Abstract
In this article, we report that carpel specification in the Oryza sativa (rice) flower is regulated by the floral homeotic gene DROOPING LEAF (DL) that is distinct from the well-known ABC genes. Severe loss-of-function mutations of DL cause complete homeotic transformation of carpels into stamens. Molecular cloning reveals that DL is a member of the YABBY gene family and is closely related to the CRABS CLAW (CRC) gene of Arabidopsis thaliana. DL is expressed in the presumptive region (carpel anlagen), where carpel primordia would initiate, and in carpel primordia. These results suggest that carpel specification is regulated by DL in rice flower development. Whereas CRC plays only a partial role in carpel identity, DL may have been recruited to have the more essential function of specifying carpels during the evolution of rice. We also show that DL interacts antagonistically with class B genes and controls floral meristem determinacy. In addition, severe and weak dl alleles fail to form a midrib in the leaf. The phenotypic analysis of dl mutants, together with analyses of the spatial expression patterns and ectopic expression of DL, demonstrate that DL regulates midrib formation by promoting cell proliferation in the central region of the rice leaf.
INTRODUCTION
The specification of each type of floral organ is a key process in flower development. Molecular and genetic studies in two model eudicots, Arabidopsis thaliana (Arabidopsis) and Antirrhinum majus (snapdragon), have established the ABC model for the determination of floral organ identities (Bowman et al., 1991; Coen and Meyerowitz, 1991). This model proposes that each class of floral homeotic gene, termed A, B, and C, works in two adjacent whorls, and combinatorial activities of these genes specify four types of floral organ: sepals, petals, stamens, and carpels.
AGAMOUS (AG) plays a central role in specifying carpel identity in the Arabidopsis flower (Yanofsky et al., 1990; Bowman et al., 1991; Bowman et al., 1999). Loss-of-function mutations in AG result in homeotic transformation of stamens into petals in whorl 3 and of carpels into reiterating ag flowers in whorl 4. Thus, AG has functions that specify stamen and carpel identity, repress the activity of class A genes in the inner two whorls, and control floral meristem determinacy. It has been suggested, however, that carpel identity also is regulated in an AG-independent pathway because mutants lacking AG function, such as the ap2 ag double mutant, retain carpeloid properties (Bowman et al., 1991). Further genetic studies have revealed that CRABS CLAW (CRC) and SPATULA (SPT) act redundantly in this pathway to specify carpels and are negatively regulated by class A and class B genes (Alvarez and Smyth, 1999; Bowman and Smyth, 1999). CRC is a member of the YABBY family of genes that encode plant-specific transcription factors with a zinc-finger domain and a helix-loop-helix domain (named the YABBY domain). In addition to CRC and SPT, the SHATTERPROOF MADS box genes partially contribute to carpel development (Pinyopich et al., 2003).
Although much progress has been made in our understanding of the molecular mechanisms of flower development in Arabidopsis, molecular developmental studies are incomplete in monocots. In the Poaceae (grass) family, flower developmental studies have focused on Zea mays (maize) and Oryza sativa (rice) (Schmidt and Ambrose, 1998; Goto et al., 2001) because of the availability of mutants, molecular tools, and information from genome analysis. In addition, grasses bear unique flowers that are distinct from those of dicots. The grass flowers have stamens and carpels like dicot flowers but lack obvious sepals and petals. Lodicules, which are specific to grass flowers and correspond to petals in dicots, are formed in the outer whorl of stamens. These three floral organs are subtended by palea and lemma. The grass carpels constitute a simple structure composed of stigmas, styles, and an ovary, in which a single ovule develops. The carpels do not differentiate to form transmitting tissues and septa as they do in Arabidopsis.
Mutations in the silky1 (si1) gene in maize and in the SUPERWOMAN1 (SPW1) gene in rice cause homeotic transformation of stamens and lodicules into carpels and palea/lemma-like structures, respectively. (Ambrose et al., 2000; Nagasawa et al., 2003). These phenotypes are similar to those of class B mutants in Arabidopsis and A. majus. Isolation of the causative genes of these mutations revealed that both si1 and SPW1 encode MADS box genes that are orthologous to Arabidopsis APETALA3 ([AP3]; Jack et al., 1992) and are expressed in the same domains in developing flowers as AP3. These results suggest that the function of class B genes and the mechanisms that specify petals/lodicules and stamens are conserved between dicots and monocots. By contrast, a loss-of-function mutation in ZAG1, an ortholog of AG, is not associated with any homeotic change in the maize flower, whereas the zag1 flower, like the ag flower in Arabidopsis, lacks floral determinacy (Mena et al., 1996). The lack of homeotic changes in zag1 is hypothesized to be attributable to the fact that this mutation is masked by the redundant functions of ZMM2, a paralog of ZAG1. Thus, the genetic mechanism that specifies carpel identity in grass species remains unclear.
Leaf morphologies also diverge in eudicots and grass species. Grass leaves have parallel venation, whereas most dicot leaves have reticulate venation. Rice leaves have three types of vein arranged in parallel: the central vein and the lateral large and small veins. The central vein comprises a strong structure, called the midrib, which serves to keep the leaves upright. Although some reports have described mutants lacking a midrib (Rao et al., 1988; Fladung et al., 1991), the genes controlling midrib formation have not been described in detail.
We found previously that severe mutations in the DROOPING LEAF (DL) locus in rice cause homeotic mutation of carpels into stamens in flowers and failure of midrib formation in leaves (Nagasawa et al., 2003). In this report, we describe isolation of the DL gene and its function in flower development and midrib formation. Our results show that DL is a member of the YABBY gene family and encodes a putative transcription factor that contains zinc-finger and helix-loop-helix domains. Phenotype and expression analyses indicate that DL is necessary for carpel specification and is negatively regulated by class B genes in rice. Among the Arabidopsis YABBY genes, DL is most similar to CRC, which has a partial role in carpel identity. Our results suggest that genes in the DL/CRC subfamily act in carpel development in both monocots and dicots, but their contribution to carpel specification may have been more crucial in grass flower development during the evolution of grasses. Finally, we show that DL regulates midrib formation by specifying cell fate to promote cell proliferation specifically in the central region of the leaf.
RESULTS
DL Regulates Carpel Identity and Floral Meristem Determinacy in the Rice Flower
Rice flowers have two lodicules, six stamens, and one pistil, which is presumably formed from three carpels, and these floral organs are subtended by palea and lemma (Figure 1A). In this article, we refer to the regions where lodicules, stamens, and a pistil develop in rice as whorl 2, whorl 3, and whorl 4, respectively, in line with the definition of regions in Arabidopsis.
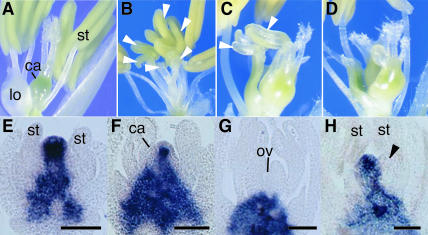
Figure 1.
