the C terminus with GUS. Despite the strong ABA resistance of mutant seeds, ABI8-GUS was undetectable histochemically in developing or mature seeds. However, fluorometric assays of extracts from mature seeds of independent transgenic lines showed that GUS activity ranged from 10 to 70 units/h/seed above the background detected in nontransgenic control seeds. Furthermore, stratification and subsequent incubation on media containing 1 μM ABA was sufficient to both delay germination and induce ABI8-GUS accumulation near the radicle tip (Figure 1 in supplemental data online), consistent with a role for ABI8 in inhibiting the radicle growth required for germination.
Figure 8.
ABI8 Expression.
(A) RNA gel blot analyses of ABI8 transcript accumulation in assorted tissues and genotypes. ABA response in different genotypes was compared in 2-week-old plants exposed to 50 μM or no ABA for the last 2 d of culture. Col, Columbia ecotype; Ler, Landsberg erecta ecotype.
(B) Localization of GUS activity derived from an ABI8-GUS fusion, including the entire ABI8 coding sequence, under control of the ABI8 promoter. Left, entire plant and root tip; right, cross-section through root tip of 2-week-old plant and high-magnification view of a single cell showing the punctate staining pattern.
(C) RNA gel blot analyses of ABI8 and ABI8-GUS transcript accumulation in transgenic lines. R, roots; S, shoots.
Although green fluorescent protein (GFP)-KOB1 expressed under control of the 35S promoter of Cauliflower mosaic virus recently was shown to localize to the plasma membrane of elongated root cells (Pagant et al., 2002), we found that the ABI8-GUS fusion protein expressed under control of its own promoter is localized primarily to the elongation zone of roots, where it accumulates in a punctate pattern within the cytoplasm (Figure 8B). A similar cytoplasmic localization of the GFP-KOB1 fusion was observed in the cell division zone of the root, indicating that membrane association is not obligate. Furthermore, ABI8-GUS transgenes exhibiting this localization pattern are sufficient to complement the mutation (Table 3), indicating that there may be substantial post-transcriptional control of ABI8 accumulation, such that minor variations in transcript levels may not be functionally significant. Consistent with the possibility of post-transcriptional control, RNA gel blot analyses indicate that the transgene is strongly expressed in both roots and shoots (Figure 8C) even though the fusion protein is undetectable in most tissues (Figure 8B), even by fluorometric assays (data not shown). The high abundance of transgene transcripts seen in Figure 8C might reflect the presence of multiple fusion genes in the transgenic lines but does not result in high-level protein accumulation.
Table 3.
Complementation of abi8 in ABI8-GUS Transgenic Lines
| Genotype | Germination (%) | (n) | Kanamycin R (%) | (n) |
|---|---|---|---|---|
| abi8/+ | 25 | (271) | NAa | |
| abi8/+ ABI8-GUS/ø | 4.9 | (144) | 77 | (81) |
| abi8/+ ABI8-GUS | 0 | (208) | 100 | (200) |
The abi8 phenotype was assayed by measuring germination and hypocotyl swelling after 7 d of incubation on 3 μM ABA. Presence, number of insertion sites, and homozygosity of transgenes was assayed by scoring kanamycin resistance (kanamycin R). The results are expressed as a percentage of the total number of seeds plated (n).
NA, not applicable.
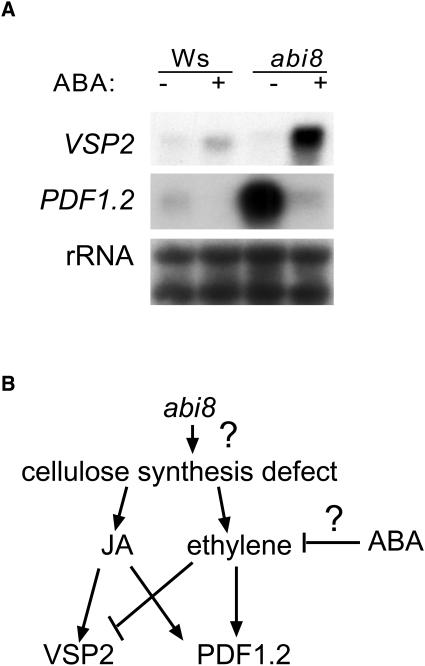
Interactions with Stress Signaling
The discovery that abi8 was allelic to a cellulose-deficient mutant raised the possibility that some of the signaling defects also might be derived from the alterations in cellulose synthesis capacity. Recent studies have shown that defects in cellulose synthesis, induced either genetically or chemically, can lead to overproduction of jasmonate (JA) and ethylene and activate stress responses dependent on these hormones (Ellis et al., 2002). To determine whether the cell wall defects of the abi8 mutant also affect this aspect of stress signaling, we tested abi8 effects on expression of two genes regulated by JA, ethylene, and cellulose deficiency: VSP2 and PDF1.2. VSP2 encodes a vegetative storage protein, whereas PDF1.2 encodes a plant defensin involved in fungal resistance. Expression of both genes is induced, to different degrees, by either JA or disruption of cellulose synthesis. PDF1.2 expression also is induced by ethylene, but VSP2 is repressed by ethylene (Ellis et al., 2002). RNA gel blot analyses indicated that ABA acts antagonistically to ethylene, in that it slightly induced VSP2 but repressed PDF1.2 expression in Wassilewskija (Ws) seedlings (Figure 9). By contrast, abi8 seedlings strongly expressed PDF1.2 in the absence of ABA but still could repress PDF1.2 and hyperinduced VSP2 in response to ABA. These results suggest that ABA regulation of VSP and PDF1.2 expression was functional or even enhanced in the abi8 mutant. Although the high basal level of PDF1.2 expression in the mutant might be explained by cell wall–based signaling, VSP expression was not similarly altered, again demonstrating that not all aspects of the abi8 phenotype can be ascribed to cellulose deficiency alone.
Figure 9.
Interactions Affecting Expression of Genes Induced by Cellulose Deficiency.
(A) RNA gel blot analyses of ABA effects on gene expression in 2-week-old plants. Blots were hybridized to indicated probes; loading uniformity was assayed by hybridization to rRNA.
(B) Proposed model of signaling interactions regulating VSP2 and PDF1.2 expression.
