Abstract
The Cdc20 and Cdh1/Fzr proteins are the substrate-specific activators of the anaphase-promoting complex (APC). In Medicago truncatula, the MtCcs52A and MtCcs52B proteins represent two subgroups of the Cdh1-type activators, which display differences in their cell cycle regulation, structure, and function. The ccs52A transcripts are present in all phases of the cell cycle. By contrast, expression of ccs52B is restricted to late G2-phase and M-phase, and its induced overexpression in BY2 cells inhibited mitosis. MtCcs52A is active in Schizosaccharomyces pombe and binds to the S. pombe APC, whereas MtCcs52B does not because of differences in the N-terminal region. We identified a new functional domain, the Cdh1-specific motif conserved in the Cdh1 proteins that, in addition to the C-box and the terminal Ile and Arg residues, was essential for the activity and required for efficient binding to the APC. Moreover, we demonstrate that cyclin-dependent kinase phosphorylation sites adjacent to the C-box may regulate the interaction with the APC. In the different plant organs, the expression of Mtccs52A and Mtccs52B displayed differences and indicated the involvement of the APC in differentiation processes.
INTRODUCTION
Controlled protein degradation mediated by ubiquitin-dependent proteolysis is a key mechanism in eukaryotes that regulates many fundamental cellular processes, including cell cycle (reviewed in Harper et al., 2002). Covalent attachment of ubiquitin chains to Lys residues on target proteins leads to their recognition and degradation by the 26S proteasome. Ubiquitination involves successive activities of the ubiquitin-activating (E1), ubiquitin-conjugating (E2), and ubiquitin ligase (E3) enzymes. The selection and specific timing of polyubiquitination of the target proteins are conferred by different E3 ubiquitin ligases. In the cell cycle, two structurally related E3 ubiquitin ligases, the anaphase-promoting complex (APC) and the Skp1/Cullin/F-box complex, have essential and complementary functions by temporally controlled degradation of various cell cycle proteins, which ensures the irreversible nature of the cell cycle (Peters, 1998).
APC is a multiprotein complex, composed of 11 to13 subunits, which is active from metaphase until S-phase (reviewed in Harper et al., 2002; Peters, 2002). The APC is essential for the regulation of metaphase-anaphase transition and exit from mitosis by ordered destruction of mitotic regulators, including securin, an inhibitor of chromosome separation, cyclin A, cyclin B, and many of the mitotic regulatory kinases (Harper et al., 2002). APC has also been linked to the control of DNA replication by degrading Cdc6 (Petersen et al., 2000), geminin (McGarry and Kirschner, 1998), and ribonucleotid reductase R2 (Chabes et al., 2003), as well as to the regulation of endoreduplication cycles (Sigrist and Lehner, 1997; Cebolla et al., 1999; Kashevsky et al., 2002; Vinardell et al., 2003).
Two subunits, APC2 and APC11, together with E2 ubiquitin-conjugating enzymes, are sufficient for ubiquitination, though this reconstituted ubiquitin ligase activity did not possess substrate specificity (Tang et al., 2001). This is defined by binding of either of the two adaptor proteins, Cdc20 (also known as Slp1, Fzy, and p55CDC) or Cdh1 (also known as Hct1, Ste9/Srw1, and Fzr), which also control stage-specific activation of the APC (Harper et al., 2002; Peters, 2002). All of these proteins contain seven WD40 repeats spanning the middle part to the C-terminus, as well as a consensus DR(F/Y)IPxR motif, called C-box in the N-terminal region, and C-terminal Ile and Arg residues (IR motif) that are involved in APC binding (Schwab et al., 2001; Vodermaier et al., 2003). Both the Cdc20 and the Cdh1 proteins contain cyclin-dependent kinase (CDK) phosphorylation sites, varying in their numbers and positions as well as in their effects on activity. Cdh1 is inactivated by hyperphosphorylation, which prevents its association with the APC (Zachariae et al., 1998; Jaspersen et al., 1999; Kramer et al., 2000), triggers its proteolysis (Blanco et al., 2000), and leads to its translocation from the nucleus to the cytoplasm (Jaquenoud et al., 2002; Zhou et al., 2003a).
These APC activators can interact with many different proteins either via their WD40 repeats or the N-terminal region. Mitotic cyclins, containing a degradation signal called destruction box (D-box, RxxLxxxxN) (Glotzer et al., 1991), are recognized as APC substrates by both the Cdc20 and Cdh1 proteins. Binding of cyclin A to mammalian Cdh1 depended on a conserved cyclin binding RVL motif within the WD40 domain that was conserved in the Cdh1 WD40 domain in diverse organisms (Sorensen et al., 2001). In addition to D-box proteins, Cdh1 interacts with a wider range of APC substrates that contain KEN- (Pfleger and Kirschner, 2000), A- (Littlepage and Ruderman, 2002), or GxEN-boxes (Castro et al., 2003).
In Saccharomyces cerevisiae (budding yeast), the cell cycle regulation of the cdc20 and cdh1 genes is different. The cdc20 RNA and Cdc20 protein are present only during late S-phase and mitosis, whereas the levels of the Cdh1 RNA and protein are constant throughout the cell cycle (Prinz et al., 1998). The activity of Cdh1 is regulated by cyclin A–dependent kinases, which decrease APCCdh1 activity during S and G2 by phosphorylation of Cdh1, allowing the accumulation of mitotic cyclins (Zachariae et al., 1998; Huang et al., 2001; Yeong et al., 2001). Increasing activity of cyclin B–dependent kinases results in the phosphorylation of the APC subunits, which is a prerequisite for the activation of APC by Cdc20 (reviewed in Zachariae and Nasmyth, 1999). APCCdc20 results in the proteolysis of mitotic cyclins and consequently reactivation of Cdh1. Because Cdc20 contains a KEN-box, it is recognized and destroyed by APCCdh1 (Prinz et al., 1998), which remains active until the S-phase (Hagting et al., 2002; Raff et al., 2002).
Most of our knowledge on APC function derives from studies on proliferating cells, though several pieces of evidence are accumulating for the implication of APC outside the cell cycle in differentiated cells. Although the expression of cdc20 seems to be restricted to proliferating cells, cdh1 transcripts are present both in mitotic and postmitotic cells. In the nuclei of postmitotic terminally differentiated neurons, both Cdh1 and APC are ubiquitously expressed (Gieffers et al., 1999), and SnoN, a negative regulator of the TGFβ signaling pathway, is degraded by APCCdh1 (Stroschein et al., 2001; Wan et al., 2001). In Homo sapiens cells, two alternatively spliced forms of cdh1 are present (Zhou et al., 2003b), whereas in Gallus domesticus (chickens) four different cdh1 genes were found, probably with specific roles in the various organs (Wan and Kirschner, 2001).
In plants, D-box–dependent degradation of mitotic cyclins by the 26S proteasome (Genschik et al., 1998) as well as the presence of the APC subunits in Arabidopsis thaliana (Blilou et al., 2002; Capron et al., 2003) indicated that the function of the APC must be conserved in plants as well.
Previously, we have identified a cell cycle switch gene, ccs52 from Medicago sativa encoding a plant homolog of the Cdh1/Fzr/Srw1 APC activators. Induced overexpression of ccs52 in Schizosaccharomyces pombe (fission yeast) elicited mitotic cyclin degradation, resulting in growth inhibition, cell elongation, and repeated endoreduplication cycles (duplication of the genome without mitosis) (Cebolla et al., 1999). In M. truncatula, this gene was dispensable for the mitotic cycles but essential for nodule differentiation (Vinardell et al., 2003). The protein was nuclear and present in all endoreduplication competent cells, indicating that it might activate the APC constitutively during the endocycles.
Our recent work indicated that the model legume M. truncatula has two distinct members of the Cdh1-type APC activators, MtCcs52A and MtCcs52B (Vinardell et al., 2003). Here, we show that MtCcs52A and MtCcs52B are differentially regulated during cell cycle and plant development and display differences in their APC interactions. Whereas MtCcs52A appears to be an ortholog of the yeast and animal Cdh1-type proteins, MtCcs52B was inactive in yeast and likely interacts specifically with plant APCs. By creating of a series of mutations in MtCcs52A, we have identified a Cdh1-specific motif (CSM) that, in addition to the C-box and the C-terminal IR residues (Schwab et al., 2001), is indispensable for APC interaction.
