Abstract
The microRNA-200 family restricts epithelial-mesenchymal transition (EMT) and metastasis in tumor cell lines derived from mice that develop metastatic lung adenocarcinoma. To determine the mechanisms responsible for EMT and metastasis regulated by this microRNA, we conducted a global LC-MS/MS analysis to compare metastatic and non-metastatic murine lung adenocarcinoma cells which had undergone EMT due to loss of miR-200. An analysis of syngeneic tumors generated by these cells identified multiple novel proteins linked to metastasis. In particular, the analysis of conditioned media, cell surface proteins, and whole cell lysates from metastatic and non-metastatic cells revealed large scale modifications in the tumor microenvironment. Specific increases were documented in extracellular matrix proteins, peptidases, and changes in distribution of cell adhesion proteins in the metastatic cell lines. Integrating proteomic data from three sub-proteomes, we defined constituents of a multilayer protein network that both regulated and mediated the effects of transforming growth factor TGFβ. Lastly, we identified extracellular matrix proteins and peptidases that were directly regulated by miR-200. Taken together, our results reveal how expression of miR-200 alters the tumor microenvironment to inhibit the processes of EMT and metastasis.
Keywords: Proteomics, EMT, metastasis
Introduction
The process of epithelial-to-mesenchymal transition (EMT), characterized by loss of intercellular adhesion and polarity, cytoskeletal reorganization that enhances cell motility, and degradation of the basement membrane has been associated with tumor progression and metastasis (1). Diverse signaling pathways regulate EMT; transforming growth factor beta (TGFβ) and RAS are capable of inducing EMT in most epithelial cell lines, while other pathways such as WNT/beta-catenin, NOTCH, NFK-β have also been shown to regulate EMT (2). Induction of EMT functions in particular through down-regulation of the epithelial adhesion protein E-cadherin (CDH1), and direct repression of Cdh1 has been shown to be under the control of transcriptional regulators ZEB1, ZEB2, TWIST1, SNAIL and SLUG, which also regulate a large number of other epithelial-related genes (3).
The importance of non-coding microRNAs (miRNAs) in tumor development and progression has become increasingly evident. Several miRNAs have been identified as either oncogenes (miR-17–92, miR-155, miR-21) or tumor suppressors (miR-15a, miR-16a, let-7) and some human tumor types can be classified by miRNA signatures (4). The miR-200 family of miRNAs consists of five members (miR-200b, 200a, 429 and miR-200c, 141) that have been demonstrated to have a role in EMT in both normal and malignant cells through double-negative feedback regulation with the ZEB transcription factors and regulation of Cdh1 and Vimentin (Vim) expression (5). This microRNA family has also been demonstrated to have pleiotropic effects, including regulation of stem cell factors and features, indicative of their importance for tissue homeostasis.
We recently demonstrated the importance of miR-200 in EMT and metastasis in a study of metastatic and non-metastatic tumors from a (Kras, p53) murine lung adenocarcinoma model (6). This genetic model has biological features and a global metastatic expression profile that is predictive of poor outcome in early-stage lung cancer (7, 8). Cell lines with high or low metastatic potential were established from these mutant Kras and p53 lung adenocarcinoma tumors, and metastatic tumors displayed a high degree of plasticity, exhibiting characteristics of EMT in tumors and 2D-culture (notably in response to EMT-inducing factors such as TGFβ), but re-expressing epithelial markers and organizing into normal epithelial structures in laminin-rich 3D Matrigel culture. MicroRNA profiling of tumors with high metastatic potential revealed loss of miR-200 as a likely regulator of metastatic potential and overexpression of the miR-200b locus in highly metastatic cells eliminated their ability to undergo EMT and metastasize.
In this work we have performed an in-depth comparative proteomic analysis of cells and tumor tissue derived from lung adenocarcinoma tumors that have undergone EMT and have a high metastatic potential to identify proteins involved in biological pathways related to metastasis (6). Analysis of whole cell lysates, cell surface proteins, and conditioned media identified novel proteins associated with EMT and provides evidence of a complex network of proteins regulating TGFβ. Reverted cells locked in an epithelial state as a result of restoration of miR-200 displayed changes in a multitude of extracellular matrix and cell adhesion proteins, suggesting miR-200 alters the microenvironment and the way in which cells interact with it.
Materials and Methods
Culture and Isotopic Labeling of Cells
Parental wildtype cell lines (393P and 344SQ) derived from lung adenocarcinomas in KrasG12D/p53R172HΔG mice and their derivatives stably expressing a control vector or vector with the miR-200B-200A-429 locus (344SQ_vector and 344SQ_200B) have been previously described (Gibbons, G&D, 2009). The cells were cultured in RPMI media (AthenaES, Baltimore, MD) containing 10% dialyzed fetal bovine serum (FBS) (Gibco) and 13C-lysine or 13C-L-lysine and 13C-L-arginine (Cambridge Isotope Labs) instead of the unlabeled amino acids, for 7–8 passages as previously described (9). The same batch of cells was used for preparation of whole cell lysates, conditioned media and extraction of cell surface proteins. The secreted proteins were obtained by gently washing the cells 3–4 times in PBS prior to addition of media without FBS, followed by growth for 24 hour. During this time, cell viability was confirmed by microscopic observation and cell counting after trypan blue staining. The conditioned media was harvested and cell debris removed by centrifugation at 5000 xg for 10 minutes followed by filtration through a 0.22 μm filter. Total cell lysates were obtained by gently washing ~2×107 cells with PBS, followed by harvesting them in 1 ml (per plate) of PBS containing 1% (w/v) octyl-glucoside (OG) and protease inhibitors (complete protease inhibitor cocktail, Roche Diagnostics, Germany).
Tumors
Syngeneic tumors from the wildtype 393P and 344SQ cells (3 of each tumor type) were generated by subcutaneous injection as previously described (6). At necropsy the tumors were flash-frozen in liquid nitrogen and stored at −80°C until subsequent processing for RNA or protein. For LC-MS/MS analysis, tumors were homogenized on liquid nitrogen and lysed in 8M urea and 1% OG in 0.1M Tris-HCl at 2 ml/gram of tumor.
Isolation of cell surface proteins
Cell surface proteins from the four cell lines, differentially-labeled with heavy or light amino acids, ~2×108 of each, were biotinylated in the culture plate after gentle washing 5 times with PBS. After a 10 minute biotinylation reaction with 10 ml (per plate) of 0.25 mg/ml Sulfo-NHS-SS-Biotin in PBS at room temperature, the reaction was quenched with 15 ml of 10 mM Lysine in PBS. Protein was extracted in 1 ml (per plate) of PBS containing 2% NP-40 and complete protease inhibitor cocktail. Biotinylated proteins were isolated by affinity chromatography using 1 ml of UltraLink Immobilized Neutravidin (Pierce). Proteins bound to the column were recovered by overnight incubation with a solution of 2% octyl-glucoside (OG) and 1mg/ml DTTT in 0.1M Tris-HCl.
Fractionation and mass spectrometry of samples
Data analysis
Enrichment analysis for Gene Ontology terms was performed on the differentially expressed proteins by Database for Annotation, Visualization and Integrated Discovery (DAVID) (10, 11). A 5-fold increase in protein enrichment in the media compared to the whole cell lysates was used as a cutoff to identify proteins that were likely to be secreted or shed, while a 2-fold increase in proteins of the cell surface compared to whole cell lysate was established for cell surface proteins. The TGFβ interacting networks were generated through the use of Ingenuity Pathway Analysis (Ingenuity Systems®). Protein interaction network analysis used the entire set of human protein-protein interactions cataloged in Entrez Gene (downloaded July 2009). Homologene was used to map between mouse genes and human orthologs. Graphical visualization of networks was generated using Cytoscape (12).
Western blot analysis
Cell lysates were prepared by extracting protein with RIPA buffer. For conditioned media, cells were grown for 48 hours in RPMI 1640 with 0.1% FBS, media removed, centrifuged, and filtered through a 0.22 uM filter. Membranes were blocked in 5% non-fat dried milk and incubated overnight at 4°C with appropriate primary antibodies (PDLIM5 (Novus Biologicals), CSRP2 and ETS-1 (Santa Cruz Biotechnology), B-Actin (Sigma-Aldrich), GAPDH (Abcam), CDH1 (BD Biosciences)).