Flower Phenotypes and Expression Patterns of OSH1.
(A) Wild-type flower.
(B) Complete transformation of carpels into stamens in the dl-sup1 flower.
(C) Partial transformation of carpels in the intermediate dl-3 flower.
(D) Multiple carpels produced in the dl-3 flower.
(E) to (G) Spatial expression patterns of OSH1 in the wild-type flower.
(H) Spatial expression patterns of OSH1 in dl-sup1. Arrowheads indicate ectopic stamens. ca, carpel; lo, lodicule; ov, ovule; st, stamen. Bars = 20 μm.
Here, we isolated and analyzed five new dl alleles in addition to four previously described dl alleles (Nagasawa et al., 2003). Three severe alleles, dl-sup3, dl-sup4, and dl-sup5, showed homeotic transformation of carpels into stamens (Figure 1B) and a variation in the number of ectopic stamens from two to seven. No vestigial carpel-like or ovule-like tissues were observed. An intermediate allele, dl-3, showed pleiotropic abnormalities, such as partial transformation of carpels (Figure 1C), the production of multiple carpels (Figure 1D), the formation of cell clusters, and an increase in the number of styles (data not shown). No phenotypic change was observed in other floral organs in any of the alleles, suggesting that mutations of DL affect only the innermost whorl. These results indicate that DL is necessary for the specification of carpel identity.
The indeterminate formation of ectopic stamens in the severe dl alleles and the production of multiple carpels and clusters of undifferentiated cells in the intermediate dl allele suggested that DL might regulate floral meristem determinacy. To test this hypothesis, we examined the spatial expression pattern of the class I knox gene OSH1, which is a molecular marker of meristematic indeterminate cells in rice (Sato et al., 1996). In the wild-type flower, OSH1 was expressed in the floral meristem and receptacle in the early stages of development (Figure 1E). The domain expressing OSH1 in the meristem became smaller as carpel primordia arose (Figure 1F) and completely disappeared when carpels developed in the fourth whorl (Figure 1G). By contrast, OSH1 expression in the dl-sup1 flower was maintained in the central region even after ectopic stamens were produced (Figure 1H). These results indicate that DL is required for regulating floral meristem determinacy in addition to specifying carpel identity.
DL Is Required for Midrib Formation in Leaves
The dl mutations cause defects in midrib formation in leaves, resulting in drooping leaf phenotypes (Nagasawa et al., 2003). Here, we examined the defects in midrib structure in these mutants in detail. The midrib in wild-type rice leaves had two structural characteristics. First, it had two large locules, called clear cells (Figure 2A), which are thought to be formed by programmed cell death after the accumulation of a large number of cells in the central region of the leaf (see below) and which constituted hollow thin cylinders aligned from proximal to distal in the leaves. Second, it formed a small vascular bundle at the adaxial side opposite the central vascular bundle (Figures 2A and 2C). In the dl mutants, the central vein lacked both clear cells and the adaxial small vascular bundle (Figure 2E). In addition, the structure of the central vein in the dl mutants resembled that of the lateral large veins (Figures 2D and 2E).
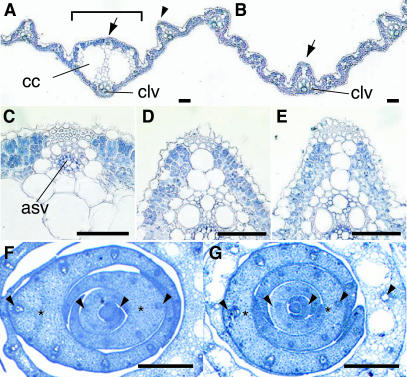
Figure 2.
Leaf Phenotypes.
(A) and (B) Cross-section of the leaf blade of the wild type (A) and dl-sup1 (B). The bracket indicates the midrib structure.
(C) to (E) Enlarged views of the inner structure at the adaxial sides of the wild-type midrib (C), indicated by the arrow in (A); the wild-type lateral vein (D), indicated by the arrowhead in (A); and the dl-sup1 central large vein (E), indicated by the arrow in (B).
(F) and (G) Cross-section of the shoot apex of the wild type (F) and dl-sup1 (G). Arrowheads indicate the central vascular bundles, and asterisks indicate the central regions of the leaves that differ in the wild type and dl-sup1. asv, adaxial small vascular bundle; cc, clear cells; clv, central large vascular bundle. Bars = 100 μm.
To determine why the midrib was missing, we analyzed the developmental patterns of leaves in the early stages. In the wild type, the central vascular bundle was initiated at the end of plastchron1 (P1). During P3 and P4, the cells in the central region of the leaf proliferated along the adaxial-abaxial axis, and a large number of cells accumulated adaxial to the central vascular bundle (Figure 2F). The midrib structure was differentiated from the large number of proliferated cells in the central region after the P5 stage. By contrast, in dl-sup1 mutants, the specific cell proliferation in the central region did not occur while the central vascular bundle was being formed (Figure 2G). The number of cell files along the adaxial-abaxial axis in the center of the dl-sup1 leaf was reduced to approximately one-third of the number in the wild-type leaf. A failure to accumulate enough cells in the central region seems to be the primary cause of leaves lacking midribs. Taken together, these observations suggest that DL regulates midrib formation by inducing cell proliferation in the central region of the leaf.
Isolation of DL
To determine the molecular functions of DL, we first tried to isolate the DL gene by a map-based cloning strategy (see supplemental data online). The dl locus was mapped to restriction fragment length polymorphism (RFLP) markers on the short arm of rice chromosome 3. A BAC contig encompassing ∼300 kb was constructed by screening using the closest marker C316 and subsequent chromosome walking. Next, we narrowed down the putative DL region using the mutant line fm28, which is supposed to have chromosomal deletion involving the dl and leafy hull sterile loci (Yoshimura et al., 1997), which are tightly linked together. The deleted region in fm28 was mapped by DNA gel blot analysis using DNA fragments arbitrarily isolated from a BAC clone (2E1213), and the deletion was found to be <40 kb. A λ contig covering the whole deletion then was constructed.
While these experiments were in progress, a new dl allele (dl-sup3) was isolated from R2 progenies that were derived from rice plants regenerated through tissue culture. Because the retrotransposon TOS17 is known to be mobilized and to cause somaclonal mutations during tissue culture (Hirochika et al., 1996), we examined whether the mutation in dl-sup3 was caused by the insertion of TOS17. DNA gel blot analysis using a probe for TOS17 indicated that one of the bands cosegregated with the drooping leaf phenotype (data not shown). Subsequent DNA gel blot analysis using a probe for the DNA sequence flanking the TOS17 in this band showed that the sequence cosegregated with the dl phenotype and genotype (Figure 3A). The TOS17 insert was found to be located in a region of ∼40 kb, which corresponded to the region found to contain the dl locus in our map-based approach. Thus, the results of our two approaches for isolating the DL gene were in agreement.
