Abstract
The pale aleurone color1 (pac1) locus, required for anthocyanin pigment in the aleurone and scutellum of the Zea mays (maize) seed, was cloned using Mutator transposon tagging. pac1 encodes a WD40 repeat protein closely related to anthocyanin regulatory proteins ANTHOCYANIN11 (AN11) (Petunia hybrida [petunia]) and TRANSPARENT TESTA GLABRA1 (TTG1) (Arabidopsis thaliana). Introduction of a 35S-Pac1 transgene into A. thaliana complemented multiple ttg1 mutant phenotypes, including ones nonexistent in Z. mays. Hybridization of Z. mays genomic BAC clones with the pac1 sequence identified an additional related gene, mp1. PAC1 and MP1 deduced protein sequences were used as queries to build a phylogenetic tree of homologous WD40 repeat proteins, revealing an ancestral gene duplication leading to two clades in plants, the PAC1 clade and the MP1 clade. Subsequent duplications within each clade have led to additional WD40 repeat proteins in particular species, with all mutants defective in anthocyanin expression contained in the PAC1 clade. Substantial differences in pac1, an11, and ttg1 mutant phenotypes suggest the evolutionary divergence of regulatory mechanisms for several traits that cannot be ascribed solely to divergence of the dicot and monocot protein sequences.
INTRODUCTION
Flavonoids are involved in many important processes, including auxin transport (Brown et al., 2001; Peer et al., 2001), attraction of pollinators (Mol et al., 1998), defense against predators (Dooner et al., 1991; Grotewold et al., 1998), and protection against UV damage (Stapleton and Walbot, 1994). The visible yet dispensable nature of many flavonoid molecules, especially anthocyanins, has enabled the identification of many biosynthetic genes and their regulators, providing an excellent model for investigations of gene regulation across a wide variety of plants. The flavonoid pathways have both similarities and differences in regulation in different species.
The anthocyanin pathway in Zea mays (maize) is one of the most thoroughly investigated branches of flavonoid metabolism. The products of the pathway are red and purple pigments that are easily scored and not required for Z. mays growth and reproduction, properties that have aided in the identification of many anthocyanin biosynthetic genes and several regulators of the pathway (reviewed in Dooner et al., 1991). Two classes of regulatory genes encode basic helix-loop-helix (bHLH) and MYB transcription factors (Paz-Ares et al., 1987; Chandler et al., 1989; Ludwig et al., 1989; Cone et al., 1993). The bHLH (BOOSTER1 [B] or RED1 [R]) and MYB (PURPLE PLANT1 [Pl1] or COLORED ALEURONE1 [C1]) proteins coordinately activate transcription of the biosynthetic genes through protein–protein interactions (Goff et al., 1992). The multiple members of each class are functionally redundant, and typically only one of the b or r genes and one of the c1 or pl1 genes are expressed in any given tissue. In aleurone, b and r alleles expressed in the seed (Styles et al., 1973) act in combination with c1, which directly binds to the promoters of the biosynthetic genes, activating transcription (Sainz et al., 1997; Lesnick and Chandler, 1998).
Comparison of anthocyanin regulation in several dicot species (Petunia hybrida [petunia], Antirrhinum majus [snapdragon], and Arabidopsis thaliana) with that in Z. mays reveals similarities and interesting differences. In both Z. mays and dicots, conserved enzymatic genes are activated in groups or modules, with each group or module responsible for flux through specific branches of the flavonoid pathways leading to anthocyanin or proanthocyanidin pigmentation. These groups or modules are regulated in each species by similar sets of regulatory proteins, including bHLH and MYB proteins. Differences include the presence of species-specific enzymes at certain positions in the pathway and differences in the grouping of genes into modules. In dicots, anthocyanin biosynthetic genes are divided into early and late biosynthetic genes (Martin and Gerats, 1993). By contrast, many of the functionally analogous early and late biosynthetic genes in Z. mays are coordinately controlled as a single module with no division between early and late biosynthetic gene regulation (reviewed in Mol et al., 1998). Z. mays and dicots also may differ with respect to regulation of the regulatory genes. In dicots, some regulatory proteins are capable of increasing the RNA levels of some of the other regulatory genes. Several examples of this have been reported, including the activation of the TRANSPARENT TESTA8 (TT8) bHLH gene by the TRANSPARENT TESTA GLABRA1 (TTG1) protein in A. thaliana and the induction of TT8 expression by ectopic expression of the TT2 MYB gene (Nesi et al., 2000, 2001; Spelt et al., 2000). This contrasts significantly with Z. mays, in which the identified regulators of the biosynthetic genes are not under each other's control (Goff et al., 1990). Significant differences between the regulatory programs in dicots and monocots also can be illustrated in comparisons of regulatory genes in one species with homologous genes in another species, combined with a consideration of whether their functions are identical. As an example, the bHLH encoding anthocyanin1 (an1) and TT8 genes in dicots (Nesi et al., 2000; Spelt et al., 2000) share stronger similarity with the Z. mays intensifier1 (in1) gene (Burr et al., 1996) than with the Z. mays b and r genes. This is significant because an1 and TT8 are positive activators of the pathway, as are b and r, but an1 and TT8 are more closely related by sequence to in1, which functions as a negative regulator. Thus, evolution of the modules of flavonoid biosynthetic genes and the regulation of these modules has taken significantly different routes in the dicot and monocot species investigated. The result is that the regulatory framework, including the hierarchical organization of the regulators' functions, cannot be predicted solely from homology.
In a screen for new regulators of the Z. mays anthocyanin pathway (Selinger and Chandler, 1999), the pale aleurone color1 (pac1) locus was identified by a mutation that resulted in pale aleurone color. Determination of the RNA levels of several of the biosynthetic genes established that pac1 is required for normal RNA levels of the biosynthetic genes. By contrast, pac1 is not required for normal RNA levels of the regulatory genes b and c1. Either B or R bHLH proteins require PAC1 for full activation of the biosynthetic genes. Different alleles of b and r that confer different tissue-specific patterns of pigmentation require pac1 function in the aleurone and scutellum but not in husk, sheath, tassel, and anthers (Selinger and Chandler, 1999).
We report the cloning and characterization of the locus encoding the PAC1 protein. The PAC1 sequence is most similar to AN11 in P. hybrida and TTG1 in A. thaliana (de Vetten et al., 1997; Walker et al., 1999). All three encode WD40 repeat proteins involved in regulation of anthocyanin pigmentation, although the precise means by which this regulation is accomplished varies among species. In addition to anthocyanin production, other species-specific traits require the function of this class of WD40 repeat proteins. To begin to understand the evolutionary context for species-specific differences in anthocyanin gene regulation, we complemented the ttg1 mutant phenotypes using a 35S-Pac1 transgene, further investigated pac1 mutant phenotypes in Z. mays, isolated another gene encoding a related WD40 repeat protein from Z. mays (mp1), and characterized the phylogenetic relationships among members of this WD40 repeat protein family.
RESULTS
Identification and Characterization of a Mu1 Insertion Cosegregating with the pac1 Mutant Phenotype
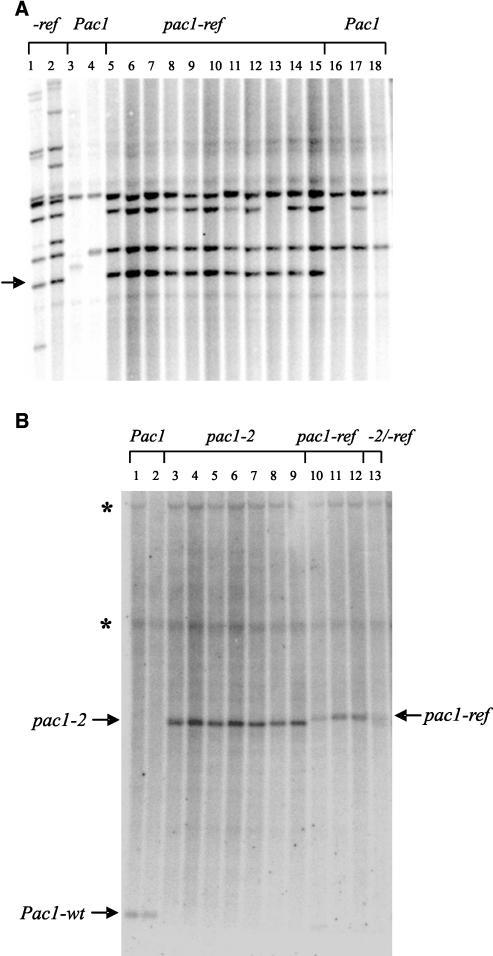
The pac1-ref allele was isolated in a screen for pale aleurone color mutants in a line that contained active Mutator (Mu) transposons (Selinger and Chandler, 1999). When crossed with an active Mu line, dark purple sectors (spots) in pac1-ref aleurones were observed, suggesting the presence of a Mu transposable element in the pac1-ref allele that was capable of excision and restoration of pac1 gene activity. By contrast, no spots were observed in progeny from crosses between pac1-ref and active Activator and Suppressor-mutator lines. A cosegregation analysis of Mu elements and the pac1-ref phenotype was used to clone the pac1 gene as follows. pac1-ref plants that had lost Mu activity (assessed as an absence of spotting) were outcrossed for several generations to non-Mu plants to dilute the number of Mu elements. One of the resulting plants (heterozygous for pac1-ref and wild-type alleles) was self-pollinated and displayed the expected 3:1 purple:pale seed phenotype. A linked marker and the pac1-ref anthocyanin phenotype were used to identify homozygous wild-type and homozygous pac1-ref progeny (see Methods). DNA gel blot analysis using Mu element probes revealed a Mu1 hybridizing band cosegregating with the pac1-ref phenotype in 11 plants (Figure 1A) and absent in three homozygous wild-type siblings. Sequence flanking this element was obtained using inverse PCR (see Methods), resulting in the sequencing of 1588 bp, including 199 bp of Mu1 sequence, 1380 bp of the putative pac1 locus, and a 9-bp duplication at the site of Mu1 insertion. To confirm that the putative pac1 sequence recovered was adjacent to the cosegregating Mu1 element and not some other Mu1 element, a probe was derived and designated SacI/SmaI 700 (Figure 2). DNA gel blot analysis revealed colocalization of this probe and the cosegregating Mu1 band and an additional 1380-bp band in wild-type plants (data not shown).