Results
Protein and mRNA profiles of metastatic and non-metastatic tumors
We first performed a comparative proteomic analysis of 344SQ (metastatic) and 393P (non-metastatic) tumors described previously in Gibbons et al. (3 tumors of each type) (6, 7). Tumor lysates were reciprocally labeled with both heavy and light acrylamide, allowing for comparisons of independent heavy/light and light/heavy metastatic vs. non-metastatic tumors, followed by reverse phase fractionation of lysate proteins and LC/MS-MS analysis of peptide digests from each fraction (9). 1261 proteins were quantified in both reciprocally labeled experiments, of which 80 had increased ratios in metastatic vs. non-metastatic and 59 had decreased ratios at a threshold of >1.5 fold change in both labelings (Supplementary Table 1A and B). Among the most highly enriched Gene Ontology categories for the proteins upregulated in metastatic tumors were “response to wounding”, “growth factor binding”, “calcium metal binding” and “extracellular space”, while downregulated proteins were enriched for “antigen processing and presentation” and “plasma membrane”. We observed upregulation of multiple markers associated with mesenchymal cell function or recruitment including CD73, PLAUR, clusterin, fibulin 2, integrin alpha-2, CXCL7, IGFBP3, and LTBP1. We further identified multiple proteins that have not previously been shown to play a role in metastasis including cell adhesion proteins LGALS2 and LRG1, and chitinase CHI3L4.
We next compared the proteomic findings with mRNA expression data previously obtained for the same tumors (6). Overall, there was significant positive correlation between mRNA and protein expression in tumors for both upregulated (p= 3.80E-12) and downregulated (p= 8.03E-12) proteins. However a large number of the differentially expressed proteins were not concordantly expressed at the transcript and protein levels, notably including insulin growth factor binding proteins 3, 4 and 7; clusterin, nucleophosmin, fetuin A and fibrinogen A and B.
Differential protein expression in metastatic and non-metastatic cell lines
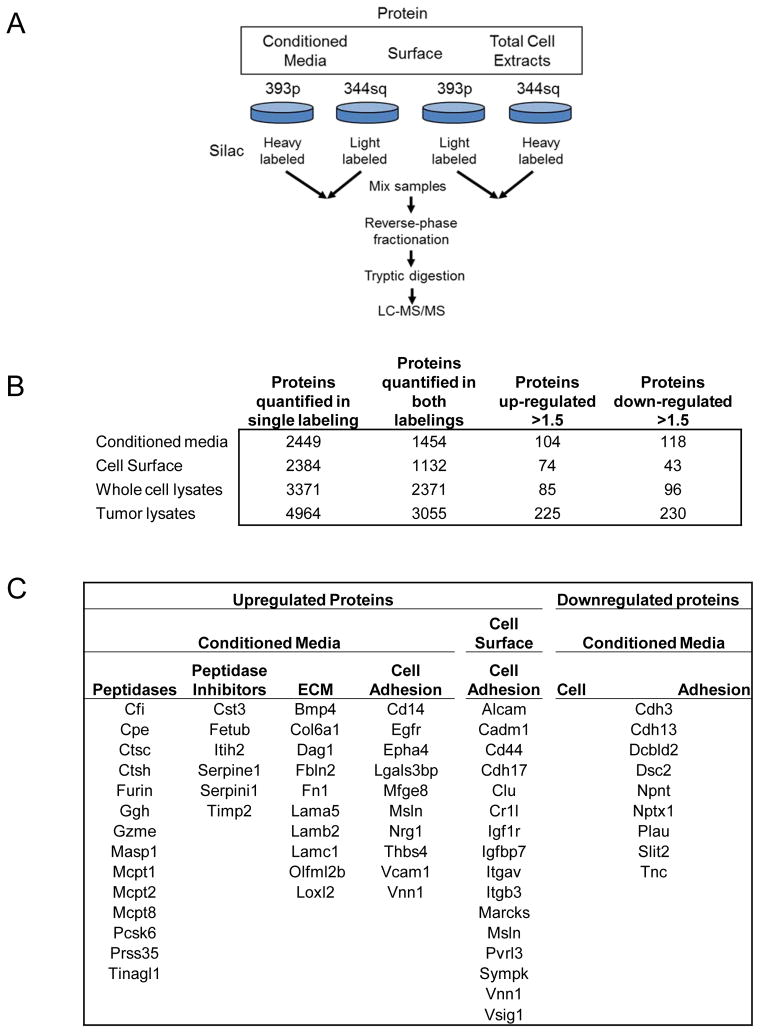
To gain a deeper understanding of the contribution of cell surface and extracellular proteins to metastasis, we performed an in-depth proteomic analysis comparing conditioned media, cell surface proteins, and whole cell lysates of metastatic (344SQ) and non-metastatic (393P) cell lines. Comparative analysis was performed using reciprocal stable isotope labeling by amino acids in cell culture (SILAC), whereby each cell line was grown in both heavy (13C-lysine or 13C-lysine + 13C-argine) and light media (allowing for comparisons of independent heavy/light and light/heavy experiments), followed by reverse phase fractionation of samples and LC/MS-MS protein analysis (9) (Figure 1A). To identify proteins differentially regulated between the metastatic and non-metastatic cell lines, we established a threshold of >1.5 fold change in both of the heavy and light SILAC labeling experiments to eliminate preferential labeling bias or contamination from trace fetal bovine serum in the media (FBS) (Figure 1B, Supplementary Tables S2A and S2B). 656, 543, and 1,299 proteins unique to each compartment were identified in the conditioned media, cell surface, and whole cell lysate (WCL), respectively. Analysis of all 225 upregulated proteins in the metastatic cells revealed Gene Ontology functions associated with immune and inflammatory response, cell adhesion, extracellular matrix, and protease activity (Supplementary Table S3). An increase in the percentage of plasma membrane proteins was found in the conditioned media was also observed, indicative of increased protein shedding from the cell surface. The Gene Ontology categories for proteins downregulated in the 344SQ cells were primarily associated with cytoskeletal regulation, cell-cell adhesion and RNA processing. Overall, substantial changes were observed in the secreted and surface protein fractions, pertaining to components of the cellular microenvironment including ECM components, peptidase and peptidase inhibitor activities and to proteins mediating cellular interactions with the microenvironment (Figure 1C). Peptidases consisting of mast cell proteases MCPT-1, 2, and 8, CFI, PCSK6 and PRSS35 were among the top 10 most highly upregulated proteins in the conditioned media of metastatic cells. Interestingly, cell adhesion molecules were also enriched by Gene Ontology analysis among both the up- and downregulated proteins in the conditioned media and the upregulated proteins on the cell surface. Differentially upregulated proteins in the whole cell lysates were primarily associated with metabolic processes, particularly glutathione metabolism and oxidoreductase activity with upregulation of five glutathione-S-transferases: A2, A4, M1, M2, and M7 in the whole cell lysates of 344SQ cells, while downregulated proteins are enriched for cytoskeletal and actin-related proteins.
Figure 1. Protein quantifications and cellular localization.
A. Overview of the experimental design for mass spectrometry analysis of conditioned media, cell surface proteins and whole cell lysate. B. The number of identified or quantified proteins in each analysis. C. Differentially regulated proteins related to the microenvironment and cellular interaction with it.
