Abstract
In Arabidopsis thaliana, the calcium binding protein Salt Overly Sensitive3 (SOS3) interacts with and activates the protein kinase SOS2, which in turn activates the plasma membrane Na+/H+ antiporter SOS1 to bring about sodium ion homeostasis and salt tolerance. Constitutively active alleles of SOS2 can be constructed in vitro by changing Thr168 to Asp in the activation loop of the kinase catalytic domain and/or by removing the autoinhibitory FISL motif from the C-terminal regulatory domain. We expressed various activated forms of SOS2 in Saccharomyces cerevisiae (yeast) and in A. thaliana and evaluated the salt tolerance of the transgenic organisms. Experiments in which the activated SOS2 alleles were coexpressed with SOS1 in S. cerevisiae showed that the kinase activity of SOS2 is partially sufficient for SOS1 activation in vivo, and higher kinase activity leads to greater SOS1 activation. Coexpression of SOS3 with SOS2 forms that retained the FISL motif resulted in more dramatic increases in salt tolerance. In planta assays showed that the Thr168-to-Asp–activated mutant SOS2 partially rescued the salt hypersensitivity in sos2 and sos3 mutant plants. By contrast, SOS2 lacking only the FISL domain suppressed the sos2 but not the sos3 mutation, whereas truncated forms in which the C terminus had been removed could not restore the growth of either sos2 or sos3 plants. Expression of some of the activated SOS2 proteins in wild-type A. thaliana conferred increased salt tolerance. These studies demonstrate that the protein kinase activity of SOS2 is partially sufficient for activation of SOS1 and for salt tolerance in vivo and in planta and that the kinase activity of SOS2 is limiting for plant salt tolerance. The results also reveal an essential in planta role for the SOS2 C-terminal regulatory domain in salt tolerance.
INTRODUCTION
Soil salinity is a serious environmental stress limiting plant productivity. Sodium ions (Na+), which are abundant in saline soils, are cytotoxic in plants when they accumulate to high concentrations. Na+ enters plant cells through transporters such as HKT1 (Rus et al., 2001) and nonselective cation channels (Amtmann and Sanders, 1999). To prevent Na+ buildup in the cytoplasm, plant cells employ Na+/H+ antiporters at the plasma membrane and tonoplast to transport Na+ into the apoplast and vacuole, respectively (Apse et al., 1999; Qiu et al., 2002). Overexpression of the Arabidopsis thaliana plasma membrane Na+/H+ antiporter Salt Overly Sensitive1 (SOS1) or the vacuolar Na+/H+ antiporter AtNHX1 improves salt tolerance in transgenic plants (Apse et al., 1999; Zhang and Blumwald, 2001; Zhang et al., 2001; Shi et al., 2003). Enhanced salt tolerance can also be achieved by overexpression of the vacuolar H+-pyrophosphatase AVP1, which generates the driving force for Na+ transport into the vacuole (Gaxiola et al., 2001).
Recently, a regulatory pathway for ion homeostasis and salt tolerance was identified in A. thaliana (Zhu, 2000, 2002). Salt stress is known to elicit a rapid increase in the free calcium concentration in the cytoplasm (Knight et al., 1997). SOS3, a myristoylated calcium binding protein, is proposed to sense this calcium signal (Liu and Zhu, 1998; Ishitani et al., 2000). SOS3 physically interacts with the protein kinase SOS2 and activates the substrate phosphorylation activity of SOS2 in a calcium-dependent manner (Halfter et al., 2000; Liu et al., 2000). SOS3 also recruits SOS2 to the plasma membrane, where the SOS3-SOS2 protein kinase complex phosphorylates SOS1 to stimulate its Na+/H+ antiport activity (Qiu et al., 2002; Quintero et al., 2002). Loss-of-function mutations in SOS3, SOS2, or SOS1 cause hypersensitivity to Na+ (Zhu et al., 1998).
SOS2 has a highly conserved N-terminal catalytic domain similar to that of Saccharomyces cerevisiae SNF1 and animal AMPK (Liu et al., 2000). Within the SOS2 protein, the N-terminal catalytic region interacts with the C-terminal regulatory domain (Guo et al., 2001). SOS3 interacts with the FISL motif in the C-terminal region of SOS2 (Guo et al., 2001), which serves as an autoinhibitory domain. A constitutively active SOS2 kinase, T/DSOS2, can be engineered by a Thr168-to-Asp change (to mimic phosphorylation by an upstream kinase) in the putative activation loop. The kinase activity of T/DSOS2 is independent of SOS3 and calcium (Guo et al., 2001). Constitutively active forms of SOS2 can also be created by removing the FISL motif (SOS2DF) or the entire C-terminal regulatory domain (SOS2/308) (Guo et al., 2001; Qiu et al., 2002). The activation loop mutation and the autoinhibitory domain deletions have a synergistic effect on the kinase activity of SOS2, and superactive SOS2 kinases T/DSOS2/308 or T/DSOS2/DF can be created when the two changes are combined (Guo et al., 2001; Qiu et al., 2002). We have shown that T/DSOS2/DF could activate the transport activity of SOS1 in vitro, whereas the wild-type SOS2 protein could not (Qiu et al., 2002). However, whether these active forms of SOS2 can function in vivo is not known.
In this study, we expressed various activated SOS2 proteins in S. cerevisiae and A. thaliana, with the aim of determining if the protein kinase activity of SOS2 is sufficient for activation of the SOS1 plasma membrane Na+/H+ antiporter in vivo and in planta, and identifying domains in the SOS2 protein that are important for its in planta function. We also investigated if the kinase activity of SOS2 is limiting for plant salt tolerance to evaluate the potential of using the activated SOS2 mutant alleles for improving the ability of plants to grow in saline soils.
RESULTS
Changes in the SOS2 Protein Produce Constitutively Active Kinases
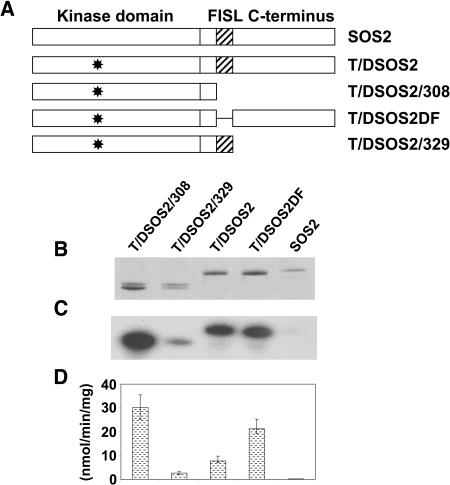
Based on its inability to autophosphorylate or phosphorylate a peptide substrate, SOS2 appears to be an inactive kinase. The calcium binding protein SOS3 has been shown to interact with and activate SOS2 in vitro in the presence of calcium (Halfter et al., 2000). We have previously shown that SOS2 kinases that are active in the absence of SOS3 and calcium (constitutively active SOS2) could be produced either by exchange of Thr168 in the activation loop to the acidic residue Asp (T/DSOS2) or by deletion of the FISL motif in the C-terminal regulatory domain of the SOS2 protein (SOS2DF) (Guo et al., 2001; Qiu et al., 2002), and that a superactive SOS2 kinase could be generated by combining these two changes (T/DSOS2DF) (Qiu et al., 2002). In this study, additional changes were made to the SOS2 kinase to allow us to develop a series of SOS2 proteins for studies of SOS2 structure and function. The FISL motif and C-terminal 117 amino acids or the C-terminal 117 amino acids were removed in the glutathione S-transferase (GST)-T/DSOS2/308 and GST-T/DSOS2/329 constructs, respectively (Figure 1A). These proteins were assayed for autophosphorylation or their ability to phosphorylate a peptide substrate, and their activities compared with those of the wild-type SOS2 protein, T/DSOS2, or T/DSOS2DF. T/DSOS2/308 had the strongest activities, followed by T/DSOS2DF, T/DSOS2, T/DSOS2/329, and SOS2 (Figures 1B to 1D). These kinase constructs served as the basis of the following transgenic studies in S. cerevisiae and A. thaliana.
Figure 1.
Active SOS2 Kinases.
Model of the domains of the wild-type and altered SOS2 protein kinases (A) is shown. The kinase activities (autophosphorylation and phosphorylation of an in vitro substrate) of altered forms of SOS2 (GST fusion proteins of T/DSOS2, T/DSOS2/308, T/DSOS2DF, and T/DSOS2/329) were evaluated. After the autophosphorylation assays, protein was separated by SDS-PAGE, and the gel was stained with Coomassie blue (B) and exposed to x-ray film (C). The ability of the same GST-SOS2 fusion proteins to phosphorylate the peptide substrate p3 (400 pmol per assay) was determined (D).