DISCUSSION
At least five independent mutations in ABI8/ELD1/KOB1 genes have been identified in screens for ABA response defects or dwarf plants (Cheng et al., 2000; Pagant et al., 2002; K. Lertpiriyapong and Z.R. Sung, personal communication). The kob1-1 and kob1-2 alleles had a T-DNA insertion in an intron and a defect affecting splicing, respectively, but still were capable of making a small amount of properly spliced transcript. By contrast, the abi8 allele results in loss of >80% of the C-terminal portion of the protein and is likely to be a biochemical null mutant. The eld1 alleles also are nulls (K. Lertpiriyapong and Z.R. Sung, personal communication). All of the mutants are morphologically similar, in that they are sterile and have severely stunted growth because of reduced cell elongation that is not rescued by treatment with any known hormones or inhibitors of hormone synthesis/response. However, the kob1-1 allele is less dwarfed than the other reported mutants. Because of the distinct nature of the screens, characterization of the various alleles has emphasized different aspects of their physiological and molecular defects.
Growth Defects in abi8/eld1/kob1 Mutants
The severe stunting of eld1 root growth was shown to reflect terminal differentiation of the root apical meristem and cessation of cell division at the root tip, with only occasional activation of cell division in newly emerging lateral roots (Cheng et al., 2000). In this regard, it is noteworthy that the eld1 mutant roots have reduced expression of cdc2a (Cheng et al., 2000), a cell cycle marker that reflects competence for cell division and whose activity is inhibited by the ABA-induced INHIBITOR OF CYCLIN DEPENDENT KINASE1 (Wang et al., 1998). In this respect, the mutants appear to display a constitutive ABA response. Both eld1 and kob1 mutants were described as having a greatly reduced zone of elongation, such that differentiation of root hairs was observed almost all of the way to the root tips; abi8 mutants also display this characteristic. All of the mutants have small cuboidal root and hypocotyl cells; the reduced cell elongation is correlated with an ∼45 to 60% decrease in cellulose synthesis in the kob1 mutants, leading Pagant et al. (2002) to suggest that reduced cellulose synthesis is a primary effect of the kob1 mutations.
Although the eld1 stele was characterized as disrupted by somewhat randomly arrayed vascular differentiation, which also is observed in abi8 mutants, this was not described for the kob1 mutants possibly because they are weaker alleles. Growth of abi8 mutant roots also arrests on standard growth media (minimal nutrient salts or Murashige and Skoog salts with 1% sucrose), but this arrest is prevented by inclusion of Glc (0.5 to 4%) in the medium. This result indicates that the mutation does not force the roots to arrest but may alter their ability to produce or respond to signals promoting growth. Microscopic examination of abi8 roots and hypocotyls showed that the improved root growth on Glc reflected maintenance of the root apical meristem and improved vascular development but that the mutant cells still were much shorter than those of the wild type. These results suggest that the cellulose synthesis defect still inhibits cell elongation, but tissue differentiation and overall morphology are subject to additional regulation.
Both eld1 and kob1 mutants were shown to have ectopic accumulat.ion of wall components, such as suberin and lignin, throughout the hypocotyl and in the cotyledons, reflected in waxy white patches on the cotyledons (Cheng et al., 2000; Pagant et al., 2002). The abi8 mutant shows similar waxy patches when grown under conditions that lead to growth arrest but not when growth is partially rescued by inclusion of at least 1% Glc (Figure 3), indicating that accumulation of these secondary products also may be a secondary effect of the mutation.
ABI8/ELD1/KOB1 Affect ABA and Glc Signaling
In addition to observing the growth defects described above, characterization from the perspective that abi8 represents an ABA response mutant has uncovered defects in ABA signaling in germination, stomatal regulation, and regulation of gene expression. However, this is not a complete lack of response because some ABA-regulated genes show normal ABA response, whereas others become constitutively expressed and/or hyperresponsive. Similarly specific but distinct effects on ABA-regulated gene expression have been described for many other ABA response mutants, including all of the other abi mutants and abh1 (Hugouvieux et al., 2001; Finkelstein et al., 2002; Hoth et al., 2002; Suzuki et al., 2003). Furthermore, some of these previously documented specificities also appear contradictory, including opposing effects of abi3 and abi5 mutations on expression of specific genes expressed late in embryogenesis (Parcy et al., 1994; Finkelstein and Lynch, 2000a) or decreased expression of several ABA-inducible genes in the ABA-hypersensitive abh1 mutant (Hugouvieux et al., 2001). Thus, abi8 mutants resemble many other ABA response mutants in displaying differential defects in ABA response. Our studies also have shown that abi8 growth is not only dependent on low concentrations of Glc but also is resistant to the inhibitory effects of high Glc, suggesting a defect in sugar signaling and/or transport. Whereas other ABA response or biosynthetic mutants have been found to be resistant to high Glc, this mutant differs from the others in that it also is Glc dependent. It also appears to act in a distinct pathway from the other ABI loci implicated in Glc response.
The fact that the impaired growth is far more severe than that observed in other wilty mutants with defects in ABA biosynthesis (aba) or signaling (abi1) is consistent with the observation that the abi8 mutation disrupts additional processes required for growth, such as cellulose synthesis, vascular differentiation, and expression of invertases or sucrose synthases, that would affect the osmotic potential of elongating root cells. These results suggest that ABA and/or Glc signaling regulates expression of these sugar-mobilizing enzymes. Although Glc regulation of these enzymes has been described in many other plant species (Koch, 1996), ABA regulation is not well characterized. These enzymes are generally encoded by multigene families, with specific family members exhibiting opposite responses to sugars, such that some are induced by feast and others by famine conditions (Koch, 1996), and it is not clear which of the A. thaliana genes are regulated by Glc and/or ABA. BLAST analyses indicate that all of these sugar-mobilizing enzymes belong to multigene families with six to nine members per family in A. thaliana, only a few of which have been analyzed extensively (Schwebel-Dugue et al., 1994; Mercier and Gogarten, 1995; Haouazine-Takvorian et al., 1997; Tymowska-Lalanne and Kreis, 1998). Our studies did not attempt to discriminate among these multitudes of family members but clearly showed that the most abundantly expressed member(s) were dependent on ABI8 for normal expression.