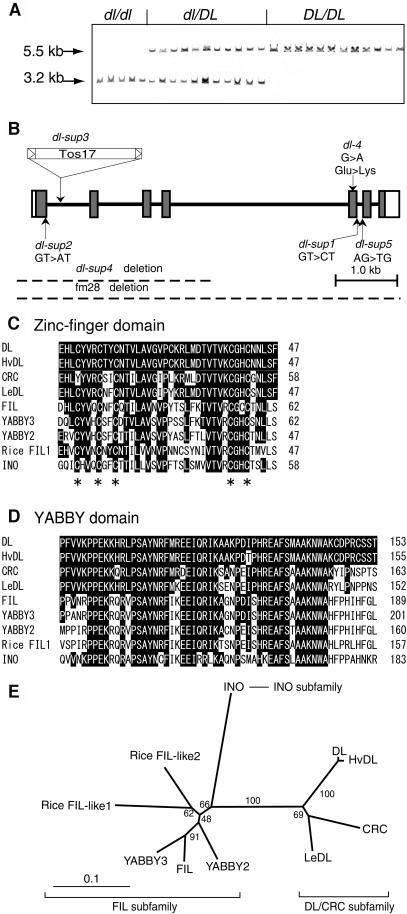
Figure 3.
Gene Isolation and Structural Features of DL.
(A) Cosegregation of the TOS17 insertion with DL genotypes. The genotypes, which were determined by the drooping leaf phenotype of the next generation (R3), are shown above the lanes. The 5.5-kb XbaI band corresponds to the wild-type allele, whereas the 3.2-kb band is produced by the insertion of TOS17, which contains an internal XbaI site.
(B) Genomic structure of DL and mutations in the six dl alleles. Boxes indicate exons and thick lines indicate introns. The coding regions are shown by shaded boxes.
(C) Zinc-finger domain. The five conserved Cys residues are indicated with asterisks.
(D) YABBY domain. The regions containing both domains have been extended slightly beyond the original definition (Bowman and Smyth, 1999) through the accumulation of more YABBY sequences. Amino acids identical to those of DL are indicated with black boxes.
(E) Phylogeny of the YABBY gene family. The tree was constructed by the neighbor-joining method (Saitou and Nei, 1987). Numbers denote bootstrap values. Amino acid sequences of HvDL (Hordeum vulgare [barley] DL homolog) and LeDL (Lycopersicon esculentum [tomato] DL homolog) were deduced from the following EST sequences: HvDL (BG369278, AL509850) and LeDL (AI485831, AI483816). Rice FIL-like proteins have been described by Sawa et al. (1999).
Sequence analysis of both the genomic DNA and the cDNA revealed that the putative DL gene covered ∼10 kb and comprised seven exons and six introns (Figure 3B). To verify whether this gene was DL, a construct containing the genomic sequence of the candidate gene was transformed into the dl-1 plant. Leaves in the transgenic plants stood upright, and the midrib was formed as in the wild type (see supplemental data online). The slight phenotypic alterations in the pistil observed in this intermediate allele also were rescued by the transgene (data not shown).
Next, we determined the DNA sequences of this candidate gene for each of the dl mutant alleles. In all severe alleles, the mutations detected were associated with serious defects at the molecular level: nucleotide substitution occurs at an RNA splicing site in dl-sup1, dl-sup2, and dl-sup5; the DNA sequence, including exon 1 to exon 4, is deleted in dl-sup4; and TOS17 is inserted into the first intron of dl-sup (Figure 3B). In the weak dl-4 allele, a single base change causes an amino acid replacement from Glu (118) to Lys in the YABBY domain (Figure 3B; see also supplemental data online).
Taken together, these results clearly indicate that the candidate gene that we isolated is derived from the dl locus.
DL Is a Member of the YABBY Gene Family and Is Closely Related to Arabidopsis CRC
The longest DL cDNA isolated was 1037 bp and encodes a putative protein of 194 amino acids (see supplemental data online). Database analysis revealed that DL is a member of the YABBY gene family, which is specific to plant genomes. In Arabidopsis, this family includes developmental genes such as CRC (Bowman and Smyth, 1999), FILAMENTOUS FLOWER (FIL) (Sawa et al., 1999), and INNER NO OUTER (INO) (Villanueva et al., 1999). Like other YABBY genes, DL encodes a protein containing two distinct domains: a zinc-finger domain in the N-terminal region and a C-terminal YABBY domain, which may form a helix-loop-helix structure and shares low similarity with the HMG box (Bowman and Smyth, 1999).
The amino acid identities between DL and other YABBY proteins (except HvDL, which belongs to the same grass family as rice) were found to vary from 48.6 to 87.5% (mean 65.0%) in the zinc-finger domain and from 62.7 to 83.6% (mean 74.6%) in the YABBY domains (Figures 3C and 3D). Although the overall homology in the zinc-finger domain was lower than that in the YABBY domain, five Cys residues were found to be conserved among nine proteins examined, except for the first Cys in CRC (Figure 3C). No obvious homology was observed in the central region or outside of the two domains.
A phylogenic tree constructed from a comparison of the two domains indicated that the YABBY genes are divided into three subfamilies: the DL/CRC subfamily, including DL and Arabidopsis CRC; the FIL subfamily, including almost all YABBY genes in Arabidopsis and rice (only one EST of rice is shown); and the INO subfamily, including only INO (Figure 3E). Some amino acid sequences, such as SF in the C-terminal region of the zinc-finger domain and PFVVK in the N-terminal region of the YABBY domain, were found to be specific to proteins in the DL/CRC subfamily. Thus, the phylogenic analysis and the amino acid comparison together indicate that CRC is the YABBY gene in Arabidopsis that is most similar to DL in rice.
Temporal and Spatial Expression Patterns of DL
We analyzed the spatial and temporal expression patterns of DL during flower and leaf development by in situ hybridization. In the wild-type flower, a bulge corresponding to the carpel primordia first arose at the flower meristem near the lemma (Figure 4A), and then the primordia developed from the flank of the meristem toward the opposite side, enclosing the meristem (Figures 4B and 4C). The meristem remained morphologically undifferentiated and later developed into the ovule. Thus, in rice flower development, the central region of the flower meristem is not consumed by carpel primordia. This contrasts with flower development in Arabidopsis, in which carpel primordia develop at the expense of the floral meristem.