The Protein Kinase Activity of SOS2 Is Partially Sufficient for Salt Tolerance in Vivo in a Heterologous System
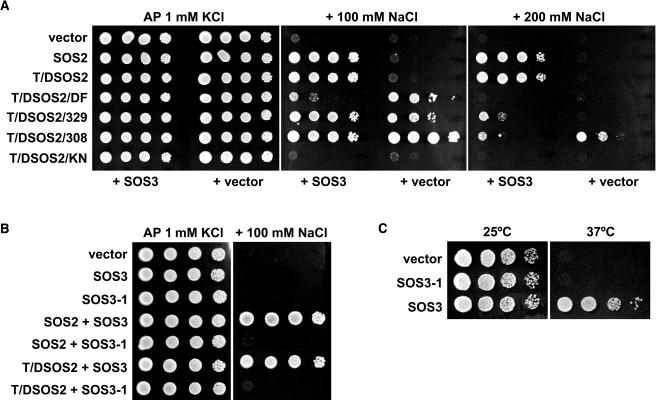
Recently, the A. thaliana SOS regulatory pathway has been reconstituted in S. cerevisiae (Quintero et al., 2002), providing an in vivo system for studies of SOS2 structure–function relationships. To determine if the kinase activity of SOS2 correlates with activation of SOS1, wild-type and constitutively active SOS2 kinases were introduced into S. cerevisiae strain YP890, in which the endogenous S. cerevisiae Na+ transporters (Na+ efflux proteins ENA1-4 and NHA1 and the vacuolar Na+/H+ exchanger NHX1) had been removed and the A. thaliana SOS1 gene was constitutively expressed from a chromosomal insertion. The transformed S. cerevisiae strains were grown on Arg phosphate (AP) medium containing 1 mM KCl and various concentrations of NaCl, and the results are shown in Figure 2. The low basal activity of SOS1 and the moderate level of expression achieved in strain YP890 failed to support cell growth above 50 mM NaCl (data not shown). The salt tolerance of YP890 was not substantially enhanced by the expression of the wild-type SOS2 (Figure 2A) but was dramatically increased by the coexpression of the SOS2-SOS3 kinase complex. There was no further increase in salt tolerance when T/DSOS2, bearing the Thr168-to-Asp mutation that mimicked the phosphorylated state of SOS2 (Gong et al., 2002), was expressed in place of the wild-type SOS2 (Figure 2). By contrast, a Lys40-to-Asn mutation in the catalytic site required for phosphotransfer activity (Gong et al., 2002) produced an inactive kinase (T/DSOS2/KN), even in the presence of SOS3. Expression T/DSOS2/308, with a truncation that removed the entire autoinhibitory C-terminal part of SOS2, strongly enhanced the ability of S. cerevisiae to grow in NaCl in the absence of SOS3, and coexpression of SOS3 failed to increase salt tolerance further because of lack of interaction between these two proteins. Deletion of the FISL motif (T/DSOS2DF) partially released SOS2 from autoinhibition, as did the truncation in T/DSOS2/329 that removed the last 117 C-terminal amino acids but retained the FISL domain. However, although SOS3 cooperated with T/DSOS2/329 through the FISL domain to activate SOS1, coexpression of SOS3 had no effect on T/DSOS2DF (Figure 2A). The greater salt tolerance imparted by T/DSOS2/308 relative to T/DSOS2DF and T/DSOS2/329 in the absence of SOS3, together with data shown in Figure 1, indicate that the entire C-terminal part of SOS2 may contribute to autoinhibition of the kinase activity. In the presence of SOS3, both T/DSOS2/308 and T/DSOS2/329 performed similarly regarding SOS1 activation (Figure 2A), despite their significantly different kinase and autophosphorylation activities in vitro (Figure 1). Together, these results demonstrate that the kinase activity determined in vitro correlates well with functionality of SOS2 in vivo and in the absence of SOS3, but they also illustrate that the capacity for binding SOS3 and recruitment to the plasma membrane is critical for the competence of SOS2 for SOS1 activation. On the other hand, none of the SOS2 kinases activated through protein truncation could increase the salt tolerance of S. cerevisiae to the same level achieved when SOS1 was coexpressed with SOS2 and SOS3 proteins retaining structural integrity, indicating that interaction between the full-length polypeptides is optimal for function.
Figure 2.
Competence of Various Forms of the SOS2 Kinase and the Ancillary Protein SOS3 to Increase the Salt Tolerance of S. cerevisiae Expressing SOS1.
(A) Wild-type SOS2, activated SOS2 kinases (T/DSOS2, T/DSOS2/DF, T/DSOS2/329, and T/DSOS2/308), and inactive SOS2 mutant (T/DSOS2/KN) were cotransformed with or without SOS3 into S. cerevisiae strain YP890 cells. Transformants were grown overnight in liquid AP medium with 1 mM KCl, and 5 μL of serial decimal dilutions were spotted onto plates containing AP medium with 1 mM KCl or supplemented with 100 or 200 mM NaCl. Plates were incubated at 28°C and photographed after 4 d.
(B) Wild-type SOS2 and activated kinase T/DSOS2 were coexpressed with wild-type SOS3 or mutant SOS3-1 in YP890 cells. Salt tolerance that resulted from the combination of these proteins was determined as indicated above.
(C) Cdc25-2 cells carrying plasmid pSRS2-1 for the expression of an hSos:SOS2 chimera were transformed to produce wild-type SOS3 or mutant SOS3-1 proteins, or with an empty vector. Cells were grown overnight at 25°C in selective medium and then spotted on duplicate YPD plates that were incubated for 2 d at either 25°C or 37°C. Growth at 37°C indicates targeting of the SOS2 kinase to the plasma membrane.
The sos3-1 mutation of A. thaliana causes deletion of three conserved amino acids in a central EF hand (Liu and Zhu, 1998). Although we have shown previously that the sos3-1 mutation drastically reduces the capacity of SOS3 to activate SOS2 in vitro (Ishitani et al., 2000), we tested if the SOS3-1 polypeptide could still interact in vivo with activated SOS2 proteins retaining the FISL motif and recruit them to the plasma membrane. Alleles SOS2 and T/DSOS2 were coexpressed with the cDNA of sos3-1 in YP890 cells. As shown in Figure 2B, the SOS3-1 mutant polypeptide failed to mediate activation of SOS1 by SOS2 or T/DSOS2. In addition, using the SOS Recruitment System (SRS) to monitor targeting of SOS2 to the plasma membrane (Quintero et al., 2002), we determined that SOS3-1 was unable to recruit SOS2 or T/DSOS2 to the plasma membrane (Figure 2C).
The Protein Kinase Activity of SOS2 Is Partially Sufficient for Salt Tolerance in Planta
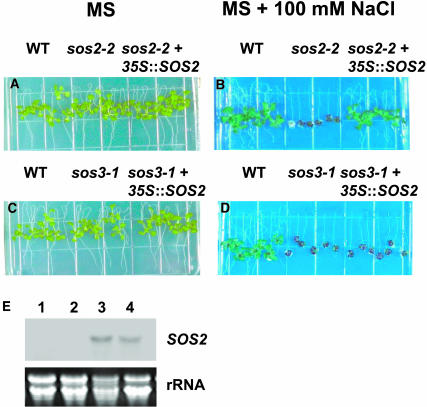
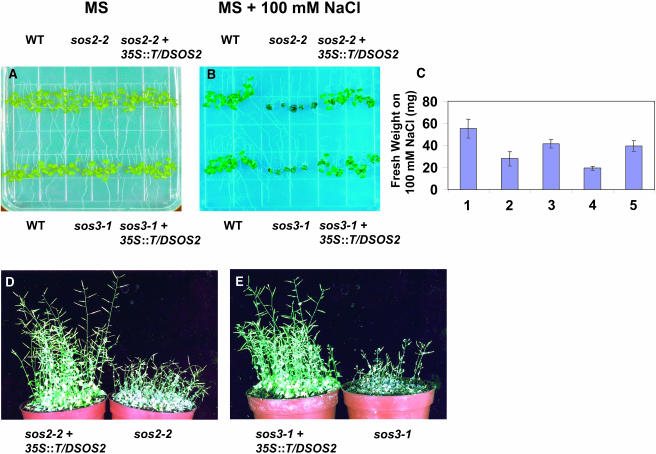
To determine if the protein kinase activity of SOS2 is sufficient for salt tolerance in planta, the wild-type and the constitutively active forms of SOS2 were expressed under the 35S promoter of Cauliflower mosaic virus (CaMV) in the sos2 and sos3 mutants of A. thaliana. Five-day-old T2 transgenic plants expressing 35S:SOS2 (germinated on MS medium without salt) were transferred to plates with either MS medium or MS medium with 100 mM NaCl. Three of twelve independent T2 transgenic lines in the sos2-2 background evaluated had salt tolerance nearly restored to levels equivalent to that of the wild type. By contrast, none of the 24 independent transgenic lines in the sos3-1 background evaluated showed any increased salt tolerance relative to the sos3-1 mutant.