Investigation of stomatal regulation revealed that the mutant fails to close its stomata in response to ABA or darkness and only weakly responds to H2O2. The partial response to H2O2 suggests that abi8 mutants may have defects in ABA-induced production of reactive oxygen species, but this does not explain their complete lack of response to ABA or the failure to respond to darkness. Another possibility is that the defect in dark response may be another aspect of the constitutively photomorphogenic phenotype described for the eld1 allele (Cheng et al., 2000). In this regard, it is noteworthy that genetic interactions affecting photomorphogenesis have been demonstrated between abi3 and de-etiolated1 mutants (Rohde et al., 2000) and that abi fusca3 and abi leafy cotyledon1 digenic mutants reveal cryptic effects resulting in a mildly deetiolated phenotype (Nambara et al., 2000; Brocard-Gifford et al., 2003). Additional interactions between darkness and ABA accumulation and signaling have been demonstrated in wild-type plants (Weatherwax et al., 1996). Although the defects in stomatal regulation also might reflect unidentified defects in guard cell structure (e.g., possibly affecting the arrangement of cellulose microfibrils that determines guard cell turgor and shape), abi8 guard cells look surprisingly normal, in that they are approximately the same size as those of wild-type plants (data not shown). Furthermore, the partial response to H2O2 is not consistent with a solely structural defect, as observed when disruption of pectic cross-linking results in locking of stomatal aperture (Jones et al., 2003).
Interactions with Other Signaling Loci
In addition to describing multiple defects in response to ABA and Glc, our genetic studies have demonstrated epistatic or additive relationships between abi8 and several loci known to affect ABA, and in some cases Glc or ethylene, signaling. At least a dozen loci have been identified that disrupt response to at least two of these signals. In general, ABA and Glc responsiveness are correlated, whereas ethylene acts antagonistically to these signals at germination and in early seedling growth. The loci shown to be associated with both ABA and sugar response have been limited so far to those encoding ABA biosynthetic enzymes (ABA1, ABA2/GIN1/ISI1/SIS4, and ABA3/GIN5/LOS5), the ABI transcription factors (ABI3, ABI4/GIN6/ISI3/SIS5/SUN6, and ABI5) and related proteins (ABF3 and ABF4), and EIN2/ERA3 and CTR1 (reviewed in Leon and Sheen, 2003). Among these, the ABI transcription factors appear to function in the same signaling pathway mediating ABA response (Finkelstein et al., 2002), and expression of all three is induced by Glc in an apparently hexokinase-dependent manner (Leon and Sheen, 2003). The EIN2/ERA3 and CTR1 loci also have been shown to function in a single pathway, in this case, mediating ethylene response via effects on EIN3 and resulting ethylene-dependent gene expression (Wang et al., 2002). This pathway recently was shown to be modulated by hexokinase-dependent degradation of EIN3 (Yanagisawa et al., 2003). Although ctr1 alleles have been isolated as enhancers of ABA resistance in abi1-1 mutants (Ghassemian et al., 2000), similar to the interactions reported for abi1-1 and abi transcription factor mutations (reviewed in Finkelstein and Rock, 2002), genetic studies testing potential interactions among the ABI transcription factors and the products of these ethylene response loci have not been reported.
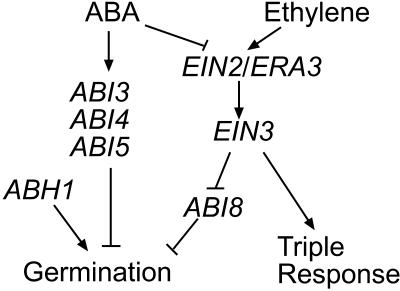
Our studies place ABI8/ELD1/KOB1 function downstream of EIN2 and EIN3 and possibly in a separate pathway from those requiring action of ABH1 and the ABI transcription factors in regulating ABA effects on germination (Figure 10). Epistasis analyses among abh1, era1, and several abi mutants indicated that ABH1 also acts in a distinct pathway from ERA1 and the ABI transcription factors (Hugouvieux et al., 2001; Brady et al., 2003). However, the additive effects of abi8 and ein mutations on Glc and ethylene response indicate that these loci mediated response to these signals via distinct pathways, suggesting that ABI8 defines a novel signaling pathway affecting Glc response. The consistently dwarfed phenotype of all of the lines carrying the abi8 mutation might reflect defects in cellulose synthesis that are not affected by any of the other loci tested in the double mutant or transgenic lines.
Figure 10.
Proposed Model of Genetic Interactions between ABI8 and Some Other ABA Response Loci Affecting Germination.
The discrepancy between transcript and protein accumulation indicates that comparisons of ABI8 transcript levels in various mutants may not be a good indicator of ABI8 activity and that ABI8 accumulation is likely to be subject to control at the level of translation and/or stability. Furthermore, differences in ABI8/ELD1/KOB1 protein accumulation or localization may be mediated by additional unidentified factors. For example, tissue-specific interactions with other protein(s) might explain why the 35S:GFP-KOB1 product was cytoplasmic in the root tip but localized to the plasma membrane elsewhere (Pagant et al., 2002). Even though these putative additional factors have not been identified yet, the observed genetic interactions and physiological responses clearly place ABI8/ELD1/KOB1 in an ABA- and Glc-regulated signaling network, suggesting that many of the growth defects may reflect signaling defects and not direct effects on cellulose biosynthesis. Furthermore, this suggests that ABA and/or Glc signaling may interact with several processes with which these signals had not been associated previously, including promotion of cellulose biosynthesis and organizing vascular differentiation. Although it may seem contradictory to ascribe the severity of the phenotype to signaling defects that are not observed in the aba or other abi mutants, this might be explained by the substantial redundancy in both ABA biosynthetic and signaling pathways. Alternatively, ABI8/ELD1/KOB1 may integrate stress signaling in response to ABA, ethylene, JA, Glc, and cell wall–derived compounds.
Potential Function of ABI8/ELD1/KOB1
The cloning and molecular analyses of ABI8/ELD1/KOB1 have raised many new questions. The predicted amino acid sequence reveals no domains of known biochemical function, and the genetic analyses place its action in a network of signaling factors. Subcellular localization of an overexpressed GFP-KOB1 fusion showed that it was in the plasma membrane of cells in the root elongation zone but showed a punctate intracellular distribution in the cell division zone at the root tip (Pagant et al., 2002). By contrast, accumulation of a complementing ABI8-GUS fusion expressed under control of the ABI8 promoter was limited to the root elongation zone and the more terminal portions of the zone of differentiation, where it was concentrated in punctate patches in the cytoplasm similar to those described for KOB1 in the root tip. These results raise the question of whether the observed plasma membrane localization of 35S:GFP-KOB1 is functionally significant or a default localization for an ectopically expressed protein with a lipid attachment site. The punctate distribution displayed by both the physiologically functional ABI8-GUS fusion and the root tip–localized GFP-KOB1 may depend on an interaction with an anchoring cytoplasmic partner that is absent throughout most of the root.