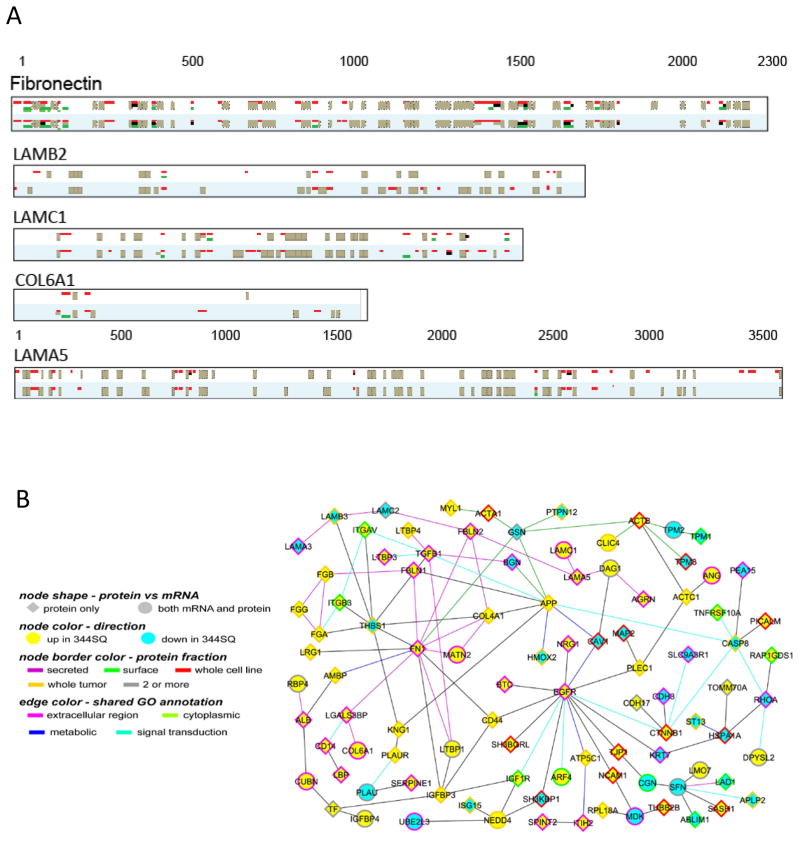
Extracellular matrix proteins COL6A1, LAMA5, LAMB2, LAMC2, fibronectin were all upregulated in 344SQ cells as well as ECM-related proteins LOXL2 which has been shown to stiffen ECM. Peptide analysis of the structural proteins COL6A1, LAMA5, LAMB2, LAMC2 and fibronectin revealed secretion of whole proteins, rather than protein fragments produced by proteolysis, providing evidence in support of tumor cells shaping their own microenvironment (Figure 2A). Upregulation of intact fibronectin in the 344SQ metastatic cells was accompanied by down-regulation of an N-terminal fragment that contains the domains for fibrinogen and collagen binding and inhibits fibronectin fibril formation (13).
Figure 2.
A. Peptide coverage of extracellular matrix proteins. Peptides upregulated in 344sq are red, downregulated peptides green, unchanged peptides black, and non-quantified peptides gray. For each protein, the top panel is labeling mix 1 and the bottom is mix 2. B. Protein-protein interaction network of differentially expressed proteins and genes (mRNAs) in 344SQ cells. Graph is comprised of proteins differentially expressed in any one of the four fractions (fold change>1.5, both duplicates). Nodes, proteins; circle nodes, proteins also differentially expressed at mRNA transcript level (P<0.01, t-test); yellow/blue, overexpression/underexpression in 344SQ, respectively. A line between two nodes signifies that the corresponding proteins can physically interact (according to the literature). Node border color, protein fraction showing differential patterns. Colored edges (other than gray), a common gene ontology term annotation shared by both of the connected proteins.
Overall, substantial concordance was observed between protein expression in cell lines and tumors with respect to metastatic status, with 17 and 16 upregulated or downregulated in common, respectively, and only 3 protein with discordant findings between the two datasets (Table 1A and 1B). The concordance observed between the cell lines and tumors is indicative of the contribution of tumor cells to the tumor proteome. Moreover, we observed significant correlation between tumor mRNA expression and proteins in the conditioned media (p=3.80E-12 for upregulated and p=8.03E-12 for downregulated proteins), cell surface (p= 2.29E-08 for upregulated and 3.01E-09 for downregulated proteins), and whole cell lysates (p=4.67E-16 for upregulated and p=5.80E-21 for downregulated proteins). As a tool for better understanding and illustrating the comparison and complementation of the protein and mRNA changes, we integrated our top differential proteins with the public Entrez Gene database of protein-protein interactions to generate a protein interaction network, in which we labeled those proteins showing corresponding differential changes at the mRNA level (Figure 2B).
Table 1.
Overlap between differentially regulated proteins in tumor lysates and cell lines
| A. Proteins upregulated in metastatic 344SQ tumors | ||||||
|---|---|---|---|---|---|---|
| Protein | Protein
|
mRNA
|
||||
| Conditioned media | Cell surface | Whole cell lysates | Tumor lysates | ratio | p-value | |
|
|
|
|||||
| Acat1 | NA | 1.79 | 1.90 | 1.80 | 1.59 | 4.4E-03 |
| Anxa10 | NA | NA | 2.39 | 5.54 | 16.10 | 5.1E-05 |
| Cdh17 | NC | 2.73 | NC | 1.99 | 0.98 | 9.5E-01 |
| Ckb | ▲ | NA | 2.73 | 2.47 | 2.78 | 7.3E-03 |
| Clu | 8.70 | 5.18 | 2.47 | 4.84 | 1.17 | 5.3E-02 |
| Fbln2 | 7.16 | NC | 2.14 | 2.03 | 0.54 | 9.4E-03 |
| Igfbp4 | 2.14 | NA | ▼ | 2.15 | 1.23 | 5.9E-04 |
| Igfbp7 | 12.92 | 5.10 | 8.97 | 3.02 | 1.23 | 1.4E-02 |
| LOC677317 | ▲ | NA | 2.56 | 2.71 | 1.59 | 1.3E-01 |
| Mcpt2 | 34.61 | NA | NA | 3.29 | 5.86 | 6.2E-04 |
| Msln | 3.97 | 12.10 | 2.94 | 1.98 | 1.44 | 7.1E-03 |
| Nt5e | NA | NA | 3.09 | 3.61 | 2.26 | 6.3E-04 |
| Procr | ▲ | ▲ | 3.29 | 2.74 | 1.24 | 3.2E-01 |
| Pxdn | 3.58 | NA | NA | 3.27 | 1.54 | 1.9E-02 |
| Rbp4 | 3.62 | NA | NA | 3.04 | 7.49 | 4.3E-05 |
| Sftpb | 18.59 | 13.68 | 8.48 | 4.39 | 46.29 | 3.1E-05 |
| Tspan8 | NC | NC | 3.46 | 3.22 | 2.56 | 2.6E-03 |
| B. Proteins downregulated in metastatic 344SQ tumors | ||||||
|---|---|---|---|---|---|---|
| Protein | Protein
|
mRNA
|
||||
| Conditioned media | Cell surface | Whole cell lysates | Tumor lysates | ratio | p-value | |
|
|
|
|||||
| Anxa6 | ▲ | NC | 0.59 | 0.58 | 0.49 | 4.9E-03 |
| Cald1 | ▼ | 0.43 | NC | 0.56 | 0.85 | 7.8E-01 |
| Capg | ▼ | NA | 0.52 | 0.37 | 0.55 | 3.1E-03 |
| Cda | NA | ▼ | 0.31 | 0.42 | 0.45 | 3.2E-06 |
| Crip1 | 0.57 | NC | NC | 0.44 | 0.84 | 3.0E-02 |
| H2-K1 | 0.41 | NA | NA | 0.42 | 0.72 | 1.3E-01 |
| Ifi204 | NA | 0.56 | 0.21 | 0.28 | 0.89 | 6.3E-01 |
| Lamc2 | ▼ | 0.50 | NA | 0.27 | 0.81 | 7.3E-02 |
| Ly6a | 0.20 | NA | 0.23 | 0.33 | 0.42 | 3.8E-03 |
| Raly | NA | NA | 0.39 | 0.39 | 0.67 | 1.5E-02 |
| S100a14 | NA | 0.30 | 0.15 | 0.09 | 0.01 | 4.9E-08 |
| Sfn | ▼ | NC | 0.28 | 0.10 | 0.44 | 8.6E-04 |
| Sh3bgrl2 | 0.44 | NA | 0.43 | 0.64 | 1.00 | 9.8E-01 |
| Tacstd1 | NC | NC | 0.15 | 0.21 | 0.08 | 1.7E-04 |
| Uchl1 | NA | NA | 0.25 | 0.53 | 0.82 | 3.0E-01 |
| Zyx | 0.53 | NA | NC | 0.61 | 0.51 | 2.9E-04 |
Values are weighted means of indepdendent and reciprocally labeled replicates. Proteins up or downregulated, but less than 1.5 are indicated by “▲” or “▼”. “NC” indicates the replicates are not concordant. “NA” indicates the protein was not quantified.