One representative T3 homozygous 35S:SOS2 line in the sos3-1 and sos2-2 backgrounds was evaluated for SOS2 transcript accumulation and growth in salt (Figure 3). RNA analysis indicated that the transgenic plants accumulated high levels of SOS2 mRNA from the transgene because the endogenous SOS2 expression was extremely low (Figure 3E) and could only be seen with prolonged exposure of the blot (data not shown). The results show that ectopic expression of SOS2 under the CaMV 35S promoter could rescue the sos2-2 phenotype (Figure 3B). As expected, the ectopic expression of SOS2 did not rescue the sos3-1 salt-hypersensitive phenotype (Figure 3D), confirming that the wild-type SOS2 protein must be activated by SOS3 in vivo for function in A. thaliana.
Figure 3.
Expression of SOS2 Complements the sos2-2 Salt-Sensitive Phenotype but Not the sos3-1 Salt-Sensitive Phenotype.
Five-day-old seedlings grown on MS agar medium were transferred to MS agar medium without NaCl ([A] and [C]) or with 100 mM NaCl ([B] and [D]); photographs were taken 10 d after transfer. SOS2 transcript levels in sos2-2, sos3-1, and 35S:SOS2 transgenic lines (E) are shown. RNA gel blot analysis was performed with total RNA extracted from sos2-2 (1), sos3-1 (2), sos2-2 (3), and sos3-1 (4) transgenic plants grown in the absence of NaCl. 25S rRNA (ethidium bromide stained) was used as a loading control. WT, wild type.
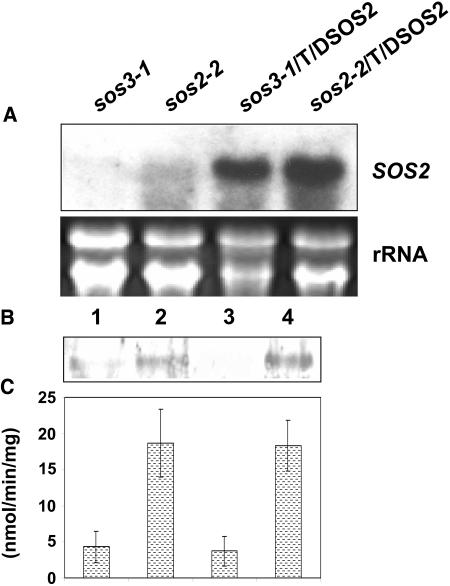
Expression of active T/DSOS2 kinase in sos2-2 and sos3-1 resulted in 5 of 12 T2 sos2-2 transgenic lines and 4 of 12 T2 sos3-1 transgenic lines, in which the shoot sensitivity but not the root sensitivity of the mutant phenotype was rescued. RNA analysis demonstrated that the sos2-2 and sos3-1 transgenic plants accumulated a high level of T/DSOS2 transcript (Figure 4A). SOS2 transcript and protein are in low abundance in A. thaliana and even when SOS2 transcript levels were higher because of the strong CaMV 35S promoter, SOS2 protein levels were still virtually undetectable using our SOS2 antisera (data not shown). Therefore, to analyze the levels of T/DSOS2 protein in the transgenic plants, total proteins were extracted from mutants and transgenic plants, and SOS2 protein (from both endogenous SOS2 and 35S:T/DSOS2) was enriched based on its binding to SOS3. The proteins were loaded onto a column containing GST-SOS3 fusion protein that was bound to glutathione-Sepharose beads. The resulting GST-SOS3-SOS2 or T/DSOS2 complexes were used for either immunoblot analysis or peptide phosphorylation assays. As shown in Figure 4B, expression of T/DSOS2 in either sos2-2 or sos3-1 resulted in the accumulation of T/DSOS2 protein at higher levels than the preexisting SOS2 protein levels in the sos2-2 and sos3-1 mutants. Based on phosphorylation of the p3 peptide, T/DSOS2 kinase activity from both the sos2-2 and sos3-1 transgenic plants was approximately four times higher than in the corresponding mutants (Figure 4C). Because several PKS (SOS2-like protein kinases) proteins also interact with SOS3 (Guo et al., 2001), the kinase activities from the untransformed mutants may not represent the activity of only SOS2.
Figure 4.
Expression of T/DSOS2 in sos2-2 and sos3-1.
RNA gel blot analysis of T/DSOS2 expression in sos3-1, sos2-2 or sos3-1 and sos2-2 transgenic lines grown in the absence of NaCl (A). 25S rRNA (ethidium bromide stained) was used as a loading control. Total protein was extracted from mutant and transgenic plants and incubated with GST-SOS3 coupled to glutathione-Sepharose beads. The GST-SOS3-T/DSOS2/SOS2 complex was used for immunoblot analysis (B) with protein from sos3-1 (1) and sos2-2 (3) mutants or sos3-1 (2) and sos2-2 (4) transgenic lines. Proteins were probed with anti-SOS2 antibody. The GST-SOS3-T/DSOS2/SOS2 complex was also used for peptide phosphorylation assays (C) with protein from sos3-1 (1), the sos3-1 transgenic line (2), sos2-2 (3), and the sos2-2 transgenic line (4).
Five-day-old seedlings of wild-type, mutant, and T/DSOS2 transgenic plants were transferred to either MS medium or MS medium containing 100 mM NaCl. No significant differences in plant growth were observed on MS medium (Figure 5A). When the plants were grown on medium containing 100 mM NaCl, the growth of the wild-type plants was retarded but root bending was largely unaffected, whereas growth of sos2-2 and sos3-1 was severely inhibited (Figure 5B) and plants died within 2 weeks (data not shown). Expression of T/DSOS2 in sos2-2 was able to partially rescue the shoot salt hypersensitivity but not the root salt hypersensitivity (Figure 5B). These results suggest that in the shoot, ectopic expression of T/DSOS2 partially restored salt tolerance in the sos2-2 background. Expression of T/DSOS2 in sos3-1 was also able to partially rescue the shoot salt hypersensitivity but not the root salt hypersensitivity (Figures 5B and 5C), suggesting that in the shoot, addition of the active kinase partially bypassed the requirement for SOS3.
Figure 5.
Expression of T/DSOS2 Partially Rescues the sos2-2 and sos3-1 Salt-Hypersensitive Phenotypes.
Five-day-old seedlings grown on MS agar medium were transferred to MS agar medium without NaCl (A) or with 100 mM NaCl (B); photographs were taken 10 d after transfer. Fresh weight (C) (in milligrams) of five plants of the wild type (1), sos3-1 (2), sos3-1 transgenic line (3), sos2-2 (4), and sos2-2 transgenic line (5) 2 weeks after transfer to MS + 100 mM NaCl (mean ± SE of three replicate experiments). Growth of sos2-2 and a sos2-2 transgenic line (D) and sos3-1 and a sos3-1 transgenic line (E) in soil in which the NaCl levels were increased by 50 mM every 4 d until a final concentration of 200 mM was reached. Photographs were taken after 15 d in 200 mM NaCl.
No differences in either vegetative or reproductive growth were seen when mutant and transgenic plants grown in soil were watered with 0.05 × MS nutrients in the absence of NaCl (data not shown). However, when the plants were treated with NaCl, sos2-2 and sos3-1 lost vigor faster, and both vegetative and reproductive growth decreased (Figures 5D and 5E). Expression of T/DSOS2 improved the growth of the mutants under NaCl stress (Figures 5D and 5E), although it did not restore salt tolerance to wild-type levels (data not shown).
The Protein Kinase Activity of SOS2 May Be Lmiting for Salt Tolerance in Arabidopsis thaliana
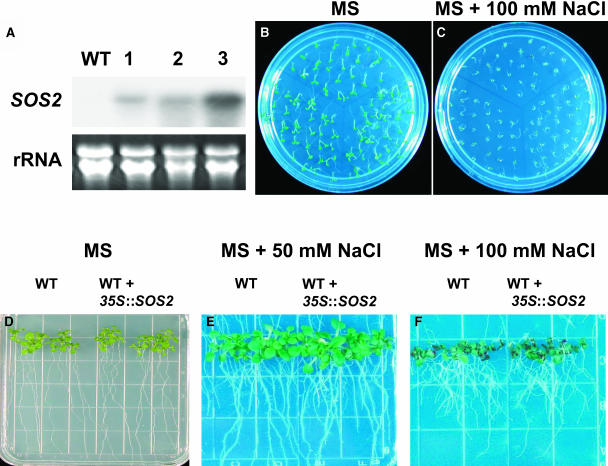
To determine if levels of SOS2 protein are limiting in vivo and if increasing active SOS2 levels lead to improved salt tolerance in planta, SOS2 and T/DSOS2 were expressed in wild-type plants under the control of the CaMV 35S promoter. Of 24 T2 35S:SOS2 transgenic lines evaluated, all had levels of salt tolerance similar to that in the untransformed wild type. The levels of SOS2 transcript were determined in three T3 homozygous 35S:SOS2 lines and strong expression was detected in all the transgenic plants (Figure 6A). The salt tolerance of two of these lines was subsequently evaluated during germination (Figures 6B and 6C) and seedling growth (Figures 6D to 6F); responses to salt at both stages were similar to those in the wild type. The lack of enhancement of salt tolerance in plants overexpressing wild-type SOS2 indicates that SOS2 protein levels are not limiting in A. thaliana in vivo.
