Abstract
A PCR-based genomic scan has been undertaken to estimate the extent and ratio of maternally versus paternally methylated DNA regions in endosperm, embryo, and leaf of Zea mays (maize). Analysis of several inbred lines and their reciprocal crosses identified a large number of conserved, differentially methylated DNA regions (DMRs) that were specific to the endosperm. DMRs were hypomethylated at specific methylation-sensitive restriction sites upon maternal transmission, whereas upon paternal transmission, the methylation levels were similar to those observed in embryo and leaf. Maternal hypomethylation was extensive and offers a likely explanation for the 13% reduction in methyl-cytosine content of the endosperm compared with leaf tissue. DMRs showed identity to expressed genic regions, were observed early after fertilization, and maintained at a later stage of endosperm development. The implications of extensive maternal hypomethylation with respect to endosperm development and epigenetic reprogramming will be discussed.
INTRODUCTION
Genomic DNA of many eukaryotes can be modified by the covalent attachment of a methyl group to the 5-carbon position of cytosine residues. In plants, DNA methylation is mainly restricted to the symmetrical sequences 5′-CpG-3′ or 5′-CpNpG-3′ but also can occur in a nonsymmetrical context (Gruenbaum et al., 1981; Meyer et al., 1994). It is widely recognized that DNA methylation has the ability to influence gene activity mainly by repressing gene expression (reviewed in Bird and Wolffe, 1999). Methylation can act directly by blocking or reducing the binding of transcription factors to target DNA sequences (Prendergast and Ziff, 1991) or indirectly by inducing a condensed chromatin structure (Kass et al., 1997). In the latter case, repression is mediated by proteins that specifically bind to methylated residues, which in turn recruit chromatin-remodeling factors (Jones et al., 1998; Nan et al., 1998). Both mechanisms are likely to occur in plants because methylation affects the accessibility of several plant proteins to their target sequences (Gierl et al., 1988; Stäiger et al., 1989; Inamdar et al., 1991; Sturaro and Viotti, 2001), and proteins with affinity for methylated sequences also have been isolated (Zhang et al., 1989; Ehrlich, 1993).
Imprinting refers to the differential expression of a gene depending on the sex of the parent that transmits it. The regulatory mechanism of imprinted gene expression implies that a cell can discriminate between genetically identical alleles and determine which is to be transcribed. Therefore, it is presumed that parental alleles are differentially marked by an epigenetic imprint, which can modulate gene activity without imposing an irreversible change on nucleotide sequence. The finding that the vast majority of mammalian imprinted genes exhibit parental differences in methylation (i.e., differentially methylated regions [DMRs]) supports this idea (reviewed in Sleutels and Barlow, 2002). In plants, DMRs have been identified in the α-tubulin (tubα3 and tubα4) and seed storage protein genes (α-zeins) of Zea mays (maize) (Bianchi and Viotti, 1988; Lund et al., 1995a, 1995b). In both cases, DMRs were specific to the endosperm and not found in the embryo or seedling. Despite only sporadic cases of genes exhibiting DMRs in plants, current evidence suggests that methylation plays a role in several parent-of-origin effects observed during seed development (Vielle-Calzada et al., 1999; Adams et al., 2000; Luo et al., 2000; Vinkenoog et al., 2000; Yadegari et al., 2000; Bushell et al., 2003). In angiosperms, seed development is initiated by a double fertilization event in which two sperm nuclei of the pollen grain are delivered into the embryo sac. One sperm nucleus fuses to the egg to produce the embryo, while a second nucleus fuses with two nuclei of the central cell to form the triploid endosperm.
To date, only genes that are preferentially expressed upon maternal transmission have been identified in Arabidopsis thaliana and Z. mays endosperm (reviewed in Baroux et al., 2002). In A. thaliana, these include genes that are important in early seed development: MEDEA (MEA), FERTILIZATION-INDEPENDENT SEED2 (FIS2), and FERTILIZATION-INDEPENDENT ENDOSPERM (FIE) (Ohad et al., 1996; Chaudhury et al., 1997; Grossniklaus et al., 1998). By reverse transcriptase PCR and/or using a β-glucuronidase reporter, each gene shows evidence of maternal expression in the endosperm, but the duration of the paternal lack of activity is distinctive. MEA and FIS2 show prolonged paternal repression compared with FIE, which shows paternal allele activity early in endosperm development (Kinoshita et al., 1999; Vielle-Calzada et al., 1999, 2000; Luo et al., 2000; Vinkenoog et al., 2000; Yadegari et al., 2000). Similar differences in the extent of paternal repression have been observed in the duplicated fie1 and fie2 in Z. mays endosperm development (Danilevskaya et al., 2003). In addition, genetic and molecular evidence shows preferential maternal expression of genes that are activated later in endosperm development, such as the R locus, which is involved in aleurone pigmentation (Kermicle, 1970); the locus dzr1, which regulates the accumulation of a 10-kD zein protein (Chaudhuri and Messing, 1994); the α-zein genes (Lund et al., 1995a); and no-apical-meristem related protein1 (Guo et al., 2003).
Because methylation plays a role in the parent-of-origin effects observed in seed development, a PCR-based approach to identify DMRs in embryo, endosperm, and young leaf tissues of Z. mays was undertaken. The results showed that Z. mays endosperm is characterized by a high degree of maternal hypomethylation. The potential role of unidirectional hypomethylation in seed development is discussed.
RESULTS
Identification of DMRs in Z. mays Endosperm
Methylation-sensitive amplified polymorphism (MSAP) was undertaken to estimate the extent of DMRs in Z. mays endosperm. This technique is a modification of amplified fragment length polymorphism (AFLP), a procedure that is based on random amplification of restriction fragments typically generated by digestion of genomic DNA with the EcoRI and MseI restriction enzymes (Vos et al., 1995). In MSAP, MseI is replaced by an enzyme sensitive to cytosine methylation, such as HpaII (Reina-Lopez et al., 1997). After digestion of genomic DNA, adaptors are attached to restriction sites that have been successfully digested. It follows that the products of the MSAP ligation reaction consist of DNA fragments that are flanked by hypomethylated HpaII and/or EcoRI sites. Thereafter, two consecutive PCR reactions, a preamplification and a selective amplification, are performed to enrich a subpopulation of the restriction fragments. The primers employed are complementary to the core sequence of adaptors and recognition sites of the restriction enzymes, and the number of nucleotides added to their 3′ termini determines their selectivity. Typically, the number of selective nucleotides is increased in the selective amplification in which one of the two primers is radioactively labeled. This enables the visualization of a subset of restriction fragments by autoradiography after separation by acrylamide gel electrophoresis.