Figure 1.
DNA Gel Blot Analysis of pac1 Mutant Alleles.
DNA samples for both blots were digested with EcoRI and HindIII.
(A) Identification of a 2.8-kb Mu1 hybridizing DNA fragment cosegregating with the pac1-ref mutant allele (arrow). Lanes 1 and 2, plants known to be pac1-ref; lanes 3 and 4, plants known to be Pac1; lanes 5 to 15, pac1-ref in family used for inverse PCR; lanes 16 to 18, Pac1 siblings of pac1-ref. An internal Mu1 fragment was used as the probe.
(B) Identification of the insertion in the pac1-2 allele. Lanes 1 and 2, Pac1; lanes 3 to 9, pac1-2; lanes 10 to 12, pac1-ref; lane 13, pac1-2/pac1-ref. Hybridization was with the SacI/SmaI 700 probe and DNA flanking the Mu1 insertion (Figure 2). Asterisks indicate fragments cross-hybridizing with the SacI/SmaI 700 probe that are not from the pac1 locus.
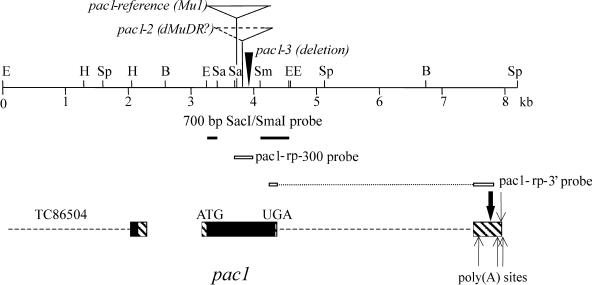
Figure 2.
The pac1 Locus and Probes.
All open reading frames in this genomic fragment are indicated by closed boxes, untranslated regions by hatched boxes, and introns by dashed lines. The pac1 transcription unit spans two exons separated by a 3017-bp intron (dashed line). Large and small arrows indicate relative abundance of sequenced poly(A) sites. The location of Mu insertions in the pac1-ref and pac1-2 alleles and the pac1-3 deletion are depicted at the top of the figure. The 9-bp duplication and Mu1 insertion for pac1-ref begins at codon 165. Sequence from pac1-2 indicates a Mu insertion at codon 192. The pac1-3 deletion begins at codon 203. The 700-bp SacI/SmaI probe was obtained by inverse PCR and is indicated by solid lines. Open boxes indicate probes for RNase protections. Part of a small putative zinc finger protein is located <1000 bp upstream of the pac1 initiator ATG site. Transcripts for this upstream gene are described in TIGR contig TC86504. B, BamHI; E, EcoRI; H, HindIII; Sa, SacI; Sm, SmaI; Sp, SpeI.
The pac1-2 and pac1-3 Alleles Confirm the Identification of the pac1 Locus
Two new alleles of pac1 (pac1-2 and pac1-3) were isolated by crossing pac1-ref plants to plants from an active Mu line and screening the resulting seed for the pale aleurone phenotype. Subsequent crosses were used to segregate these new alleles away from the pac1-ref allele. The pac1-2 allele displayed a spotted phenotype in active Mu lines, suggesting excision of an element and restoration of wild-type gene activity. To determine unequivocally that the Mu1 cosegregating sequence cloned from the pac1-ref allele identified the pac1 locus, DNA gel blot analysis was performed using the SacI/SmaI 700 probe and DNA from plants with the pac1-2 allele. This experiment revealed an ∼2700-bp band consistent with a 1300-bp insertion within the same 1380-bp fragment as the insertion associated with the pac1-ref allele (Figure 1B). The other new allele, pac1-3, displayed a pale aleurone phenotype with no detectable spotting in the presence of active Mu. DNA gel blot analysis using the SacI/SmaI 700 probe and DNA from plants with the pac1-3 allele revealed a hybridizing band slightly smaller than that of the wild type (data not shown).
Regions of the pac1-2 and pac1-3 alleles were PCR amplified and sequenced (see Methods). Sequence from the pac1-2 allele revealed a Mu element insertion within the same open reading frame (ORF) interrupted in the pac1-ref allele. The Mu element in the pac1-2 allele had characteristics of a MuDR element with a significant internal deletion, and it therefore was designated dMuDR (see Methods). Amplification of the pac1-3 allele revealed a 25-bp deletion within the pac1 ORF resulting in a frameshift starting at putative amino acid 203, altering the next 20 amino acids and introducing a premature stop codon. The locations of the Mu element insertions in the pac1-ref and pac1-2 alleles and the deletion in the pac1-3 allele are indicated in Figure 2. Taken together, characterization of these mutants firmly establishes the identification of the pac1 locus.
Analysis of pac1 Genomic Structure and mRNA Expression
To characterize a wild-type allele of the pac1 gene, BAC clones containing the gene were identified in the Clemson University Genomics Institute (CUGI) B73 genomic BAC library (see Methods). Comparison of fragments produced by restriction digestion and restriction mapping of wild-type genomic DNA revealed that the BAC insert chosen for sequencing had the same restriction map as pac1 wild-type genomic DNA. The resulting pac1 sequence was deposited to GenBank (accession number AY115485).
An ORF of 353 amino acids within a single exon spans the region containing the two Mu insertions and the pac1-3 deletion (Figure 2). To further characterize the gene's structure, 3′ rapid amplification of cDNA ends (RACE) was performed using RNA isolated from immature tassel (see Methods). Thirteen 3′ RACE clones were isolated, sequenced, and compared with the genomic locus, revealing an intron in the genomic sequence starting a few base pair 3′ of the ORF's stop codon as well as five alternative poly(A) addition sites (indicated by arrows in Figure 2). The detection of another gene in the genomic sequence within 1 kb upstream of the pac1 start codon and the lack of significant ORFs in the sequences between the two coding regions suggest the identification of the complete pac1 ORF.
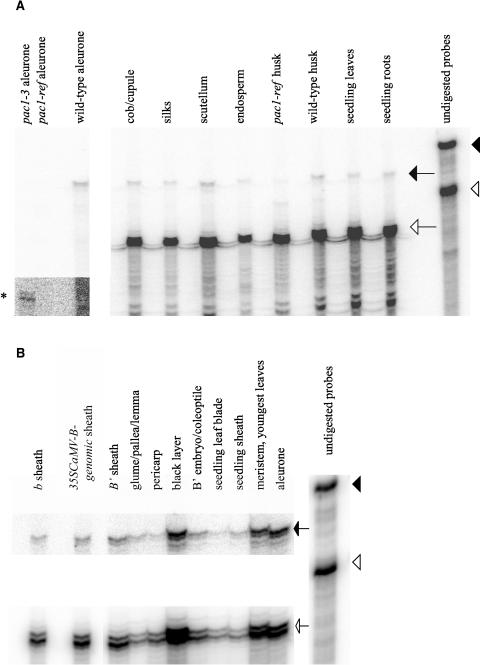
To determine which tissues express pac1, RNase protection experiments were performed using the pac1-rp-300 probe (see Methods; Figure 2) and actin1 as an internal control. Low levels of pac1 transcript were detected in RNA from all wild-type tissues tested (Figures 3A and 3B; Methods). The expression levels varied no more than threefold in all tissues, and the amount of signal resulting from our protections was roughly one-third to one-half of that seen in protections with b and c1 (data not shown), suggesting that pac1 is expressed more ubiquitously but at a lower level than either the bHLH or MYB transcription factors. Similar results were observed (data not shown) using the pac1-rp-3′ probe, which spans the splice site of the two pac1 exons (see Methods; Figure 2).
Figure 3.
RNase Protections Using the pac1-rp-300 Probe.
Arrowheads indicate undigested probes, and arrows indicate probes protected by sample RNA (closed, pac1; open; actin1).
(A) The pac1-rp-300 probe was protected in the absence of the probe for actin1 for the first three samples, which are from aleurones of pac1-3, pac1-ref, and a wild-type control. The remaining samples include both actin1 and pac1 probes. The asterisk in the panel at left, which contains a longer exposure, shows the expected protected product for the pac1-3 allele. The pac1-ref and wild-type husks were from plants segregating in the same family. All other plant parts were from homozygous wild-type individuals.
(B) All samples were from Pac1 wild-type individuals. The levels were adjusted upward in Adobe Photoshop for the pac1 protected probe because the signal was weak compared with actin1.
Examination of transcript levels from mutant pac1 alleles revealed substantial differences when compared with the wild type. When using the pac1-rp-300 probe (Figure 3A), which spans the locations of the insertions and the deletion, a smaller 180-base fragment (asterisk in Figure 3A) was observed when pac1-3 RNA was used, consistent with internal cleavage to a smaller product because of a lack of complementary sequence of the probe and the pac1-3 transcript in the region of the pac1-3 deletion. pac1-ref (Figure 3A) and pac1-2 (data not shown) RNA protected substantially less pac1-rp-300 probe than the wild type and did not reveal any significant smaller products, suggesting that the region covered by the pac1-rp-300 probe is either not well transcribed or is substantially altered for these two alleles. We favor the latter explanation because levels of pac1 RNA for all three alleles did not differ from those of the wild type when the pac1-rp-3′ probe was used (data not shown). Occasionally, a trace of protected RNA that is similar in size to the wild type is observed in pac1-ref and pac1-2 samples (signal normalized to actin1 for pac1-ref husk in Figure 3A is 13% of that of wild-type sibling husk), suggesting that in a minority of transcripts in at least some tissues, the Mu elements may be spliced out or that the samples may contain sectors of tissue with somatic excisions of the Mu elements.