RESULTS
Mtccs52A and Mtccs52B Encode Cdh1-Type APC Activators in M. truncatula
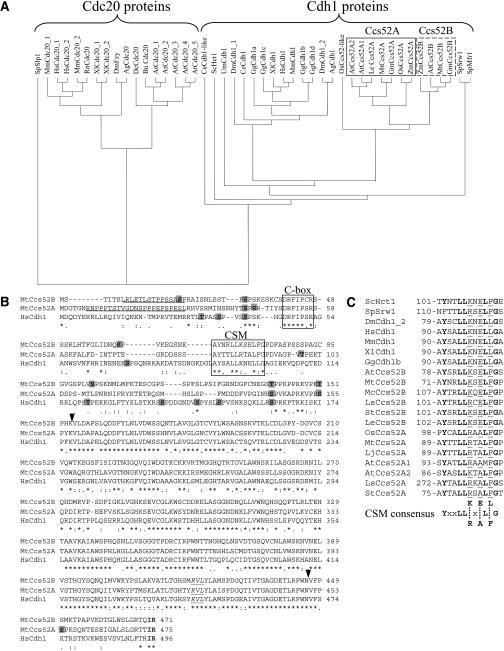
The Mtccs52A and Mtccs52B cDNAs were isolated from M. truncatula nodule and root libraries, respectively (Vinardell et al., 2003). Phylogenetic analysis of known and putative APC activators, originating from homologous cDNAs or genome sequences, grouped the Cdc20 and Cdh1 proteins separately and showed clearly that both the MtCcs52A and MtCcs52B proteins belong to the Cdh1 group of APC activators (Figure 1A). Within this group, animal and plant proteins were on different branches, and strikingly, the A-type and B-type Ccs52 proteins represented two subgroups of the Cdh1-type proteins in plants. MtCcs52A and MtCcs52B shared 63% identity. MtCcs52A was more homologous to the Glycine max protein GmBG044933 (93% identity) or to AtCcs52A1 (At4g22910) (79.7%) and AtCcs52A2 (At4g11920) (77.2%) from A. thaliana than to MtCcs52B. Likewise, MtCcs52B was more related to GmAI736659 (92% identity) and AtCcs52B (At5g13840) (83.3%) than to MtCcs52A.
Figure 1.
The Structure and Conserved Motifs of the M. truncatula Ccs52A and Ccs52B Proteins and Their Relatedness to Other APC Activators.
(A) Phylogenetic tree of APC activators classifies the Cdc20 and Cdh1/Ccs52 proteins in separate groups. BLASTX and TBLASTX were performed with Mtccs52A and Mtccs52B cDNA. Translated sequences were aligned with CLUSTAL W at EMBL-EBI and visualized using the TreeView program.
(B) Comparison of MtCcs52A and MtCcs52B to the H. sapiens Cdh1 protein. Putative PEST motifs are underlined; S and T residues of potential CDK phosphorylation sites are indicated on shaded background; the conserved terminal IR residues are in bold; the C-box and CSM sequences are shown in frames; and the conserved cyclin binding RVL motif is in italic and underlined. Arrowheads indicate the start and the end of the WD40 repeat region.
(C) The CSM is conserved in the Cdh1 proteins. A consensus sequence is deduced, where identical residues are in bold and x indicates any amino acid. Positions of the CSMs are given. Ag, Anopheles gambiae; At, A. thaliana; Bn, Brassica napus; Ce, Caenorhabditis elegans; Dc, Daucus carota; Dm, D. melanogaster; Gg, Gallus gallus; Gm, G. max; Hs, H. sapiens; Le, Lycopersicon esculentum; Lj, Lotus japonicus; Ls, Lactuca sativa; Mc, Mesembryanthemum crystallinum; Mm, Mus musculus; Mt, M. truncatula; Os, Oryza sativa; Rn, Rattus norvegicus; Sc, S. cerevisiae; Sp, S. pombe; St, Solanum tuberosum; Um, Ustilago maydis; Xl, Xenopus laevis; Zm, Zea mays.
The overall structures of the MtCcs52A and MtCcs52B proteins were similar to each other and to the H. sapiens Cdh1 protein (Figure 1B), but there were also marked differences. The N-terminal region in front of the C-box was longer in MtCcs52A than in MtCcs52B; however, their final sizes were similar (475 and 471 amino acids, respectively) and slightly smaller than that in the HsCdh1 protein (496 amino acids). A PEST motif, responsible for rapid turnover of proteins (Rogers et al., 1986), was predicted at the N terminus of MtCcs52A (PESTfind score 18.67) and MtCcs52B (PESTfind score 7.02), whereas this motif is absent in the H. sapiens and animal Cdh1 proteins. The C-box sequence was identical in MtCcs52A and HsCdh1 (DRFIPSR) but differed in MtCcs52B and in all other B-type plant proteins at the sixth position, where instead of Ser a Cys residue was present. Both Medicago proteins, like the H. sapiens and other Cdh1 proteins, terminated with Ile and Arg residues and contained the consensus cyclin binding RVL motif in the WD40 repeat region. MtCcs52A contained six putative CDK phosphorylation sites, whereas MtCcs52B had seven and HsCdh1 had nine. Two phosphorylation sites were conserved in all the three proteins: one in front of the C-box, the other before the WD40 repeats. In the MtCcs52A and MtCcs52B proteins, a third phosphorylation site was conserved at positions 155 and 151, respectively.
Downstream of the C-box, similarity was found along 12 residues in the MtCcs52A, MtCcs52B, and HsCdh1 proteins starting at positions 89, 71, and 90, respectively. Because this sequence was absent in the Cdc20 proteins and a consensus YxxLL(K/R)x(E/A)L(F/L)G was present in the Cdh1 proteins (Figure 1C), that was designated as CSM. All plant Ccs52 proteins contained Phe at the 10th position. At the eighth position, an Ala residue was present in the Ccs52A proteins, whereas Glu was present in the Ccs52B proteins. At the sixth position, Arg was preferentially present in the A-type proteins, and Lys was present in the B-type proteins. Moreover, in the Ccs52B proteins the CSM contained an Arg residue at the third position.
The ccs52A and ccs52B Genes Are Differently Regulated during the Cell Cycle
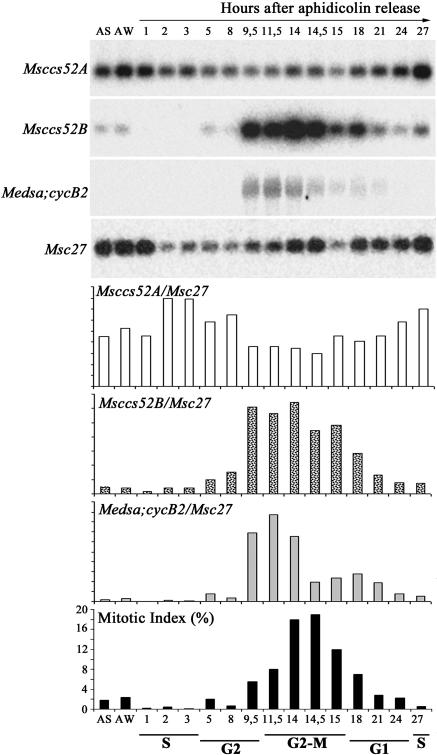
The temporal control of APC activation is determined in part by the presence and availability of the APC activators. Their regulation therefore might contribute to APC activation and cell cycle control. Because M. truncatula cell cultures cannot be efficiently synchronized and the M. sativa and M. truncatula genomes are highly homologous, cell cycle regulation of the ccs52A and ccs52B genes was studied in the M. sativa A2 cell culture that can be efficiently synchronized with aphidicolin and propyzamide treatments (Roudier et al., 2000). The ccs52 genes exhibit high conservation in these two Medicago species. In the case of ccs52A, the two genes share 98% identity in the coding sequences and 91% identity in the promoter regions (our unpublished data), suggesting orthologous function and regulation. In our previous work (Vinardell et al., 2003), an identical expression pattern of the Mt/Msccs52A genes has been confirmed during root and nodule development by in situ hybridization. The Mt/Msccs52A and Mt/Msccs52B genes could be amplified with the same primers, and their expression patterns were studied by reverse transcription (RT)–PCR. The ccs52A transcripts were present in all cell cycle phases, with a slight decrease in G2 and G2/M and a small increase during G1 until the beginning of S-phase, a pattern similar to that described for cdh1 (Prinz et al., 1998). Unexpectedly, expression of ccs52B was strongly regulated during the cell cycle. It was induced in late G2 with maximal transcript levels during G2/M-phase and M-phase, which then gradually decreased and disappeared during the G1-phase (Figure 2). This expression profile was surprisingly similar to that of the mitotic cyclin Medsa;cycB2. Thus, expression of the Msccs52A and Msccs52B genes during the cell cycle displayed marked differences, suggesting distinct functions for these APC activators.
Figure 2.
Differential Regulation of the ccs52 Genes during the Cell Cycle.