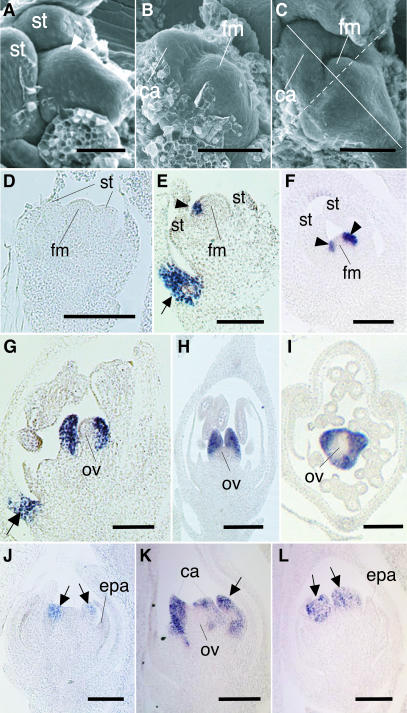
Figure 4.
Scanning Electron Micrographs of Carpel Development and in Situ Localization of DL Transcripts.
(A) Scanning electron micrograph of the wild-type floral meristem just before carpel initiation.
(B) and (C) Scanning electron micrographs of carpel primordia and the flower meristem in the wild-type flower.
(D) to (I) Localization of DL transcripts in the wild-type flower ([D] to [H]) longitudinal sections; [I] transverse section). Sectioned planes are indicated in (C) by a solid line (for [E] and [G]) and by a dashed line (for [D], [F], and [H]).
(J) to (L) Localization of DL transcripts in the spw1 flower. The flower stage shown in (J) is the same as that shown in (D).
Arrowheads in (A), (E), and (F) indicate the carpel anlagen. Arrows in (E) and (G) are DL signals in the region that correspond to the midrib in the lemma. Arrows in (J) to (L) indicate ectopic carpels in whorl 3. ca, carpel; epa, ectopic palea-like organ; fm, floral meristem; ov, ovule; st, stamen. Bars = 20 μm.
DL transcripts were not detected from the early stages of flower development to the stage when the stamen primordia arose (Figure 4D). DL expression was detected first in a few cells in the flower meristem at the lemma side (Figure 4E) and then in cells at the flank of the meristem (Figure 4F). These DL expression domains corresponded to the presumptive region (carpel anlagen), where carpel primordia would initiate. Shortly after this expression, carpel primordia began to form. At this stage, DL was expressed specifically and uniformly in carpel primordia (Figures 4G to 4I). No expression was detected in the flower meristem (Figures 4E and 4F) or the ovule primordium (Figures 4G to 4I). By contrast, expression of DL was not observed in dl-sup1 mutants throughout flower development (data not shown). Thus, the expression domains of DL are restricted to the carpel anlagen and the carpel primordia, and these expression patterns of DL are consistent with its specific role in carpel development.
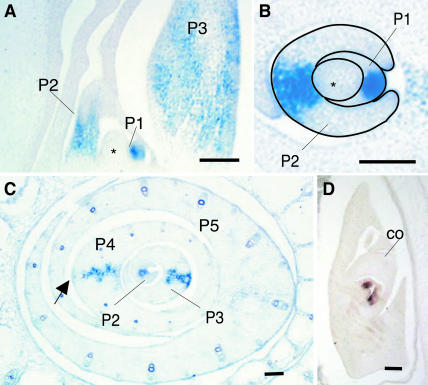
During leaf development, DL transcripts were detected first in the central region of the P1 primordia (Figures 5A and 5B). As the wild-type leaf grows, the central region becomes thicker through cell proliferation, and at this stage, DL expression was restricted to several cell arrays in the central region (Figure 5C). During the P1 to P3 stages, the DL expression domain spanned from the adaxial to the abaxial epidermal tissues. At the P4 stage, DL expression disappeared from the abaxial mesophyll, which was almost mature, but was maintained in the central region abaxial to the central vascular bundle (Figure 5C). The expression of DL was hardly detectable in the P5 leaves (Figure 5C). The disappearance of DL transcripts was confirmed by longitudinal sections, which contained all parts of the P5 leaves (data not shown). DL expression also was detected in the central region of the leaf primordia in the embryo (Figure 5D) and in the region that corresponds to the midrib in the lemma (Figures 4E and 4G). Thus, the expression of DL is consistent with its specific role in midrib development.
Figure 5.
Localization of DL Transcripts in Developing Wild-Type Leaves and an Embryo.
(A) to (C) DL expression in leaves. Longitudinal (A) and transverse ([B] and [C]) section of a vegetative shoot apex. P1 and P2 leaf primordia and the shoot apical meristem are outlined in (B). The section of (C) was taken 100 μm above that shown in (B).
(D) DL expression in an embryo 10 d after pollination, when two leaves had initiated.
Plastochron numbers are indicated as P1 to P5. Asterisks indicate the shoot apical meristem. The arrow shows the abaxial mesophyll. co, coleoptyle. Bars = 20 μm.
DL Expression Is Negatively Regulated by SPW1
Loss-of-function mutations of SPW1 cause homeotic conversion of lodicules and stamens into palea-like organs and carpels, respectively (Nagasawa et al., 2003). To investigate how DL genetically interacts with SPW1, we examined the DL expression pattern in the spw1-1 flower. DL transcripts first were detected ectopically in the primordia in whorl 3, which developed ectopic carpels (Figure 4J). The timing of this expression was earlier than that observed in the wild type (cf. Figure 4D). Expression of DL continued during the development of both the ectopic carpels and the original carpels (Figures 4K and 4L). These observations indicate that DL is involved in the specification of the ectopic carpels in the spw1 mutant and that the expression of DL in whorl 3 may be regulated negatively by SPW1 in the wild type. No expression was observed in the ectopic palea-like organs in whorl 2 in the spw1 mutant (Figures 4J and 4L).
Ectopic Expression of DL
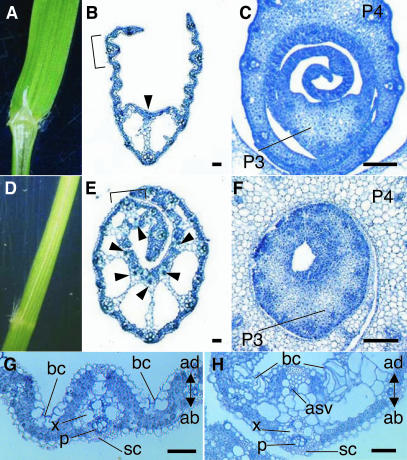
To gain further insight into the functions of DL, we produced transgenic rice plants that overexpressed DL under the control of the constitutive rice actin promoter. ACTIN:DL plants showed aberrant seedling phenotypes and died after producing four to eight leaves. Leaf blades in ACTIN:DL plants curled toward the inside, forming a cylinder-like structure. New leaves were prevented from emerging from these cylindrical leaves (Figure 6D). Transverse sections showed that the leaf blades were thickened, and midrib-like structures were formed in the lateral regions as well as in the central region in ACTIN:DL lines (Figure 6E). Clear cells were ectopically formed in the lateral regions, and the small adaxial vascular bundle that is specific to the midrib in the wild-type leaf also developed at the adaxial side opposite the large vascular bundle in the ACTIN:DL leaf. The leaf blades became gradually thinner toward the marginal regions. Analysis of earlier developmental stages of the leaves showed that cells proliferated along the adaxial-abaxial axis not only in the central regions but also in the lateral regions in ACTIN:DL lines (Figure 6F). Taken together, these observations support the idea that DL regulates midrib formation by promoting cell proliferation in the central region of wild-type leaves.