The PAC1 Protein Is a Homolog of AN11 and TTG1
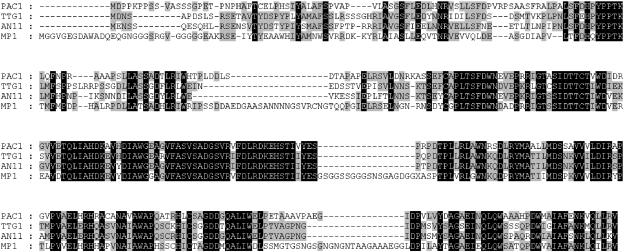
In a Basic Local Alignment Search Tool (BLASTp) search (Altschul et al., 1997) using the PAC1 sequence as a query, TTG1 (A. thaliana) and AN11 (P. hybrida) were identified as the strongest hits, each with e-110 expect values. Significantly, AN11 and TTG1 are involved in the expression of several traits in their respective plants, including anthocyanin production. Figure 4 shows an alignment of PAC1, AN11, and TTG1. All three are well conserved, with the noticeable exception of the most N-terminal portion of PAC1. The most similar Z. mays protein identified to date, MP1, which is discussed in detail later, shares less homology with PAC1 than do AN11 and TTG1. In summary, PAC1, a regulator of anthocyanins in Z. mays, is a homolog of proteins previously identified as regulators of the anthocyanin pathway in P. hybrida (de Vetten et al., 1997) and A. thaliana (Walker et al., 1999).
Figure 4.
Alignment of PAC1, TTG1, AN11, and MP1.
Amino acids conserved in all four proteins are indicated in black. Gray indicates identity in two or three of the proteins.
The PAC1 Protein Complements ttg1-1 and ttg1-13 Phenotypes in A. thaliana
Because of the high similarity of PAC1 with TTG1 and the involvement of both proteins in anthocyanin pigmentation, the ability of PAC1 to complement ttg1 mutants was investigated. Of particular interest was whether the Z. mays PAC1 protein could complement not only ttg1 defects in anthocyanin production but also other ttg1 defects as well. A PAC1 expression vector, designated 35S-Pac1, which included a gene encoding resistance for the herbicide Basta, was transformed into plants homozygous for one of two ttg1 mutant alleles. Both ttg1 alleles result in severe defects in trichome differentiation, anthocyanin pigmentation, seed coat pigmentation, and seed coat mucilage (Koorneef, 1981; Larkin et al., 1999). The molecular basis of the mutant phenotypes are a premature stop codon 25 amino acids from the C terminus of the protein in ttg1-1 (Walker et al., 1999) and a deletion of the entire coding region in ttg1-13 (Larkin et al., 1999). Figure 5 and Table 1 show the results of these complementation experiments. Significantly, and presented in more detail below, when any ttg1 mutant phenotype was complemented, complementation of all of the other tested ttg1 mutant phenotypes was observed.
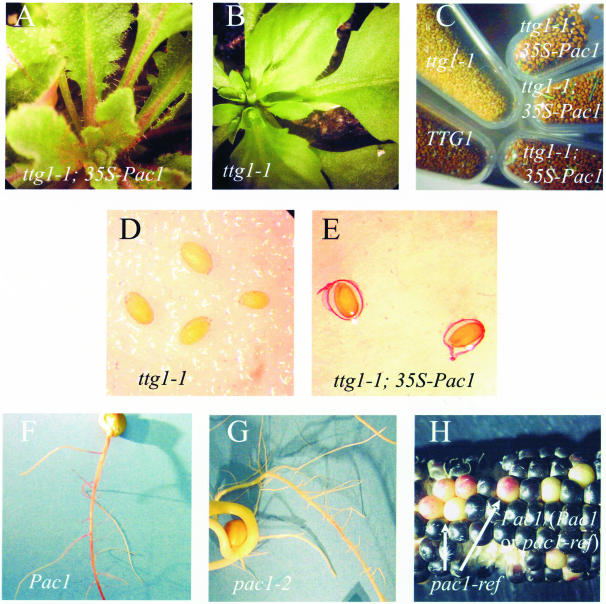
Figure 5.
Phenotypes of 35S-Pac1 Complemented ttg1-1 Mutants and pac1 Mutants.
(A) and (B) Anthocyanin, trichome, and leaf phenotypes are complemented by the 35S-Pac1 transgene. A ttg1-1 plant containing the 35S-Pac1 transgene (A) and a nontransgenic sibling (B).
(C) The 35S-Pac1 transgene restores wild-type seed pigmentation to ttg1-1 mutants. Mature seed phenotypes of wild-type (TTG1) Columbia, ttg1-1 in the Columbia background (yellow seed), and three independent ttg1-1 lines containing the 35S-Pac1 transgene.
(D) and (E) Mucilage production is restored by the 35S-Pac1 transgene. Ruthenium red staining of ttg1-1 Columbia (D) and ttg1-1; 35S-Pac1 seeds (E).
(F) and (G) Root phenotypes in wild-type (F) and pac1-2 (G) sibling seedlings expressing the anthocyanin regulatory gene r. Plant in (F) received a root anthocyanin score of 8. Plant in (G) was from the most darkly pigmented seed in the pac1-2 category and received a root anthocyanin score of 4.
(H) in1; pr1; R ear segregating Pac1/(Pac1 or pac1-ref) and pac1-ref kernels. All pac1-ref kernels were pale.
Table 1.
Phenotypes of Primary 35S-Pac1 Transformants while under Basta Herbicide Selection
| Allele (Background)a | Number of Seedlings with Trichomes (Total Scored)bc | Number of Plants with Trichomes Yielding Brown Seed and Mature Plant Anthocyanins (Total Scored) |
|---|---|---|
| ttg1-1 (Columbia) | 26 (39) | 6 (6) |
| ttg1-1 (Landsberg erecta) | 13 (15) | 3 (3) |
| ttg1-13 | 12 (16) | 1 (1) |
The ttg1-13 allele was partially introgressed from the RLD ecotype into Columbia.
Number of trichomes per first true leaf ranged from 4 to 38 trichomes with an average of 17.7 trichomes (sd of 7.3, n = 30 plants, all alleles pooled). No trichomes were seen in several hundred nontransformed ttg1 mutants.
All plants lacking trichomes as seedlings lacked anthocyanins as mature plants. The four such plants that were followed to silique shatter yielded yellow seed, consistent with no proanthocyanidin synthesis.
To determine if the 35S-Pac1 transgene complements the ttg1 defect in trichomes, all primary transformant (Basta-resistant) seedlings were screened for trichomes. Trichomes were observed in 72% of the resistant seedlings (Figures 5A and 5B, Table 1), whereas no trichomes were observed in any nontransformed controls (ttg1-1 and ttg1-13 parental lines and nontransgenic mutant lines resulting from the floral dip). To determine if anthocyanin defects in ttg1 mutants were complemented, mature plants were examined (Table 1). Ten out of fourteen mature Basta-resistant plants had increased anthocyanin pigments, whereas four showed no complementation. The four anthocyanin noncomplemented plants also lacked trichomes and, importantly, yielded only Basta-sensitive progeny that were not complemented for any ttg1 phenotype, suggesting that these four plants either escaped initial Basta selection and were not transformed or were subsequently silenced for transgene activity.
Examination of the seed phenotypes of 35S-Pac1 ttg1 plants showed that all 10 primary regenerant Basta-resistant plants yielded brown seeds upon self-pollination (Figure 5C), indicating complementation of the ttg1 proanthocyanidin defect in the maternally derived seed coat (Table 1, Figure 5C). Similarly, seeds from Basta-resistant plants were positive for the presence of mucilage on the seed coat using ruthenium red staining (Figure 5E). By contrast, little to no mucilage was detected in the seed coats of nontransformed ttg1 mutants (Figure 5D), and control seeds were yellow, indicating a lack of proanthocyanidins in all ttg1 mutant controls (Figure 5C).
To examine the transmission of complemented phenotypes resulting from expression of 35S-Pac1 in ttg1 plants, we planted seed obtained from self-pollination of 9 of the 10 complemented lines grown under nonselective conditions. In all lines, when one phenotype was complemented, all phenotypes were complemented: anthocyanins, trichomes, proanthocyanidins, and seed coat mucilage (root phenotypes associated with ttg1 mutants are difficult to score, and complementation of the ttg1 root defects therefore was not examined rigorously).
pac1 Affects Anthocyanin Regulation in Z. mays Seedling Roots but Does Not Affect Root Morphology or Trichomes
The observations that pac1 was expressed in all Z. mays tissues tested prompted a further examination of pac1 mutant phenotypes in a variety of Z. mays tissues. Consistent with what was previously reported (Selinger and Chandler, 1999), anthocyanin pigmentation of above ground tissues, mature husk, sheath, tassel, or anthers was not obviously affected in comparisons of pac1 and wild-type plants. However, in roots, a noticeable reduction in anthocyanin pigmentation was observed in pac1 mutants. To score this effect, a 1 to 8 root color score was devised, in which a score of 1 corresponded to the absence of anthocyanins, and a score of 8 corresponded to the highest anthocyanin levels observed in this experiment (for examples of root color scores of 8 and 4, see Figures 5F and 5G, respectively). The roots of 11 mutant pac1-2 plants had an average color score of 2.1 (sd = 1.1, range: 1 to 5). By contrast, nine wild-type siblings had an average color score of 7.2 (sd = 2.1, range: 4 to 8), indicating that pac1 function is required for strong activation of the anthocyanin pathway in roots.