The discrepancy between the extremely limited vegetative expression pattern of the ABI8-GUS fusion protein and its ability to complement the mutation's effects in tissues throughout the plant body suggests that ABI8 effects on shoot tissue may depend on either very low levels of protein or a long-distance signaling mechanism that preconditions shoots to respond to ABA and/or other signals. Fluorometric assays of ABI8-GUS activity in mature seeds demonstrated that it accumulates to levels one to three orders of magnitude lower than those observed for promoter fusions with the ABI transcription factors (Söderman et al., 2000; Brocard et al., 2002; R.R. Finkelstein, unpublished data). However, ABI8-GUS activity increased significantly in seeds stratified in the presence of ABA, consistent with a role for ABI8 in maintaining developmental arrest in imbibing seeds. This expression was strongest in the radicle tip, where it might be involved in preventing production of hydrolases needed for emergence and/or reactivation of cell cycling in postgerminative growth.
As suggested with respect to cev1, a cellulose synthase mutant, disruption of cell wall synthesis may cause release of cell wall–derived compounds, such as oligosaccharides, that may act as long-distance signals themselves. Alternatively, ABI8 might aid transport of water and available signals simply by promoting vascular differentiation. However, several lines of evidence indicate that it is unlikely that the defects of abi8/eld1/kob1 mutants can be explained fully in terms of effects on root and hypocotyl vascular development and the resulting capacity for long-distance transport. First, even isolated leaves of abi8 plants with rescued vascular development have altered stomatal response to applied ABA. In addition, these rescued plants display aberrant ABA regulation of gene expression. Furthermore, vascular development is disrupted in mutant tissues in which ABI8 appears to be not usually expressed, based on ABI8-GUS activity (e.g., the hypocotyls). ABI8 accumulation also may be transiently induced in specific tissues in response to as yet unidentified signals. In summary, the abi8/eld1/kob1 mutants provide evidence linking ABA and/or Glc signaling to promotion of cellulose biosynthesis and organizing vascular differentiation and provide an opportunity to decipher the function of a novel essential protein and possibly a novel signaling mechanism.
METHODS
Plant Material
The abi8 mutant was isolated from a T-DNA insertion line collection in the Ws background produced by Feldmann (1991) and made available by agreement with DuPont. The abh1, ein2-1, and ein3-1 mutants are described by Roman et al. (1995) and Hugouvieux et al. (2001). The 35S:ABI lines are described by Parcy et al. (1994) (2x35S:ABI3, isolate C7A19) and Brocard et al. (2002) (35S:ABI5, isolate 2A4). The ABI4:GUS fusion includes 3 kb of upstream genomic sequence and four codons of the ABI4 sequence in a translational fusion, as described by Söderman et al. (2000). The RAB18:GUS and Dc3:GUS fusions are described by Ghassemian et al. (2000) and Chak et al. (2000), respectively. The ABI8-GUS fusion was constructed by engineering a SacII site into the final codon of ABI8 and then fusing a 4.3-kb fragment containing 1.3 kb of upstream sequence and the entire ABI8 coding sequence, including introns into pBI101.3. The transgene was introduced into abi8/+ plants by Agrobacterium tumefaciens–mediated floral-dip transformation (Clough and Bent, 1998). Primary transformants were selected for growth in the presence of 40 μg/mL of kanamycin; their progeny were tested for segregation of kanamycin resistance and the abi8 phenotype.
For RNA isolation from vegetative tissues, plants were grown 12 d on 0.5 × MS nutrients (Sigma, St. Louis, MO) supplemented with 1% sucrose and 0.55% agar and then transferred to fresh media with or without 50 μM ABA for 2 additional d before harvest. For RNA isolation from leaves, flowers, and siliques, plants were grown in soil in continuous light at 22°C. Leaves were harvested during rosette stage, and flowers include open flowers and buds. Siliques were harvested in pools corresponding to four developmental stages: early embryogenesis (1 to 5 d post anthesis [dpa]), maturation (8 to 11 dpa), postabscission (12 to 16 dpa), and late embryogenesis (17 to 21 dpa). For RNA isolation from roots and shoots, 2-d-old plants were transferred into Gamborg B5 salts (Gamborg, 1968) supplemented with 1% Glc and grown hydroponically for 3 weeks under agitation in continuous light at 22°C before harvest. All growing tissues were weighed, flash frozen in liquid nitrogen, and stored at −70°C until extraction. Dry seed were stored at room temperature.
Positional Cloning
After preliminary linkage analysis indicated that the abi8 mutation was near the top of chromosome 3, plants heterozygous for the abi8 mutation were outcrossed to Landsberg marker lines carrying the hy2 and glabrous1 mutations. Mapping populations were produced by identifying F2 families segregating abi8 and then scoring the resulting F3 families at abi8 and hy2 to identify recombinants. Additional recombinants were identified by screening F2 individuals for recombinations between two flanking markers scorable by PCR: the cleaved amplified polymorphic sequence (CAPS) marker 17D8L and the simple sequence length polymorphism marker nga126 (Bell and Ecker, 1994). Fine-mapping within these recombinant populations made use of additional CAPS markers, restriction fragment length polymorphisms, and single nucleotide polymorphisms. Reaction and cycling conditions for CAPS markers were as described by Konieczny and Ausubel (1993).
Double Mutant Construction
To construct double mutants, abi8/+ plants were crossed with abh1, ein2, or ein3 plants. Progeny of abi8/+ abh1 double mutants were identified on the basis of 100% Basta resistance and abi8 segregation. Seed of abi8/+ ein individuals were identified on the basis of their phenotype on ACC in the dark (75% of plants with long hypocotyl, no plant showing ethylene triple response) and abi8 segregation. Homozygous double mutants were not fertile. To introduce transgenes into the abi8 background, transgenic lines were crossed onto abi8/+ individuals, which were identified at seed maturity by segregation of the abi8 mutation in the selfed progeny. Transgenic F1s were selected on the basis of kanamycin resistance, and their progeny were tested for segregation of the abi8 allele. Kanamycin-resistant F2s were selected and their progeny scored to identify homozygous transgenic lines segregating the abi8 mutation.