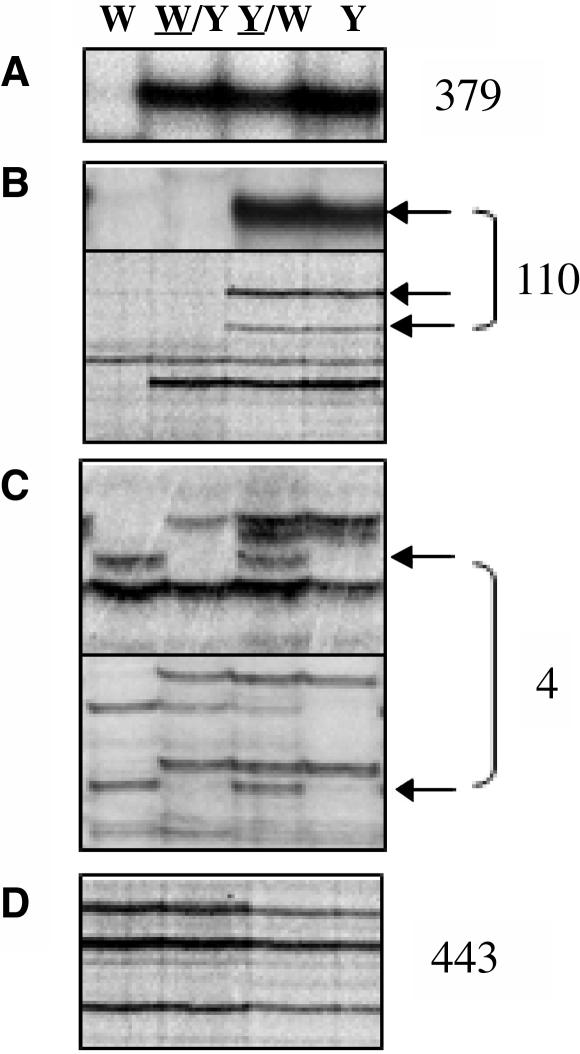
Because DMRs have been identified previously in the zein and tubulin genes by DNA gel blot analysis using the inbred lines W64A, A69Y, and their reciprocal crosses W64A/A69Y and A69Y/W64A (the seed parent of the cross is underlined) (Lund et al., 1995a, 1995b), the same materials were chosen for analysis by MSAP. DNA was extracted from endosperms harvested at 15 d after pollination (DAP) and digested with the EcoRI and HpaII restriction enzymes. MSAP analysis was conducted using 24 selective primer combinations, and the HpaII primer was radioactively labeled in the selective PCR amplification. Because of the triploid nature of the endosperm, an inbred line harbors three copies of a given restriction fragment, two copies from the maternal genome complements, and one copy from the paternal genome complement, whereas the reciprocal crosses W64A/A69Y and A69Y/W64A (where W64A is the seed and pollen parent, respectively) carry two maternal copies or one paternal copy, respectively. By comparing MSAP profiles of the inbred lines and their reciprocal F1 hybrids, the parental methylation status of polymorphic restriction fragments could be deduced. For example, a fragment specific to the A69Y inbred line that was detected upon maternal and paternal transmission (in the reciprocal crosses A69Y/W64A and W64A/A69Y, respectively) represented a DNA region without parental differences in methylation (Figure 1A, a normal polymorphic profile). A fragment that was only present when a particular inbred line was the seed or pollen donor could represent a DMR (Figures 1B and 1C, maternal and paternal profiles, respectively). By contrast, parental differences in methylation could not be determined for restriction fragments that were conserved between the inbred lines because their parental alleles were indistinguishable (Figure 1D, monomorphic profiles). Out of 936 profiles identified, 379 exhibited a normal polymorphic profile, 110 exhibited a maternal profile, 4 exhibited a paternal profile, and 443 exhibited a monomorphic profile. Maternal profiles accounted for 22.3% of polymorphic profiles (or 11.8% of total profiles, defined as the sum of monomorphic and polymorphic profiles), whereas only 0.8% of polymorphic profiles (or 0.4% of total profiles) represented paternal profiles.
Figure 1.
Identification of DMRs in Endosperm DNA by MSAP.
Representative MSAP profiles of an EcoRI/HpaII digest of endosperm DNA extracted from the inbred lines W64A (W) and A69Y (Y) and the reciprocal crosses W64/A69Y (W/Y) and A69Y/W64 (Y/W) (the seed parent of the reciprocal cross is underlined) harvested at 15 DAP. The sum of profiles scored of both inbred lines using 24 selective primer combinations is indicated at the right of each panel.
(A) An MSAP restriction fragment specific to the A69Y inbred line that did not show parental differences in methylation (a normal polymorphic profile).
(B) MSAP restriction fragments that only were detected when the A69Y inbred line was the seed parent (maternal profiles are indicated by arrows).
(C) MSAP restriction fragments that only were detected when the W64A inbred line was the pollen parent (paternal profiles are indicated by arrows).
(D) Monomorphic profiles.
Although the most likely explanation for the parent-of-origin profiles was that they represented DMRs, similar profiles could result from lack of homozygosity of the inbred lines employed, contamination of pericarp tissue, or preferential elimination/recombination of specific paternal chromosomal fragments, or they could represent maternally inherited organelle DNA. To exclude such possibilities, samples also were analyzed by AFLP, substituting the HpaII restriction enzyme with MseI, which is insensitive to methylation. Twelve selective primer combinations were employed, and the EcoRI primer was labeled in the selective PCR reaction. In contrast with MSAP, only 0.2% of total profiles exhibited a parent-of-origin effect (data not shown), indicating that the parental profiles observed in Figures 1B and 1C probably represented DMRs. We define the profiles as maternal methylation profiles (MMPs) and paternal methylation profiles (PMPs), respectively.
DMRs Are Conserved in Different Genetic Backgrounds and during Endosperm Development
DMRs specific to the W64A or A69Y inbred lines were identified only when they were distinguishable as genotype-specific allelic fragments. To scan the Z. mays genome for new DMRs, additional reciprocal hybrids generated from W64A, A69Y, W23, B73, and Mo17 inbred lines were analyzed by MSAP. The profiles of each pair of inbred lines and their F1 reciprocals were compared and DMRs scored in autoradiograms of 20 selective primer combinations.
Most DMRs proved to be genetic background independent. For example, 48 (22/46) and 51% (29/57) of MMPs present in the A69Y or W64A inbred lines also were found when either inbred line was crossed to the W23 inbred line (Figure 2A, top and bottom panels, respectively). Other MMPs, initially identified by comparing W64A with the A69Y inbred line also were evident for the W23 inbred line (Figure 2B, top and bottom panels, respectively). As expected, these conserved MMPs showed monomorphic profiles in the crosses between W23 and W64A or between W23 and A69Y inbred lines (Figure 2B, top and bottom panels, respectively). The frequencies of DMRs were found to vary from 12 to 14% in the different inbred lines studied (data not shown).
Figure 2.
Analysis of DMRs in Different Z. mays Inbred Lines by MSAP.
Representative MSAP profiles of an EcoRI/HpaII digest of DNA extracted from the inbred lines W64A, A69Y, and W23 (23) and their respective reciprocal crosses harvested at 15 DAP. Other abbreviations are as in Figure 1.