Because TTG1 function in A. thaliana is required for normal root (Galway et al., 1994) and trichome development, possible effects of the pac1 mutant on Z. mays root and trichome development were investigated. Z. mays pac1-2 and wild-type siblings showed no differences in root morphology or abundance of root hairs (data not shown). Likewise, no pac1 mutant effects were demonstrated in pac1-ref and wild-type siblings with respect to architecture and spacing of the two trichome classes in Z. mays, prickle hairs and macrohairs, on mature leaf blades. In summary, pac1 mutant phenotypes in Z. mays differ significantly from ttg1 mutant root and trichome phenotypes in A. thaliana, in that pac1 mutants do not display defects in root and trichome development that are characteristic of ttg1 mutants.
In addition to effects on anthocyanin pigmentation, significant pac1 effects on height were noticed with all three pac1 mutant alleles. We examined each mutant allele relative to their wild-type siblings and each showed a significant difference (data not shown). The pooled data demonstrated that wild-type plants were 17 cm taller on average (n = 55; mean, 139 cm; se = 13) than pac1 mutant plants (n = 55; mean, 122 cm; se = 15; t test, P < 0.001). There was no significant corresponding decrease in the number of leaves (means: the wild type, 17.5 with se of 1; mutant, 17.4 with se of 1.2). Thus, the wild-type function of pac1 is required for normal stature with the loss of function leading to shorter plants. This difference is fully recessive because heterozygous individuals had equivalent heights as wild-type individuals (27 confirmed heterozygotes had a mean of 141 cm with a se of 7, and 11 homozygotes had a mean of 140 cm with a se of 9).
Genetic Interactions of pac1 with Other Z. mays Anthocyanin Regulatory Loci
The bHLH and MYB proteins encoded by b and c1 loci are required for activation of the anthocyanin pathway in Z. mays. It was demonstrated previously (Selinger and Chandler, 1999) and verified (this study, data not shown) that b and c1 RNA levels are similar in pac1-ref and wild-type individuals, suggesting that pac1 does not influence the expression of either of these genes at the transcriptional or RNA levels. To test for the possibility that pac1 expression is influenced by b and/or c1 expression levels, RNase protections were performed using the pac1-rp-300 probe and RNA from adult sheaths, a tissue with no c1 expression (Cone et al., 1993). pac1 RNA levels were assayed in sheaths of plants with a nonfunctional allele of b producing no anthocyanin pigmentation (b), a functional allele of b producing a moderate amount of anthocyanin pigmentation (B′), and a strongly expressed functional transgenic allele of b producing intense anthocyanin pigmentation (35SCaMV-B-I-genomic; C.C. Carey and V.L. Chandler, unpublished data). There was no change in pac1 RNA levels in response to increasing b expression, indicating that pac1 expression is not under the control of b (Figure 3B). Although expression of pac1 mRNA does not require expression of c1, we have not formally ruled out the possibility that the MYB gene pl1 may regulate pac1 in tissues such as sheath and leaf in place of c1. However, because most pl1 alleles are not expressed in the aleurone, there is no absolute requirement of pac1 for pl1 expression. In conclusion, pac1 regulates neither b nor c1 at the RNA level, and pac1 expression does not require b or the exclusive action of c1 or pl1.
The in1 gene (Burr et al., 1996) encodes a bHLH protein in Z. mays with the most similarity to TT8, the bHLH protein in A. thaliana thought to function like Z. mays B and R in activation of anthocyanin and proanthocyanidin biosynthetic genes. The fact that in1 mutations result in very intense pigmentation in the aleurone suggests that IN1 acts as a repressor (Burr et al., 1996) or diverts metabolic precursors into an alternative pathway (Nesi et al., 2000). Because of the established relationship of some of the bHLH regulators with WD40 repeat proteins, we examined the genetic interaction of pac1 and in1 by creating a line that was homozygous R; in1; pr1 (red aleurone, a locus encoding flavonoid 3′-hydroxylase function [Larson et al., 1986]) and heterozygous for pac1-ref. Two ears resulting from self-pollination of these plants displayed one-quarter pale and spotted kernels (consistent with somatic excision of the Mu1 element in the pac1-ref allele) and three-quarters dark kernels (Figure 5H). DNA gel blot analysis verified that all pale seeds tested were homozygous for the pac1-ref allele, and purple seeds were either heterozygous or wild-type for pac1 (Methods). Therefore, the function of PAC1 cannot simply be the removal of the IN1 putative repressor molecule to allow full function of R, otherwise all in1 mutant seed would have been dark, regardless of pac1 genotype. Rather, pac1 is epistatic to in1 and PAC1 function is required for the activation of the anthocyanin pathway in aleurones in the absence of IN1 function.
Identification and Characteristics of the WD40 Repeat Gene Family Related by Sequence to the Plant Anthocyanin Regulators
It is intriguing that pac1 mutants only affect anthocyanin pigmentation in a subset of tissues, given the homology of PAC1 with AN11 and TTG1 and that mutations in the an11 and TTG1 genes eliminate anthocyanin pigmentation entirely. Likewise, it is interesting that pac1 mutants do not affect traits such as root hairs and trichomes as might have been predicted given the homology of PAC1 with TTG1. One explanation for these observations is the presence of Z. mays genes with shared, partially redundant functions. Consistently, DNA gel blot analysis using SacI/SmaI 700 revealed DNA fragments that hybridized but that were independent of the pac1 locus (Figure 1B, fragments marked with asterisks). In addition, two clones were identified from the CUGI B73 BAC library, which contained an ORF of the WD40 repeat protein family identical to a sequence obtained from an EST database and previously named MP1 (Hernandez et al., 2000). Consistent with there being two separate groups of related WD40 repeat proteins, the MP1 protein is more similar in sequence to ATAN11A and ATAN11B (de Vetten et al., 1997) (68% identity) from A. thaliana than to PAC1 (58% identity) or TTG1 and AN11. Using mp1 DNA sequence as a probe for stringent DNA gel blot analysis, no hybridization was observed to pac1 nor to the fragments that cross-hybridized with SacI/SmaI 700, suggesting that the two other fragments identified with pac1 probes using DNA gel blot analysis may represent other genes.
Sequence analysis and phylogenetic trees of PAC1 and MP1 proteins and their homologs in several species could shed light on the evolutionary history of these WD40 repeat proteins and set the foundation for studies addressing the redundant or divergent functions of these proteins. To that end, sequences of the two Z. mays proteins PAC1 and MP1 were used to identify homologs in GenBank, the Plant Genome Database, and the Index for Genomic Research (TIGR) gene indices from all plant species and a representative sampling of other eukaryotes. In all cases, proteins identified as homologs were more similar to PAC1 and MP1 than to other WD40 repeat proteins in their respective genomes. As a general rule, homologous sequences had TBLASTN expect values of e-25 and higher significance, whereas several more distantly related WD40 repeat sequences were excluded at expect values of e-10 and lower significance.
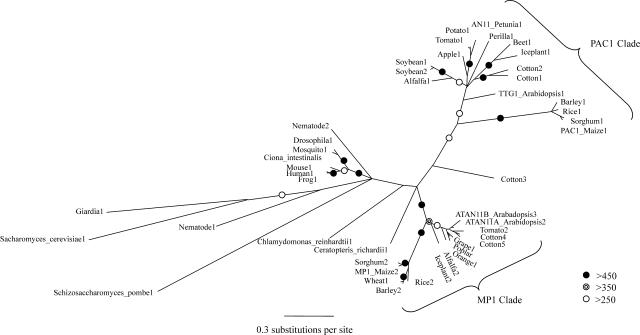
Seventy-five deduced protein sequences, including those of AN11 and TTG1, were assembled into a multiple alignment (see supplemental data online). The multiple alignment, which included 30 full-length sequences, suggested that there are at least two classes of this WD40 repeat protein family in plants. Ninety-seven amino acids of the N-terminal sequence of 46 proteins (31 species) were used to generate the representative phylogeny shown in Figure 6 (see Methods). The tree shows three well-supported clades, with two of the clades consisting solely of plant proteins and the third consisting of the animal proteins except nematode. The PAC1 clade includes PAC1, AN11, and TTG1, for which anthocyanin mutant phenotypes have been identified, whereas the MP1 clade includes MP1, ATAN11A, and ATAN11B, for which no mutant phenotypes have been described. Thus, the phylogenies and multiple alignments support a scenario in which an ancestral gene duplication in a plant ancestor led to the PAC1 clade (containing known anthocyanin regulators) and the MP1 clade of proteins. In several plant species, there were subsequent duplication events in either or both clades. Chlamydomonas reinhardtii and Ceratopteris richardii (fern) appear to have sequences shared with both the MP1 and PAC1 clades, such that they are classified into neither clade.
Figure 6.
Phylogenetic Relationships of PAC1 and MP1 Homologs.
Three major clades of PAC1 and MP1 homologs are apparent. The PAC1 clade consists of PAC1, AN11, and TTG1. The MP1 clade contains MP1, ATAN11A, and ATAN11B. A third clade consists of animal species within a grouping of non-plant species. The consensus unrooted tree is depicted, with leaf and branch lengths representing distance derived from maximum likelihood analysis. Leaves that were minimal in length have been exaggerated to emphasize position on branches. Five hundred trees were generated by bootstrapping. Number of trees supporting each branch point is as presented in the key, with the exception that branch points with <250 supporting trees are unmarked. The bar indicates a length representing 0.3 amino acid substitutions per site.
In contrast with plants, there are generally one or two homologous genes in most non-plant species, with a well supported clade within the animals. Nematode, with three loci that are quite divergent from the other animal family members, had the most representatives of this WD40 repeat subfamily. No functional assignment in any of the non-plant species has been made nor could any function be inferred from investigating coexpression patterns of the Saccharomyces cerevisiae homolog with other transcripts in publicly available microarray databases or from investigating protein interaction databases. In summary, several completely sequenced genomes in non-plant species were searched, revealing that fewer of these WD40 protein family members are found in non-plant species and that these form a group distinct from the two clades in plants.