Transcript levels of the Msccs52A, Msccs52B, Medsa;cycB2, and the constitutively expressed Msc27 gene were analyzed by RT-PCR in asynchronously growing (AS) and synchronized M. sativa A2 cell suspensions according to Roudier et al. (2000). Msccs52A, Msccs52B, and Medsa;cycB2 signals normalized with that of Msc27, and mitotic indexes are shown. AW, aphidicolin-treated cells after wash.
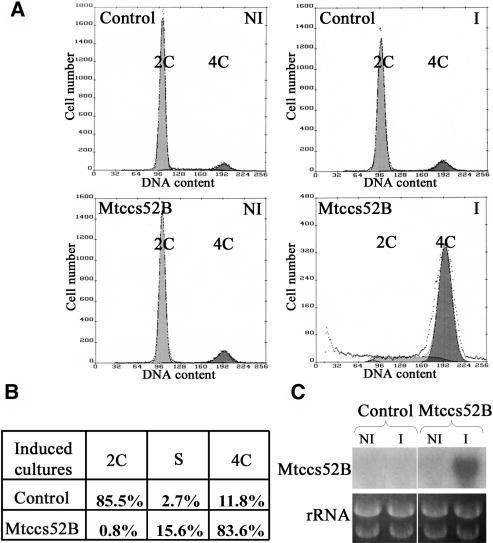
To gain more information about the cell cycle function of Mtccs52B, the Nicotiana tabacum cell line BY2-tetR17 expressing the tetracycline repressor (David and Perrot-Rechenmann, 2001) was stably transformed with the Mtccs52B cDNA under the control of the tetracycline-inducible promoter (Gatz et al., 1992). The reason for using a heterologous system was that no inducible gene expression systems or stable transformation protocols have been worked out for M. sativa and M. truncatula cell cultures. The effect of Mtccs52B expression on cell cycle was studied by flow cytometry, measuring the numbers of the 2C, S, and 4C nuclei in noninduced and induced cultures compared with those transformed with the empty vector (Figure 3). Although the control and the noninduced Mtccs52B cultures contained predominantly 2C cells, strong expression of Mtccs52B resulted in a dramatic increase in the number of 4C cells (83.6%), and the lack of 2C cells suggested that induced overexpression of Mtccs52B may block mitosis and reentering of the cells into G1-phase. Parallel transformations of BY2 cells with Mtccs52A did not result in the formation of transgenic calli, suggesting that leaky expression of Mtccs52A inhibited cell proliferation.
Figure 3.
Induced Expression of Mtccs52B Arrests BY2-Rtet17 Cells with 4C DNA Content.
(A) Flow cytometry analysis of nuclear DNA content of BY2-Rtet17 cells transformed with pBinOP (control) or pBinOP-MtCcs52B cDNA. Cultures were induced with tetracycline for 72 h. C, haploid DNA content; I, induced cultures; NI, noninduced cultures.
(B) Distribution of 2C, S, and 4C cells in the induced cultures from (A).
(C) RNA gel blot analysis of Mtccs52B in noninduced and induced cultures.
Mtccs52A and Mtccs52B Are Not Functional Homologs in S. pombe
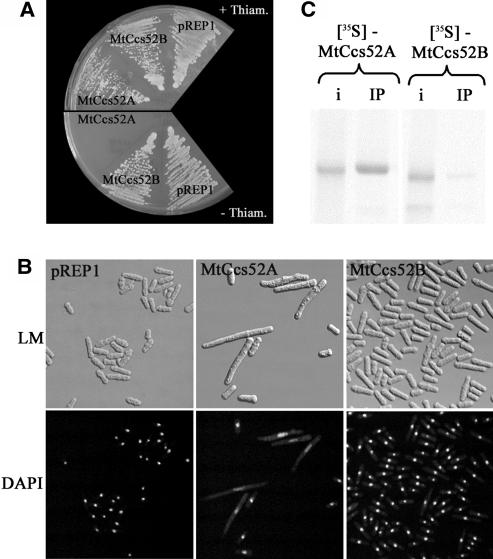
Our previous work (Cebolla et al., 1999) showed that the plant protein MsCcs52A was an ortholog of the S. pombe Srw1/Ste9. Because induced overexpression of Msccs52A elicited a severe phenotype manifested by growth inhibition and extreme elongation of the yeast cells (Cebolla et al., 1999), we investigated whether MtCcs52A and MtCcs52B have redundant or distinct functions in S. pombe. Both the Mtccs52A and Mtccs52B cDNAs were cloned in the pRep1 yeast vector and expressed from the nmt1 promoter, allowing gene expression in the absence of thiamine. Overexpression of Mtccs52A in S. pombe also resulted in growth arrest and cell enlargement (Figures 4A and 4B). Surprisingly, overexpression of Mtccs52B did not provoke any visible phenotypes, and both the growth and the morphology of the cells, observed with light microscopy and 4′,6-diamidino-2-phenylindole (DAPI) staining under UV light, were similar to the control S. pombe strain that was transformed with the empty vector (Figures 4A and 4B). These data indicated that the activity of MtCcs52A and MtCcs52B proteins, at least in yeast, is different.
Figure 4.
MtCcs52A and MtCcs52B Act Differently in S. pombe.
(A) Growth of S. pombe transformed with pREP1-MtCcs52A, pREP1-MtCcs52B, or the empty vector pREP1 in the presence and absence of thiamine.
(B) Phenotype of strains from (A) induced withdrawal of thiamine and observed with light microscopy or with UV light after DAPI staining of the nuclei. DAPI, UV light observation of DAPI-stained cells; LM, light microscopy.
(C) Binding of the in vitro translated [35S]Met-labeled MtCcs52A and MtCcs52B proteins to the S. pombe APC immunoprecipitated via Myc-lid1. i, 10% of the input of the [35S]Met-labeled proteins; IP, immunoprecipitation.
MtCcs52A, but Not MtCcs52B, Can Interact with the S. pombe APC in Vitro
One possibility to explain this difference is that MtCcs52A, but not MtCcs52B, is capable of binding to the yeast APC. To test this hypothesis, we performed in vitro binding assays between immunoprecipitated APC complex and in vitro translated [35S]Met-labeled MtCcs52A and MtCcs52B proteins (Figure 4C). Although >10% of the input MtCcs52A protein bound to the APC, MtCcs52B was practically absent. The lack of efficient interaction between MtCcs52B and the yeast APC may therefore explain why overexpression of Mtccs52B had no phenotype in S. pombe.
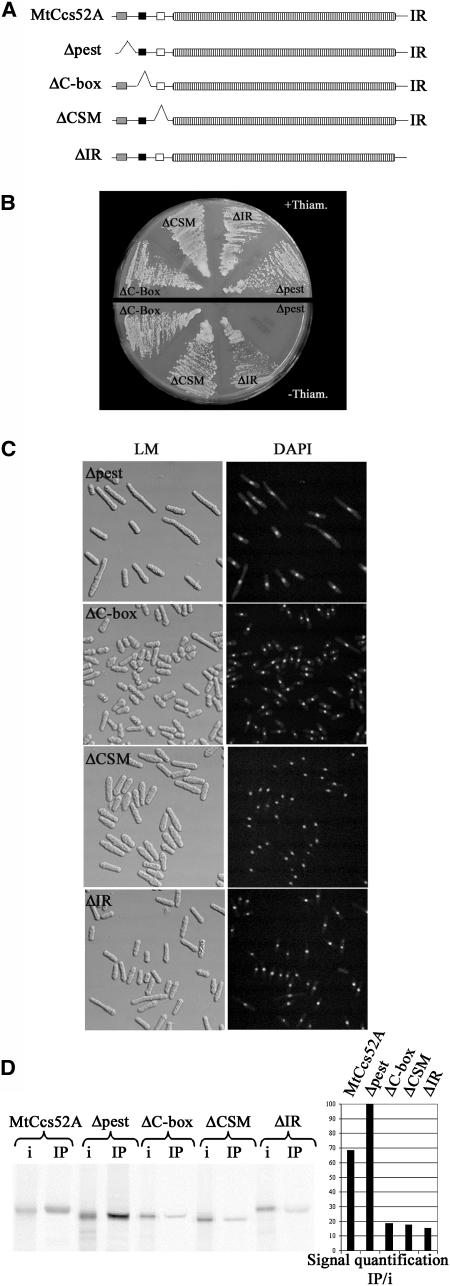
The PEST Motif Is Dispensable but the CSM, in Addition to the C-Box and the IR Tail, Is Required for Binding of MtCcs52A to the APC
Using the in vivo functional assay in S. pombe and the in vitro APC interaction test, we performed structure-function studies on the MtCcs52A protein. A set of deletion derivatives was created, which eliminated the putative PEST motif (deletion of 24 amino acids), the C-box (deletion of 20 amino acids), the CSM (deletion of 43 amino acids), and the IR tail (replacing I474 with a stop codon) (Figure 5A). These constructs were introduced into S. pombe and overexpressed from the nmt1 promoter. All mutations, except the Δpest, resulted in the inactivation of MtCcs52A and allowed growth of S. pombe in the absence of thiamine (Figure 5B). Induced expression of Δpest provoked the same phenotype as the wild-type MtCcs52A protein. Overexpression of ΔIR occasionally produced a few elongated cells. All the other mutant derivatives had no effect on cell size, although their shape was somewhat abnormal (Figure 5C). Consistent with these results, the Δpest derivative interacted robustly with the yeast APC; its binding was even more efficient (100%) than that of the wild-type protein (∼70%) (Figure 5D). This suggests that the absence of the PEST motif might stabilize MtCcs52A and the formation of the MtCcs52A-APC complexes. By contrast, all the other deletions in the C-box, CSM, or the IR tail reduced the interaction of MtCcs52A with the APC (Figure 5D). The signal intensities of the APC-bound ΔC-box, ΔCSM, and IR tail mutant proteins were <20%. Even if the intensity of detection is not linearly proportional with the amount of the detected proteins, this reduction appears to be significant and correlated with the loss of activity. These data showed that the consensus CSM represents a novel functional domain of the Cdh1-type APC activators, which, in addition to the C-box and the IR motif, is important for APC interaction.