Figure 6.
Expression of SOS2 Does Not Increase Salt Tolerance in A. thaliana.
SOS2 transcript levels in the wild type and three 35S:SOS2 transgenic lines grown in the absence of NaCl (A) are shown. 25S rRNA (ethidium bromide stained) was used as a loading control. Seeds from the wild type (top) and two 35S:SOS2 transgenic lines (bottom) were germinated on MS medium (B) or MS + 100 mM NaCl (C); photographs were taken 5 (left panel) and 10 d (right panel) after germination. Five-day-old seedlings grown on MS agar medium were transferred to MS agar without NaCl (D) or with 50 (E) or 100 mM NaCl (F); photographs were taken 10 d after transfer.
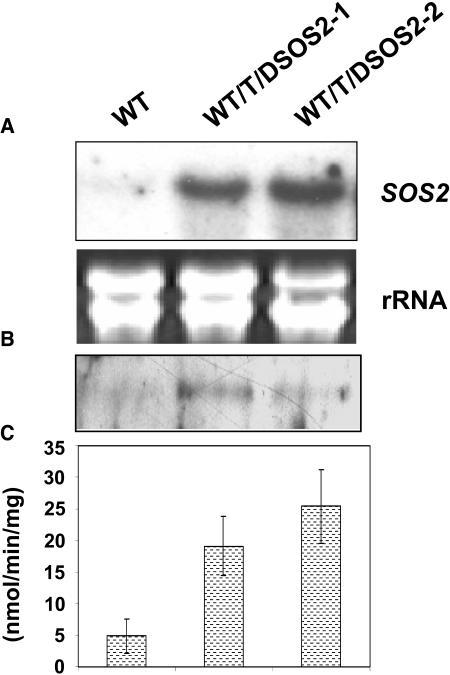
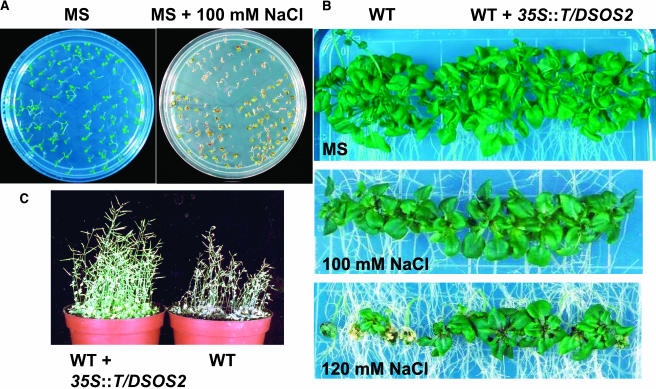
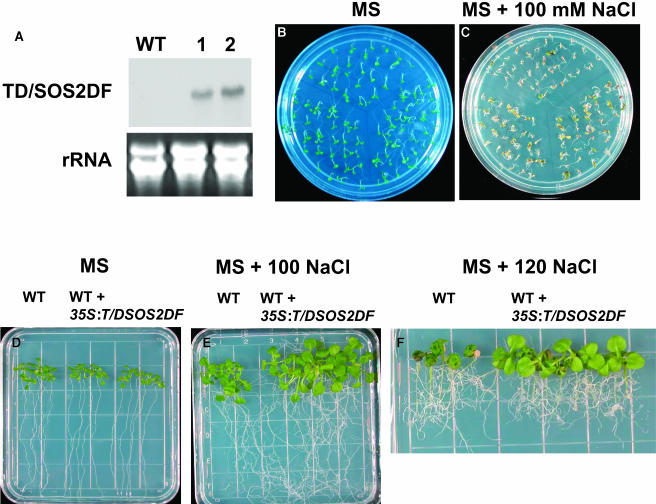
Exchange of Thr168 in the activation loop of the SOS2 protein with Asp mimics the phosphorylation of Thr168 by an unknown upstream kinase and leads to activation of SOS2 (Guo et al., 2001). When T/DSOS2 was expressed in wild-type plants, 7 of 34 T2 transgenic lines evaluated showed increased salt tolerance compared with untransformed wild-type plants. Two of the seven T3 homozygous 35S:T/DSOS2 lines were analyzed for T/DSOS2 transcript and protein accumulation and salt tolerance. The transgenic plants accumulated high levels of T/DSOS2 transcript and protein (Figures 7A and 7B). T/DSOS2 kinase activity from the transgenic plants was enhanced four to five times over the kinase activity levels in the wild type (Figure 7C). No difference was seen when seeds from transgenic or wild-type plants were germinated on MS medium without salt (Figure 8A, left panel). However, seeds from the transgenic lines showed more rapid germination on MS medium containing 100 mM NaCl (Figure 8A, right panel), and seedling development proceeded further in salt (green cotyledons developed) in the transgenic plants. Growth of wild-type and transgenic seedlings in the absence of salt was similar (Figure 8B, top panel). However, when seedlings were transferred to medium with NaCl, the transgenic plants showed significantly less growth inhibition, which was especially evident at 120 mM NaCl (Figure 8B, middle and bottom panels).
Figure 7.
Expression of T/DSOS2 in A. thaliana.
T/DSOS2 transcript levels in the wild-type and WT/T/DSOS2 transgenic lines (A). RNA gel blot analysis with total RNA extracted from the wild type and two WT/T/DSOS2 lines grown in the absence of NaCl. 25S rRNA (ethidium bromide stained) was used as a loading control. Total protein was extracted from the wild-type and transgenic plants and incubated with GST-SOS3 coupled to glutathione-Sepharose beads. The GST-SOS3-T/DSOS2/SOS2 protein complex was used for immunoblot analysis with anti-SOS2 antibody (B) and peptide phosphorylation assays (C).
Figure 8.
Expression of T/DSOS2 Increases Salt Tolerance in A. thaliana.
(A) Seeds from the wild type (top) and two WT/T/DSOS2 transgenic lines (bottom) were germinated on MS medium (left panel) or MS + 100 mM NaCl (right panel); photographs were taken 5 (left panel) and 10 d (right panel) after germination.
(B) Five-day-old seedlings from the wild type or WT/T/DSOS2 transgenic lines grown on MS agar were transferred to MS agar without NaCl (top panel), with 100 (middle panel), or 120 mM NaCl (bottom panel); photographs were taken 15 d after transfer.
(C) Wild-type and transgenic (WT + 35S:T/DSOS2) plants were grown in soil in which the NaCl levels were increased by 50 mM every 4 d until a final concentration of 200 mM was reached. Photographs were taken after 15 d in 200 mM NaCl.
To test the salt tolerance of the plants when grown in soil, wild-type and transgenic seeds were germinated in soil and watered with 0.05 × MS nutrients. After 3 weeks, the plants were treated with NaCl by progressively increasing the NaCl concentration 50 mM every 4 d until a final concentration of 200 was reached (Shi et al., 2003). The transgenic plants showed improved vegetative and reproductive growth in soil with 200 mM NaCl when compared with growth of the wild-type plants (Figure 8C); no difference was found when plants were grown without NaCl (data not shown). The increased salt tolerance of the plants expressing the T/DSOS2 kinase suggests that levels of activated kinase may be limiting in A. thaliana in vivo and that increasing active SOS2 levels in planta can lead to improved salt tolerance.