DISCUSSION
WD40 Anthocyanin Regulatory Proteins Are More Conserved Than Other Classes of Anthocyanin Regulators
The 353–amino acid PAC1 protein has high similarity to WD40 repeat proteins encoded by AN11 and TTG1 both in terms of sequence and with respect to common function because they are all regulators of the anthocyanin pathway. This high degree of conservation between species contrasts with what is observed with the other two classes of regulators (bHLH and MYB). With respect to amino acid sequence, PAC1 shares 62% identity with AN11 and TTG1 as well as with the proposed anthocyanin regulator PFWD in Perilla frutescens (Sompornpailin et al., 2002). By contrast, only 30% identity is evident in pair-wise comparisons of any of the following bHLH homologs: B/R in Z. mays (Chandler et al., 1989; Ludwig et al., 1989), IN1 in Z. mays (Burr et al., 1996), TT8 in A. thaliana (Nesi et al., 2000), GLABRA3 and MYC146 in A. thaliana (Payne et al., 2000), AN1 in P. hybrida (Spelt et al., 2000), JAF13 in P. hybrida (Quattrocchio et al., 1998), and DELILAH in A. majus (Goodrich et al., 1992). Given the number of developmental programs in which PAC1, AN11, and TTG1 are implicated, it is likely that a large number of interacting partners combined with the pressures of conserving the seven-bladed propeller structure contributes to the high degree of conservation among these three proteins. Modeling of protein secondary structure and a consideration of the location of ttg1 point mutations suggest that these proteins can form seven-bladed propeller structures typical of other WD40 repeat proteins (see supplemental data online).
A pac1 Transgene Functionally Complements ttg1 Mutants
The 35S-Pac1 transgene complements all ttg1 mutant phenotypes, including those resulting from developmental programs that differ significantly between Z. mays and A. thaliana (trichomes) and are nonexistent in Z. mays (seed coat mucilage), demonstrating that PAC1 contains most if not all functional competencies characteristic of TTG1 in spite of the evolutionary distance between monocots and dicots. One interpretation of these results is that there are no distinct domains within these WD40 repeat proteins responsible for function in distinct pathways. Consistent with this, Larkin et al. (1999) showed that all weak and strong mutant alleles of ttg1 affected trichome, root, and anthocyanin phenotypes. Complementation of ttg1 mutants has been observed using an an11 transgene (C.T. Payne, F. Zhang, and A.M. Lloyd, unpublished data), and expression of PFWD from P. frutescens (Sompornpailin et al., 2002) in wild-type A. thaliana plants results in supernumerary trichomes and enhanced anthocyanin pigmentation, suggesting a role for PFWD in regulation of these traits in P. frutescens. As a result, sequence comparison between PAC1 clade proteins should facilitate determination of residues structurally required for function in all pathways.
Is a Protein Homologous to PAC1 Required for Z. mays Trichome, Root Hair, and Tissue-Specific Regulation of the Anthocyanins?
In Z. mays, the most obvious phenotypes resulting from the pac1 mutations are reduced anthoycanin pigmentation in a subset of normally pigmented tissues. Importantly, this is in contrast with A. thaliana ttg1 mutants, in which anthocyanins are eliminated from all tissues throughout the plant. Shortened stature is observed in plants with pac1 mutant alleles, whereas no such effect is reported in ttg1 or an11 mutants in A. thaliana or P. hybrida. Significant and additional contrasts include the A. thaliana ttg1 mutant effects on proanthocyanidins, trichomes, seed-coat mucilage, and root hair development and the absence of these effects in Z. mays pac1 mutants. In P. hybrida, an11 mutants affect anthocyanins, proanthocyanidins, seed-coat cell morphology, and flower pH, but trichome and root development are unaffected. Thus, many different processes are variously affected by these mutations in different species, with pertubation of anthocyanin pigmentation being the common phenotype. This raises a central evolutionary question: Does the diversification of the nonanthocyanin functions result from evolution of the protein sequences, or does the diversification of other functions reflect evolution at another level, such as the presence or absence of redundantly functioning proteins and/or a difference in the regulatory networks controlling these processes in different species?
Based on the ability of the pac1 transgene to complement the ttg1 mutant phenotypes, it appears that the differences in nonanthocyanin functions among these species is not the result of differences at the protein sequence level but is the result of differences in organization of the genome or in the mechanisms of regulation of these traits. Gene duplications resulting in redundantly functioning WD40 repeat proteins with different tissue-specific expression patterns within the same organism could explain the differences in phenotypes observed in Z. mays, P. hybrida, and A. thaliana. The observation that an ancestral duplication led to PAC1 and MP1 clade proteins suggests that proteins from each class still may share common function, in which case, MP1 may functionally replace or be primarily responsible for normal anthocyanin pigmentation in some tissues and also may be responsible for trichome and root development.
Evidence arguing against PAC1 and MP1 clades retaining some common functionality is that several A. thaliana programs are apparently affected when ttg1 is mutant, despite the presence and presumed function of two MP1 clade proteins (ATAN11A and ATAN11B). ESTs representing at least one of these proteins are as widespread and as abundant as ESTs representing TTG1, suggesting that MP1 and PAC1 clade proteins may not be functionally equivalent in A. thaliana. Evidence arguing that PAC1 and MP1 clade proteins may retain the capacity for common functionality is transient activation experiments in P. hybrida, in which expression of a luciferase reporter driven by an anthocyanin biosynthetic gene promoter was enhanced by coexpression of ATAN11A and even the Homo sapiens (human) homolog from the non-plant clade (de Vetten et al., 1999). Our observation of some, albeit reduced, anthocyanin pigmentation in pac1 mutant tissues, such as roots, suggests that different tissues in Z. mays vary quantitatively in their requirement for PAC1 protein function. Again, this would be in marked contrast with A. thaliana, in which all tissues appear to absolutely require TTG1 protein function for anthocyanin pigmentation. We consider three possibilities to explain these differences. First, MP1 may regulate the anthocyanin pathway in some Z. mays tissues, such as husk and sheath, and may function in the regulation of trichome developmental programs. Second, it is possible that an as yet unsequenced PAC1 homolog in Z. mays may serve one or more of these functions. Third, it is possible that regulation of anthocyanins in husk, sheath, and anthers is distinct from regulation in Z. mays aleurones, scutella, and roots, such that a WD40 repeat protein is not required. Clearly, the identification of other homologs and an understanding of MP1 function will be required to elucidate requirements for WD40 repeat protein function.
Models for WD40 Repeat Protein Interactions with bHLH and MYB Proteins
The bHLH and MYB proteins in the Z. mays anthocyanin pathway were shown to physically interact several years ago (Goff et al., 1992). When Lloyd et al. (1992) showed that the Z. mays r gene could complement ttg1 mutant defects, they suggested that TTG1 may encode or activate a bHLH protein in A. thaliana. The cloning of genes encoding AN11 and TTG1 (de Vetten et al., 1997; Walker et al., 1999) and other regulatory proteins established that genetic interactions between bHLH, MYB, and WD40 repeat proteins are required for anthocyanin regulation and trichome development. In addition to the physical interaction between bHLH and MYB proteins demonstrated by Goff et al. (1992), bHLH and MYB physical interactions have been found between regulators of A. thaliana trichome development (Payne et al., 2000) as well as between proteins purported to be regulators of the anthocyanin pathway in P. frutescens (Sompornpailin et al., 2002). In A. thaliana and P. frutescens, interactions between bHLH and the PAC1 clade also were found. Thus, a reasonable hypothesis is that a triad of MYB, bHLH, and WD40 proteins form a complex required for regulation of several additional traits, including root hair development and seed coat mucilage. A simplistic view of this complex would entail a bHLH at the center of this complex that interacts with a WD40 repeat protein on one side and a MYB protein on the other side. Although this interpretation may be correct, there are several complexities. First, in A. thaliana siliques, the expression of the anthocyanin and proanthocyanidin regulatory bHLH, TT8, requires the function of TTG1 (Nesi et al., 2000). Therefore, TTG1 appears to function upstream of the bHLH. Second, in P. hybrida, the AN2 and AN4 putative MYB proteins activate accumulation of the bHLH an1 RNA (Spelt et al., 2000). Therefore, in these dicots, some of the regulatory genes appear to be under the regulation of other regulatory proteins. One possibility is that once each protein is expressed, it is capable of interacting with the others. There could be benefits for the evolution of such a feedforward loop in which the formation of the regulatory triad could additionally upregulate the expression of the regulated regulator (TT8 or an1), thereby helping to lock in expression of the anthocyanin pathway. In Z. mays, it is striking that we have tested for and found no evidence that any of the regulators regulate each other. The Z. mays bHLH, MYB, and PAC1 clade WD40 repeat proteins appear to be regulated independently, and expression of all three are required for anthocyanin pigmentation in Z. mays kernels and roots. Thus, although a general role for the bHLH, MYB, and WD40 repeat proteins as regulators of the anthocyanin pathway has been conserved in monocots and dicots, the mechanisms of regulation of these regulators has been reorganized.
In summary, PAC1 clade WD40 repeat proteins in Z. mays, P. hybrida, A. thaliana, and possibly P. frutescens are required in their respective organisms not only for anthocyanin pigmentation but also for expression of several different traits, with the exact traits varying by species. The PAC1 protein functionally complements ttg1 mutant phenotypes in A. thaliana, demonstrating that PAC1 clade proteins have likely retained all of their functional competency since the divergence of monocots and dicots. This result suggests that differences in the regulation of anthocyanin and proanthocyanidin pathways in Z. mays, P. hybrida, and A. thaliana include not only differences in the organization of the biosynthetic genes that are regulated by these bHLH, WD40 repeat, and MYB proteins but also differences in the regulation of the regulators of these genes. Finally, mutants of the PAC1 clade WD40 repeat proteins (PAC1, TTG1, and AN11) suggest the primary importance of this clade for anthocyanin regulation.