Protein components of the TGFβ network in metastatic tumors and cell line compartments
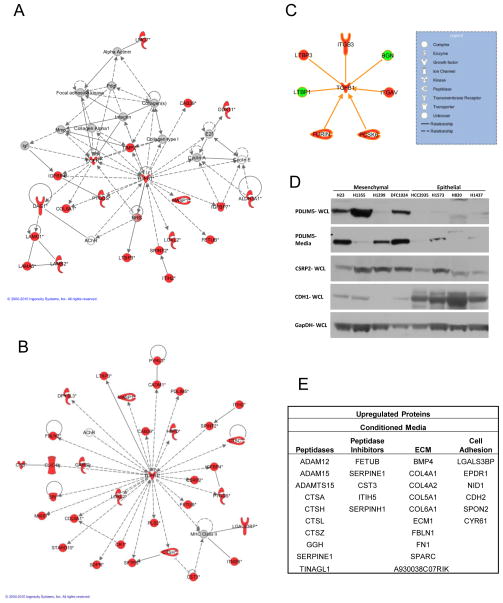
We next used Ingenuity Pathway Analysis (IPA) tools to identify potential regulatory pathways accounting for the differences between the metastatic and non-metastatic cells. Analysis of differentially regulated proteins from the conditioned media, cell surface, and whole cell lysates revealed regulatory nodes associated with NFκB, fibronectin and p38 MAPK (Supplementary Figure S1). Networks with TGFβ-1 were also identified in the individual compartments, particularly the conditioned media (Figure 3A, see Supplementary Figure S2 for TGFβ containing networks from all sub-proteomes). However, analysis of the combined upregulated proteins by IPA identified TGFβ-1 as the central regulatory node in the most highly significant network, with an IPA significance score of 64 versus 33 for the second network generated (Figure 3B). This finding highlights the power of data synthesis from multiple cellular compartments to enhance the ability to find master regulators, as TGFβ-1 was identified as one of several regulatory proteins in the individual analyses but was shown to be the dominant regulator in the analysis of combined protein compartments. Furthermore, we observed evidence of a multilayer regulation of TGFβ-1 in the differential expression of TGFβ-1 regulatory proteins. Proteases PCSK6 (also known as PACE4) and FURIN activate TGFβ-1 through proteolytic cleavage, TGFβ latency complex protein LTBP3 and Integrins αV and β3, which have been shown to be involved in TGFβ-1 activation, were all upregulated in 344SQ cells while the TGFβ latency complex protein, LTBP1 and the TGFβ-1 binding proteins BGN were downregulated (Figure 3C) (14, 15). We further identified isoform differences in the latent transforming growth factor binding proteins (LTBP’s). An N-terminal peptide corresponding to cleavage at the LTBP3 hinge region which elutes out earlier by reverse phase HPLC is reduced in metastatic 344SQ cells while there is upregulation of the full length protein. The LTBP1 proteins are downregulated in 344SQ cells and also show likely downregulation of an N-terminal product, although it is larger than the cleavage product resulting from processing of LTBP1 at the hinge region (16).
Figure 3. TGFβ signaling in metastatic cells.
A. The most significant network from Ingenuity Pathway Analysis for upregulated proteins in the conditioned media shows evidence of TGFβ-1 regulation. B. Network analysis of upregulated proteins combined from conditioned media, cell surface and total cell extracts reveals a stronger and more significant node than from the individual compartments C. Proteins directly interacting with and regulating TGFβ-1. Network objects colored red indicate upregulation and green objects indicate downregulation. D. Western blots comparing protein expression in epithelial and mesenchymal NSCLC cell lines. E. Microenvironment related proteins downregulated with miR-200 expression.
Identification of novel metastasis associated proteins
Integrated data analysis of lysate, conditioned media and cell surface components for proteins associated with metastasis yielded several novel proteins. LRRC8 is a known endoplasmic reticulum protein which we found to be upregulated on the surface of metastatic cells. Cytoskeletal protein PDLIM5 was also upregulated on the surface of metastatic cells and showed evidence of cleavage, with the first half and the second half of the protein eluting out in different fractions. Another lim containing protein, CSRP2, was upregulated and occurred as an intact protein in the cell surface fraction. Many novel secreted proteins were also upregulated. Fibulin 2, previously suggested to be a tumor suppressor was upregulated 7-fold, MASP1 and peroxidasin were upregulated 3-fold. In addition several cytoplasmic and nuclear proteins were found to be secreted by metastatic cells including ST3GAL4, ST6GAL1, SIL1, and SDF4. Expression of several proteins upregulated in 344SQ cells was evaluated in human non-small cell lung cancer cell lines (NSCLC) with mesenchymal or epithelial features. CSRP2 and PDLIM5 were expressed at higher levels in cells with mesenchymal features than more epithelial cell lines. PDLIM5 was further upregulated in conditioned media from mesenchymal NSCLC cell lines (Figure 3D). These findings demonstrate the applicability of findings in the mouse model to human lung cancer.
Identification of miR-200 regulated proteins
miR-200 regulates EMT and metastasis at least in part through a negative regulatory loop with the Zeb1/2 family of transcriptional repressors (6, 17). As one of the primary biological differences between the metastatic 344SQ cells and the non-metastatic 393P cells is the expression of the miR-200 family members, we investigated the effect of miR-200 on protein expression. For this analysis, 344SQ cell lines (which normally have low miR-200 expression) with stable miR200b-200a-429 (344SQ_miR-200) expression or a vector control (344SQ_vector) were established and their protein constituents analyzed by LC-MS/MS. We identified 193 upregulated proteins and 179 downregulated proteins in the combined sub-proteomes in 344SQ_miR-200 cells (Supplementary Tables S4A, S4B and S4C). Gene Ontology analysis of differentially regulated proteins revealed similar findings to the 344SQ/393P analysis, as categories associated with peptidase activity, cell adhesion and extracellular matrix among the proteins downregulated with miR-200 restoration, while proteins upregulated with miR-200 restoration were associated primarily with cytoskeletal regulation and cell adhesion (Supplementary Tables S5).
Restoration of miR-200 expression affects the microenvironment through protein shedding and secretion
To ascertain changes in cellular functions after restoration of miR-200 expression, we assessed the differentially regulated proteins from conditioned media, cell surface, and whole cell lysates. The most striking effect of miR-200 expression was a change in protein constituents in the media resulting from protein secretion and shedding with downregulation of extracellular matrix, peptidases and cell adhesion proteins in the conditioned media from the 344SQ_miR-200 cells (Figure 3E). Twenty-two proteins upregulated in the 344SQ/393P conditioned media were downregulated in conditioned media from 344SQ_miR-200 cells, suggesting direct regulation by miR-200 (Table 2A). Furthermore, there was significant correlation between protein and mRNA expression in the downregulated proteins from the 344SQ_miR-200/vector control comparison (p=6.38E-12), supporting the role miR-200 plays in altering the cellular microenvironment. To validate proteins regulated by miR-200, we analyzed expression of proteins downregulated after miR-200 expression in 344SQ cells in a set of human NSCLC cell lines for which we have microRNA expression data. Several proteins in each compartment correlated with miR-200 family expression at both the RNA and protein level in NSCLC human cell lines (Table 2B), including known miR-200 targets such as CDH1, but also EPS8L2, PLS1, LSR and others.