Enhancement of SOS1 Activity in Vivo by Constitutively Active SOS2
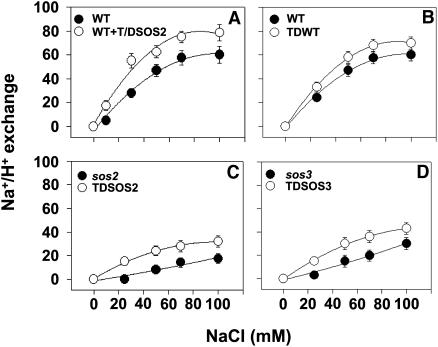
Previous studies have shown that active SOS2 protein stimulates the Na+/H+ antiport activity of SOS1 in vitro (Qiu et al., 2002), suggesting that SOS2 directly regulates the activity of SOS1. To determine if in vivo SOS2 kinase activity is sufficient to regulate SOS1 activity and if SOS1 activation might contribute to the improved salt tolerance conferred by T/DSOS2, we measured SOS1 transport activity in the 35S:T/DSOS2 transgenic plants and the untransformed wild-type, sos2-2, and sos3-1 control plants. For these studies, highly purified plasma membrane vesicles were isolated from wild-type, sos2-2, sos3-1, and their T/DSOS2 transgenic plants after treatment with 250 mM NaCl for 3 d. When T/DSOS2 protein was added in vitro to plasma membrane vesicles isolated from untransformed wild-type plants, Na+/H+-exchange activity increased with increasing NaCl concentration and was higher than activity in the absence of T/DSOS2 protein at all NaCl concentrations (Figure 9A). A maximum stimulation of activity of 40% relative to activity without added protein was measured with 100 mM NaCl. Na+/H+ exchange activity, measured in vesicles isolated from T/DSOS2 transgenics of the wild-type, sos2, and sos3 plants, was higher than in the respective untransformed controls (Figures 9B to 9D); however, the exchange activity of the sos2-2 and sos3-1 transgenic lines was restored to only half to two-thirds of the levels of activity measured in the untransformed wild type, in agreement with the partial suppression of their salt sensitivity (Figure 5). These results demonstrate that expression of the active kinase T/DSOS2 enhanced SOS1 activity in vivo in the transgenic plants. These measurements also provide further evidence that more than SOS2 is required for full SOS1 activity and salt tolerance in vivo. Besides activating SOS1, expression of the active kinase may also enhance salt tolerance through other mechanisms (e.g., enhancement of vacuolar Na+/H+ antiport activity) because SOS2 has been shown to be a regulator of vacuolar Na+/H+ antiporters (Qiu et al., 2004).
Figure 9.
Active T/DSOS2 Increases Plasma Membrane Na+/H+-Exchange Activity in Vitro and in Vivo.
When added in vitro, T/DSOS2 protein stimulates plasma membrane Na+/H+-exchange (antiport) activity in vesicles isolated from wild-type plants (A). Transport assays were performed as described in Methods. The pH gradient (ΔpH) was formed in the absence (closed circle) or presence (open circle) of T/DSOS2 protein. When ΔpH reached steady state, NaCl was added over a range of final concentrations (0 to 100 mM), and the initial rates of dissipation (Na+/H+ exchange) were measured. When compared with activity in the wild type, sos2, and sos3, plasma membrane Na+/H+-exchange activity is higher in the wild-type (B), sos2 (C), and sos3 (D) plants overexpressing T/DSOS2. Assays were performed using vesicles isolated from the parental (closed circle, [B] to [D]) and transgenic (open circle, [B] to [D]) plants. When ΔpH reached steady state, NaCl was added over a range of final concentrations (0 to 100 mM), and the initial rates of dissipation were measured. Units of Na+/H+ exchange are Δ%F mg−1 protein min−1. Data in (A) to (D) represent mean ± SE of at least three replicate experiments. Each replicate experiment was performed using independent membrane preparations.
The C-Terminal Region of SOS2 Is Required for Function in Planta
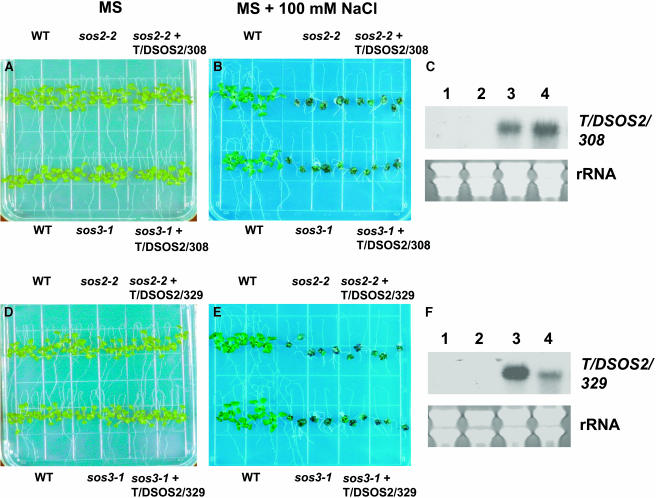
The above experiments showed that the active kinase T/DSOS2 could enhance SOS1 activity and salt tolerance when expressed either in wild-type, sos2, or sos3 plants. Because T/DSOS2/308 (with the Thr168-to-Asp change and in which the FISL domain and C-terminal 117 amino acids were removed) exhibited the highest protein kinase activity in vitro (Figure 1) and was the most competent for activation of SOS1 in S. cerevisiae in the absence of SOS3 (Figure 2), T/DSOS2/308 was expressed in the sos2-2 or sos3-1 mutants under the CaMV 35S promoter. Twenty-four independent T2 transgenic lines from each transformation were tested for growth in salt; none had salt tolerance that was greater than that of either the sos2-2 or sos3-1 mutant. One representative T3 homozygous line from expression of T/DSOS2/308 in sos2-2 (Figures 10A and 10B, top) and sos3-1 (Figures 10A and 10B, bottom) is shown. Although the transgene was expressed at high levels in the transgenic plants (Figure 10C), salt tolerance was not enhanced. These results suggest that the FISL motif and/or the C-terminal 117 amino acids are required for salt tolerance in planta.
Figure 10.
Expression of T/DSOS2/308 or T/DSOS2/329 Does Not Complement the sos2-2 and sos3-1 Salt-Sensitive Phenotypes.
Five-day-old seedlings grown on MS agar medium were transferred to MS agar medium without NaCl ([A] and [D]) or with 100 mM NaCl ([B] and [E]); photographs were taken 10 d after transfer. T/DSOS2/308 (C) or T/DSOS2/329 (F) transcript levels in sos2-2 (1), sos3-1 (2), or sos2-2 (3) and sos3-1 (4) transgenic lines grown in the absence of NaCl. 25S rRNA (ethidium bromide stained) was used as a loading control.
Compared with T/DSOS2/308, T/DSOS2/329 (with the Thr168-to-Asp change and in which the C-terminal 117 amino acids were removed) contains the FISL motif but is not as active because the FISL motif is inhibitory to SOS2 activity (Figures 1 and 2). When 35S:T/DSOS2/329 was expressed in the sos2-2 or sos3-1 mutants, salt tolerance was not restored. One representative T3 homozygous line from expression of T/DSOS2/329 in sos2-2 (Figures 10D and 10E, top) and sos3-1 (Figures 10D and 10E, bottom) is also shown. As with T/DSOS2/308, expression of the transgenes was high in the transgenic plants (Figure 10F), but salt tolerance was not enhanced. The data from the analysis of the sos2-2 and sos3-1 transgenic lines expressing T/DSOS2/329 suggest that adding back the FISL motif is not sufficient to restore the function to the active T/DSOS2/308 kinase in planta. Together with the data from the T/DSOS2/308 expressing transgenic plants, the results reveal a critical role for the C-terminal region of SOS2 in salt tolerance in planta.
To further examine the role of the FISL motif and the C-terminal 117 residues, T/DSOS2DF (with the Thr168-to-Asp change and in which the FISL domain was removed) was expressed in the wild-type A. thaliana and the sos2-2 or sos3-1 backgrounds. When T/DSOS2DF was expressed in the wild-type plants, 4 of 12 of T2 transgenic lines evaluated were more salt tolerant than the untransformed wild type. The levels of T/DSOS2DF transcript were determined in two T3 homozygous lines, and high accumulation in both was detected (Figure 11A). When these plants were evaluated for salt tolerance during germination and seedling growth, no significant differences in germination were detected on medium without salt (Figure 11B). By contrast, the transgenic plants had faster germination and improved seedling development on MS medium containing 100 mM NaCl (Figure 11C). When 5-d-old seedlings were transferred to MS medium, the growth of wild-type and transgenic plants was similar (Figure 11D). When seedlings were transferred to MS medium containing 100 mM or 120 mM NaCl, growth of the transgenic plants was less inhibited by NaCl (Figures 11E and 11F).
Figure 11.
Expression of T/DSOS2DF Increases Salt Tolerance in A. thaliana.
T/DSOS2DF transcript levels in the wild type and two transgenic lines grown in the absence of NaCl (A). 25S rRNA (ethidium bromide stained) was used as a loading control. Seeds from the wild type (top) and two transgenic lines (bottom) were germinated on MS medium (B) or MS + 100 mM NaCl (C); photographs were taken 5 (left panel) and 10 d (right panel) after germination. Five-day-old seedlings grown on MS agar medium were transferred to MS agar without NaCl (D) or with 100 (E) or 120 mM NaCl (F); photographs were taken 10 (D) and 15 d ([E] and [F]) after transfer.
We attempted to enrich the T/DSOS2DF protein by incubating total protein extracts (from a transgenic line with increased salt tolerance) with GST-SOS3 on glutathione-Sepharose beads. However, T/DSOS2DF protein could not be detected by immunoblot analysis (data not shown), and no T/DSOS2DF kinase activity was detected in peptide phosphorylation assays (data not shown), indicating that T/DSOS2DF did not interact with SOS3 and further supporting previous interaction studies suggesting that the FISL motif is required for SOS2/SOS3 interaction.