METHODS
Genetic Nomenclature
Genes are indicated by lower case italic letters. Dominant wild-type Z. mays alleles are indicated by italic letters with the first letter capitalized. Lower case italic letters indicate recessive Z. mays alleles. Homozygous alleles are indicated with a single designation, for example, pac1-ref refers to an individual homozygous for the pac1-ref allele. Pac1/(Pac1 or pac1-ref) indicates a wild-type allele of pac1 in combination with either a wild-type or mutant allele of pac1. Proteins are indicated by non-italic capital letters. In A. thaliana, TTG1 refers to a wild-type allele, and ttg1-1 and ttg1-13 refer to recessive mutant alleles.
Origin and Analysis of the pac1-ref, pac1-2, and pac1-3 Alleles
Isolation of pac1-ref stocks was as described by Selinger and Chandler (1999). Seed used for inverse PCR cloning of pac1-ref was from a self-pollinated pac1-ref/Pac1; in1; R-r; b plant. Certain alleles of r or b are required to see pac1 phenotypes in the aleurone. In this work, observation of the pac1 phenotype was facilitated by either the R-r allele of r, the B-Peru allele of b, or both. We isolated the pac1-2 allele using a directed tagging scheme as follows: b; R-r; pac1-ref plants were crossed by B-Peru; r-r; Mu plants, resulting in the identification of a pale aleurone kernel heterozygous for the newly identified pac1-2 allele and the pac1-ref allele. Similarly, we isolated the pac1-3 allele using a directed tagging scheme as follows: B-Peru/B-I; pac1-ref plants were crossed to B-Peru; Mu plants, resulting in the identification of a pale aleurone kernel heterozygous for the newly identified pac1-3 allele and the pac1-ref allele. The pac1-2 and pac1-3 alleles were segregated from the pac1-ref allele by a series of outcrosses and self-pollinations and confirmed by DNA gel blot analysis using the 700-bp SacI/SmaI probe (Figure 2).
Mapping the pac1 Locus
The pac1 locus was previously mapped to the distal end of the long arm of chromosome 5 (Selinger and Chandler, 1999). To facilitate cloning of the pac1 locus, a Pac1/pac1-ref plant derived from a heterogeneous background was self-pollinated and progeny plants used for mapping. We identified a linked molecular marker for distinguishing pac1-ref and Pac1 alleles by screening several simple sequence repeat (SSR) markers on chromosome 5. Parental pac1-ref and Pac1 lines displayed a size polymorphism (140 and 170 bp, respectively) using the SSR primers Phi087 (forward, 5′-GAGAGGAGGTGTTGTTTGACACAC-3′) and Phi087 (reverse, 5′-ACAACCGGACAAGTCAGCAGATTG-3′) (Maize Genetics and Genomics Database, http://www.maizegdb.org). Plants from pale and dark seeds in the mapping population were screened for linkage with pac1. All 25 plants from pale seed were positive only for the 140-bp product, suggesting a distance of <3.9 centimorgan from the phi087 SSR marker. Ten plants from dark seed were positive for both the 140- and 170-bp products, and three plants only produced the 170-bp product. Subsequent probing with Mu1 (described below and in Results) confirmed the presence of the pac1-ref cosegregating band in 11 plants from the pale seed and its absence in the three wild-type plants, from which only the 170-bp Phi087 PCR product was obtained.
Cosegregation Analysis and PCR Identification of the pac1-ref, pac1-2, and pac1-3 Alleles
DNA samples from plants homozygous for pac1-ref or Pac1 were digested with EcoRI and HindIII, blotted, and then probed with a Mu1-specific sequence from plasmid pA/B5 (Talbert et al., 1989). To recover DNA flanking the Mu1 element, pac1-ref DNA was digested with EcoRI and self-ligated under very dilute conditions to obtain circular products. Inverse PCR of the pac1-ref locus made use of primers vc141 (5′-CGCACGGGAACGGTAAACGGGGACAGAAAAC-3′) and vc143 (5′-TGACAGAGACACGAGACGAAACAAGCTGAAGG-3′) within the Mu1 element, resulting in amplification across the terminal inverted repeats into the DNA flanking the element. PCR products were used as templates for a second reaction with primers vc140 (5′-AGTTTGGCTGTCGCGTGCGTCTCCAAAACAG-3′) and vc144 (5′-AGTTTGGCTGTCGCGTACGTCTCTAAAACAG-3′), which are nested toward the ends of the Mu1 element relative to vc141 and vc143. A 1588-bp product was present in homozygous mutant samples and absent in homozygous wild-type control samples. GenBank accession numbers AY442344 and AY442345 for pac1-ref were derived from this product. Part of this 1588-bp product was designated the SacI/SmaI 700 probe and used for hybridization to DNA gel blots. The pac1-2 allele was amplified from pac1-2 genomic DNA using a primer common for many Mu element termini (rm005, 5′-CATTTCGTCGAATCCCCTTCC-3′) and a primer in the pac1 locus (vc293, 5′-GGTACAGCCGCCGCCGTCTCAG-3′). The sequence obtained from the pac1-2 locus (GenBank accession number AY442262) included 28 bp of sequence best matching the ends of MuDR, and DNA gel blot analysis revealed a BamHI site in the vicinity of the insertion. The size (∼1400 bp) of the band visualized with DNA gel blot analysis using pac1-2 DNA, SacI/SmaI 700 probe, and EcoRI/HindIII enzymes suggests that the pac1-2 allele contains a MuDR element with a significant internal deletion that preserved one of the BamHI sites in MuDR. The pac1-3 allele (GenBank accession number AY442261) was amplified from pac1-3 genomic DNA using two primers in the pac1 locus: vc181 (5′-CAACCGCAAGGCCTCCTCCGAGTTCTG-3′) and vc189 (5′-GTACACTAGCACAGGATCAATCC-3′).
Identification and Sequencing of BAC Clones Containing the pac1 and mp1 Loci
BAC filters from the CUGI Zea mays National Science Foundation B73 library were hybridized according to directions (http://www.genome.clemson.edu/groups/bac/protocols/bacmanual.html) with the 700-bp SacI/SmaI probe. We ordered several of the strongest hybridizing clones and screened them by PCR amplification for the pac1 locus using primers vc178 (5′-CGCCTTTGAGAACAAGGTCCAG-3′) and vc182 (5′-AGAGCTGGAATTAAAGTTCAATTCCTTGC-3′). Positive clones included ZMMBBb0174B16, ZMMBBb0176H10, ZMMBBb0213M14, ZMMBBb0231M22, ZMMBBb0124P19, ZMMBBb0156C21, and ZMMBBb0227P08. Three overlapping subclones from clone ZMMBBb0124P19 were isolated, sequenced, and assembled into a contig of 8239 bp (GenBank accession number AY115485). To identify potential homologs of pac1, we examined additional BAC clones hybridizing to the 700-bp SacI/SmaI probe using restriction mapping and DNA gel blot analysis, hybridizing sequentially with the 5′ and 3′ halves of the pac1 coding region. Several clones, which hybridized strongly to both halves of the coding region, were shown by restriction mapping not to be pac1. To determine if these BAC clones contained genes homologous to pac1, they were subjected to additional restriction mapping, subcloning, and sequencing. Clone ZMMBBb099L23, which was subcloned and sequenced, and clone ZMMBBb0139C12, which was shown to be overlapping with ZMBBb099L23 based on its restriction pattern, both contain the mp1 locus (GenBank accession number AY339884), a gene previously identified by Hernandez et al. (2000) as having homology with an11. Erich Grotewold provided us with the mp1 EST sequence, enabling verification that the genomic clones represented the same gene as mp1.
RNA Analysis
Total RNA was prepared using TRIzol reagent (Invitrogen Life Technologies, San Diego, CA) for husk, sheath (after pollination), and immature tassel (2 cm in length) or the endosperm RNA isolation method described by Wessler (1994) for aleurone, scutellum, black layer, endosperm, and pericarp all at 23 to 25 d after pollination. RNA from freshly emerged silks, pallea/lemma, and cob/cupules from the same ear and 3-week-old seedling root, leaf, sheath, and inner leaf/meristem also were prepared using the endosperm RNA isolation method. 3′ RACE of immature tassel and aleurone total RNA was done according to instructions in the 5′/3′ RACE kit from Boehringer Mannheim (Roche Diagnostics, Indianapolis, IN) using pac1 primer vc177 (5′-TGATCTGGGAACTGCCTGAGAC-3′) or vc178 (5′-CGCCTTTGAGAACAAGGTCCAG-3′). 3′ RACE was more successful with immature tassel than with aleurone and was therefore the source of all 3′ RACE clones. RNase protections were as described by Selinger and Chandler (1999) using antisense probes designed against the pac1 cDNA. The pac1-rp-3′ probe resulted from the 3′ RACE reactions using the pac1 primer vc177 and therefore spans the splice site for the two pac1 exons and ends at the most common poly(A) site. The pac1-rp-300 probe, which spans the locations of all three pac1 mutations, was generated by PCR from wild-type DNA using primers vc554 (5′-TCACCTCCTTCGATTGGAAC-3′) and vc555 (5′-GCCATATAGCGGAGGTCAGA-3′). In all protections, 5 to 10 ug of total RNA and the internal controls actin1 (Selinger and Chandler, 1999) or ubiquitin2 (Dorweiler et al., 2000) were used.