Figure 5.
Binding of MtCcs52A to the APC Requires the C-Box, CSM, and IR Motifs but Not the PEST Motif.
(A) Schematic representation of the mutations in the MtCcs52A protein.
(B) Growth of S. pombe transformed with pREP1-Δpest, pREP1-ΔC-box, pREP1-ΔCSM, or pREP1-ΔIR in the presence and absence of thiamine.
(C) Phenotype of induced cells from (B) as in Figure 3B.
(D) Immunoprecipitation of APC via Myc-lid1 results in efficient binding of the [35S]Met-labeled MtCcs52A and Δpest but not the ΔC-box, ΔCSM, and ΔIR proteins. IP signals were normalized according to their respective input signals. The binding efficiency of proteins is presented relative to the maximum value (Δpest = 100%).
Phosphorylation in the Vicinity of the C-Box May Negatively Regulate Binding of MtCcs52A to APC
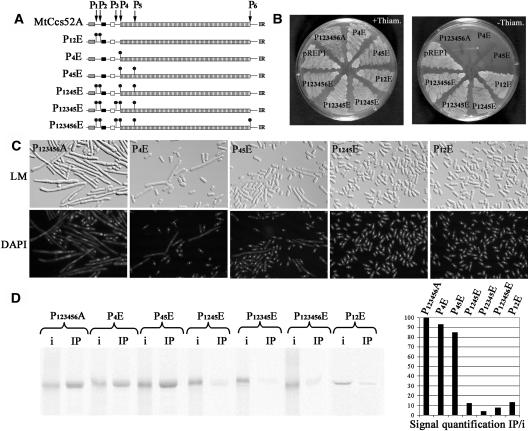
In MtCcs52A, there are six putative CDK phosphorylation sites, at positions 43 (P1), 45 (P2), 99 (P3), 144 (P4), 155 (P5), and 454 (P6) (Figure 6A). To study how they might affect activity, the Ser and Thr residues of the putative CDK phosphorylation sites were replaced either by Ala, creating a nonphosphorylable version of the protein, or by Glu, providing a negative charge and mimicking phosphorylation. When a series of Ala mutations was made, affecting single, multiple (data not shown), or all the six phosphorylation sites (P123456A), MtCcs52A remained active and elicited growth arrest (Figures 6B and 6C). The P123456A form of MtCcs52A appeared to be even more active than the wild-type protein, provoking more efficiently the elongated yeast phenotype (cf. Figures 4B and 6C). This nonphosphorylated P123456A-MtCcs52A also formed a complex with the APC in vitro (cf. Figures 5D and 6D).
Figure 6.
A Nonphosphorylable Form of MtCcs52A Interacts with the Yeast APC, but Phosphomimetic Amino Acids in the Vicinity of the C-Box Inhibit This Binding.
(A) Schematic representation of the different mutations in the MtCcs52A protein mimicking phosphorylation.
(B) Growth of S. pombe transformed with pREP1-P123456A, pREP1-P12E, pREP1-P4E, pREP1-P45E, pREP1-P1245E, pREP1-P12345E, pREP1-P123456E, or with the empty vector pREP1 in the presence and absence of thiamine.
(C) Phenotype of strains expressing pREP1-P123456A, pREP1-P4E, pREP1-P45E, pREP1-P1245E, and pREP1-P12E.
(D) Immunoprecipitated APC binds efficiently P1234566A, P4E, and P45E but not the P12E, P1245E, P12345E, and P123456E proteins. IP signals were quantified as in Figure 4.
When the P4 site or both the P4 and P5 sites were mutated to Glu, the P4E and P45E proteins remained active, inhibiting yeast growth (Figure 6B), and induced the formation of elongated cells, albeit less efficiently than the wild type (Figure 6C). Both the P4E and P45E proteins were also able to efficiently interact with the APC (Figure 6D). The mutation P1245E, affecting four CDK phosphorylation sites, resulted in the inactivation of the protein; the yeast strain expressing P1245E was able to grow, and the cells were normal. Consistent with the yeast phenotype, P1245E and consequently also P12345E and P123456E exhibited extremely reduced binding to the APC (Figure 6D). Compared with P45E, P1245E had a drastic effect on activity, which suggested that the P12 sites (S43 and S45) might have particular importance. Therefore, P12E was constructed, which resulted in the inactivation of the protein and drastically reduced the interaction with APC (Figure 6D). This demonstrated that the introduced negative charge mimicking phosphorylation of MtCcs52A in the close vicinity of the C-box may negatively regulate its interaction with the APC.
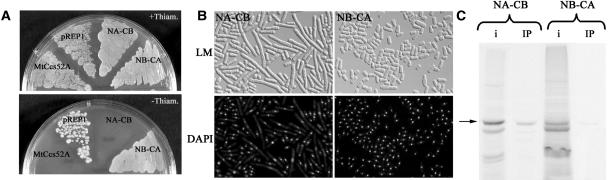
A Chimeric Protein Containing the N-Terminal Part of MtCcs52A Fused with the C-Terminal Region of MtCcs52B Is Functional in Yeast
The MtCcs52A and MtCcs52B proteins share 63% identity, with differences both at the N-terminal and the WD40 repeat regions. Although both of them contain the three functional domains, the C-box, the CSM, and the IR, MtCcs52B was unable to interact with the yeast APC. Because the C-box and the CSM sequences are not identical in the two proteins and there are differences in the neighboring sequences, it was possible that the interaction with the S. pombe APC might depend on the N-terminal region. To test this hypothesis, two chimeras were constructed, fusing either the N-terminal part of MtCcs52B with the WD40 domains of MtCcs52A (NB-CA hybrid protein) or the N-terminal part of MtCcs52A to the WD40 domains of MtCcs52B (NA-CB). The N-terminal part of MtCcs52B rendered the NB-CA fusion protein inactive in yeast, whereas NA-CB, with the N-terminal part of MtCcs52A, was active, resulting in growth inhibition and elongation of cells (Figures 7A and 7B). Consistent with these results, NA-CB interacted in vitro with the yeast APC, whereas binding of NB-CA to the APC was not significant (Figure 7C). This result confirmed that the N-terminal part of MtCcs52A determines the specificity of the interaction with the yeast APC, whereas the C-terminal part of MtCcs52B contributes with its IR tail to the APC binding. Moreover, the activity of the NA-CB hybrid protein implies that NA-CB was able to recruit at least one of the yeast APC substrates, the Cdc13 mitotic cyclin, probably via the conserved cyclin binding motif present in the CB part of the hybrid protein.
Figure 7.
The N-Terminal Part of MtCcs52A Is Essential for Its Activity in Yeast.
(A) Growth of S. pombe transformed with pREP1, pREP1-MtCcs52A, or with the pREP1-NA-CB and pREP1-NB-CA chimeras in the presence and absence of thiamine.
(B) Morphology of the strains expressing pREP1-NA-CB and pREP1-NB-CA in the absence of thiamine.
(C) NA-CB but not the NB-CA protein interacts efficiently with the S. pombe APC. Arrow indicates the correct size of the chimeric proteins, whereas the bands with higher mobility correspond probably to degradation products.