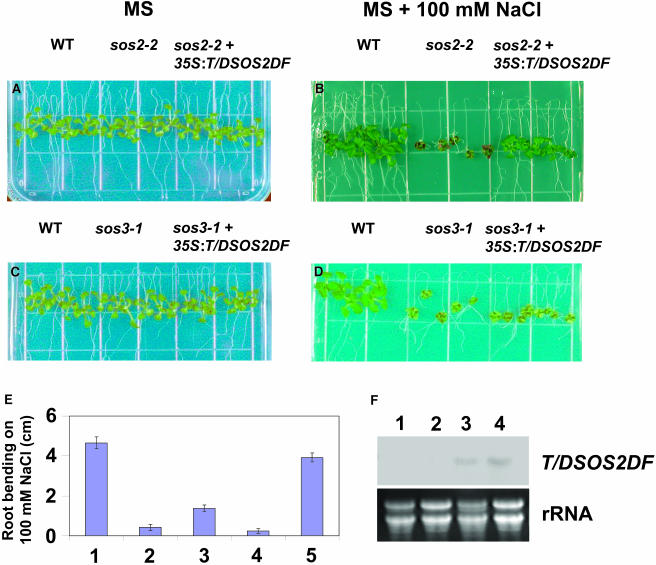
When T/DSOS2DF was expressed in the sos2-2 and sos3-1 backgrounds, 3 of 12 T2 sos2-2 transgenic lines had salt tolerance that was restored almost to wild-type levels. However, of the 24 T2 sos3-1 transgenic lines evaluated, all had the sos3-1 phenotype with only slight root bending. Representative T3 homozygous sos2-2 and sos3-1 transgenic lines are shown in Figure 12. The T/DSOS2DF transcript was detected in both transgenic lines (Figure 12F). When 5-d-old seedlings from the wild-type, sos2-2, sos3-1, and transgenic sos2-2 or sos3-1 lines were transferred to MS medium without salt, no significant differences in growth were found (Figures 12A and 12C). However, when the seedlings were transferred to MS medium containing 100 mM NaCl, sos2-2 plants died within 2 weeks, whereas the phenotype of the sos2-2 transgenic plants was similar to the wild type but with slightly smaller shoots and fewer lateral roots (Figure 12B). Expression of T/DSOS2DF in sos3-1 led to a slight increase in root elongation relative to sos3-1 when plants were grown on 100 mM NaCl (Figures 12D and 12E); however, both sos3-1 and the sos3-1 transgenic lines were unable to survive on this medium for >2 weeks.
Figure 12.
Expression of T/DSOS2DF Rescues the sos2-2 Salt-Hypersensitive Phenotype but Not the sos3-1 Salt-Hypersensitive Phenotype.
Five-day-old seedlings grown on MS agar medium were transferred to MS agar medium without NaCl ([A] and [C]) or with 100 mM NaCl ([B] and [D]); photographs were taken 10 d after transfer. Root growth (E) (in centimeters) of the wild type (1), sos3-1 (2), a sos3-1 transgenic line (3), sos2-2 (4), and a sos2-2 transgenic line (5) grown on MS medium + 100 mM NaCl for 2 weeks (mean ± SE of three replicate experiments). T/DSOS2DF transcript levels (F) are shown. RNA gel blot analysis with total RNA extracted from sos2-2 (1), sos3-1 (2), a sos2-2 transgenic line (3), and a sos3-1 transgenic line (4) grown in the absence of NaCl. 25S rRNA (ethidium bromide stained) was used as a loading control.
The results with the 35S:T/DSOS2DF transgenic plants demonstrate that the 117 residues C terminal to the FISL motif are necessary and sufficient for the in planta function of the active SOS2 kinase proteins in wild-type and sos2-2 mutant plants. However, function of the active kinase in sos3-1 mutant plants appears to require the FISL motif as well. Improved salt tolerance in the wild-type transgenic plants provides further support that the kinase activity of SOS2 is limiting in vivo, and increasing this activity can be beneficial for salt tolerance.
DISCUSSION
Genetic analysis of sos1, sos2, and sos3 mutants suggested that SOS1, SOS2, and SOS3 function in the same pathway for Na+ homeostasis in A. thaliana (Zhu et al., 1998). SOS2 is activated by its interacting protein SOS3 in a calcium-dependent manner (Halfter et al., 2000). When expressed in S. cerevisiae, the SOS3-SOS2 complex phosphorylates and activates SOS1 to enhance Na+ efflux and salt tolerance (Quintero et al., 2002). The Na+/H+ exchange activity of SOS1 is substantially diminished in sos2 and sos3 mutant plants, and in vitro addition of the activated form of SOS2, T/DSOS2DF, rescues the exchange activity in not only sos2 but also sos3 plasma membrane vesicles (Qiu et al., 2002). Therefore, the requirement of SOS3 in vitro for SOS1 activation can be bypassed by the activated SOS2 protein. Results presented here demonstrate that in S. cerevisiae, the requirement of SOS3 in salt tolerance can also be partially bypassed in vivo by the activated forms of SOS2. However in planta, only the activated form of SOS2 that retains structural integrity (i.e., T/DSOS2) can bypass the requirement for SOS3. These results show that data obtained in vitro and even in vivo from a heterologous system only partially reflect what happens in planta. The in planta experiments thus reveal new functions of the regulatory proteins and their essential structural domains.
The activity and functionality of the different forms of SOS2 in vitro, in S. cerevisiae, and in wild-type and mutant A. thaliana is summarized in Table 1. In S. cerevisiae, the effect of the kinase forms on SOS1 activation and salt tolerance is largely correlated with their in vitro kinase activities when both are measured in the absence of SOS3. For example, T/DSOS2/308 is most active in vitro and is also most effective in enhancing the salt tolerance of the S. cerevisiae cells not expressing SOS3, whereas wild-type SOS2 is essentially inactive in both assays. By contrast, the ability of SOS2 variants to activate SOS1 in vivo is also dependent on their ability to interact with the ancillary protein SOS3 through the FISL motif. Thus, activated forms T/DSOS2 and T/DSOS2/329, both containing the FISL motif, conveyed greater salt tolerance in the presence of SOS3, whereas T/DSOS2DF and T/DSOS2/308 did not. The results demonstrate that localization of activated SOS2 to the membrane via its interaction with SOS3 enhances but is not necessary for activation of SOS1. Structural integrity of SOS2 is also important because SOS2-SOS3 and T/DSOS2-SOS3 complexes yielded maximal activation of SOS1 and salt tolerance above 100 mM NaCl (Figure 2 and data not shown). Although T/DSOS2/308, T/DSOS2/DF, and T/DSOS2/329 are all more active in vitro and result in limited independence from SOS3 in S. cerevisiae cells, they are unable to bypass the SOS3 deficiency in planta. Surprisingly, only the T/DSOS2 form was able to partially rescue the sos3-1 mutant phenotype when expressed in A. thaliana, despite the strict dependence of T/DSOS2 on a functional SOS3 protein in S. cerevisiae. The sos3-1 mutation causes a deletion of three amino acids in one of the EF hands of SOS3 that reduces but does not eliminate the calcium binding of SOS3 (Liu and Zhu, 1998; Ishitani et al., 2000). It is therefore possible that this mutant form of SOS3 is still partially functional. Because T/DSOS2DF, which varies from T/DSOS2 only in the removal of the FISL motif, did not suppress the sos3-1 mutation whereas T/DSOS2 did, it was possible that the mutant polypeptide SOS3-1 could still bind to T/DSOS2 and target the activated kinase to the plasma membrane for the phosphorylation of SOS1. However, previous studies have shown that the mutant SOS3-1 protein does not interact with SOS2 in an S. cerevisiae two-hybrid assay (Ishitani et al., 2000), and we have shown here that SOS3-1 is unable to recruit SOS2 or T/DSOS2 to the plasma membrane (Figures 2B and 2C). Alternatively, T/DSOS2 could interact with another SOS3-like calcium binding protein (SCaBP) and be targeted to the plasma membrane in the absence of SOS3. If so, it would also explain why T/DSOS2DF partially rescued the sos2-2 mutant phenotype but could not suppress the sos3-1 mutation because deletion of the FISL motif eliminates interaction with SOS3 and other SCaBPs. The sos2-2 mutation results in a truncated protein containing the kinase catalytic domain (Liu et al., 2000). It cannot be ruled out that the truncated protein in the mutant may influence the results in planta. It should also be noted that SOS2 physically interacts with other proteins that, directly or indirectly, may help recruit T/DSOS2 to membranes in a SOS3-independent manner. For instance, it has been recently shown that SOS2 binds to ABI2, a protein phosphatase 2C involved in abscisic acid and stress signaling (Ohta et al., 2003). Moreover, it is reasonable to expect that SOS2, besides activating SOS1, may fulfill additional roles leading to plant salt tolerance that could be independent of its interaction with SCaBPs and/or targeting to the plasma membrane. A better knowledge of the various functional domains of SOS2 and SOS3 and related proteins will be needed to fully understand the complexity of this pathway. Nevertheless, the observations here collectively reveal a requirement for the C-terminal regulatory region of SOS2 for salt tolerance in planta.