Table 2.
miR-200 regulation of proteins
| A. Secreted proteins regulated by miR-200 | ||||||
|---|---|---|---|---|---|---|
| Gene | Protein
|
mRNA
|
||||
| 344SQ/393P
|
344SQ_miR-200/344SQ_control
|
344SQ/393P
|
344SQ_miR-200/344SQ_control
|
|||
| ratio | ratio | ratio | p-value | ratio | p-value | |
|
|
|
|
|
|||
| B4galt5 | 4.60 | 0.39 | 1.20 | 3.35E-01 | 1.29 | 1.48E-01 |
| Bmp4 | 2.90 | 0.43 | 4.88 | 5.13E-03 | 0.17 | 7.89E-03 |
| Col6a1 | 3.47 | 0.57 | 1.97 | 3.98E-03 | 0.71 | 6.40E-02 |
| Cst3 | 1.76 | 0.49 | 1.05 | 5.39E-01 | 0.46 | 2.79E-04 |
| Ctsc | 3.74 | 0.48 | 1.27 | 2.83E-01 | 1.23 | 2.79E-01 |
| Ctsh | 5.36 | 0.28 | 1.83 | 9.79E-03 | 0.26 | 1.15E-02 |
| Egfr | 2.84 | 0.39 | 1.24 | 5.72E-02 | 1.16 | 5.88E-02 |
| Fetub | 2.98 | 0.47 | 0.87 | 3.34E-02 | 0.87 | 2.69E-01 |
| Fn1 | 1.73 | 0.48 | 0.88 | 9.23E-02 | 2.15 | 7.12E-02 |
| Ggh | 2.91 | 0.59 | 0.69 | 4.85E-01 | 1.01 | 9.24E-01 |
| Gnptg | 2.26 | 0.39 | 1.09 | 5.20E-01 | 0.78 | 2.70E-01 |
| Lgals3bp | 6.52 | 0.42 | 1.88 | 1.04E-02 | 0.20 | 2.26E-03 |
| Loxl2 | 4.34 | 0.36 | 0.87 | 7.84E-01 | 0.67 | 1.77E-01 |
| Man2b1 | 2.59 | 0.44 | 0.92 | 2.46E-01 | 1.04 | 4.62E-01 |
| Manba | 2.67 | 0.39 | 1.20 | 3.73E-01 | 0.59 | 2.39E-01 |
| Mcpt2 | 34.61 | 0.10 | 5.86 | 6.16E-04 | 0.01 | 6.88E-05 |
| Mcpt8 | 12.11 | 0.58 | 3.97 | 2.22E-03 | 0.64 | 8.32E-02 |
| Mfge8 | 5.06 | 0.61 | 0.99 | 5.92E-01 | 0.86 | 4.23E-01 |
| Prl2c4 | 2.60 | 0.25 | 0.37 | 2.57E-02 | 0.36 | 3.29E-02 |
| Serpine1 | 6.95 | 0.25 | 1.13 | 1.09E-01 | 0.68 | 3.56E-01 |
| Sftpb | 18.59 | 0.08 | 46.29 | 3.10E-05 | 0.02 | 1.06E-05 |
| Tinagl1 | 4.04 | 0.52 | 1.24 | 2.09E-02 | 0.89 | 2.43E-01 |
| B. Protein and gene expression correlation with miR-200 family members in human NSCLC cell lines | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Conditioned Media | 344SQ_miR-200/344SQ_control
|
Protein
|
mRNA
|
||||||||
| protein ratio | miR-200a | miR-200b | miR-429 | miR-200c | miR-141 | miR-200a | miR-200b | miR-429 | miR-200c | miR-141 | |
|
|
|
|
|||||||||
| CDH1 | 1.67 | 0.59 | 0.56 | 0.37 | 0.7 | 0.7 | 0.71 | 0.74 | 0.7 | 0.92 | 0.92 |
| EPS8L2 | 2.05 | 0.53 | 0.56 | 0.39 | 0.59 | 0.63 | 0.56 | 0.51 | 0.44 | 0.22 | 0.22 |
| IRF2BP2 | 3.07 | 0.14 | 0.41 | 0.36 | 0.18 | 0.16 | 0.34 | 0.46 | 0.39 | 0.4 | 0.42 |
| KRT7 | 2.52 | 0.63 | 0.59 | 0.53 | 0.53 | 0.55 | 0.55 | 0.52 | 0.48 | 0.49 | 0.48 |
| KRT8 | 1.78 | 0.23 | 0.49 | 0.27 | 0.44 | 0.49 | 0.32 | 0.31 | 0.26 | 0.48 | 0.46 |
| KRT19 | 2.24 | 0.6 | 0.59 | 0.45 | 0.58 | 0.59 | 0.65 | 0.59 | 0.52 | 0.76 | 0.76 |
| Cell Surface | |||||||||||
|
|
|
|
|||||||||
| Atp1b1 | 1.65 | 0.39 | 0.45 | 0.45 | 0.39 | 0.42 | 0.4 | 0.44 | 0.38 | 0.5 | 0.5 |
| F3 | 1.64 | 0.49 | 0.45 | 0.32 | 0.42 | 0.4 | 0.67 | 0.66 | 0.61 | 0.57 | 0.56 |
| F11R | 1.71 | 0.35 | 0.44 | 0.24 | 0.31 | 0.34 | 0.54 | 0.61 | 0.57 | 0.57 | 0.58 |
| LSR | 1.57 | 0.45 | 0.43 | 0.42 | 0.43 | 0.4 | 0.42 | 0.38 | 0.35 | 0.53 | 0.53 |
| SDCBP2 | 1.89 | 0.38 | 0.3 | 0.21 | 0.41 | 0.42 | 0.6 | 0.62 | 0.5 | 0.62 | 0.63 |
| Whole Cell lysate | |||||||||||
|
|
|
|
|||||||||
| EPS8L2 | 1.65 | 0.55 | 0.57 | 0.37 | 0.57 | 0.58 | 0.56 | 0.51 | 0.44 | 0.22 | 0.22 |
| GOLGA2 | 1.63 | 0.35 | 0.26 | 0.26 | 0.44 | 0.4 | 0.39 | 0.44 | 0.38 | 0.59 | 0.59 |
| PLS1 | 1.60 | 0.44 | 0.55 | 0.36 | 0.44 | 0.46 | 0.63 | 0.62 | 0.62 | 0.5 | 0.49 |
Table 1A. Overlap between proteins upregulated in 344SQ cells and downregulated after re-expression of miR-200. Values for proteins are weighted averages of reciprocal labeling experiments. B. Correlation coefficients with miR-200 family members in 38 human NSCLC cell lines. Spearman and Pearson correlation coefficients are listed for protein and mRNA expression, respectively, with r values having a p<0.01 in bold.
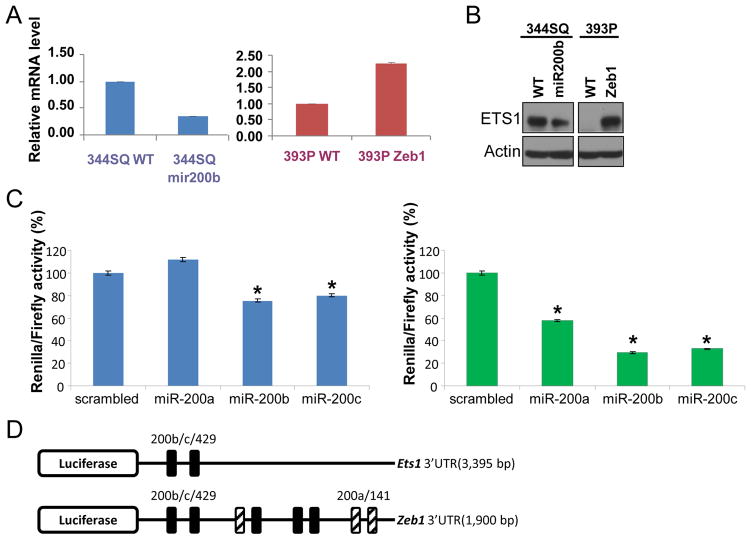
We have previously shown that miR-200 expression alters many genes at the expression level through an indirect effect. Though restoring miR-200 expression in 344SQ cells reverted the epithelial-to-mesenchymal transition, the small number of overlapping proteins upregulated in 344SQ cells and downregulated after miR-200 expression is restored suggests the occurrence of regulatory mechanisms other than direct inhibition of miR-200. To elucidate alternative mechanisms for regulation of genes associated with EMT, the publicly available software package Amadeus was used to search for common DNA motifs in promoter sequences from differentially regulated genes (Supplementary Table S6) (18). One transcription factor identified in upregulated proteins is the oncogene C-ets-1, a member of the ETS family that has been shown to be upregulated in invasive cancers and to be an effector of TGFβ induced EMT, by upregulating Zeb1 (19, 20). ZEB1 DNA binding elements were enriched in both the downregulated mRNA and protein datasets (Suplementary Table S7). Zeb1 is a validated target of miR-200 that was previously demonstrated to be upregulated in 344SQ cells, while C-ets-1 was recently demonstrated to be a direct miR-200 target in human endothelial cells. Expression of C-ets-1 was regulated by miR-200 at both the mRNA and protein level (Figure 4A and 4B). Activity of a luciferase reporter containing the 3′UTR for C-ets-1 was directly regulated by co-transfection of miR-200B and C, but not miR-200A, as predicted from the seed sequence sites found in the 3′ UTR (Figure 4C and 4D). This is in contrast to the 3′ UTR for Zeb1, which has documented sites for both of the miR-200 family seed sequences. One other transcription factor, AP-2REP, was also identified in both the downregulated mRNA and protein data sets and has been shown to be amplified in invasive gastric cancer and salivary tumors (21). ATF transcription factors, known to mediate and regulate effects of TGFβ were also enriched in the upregulated mRNA’s. Interestingly, binding sites for ETS2, which shares overlapping function with ETS1 during mouse development, was identified in the downregulated mRNA, suggesting differential roles for ETS1 and ETS2 during tumor progression, a finding supported by transcript analysis in human lung cancer cell lines (data not shown).