Germination Assays
Germination assays were performed with seeds that were surface sterilized in 5% hypochlorite and 0.02% Triton X-100 and then rinsed several times with sterile water before plating on minimal medium (Haughn and Somerville, 1986) containing 0.7% (w/v) agar supplemented with different concentrations of ABA, Glc, or sorbitol. The dishes were incubated for 3 d at 4°C to break any residual dormancy and then transferred to 22°C in continuous light (50 to 70 μE·m−2·s−1); germination was scored after 4 to 9 d.
Stomatal Aperture Measurements
Stomatal apertures were measured essentially as described previously by Kwak et al. (2002), except that the pH of the opening solution (10 mM Mes-Tris, 5 mM KCl, and 50 μM CaCl2) was decreased from 6.15 to 5.6 to promote ABA uptake. Leaves were incubated 3 h in light to promote stomatal opening and then for an additional 3 h with or without 10 μM ABA or 100 μM H2O2. For testing photoregulation, leaves were incubated continuously in the dark. After these treatments, leaves were fragmented in a Waring blender, and epidermal fragments were quickly mounted for photomicrography. Each experiment was replicated three times. Average stomatal aperture ratios (pore width:length) were determined from measurements of 59 to 225 stomata per treatment; statistical analyses were conducted using Student's t test, assuming an unequal variance.
RNA Gel Blot Analysis
RNA was isolated from immature siliques as previously described (Söderman et al., 2000). RNA was extracted from vegetative tissues by either a hot-phenol extraction procedure or Plant RNeasy (Qiagen, Valencia, CA) minipreps (Söderman et al., 2000). RNA concentrations were estimated based on absorbance at 260 and 280 nm.
Total RNA (5 to 10 μg per lane) was size fractionated on MOPS [3-(N-morpholino)-propanesulfonic acid]-formaldehyde gels and then transferred to nylon membranes (Osmonics, Westborough, MA) using 20 × SSPE (1× SSPE is 0.115 M NaCl, 10 mM sodium phosphate, and 1mM EDTA, pH 7.4) as blotting buffer. RNA was bound to the filters by UV cross-linking (120 mJ cm−2 at 254 nm). Uniformity of loading and transfer was assayed qualitatively by methylene blue staining and hybridization to an rDNA probe. Transcripts from ABA-inducible genes were detected by hybridization to cDNA clones as described by Söderman et al. (2000), labeled by random priming to a specific activity of 108 cpm μg−1. The cor6.6, cor15a, and cor78 cDNA clones are described by Hajela et al. (1990); osmotin (242K20T7), dehydrin (ATTS0613), cytoplasmic invertase (158A15T7), and sucrose synthase (182C20T7) cDNA clones are ESTs provided by the ABRC; and the RAB18 cDNA was an EST (PAP023) provided by M. Delseny. The RBCS (At5g38430), CAB (At5g01530), PDF1.2 (At5g44420), and VSP2 (At5g24770) transcripts were detected by hybridization to PCR fragments amplified from Ws genomic DNA. Hybridization conditions were 50% formamide, 5 × SSPE, 5 × Denhardt's solution (1× Denhardt's solution is 0.02% Ficoll, 0.02% polyvinylpyrrolidone, and 0.02% BSA), 0.1% SDS, and 200 mg mL−1 of DNA at 43°C for 16 to 24 h in a Hyb-Aid rotisserie oven. Filters were washed twice at 60°C in 2 × SSC (1× SSC is 0.15 M NaCl and 0.015 M sodium citrate) and 0.1% SDS and once at 60°C in 0.2 × SSC and 0.1% SDS for 30 to 60 min. ABI8 transcripts were detected by hybridization to a random-priming labeled cDNA clone in 7% SDS, 0.5 M sodium phosphate, pH 7.2, 1 mM EDTA, and 1% BSA at 65°C for 16 to 24 h (Church and Gilbert, 1984); the final wash for these filters was 40 mM Na phosphate buffer, pH 7.2, 1% SDS, and 1 mM EDTA at 60 to 65°C.
Microscopy
For comparisons of root differentiation, 7- or 11-d-old wild-type and abi8 seedlings, grown on minimal medium or minimal medium supplemented with 1% Glc, were fixed for 24 h in a 4% formalin/5% acetic acid/50% alcohol solution, followed by several rinses in 1 × PBS. Seedlings were stained with 10 μg mL−1 propidium iodide for 1 h with gentle agitation, followed by several rinses in 1 × PBS. Whole seedlings were mounted in glycerol or wintergreen oil and viewed on a Bio-Rad 1024 laser scanning confocal microscope (Hercules, CA). Images were processed with Adobe Photoshop (Mountain View, CA).
For GUS localization, 15-d-old seedlings were infiltrated with 5-bromo-4-chloro-3-indolyl-β-glucuronic acid for 10 min, followed by an overnight incubation in the dark at 37°C, as described by Brocard et al. (2002). Seedlings then were fixed for 4 h at 4°C in 4% glutaraldehyde and 0.05 M sodium cocadylate buffer, pH 7.0, with a change of fresh fixative after the first 2 h. Seedlings then were dehydrated in a graded series of ethanol and propylene oxide, gradually infiltrated with Spurr's resin, and finally embedded in 100% resin and incubated at 65°C for several days. Three-micrometer-thick sections were cut using the LKB Bromma 2088 Ultrotome V (Leica Instruments, Bannockburn, IL). Sections were attached to glass slides with heat and viewed on an Olympus BX51 microscope (Tokyo, Japan). Images were taken with an Optronics Magnafire camera and processed with Adobe Photoshop.
Seedling anatomy was compared in 11-d-old wild-type and abi8 seedlings grown on minimal medium or minimal medium supplemented with 1% Glc. Seedlings were fixed as described above with the addition of a postfixation treatment in 2% osmium tetroxide and 0.05 M sodium cacodylate buffer for 1 h at room temperature before the seedlings were dehydrated. Two- to three-micrometer-thick sections were cut using the LKB Bromma 2088 Ultrotome V. Cross-sections were attached to glass slides with heat, stained with 0.5% toluidine blue O, and then viewed with an Olympus BX51 microscope. Images were taken with an Optronics Magnafire or Microfire camera and processed with Adobe Photoshop.