Table 1.
Summary of the in Vitro and in Vivo Activities of the Wild-Type and Activated Forms of SOS2
|
S. cerevisiae Growth (in Salt)
|
A. thaliana Growth (in Salt)
|
||||||
|---|---|---|---|---|---|---|---|
| Autophosphorylation | Peptide Phosphorylation | +SOS3 | −SOS3 | sos2 | sos3 | WT | |
| SOS2 | − | − | ++++ | − | + | − | − |
| T/DSOS2 | ++ | ++ | ++++ | − | + | + | + |
| T/DSOS2DF | ++ | +++ | + | + | + | − | + |
| T/DSOS2/329 | + | + | ++ | + | − | − | ND |
| T/DSOS2/308 | +++ | ++++ | ++ | ++ | − | − | ND |
The level of phosphorylation activity in vitro and relative salt tolerance in vivo are indicated for each SOS2 variant by the number of + symbols. Minus symbol indicates undetectable phosphorylation or no growth. ND, not determined; WT, wild-type form of SOS2.
Another unexpected observation is that T/DSOS2 partially rescues the salt hypersensitivity in the shoot but not the root in sos2 and sos3 mutants. The lack of effect in the root is not likely explained by the use of the CaMV 35S promoter because the wild-type SOS2 expressed under the same promoter does rescue the sos2 mutant in both the shoot and root. A root-specific regulation of SOS2 may occur through its activation loop, and the T/D mutation may interfere with such a regulation. Although the hypothetical upstream protein kinase(s) for SOS2 has not been identified, it is conceivable that there might be a root-specific isoform of such a kinase. On the other hand, expression of T/DSOS2DF can rescue the sos2 mutant phenotype. Thus, if the hypothetical root-specific upstream kinase is responsible for the inactivity of T/DSOS2 in the root, it must not have an effect on T/DSOS2DF.
Regulatory genes are often considered superior targets of biotechnological applications for plant improvement because they control many downstream effector genes. For example, ectopic expression of the CBF/DREB1A family of transcription factors and the MAPKKK ANP1 have been shown to substantially improve plant tolerance to various abiotic stresses (Jaglo-Ottosen et al., 1998; Gilmour et al., 2000; Kovtun et al., 2000). SOS2 is a key regulator of ion transporters (Zhu, 2002), some of which have been shown to confer increased salt tolerance when overexpressed in transgenic plants (Apse et al., 1999; Shi et al., 2003). In this study, we evaluated the feasibility of using SOS2 to improve plant salt tolerance. Overexpression of the wild-type SOS2 did not confer any increased salt tolerance in transgenic A. thaliana. However, ectopic expression of the activated forms T/DSOS2 and T/DSOS2/DF led to measurable enhancement in salt tolerance in transgenic A. thaliana. These results raise the hope that by exploring various versions of the protein kinase, an effective allele may be identified that might become useful even in field conditions.
METHODS
Preparation of Active SOS2 Kinase Expression Plasmids and Plant Transformation
For expression of constitutively active SOS2 kinase in A. thaliana, DNA fragments of T/DSOS2, T/DSOS2/308, T/DSOS2/329, and T/DSOS2DF were digested from their GST fusion constructs (Guo et al., 2001) with BamHI and EcoRI and cloned into a binary vector (pCAMBIA1027) under the control of the CaMV 35S promoter. The plasmids were introduced into Agrobacterium tumefaciens strain GV3101 by electroporation and then transferred into wild-type (A. thaliana Columbia ecotype), sos2-2, or sos3-1 mutant plants by vacuum infiltration. Hygromycin-resistant transgenic T2 and T3 plants were tested for growth in salt.
Growth Measurements
Seeds of wild-type, sos2-2, sos3-1, and transgenic plants were surface-sterilized in 7% (w/v) hypochlorite and 0.01% (w/v) Triton X-100 and then rinsed five times with sterile water. The seeds were sown on an MS nutrient medium (JRH Biosciences, Lenexa, KS) containing 0.6% agar and the indicated NaCl concentrations. The seeds were stratified at 4°C for 3 d and then transferred to 22°C under continuous light for measurements of germination and growth.
For seedling growth in salt, 5-d-old seedlings of wild-type, sos2-2, sos3-1, and transgenic plants were transferred to MS medium containing 1.2% agar and the indicated NaCl concentrations. Growth was monitored using a root bending assay (Zhu et al., 1998). Plant salt tolerance in soil was assayed as described in Shi et al. (2003).
RNA Analysis
Total RNA was extracted from 2-week-old seedlings, and 40 μg of each sample was used for RNA analysis as described (Guo et al., 2001).
Immunoblot Analysis and Kinase Assays
Total proteins (5 g from 10-d-old seedlings) were extracted at 4°C from wild-type, sos2-2, sos3-1, and transgenic plants in 10 mL 1 × PBS buffer (137 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4, and 1.4 mM NaH2PO4, pH 7.4) with 5 mM dithiothreitol, 2 μg aprotinin mL−1, 2 μg leupeptin mL−1, and 2 mM phenylmethanesulfonyl fluoride. To isolate sufficient amounts of T/DSOS2 protein, GST-SOS3 fusion protein (Halfter et al., 2000) was first purified using glutathione-Sepharose beads (Amersham Pharmacia Biotech, Uppsala, Sweden). Total A. thaliana proteins were then incubated with 100 μL of GST-SOS3 coupled to the Sepharose beads for 2 h at 4°C. The GST-SOS3 beads-T/DSOS2 protein complex was washed three times with 1 × PBS buffer. Ten microliters of the protein complex were used for either immunoblot analysis or protein kinase assays.
For immunoblot analysis, 3 μL of 3 × protein loading buffer (200 mM Tris-HCl, pH 6.8, 8% SDS, 30% glycerol, 1.5% β-mercaptoethanol, and 0.3% bromophenol blue) were added to 10 μL protein, and the samples were boiled for 5 min. The samples were run on a 10% SDS-PAGE gel, and the proteins were transferred to a pure nitrocellulose membrane (Bio-Rad Laboratories, Hercules, CA) at 80 V for 60 min. The membrane was blocked overnight at 4°C in 1 × PBS buffer with 5% fat-free milk, rinsed one time with 1 × PBS, and incubated with SOS2 antibodies (diluted 1:1000) for 3 h at room temperature. After three washes with 1 × PBS buffer, the membrane was incubated with anti-rabbit IgG secondary antibody (Amersham Biosciences, Piscataway,NJ) diluted 1:2500 for 1 h at room temperature. The membrane was then washed five times with 1×PBS and the immunoreactive bands detected using the chemiluminescent ECL detection substrate (Amersham Biosciences).
Ten microliters of SOS3-T/DSOS2 beads were used for p3 peptide phosphorylation assays as described by Halfter et al. (2000).
Na+/H+ Exchange
Plasma membrane vesicles were isolated using aqueous two-phase partitioning as described (Qiu and Su, 1998; Qiu et al., 2002). Na+/H+ exchange (antiport) activity was measured as a Na+-induced dissipation of the pH gradient (ΔpH, i.e., a Na+-induced increase in quinacrine fluorescence; Qiu et al., 2002). When ΔpH reached steady state, NaCl was added to initiate Na+ transport. To determine initial rates of Na+/H+ exchange (change in fluorescence per minute; Δ%F min−1), changes in relative fluorescence were measured during the first 15 s after addition of Na+. Specific activity was calculated by dividing the initial rate by the mass of plasma membrane protein in the reaction (Δ%F mg−1 protein min−1). To determine whether T/DSOS2 activated SOS1 in vitro, 200 ng of T/DSOS2 protein was preincubated with wild-type membrane vesicles for 7 min at room temperature before the antiport activity assays.