Figure 4. ETS-1 regulation by miR-200.
A. Quantitative RT-PCR analysis of ETS-1 mRNA levels in 344SQ or 393P cells stably transfected with an empty vector control (WT), the miR-200b/a/429 locus (200b), or Zeb1 normalized on the basis of L32 ribosomal protein mRNA levels and expressed as mean values of triplicate cultures relative to control transfectants, which were set at 1.0. B. Western blot analysis of the same cell lines. C. 344SQ cells were transiently co-transfected with the indicated pre-miRs or scrambled oligomer (10 nM) and reporter plasmids (500 ng) that are linked to the full-length 3′-UTR of ETS-1 (left) or Zeb1 (right). Results were normalized on the basis of renilla luciferase and expressed as the mean values of triplicate wells. * p<0.05. D. Map of 3′UTR’s for luciferase assays showing miR-200 target sites.
Discussion
In this work we have performed an in-depth systems analysis of metastatic lung tumors which spontaneously undergo EMT, identifying changes in the deposition of extracellular matrix, protease function and cell adhesion. While prior studies of EMT using proteomics have been primarily based on total lysates from cancer cell lines, we expanded on previous findings with analysis of tumors and sub-proteomes from primary cell lines (22–24). Our initial proteomic analysis comparing metastatic and non-metastasizing primary cell lines grown as syngenic murine tumors revealed a large number of proteins (80 up and 59 down) potentially involved in tumor progression, while analysis by LC-MS/MS of cultured cell lines labeled in vitro enabled protein identification and quantitation of intracellular, cell surface, and secreted/shed proteins, greatly increasing the total number of differentially regulated proteins and providing insight into protein processing. Integration of data from the sub-proteomes enabled identification of relevant biological functions such as changes in extracellular matrix and cell adhesion and pertinent regulatory networks, notably regulators of the metastatic driver TGFβ-1. Findings included increased expression of the proteolytic activators FURIN and PCSK6 in the conditioned media, as well as differential regulation of latent transforming binding proteins.
Extracellular matrix regulation of cell behavior is transmitted through cell surface receptors, embedded cytokines and growth, as well as by post-translational modification and matrix stiffness. Numerous changes were observed in extracellular matrix proteins, proteases and cell adhesion in the metastatic 344SQ cells, including upregulation of full length Lamins A5, B2 and C1, Collagen 6A1, fibronectin, Loxl2, a protein involved in the stiffening of collagen, and biglycan, a collagen binding partner. While the classical model of tumor progression includes degradation of the extracellular matrix for cells to invade through the basement membrane, we observed increased production of particular full-length extracellular matrix proteins without evidence of degradation. These effects are in part driven by miR-200 expression as there was significant downregulation of ECM proteins after miR-200 re-expression. Interestingly, while there was an increase in laminins (Laminin Alpha 5, Beta2 and Gamma 1) secreted by the 344SQ cells, collagens (Collagens 4A1, 4A2, 5A1, 6A1) were the primary ECM structural component reduced with miR-200 restoration. Increased extracellular matrix production has been observed in several cancer types; oral squamous cell carcinomas, colorectal cancer, breast cancer (25, 26). The apparent switch from laminins to collagens, along with expression of fibronectin and lysyl oxidase homolog 2, stiffens the matrix, a finding which has been shown to aid in tumor progression in several tumor models, but not so far in lung cancer (reviewed in (27)). This downregulation of collagens and matrix stiffening proteins with miR-200 expression implies a specific novel role for miR-200 in collagen production. Other microRNAs have previously been demonstrated to play roles in fibrosis through regulation of ECM proteins; miR-29 family members downregulate collagens and fibrillins in hepatic and cardiac fibroblasts, while mir-21 mediates pulmonary fibrosis, but this is the first evidence of microRNA effects on the microenvironment during tumorigenesis (28, 29). Recent work by Korpal et al. identified Sec23a as a miR-200 target important for mediating the secretion of metastasis-related proteins in breast cancer cell line(30). Many of the proteins they identified as being miR-200 or Sec23a-dependent were also identified in our study, such as IGFBP4, Tinagl1, and Ltbp3, although the total number of differentially-regulatd proteins in our study was greater, likey due to the higher resolution provided by more extensive fractionation. Sec23a itself was upregulated in the mRNA expression analysis, but only in one of the two 344SQ/393P proteomic analyses, not meeting our stringent threshold for upregulation.
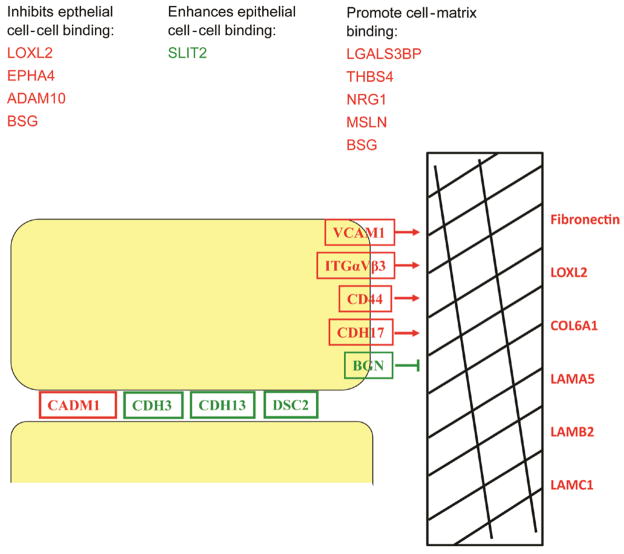
In addition to ECM structural proteins, we also observed differential regulationof peptidases and cell adhesion proteins in 344SQ cells and downregulation of peptidases with miR-200 restoration. Of the 22 proteins in the 344SQ/393P conditioned media that appear to be directly regulated by miR-200, 8 were peptidases or peptidase inhibitors. Expression and processing of cell adhesion molecules were also modified in 344SQ cells and closer investigation of the specific cellular adhesion proteins reveals a shift from proteins functioning in cell-cell adhesion and epithelial phenotype to cell-matrix adhesion and mesenchymal phenotype (Figure 5). Cell surface adhesion molecules CDH17, Integrins αV and β3, CD44, all directly bind cells to extracellular matrix while secreted LGALS3BP, Neuregulin and THBS4 increase cell-matrix adhesion (31, 32) and Adam10, Loxl2, and EPHA4 inhibit cell-cell adhesion (33–35). Adhesion proteins downregulated in the media, such as Cadherins 3, 13, Desmocollin 2 and SLIT2 promote epithelial cell-cell adhesion and loss is associated with increases in invasion and tumor progression (36–38). The changes in extracellular matrix, peptidases and cell adhesion proteins represent remodeling of the microenvironment and the cell surface after or coincident with EMT.