Mtccs52A and Mtccs52B Have Distinct Expression Patterns in Plant Organs
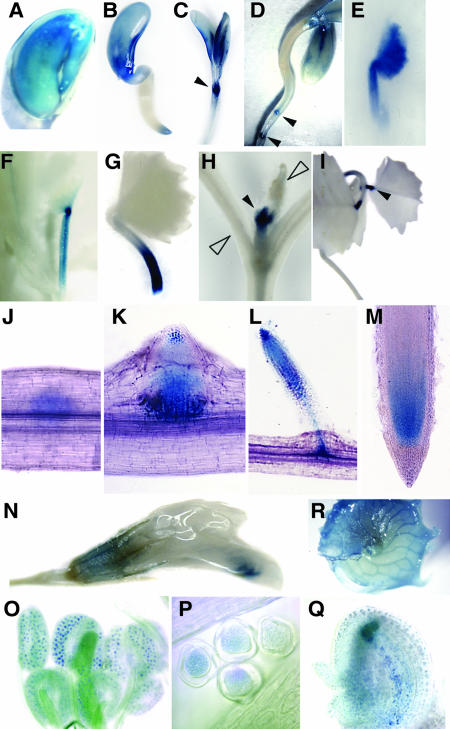
Our previous study revealed expression of Mtccs52B in the nodulation-competent root zone but not in the nodules, whereas Mtccs52A was induced in the nodule primordium, and its expression was linked to differentiation and endoreduplication of nodule cells (Vinardell et al., 2003). Because little is known about the spatial and temporal expression pattern of the cdh1 genes in higher organisms, we looked more generally at Ccs52A expression during plant development, with the help of transgenic M. truncatula expressing the uidA reporter gene coding for β-glucuronidase (GUS) from the Mtccs52A promoter (Figure 8). In the embryo, GUS activity was detectable both in the radicle and in the cotyledons (Figure 8A), indicating expression of Mtccs52A in these regions. In the growing seedlings, the expression was maintained in the cotyledons, but in the root it was restricted to the root apex (Figure 8B). Later, the GUS staining progressively disappeared from the cotyledons, and coincidently expression of Mtccs52A was induced by initiation of the first leaf in the shoot apex (Figure 8C). Simultaneously in the root, Mtccs52A expression was detectable at the initiation of the lateral root primordia (Figure 8D). Expression of Mtccs52A during leaf development was transient. The intense blue staining observed in the tiny leaflet (Figure 8E) disappeared rapidly, whereas the staining persisted in the petiole (Figure 8F), but then it also vanished progressively (Figure 8G). At this stage, Mtccs52A expression was completely switched off both in the cotyledons and the first leaflet, and it was only detectable at the shoot apex, where development of additional leaves started (Figure 8H). In the trifoliate leaves, the GUS staining was only detectable at the petiole–leaf junctions (Figure 8I). During lateral root development, Mtccs52A was induced very early, before the emergence of the lateral root primordium (Figure 8J). At the outgrowth of the differentiating lateral root primordium, the staining concentrated to the apical and to the basal regions (Figure 8K). With the growth of the lateral root, the expression was maintained in the apical region and was transiently present at the junction of the main and lateral roots in the vascular tissues (Figure 8L). Later, the expression was in the meristematic region and in the differentiating cell files as in the primary root (Figure 8M). In the flower, weak blue color was detectable in the sepal and stronger staining in the anther (Figures 8N and 8O), where Mtccs52A expression was localized in the pollen grains (Figure 8P). After pollination, Mtccs52A was expressed in the immature seed (Figure 8Q). In the seedpod, mainly the vascular bundles were stained (Figure 8R). These expression data showed clearly that Mtccs52A is involved in different stages of plant development. The mostly transient Mtccs52A expression during the initiation and differentiation of the organs also indicated that MtCcs52A, in addition to cell cycle exit, might contribute to the determination of cell fate by destruction of specific target proteins.
Figure 8.
Expression of Mtccs52A during the Development of Transgenic M. truncatula Plants Harboring the Mtccs52A Promoter-uidA Fusion.
GUS staining is shown in the embryo (A), developing seedling (B), shoot apex with opening cotyledons (C), at the initiation of secondary roots (D), during leaf development ([E] to [H]), in differentiated leaves (I), during successive stages of lateral root development ([J] to [M]), in flower (N), in pollen sac (O), in pollen grain (P), in developing embryo (Q), and in the seedpod (R). Closed arrowheads point to the induction of the Mtccs52A expression, whereas the open arrowheads point to the decay of expression.
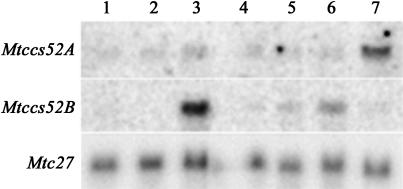
As no transgenic lines were available with Mtccs52B promoter-reporter gene fusion, the expression of Mtccs52B in different organs was studied by RNA gel blot hybridization and compared with that of Mtccs52A using specific probes for each gene (Figure 9). This hybridization confirmed the expression of Mtccs52A in the developing organs that was the most pronounced in root nodules. The highest expression level of Mtccs52B was in the shoot apex, but the transcripts were also present in the root, especially in the root tip and in the hypocotyls, whereas they were not detectable in the cotyledons and likely absent during leaf development. These data suggest that both Mtccs52A and Mtccs52B are involved in plant development, probably contributing differently to the development of various organs.
Figure 9.
Expression of Mtccs52A and Mtccs52B Genes Detected by RNA Gel Blot Analysis in Different M. truncatula Organs.
Lanes correspond to 15μg total RNA from young leaves (1), cotyledons (2), shoot apices (3), hypocotyls (4), roots (5), root tips (6), and nodules (7).
DISCUSSION
Ccs52A and Ccs52B Represent Two Distinct Classes of Cdh1-Type APC Activators in Plants
We have demonstrated that plants have two classes of the Cdh1-type APC activators, the Ccs52A and Ccs52B proteins that are differentially regulated and might have distinct functions during cell cycle and plant development. The overall structures of the MtCcs52A and MtCcs52B proteins are similar (63% identical), however each of them is more related to the same type of proteins from other species than to each other, thereby forming two subgroups of the Ccs52 proteins.
The cell cycle control and developmental regulation of the ccs52A and ccs52B genes exhibited distinct features. During the cell cycle, ccs52A expression was constitutive, however both the transcript levels and the amounts of the Ccs52A protein (data not shown) were somewhat reduced during G2 and G2/M. Expression of ccs52B was surprisingly similar to that of cyclin B. This expression pattern has not been described for the cdh1 genes, and this was also different from the reported expression profile of cdc20 from late S-phase to M-phase (Prinz et al., 1998), suggesting the involvement of MtCcs52B in late G2-phase and M-phase functions. Induced expression of Mtccs52B arrested BY2-Rtet17 cells with 4C DNA content, probably because of unscheduled degradation of certain mitotic regulator(s). This hypothesis is reinforced by identification of Cc52B interacting proteins involved in M-phase control and exit from mitosis (our unpublished data).
Expression of both genes was detected in various plant organs; however, their transcript levels differed in most organs. The spatial and temporal expression pattern of Mtccs52A, followed by the Mtccs52A promoter–driven GUS activity in transgenic M. truncatula, indicated roles for MtCcs52A in pollen grains, seedpods, and the embryos and during primary and lateral root and leaf development. By RNA gel blot analysis, expression pattern of Mtccs52A and Mtccs52B displayed differences in the tested organs. These data, together with the identification of Ccs52 interacting partners (our unpublished data), indicate nonredundant roles for Ccs52A and Ccs52B proteins. This is further supported by overexpression of the Mtccs52 genes in S. pombe, in which Mtccs52A was active and Mtccs52B was inactive.
APC Interaction Requires CSM, a Novel Functional Domain in the Cdh1-Type Proteins and Depends on the N-Terminal Regions
Previous work on the S. cerevisiae Hct1 protein demonstrated the importance of the C-box and the terminal IR motif in the APC interaction (Schwab et al., 2001). Because the deletion of 74 amino acids from the N terminus, including the C-box, abolished the binding of Hct1, it was concluded that this conserved, seven amino acid–long motif was required for APC interaction. We observed the conservation of an additional motif that was exclusively present in the Cdh1-type proteins and studied the significance of this CSM sequence in the Ccs52A protein by deletion analysis. We showed that the lack of CSM significantly decreased the binding of Ccs52A to the APC and inactivated the protein. Similarly, deletion of the C-box or mutation of the IR motif reduced the interaction with APC at the same extent. Therefore, all of these three motifs, the C-box, CSM, and IR, are needed for binding of Cdh1-type activators to the APC. During the preparation of this article, Vodermaier et al. (2003) reported that the TPR domains in the H. sapiens APC3 and APC7 subunits bind the C-terminal IR tail of the Cdh1 proteins. Similar to our results, mutations in the C-box and in the IR tail of the H. sapiens Cdh1 decreased the binding, however even the combination of the two mutations did not abolish completely the interaction with the APC. This is in-line with our finding that a third motif, the CSM, is also necessary for the binding of the Cdh1-type proteins to the APC. Therefore, the remaining residual but significant binding of the H. sapiens Cdh1 in the absence of both the C-box and the IR tail is likely mediated by CSM.