Figure 6.
Phenotypes of ACTIN:DL Plants.
(A) and (D) Morphology of leaf blades.
(B) and (E) Transverse sections of the leaf blades of seedlings.
(C) and (F) Transverse sections of developing (P3 to P4) leaf blades.
(G) and (H) Enlarged view of the regions shown by the brackets in (B) and (E), respectively.
(A), (B), (C), and (G) Wild-type.
(D), (E), (F), and (H) ACTIN:DL.
Arrowheads indicate adaxial small vascular bundles. ab, abaxial; ad, adaxial; asv, adaxial small vascular bundle; bc, bulliform cells; p, phloem; sc, sclerenchyma; x, xylem. Bars = 200 μm.
No alterations in adaxial-abaxial identity were observed in the arrangement of the leaf inner structure (Figures 6G and 6H). Bulliform cells, which are responsible for the leaf rolling that occurs under drought conditions in rice, were deposited in the adaxial side of the leaf blade in ACTIN:DL lines as they are in the wild type. The sclerenchyma was formed abaxial to the larger vascular bundle in ACTIN:DL lines as it is in the wild type. Similarly, the arrangement of xylem and phloem in ACTIN:DL lines was the same as that in the wild type. These observations suggest that DL is not sufficient for the control of dorsoventrality.
DISCUSSION
We have isolated the rice DL gene by two alternative strategies: map-based cloning and transposon tagging. DL encodes a putative transcriptional regulator that contains zinc-finger and YABBY domains and is most similar to CRC among the Arabidopsis YABBY genes. Analyses of phenotypes and special expression patterns strongly suggest that DL is involved in carpel specification in the flower and midrib development in the leaf.
DL Functions in Flower Development
A severe or intermediate mutation in DL causes complete or partial homeotic conversion of carpels into stamens without affecting the identities of other floral organs, suggesting that DL functions only in whorl 4. The expression of DL is detected first in the presumptive region, where carpels initiate, and then specifically expressed in carpel primordia. Thus, the spatial and temporal patterns of DL expression in carpel development are correlated with floral phenotypes. DL also is expressed in ectopic carpel primordia in whorl 3 of the spw1 flower, in which stamens are homeotically transformed into carpels, suggesting that DL is involved in the development of these ectopic carpels. In fact, neither the original carpels nor the ectopic carpels observed in spw1 were initiated in dl-sup1 spw1-1 double mutants (Nagasawa et al., 2003). These results clearly indicate that DL is a floral homeotic gene that regulates carpel specification in rice.
The flowers in severe loss-of-function dl mutants seemingly resemble those in superman (sup) mutants of Arabidopsis (Bowman et al., 1992; Sakai et al., 1995) because the number of stamens is increased in both types of plant. A detailed comparison of the mutant phenotypes suggests, however, that the functions of DL and SUP differ (Nagasawa et al., 2003). The spatial expression patterns of DL also indicate that the functions of the two genes are distinct. First, DL is specifically expressed in carpel anlagen and carpel primordia, whereas SUP is expressed in the inner part of whorl 3 (Sakai et al., 1995). Second, DL is ectopically expressed in whorl 3 of the spw1 flower, whereas the spatial expression pattern of SUP is not altered in the ap3 flower, although its expression level is decreased (Sakai et al., 2000). Neither carpels nor stamens are developed in the dl-sup1 spw1-1 double mutant (Nagasawa et al., 2003), whereas the ap3 sup double mutant shows a phenotype similar to that of the ap3 single mutant (Sakai et al., 2000). Arabidopsis SUP is thought to be involved in maintaining the boundary between whorl 3 and whorl 4, and defects in this gene cause extra stamens to form in whorl 3 at the expense of carpel development (Sakai et al., 1995). By contrast, the severe phenotypes of dl mutants may be explained by homeotic transformation of carpels into stamens, together with loss of floral meristem determinacy.
Homeotic transformation of stamens into carpels in spw1 mutants suggests that DL is regulated negatively by SPW1 in whorl 3 in the wild type (Nagasawa et al., 2003). Here, we have clearly proved this hypothesis by showing that DL is expressed in the ectopic carpel primordia in whorl 3 of the spw1 flower. Conversely, homeotic transformation of carpels into stamens, coupled with the spatial expression pattern of SPW1 in severe dl mutants, indicates that SPW1 is negatively controlled by DL in whorl 4 in the wild type (Nagasawa et al., 2003). Thus, DL and SPW1 antagonistically regulate each other. Further molecular studies may be required to address the question of whether this mutual negative regulation is direct or indirect. Although DL transcripts were detected in whorl 3, they were not detected in whorl 2 in the spw1 flower, suggesting that DL is repressed by class A genes or requires the function of class C genes.
It is of great interest to know how class C genes are involved in carpel development in rice. In maize, a loss-of-function mutation of ZAG1 does not affect floral organ identity on its own (Mena et al., 1996) but causes loss of carpel identity when coupled with a mutation in si1 (Ambrose et al., 2000). Moreover, it has been suggested that the ZAG1 paralog ZMM2 acts redundantly in carpel identity (Mena et al., 1996). In rice, antisense suppression of the AG ortholog OsMADS3 produces flowers with abnormal carpels but does not cause clear homeotic change in carpels (Kang et al., 1998). By contrast, two other functions of class C genes—namely, the negative regulation of class A genes and stamen specification in conjunction with class B genes—have been demonstrated by the results from antisense suppression and ectopic expression of OsMADS3 (Kang et al., 1998; Kyozuka and Shimamoto, 2002). We have found that rice has another AG ortholog in addition to OsMADS3 (T. Yamaguchi and H.-Y. Hirano, unpublished data). Therefore, to reveal the contribution of class C genes to carpel specification in rice, an approach based on the knockout of both class C genes will be required.
We also have shown that DL regulates determinacy of the floral meristem. This was predicted from the phenotypes of the dl mutants and confirmed by the expression pattern of OSH1. Studies of zag1 mutants have demonstrated that this class C gene in the grass family, like AG in Arabidopsis, is involved in the control of floral determinacy as is the indeterminate floral apex1 gene (Mena et al., 1996; Laudencia-Chingcuanco and Hake, 2002). The regulation of floral meristem determinacy and the interaction between DL and these genes will be interesting subjects for future studies of flower development in rice. It also is of great interest to note that some genes, such as DL and AG, are involved in the regulation of both carpel identity and floral meristem determinacy. Therefore, the two functions may be closely associated with each other.