Figure 5. Regulation of cell adhesion proteins in 344SQ metastatic cells.
Cell adhesion and ECM proteins are differentially regulated in metastatic cells. Changes of protein expression observed in 344SQ cells reveals enhanced binding to extracellular matrix and decreased cell-cell adhesion along with upregulation of extracellular matrix. Upregulated proteins are labeled in red and downregulated proteins in green.
In order for a tumor to progress, tumor cells must be capable of self-renewal, likely through cancer stem cells. The stem cell marker CD44 was upregulated in 344SQ cells along with mesenchymal stem cell markers CD9 and CD106 (VCAM1) (39, 40). We also observed upregulation of two aldehyde dehydrogenases (ALDH3A1 and ALDH1A7); aldehyde dehydrogenase activity and ALDH3A1 in particular have been suggested as markers of cancer stem cells in several cancer types (41). An increase in glutathione transferase expression has also been correlated with CD133 expression in NSCLC tumor samples and we have recently identified a metastatic subpopulation of cells in this model to be CD133+ (42). We further observe upregulation of five glutathione S-transferases (GSTM1, GSTA2, GSTM2, GSTA4, GSTM7) in metastatic 344SQ cells, suggesting glutathione S-transferase activity plays a plays an important role in metastasis and may be another potential marker for cancer stem cells. Reactive oxygen species (ROS) play a role in many aspects of metastasis, including cell adhesion, motility, cell death and other cancer-related pathways and glutathione scavenging of reactive species and free radicals is a mechanism by which cells prevent the damage of ROS, with Glutathione-S-transferases (GST’s) catalyzing the binding of reduced glutathione to both endogenous and exogenous reactive species, reducing the toxicity of these molecules(43).
In our proteomics analysis, we observed evidence of transcriptional regulation in 8 of the 22 proteins downregulated in conditioned media after miR-200 overexpression. Furthermore, comparison of the 22 gene list with Targetscan and Pictar predicted miR-200 regulated genes reveals only one miR-200 predicted gene on the list, fibronectin. We cannot ascertain from this data whether the affected genes in this list are direct targets of miR-200, but previous analysis of microRNA regulation also revealed large numbers of affected genes that do not have 3′ UTR consensus sequences for miR-200 regulation (44, 45). miR-200 functions in a feedback loop with the transcriptional repressors Zeb1 and Zeb2 to regulate the epithelial-to-mesenchymal transition through miR-200 active sites in the 3′ untranslated region (UTR) of Zeb, while ZEB1 and ZEB2 are also capable of repressing transcription of miR-200 family members (46). ZEB1 was found to be overexpressed in metastatic 344SQ cells and may be responsible for changes in gene expression that are not accounted for by miR-200. The identification of ZEB1 binding domains in the promoters of downregulated proteins and transcripts raises the possibility that the miR-200 family’s effect is a combination of direct suppression and regulation of multiple transcription factors, such as Zeb1 and c-ets-1. Promoter analysis of differentially regulated genes further revealed other potential transcription factors with a role in EMT such as Ap-2rep. Further study is required to elucidate the combined role for these factors in EMT and metastasis.
Supplementary Material
Acknowledgments
Grant Support
Funding was provided by the NCI Mouse Models of Human Cancer Consortium and NIH R01 CA157450. D.L.G. was supported by a Young Investigator Award from The ASCO Cancer Foundation, an International Association for the Study of Lung Cancer (IASLC) Fellow Grant, and NCI K08 CA151651. J.M.K. is the Elza A. and Ina Shackelford Freeman Endowed Professor in Lung Cancer.
Reference List
- 1.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–90. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 2.Siegel PM, Shu W, Massague J. Mad upregulation and Id2 repression accompany transforming growth factor (TGF)-beta-mediated epithelial cell growth suppression. J Biol Chem. 2003;278:35444–50. doi: 10.1074/jbc.M301413200. [DOI] [PubMed] [Google Scholar]
- 3.Huber MA, Kraut N, Beug H. Molecular requirements for epithelial-mesenchymal transition during tumor progression. Curr Opin Cell Biol. 2005;17:548–58. doi: 10.1016/j.ceb.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 4.Bracken CP, Gregory PA, Khew-Goodall Y, Goodall GJ. The role of microRNAs in metastasis and epithelial-mesenchymal transition. Cell Mol Life Sci. 2009;66:1682–99. doi: 10.1007/s00018-009-8750-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Park SM, Gaur AB, Lengyel E, Peter ME. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 2008;22:894–907. doi: 10.1101/gad.1640608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gibbons DL, Lin W, Creighton CJ, Rizvi ZH, Gregory PA, Goodall GJ, et al. Contextual extracellular cues promote tumor cell EMT and metastasis by regulating miR-200 family expression. Genes Dev. 2009;23:2140–51. doi: 10.1101/gad.1820209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zheng S, El-Naggar AK, Kim ES, Kurie JM, Lozano G. A genetic mouse model for metastatic lung cancer with gender differences in survival. Oncogene. 2007;26:6896–904. doi: 10.1038/sj.onc.1210493. [DOI] [PubMed] [Google Scholar]
- 8.Gibbons DL, Lin W, Creighton CJ, Zheng S, Berel D, Yang Y, et al. Expression signatures of metastatic capacity in a genetic mouse model of lung adenocarcinoma. PLoS One. 2009;4:e5401. doi: 10.1371/journal.pone.0005401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Faca VM, Ventura AP, Fitzgibbon MP, Pereira-Faca SR, Pitteri SJ, Green AE, et al. Proteomic analysis of ovarian cancer cells reveals dynamic processes of protein secretion and shedding of extra-cellular domains. PLoS One. 2008;3:e2425. doi: 10.1371/journal.pone.0002425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome biology. 2003;4:P3. [PubMed] [Google Scholar]
- 11.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature protocols. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 12.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome research. 2003;13:2498–504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saulnier R, Bhardwaj B, Klassen J, Leopold D, Rahimi N, Tremblay E, et al. Fibronectin fibrils and growth factors stimulate anchorage-independent growth of a murine mammary carcinoma. Experimental cell research. 1996;222:360–9. doi: 10.1006/excr.1996.0045. [DOI] [PubMed] [Google Scholar]
- 14.Constam DB, Robertson EJ. Tissue-specific requirements for the proprotein convertase furin/SPC1 during embryonic turning and heart looping. Development (Cambridge, England) 2000;127:245–54. doi: 10.1242/dev.127.2.245. [DOI] [PubMed] [Google Scholar]
- 15.Dubois CM, Laprise MH, Blanchette F, Gentry LE, Leduc R. Processing of transforming growth factor beta 1 precursor by human furin convertase. J Biol Chem. 1995;270:10618–24. doi: 10.1074/jbc.270.18.10618. [DOI] [PubMed] [Google Scholar]
- 16.Taipale J, Miyazono K, Heldin CH, Keski-Oja J. Latent transforming growth factor-beta 1 associates to fibroblast extracellular matrix via latent TGF-beta binding protein. The Journal of cell biology. 1994;124:171–81. doi: 10.1083/jcb.124.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gregory PA, Bracken CP, Bert AG, Goodall GJ. MicroRNAs as regulators of epithelial-mesenchymal transition. Cell Cycle. 2008;7:3112–8. doi: 10.4161/cc.7.20.6851. [DOI] [PubMed] [Google Scholar]