Quantification of Soluble Sugars and Starch
Starch and soluble sugar (sucrose, fructose, and Glc) levels in 7-d-old plants grown on minimal medium were determined as previously described by Chia et al. (2000). After extraction of soluble sugars, the extract was divided into three fractions for parallel determinations of sucrose, fructose, and Glc levels. Sucrose was digested with 400 units invertase and 1 unit phosphoglucoisomerase, followed by measurement of released Glc by the Infinity Glc reagent (Sigma). Fructose and Glc levels were determined by digestion of extracted soluble sugars with 1 unit phosphoglucoisomerase or no enzyme, respectively, followed by measurement of released Glc by the Infinity Glc reagent. Control experiments indicated that the Glc reagent was not contaminated with phosphoglucoisomerase, such that the measured Glc levels did not include any contribution from endogenous fructose levels (data not shown).
Supplementary Material
Acknowledgments
We thank P. Scolnik and T. Caspar for providing the opportunity to screen DuPont's T-DNA insertion collection, M. Thomashow for the cDNAs of cor15a, cor47, and cor78, and M. Delseny for the RAB18 cDNA. We thank the ABRC team at Ohio State University for efficient distribution of clones and seed stocks, including the BAC clones used in the positional cloning, the RAB28, osmotin, dehydrin, invertase, and sucrose synthase cDNAs, and the T-DNA insertion lines from the SALK SIGnAL collection. We also thank J. Schroeder and his lab for gift of abh1 and for valuable discussion about the stomatal experiments, P. McCourt for gifts of ein2-1, ein3, and RAB18:GUS, T. Thomas for gift of Dc3:GUS, and J. Giraudat and F. Parcy for gifts of 35S:ABI3 and ABI3:GUS transgenic lines. We thank B. Matsumoto for microscopy advice and assistance and J. Cooper for critical reading and helpful discussion of this manuscript. This work was partially supported by USDA Grant 97-35304-4875. I.B.-G., M.E.G., and T.J.L. also were partially supported by National Science Foundation Grant IBN-9982779. Funding for the SIGnAL indexed insertion mutant collection was provided by the National Science Foundation.
On-line version contains Web-only data.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Ruth R. Finkelstein (finkelst@lifesci.ucsb.edu).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.018077.
References
- Alonso, J., Hirayama, T., Roman, G., Nourizadeh, S., and Ecker, J. (1999). EIN2, a bifunctional transducer of ethylene and stress responses in Arabidopsis. Science 284, 2148–2152. [DOI] [PubMed] [Google Scholar]
- Arenas-Huertero, F., Arroyo, A., Zhou, L., Sheen, J., and Leon, P. (2000). Analysis of Arabidopsis glucose insensitive mutants, gin5 and gin6, reveals a central role of the plant hormone ABA in the regulation of plant vegetative development by sugar. Genes Dev. 14, 2085–2096. [PMC free article] [PubMed] [Google Scholar]
- Beaudoin, N., Serizet, C., Gosti, F., and Giraudat, J. (2000). Interactions between abscisic acid and ethylene signaling cascades. Plant Cell 12, 1103–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell, C.J., and Ecker, J.R. (1994). Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics 19, 137–144. [DOI] [PubMed] [Google Scholar]
- Berger, S., Bell, E., and Mullet, J. (1996). Two methyl jasmonate-insensitive mutants show altered expression of AtVsp in response to methyl jasmonate and wounding. Plant Physiol. 111, 525–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady, S., Sarkar, S., Bonetta, D., and McCourt, P. (2003). The ABSCISIC ACID INSENSITIVE 3 (ABI3) gene is modulated by farnesylation and is involved in auxin signaling and lateral root development in Arabidopsis. Plant J. 34, 67–75. [DOI] [PubMed] [Google Scholar]
- Brocard, I., Lynch, T., and Finkelstein, R. (2002). Regulation and role of the Arabidopsis ABA-insensitive (ABI)5 gene in ABA, sugar and stress response. Plant Physiol. 129, 1533–1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brocard-Gifford, I., Lynch, T., and Finkelstein, R. (2003). Regulatory networks in seeds integrating developmental, ABA, sugar and light signaling. Plant Physiol. 131, 78–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cary, A., Liu, W., and Howell, S. (1995). Cytokinin action is coupled to ethylene in its effects on the inhibition of root and hypocotyl elongation in Arabidopsis thaliana seedlings. Plant Physiol. 107, 1075–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chak, R.K.F., Thomas, T.L., Quatrano, R.S., and Rock, C.D. (2000). The genes ABI1 and ABI2 are involved in abscisic acid- and drought-inducible expression of the Daucus carota L. Dc3 promoter in guard cells of transgenic Arabidopsis thaliana (L.) Heynh. Planta 210, 875–883. [DOI] [PubMed] [Google Scholar]
- Chao, Q., Rothenberg, M., Solano, R., Roman, G., Terzaghi, W., and Ecker, J. (1997). Activation of the ethylene gas response pathway in Arabidopsis by the nuclear protein ETHYLENE-INSENSITIVE3 and related proteins. Cell 89, 1133–1144. [DOI] [PubMed] [Google Scholar]
- Cheng, J.-C., Lertpiriyapong, K., Wang, S., and Sung, Z.R. (2000). The role of the Arabidopsis ELD1 gene in cell development and photomorphogenesis in darkness. Plant Physiol. 123, 509–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, W.-H., Endo, A., Zhou, L., Penney, J., Chen, H.-C., Arroyo, A., Leon, P., Nambara, E., Asami, T., Seo, M., Koshiba, T., and Sheen, J. (2002). A unique short-chain dehydrogenase/reductase in Arabidopsis glucose signaling and abscisic acid biosynthesis and functions. Plant Cell 14, 2723–2743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chia, D.W., Yoder, T.J., Reiter, W.-D., and Gibson, S.I. (2000). Fumaric acid: An overlooked form of fixed carbon in Arabidopsis and other plant species. Planta 211, 743–751. [DOI] [PubMed] [Google Scholar]
- Church, G., and Gilbert, W. (1984). Genomic sequencing. Proc. Natl. Acad. Sci. USA 81, 1991–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S., and Bent, A. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Coruzzi, G.M., and Zhou, L. (2001). Carbon and nitrogen sensing and signaling in plants: Emerging ‘matrix effects’. Curr. Opin. Plant Biol. 4, 247–253. [DOI] [PubMed] [Google Scholar]
- Eckardt, N.A. (2002). Abscisic acid biosynthesis gene underscores the complexity of sugar, stress, and hormone interactions. Plant Cell 14, 2645–2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis, C., Karafyllidis, I., Wasternack, C., and Turner, J. (2002). The Arabidopsis mutant cev1 links cell wall signaling to jasmonate and ethylene responses. Plant Cell 14, 1557–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldmann, K. (1991). T-DNA insertion mutagenesis in Arabidopsis: Mutational spectrum. Plant J. 1, 71–82. [Google Scholar]
- Finkelstein, R., and Lynch, T. (2000. a). The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell 12, 599–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein, R., and Lynch, T. (2000. b). Abscisic acid inhibition of radicle emergence but not seedling growth is suppressed by sugars. Plant Physiol. 122, 1179–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein, R., Gampala, S., and Rock, C. (2002). Abscisic acid signaling in seeds and seedlings. Plant Cell 14 (suppl.), S15–S45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein, R., and Gibson, S.I. (2002). ABA and sugar interactions regulating development: “Cross-talk” or “voices in a crowd”? Curr. Opin. Plant Biol. 5, 26–32. [DOI] [PubMed] [Google Scholar]
- Finkelstein, R., and Rock, C. (2002). Abscisic acid biosynthesis and signaling. In The Arabidopsis Book, C.R. Somerville and E.M. Meyerowitz, eds (Rockville, MD: American Society of Plant Biologists).