Yeast Growth
The S. cerevisiae strain YP890 is a derivative of AXT3K (Δena1:HIS3:ena4, nha1:LEU2, and nhx1:KanMX) (Quintero et al., 2002), in which a PGK1:SOS1:CYC1 expression cassette was inserted at the 3′ untranslated region of the chromosomal gene CYC1. The chromosomal placement of the transgene and the use of the PGK1 promoter provide moderate and constitutive expression of the A. thaliana SOS1 protein in YP890 cells. The plasmids that contain either wild-type SOS2, activated forms of SOS2 (T/DSOS2, T/DSOS2/308, T/DSOS2/329, and T/DSOS2/DF), or the inactive SOS2 mutant bearing substitution Lys40 to Asn (T/DSOS2/KN) were made by inserting BamHI-EcoRI fragments from pGEX-SOS2 derivatives (Guo et al. 2001) into the BamHI-EcoRI sites of the p414GPD vector. The cDNAs of wild-type SOS3 and mutant sos3-1 were cloned into the XbaI-XhoI sites of the expression vector pYPGE15. Transformation of S. cerevisiae was performed using a standard lithium–polyethylene glycol method. The ability of S. cerevisiae to grow in salt was tested on AP medium, which is essentially free of alkali cations. Strains were cultured overnight in liquid AP medium supplemented with 1 mM KCl. After harvest, cells were resuspended and diluted decimally in distilled water. Aliquots (5 μL) were spotted onto AP plates supplemented with 1 mM KCl and various concentrations of NaCl, as noted, and grown for 3 to 4 d at 28°C.
SRS
Plasmid pSRS2-1 containing the gene fusion hSos:SOS2 was used to monitor plasma membrane targeting of SOS2 (Quintero et al., 2002). SOS3 and sos3-1 were expressed using the vector plasmid pYPGE15 as described above. All plasmids used for SRS were transformed into the S. cerevisiae strain Cdc25-2 (Matα, cdc25-2, ura3, lys2, leu2, trp1, his3 , and ade101), which is conditional lethal at 37°C unless the fusion protein hSos:SOS2 reaches the plasma membrane (Aronheim et al., 1997). Cell viability at 37°C was determined in YPD plates (1% yeast extract, 2% peptone, and 2% glucose).
Acknowledgments
This work was supported by National Institutes of Health Grant R01GM59138 to J.-K.Z., U.S. Department of Energy Grant DE-FG03-93ER20120 to K.S.S., and the Southwest Consortium on Plant Genetics and Water Resources to J.-K.Z. and K.S.S. F.J.Q. and J.M.P. were supported by Grant BIO2000-0398 from the Spanish Ministry of Science and Technology and Grant CVI-148 from Junta de Andalucía.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Yan Guo (guoyan@nibs.ac.cn).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.019174.
References
- Amtmann, A., and Sanders, D. (1999). Mechanisms of Na+ uptake by plant cells. Adv. Bot. Res. 29, 76–112. [Google Scholar]
- Apse, M.P., Aharon, G.S., Snedden, W.A., and Blumwald, E. (1999). Salt tolerance conferred by overexpression of a vacuolar Na+/H+ antiport in Arabidopsis. Science 285, 1256–1258. [DOI] [PubMed] [Google Scholar]
- Aronheim, A., Zandi, E., Hennemann, H., Elledge, S.J., and Karin, M. (1997). Isolation of an AP-1 repressor by a novel method for detecting protein-protein interactions. Mol. Cell. Biol. 17, 3094–3102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaxiola, R.A., Li, J., Undurraga, S., Dang, L.M., Allen, G.J., Alper, S.L., and Fink, G.R. (2001). Drought- and salt-tolerant plants result from overexpression of the AVP1 H+-pump. Proc. Natl. Acad. Sci. USA 98, 11444–11449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour, S.J., Sebolt, A.M., Salazar, M.P., Everard, J.D., and Thomashow, M.F. (2000). Overexpression of the Arabidopsis CBF3 transcriptional activator mimics multiple biochemical changes associated with cold acclimation. Plant Physiol. 124, 1854–1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong, D., Guo, Y., Jagendorf, A.T., and Zhu, J.K. (2002). Biochemical characterization of the Arabidopsis protein kinase SOS2 that functions in salt tolerance. Plant Physiol. 130, 256–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, Y., Halfter, U., Ishitani, M., and Zhu, J.-K. (2001). Molecular characterization of functional domains in the protein kinase SOS2 that is required for plant salt tolerance. Plant Cell 13, 1383–1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halfter, U., Ishitani, M., and Zhu, J.-K. (2000). The Arabidopsis SOS2 protein kinase physically interacts with and is activated by the calcium-binding protein SOS3. Proc. Natl. Acad. Sci. USA 97, 3735–3740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishitani, M., Liu, J., Halfter, U., Kim, C.-S., Shi, W., and Zhu, J.-K. (2000). SOS3 function in plant salt tolerance requires N-myristoylation and calcium-binding. Plant Cell 12, 1667–1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaglo-Ottosen, K.R., Gilmour, S.J., Zarka, D.G., Schabenberger, O., and Thomashow, M.F. (1998). Arabidopsis CBF1 overexpression induces COR genes and enhances freezing tolerance. Science 280, 104–106. [DOI] [PubMed] [Google Scholar]
- Knight, H., Trewavas, A.J., and Knight, M.R. (1997). Calcium signaling in Arabidopsis thaliana responding to drought and salinity. Plant J. 12, 1067–1078. [DOI] [PubMed] [Google Scholar]
- Kovtun, Y., Chiu, W.L., Tena, G., and Sheen, J. (2000). Functional analysis of oxidative stress-activated mitogen-activated protein kinase cascade in plants. Proc. Natl. Acad. Sci. USA 97, 2940–2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J., Ishitani, M., Halfter, U., Kim, C.-S., and Zhu, J.-K. (2000). The Arabidopsis thaliana SOS2 gene encodes a protein kinase that is required for salt tolerance. Proc. Natl. Acad. Sci. USA 97, 3730–3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J., and Zhu, J.-K. (1998). A calcium sensor homolog required for plant salt tolerance. Science 280, 1943–1945. [DOI] [PubMed] [Google Scholar]
- Ohta, M., Guo, Y., Halfter, U., and Zhu, J.K. (2003). A novel domain in the protein kinase SOS2 mediates interaction with the protein phosphatase 2C ABI2. Proc. Natl. Acad. Sci. USA 100, 11771–11776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu, Q.S., Guo, Y., Dietrich, M.A., Schumaker, K.S., and Zhu, J.-K. (2002). Regulation of SOS1, a plasma membrane Na+/H+ exchanger in Arabidopsis thaliana, by SOS2 and SOS3. Proc. Natl. Acad. Sci. USA 99, 8436–8441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu, Q.S., Guo, Y., Quintero, F.J., Pardo, J.M., Schumaker, K.S., and Zhu, J.-K. (2004). Regulation of vacuolar Na+/H+ exchange in Arabidopsis thaliana by the SOS pathway. J. Biol. Chem. 279, 207–215. [DOI] [PubMed] [Google Scholar]
- Qiu, Q.S., and Su, X.F. (1998). The influence of extracellular-side Ca2+ on the activity of the plasma membrane H+-ATPase from wheat roots. Aust. J. Plant Physiol. 25, 923–928. [Google Scholar]
- Quintero, F.J., Ohta, M., Shi, H., Zhu, J.-K., and Pardo, J.M. (2002). Reconstitution in yeast of the Arabidopsis SOS signaling pathway for Na+ homeostasis. Proc. Natl. Acad. Sci. USA 99, 9061–9066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rus, A., Yokoi, S., Sharkhuu, A., Reddy, M., Lee, B.H., Matsumoto, T.K., Koiwa, H., Zhu, J.K., Bressan, R.A., and Hasegawa, P.M. (2001). AtHKT1 is a salt tolerance determinant that controls Na(+) entry into plant roots. Proc. Natl. Acad. Sci. USA 98, 14150–14155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, H., Lee, B., Wu, S.-J., and Zhu, J.-K. (2003). Overexpression of a plasma membrane Na+/H+ antiporter improves salt tolerance in Arabidopsis. Nature Biotechnol. 21, 81–85. [DOI] [PubMed] [Google Scholar]
- Zhang, H.X., and Blumwald, E. (2001). Transgenic salt-tolerant tomato plants accumulate salt in foliage but not in fruit. Nature Biotechnol. 19, 765–768. [DOI] [PubMed] [Google Scholar]
- Zhang, H.X., Hodson, J.N., Williams, J.P., and Blumwald, E. (2001). Engineering salt-tolerant Brassica plants: Characterization of yield and seed oil quality in transgenic plants with increased vacuolar sodium accumulation. Proc. Natl. Acad. Sci. USA 98, 12832–12836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, J.-K. (2000). Cell signaling under salt, water and cold stresses. Curr. Opin. Plant Biol. 4, 401–406. [DOI] [PubMed] [Google Scholar]
- Zhu, J.-K. (2002). Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 53, 247–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, J.-K., Xiong, L., Ishitani, M., Liu, J., Lee, H., Stevenson, B., and Shi, W. (1998). Identification of genes important for environmental stress tolerance in plants. In Breeding and Biotechnology of Environmental Stress in Rice, Y. Sato, ed (Sapporo, Japan: Hokkaido National Agricultural Experiment Station), pp. 105–113.