DL Function in Midrib Formation
In developing leaves, before the formation of a midrib structure, the central region adaxial to the central vascular bundle is thickened by cell proliferation. This proliferation provides enough cells to form a midrib structure. In dl mutants, this specific cell proliferation seems not to occur because the number of cell files in the center of the leaf is reduced and no obvious thickening of the central region is observed. In the wild type, DL starts to be strongly expressed in the central region of the leaf before thickening and continues to be expressed during thickening. Therefore, the function of DL may be to specify cell fate to promote the specific proliferation of cells in the center of the leaf.
Our results from the overexpression of DL strongly support this hypothesis. In ACTIN:DL plants, the lateral regions of the leaf also became thick from vigorous cell proliferation, resulting in the formation of midrib-like structures in these regions. Thus, we conclude that the primary function of DL in leaf development is to specify the midrib by promoting cell proliferation specifically along the adaxial-abaxial axis. The leaf thickness was reduced toward the peripheral region and midrib-like structures did not form in the leaf margins in ACTIN:DL plants, suggesting that the function of DL is regulated negatively by unknown factors in the peripheral and marginal regions. Furthermore, as DL is expressed and functions to form the midrib only in the central region of the leaf in the wild type, it may be associated with the central lateral properties of the leaf.
Evolutionary Implications of DL Function
Phylogenic analysis revealed that DL is most similar to CRC among Arabidopsis YABBY genes. We have isolated seven additional rice YABBY genes, which are classified into the FIL or INO subfamilies (K. Harada, A. Takamura, T. Yamaguchi, and H.-Y. Hirano, unpublished data). We could not identify any other YABBY genes in the sequence database of the rice genome, for which sequencing has been almost completed. Because all of the Arabidopsis YABBY genes have been described (Bowman and Smyth, 1999; Sawa et al., 1999; Siegfried et al., 1999; Villanueva et al., 1999), these analyses confirm that DL and CRC are orthologs.
Our studies indicate that DL has three functions in rice flower development: specification of carpel identity, control of floral meristem determinacy, and antagonistic regulation with class B genes. In Arabidopsis, genetic and molecular analyses have revealed that CRC is involved in an AG-independent pathway of carpel specification and in control of floral determinacy (Alvarez and Smyth, 1999). CRC also is negatively regulated by class B genes (Alvarez and Smyth, 1999; Bowman and Smyth, 1999). In addition, the formation of ectopic stamen interior to the carpels in crc/crc ag/+ plants suggests that CRC represses class B genes (Alvarez and Smyth, 1999). From these observations, we conclude that the functions of the DL/CRC subfamily are fundamentally conserved between rice and Arabidopsis.
Unlike severe loss-of-function mutations of DL, however, loss-of-function mutations of CRC do not result in homeotic transformation of carpels (Alvarez and Smyth, 1999; Bowman and Smyth, 1999). DL may have been recruited to acquire critical functions to specify carpel identity during grass evolution (see below). Alternatively, genes in the DL/CRC subfamily of YABBY genes may contribute differently to carpel specification, depending on the plant species. It is possible that the diverse contribution of DL/CRC may be associated with differences in carpel architecture. The Arabidopsis carpel, especially the ovary, has a complex structure consisting of various differentiated tissues, such as septa, abaxial repla, transmitting tissues, and placenta (Bowman et al., 1999), whereas the rice pistil is simple because its ovary does not contain these differentiated tissues and encloses a single ovule. It is plausible that the crc mutation does not cause serious defects in carpel development in Arabidopsis because many genes coordinately specify the complex differentiation of the carpel (Alvarez and Smyth, 1999; Bowman and Smyth, 1999; Eshed et al., 1999; Kuusk et al., 2002; Pinyopich et al., 2003). By contrast, loss of the function of DL may be critical in rice because it is thought that only a few genes regulate carpel development. In either case, we conclude that DL has a predominant role in carpel specification in rice.
Defects in both carpel identity and midrib formation have been reported in mutants of two other grass species, Pennisetum americanum (pearl millet) and Panicum aestivum (Rao et al., 1988; Fladung et al., 1991). Because the phenotypes of these mutants are quite similar to those of the rice dl mutants, these mutations may be caused by DL orthologs in each plant. This suggests that genes in the DL/CRC subfamily may have been recruited to play an essential role in carpel specification and midrib formation during grass evolution. As crc mutants do not show any phenotypic changes in the leaf and CRC is not expressed in the leaf (Alvarez and Smyth, 1999; Bowman and Smyth, 1999), the DL function that controls midrib formation may have been acquired in the monocot lineage or during grass evolution.
Our results have shown that DL is a member of the YABBY gene family. In Arabidopsis, the function of YABBY genes is thought to be to promote abaxial cell fate in the lateral organs (Eshed et al., 1999; Sawa et al., 1999; Siegfried et al., 1999; Bowman, 2000), except for CRC, which plays a role in nectary development in addition to its role in regulating abaxial cell fate in carpels (Alvarez and Smyth, 1999; Bowman and Smyth, 1999). It is unlikely, however, that DL is involved in abaxial specification in rice. The phenotypes of dl mutants were not associated with abaxial identity, and no expression of DL was observed on the abaxial side in either the carpel or the leaf. Overexpression of DL did not cause structural alterations associated with dorsoventrality of the leaf. The YABBY genes seem to have evolved recently during plant evolution because database analysis indicates that no YABBY-like genes have been detected in animal and fungal genomes. Therefore, it is plausible that the YABBY genes have acquired their diversified functions during the evolution of monocots and dicots. Furthermore, because DL/CRC and INO regulate development of the carpel and outer integument, respectively (Alvarez and Smyth, 1999; Bowman and Smyth, 1999; Villanueva et al. 1999), both of which are characteristic of angiosperms, elucidation of the YABBY gene function in various angiosperms, including primitive species, may aid our understanding of the evolution of body plans in angiosperms and the mechanisms by which they are determined.
METHODS
Materials
The rice strains used in this study were O. sativa spp japonica. Taichung 65 (T65) was used for the wild type in cDNA isolation, histological observation, and in situ hybridization. Mutants dl-1, dl-2, dl-sup1, and dl-sup2 were described by Nagasawa et al. (2003). dl-sup3, dl-sup4, and dl-sup5 were isolated from the R2 population of plants (Nipponbare) regenerated by the method of Hirochika et al. (1996). dl-3 and dl-4 were obtained from an M2 population of Kinmaze that had been chemically mutagenized with N-methyl-N-nitrosourea. A chromosomal deletion line, fm28, was isolated from the M2 population (Nihon-masari), which was produced by γ-ray irradiation.