- Fujita, H., and Syono, K. (1996). Genetic analysis of the effects of polar auxin transport inhibitors on root growth in Arabidopsis thaliana. Plant Cell Physiol. 37, 1094–1101. [DOI] [PubMed] [Google Scholar]
- Gamborg, O. (1968). Nutrient requirements of suspension cultures of soybean root cells. Exp. Cell Res. 50, 151–158. [DOI] [PubMed] [Google Scholar]
- Gazzarrini, S., and McCourt, P. (2001). Genetic interactions between ABA, ethylene and sugar signaling pathways. Curr. Opin. Plant Biol. 4, 387–391. [DOI] [PubMed] [Google Scholar]
- Ghassemian, M., Nambara, E., Cutler, S., Kawaide, H., Kamiya, Y., and McCourt, P. (2000). Regulation of abscisic acid signaling by the ethylene response pathway in Arabidopsis. Plant Cell 12, 1117–1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson, S.I. (2000). Plant sugar-response pathways. Part of a complex regulatory web. Plant Physiol. 124, 1532–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajela, R., Horvath, D., Gilmour, S., and Thomashow, M. (1990). Molecular cloning and expression of cor (cold-regulated) genes in Arabidopsis thaliana. Plant Physiol. 93, 1246–1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haouazine-Takvorian, N., Tymowska-Lalanne, Z., Takvorian, A., Tregear, J., Lejeune, B., Lecharny, A., and Kreis, M. (1997). Characterization of two members of the Arabidopsis thaliana gene family, At-β-fruct3 and At-β-fruct4, coding for vacuolar invertases. Gene 197, 239–251. [DOI] [PubMed] [Google Scholar]
- Haughn, G., and Somerville, C. (1986). Sulfonylurea-resistant mutants of Arabidopsis thaliana. Mol. Gen. Genet. 204, 430–434. [Google Scholar]
- Himmelbach, A., Yang, Y., and Grill, E. (2003). Relay and control of abscisic acid signaling. Curr. Opin. Plant Biol. 6, 470–479. [DOI] [PubMed] [Google Scholar]
- Hoth, S., Morgante, M., Sanchez, J.-P., Hanafey, M., Tingey, S., and Chua, N.-H. (2002). Genome-wide gene expression profiling in Arabidopsis thaliana reveals new targets of abscisic acid and largely impaired gene regulation in the abi1-1 mutant. J. Cell Sci. 115, 4891–4900. [DOI] [PubMed] [Google Scholar]
- Hugouvieux, V., Kwak, J., and Schroeder, J. (2001). A mRNA cap binding protein, ABH1, modulates early abscisic acid signal transduction in Arabidopsis. Cell 106, 477–487. [DOI] [PubMed] [Google Scholar]
- Huijser, C., Kortstee, A., Pego, J., Weisbeek, P., Wisman, E., and Smeekens, S. (2000). The Arabidopsis SUCROSE UNCOUPLED-6 gene is identical to ABSCISIC ACID INSENSITIVE-4: Involvement of abscisic acid in sugar responses. Plant J. 23, 577–585. [DOI] [PubMed] [Google Scholar]
- Ishitani, M., Xiong, L., Stevenson, B., and Zhu, J.-K. (1997). Genetic analysis of osmotic and cold stress signal transduction in Arabidopsis: Interactions and convergence of abscisic acid-dependent and abscisic acid-independent pathways. Plant Cell 9, 1935–1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, L., Milne, J., Ashford, D., and McQueen-Mason, S. (2003). Cell wall arabinan is essential for guard cell function. Proc. Natl. Acad. Sci. USA 100, 11783–11788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch, K.E. (1996). Carbohydrate-modulated gene expression in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47, 509–540. [DOI] [PubMed] [Google Scholar]
- Konieczny, A., and Ausubel, F.A. (1993). A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers. Plant J. 4, 403–410. [DOI] [PubMed] [Google Scholar]
- Kuhn, J., and Schroeder, J. (2003). Impacts of altered RNA metabolism on abscisic acid signaling. Curr. Opin. Plant Biol. 6, 463–469. [DOI] [PubMed] [Google Scholar]
- Kwak, J.M., Moon, J.-H., Murata, Y., Kuchitsu, K., Leonhardt, N., DeLong, A., and Schroeder, J.I. (2002). Disruption of a guard cell-expressed protein phosphatase 2A regulatory subunit, RCN1, confers abscisic acid insensitivity in Arabidopsis. Plant Cell 14, 2849–2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laby, R., Kincaid, M., Kim, D., and Gibson, S. (2000). The Arabidopsis sugar-insensitive mutants sis4 and sis5 are defective in abscisic acid synthesis and response. Plant J. 23, 587–596. [DOI] [PubMed] [Google Scholar]
- Leon, P., and Sheen, J. (2003). Sugar and hormone connections. Trends Plant Sci. 8, 110–116. [DOI] [PubMed] [Google Scholar]
- Leung, J., and Giraudat, J. (1998). Abscisic acid signal transduction. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49, 199–222. [DOI] [PubMed] [Google Scholar]
- Lovegrove, A., and Hooley, R. (2000). Gibberellin and abscisic acid signalling in aleurone. Trends Plant Sci. 5, 102–110. [DOI] [PubMed] [Google Scholar]
- McCourt, P. (1999). Genetic analysis of hormone signaling. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 219–243. [DOI] [PubMed] [Google Scholar]
- Mercier, R., and Gogarten, J. (1995). A second cell wall acid invertase gene in Arabidopsis thaliana. Plant Physiol. 107, 659–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nambara, E., Hayama, R., Tsuchiya, Y., Nishimura, M., Kawaide, H., Kamiya, Y., and Naito, S. (2000). The role of ABI3 and FUS3 loci in Arabidopsis thaliana on phase transition from late embryo development to germination. Dev. Biol. 220, 412–423. [DOI] [PubMed] [Google Scholar]