(A) MMPs specific to the W23 and A69Y inbred lines (top panel) or to the W64A inbred line (bottom panel) are indicated by arrows.
(B) An MMP that was conserved between the W64 and W23 inbred lines (top panel) and an MMP that was conserved between the A69Y and W23 inbred lines (bottom panel).
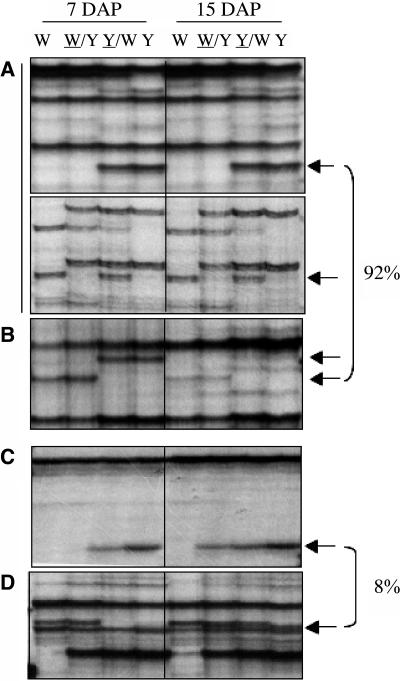
The stability of DMRs during endosperm development was studied to determine if endoreduplication affected the generation or stability of DMRs. Endoreduplication is a maternally controlled process by which DNA replication is not followed by cell division (Kowles et al., 1997; Dilkes et al., 2002; Leblanc et al., 2002). Because this process initiates at ∼9 DAP, DNA was extracted from endosperms harvested at 7 DAP and analyzed by MSAP. The restriction fragment profiles from 16 selective primer combinations were compared with those previously obtained from endosperm DNA harvested at 15 DAP (Figure 3). The majority (92%) of DMRs were maintained during endosperm development (Figures 3A and 3B), although 4% showed a decrease in signal from 7 to 15 DAP (Figure 3B). Only 8% of DMRs exhibited changes during endosperm development. These regions exhibited an MMP at 7 DAP but either a polymorphic or monomorphic profile at 15 DAP (Figures 3C and 3D, respectively). The results indicated that all DMRs were present early in endosperm development and that these methylation patterns were faithfully maintained during development.
Figure 3.
Stability of DMRs during Endosperm Development by MSAP.
Representative MSAP profiles of an EcoRI/HpaII digest of DNA extracted from endosperms harvested at 7 and 15 DAP from the W64A and A69Y inbred lines and their reciprocal crosses. The percentage of DMRs with a given profile is indicated at the right. This percentage was calculated from profiles scored of two inbred lines with 16 selective primer combinations. Arrows indicate DMRs, and abbreviations are as in Figure 1.
(A) An MMP and a PMP that were stable at the two developmental stages studied (top and bottom panels, respectively).
(B) An MMP that was stable at the two developmental stages, albeit with a lower intensity at 15 DAP.
(C) A restriction fragment that showed an MMP at 7 DAP but a normal polymorphic profile at 15 DAP.
(D) A restriction fragment that showed an MMP at 7 DAP but a monomorphic profile at 15 DAP.
DMRs Are Specifically Hypomethylated in the Endosperm
Numerous lines of evidence suggest that imprinting is central to endosperm development, with mainly indirect consequences on embryo growth (Kermicle, 1970; Lin, 1984; Haig and Westoby, 1991). If methylation is a component of the imprinting process, DMRs would be expected to be endosperm-specific. To investigate this, embryo and leaf DNAs of the W64A and A69Y inbred lines and their reciprocal F1 hybrids were subjected to MSAP. Based on 24 selective primer combinations, the endosperm had the largest number of total and tissue-specific profiles (i.e., profiles that were absent in the two other tissues) compared with leaf and embryo tissues, and 91% of endosperm-specific polymorphic profiles represented DMRs (Table 1).
Table 1.
MSAP of Embryo, Leaf, and Endosperm Tissues
| Tissue-Specific Profilesc
|
|||||
|---|---|---|---|---|---|
| Tissuea | Total | Totalb | Monomorphic | Polymorphic | DMR/Polymorphic (%)d |
| Embryo | 766 | 9 | 6 | 3 | 0 |
| Leaf | 768 | 10 | 5 | 5 | 0 |
| Endosperm | 936 | 178 | 60 | 118 | 91 |
DNA was extracted from embryos and endosperms harvested 15 DAP or from 2-week-old leaves of the inbred lines W64A, A69Y, and their reciprocal F1 hybrids.
Total profiles refer to the sum of monomorphic and polymorphic profiles of both inbred lines.
Tissue-specific refers to profiles only present in the tissue indicated; the values refer to the sum of both inbred lines analyzed by 24 selective primer combinations.
DMR refers to an MMP or a PMP.
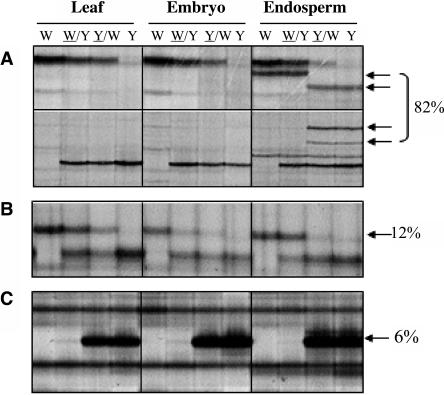
Comparisons of MSAP profiles between tissues showed that 82% of DMRs were completely absent in both embryo and leaf tissues (Figure 4A). Twelve percent of fragments that exhibited an MMP in the endosperm were detected in both leaf and embryo, but none showed an MMP in these tissues (Figure 4B). This indicated that the majority of endosperm-specific DMRs were methylated both maternally and paternally in leaf and embryo. To confirm this assumption, the total methyl-cytosine content of leaf and endosperm DNA was analyzed by high performance capillary electrophoresis (HPCE). Indeed, the results showed that the endosperm exhibited a 13% reduction in total methylation compared with leaf DNA (Table 2).
Figure 4.
Analysis of DMRs in Leaf, Embryo, and Endosperm Tissues by MSAP.
Representative MSAP profiles of an EcoRI/HpaII digest of DNA extracted from 2-week-old leaf tissues and from embryos and endosperm harvested at 15 DAP of the inbred lines W64A, A69Y, and their reciprocal crosses. The percentage of MMPs that exhibited each profile is indicated at the right. Arrows indicate MMPs, and abbreviations are as in Figure 1.
(A) Restriction fragments that showed an MMP specific to the endosperm.
(B) A restriction fragment that was detected in all tissues but only showed an MMP in the endosperm.
(C) A restriction fragment that showed an MMP in all tissues (a common MMP).
Table 2.