Isolation of DL
The dl locus has been mapped to the short arm of rice chromosome 3 (Yoshimura et al., 1997). The dl mutant (Japonica) was crossed with Kasalath (Indica), and F2 plants with the drooping leaf phenotype were selected and used for the following experiments. The dl locus was mapped among RFLP markers, and the marker C316 was found to be closest to this locus (no recombinant was found in 304 chromosomes). Next, we constructed a BAC contig by screening of a BAC library (Nakamura et al., 1997) with C316 and subsequent chromosomal walking. This BAC contig covered ∼300 kb. Sequences at both ends of the BAC contig showed recombination with the dl locus (R176, one recombinant; L1L, four recombinants), indicating that the dl locus is contained in this contig. Next, we used a mutant line, fm28, which probably has a chromosomal deletion that includes dl. The deleted region in fm28 was mapped by DNA gel blot analysis using DNA fragments arbitrarily isolated from a BAC clone (2E1213), and the size of the deletion was found to be <40 kb. The DNA (15 to 18 kb) derived from the BAC clone 2E1213 was subcloned into λDASH (Stratagene, La Jolla, CA), and a λ contig covering the deleted region was constructed.
A new mutant with drooping leaf was identified in the R2 population of regenerated rice plants through tissue culture. Genetic complementation analysis showed that this new mutant was allelic to other dl mutants, such as dl-1. The genotypes of 28 R2 plants, determined by the phenotype of the next generation, were DL/DL (12 plants), DL/dl (11 plants), and dl/dl (5 plants). DNA gel blot analysis using the retrotransposon TOS17 as a probe detected a 3.2-kb band in all plants with the dl/dl and DL/dl genotypes but was not detected in plants with DL/DL. DNA fragments specific to the dl/dl and DL/dl genotypes were isolated from a gel, and a DNA fragment (∼500 bp) flanking TOS17 was amplified by thermal asymmetric interlaced PCR. This fragment was used as a probe for subsequent DNA gel blot analysis.
Isolation of DL cDNA and DNA Sequencing
A cDNA library was constructed from poly(A)+ RNA isolated from inflorescences containing developing flowers at early stages using λZAPII (Stratagene). The cDNA library was screened with a 4.0-kb DNA fragment flanking TOS17. After a second screening, five positive clones were obtained out of 5 × 105 clones. All five clones are identical, and the longest cDNA was 1019 bp. The λDNA (D14) including the DL genomic DNA was subcloned into pBluescript SK+ (Stratagene). The nucleotide sequence was determined with a DYE primer cycle sequencing ready reaction kit (Applied Biosystems, Foster City, CA) after making nested deletion series using a double-stranded nested deletion kit (Pharmacia, Piscataway, NJ). For identification of mutations in the dl mutants, DNA fragments including several exons were amplified with primers containing sequences complementary to universal primers (21M13 and RV) at the outside introns, and nucleotide sequences were determined by the DYE primer method.
Complementation Test and Overexpression of DL
The EcoRI fragment (thick line in Figure 1 in the supplemental data online) of λD14 was isolated, cloned into a binary vector, and used for complementation analysis. For overexpression, DL cDNA was inserted in the binary vector containing the cassette of the rice actin (Act1) promoter and the nos terminator (Sentoku et al., 2000). Agrobacterium tumefaciens–mediated transformation was performed using calli derived from scutellum according to Hiei et al. (1994).
In Situ Hybridization
A part of the DL cDNA (100 to 447) was amplified and cloned into a T-dvector and used for the synthesis of an RNA probe to avoid cross-hybridization among highly conserved sequences corresponding to the YABBY domain. In situ hybridization with digoxigenin-labeled RNA was conducted as described by Kouchi and Hata (1993). Tissues were fixed with 4% (w/v) paraformaldehyde and 0.25% glutaraldehyde in 0.1 M sodium phosphate buffer and embedded in Paraplast Plus (Oxford Labware, St. Louis, MO). Microtome sections (7 to 10 μm thick) were applied to glass slides treated with Vectabond (Vector Laboratories, Burlingame, CA).
Sequence data of the cDNA and genomic DNA of DL have been deposited with the EMBL/GenBank data libraries under accession numbers AB106553 and AB106554, respectively.
Supplementary Material
Acknowledgments
We thank the Rice Genome Project (Tsukuba, Japan) for providing RFLP marker clones. This research was supported by Grants-in-Aid for Scientific Research (09460002 and 12037205) from the Ministry of Education, Culture, Sports, Science, and Technology and a Grant-in-Aid (Bio Design Program) from the Ministry of Agriculture, Forestry, and Fisheries of Japan (to H.-Y.H.).
On-line version contains Web-only data.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Hiro-Yuki Hirano (ahirano@mail.ecc.u-tokyo.ac.jp).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.018044.
References
- Alvarez, J., and Smyth, D.R. (1999). CRABS CLAW and SPATULA, two Arabidopsis genes that control carpel development in parallel with AGAMOUS. Development 126, 2377–2386. [DOI] [PubMed] [Google Scholar]
- Ambrose, B.A., Lerner, D.R., Ciceri, P., Padilla, C.M., Yanofsky, M.F., and Schmidt, R.J. (2000). Molecular and genetic analyses of the silky1 gene reveal conservation in floral organ specification between eudicots and monocots. Mol. Cell 5, 569–579. [DOI] [PubMed] [Google Scholar]
- Bowman, J.L. (2000). The YABBY gene family and abaxial cell fate. Curr. Opin. Plant Biol. 3, 17–22. [DOI] [PubMed] [Google Scholar]
- Bowman, J.L., Baum, S.F., Eshed, Y., Putterill, J., and Alvarez, J. (1999). Molecular genetics of gynoecium development in Arabidopsis. Curr. Top. Dev. Biol. 45, 155–205. [DOI] [PubMed] [Google Scholar]
- Bowman, J.L., Sakai, H., Jack, T., Weigel, D., Mayer, U., and Meyerowitz, E.M. (1992). SUPERMAN, a regulator of floral homeotic genes in Arabidopsis. Development 114, 599–615. [DOI] [PubMed] [Google Scholar]
- Bowman, J.L., and Smyth, D.R. (1999). CRABS CLAW, a gene that regulates carpel and nectary development in Arabidopsis, encodes a novel protein with zinc finger and helix-loop-helix domains. Development 126, 2387–2396. [DOI] [PubMed] [Google Scholar]