In the MtCcs52A and MtCcs52B proteins, the IR tail was conserved, but the C-box and the CSM sequences displayed differences, which suggested that the inability of MtCcs52B to bind to the yeast APC might be because of these differences. By exchanging the N-terminal regions of the Ccs52A and Ccs52B proteins, we showed that the interaction with the S. pombe APC was only possible when the N-terminal region of the Ccs52A was present in the chimeric protein. In this NA-CB protein, the C-box sequence was identical to that of the yeast Srw1/Ste9 protein, whereas in MtCcs52B it differed in one amino acid. For the activity of the NA-CB hybrid protein, MtCcs52B provided the IR tail and recruited the yeast mitotic cyclin because the yeast phenotype was a result of degradation of this cyclin, as we showed previously (Cebolla et al., 1999). Thus, the lack of interaction between the Ccs52B protein and the S. pombe APC was indeed because of differences in the N-terminal sequences.
Interaction of Ccs52A with APC May Be Regulated by Conserved Phosphorylation Sites in Front of the C-Box and Might Be Influenced by Its Turnover via the PEST Motif
The Cdh1 proteins contain several CDK phosphorylation sites. In yeast and H. sapiens cells, hyperphosphorylation of Cdh1 or mutations mimicking its hyperphosphorylation prevented the binding of Cdh1 to the APC (Zachariae et al., 1998; Jaspersen et al., 1999; Blanco et al., 2000; Kramer et al., 2000). MtCcs52A contains six CDK phosphorylation sites, and by creation of a series of mutations mimicking phosphorylation, we showed that it was not the degree of phosphorylation but the phosphorylation of particular sites, namely in front of the C-box, that abolished the binding of Ccs52A to the APC. Analysis of Ala mutations, mimicking the nonphosphorylated sites in Ccs52A, did not reveal any differences among the sites, whereas mutations in all the six sites increased the activity in yeast, provoking more efficiently the formation of elongated cell types.
Ccs52A lacking the N-terminal PEST motif behaved similarly to the wild-type protein. On the other hand, more (Δpest)Ccs52A than wild-type protein could be recovered from the APC in the in vitro binding assay (Figure 5D). This may indicate that the PEST sequence is functional and regulates the turnover of the protein. Thus, the lack of this motif may stabilize Ccs52A and increase the formation of the APC(Δpest)Ccs52A complexes.
Possible Functions of the Plant Ccs52A and Ccs52B Proteins
In Drosophila melanogaster, the APC may exist in the forms of several related complexes that might be differentially localized and perform distinct functions (Huang and Raff, 2002). APCs have not been isolated from plants; however, APC subunits have been predicted in the A. thaliana genome (Capron et al., 2003). Surprisingly, Cdc27 (corresponding to the H. sapiens APC3 subunit) is encoded by two genes in A. thaliana, which leads to the formation of two functionally distinct APCs (Blilou et al., 2002) and suggests higher complexity of the APC forms in plants than in yeasts and animals. In M. truncatula, we have isolated several cDNA clones encoding putative APC components (our unpublished data), however plant APC complexes remain to be identified. It is possible that Ccs52A and Ccs52B may bind to different APC forms or subcomplexes. Because Ccs52B was unable to bind to the yeast APC, it may interact only with plant APCs or certain forms of plant APCs.
The plant Ccs52A and Ccs52B proteins may share their Cdh1 functions. Ccs52A might be critical for APC activity in all cell cycle phases, whereas Ccs52B could have specific roles in M-phase progression, alternative or complementary to those of Ccs52A. Similarly, their function during plant development could be distinct and specific. APCCcs52A was dispensable for nodule meristem formation but required for endoreduplication cycles and differentiation of nodule cells (Vinardell et al., 2003). Therefore, Ccs52A might have major roles in postmitotic, differentiating cells, in which degradation of specific APC targets could contribute to differentiation of given cell types, tissues, or organs. The Ccs52A, Ccs52B, and numerous Cdc20 proteins in plants interacting with different APC forms can thus provide a finely tuned regulation and substrate specificity of the plant APCs during cell cycle and developmental programs.
METHODS
DNA Constructs and Genetic Manipulations
PCR amplifications were made with Pfu enzyme (Promega, Madison, WI) using the oligonucleotides listed in the supplemental data online. Deletions in the Mtccs52A cloned in pBluescript SK+ (Stratagene, La Jolla, CA) were constructed by amplifying cDNAs fragments from the flanking regions with T3/T7 primers in combination with primers containing ClaI sites that were restricted with ClaI and ligated. ΔC-box deletion (Δ26 to 46) was obtained by ligation of the T3/Cla1 fragment and the Cla2/T7 fragment. ΔCSM deletion (Δ75 to 118) was generated by ligating the T3/Cla3 fragment with the Cla4/T7 fragment. Prediction of PEST motif was according to http://www.at.embnet.org/embnet/tools/bio/PESTfind/. Δpest deletion (Δ7 to 30) was created with ΔpestA and T7 oligonucleotides. Phosphorylation sites were predicted according to the consensus sequences (S/T)Px(K/R) and (S/T)P(K/R) (Nigg, 1993). Amino acid replacements in MtCcs52A were performed by site-directed mutagenesis (QuikChange site-directed mutagenesis kit; Stratagene). NA-CB and NB-CA hybrid proteins were constructed by amplifying the 5′ and 3′ regions of Mtccs52A and Mtccs52B cDNA with primers containing SalI sites that were restricted with SalI and ligated. In NA-CB, the 5′ part of Mtccs52A cDNA (corresponding to amino acids 1 to 140) was amplified with T3/ NA-L and ligated to the 3′ part of Mtccs52B cDNA (corresponding to amino acids 138 to 471) amplified with CB-R/T7. In NB-CA, the 5′ part of Mtccs52B cDNA (corresponding to amino acids 1 to 138) was amplified with T3/ NB-L and ligated to the 3′ part of Mtccs52A cDNA (amino acids 143 to 475) amplified with CA-R/T7. All mutant derivatives of the Mtccs52A were cloned in pREP1 and verified by sequencing. These pREP1 plasmids as well as the empty vector were transformed into S. pombe SP-Q01 (Stratagene) according to Agatep et al. (1998) and plated on Edinburgh minimal medium containing 50 μM thiamine. Single colonies from each transformation were grown in liquid medium in the presence of thiamine overnight, and then gene expression was induced by culturing the cells for 8 h in EMM medium without thiamine. Cells were fixed in 70% ethanol, stained with DAPI (0.5 μg/mL), and analyzed by fluorescent microscopy.
Plant Manipulations
Transgenic M. truncatula lines containing the Mtccs52A promoter-uidA transcriptional fusion and the histochemical GUS staining are described by Vinardell et al. (2003). Chlorophyll was removed from the green tissues with repeated washes in 30% ethanol and 70% acetone. Synchronization of the M. sativa A2 cell culture and RT-PCR analysis were according to Roudier et al. (2000). Msccs52A was amplified with p52GL and p52-3′ end primers (17 cycles), Msccs52B with c12P1 and c12P2 (21 cycles), Medsa;cycB2 with pro4L and pro7R (20 cycles), and Msc27 with Msc27-1 and Msc27-1 (15 cycles). Amplified cDNAs were blotted, hybridized with specific probes, and exposed on a Storm PhosphorImager (Molecular Dynamics, Sunnyvale, CA).
For induced expression of Mtccs52B in BY2 cells, the Mtccs52B cDNA was cloned in the XbaI/SalI sites of the pBinOP vector (Gatz et al., 1992) containing the Triple-Op repressible promoter. The empty vector pBinOP and pBinOP-Mtccs52B were transformed into BY2-Rtet17 cells expressing the tetracycline repressor, according to David and Perrot-Rechenmann (2001). Ten independent transgenic lines were selected and propagated in liquid cultures. Mtccs52B transcript levels were analyzed by RNA gel blot analysis after 72 h of tetracycline induction. DNA contents of nuclei were measured by flow cytometry (Roudier et al., 2000) in selected lines displaying high Mtccs52B expression.