Total Cytosine Methylation in Leaf and Endosperm
| Tissuea | Total mC (%)b | Tissue Methylation Level (%)c | Decrease in Methylation (%) |
|---|---|---|---|
| Leaf | 24.8 ± 0.71 | 100 | 0 |
| Endosperm | 21.6 ± 0.63 | 87 | 13 |
DNA was extracted from 2-week-old leaves of 12 individual plants or from endosperms of 12 individual seeds harvested 15 DAP of the W64A inbred line.
The total methyl-cytosine (mC) content was analyzed by HPCE as described by Fraga and coworkers (2000); quantifications of the relative methylation in the DNA samples were calculated as the percentage of methyl-cytosine of total cytosine (C) [i.e., mC peak area × 100/(C peak area + mC peak area)]. The mean values indicated of leaf and endosperm DNA were significantly different (P < 0.01).
The methyl-cytosine content of the endosperm was normalized to leaf DNA levels.
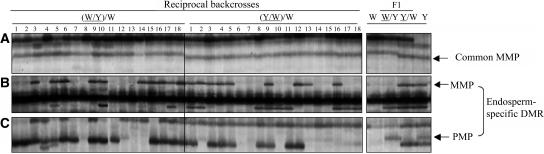
Although no DMRs specific to embryo or leaf tissues were detected, 6% (7/114) of MMPs were common to all three tissue types (Figure 4C). To elucidate if common MMPs represented genomic or organelle DNA, the seven fragments were sequenced and analyzed in reciprocal backcrosses of 18 individual endosperms by MSAP. In addition, 10 endosperm-specific DMRs (nine representative of an MMP and one representative of a PMP) were included as controls. Backcrosses were generated using the reciprocal F1 hybrids W64A/A69Y and A69Y/W64 as seed parents and the W64A inbred line as a pollen parent. Only three out of seven fragments with a common MMP had sequence identity to mitochondrial genes, but all fragments exhibited grandparental effects in the segregation analysis (Figure 5A). By contrast, endosperm-specific DMRs segregated as expected. For example, a restriction fragment that showed an endosperm-specific MMP specific to the A69Y inbred line segregated in both backcrosses (Figure 5B), whereas a fragment with a PMP specific to the A69Y inbred line was absent in both reciprocal backcrosses (Figure 5C). The results indicated that common MMPs did not represent DMRs but probably resulted from mitochondrial or chloroplast genomes.
Figure 5.
Reciprocal Backcross Analysis of DMRs in Endosperm Tissue by MSAP.
Representative MSAP profiles of an EcoRI/HpaII digest of DNA extracted from 18 individual endosperms of the reciprocal backcrosses (W64A/A69Y)/W64A and (A69Y/W64A)/W64A, indicated as (W/Y)/W and (Y/W)/W, respectively) harvested at 15 DAP. The MSAP profiles of the inbred lines W64A and A69Y and their respective reciprocal F1 hybrids are shown in the panels at right. Arrows indicate DMRs specific to the A69Y inbred line. Other abbreviations are as in Figure 1.
(A) A restriction fragment representing a common MMP that showed a grandparental effect in the reciprocal backcrosses.
(B) A restriction fragment representing an endosperm-specific MMP that segregated in both backcrosses.
(C) A restriction fragment representing an endosperm-specific PMP that was not detected upon maternal transmission.
Sequence Analysis of DMRs
To characterize the nature of endosperm-specific DMRs, fragments showing an MMP or a PMP were excised from acrylamide gels, reamplified by PCR, cloned, and sequenced. The sequences of 31 fragments with an MMP and one fragment with a PMP were successfully obtained (Table 3). The fragments ranged from 162 to 985 bp in size, the largest representing PMP2. The low success rate of obtaining consistent sequence data of the four PMP fragments identified probably reflected their size because all of them were ∼1 kb or larger. Under the experimental conditions employed, such fragments were poorly resolved and often lead to the isolation of multiple bands upon excision from acrylamide gels. Approximately 53% of DMRs revealed identity to cDNA sequences from a variety of different tissues and almost all of them showed sequence identity to unmethylated sequences from methyl-filtered DNA libraries (Rabinowicz et al., 1999). Only two DMRs (MMP5 and MMPa3) showed homology to retrotransposons, indicating that the majority of highly repetitive DNA regions were methylated on both parental genomes in the endosperm.
Table 3.
Sequence Analysis of DMRs
| DMRa | Selective Primer Combinationb | Inbredc | Lengthd | ESTe | DNA or Protein Homologye |
|---|---|---|---|---|---|
| MMP4 | ACA/CGAA | W23 | 757 | None | CC933496 |
| MMP5* | As above | W23 | 546 | BM339610 | AF078917 (centromeric retrotransposon-like repeat CentA) |
| MMM7* | ACA/AATT | W64 | 344 | BU093060 | BAB12027 (hypothetical ring finger protein, Oryza sativa) |
| MMP8 | ACA/CGAA | A69Y | 351 | None | None |
| MMP9* | As above | A69Y | 583 | BM080498 | NP_563889 (expressed protein, A. thaliana) |
| MMP11* | AGA/CGAA | W23 | 197 | AW172080 | NP_921858 (En/Spm-like transposon, O. sativa) |
| MMP21* | ACC/TAGC | W64 | 326 | CF056235 | NP_908521 (transposon Tip100, O. sativa) |
| MMP22* | AGT/TAGC | A69Y | 540 | None | None |
| MMP26* | AGA/TCCA | A69Y | 509 | None | BZ368936 |
| PMP2* | AGA/CGAA | W64 | 985 | CF041957 | AAB84331 (putative N-acetylglucosaminephosphotransferse, A. thaliana) |
| MMP43* | AGT/AATT | W64 | 665 | CD976902 | NP_190991 (RNA recognition motif containing protein, A. thaliana) |
| MPM53* | ACA/CGTT | W64 | 292 | None | BZ539615 |
| MMPa3 | ACA/AATT | B73 | 443 | CD526326 | AF479697 (putative target of liguless3/4); NP_922450 (putative gypsy-type retrotransposon) |
| MMPa4 | As above | B73 | 344 | As MMP7 | As MMP7 |
| MMPa5 | As above | Mo17 | 317 | None | CG383037 |
| MMPa6 | As above | B73 | 188 | None | BH869432 |
| MMPe3 | AGA/CGTT | Mo17 | 305 | CF049215 | X15406 (3′ noncoding region of GpaI pseudogene; NP_198800 (putative RNA-like helicase, O. sativa) |
| MMPe4 | As above | B73 | 330 | None | BZ317541 (methylation-filtered genomic sequence) |
| MMPf1 | ACC/CGTT | B73 | 227 | None | None |
| MMPh5 | AGA/CGAA | Mo17 | 294 | CA275440 (Saccharum officinarum) | CAE05494 (putative protein, O. sativa) |
| MMPh7 | As above | B73 | 162 | None | BH783657 |
| MMPi2 | ACC/CGAA | B73 | 506 | None | BH870119 |
| MMPi3 | As above | Mo17 | 494 | None | As MMPi2 |
| MMPi6 | As above | Mo17 | 268 | CF037288 | NP_914980 (putative protein, O. sativa) |
| MMPi7 | As above | Mo17 | 258 | None | NP_922823 (putative transposase, O. sativa) |
| MMPm1 | ACA/CGTT | B73 | 595 | CA399229 | None |
| MMPm2 | As above | Mo17 | 552 | CF674952 | S29330 (hypothetical En-1 transposon protein) |
| MMPn4.1 | AGA/TAGC | B73 | 313 | None | BH777106 |
| MMPn4.2 | As above | B73 | 313 | CB927767 (Sorghum bicolor) | NP_912742 (unnamed protein, O. sativa) |
| MMPn5 | As above | B73 | 233 | BG104058 (S. propinquum) | NP_908906 (putative Ser/Thr protein phosphatase, O. sativa) |
| MMPn6 | As above | B73 | 226 | CA140644 (S. officinarum) | NP_916752 (putative protein, O. sativa) |
| MMPp3 | AGT/TAGC | B73 | 554 | CD227645 (S. bicolor) | NP_199332 (cyclin family, A. thaliana) |
An MSAP fragment that showed an MMP or a PMP was excised after acrylamide gel electrophoresis, reamplified using the appropriate primer combinations, cloned, and subjected to DNA sequencing. Only fragments exhibiting identical sequences of three independent clones were included.