Complementation of ttg1 Mutants
A pac1 genomic fragment was cloned into a binary expression vector by replacing GUS in the GSA1131 vector (http://www.chromdb.org/) using NcoI and BamHI sites engineered into the pac1 transcript at the start and stop codons using primers vc280 (5′-CATGCCATGGACCCACCCAAGCCGCC-3′) and vc281 (5′-GCGGGATCCTCAGACCCTAAGAAGCTGGACC-3′). The source of the template was a subclone of BAC ZMMBBb0124P19 and, therefore, the B73 allele. The GSA1131 vector included a Basta resistance gene for herbicide-resistant selection of transformants. The Agrobacterium tumefaciens–mediated floral dip method (Clough and Bent, 1998) was used to transform the ttg1-1 and ttg1-13 lines described in Larkin et al. (1999), and the ttg1-1 stock (CS89) from Koornneef (1981) was obtained from the ABRC. The former stocks were predominantly in the Columbia background, and the latter stock was in the Landsberg erecta background. A total of 32,000 seeds were planted. The transformation efficiency was ∼0.2% for each line. Lines that were propagated segregated 60 to 81% Basta resistance with a median of 71%, suggesting that the propagated lines segregated a single transgenic locus in the expected 3:1 ratio. Trichome phenotypes were observed in 35S-Pac1 ttg1 regenerants from all three ttg1 mutant lines. Medium used for plates was MS (Sigma, St. Louis, MO) with 3% (w/v) sucrose and 4 g of phytagar. For selection, Basta was used at a concentration of 10 μg/mL. Seed was imbibed for 3 or 8 d at 4°C and then plated. Seedlings were transplanted into soil at leaf stages 4 to 8 for the first planting of transformed seed or at leaf stages 2 to 4 for the second planting of transformants. In most experiments, controls included parental mutant lines as well as seeds resulting from the A. tumefaciens–mediated floral dip that did not appear to be transformed. Together, these controls are referred to as nontransformed. Plants for all experiments were grown in an 18-h-light/6-h-dark cycle with fluorescent lighting. Ruthenium red staining of seed coat mucilage was as described by Penfield et al. (2001).
Z. mays Seedling Root Color Score
A pac1-2/Pac1 B-Peru plant heterozygous for a functional and nonfunctional r allele was self-pollinated, resulting in purple and pale spotted seeds. Fourteen purple (Pac1−) and 14 pale (pac1-2) spotted seeds were germinated in paper towels moistened with tap water and grown in the dark until roots were 15 cm in length on average, at which point they were scored for pigment and harvested for DNA. Because the r allele responsible for root pigmentation was heterozygous with an allele incapable of function in the root, progeny were screened using DNA gel blot analysis for the presence of restriction length polymorphisms indicative of the root-functioning r allele. We excluded all seedlings that did not carry the functional r allele, leaving 11 pac1-2 mutant and 9 wild-type Pac1 or Pac1/pac1-2 plants. All excluded seedlings had a root color score of 1. The pac1 genotypes were confirmed by DNA gel blot analysis.
Generation of the in1; pr1; R-r Stock Segregating pac1-ref and Pac1
An in1; pr1; R-scm (a revertant of the R-mb allele giving full seed color; Maize Genetics and Genomics Database, http://www.maizegdb.org) plant was crossed with a pac1-ref; In1(wt); R-r plant. All resulting seeds were dark. Plants derived from this seed were self-pollinated, generating red, pale, and dark seed. Red seeds were planted (pr1 Pac1/pr1 [Pac1 or pac1-ref]; In1/[In1 or in1]), and the resulting plants self-pollinated. Dark progeny seeds were planted (pr1[Pac1 or pac1-ref]/pr1[Pac1 or pac1-ref]; in1) and self-pollinated. Two of these self-pollinated plants (ds1432-4 and ds1474-1) generated ears that were segregating one-quarter pale and three-quarters dark. The phi087 SSR marker polymorphism and DNA gel blot analysis was used to determine pac1 wild-type and mutant genotypes. All assayed pale seed from one ear (ds1432-4) were pac1-ref; whereas all assayed dark seed from the same ear were heterozygous or wild-type for Pac1. Similar results were obtained with the other ear (ds1474-1).
Bioinformatics and Construction of Alignments/Phylogenies
Publicly available sequences were collected from GenBank through NCBI (http://www.ncbi.nlm.nih.gov/), from the Plant Genome Database (http://www.plantgdb.org/), and from the gene indices (Quackenbush et al., 2001) at the TIGR Gene Index Databases, Institute for Genomic Research, Rockville, MD (http://www.tigr.org/tdb/tgi) from May to July, 2003. We aligned an initial set of 33 homologs using CLUSTAL W at Baylor College of Medicine (http://searchlauncher.bcm.tmc.edu/) with the default Dayhoff percent accepted mutation weight matrix. The alignment was fine-tuned manually using GeneDoc (http://www.psc.edu/biomed/genedoc/). The remaining sequences were added, manually aligned, and have been made available in Fasta format (see supplemental data online). A region that included the first 131 amino acids of PAC1 was selected to construct the alignment used in the phylogeny, resulting in the use of 46 unique coding regions from 31 species, from which all regions of insertions and deletions were removed leaving each sequence with 97 amino acids (indicated by shading or the use of lower case letters in the PAC1 sequence in the supplemental data online). The phylogenies were constructed using the PHYLIP package of programs (http://evolution.genetics.washington.edu/phylip.html) (Felsenstein, 1989) and Protml (Adachi and Hasegawa, 1996). Fitch, Kitsch, and neighbor joining programs all were used as documented in the PHYLIP package of programs, with each giving approximately the same tree topology. For the results presented in Figure 6, 500 bootstraps were performed with neighbor joining to derive a consensus tree that was used as an input for the Protml program with default options to derive distance measurements. Bootstrap values for the consensus tree from neighbor joining are indicated along the branches for values >250. The Oryza sativa (rice) and Z. mays pac1 loci are syntenous, as determined using the Gramene Web site (http://www.gramene.org/).
Accession and Sequence Identifier Numbers
GenBank accession numbers for pac1 and mp1 are as follows: AY115485 (pac1), AY442344 and AY442345 (pac1-ref), AY442262 (pac1-2), AY442261 (pac1-3), and AY339884 (mp1).
TIGR gene indices, GenBank, and Plant Genome Database report, accession, identifier numbers, and scientific names are as below for sequences used in the phylogeny. Each is further commented on in the supplemental data online. TIGR gene indices reports: TC73929 (Barley1 Hordeum vulgare), TC66955 (Potato1 Solanum tuberosum), TC126206 (Tomato1 Lycopersicon esculentum), TC146713 (Soybean1 Glycine max), TC146716 (Soybean2 G. max), TC93713 (Alfalfa1 Medicago truncatula), TC17885 (Cotton1 Gossypium hirsutum), TC15388 (Cotton2 G. hirsutum), TC185139 (TTG1_Arabidopsis1 A. thaliana), TC5440 (Iceplant1 Mesembryanthemum crystallinum), TC16648 (Cotton3 G. hirsutum), TC21234 (Chlamydomonas_reinhardtii1 C. reinhardtii), TC10687 (Grape1 Vitis vinifera), TC89552 (Alfalfa2 M. truncatula), TC17903 (Cotton4 G. hirsutum), TC52375, TC51908, and GenBank BE592997 (Sorghum2 Sorghum bicolor), TC78949 (Barley2 H. vulgare), TC142332 (Frog1 Xenopus Laevis), TC804879 (Mouse1 Mus musculus), THC1438474 (Human1 H. sapiens), TC153935 (Drosophila1 Drosophila melanogaster), TC36782 (Mosquito1 Anopheles gambiae), TC75759 (Nematode1 Caenorhabditis elegans), TC26097 (Ciona_intestinalis1 Ciona intestinalis), and TC11950 (Saccharomyces_cerevisiae1 S. cerevisiae).
GenBank accession and GenInfo numbers: BG560471 GI 13589469 (Sorghum1 S. propinquum), AY115485 (PAC1_Maize1 Z. mays), AP004178 GI 15718435 (Rice1 O. sativa range 71305 to 72372), AB059642 GI 14270084 (Perilla1 P. frutescens), AF220203 GI 6752885 (Apple1 Malus x domestica), U94748 GI 2290531 (AN11_Petunia1 Petunia x hybrida), BQ087012 GI 20046213 (Ceratopteris_richardii1(Fern) C. richardii), U94746 GI 2290527 (ATAN11A_Arabidopsis2 A. thaliana), X97488 GI 1495264 (ATAN11B_Arabdidopsis3 A. thaliana), BE033994 GI 8329003 (Iceplant2 M. crystallinum), AF530912 GI 22324808 (Cotton5 G. hirsutum), AY339884 (MP1_Maize2 Z. mays), AP004772 GI 18844992 (Rice2 O. sativa range 96686 to 97918), CB611069 GI 29550682 (Orange1 Citrus sinensis), BF098437 GI 10904147 (Tomato2 L. esculentum), BU825070 GI 23996202 (Poplar1 Populus tremula x P. tremuloides), CAB02116.2 GI 7160713 (Nematode2 C. elegans), T39719 GI 7490148 (Schizosaccharomyces_pombe1 Schizosaccharomyces pombe), and EAA38483 GI 29246903 (Giardia1 Giardia lamblia).
Plant Genome Datebase EST contig numbers: BVSVtuc03-04-08.2097 (Beet1 Beta vulgaris) and TAtuc02-12-22.2204 (Wheat1 Triticum aestivum).
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession numbers AY115485, AY339884, AY442261, AY442262, AY442344, and AY442345.
Supplementary Material
Acknowledgments
We thank Erich Grotewold for confirmation of our identification of the genomic sequence for the mp1 gene and comments on this manuscript, Lyudmila Sidorenko for critical reading and comments on this manuscript, John Larkins for providing the A. thaliana ttg1-1 and ttg1-13 lines, and Jade Rusche for his extensive help with the A. thaliana experiments. USDA Grant 99-35301-7791 funded this work.
On-line version contains Web-only data.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Vicki L. Chandler (chandler@ag.arizona.edu).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.018796.
References
- Adachi, J., and Hasegawa, M. (1996). MOLPHY version 2.3: Programs for molecular phylogenetics based on maximum likelihood. Computer Science Monographs, No. 28. (Tokyo, Japan: Institute of Statistical Mathematics).
- Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. (1997). Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, D.E., Rashotte, A.M., Murphy, A.S., Normanly, J., Tague, B.W., Peer, W.A., Taiz, L., and Muday, G.K. (2001). Flavonoids act as negative regulators of auxin transport in vivo in arabidopsis. Plant Physiol. 126, 524–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burr, F.A., Burr, B., Scheffler, B.E., Blewitt, M., Wienand, U., and Matz, E.C. (1996). The maize repressor-like gene intensifier1 shares homology with the r1/b1 multigene family of transcription factors and exhibits missplicing. Plant Cell 8, 1249–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler, V.L., Radicella, J.P., Robbins, T.P., Chen, J., and Turks, D. (1989). Two regulatory genes of the maize anthocyanin pathway are homologous: Isolation of B utilizing R genomic sequences. Plant Cell 1, 1175–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Cone, K.C., Cocciolone, S.M., Burr, F.A., and Burr, B. (1993). Maize anthocyanin regulatory gene pl is a duplicate of c1 that functions in the plant. Plant Cell 5, 1795–1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vetten, N., Quattrocchio, F., Mol, J., and Koes, R. (1997). The an11 locus controlling flower pigmentation in petunia encodes a novel WD-repeat protein conserved in yeast, plants, and animals. Genes Dev. 11, 1422–1434. [DOI] [PubMed] [Google Scholar]
- de Vetten, N., ter Horst, J., van Schaik, H.P., de Boer, A., Mol, J., and Koes, R. (1999). A cytochrome b5 is required for full activity of flavonoid 3′, 5′-hydroxylase, a cytochrome P450 involved in the formation of blue flower colors. Proc. Natl. Acad. Sci. USA 96, 778–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooner, H.K., Robbins, T.P., and Jorgensen, R.A. (1991). Genetic and developmental control of anthocyanin biosynthesis. Annu. Rev. Genet. 25, 173–199. [DOI] [PubMed] [Google Scholar]
- Dorweiler, J.E., Carey, C.C., Kubo, K.M., Hollick, J.B., Kermicle, J.L., and Chandler, V.L. (2000). Mediator of paramutation1 is required for establishment and maintenance of paramutation at multiple maize loci. Plant Cell 12, 2101–2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein, J. (1989). PHYLIP: Phylogeny inference package (version 3.2). Cladistics 5, 164–166. [Google Scholar]
- Galway, M.E., Masucci, J.D., Lloyd, A.M., Walbot, V., Davis, R.W., and Schiefelbein, J.W. (1994). The TTG gene is required to specify epidermal cell fate and cell patterning in the Arabidopsis root. Dev. Biol. 166, 740–754. [DOI] [PubMed] [Google Scholar]
- Goff, S.A., Cone, K.C., and Chandler, V.L. (1992). Functional analysis of the transcriptional activator encoded by the maize B gene: Evidence for a direct functional interaction between two classes of regulatory proteins. Genes Dev. 6, 864–875. [DOI] [PubMed] [Google Scholar]
- Goff, S.A., Klein, T.M., Roth, B.A., Fromm, M.E., Cone, K.C., Radicella, J.P., and Chandler, V.L. (1990). Transactivation of anthocyanin biosynthetic genes following transfer of B regulatory genes into maize tissues. EMBO J. 9, 2517–2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich, J., Carpenter, R., and Coen, E.S. (1992). A common gene regulates pigmentation pattern in diverse plant species. Cell 68, 955–964. [DOI] [PubMed] [Google Scholar]
- Grotewold, E., Chamberlin, M., Snook, M., Siame, B., Butler, L., Swenson, J., Maddock, S., Clair, G.S., and Bowen, B. (1998). Engineering secondary metabolism in maize cells by ectopic expression of transcription factors. Plant Cell 10, 721–740. [PMC free article] [PubMed] [Google Scholar]
- Hernandez, J.M., Pizzirusso, M., and Grotewold, E. (2000). The maize MP1 gene encodes a WD-protein similar to An11 and TTG. Maize Genetics Cooperative Newsletter 74, 24. [Google Scholar]
- Koorneef, M. (1981). The complex syndrome of TTG mutants. Arabidopsis Information Service 18, 45–51. [Google Scholar]
- Larkin, J.C., Walker, J.D., Bolognesi-Winfield, A.C., Gray, J.C., and Walker, A.R. (1999). Allele-specific interactions between ttg and gl1 during trichome development in Arabidopsis thaliana. Genetics 151, 1591–1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson, R., Bussard, J.B., and Coe, E.H., Jr. (1986). Gene-dependent flavonoid 3′-hydroxylation in maize. Biochem. Genet. 24, 615–624. [DOI] [PubMed] [Google Scholar]
- Lesnick, M.L., and Chandler, V.L. (1998). Activation of the maize anthocyanin gene a2 is mediated by an element conserved in many anthocyanin promoters. Plant Physiol. 117, 437–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd, A.M., Walbot, V., and Davis, R.W. (1992). Arabidopsis and Nicotiana anthocyanin production activated by maize regulators R and C1. Science 258, 1773–1775. [DOI] [PubMed] [Google Scholar]
- Ludwig, S.R., Habera, L.F., Dellaporta, S.L., and Wessler, S.R. (1989). Lc, a member of the maize R gene family responsible for tissue-specific anthocyanin production, encodes a protein similar to transcriptional activators and contains the myc-homology region. Proc. Natl. Acad. Sci. USA 86, 7092–7096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, C., and Gerats, T. (1993). Control of Pigment Biosynthesis Genes during Petal Development. Plant Cell 5, 1253–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mol, J., Grotewold, E., and Koes, R. (1998). How genes paint flowers and seeds. Trends Plant Sci. 3, 212–217. [Google Scholar]
- Nesi, N., Debeaujon, I., Jond, C., Pelletier, G., Caboche, M., and Lepiniec, L. (2000). The TT8 gene encodes a basic helix-loop-helix domain protein required for expression of DFR and BAN genes in Arabidopsis siliques. Plant Cell 12, 1863–1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesi, N., Jond, C., Debeaujon, I., Caboche, M., and Lepiniec, L. (2001). The Arabidopsis TT2 gene encodes an R2R3 MYB domain protein that acts as a key determinant for proanthocyanidin accumulation in developing seed. Plant Cell 13, 2099–2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne, C.T., Zhang, F., and Lloyd, A.M. (2000). GL3 encodes a bHLH protein that regulates trichome development in arabidopsis through interaction with GL1 and TTG1. Genetics 156, 1349–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paz-Ares, J., Ghosal, D., Wienand, U., Peterson, P.A., and Saedler, H. (1987). The regulatory c1 locus of Zea mays encodes a protein with homology to myb proto-oncogene products and with structural similarities to transcriptional activators. EMBO J. 6, 3553–3558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peer, W.A., Brown, D.E., Tague, B.W., Muday, G.K., Taiz, L., and Murphy, A.S. (2001). Flavonoid accumulation patterns of transparent testa mutants of arabidopsis. Plant Physiol. 126, 536–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penfield, S., Meissner, R.C., Shoue, D.A., Carpita, N.C., and Bevan, M.W. (2001). MYB61 is required for mucilage deposition and extrusion in the Arabidopsis seed coat. Plant Cell 13, 2777–2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quackenbush, J., Cho, J., Lee, D., Liang, F., Holt, I., Karamycheva, S., Parvizi, B., Pertea, G., Sultana, R., and White, J. (2001). The TIGR Gene Indices: Analysis of gene transcript sequences in highly sampled eukaryotic species. Nucleic Acids Res. 29, 159–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quattrocchio, F., Wing, J.F., van der Woude, K., Mol, J.N., and Koes, R. (1998). Analysis of bHLH and MYB domain proteins: Species-specific regulatory differences are caused by divergent evolution of target anthocyanin genes. Plant J. 13, 475–488. [DOI] [PubMed] [Google Scholar]
- Sainz, M.B., Grotewold, E., and Chandler, V.L. (1997). Evidence for direct activation of an anthocyanin promoter by the maize C1 protein and comparison of DNA binding by related Myb domain proteins. Plant Cell 9, 611–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selinger, D.A., and Chandler, V.L. (1999). A mutation in the pale aleurone color1 gene identifies a novel regulator of the maize anthocyanin pathway. Plant Cell 11, 5–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sompornpailin, K., Makita, Y., Yamazaki, M., and Saito, K. (2002). A WD-repeat-containing putative regulatory protein in anthocyanin biosynthesis in Perilla frutescens. Plant Mol. Biol. 50, 485–495. [DOI] [PubMed] [Google Scholar]
- Spelt, C., Quattrocchio, F., Mol, J.N., and Koes, R. (2000). anthocyanin1 of petunia encodes a basic helix-loop-helix protein that directly activates transcription of structural anthocyanin genes. Plant Cell 12, 1619–1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stapleton, A.E., and Walbot, V. (1994). Flavonoids can protect maize DNA from the induction of ultraviolet radiation damage. Plant Physiol. 105, 881–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Styles, E.D., Ceska, O., and Seah, K.T. (1973). Developmental differences in action of r and b alleles in maize. Can. J. Genet. Cytol. 15, 59–72. [Google Scholar]
- Talbert, L.E., Patterson, G.I., and Chandler, V.L. (1989). Mu transposable elements are structurally diverse and distributed throughout the genus Zea. J. Mol. Evol. 29, 28–39. [DOI] [PubMed] [Google Scholar]
- Walker, A.R., Davison, P.A., Bolognesi-Winfield, A.C., James, C.M., Srinivasan, N., Blundell, T.L., Esch, J.J., Marks, M.D., and Gray, J.C. (1999). The TRANSPARENT TESTA GLABRA1 locus, which regulates trichome differentiation and anthocyanin biosynthesis in Arabidopsis, encodes a WD40 repeat protein. Plant Cell 11, 1337–1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessler, S.R. (1994). Isolation of RNA from Wx and wx Endosperms. In The Maize Handbook, M. Freeling and V. Walbot, eds (New York, NY: Springer-Verlag), pp. 545–546.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.