Binding of the MtCcs52A and MtCcs52B Proteins to the S. pombe APC
The MtCcs52A, MtCcs52B, Δpest, ΔC-box, ΔCSM, ΔIR, P12E, P4E, P45E, P1245E, P12345E, P123456E, and P1234566A proteins were in vitro translated and labeled with [35S]Met (TNT T7/T3 coupled reticulocyte lysate system; Promega). The APC was immunoprecipitated from the S. pombe strain S813 lid1:lid-9xMyc (Berry et al., 1999). Overnight culture of strain S813 was diluted to OD600 = 0.2 in complete medium and grown for 5 h. Yeast protein extracts were prepared in 50 mM Tris-HCl, pH 7.5, 150mM NaCl, 0.5% Nonidet P-40, 50 mM NaF, 1 mM Na3VO4, 1 mM PMSF, 1 mM DTT, 1 mM MgCl2, 0.5mM ATP, 10 μg/mL aprotinin, 10 μg/mL leupeptin, and 10 μg/mL pepstatin. One milligram of total yeast proteins was incubated on ice for 2 h with 3 μg of mouse monoclonal anti-Myc antibody (9E10; Roche, Basel, Switzerland) and then overnight with 20 μL of Protein G Plus Agarose (TEBU, Le Perray-en-Yvelines, France) at 4°C. After three washes with the extraction buffer, one-fourth of the in vitro translated proteins were incubated with the immunoprecipitated Myc-lid-APC for 4 h. Beads were washed five times with the extraction buffer, two times with the extraction buffer supplemented with 50 mM NaCl, then boiled in 15 μL of loading buffer, and the proteins were separated in 10% acrylamide gel.
RNA Gel Blot Analysis
Fifteen micrograms of total RNA from shoot apices, root tips, hypocotyls, cotyledons, and roots of 3-d-old, in vitro grown M. truncatula plantlets, from young leaves of 13-d-old plantlets, and from nodules were used for RNA gel blot analysis according to Roudier et al. (2000). The membrane was successively hybridized with the Mtccs52A, Mtccs52B, and Msc27 probes. Signals were analyzed by PhosphorImager (Molecular Dynamics). The Mtccs52A probe was amplified with p52GL/p52-3′ end oligonucleotides, resulting in a 297-bp PCR fragment from nucleotides 1473 to 1770 downstream of the translation initiation site. The Mtccs52B probe, amplified with c12P1/ c12P2, corresponded to a 465-bp fragment from nucleotides 1094 to 1559 downstream of the translation initiation. The Mtc27 probe, amplified with Msc27-1/Msc27-2, corresponded to a 310-bp fragment from nucleotides 87 to 397 bp downstream of the translation initiation.
Mtccs52A and Mtccs52B have been deposited in GenBank under accession numbers AF134835 and AY357299, respectively. Other accession numbers presented in Figure 1 are the following: SpSlp1, P78972; MmCdc20_1, XP 139652; HsCdc20_1, NP 001246; HsCdc20_2, A56021; MmCdc20_2, NP 075712; RnCdc20, NP 741990; XlCdc20_1, AAH42288; XlCdc20_2, AAC41376; DmFzy, NP 477501; AgCdc20, CP10238; DcCdc20, T14352; BnCdc20, AJ224078; AtCdc20_1, At4g33260; AtCdc20_2, At4g33270; AtCdc20_3, At5g27570; AtCdc20_4, At5g26900; AtCdc20_5, At5g27080; CeCdh1-like, NP 495051; ScHct1, P53197; UmCdh1, AY118173; DmCdh1_1, NP 611854; CeCdh1, NP 496075; GgCdh1a, AAL31947; GgCdh1c, AAL31949; XlCdh1, CAA74576; HsCdh1, NP 057347; MmCdh1, NP 062731; GgCdh1b, AAL31948; GgCdh1d, AAL31950; DmCdh1_2, NP 726941; AgCdh1, CP12792; OsCcs52-like, AP003298; AtCcs52A2, At4g11920; AtCcs52A1, At4g22910; LeCcs52A, AW030735; MtCcs52A, AF134835; GmCcs52A, BG044933; OsCcs52A, AAN74839; ZmCcs52A, AY112458; ZmCcs52B, AI861254; AtCcs52B, At5g13840; MtCcs52B, AY357299; GmCcs52B, AI736659; SpSrw1, O13286; SpMfr1, O94423; McCcs52B, MO15G09; LsCcs52B, TC7094; StCcs52B, TC59462; LeCcs52B, TC121578; LjCcs52A, BI420244; LsCcs52A, QGG13M0; and StCcs52A, TC62503.
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession numbers AF134835 and AY357299.
Supplementary Material
Acknowledgments
We thank F. Roudier for providing cDNAs from synchronized M. sativa A2 cells, S. Moreno for the S. pombe strain S813, C. Perrot-Rechenmann for the BY2-Rtet17 cell line, N. Mansion for her help in the photographic work, and E. Kiss for his initial contribution to the work. We are grateful to R. Karess for his critical reading of the manuscript. J.M.V. and A.C. were supported by the TMR Marie Curie fellowship program. S.T. is the recipient of a grant from the French Ministry of Research.
On-line version contains Web-only data.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Eva Kondorosi (eva.kondorosi@isv.cnrs-gif.fr).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.018952.
References
- Agatep, R., Kirkpatrick, R.D., Parchaliuk, D.L., Woods, R.A., and Gietz, R.D. (1998). Transformation of Saccharomyces cerevisiae by the lithium acetate/single-stranded carrier DNA/polyethylene glycol (LiAc/ss-DNA/PEG) protocol. Technical Tips Online, http://tto.trends.com.
- Berry, L.D., Feoktistova, A., Wright, M.D., and Gould, K.L. (1999). The Schizosaccharomyces pombe dim1(+) gene interacts with the anaphase-promoting complex or cyclosome (APC/C) component lid1(+) and is required for APC/C function. Mol. Cell. Biol. 19, 2535–2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco, M.A., Sanchez-Diaz, A., de Prada, J.M., and Moreno, S. (2000). APC(ste9/srw1) promotes degradation of mitotic cyclins in G(1) and is inhibited by cdc2 phosphorylation. EMBO J. 19, 3945–3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blilou, I., Frugier, F., Folmer, S., Serralbo, O., Willemsen, V., Wolkenfelt, H., Eloy, N.B., Ferreira, P.C., Weisbeek, P., and Scheres, B. (2002). The Arabidopsis HOBBIT gene encodes a CDC27 homolog that links the plant cell cycle to progression of cell differentiation. Genes Dev. 16, 2566–2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capron, A., Serralbo, O., Fulop, K., Frugier, F., Parmentier, Y., Dong, A., Lecureuil, A., Guerche, P., Kondorosi, E., Scheres, B., and Genschik, P. (2003). The Arabidopsis anaphase-promoting complex or cyclosome: Molecular and genetic characterization of the APC2 subunit. Plant Cell 15, 2370–2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro, A., Vigneron, S., Bernis, C., Labbe, J.C., and Lorca, T. (2003). Xkid is degraded in a D-box, KEN-box, and A-box-independent pathway. Mol. Cell. Biol. 23, 4126–4138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cebolla, A., Vinardell, J.M., Kiss, E., Olah, B., Roudier, F., Kondorosi, A., and Kondorosi, E. (1999). The mitotic inhibitor ccs52 is required for endoreduplication and ploidy-dependent cell enlargement in plants. EMBO J. 18, 4476–4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabes, A.L., Pfleger, C.M., Kirschner, M.W., and Thelander, L. (2003). Mouse ribonucleotide reductase R2 protein: A new target for anaphase-promoting complex-Cdh1-mediated proteolysis. Proc. Natl. Acad. Sci. USA 100, 3925–3929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David, K.M., and Perrot-Rechenmann, C. (2001). Characterization of a tobacco Bright Yellow 2 cell line expressing the tetracycline repressor at a high level for strict regulation of transgene expression. Plant Physiol. 125, 1548–1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatz, C., Frohberg, C., and Wendenburg, R. (1992). Stringent repression and homogeneous de-repression by tetracycline of modified CaMV 35S promoter in intact transgenic tobacco plants. Plant J. 2, 397–404. [DOI] [PubMed] [Google Scholar]
- Genschik, P., Criqui, M.C., Parmentier, Y., Derevier, A., and Fleck, J. (1998). Cell cycle-dependent proteolysis in plants: Identification of the destruction box pathway and metaphase arrest produced by the proteasome inhibitor mg132. Plant Cell 10, 2063–2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gieffers, C., Peters, B.H., Kramer, E.R., Dotti, C.G., and Peters, J.M. (1999). Expression of the CDH1-associated form of the anaphase-promoting complex in postmitotic neurons. Proc. Natl. Acad. Sci. USA 96, 11317–11322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glotzer, M., Murray, A.W., and Kirschner, M.W. (1991). Cyclin is degraded by the ubiquitin pathway. Nature 349, 132–138. [DOI] [PubMed] [Google Scholar]