Nucleotide extensions of EcoRI-00 and HpaII-00 primers (see Methods) used in the selective amplification reaction.
The inbred line that exhibited an MMP or a PMP.
Length of the fragment analyzed.
BlastN (Basic Local Alignment Search Tool) and BlastX similarity scores were considered significant for E values <0.00001. Unless indicated, EST and genomic clones were from Z. mays; the BlastX hits indicated of MMP9, PMP2, MMPa3, MMPi6, and MMPn6 were specific to the EST sequence, and the others were common to both the EST and DMR.
Refers to an MMP or a PMP analyzed by DNA gel blotting.
GpaI, glyceraldehyde-3-phosphate dehyrogenase subunit A.
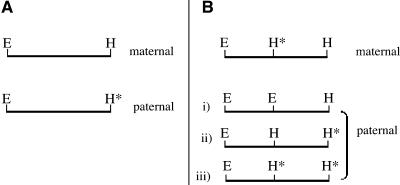
All sequenced DMRs represented EcoRI/HpaII restriction fragments, indicating that a DMR can result from differential methylation of either an HpaII site or an EcoRI site (the EcoRI restriction enzyme is sensitive to methylation of the cytosine residue of the GAATTC restriction site). However, because few DMRs were revealed by AFLP when the EcoRI primer was labeled as opposed to the HpaII selective primer, it was concluded that DMRs resulted from parental differences in methylation of an HpaII site. Seventy-five percent of sequenced MMP fragments lacked internal HpaII sites. This implied that the failure to detect these fragments upon paternal transmission resulted from paternal methylation of the external HpaII site (Figure 6A). Because 25% of the remaining MMP fragments contained an internal HpaII site (Figure 6B, i and ii), this site must have been methylated upon maternal transmission, whereas the external HpaII site was hypomethylated (Figure 6B). The lack of these fragments upon paternal transmission resulted from either paternal hypomethylation or hypermethylation of one or both HpaII sites (Figure 6B, i, ii, and iii). Assuming that the internal HpaII site of these MMP fragments was paternally hypomethylated, HpaII/EcoRI restriction fragments ranging from 120 to 555 bp in size would have been generated in the initial MSAP restriction digest. Obviously, the probability of detecting these particular PMP fragments together with their corresponding MMPs using the same primer combinations is remote, but it illustrates that paternal hypomethylation of specific HpaII sites would produce fragments within the range of effective separation by MSAP (i.e., between 50 and 1000 bp). Because only four PMP fragments ∼1 kb or larger in size were detected (corresponding to 3.5% of total DMRs), this suggests that few, if any, DNA regions were specifically hypomethylated upon paternal transmission. It follows that a PMP fragment is predicted to contain an internal HpaII site that is maternally hypomethylated. This was found to be the case for PMP2 (see below).
Figure 6.
Parental Methylation Status of a Restriction Fragment That Showed an MMP by MSAP.
An EcoRI (E)/HpaII (H) restriction fragment showing the possible methylation status of HpaII sites upon maternal versus paternal transmission. The asterisks indicate a methylated cytosine residue.
(A) A fragment without internal HpaII sites. The external HpaII site is maternally hypomethylated and paternally methylated.
(B) A fragment with an internal HpaII site. The external and internal HpaII sites are maternally hypomethylated and hypermethylated, respectively. The methylation status of this paternal fragment may be one of the following: i, hypomethylated at both HpaII sites; ii, hypomethylated at the internal HpaII site only; or iii, hypermethylated at both HpaII sites.
DNA Gel Blot Analysis of DMRs
Nine fragments showing an MMP and one showing a PMP were used as probes in DNA gel blot analysis (marked with asterisks in Table 3). Genomic DNA from endosperms harvested at 15 DAP or from 2-week-old leaf tissue was digested with the combination of EcoRI and HpaII (EH) restrictions enzymes used in MSAP or with HindIII, a restriction enzyme that is insensitive to 5′-CpG-3′ methylation. For some probes, additional restriction digests were undertaken.
DMRs without Internal HpaII Sites
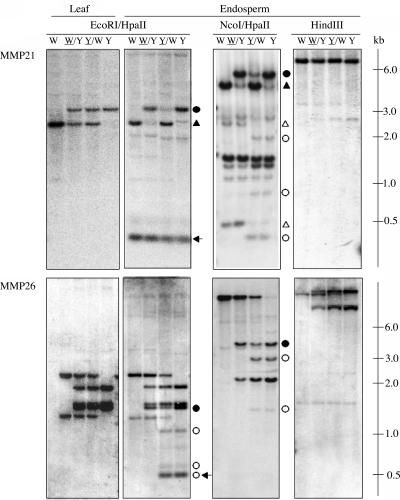
MMP21 was a W64A-specific 309-bp fragment that lacked an internal HpaII site. In an EH digest of endosperm DNA, the smallest hybridizing fragment in the W64A inbred line should represent an MMP, whereas a PMP of a larger size fragment would be expected. Hybridization to an ∼300-bp fragment was observed (Figure 7, arrow in top panel), but the parental methylation status could not be determined because a similar size fragment hybridized to the A69Y inbred line. However, ∼2.5- and 3.0-kb fragments specific to the W64A and A69Y inbred lines, respectively, showed a PMP (Figure 7, closed triangles and circles, respectively, in top panel). It was concluded that the 300-bp fragment exhibited an MMP in both inbred lines that was not resolved by DNA gel blotting. Indeed, hybridization of MMP21 to an NcoI/HpaII double digest of endosperm DNA revealed MMPs and a PMP specific to the W64A (Figure 7, open and closed triangles, respectively, in top panel) and A69Y inbred lines (Figure 7, open and closed circles, respectively, in top panel). Similar results were found for MMP26, an A69Y-specific 509-bp fragment that also lacked an internal HpaII site. Three fragments exhibited an MMP in endosperm DNA digested with EH (Figure 7, open circles in bottom panel), the smallest corresponding in size to the fragment isolated by MSAP (Figure 7, arrow in bottom panel). Both reciprocal crosses showed hybridization to an ∼1.5-kb fragment specific to the A69Y inbred line, but the intensity of hybridization was higher in the F1 hybrid in which the A69Y inbred was the pollen parent (i.e., W64A/A69Y) (Figure 7, closed circles in bottom panel). This suggested that only some copies of the 1.5-kb fragment exhibited a PMP. The specific hybridization patterns observed in MMP26 were echoed in an NcoI/HpaII double digest of endosperm DNA (Figure 7, bottom panel). In addition, neither probes showed DMRs in leaf DNA digested with EH (Figure 7, top and bottom panels at left) or in endosperm DNA digested with HindIII (Figure 7, top and bottom panels at right).