- 18.Linhart C, Halperin Y, Shamir R. Transcription factor and microRNA motif discovery: the Amadeus platform and a compendium of metazoan target sets. Genome Res. 2008;18:1180–9. doi: 10.1101/gr.076117.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dittmer J. The biology of the Ets1 proto-oncogene. Molecular cancer. 2003;2:29. doi: 10.1186/1476-4598-2-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koinuma D, Tsutsumi S, Kamimura N, Taniguchi H, Miyazawa K, Sunamura M, et al. Chromatin immunoprecipitation on microarray analysis of Smad2/3 binding sites reveals roles of ETS1 and TFAP2A in transforming growth factor beta signaling. Mol Cell Biol. 2009;29:172–86. doi: 10.1128/MCB.01038-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nakamura Y, Migita T, Hosoda F, Okada N, Gotoh M, Arai Y, et al. Kruppel-like factor 12 plays a significant role in poorly differentiated gastric cancer progression. Int J Cancer. 2009;125:1859–67. doi: 10.1002/ijc.24538. [DOI] [PubMed] [Google Scholar]
- 22.Keshamouni VG, Jagtap P, Michailidis G, Strahler JR, Kuick R, Reka AK, et al. Temporal quantitative proteomics by iTRAQ 2D-LC-MS/MS and corresponding mRNA expression analysis identify post-transcriptional modulation of actin-cytoskeleton regulators during TGF-beta-Induced epithelial-mesenchymal transition. J Proteome Res. 2009;8:35–47. doi: 10.1021/pr8006478. [DOI] [PubMed] [Google Scholar]
- 23.Mathias RA, Wang B, Ji H, Kapp EA, Moritz RL, Zhu HJ, et al. Secretome-based proteomic profiling of Ras-transformed MDCK cells reveals extracellular modulators of epithelial-mesenchymal transition. J Proteome Res. 2009;8:2827–37. doi: 10.1021/pr8010974. [DOI] [PubMed] [Google Scholar]
- 24.Zhou C, Nitschke AM, Xiong W, Zhang Q, Tang Y, Bloch M, et al. Proteomic analysis of tumor necrosis factor-alpha resistant human breast cancer cells reveals a MEK5/Erk5-mediated epithelial-mesenchymal transition phenotype. Breast Cancer Res. 2008;10:R105. doi: 10.1186/bcr2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kulasekara KK, Lukandu OM, Neppelberg E, Vintermyr OK, Johannessen AC, Costea DE. Cancer progression is associated with increased expression of basement membrane proteins in three-dimensional in vitro models of human oral cancer. Arch Oral Biol. 2009;54:924–31. doi: 10.1016/j.archoralbio.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 26.Kass L, Erler JT, Dembo M, Weaver VM. Mammary epithelial cell: influence of extracellular matrix composition and organization during development and tumorigenesis. The international journal of biochemistry & cell biology. 2007;39:1987–94. doi: 10.1016/j.biocel.2007.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kumar S, Weaver VM. Mechanics, malignancy, and metastasis: the force journey of a tumor cell. Cancer Metastasis Rev. 2009;28:113–27. doi: 10.1007/s10555-008-9173-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu G, Friggeri A, Yang Y, Milosevic J, Ding Q, Thannickal VJ, et al. miR-21 mediates fibrogenic activation of pulmonary fibroblasts and lung fibrosis. J Exp Med. 2010;207:1589–97. doi: 10.1084/jem.20100035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roderburg C, Urban GW, Bettermann K, Vucur M, Zimmermann H, Schmidt S, et al. Micro-RNA profiling reveals a role for miR-29 in human and murine liver fibrosis. Hepatology. 2011;53:209–18. doi: 10.1002/hep.23922. [DOI] [PubMed] [Google Scholar]
- 30.Korpal M, Ell BJ, Buffa FM, Ibrahim T, Blanco MA, Celia-Terrassa T, et al. Direct targeting of Sec23a by miR-200s influences cancer cell secretome and promotes metastatic colonization. Nat Med. 2011 doi: 10.1038/nm.2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ozaki Y, Kontani K, Teramoto K, Fujita T, Tezuka N, Sawai S, et al. Involvement of 90K/Mac-2 binding protein in cancer metastases by increased cellular adhesiveness in lung cancer. Oncology reports. 2004;12:1071–7. [PubMed] [Google Scholar]
- 32.Dunkle ET, Zaucke F, Clegg DO. Thrombospondin-4 and matrix three-dimensionality in axon outgrowth and adhesion in the developing retina. Exp Eye Res. 2007;84:707–17. doi: 10.1016/j.exer.2006.12.014. [DOI] [PubMed] [Google Scholar]
- 33.Puschmann TB, Turnley AM. Eph receptor tyrosine kinases regulate astrocyte cytoskeletal rearrangement and focal adhesion formation. J Neurochem. 2010;113:881–94. doi: 10.1111/j.1471-4159.2010.06655.x. [DOI] [PubMed] [Google Scholar]
- 34.Schietke R, Warnecke C, Wacker I, Schodel J, Mole DR, Campean V, et al. The lysyl oxidases LOX and LOXL2 are necessary and sufficient to repress E-cadherin in hypoxia: insights into cellular transformation processes mediated by HIF-1. J Biol Chem. 2010;285:6658–69. doi: 10.1074/jbc.M109.042424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stamenkovic I, Yu Q. Shedding light on proteolytic cleavage of CD44: the responsible sheddase and functional significance of shedding. J Invest Dermatol. 2009;129:1321–4. doi: 10.1038/jid.2009.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kuphal S, Martyn AC, Pedley J, Crowther LM, Bonazzi VF, Parsons PG, et al. H-cadherin expression reduces invasion of malignant melanoma. Pigment Cell Melanoma Res. 2009;22:296–306. doi: 10.1111/j.1755-148X.2009.00568.x. [DOI] [PubMed] [Google Scholar]
- 37.Sarrio D, Palacios J, Hergueta-Redondo M, Gomez-Lopez G, Cano A, Moreno-Bueno G. Functional characterization of E- and P-cadherin in invasive breast cancer cells. BMC Cancer. 2009;9:74. doi: 10.1186/1471-2407-9-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tseng RC, Lee SH, Hsu HS, Chen BH, Tsai WC, Tzao C, et al. SLIT2 attenuation during lung cancer progression deregulates beta-catenin and E-cadherin and associates with poor prognosis. Cancer Res. 2010;70:543–51. doi: 10.1158/0008-5472.CAN-09-2084. [DOI] [PubMed] [Google Scholar]
- 39.Halfon S, Abramov N, Grinblat B, Ginis IO. Markers distinguishing mesenchymal stem cells from fibroblasts are downregulated with passaging. Stem cells and development. doi: 10.1089/scd.2010.0040. [DOI] [PubMed] [Google Scholar]
- 40.Peng L, Ran YL, Hu H, Yu L, Liu Q, Zhou Z, et al. Secreted LOXL2 is a novel therapeutic target that promotes gastric cancer metastasis via the Src/FAK pathway. Carcinogenesis. 2009;30:1660–9. doi: 10.1093/carcin/bgp178. [DOI] [PubMed] [Google Scholar]
- 41.Moreb JS, Baker HV, Chang LJ, Amaya M, Lopez MC, Ostmark B, et al. ALDH isozymes downregulation affects cell growth, cell motility and gene expression in lung cancer cells. Molecular cancer. 2008;7:87. doi: 10.1186/1476-4598-7-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang Y, Ahn YH, Gibbons DL, Zang Y, Lin W, Thilaganathan N, et al. The Notch ligand Jagged2 promotes lung adenocarcinoma metastasis through a miR-200-dependent pathway in mice. J Clin Invest. 2011 doi: 10.1172/JCI42579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Anasagasti MJ, Alvarez A, Martin JJ, Mendoza L, Vidal-Vanaclocha F. Sinusoidal endothelium release of hydrogen peroxide enhances very late antigen-4-mediated melanoma cell adherence and tumor cytotoxicity during interleukin-1 promotion of hepatic melanoma metastasis in mice. Hepatology. 1997;25:840–6. doi: 10.1002/hep.510250410. [DOI] [PubMed] [Google Scholar]
- 44.Cheng J, Zhou L, Xie QF, Xie HY, Wei XY, Gao F, et al. The impact of miR-34a on protein output in hepatocellular carcinoma HepG2 cells. Proteomics. 2010;10:1557–72. doi: 10.1002/pmic.200900646. [DOI] [PubMed] [Google Scholar]
- 45.Burk U, Schubert J, Wellner U, Schmalhofer O, Vincan E, Spaderna S, et al. A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Rep. 2008;9:582–9. doi: 10.1038/embor.2008.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.