- Hagting, A., Den Elzen, N., Vodermaier, H.C., Waizenegger, I.C., Peters, J.M., and Pines, J. (2002). Human securin proteolysis is controlled by the spindle checkpoint and reveals when the APC/C switches from activation by Cdc20 to Cdh1. J. Cell Biol. 157, 1125–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper, J.W., Burton, J.L., and Solomon, M.J. (2002). The anaphase-promoting complex: It's not just for mitosis anymore. Genes Dev. 16, 2179–2206. [DOI] [PubMed] [Google Scholar]
- Huang, J.N., Park, I., Ellingson, E., Littlepage, L.E., and Pellman, D. (2001). Activity of the APC(Cdh1) form of the anaphase-promoting complex persists until S phase and prevents the premature expression of Cdc20p. J. Cell Biol. 154, 85–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, J.Y., and Raff, J.W. (2002). The dynamic localisation of the Drosophila APC/C: Evidence for the existence of multiple complexes that perform distinct functions and are differentially localised. J. Cell Sci. 115, 2847–2856. [DOI] [PubMed] [Google Scholar]
- Jaquenoud, M., van Drogen, F., and Peter, M. (2002). Cell cycle-dependent nuclear export of Cdh1p may contribute to the inactivation of APC/C(Cdh1). EMBO J. 21, 6515–6526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaspersen, S.L., Charles, J.F., and Morgan, D.O. (1999). Inhibitory phosphorylation of the APC regulator Hct1 is controlled by the kinase Cdc28 and the phosphatase Cdc14. Curr. Biol. 9, 227–236. [DOI] [PubMed] [Google Scholar]
- Kashevsky, H., Wallace, J.A., Reed, B.H., Lai, C., Hayashi-Hagihara, A., and Orr-Weaver, T.L. (2002). The anaphase promoting complex/cyclosome is required during development for modified cell cycles. Proc. Natl. Acad. Sci. USA 99, 11217–11222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer, E.R., Scheuringer, N., Podtelejnikov, A.V., Mann, M., and Peters, J.M. (2000). Mitotic regulation of the APC activator proteins CDC20 and CDH1. Mol. Biol. Cell 11, 1555–1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Littlepage, L.E., and Ruderman, J.V. (2002). Identification of a new APC/C recognition domain, the A box, which is required for the Cdh1-dependent destruction of the kinase Aurora-A during mitotic exit. Genes Dev. 16, 2274–2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGarry, T.J., and Kirschner, M.W. (1998). Geminin, an inhibitor of DNA replication, is degraded during mitosis. Cell 93, 1043–1053. [DOI] [PubMed] [Google Scholar]
- Nigg, E.A. (1993). Targets of cyclin-dependent protein kinases. Curr. Opin. Cell Biol. 5, 187–193. [DOI] [PubMed] [Google Scholar]
- Peters, J.M. (1998). SCF and APC: The Yin and Yang of cell cycle regulated proteolysis. Curr. Opin. Cell Biol. 10, 759–768. [DOI] [PubMed] [Google Scholar]
- Peters, J.M. (2002). The anaphase-promoting complex: Proteolysis in mitosis and beyond. Mol. Cell 9, 931–943. [DOI] [PubMed] [Google Scholar]
- Petersen, B.O., Wagener, C., Marinoni, F., Kramer, E.R., Melixetian, M., Denchi, E.L., Gieffers, C., Matteucci, C., Peters, J.M., and Helin, K. (2000). Cell cycle- and cell growth-regulated proteolysis of mammalian CDC6 is dependent on APC-CDH1. Genes Dev. 14, 2330–2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfleger, C.M., and Kirschner, M.W. (2000). The KEN box: An APC recognition signal distinct from the D box targeted by Cdh1. Genes Dev. 14, 655–665. [PMC free article] [PubMed] [Google Scholar]
- Prinz, S., Hwang, E.S., Visintin, R., and Amon, A. (1998). The regulation of Cdc20 proteolysis reveals a role for APC components Cdc23 and Cdc27 during S phase and early mitosis. Curr. Biol. 8, 750–760. [DOI] [PubMed] [Google Scholar]
- Raff, J.W., Jeffers, K., and Huang, J.Y. (2002). The roles of Fzy/Cdc20 and Fzr/Cdh1 in regulating the destruction of cyclin B in space and time. J. Cell Biol. 157, 1139–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers, S., Wells, R., and Rechsteiner, M. (1986). Amino acid sequences common to rapidly degraded proteins: The PEST hypothesis. Science 234, 364–368. [DOI] [PubMed] [Google Scholar]
- Roudier, F., Fedorova, E., Gyorgyey, J., Feher, A., Brown, S., Kondorosi, A., and Kondorosi, E. (2000). Cell cycle function of a Medicago sativa A2-type cyclin interacting with a PSTAIRE-type cyclin-dependent kinase and a retinoblastoma protein. Plant J. 23, 73–83. [DOI] [PubMed] [Google Scholar]
- Schwab, M., Neutzner, M., Mocker, D., and Seufert, W. (2001). Yeast Hct1 recognizes the mitotic cyclin Clb2 and other substrates of the ubiquitin ligase APC. EMBO J. 20, 5165–5175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigrist, S.J., and Lehner, C.F. (1997). Drosophila fizzy-related down-regulates mitotic cyclins and is required for cell proliferation arrest and entry into endocycles. Cell 90, 671–681. [DOI] [PubMed] [Google Scholar]
- Sorensen, C.S., Lukas, C., Kramer, E.R., Peters, J.M., Bartek, J., and Lukas, J. (2001). A conserved cyclin-binding domain determines functional interplay between anaphase-promoting complex-Cdh1 and cyclin A-Cdk2 during cell cycle progression. Mol. Cell. Biol. 21, 3692–3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroschein, S.L., Bonni, S., Wrana, J.L., and Luo, K. (2001). Smad3 recruits the anaphase-promoting complex for ubiquitination and degradation of SnoN. Genes Dev. 15, 2822–2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, Z., Li, B., Bharadwaj, R., Zhu, H., Ozkan, E., Hakala, K., Deisenhofer, J., and Yu, H. (2001). APC2 Cullin protein and APC11 RING protein comprise the minimal ubiquitin ligase module of the anaphase-promoting complex. Mol. Biol. Cell 12, 3839–3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinardell, J.M., Fedorova, E., Cebolla, A., Kevei, Z., Horvath, G., Kelemen, Z., Tarayre, S., Roudier, F., Mergaert, P., Kondorosi, A., and Kondorosi, E. (2003). Endoreduplication mediated by the anaphase-promoting complex activator CCS52A is required for symbiotic cell differentiation in Medicago truncatula nodules. Plant Cell 15, 2093–2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vodermaier, H.C., Gieffers, C., Maurer-Stroh, S., Eisenhaber, F., and Peters, J.M. (2003). TPR subunits of the anaphase-promoting complex mediate binding to the activator protein CDH1. Curr. Biol. 13, 1459–1468. [DOI] [PubMed] [Google Scholar]
- Wan, Y., and Kirschner, M.W. (2001). Identification of multiple CDH1 homologues in vertebrates conferring different substrate specificities. Proc. Natl. Acad. Sci. USA 98, 13066–13071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan, Y., Liu, X., and Kirschner, M.W. (2001). The anaphase-promoting complex mediates TGF-beta signaling by targeting SnoN for destruction. Mol. Cell 8, 1027–1039. [DOI] [PubMed] [Google Scholar]
- Yeong, F.M., Lim, H.H., Wang, Y., and Surana, U. (2001). Early expressed Clb proteins allow accumulation of mitotic cyclin by inactivating proteolytic machinery during S phase. Mol. Cell. Biol. 21, 5071–5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zachariae, W., and Nasmyth, K. (1999). Whose end is destruction: Cell division and the anaphase-promoting complex. Genes Dev. 13, 2039–2058. [DOI] [PubMed] [Google Scholar]
- Zachariae, W., Schwab, M., Nasmyth, K., and Seufert, W. (1998). Control of cyclin ubiquitination by CDK-regulated binding of Hct1 to the anaphase promoting complex. Science 282, 1721–1724. [DOI] [PubMed] [Google Scholar]
- Zhou, Y., Ching, Y.P., Chun, A.C., and Jin, D.Y. (2003. a). Nuclear localization of the cell cycle regulator CDH1 and its regulation by phosphorylation. J. Biol. Chem. 278, 12530–12536. [DOI] [PubMed] [Google Scholar]
- Zhou, Y., Ching, Y.P., Ng, R.W., and Jin, D.Y. (2003. b). Differential expression, localization and activity of two alternatively spliced isoforms of human APC regulator CDH1. Biochem. J. 374, 349–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.