Figure 7.
DNA Gel Blot Analysis of DMRs.
DNA extracted from endosperms harvested at 15 DAP or from 2-week-old leaves of the inbred lines W64A, A69Y, and their reciprocal hybrids was digested with the restriction enzymes indicated. DNA gel blot analysis of leaf and endosperm DNA probed with MMP21 and MMP26 that lacked internal HpaII sites (top and bottom panels, respectively). MMP21 was a 309-bp fragment that showed an MMP specific to the W64A inbred line by MSAP; MMP26 was a 509-bp fragment that exhibited an MMP specific to the A69Y inbred line by MSAP. Arrows indicate a fragment that corresponds in size to the DMR isolated by MSAP. Open and closed triangles indicate MMPs and PMPs, respectively, specific to the W64A inbred line, and open and closed circles indicate MMPs and PMPs, respectively, specific to the A69Y inbred line. Abbreviations are as in Figure 1.
DMRs with Internal HpaII Sites
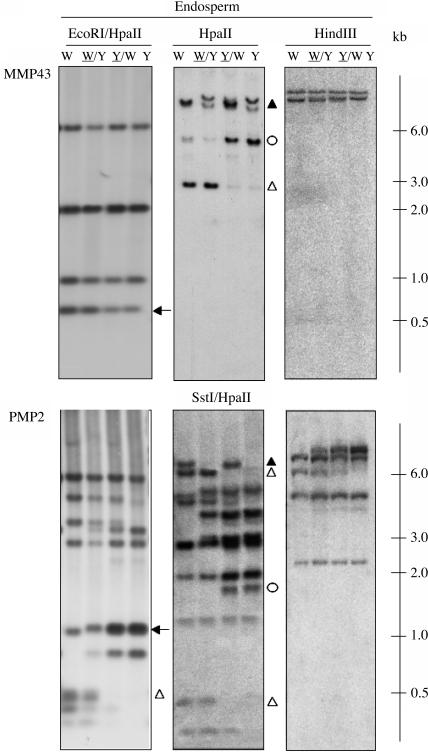
We have argued previously that DMRs, with or without internal HpaII sites, were hypomethylated upon maternal transmission. To strengthen this conclusion, MMP43, a 665-bp W64A-specific fragment that contained an internal HpaII site, was hybridized to an EH digest of endosperm DNA. If both internal and external HpaII sites of MMP43 were hypomethylated paternally (Figure 6B, i), 555- and 110-bp fragments would be expected to show PMPs, whereas a 665-bp fragment should exhibit an MMP. Because the smallest hybridizing fragment was ∼600 bp (Figure 8, arrow in top panel), the internal HpaII site of MMP43 was methylated both paternally and maternally. Because of a lack of polymorphism between the W64A and A69Y inbred lines of the 600-bp fragment, the parental methylation status of the external HpaII site could not be determined. However, upon hybridization of MMP43 to an HpaII digest of endosperm DNA, it was confirmed that the external HpaII site of MMP43 was maternally hypomethylated (Figure 8, top middle panel). Likewise, PMP2, a 985-bp W64A-specific fragment, was found to be hypomethylated upon maternal transmission. In an EH digest of endosperm DNA, hybridization to an ∼1-kb fragment in both inbred lines impeded the identification of a PMP (Figure 8, arrow in bottom panel). However, two smaller comigrating fragments, which corresponded in size to those expected if the internal and external HpaII sites of PMP2 were maternally hypomethylated (i.e., 475 and 510 bp; Figure 8, open triangle in bottom panel at left), showed an MMP. It was concluded that the internal HpaII site of PMP2 was hypomethylated upon maternal transmission. Because of the poor resolution of the PMP in the EH digest, PMP2 was hybridized to endosperm DNA restricted with SstI and HpaII, confirming the presence of DMRs in both inbred lines (Figure 8, bottom middle panel). As observed previously, neither of the probes exhibited parental profiles in endosperm DNA digested with HindIII (Figure 8, top and bottom panels at right) nor in leaf DNA digested with EH (data not shown).
Figure 8.
DNA Gel Blot Analysis of DMRs.
DNA extracted from endosperms harvested at 15 DAP or from 2-week-old leaves of the inbred lines W64A, A69Y, and their reciprocal hybrids was digested with the restriction enzymes indicated. DNA gel blot analysis of endosperm DNA probed with MMP43 and PMP2 that contained an internal HpaII site (top and bottom panels, respectively). MMP43 was a 665-bp fragment that showed an MMP specific to the W64A inbred line by MSAP; PMP2 was a 985-bp fragment that exhibited a PMP specific to the W64A inbred line by MSAP. Open and closed triangles indicate MMPs and PMPs, respectively, specific to the W64A inbred line, and open and closed circles indicate MMPs and PMPs, respectively, specific to the A69Y inbred line. Arrows indicate a fragment that corresponds in size to the DMR isolated by MSAP. Abbreviations are as in Figure 1.
In summary, the 10 DMRs analyzed were all hypomethylated in the endosperm, and DMRs were specific to the endosperm. The majority of fragments were hypomethylated upon maternal transmission, but for 4/10 probes (MMP5, MMP9, MMP11, and MMP43), the parental origin of hypomethylation could not be determined because of the lack of polymorphism between the W64A and A69Y inbred lines in the EH digest. However, extensive analysis of MMP43 showed that the failure to detect an MMP resulted from the reduced sensitivity of DNA gel blotting compared with MSAP (Figure 8, top panel). Several DMRs, identified as being specific to a particular inbred line by MSAP, were found to be differentially methylated in both inbred lines by DNA gel blotting (MMP7, MMP21, MMP43, and PMP2). For MMP26, maternal hypomethylation was partial.