- Bowman, J.L., Smyth, D.R., and Meyerowitz, E.M. (1991). Genetic interactions among floral homeotic genes of Arabidopsis. Development 112, 1–20. [DOI] [PubMed] [Google Scholar]
- Coen, E.S., and Meyerowitz, E.M. (1991). The war of the whorls: Genetic interactions controlling flower development. Nature 353, 31–37. [DOI] [PubMed] [Google Scholar]
- Eshed, Y., Baum, S.F., and Bowman, J.L. (1999). Distinct mechanisms promote polarity establishment in carpels of Arabidopsis. Cell 9, 199–209. [DOI] [PubMed] [Google Scholar]
- Fladung, M., Bossinger, G., Roeb, G.W., and Salamini, F. (1991). Correlated alterations in leaf and flower morphology and rate of leaf photosynthesis in a midribless (mbl) mutant of Panicum maximum Jacq. Planta 184, 356–361. [DOI] [PubMed] [Google Scholar]
- Goto, K., Kyozuka, J., and Bowman, J.L. (2001). Turning floral organs into leaves, leaves into floral organs. Curr. Opin. Genet. Dev. 11, 449–456. [DOI] [PubMed] [Google Scholar]
- Hiei, Y., Ohta, S., Komari, T., and Kumashiro, T. (1994). Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 6, 271–282. [DOI] [PubMed] [Google Scholar]
- Hirochika, H., Sugimoto, K., Otsuki, Y., Tsugawa, H., and Kanda, M. (1996). Retrotransposons of rice involved in mutations induced by tissue culture. Proc. Natl. Acad. Sci. USA 93, 7783–7788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack, T., Brockman, L.L., and Meyerowitz, E.M. (1992). The homeotic gene APETALA3 of Arabidopsis thaliana encodes a MADS box and is expressed in petals and stamens. Cell 68, 683–697. [DOI] [PubMed] [Google Scholar]
- Kang, H.-G., Jeon, J.-S., Lee, S., and An, G. (1998). Identification of class B and class C floral organ identity genes from rice plants. Plant Mol. Biol. 38, 1021–1029. [DOI] [PubMed] [Google Scholar]
- Kouchi, H., and Hata, S. (1993). Isolation and characterization of novel nodulin cDNAs representing genes expressed at early stages of soybean nodule development. Mol. Gen. Genet. 238, 106–119. [DOI] [PubMed] [Google Scholar]
- Kuusk, S., Sohlberg, J.J., Long, J.A., Fridborg, I., and Sundberg, E. (2002). STY1 and STY2 promote the formation of apical tissues during Arabidopsis gynoecium development. Development 129, 4707–4717. [DOI] [PubMed] [Google Scholar]
- Kyozuka, J., and Shimamoto, K. (2002). Ectopic expression of OsMADS3, a rice ortholog of AGAMOUS, caused a homeotic transformation of lodicules to stamens in transgenic rice plants. Plant Cell Physiol. 43, 130–135. [DOI] [PubMed] [Google Scholar]
- Laudencia-Chingcuanco, D., and Hake, S. (2002). The indeterminate floral apex1 gene regulates meristem determinacy and identity in the maize inflorescence. Development 129, 2629–2638. [DOI] [PubMed] [Google Scholar]
- Mena, M., Ambrose, B.A., Meeley, R.B., Briggs, S.P., Yanofsky, M.F., and Schmidt, R.J. (1996). Diversification of C-function activity in maize flower development. Science 274, 1537–1540. [DOI] [PubMed] [Google Scholar]
- Nagasawa, N., Miyoshi, M., Sano, Y., Satoh, H., Hirano, H.-Y., Sakai, H., and Nagato, Y. (2003). SUPERWOMAN 1 and DROOPING LEAF genes control floral organ identity in rice. Development 130, 705–718. [DOI] [PubMed] [Google Scholar]
- Nakamura, S., Asakawa, S., Ohmido, N., Fukui, K., Shimizu, N., and Kawasaki, S. (1997). Construction of an 800-kb contig in the near-centromeric region of the rice blast resistance gene Pi-ta-2 using a highly representative rice BAC library. Mol. Gen. Genet. 254, 611–620. [DOI] [PubMed] [Google Scholar]
- Pinyopich, A., Ditta, G.S., Savidge, B., Liljegren, S.J., Baumann, E., Wisman, E., and Yanofsky, M.F. (2003). Assessing the redundancy of MADS-box genes during carpel and ovule development. Nature 424, 85–88. [DOI] [PubMed] [Google Scholar]
- Rao, S.A., Mengesha, M.H., and Reddy, C.R. (1988). Characteristics, inheritance, and allelic relationships of midribless mutants in pearl millet. J. Hered. 79, 18–20. [Google Scholar]
- Saitou, N., and Nei, M. (1987). The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425. [DOI] [PubMed] [Google Scholar]
- Sakai, H., Krizek, B.A., Jacobsen, S.E., and Meyerowitz, E.M. (2000). Regulation of SUP expression identifies multiple regulators involved in arabidopsis floral meristem development. Plant Cell 12, 1607–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai, H., Medrano, L.J., and Meyerowitz, E.M. (1995). Role of SUPERMAN in maintaining Arabidopsis floral whorl boundaries. Nature 378, 199–203. [DOI] [PubMed] [Google Scholar]
- Sato, Y., Hong, S.K., Tagiri, A., Kitano, H., Yamamoto, N., Nagato, Y., and Matsuoka, M. (1996). A rice homeobox gene, OSH1, is expressed before organ differentiation in a specific region during early embryogenesis. Proc. Natl. Acad. Sci. USA 93, 8117–8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawa, S., Watanabe, K., Goto, K., Kanaya, E., Morita, E.H., and Okada, K. (1999). FILAMENTOUS FLOWER, a meristem and organ identity gene of Arabidopsis, encodes a protein with a zinc finger and HMG-related domains. Genes Dev. 13, 1079–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt, R.J., and Ambrose, B.A. (1998). The blooming of grass flower development. Curr. Opin. Plant Biol. 1, 60–67. [DOI] [PubMed] [Google Scholar]
- Sentoku, N., Sato, Y., and Matsuoka, M. (2000). Overexpression of rice OSH genes induces ectopic shoots on leaf sheaths of transgenic rice plants. Dev. Biol. 220, 358–364. [DOI] [PubMed] [Google Scholar]
- Siegfried, K.R., Eshed, Y., Baum, S.F., Otsuga, D., Drews, G.N., and Bowman, J.L. (1999). Members of the YABBY gene family specify abaxial cell fate in Arabidopsis. Development 126, 4117–4128. [DOI] [PubMed] [Google Scholar]
- Villanueva, J.M., Broadhvest, J., Hauser, B.A., Meister, R.J., Schneitz, K., and Gasser, C.S. (1999). INNER NO OUTER regulates abaxial- adaxial patterning in Arabidopsis ovules. Genes Dev. 13, 3160–3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanofsky, M.F., Ma, H., Bowman, J.L., Drews, G.N., Feldmann, K.A., and Meyerowitz, E.M. (1990). The protein encoded by the Arabidopsis homeotic gene agamous resembles transcription factors. Nature 346, 35–39. [DOI] [PubMed] [Google Scholar]
- Yoshimura, A., Ideta, O., and Iwata, N. (1997). Linkage map of phenotype and RFLP markers in rice. Plant Mol. Biol. 35, 49–60. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.