- Pagant, S., Bichet, A., Sugimoto, K., Lerouxel, O., Desprez, T., McCann, M., Lerouge, P., Vernhettes, S., and Hofte, H. (2002). KOBITO1 encodes a novel plasma membrane protein necessary for normal synthesis of cellulose during cell expansion in Arabidopsis. Plant Cell 14, 2001–2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parcy, F., Valon, C., Raynal, M., Gaubier-Comella, P., Delseny, M., and Giraudat, J. (1994). Regulation of gene expression programs during Arabidopsis seed development: Roles of the ABI3 locus and of endogenous abscisic acid. Plant Cell 6, 1567–1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postlethwait, S., and Nelson, O. (1957). A chronically wilted mutant of maize. Am. J. Bot. 44, 628–633. [Google Scholar]
- Rock, C., and Ng, P. (1999). Dominant Wilty mutants of Zea mays (Poaceae) are not impaired in abscisic acid perception or metabolism. Am. J. Bot. 86, 1796–1800. [PubMed] [Google Scholar]
- Rohde, A., De Rycke, R., Beeckman, T., Engler, G., Van Montagu, M., and Boerjan, W. (2000). ABI3 affects plastid differentiation in dark-grown Arabidopsis seedlings. Plant Cell 12, 35–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolland, F., Moore, B., and Sheen, J. (2002). Sugar sensing and signaling in plants. Plant Cell 14 (suppl.), S185–S205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roman, G., Lubarsky, B., Kieber, J.J., Rothenberg, M., and Ecker, J.R. (1995). Genetic analysis of ethylene signal transduction in Arabidopsis thaliana: Five novel mutant loci integrated into a stress response pathway. Genetics 139, 1393–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rook, F., Corke, F., Card, R., Munz, G., Smith, C., and Bevan, M.W. (2001). Impaired sucrose-induction mutants reveal the modulation of sugar-induced starch biosynthetic gene expression by abscisic acid signalling. Plant J. 26, 421–433. [DOI] [PubMed] [Google Scholar]
- Schwebel-Dugue, N., El Mtili, N., Krivitzky, M., Jean-Jacques, I., Williams, J., Thomas, M., Kreis, M., and Lecharny, A. (1994). Arabidopsis gene and cDNA encoding cell-wall invertase. Plant Physiol. 104, 809–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheen, J., Zhou, L., and Jang, J. (1999). Sugars as signaling molecules. Curr. Opin. Plant Biol. 2, 410–418. [DOI] [PubMed] [Google Scholar]
- Söderman, E., Brocard, I., Lynch, T., and Finkelstein, R. (2000). Regulation and function of the Arabidopsis ABA-insensitive4 (ABI4) gene in seed and ABA response signaling networks. Plant Physiol. 124, 1752–1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solano, R., Stepanova, A., Chao, Q., and Ecker, J. (1998). Nuclear events in ethylene signaling: A transcriptional cascade mediated by ETHYLENE-INSENSITIVE3 and ETHYLENE-RESPONSE-FACTOR1. Genes Dev. 12, 3703–3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steber, C., and McCourt, P. (2001). A role for brassinosteroids in germination in Arabidopsis. Plant Physiol. 125, 763–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturm, A., and Tang, G. (1999). The sucrose-cleaving enzymes of plants are crucial for development, growth and carbon partitioning. Trends Plant Sci. 4, 401–407. [DOI] [PubMed] [Google Scholar]
- Suzuki, M., Ketterling, M.G., Li, Q.-B., and McCarty, D. (2003). Viviparous1 alters global gene expression patterns through regulation of abscisic acid signaling. Plant Physiol. 132, 1664–1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tymowska-Lalanne, Z., and Kreis, M. (1998). Expression of the Arabidopsis thaliana invertase gene family. Planta 207, 259–265. [DOI] [PubMed] [Google Scholar]
- Wang, H., Qi, Q., Schorr, P., Cutler, A.J., Crosby, W., and Fowke, L.C. (1998). ICK1, a cyclin-dependent protein kinase inhibitor from Arabidopsis thaliana interacts with both Cdc2a and CycD3, and its expression is induced by abscisic acid. Plant J. 15, 501–510. [DOI] [PubMed] [Google Scholar]
- Wang, K.L.-C., Li, H., and Ecker, J.R. (2002). Ethylene biosynthesis and signaling networks. Plant Cell 14, S131–S151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weatherwax, S.C., Ong, M.S., Degenhardt, J., Bray, E.A., and Tobin, E.M. (1996). The interaction of light and abscisic acid in the regulation of plant gene expression. Plant Physiol. 111, 363–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, A., Pickett, F., Turner, J., and Estelle, M. (1990). A dominant mutation in Arabidopsis confers resistance to auxin, ethylene and abscisic acid. Mol. Gen. Genet. 222, 377–383. [DOI] [PubMed] [Google Scholar]
- Yanagisawa, S., Yoo, S., and Sheen, J. (2003). Differential regulation of EIN3 stability by glucose and ethylene signalling in plants. Nature 425, 521–525. [DOI] [PubMed] [Google Scholar]
- Ziegelhoffer, E., Medrano, L., and Meyerowitz, E. (2000). Cloning of the Arabidopsis WIGGUM gene identifies a role for farnesylation in meristem development. Proc. Natl. Acad. Sci. USA 97, 7633–7638. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.