DISCUSSION
Characteristics of DMRs
MSAP analysis of embryo, endosperm, and leaf DNA has provided clear evidence of an asymmetry in parental patterns of methylation that is specific to the endosperm. Approximately 96% of DMRs exhibited an MMP, whereas only 4% showed a PMP. Sequence and DNA gel blot analysis demonstrated that the origin of both profiles was identical; for example, DMRs resulted from maternal hypomethylation of specific HpaII sites, whereas the corresponding sites were methylated paternally. An almost identical situation was reported for specific alleles of the α-zein genes, the α-tubulin genes, and the R locus in Z. mays (Lund et al., 1995a, 1995b; Alleman and Doctor, 2000). At least 50% of DMRs were conserved between genotypes. This percentage is presumably higher because the identification of a conserved DMR relies on polymorphism between the inbred lines studied. Indeed, the differences in MMP frequencies between the inbred lines studied most probably reflect differences in their degree of polymorphism. In addition, MMPs identified as being specific to one inbred line by MSAP exhibited DMRs in both inbred lines by DNA gel blotting. The high frequency and diverse nature of DMRs is indicative of a genome-wide process that leads to parental differences in methylation in the endosperm.
The majority of endosperm-specific maternally hypomethylated DNA regions were methylated to a similar extent in both embryo and leaf. Interestingly, a similar situation has been observed in the mammalian placenta, which like the endosperm, mediates the transfer of nutrients from mother to offspring. In the placenta, general hypomethylation of both genic and repetitive DNA sequences is observed, whereas the same sequences are methylated in embryonic and adult tissues (Chapman et al., 1984; Razin et al., 1984; Rossant et al., 1986). Although it has not been determined if hypomethylation occurs preferentially in maternal or paternal DNA regions, it is interesting to note that the paternal X-chromosome is preferentially inactivated in the Mus musculus (mouse) placenta as opposed to random X-inactivation in the embryo (Takagi and Sasaki, 1975).
Epigenetic Reprogramming during Seed Development
Z. mays endosperm exhibited an ∼13% reduction in methylation compared with the embryo and leaf. Given that 91% of endosperm-specific polymorphic profiles represented DMRs, maternal hypomethylation must contribute significantly to the reduced level of 5-methylcytosine observed in the endosperm. Once established, maternal hypomethylation remained stable during development and was not modified by the pollen donor. This suggests that major methylation changes occur at an early stage of endosperm development, if not during female gametogenesis. Given that the methylation status of the paternal allele was almost identical among endosperm, embryo, and leaf tissues, it might be argued that the paternal genome does not undergo similar changes in methylation. Using an antibody against 5-methylcytosine during Nicotiana tabacum (tobacco) pollen development, the generative nucleus has been reported to show a fivefold reduction in methylation compared with the vegetative nucleus (Oakeley et al., 1997). If these differences reflect a net change in methylation, and not the loss of accessibility to DNA because of an altered chromatin structure, similar methylation changes also may occur during pollen development.
Methylation and Parent-of-Origin Effects in Endosperm Development
In A. thaliana, a genome-wide imprinting mechanism that ensures maternal control of early seed development has been proposed to occur (Vielle-Calzada et al., 2000; Baroux et al., 2002). This relates to the observation that a large number of genes are transiently silenced during early embryo and endosperm development upon pollen transmission (Vielle-Calzada et al., 2000). A similar behavior has been reported in fie2 in Z. mays (Danilevskaya et al., 2003). However, the paternal genome cannot be considered completely silenced during early seed development because several genes are transcribed upon pollen transmission in both Z. mays and A. thaliana (Springer et al., 2000; Weijers et al., 2001; Scholten et al., 2002).
It is tempting to suggest that global hypomethylation plays a role in the genome-wide maternal expression observed early in seed development either directly by silencing paternally derived genes or by acting downstream of other chromatin remodeling factors such as histone modifications. Whatever the mechanism, several observations suggest that maternal hypomethylation is a hallmark of expressed genic regions in Z. mays endosperm. An inverse correlation between methylation and gene expression is evident for the imprinted R and α-zein genes (Lund et al., 1995a, 1995b; Alleman and Doctor, 2000). In addition, global analysis of expression levels shows a higher percentage of maternal than paternal transcripts that cannot be explained by gene dosage alone (Guo et al., 2003). The observation that the size and shape of the endosperm corresponds to the maternal genotype also suggests that many processes of endosperm development are maternally regulated.
The parental conflict hypothesis proposed by Haig and Westoby (1989) is based on the concept that maternally and paternally derived genes compete over available resources in the offspring. Because the seed parent bears the cost of reproduction, genes promoting endosperm development are predicted to show preferential paternal expression, whereas growth inhibitors are maternally expressed. This hypothesis is supported by the nonreciprocal phenotypes observed in interploidy crosses in which an excess of maternal and paternal genomes is associated with small and large seeds, respectively (Lin, 1984; Haig and Westoby, 1991; Scott et al., 1998). In Z. mays, lack of the paternal contribution of specific chromosomal regions produces subnormal endosperms, indicating that these regions contain gene/genes (termed endosperm size factors [EFs]) that promote endosperm growth upon pollen transmission (Lin, 1982). However, as the small kernel phenotype is enhanced by adding extra maternal doses of the same or different EF-containing chromosomal regions, EFs also must function when maternally transmitted, albeit with an opposite effect (Birchler and Hart, 1987; Birchler, 1993). This has lead to the proposal that parent-of-origin effects result from an imbalance in dosage-sensitive factors during megagametogenesis and in their subsequent interaction with dosage-dependant functions after fertilization (Birchler, 1993). In A. thaliana, a potential role for methylation in parent-of-origin effects has been uncovered by crossing wild-type and hypomethylated plants, which show an 85% reduction in the global cytosine level because of the presence of a METHYLTRANSFERASE1 antisense (MET1 a/s) (Finnegan et al., 1996; Adams et al., 2000). For example, a cross between a MET1 a/s seed parent and a wild-type pollen parent (MET1 a/s × 2x) phenocopies the paternal excess phenotype observed by crossing a diploid seed parent to a tetraploid pollen parent. This suggests that genes regulating endosperm growth are silenced maternally by methylation. Although the lack of paternal hypomethylation found in our study runs counter to this idea, a simple explanation might be that maternally expressed genes greatly exceed paternally expressed genes in number, and only an exhaustive analysis will reveal paternally hypomethylated regions. Alternatively, paternally expressed genes are regulated by different mechanisms, or hypomethylation is not always indicative of a transcriptionally active state. In mammals, parental asymmetry in imprint acquisition has been observed because most maternally imprinted genes are silenced directly by maternal methylation, whereas the majority of paternally silenced genes are repressed indirectly via an antisense mechanism (Reik et al., 2001). In the latter case, an antisense molecule only is transcribed from the paternal allele because the maternal copy of the transcript is silenced by methylation (reviewed in Sleutels and Barlow, 2002). However, the observation that numerous aspects of endosperm development are maternally controlled and the seeming lack of paternal-specific hypomethylation suggests that seed size may result from an imbalance of dosage-sensitive genes upon maternal transmission and not from parental imprinting.
METHODS
Plant Material
Z. mays plants were grown in the field and under standard greenhouse conditions during the years 1993 to 2002. Immature seeds were collected at 7 and 15 DAP, frozen in liquid nitrogen, and stored at −80°C. Endosperms harvested at 7 DAP were dissected under a dissecting microscope and collected in a mannitol solution (750 mosM/kg H2O). The embryo and remaining sporophytic tissues were manually eliminated under an inverted microscope using glass needles.
MSAP Analysis
MSAP was performed as reported by Reina-Lopez and coworkers (1997), with the following modifications. Enzymes were provided by New England Biolabs ([NEB]; Hitchin, UK). Five hundred nanograms of genomic DNA were digested for 1 h with 5 units of HpaII using NEB buffer 1 in a 30-μL reaction volume. Restriction digest and ligation reactions were performed simultaneously for an additional 3 h in a final volume of 40 μL. The restriction-ligation mix contained 5 units of HpaII and EcoRI, 10 units of T4 DNA ligase, 5 pmole EcoRI adaptor, 50 pmole HpaII adaptor, and 1 mM ATP. The enzymes were inactivated for 15 min at 65°C, and a second digest was performed for 1 h with 5 units of HpaII and EcoRI, again followed by heat inactivation. The adaptor sequences were as follows: EcoRI, 5′-CTCGTAGACTGCGTACC-3′ and 5′-AATTGGTACGCAGTCTAC-3′; and HpaII, 5′-GATCATGAGTCCTGCT-3′ and 5′-CGAGCAGGACTCATGA-3′.
The preamplification reaction was performed with primers complementary to the core of the adaptor sequences and to the target sequences of EcoRI and HpaII. The sequences of the EcoRI (EcoRI-00) and HpaII (HpaII-00) preselective primers were 5′-AGACTGCGTACCAATTC-3′ and 5′-TCATGAGTCCTGCTCGG-3′, respectively. Two microliters of the digestion-ligation mix (diluted 1:3) were added to the preamplification mix consisting of 1 × PCR buffer (100 mM Tris HCl, 15 mM MgCl2, and 500 mM KCl, pH 8.3), 0.1 mM deoxynucleotide triphosphate (dNTP), 50 ng of EcoRI-00 primer and HpaII-00 primer, and 1 unit of Taq polymerase (Roche Biochemicals, Indianapolis, IN). PCR conditions were 72°C for 2 min and 94°C for 3 min, followed by 25 cycles of 95°C for 30 s, 56°C for 30 s, and 72°C for 1 min. The final extension was at 72°C for 4 min.
Selective primers had identical sequences to the preselective primers but included the addition of a number of nucleotides at the 3′ termini. Selective nucleotides of the EcoRI-00 primer were EcoRI-01 AGT, EcoRI-02 ACA, EcoRI-03 AGA, and EcoRI-04 ACC. The HpaII selective primers were HpaII-01 TCCA, HpaII-02 TAGC, HpaII-03 CGAA, HpaII-03A CGTT, HpaII-04 AATT, and HpaII-04A AACC. Selective PCR used 2 μL of preamplification mix (diluted 1:10) in a 10-μL reaction volume containing 1 × PCR buffer, 0.1 mM dNTP, 50 ng of EcoRI-00 primer, 50 ng of γ-33P–labeled HpaII-00 primer, and 1 unit of Taq polymerase. The HpaII primer was end-labeled by incubating 50 ng of primer with 50 μCi of [γ-33P]ATP, 10 units of polynucleotide kinase, and 5 μL of 1 × OPA buffer (100 mM Tris acetate, pH 7.5, 100 mM magnesium acetate, and 500 mM potassium acetate) (Amersham Pharmacia Biotech, Little Chalfont, UK). The reaction was incubated for 1 h at 37°C, followed by heat inactivation for 15 min at 65°C. The PCR cycle employed was the standard AFLP protocol (Vos et al., 1995).
The PCR samples were mixed 1:1 (v/v) with denaturating buffer (98% formamide, 10 mM EDTA, 0.1% bromophenol blue, and 0.1% xylene cyanol) and separated on 6% polyacrylamide sequencing gel (Bio-Rad, Hercules, CA) for 3 h at 90 W. Gels were dried and exposed to BioMax MR film (Eastman Kodak, Rochester, NY) for 1 to 4 d at −80°C. For each sample, three independent MSAP reactions were performed. When the results were reproducible, one sample was used for further analysis.
AFLP Analysis
AFLP was conducted according to Vos and coworkers (1995), and NEB provided all enzymes used in the AFLP protocol. Genomic DNA (0.5 μg) was digested for 1 h with 5 units of EcoRI and MseI in a 20-μL reaction volume. After heat inactivation (15 min at 65°C), ligation of adapters was performed for 3 h at 37°C by adding an equal volume of ligation mixture containing 1 × OPA buffer, 5 mM DTT, 5 pmole EcoRI adaptor (5′-CTCGTAGACTGCGTACC-3′, 5′-AATTGGTACGCAGTC-3′), 50 pmole MseI adaptor (5′-GACGATGAGTCCTGAG-3′, 5′-TACTCAGGACTCAT-3′), 5 units T4 ligase enzyme, and 1 mM ATP. The reaction was stopped by heat inactivation for 15 min at 65°C and diluted to 120 μL.
Preamplification used 5 μL of the diluted restriction-ligation mixture in a 20-μL reaction volume containing 1 × PCR buffer, 0.1 mM dNTP, 50 ng preselective primers, and 1 unit of Taq polymerase. Preselective primers were complementary to the core sequences of EcoRI and Mse1 adaptors, including one selective nucleotide for both EcoRI (E-01, 5′-GACTGCGTACCAATTCA-3′) and MseI (Mse-02, 5′-GATGAGTCCTGAGTAAC-3′) primers. The preamplification consisted of 25 cycles at 94°C for 30 s, 56°C for 30 s, and 72°C for 1 min, with a final extension at 72°C for 5 min. The preamplification product was diluted to 200 μL and stored at −20°C before use. Selective amplification was as described for MSAP analysis. The selective primers were identical to the AFLP preselective primers but included two additional nucleotides at the 3′ termini. The EcoRI selective primers were E31 AA and E32 AC. The selective MseI primers were M47 AA, M48 AC, M49 AG, M50 AT, M51 CA, M52 CC, M58 GT, M59 TA, and M60 TC.
Isolation, Cloning, and Sequence Analysis of DMRs
DMRs were excised from acrylamide gels, suspended in 30 μL of 0.5 × TE buffer (1× TE buffer is 10 mM Tris HCl, pH 8.0, and 1 mM EDTA) and incubated for 10 min at 65°C. One microliter of the solution was used in a standard PCR reaction with the appropriate primer combinations. The resulting fragments were cloned in pGEM-T Easy vector (Promega, Madison, WI), and three independent clones were sequenced for each DMR.
DNA Extraction and DNA Gel Blot Analysis
DNA extraction and restriction digests were performed as by Bianchi and Viotti (1988). Ten micrograms of each sample were separated on a 0.8 to 1.2% agarose gel and blotted onto a positively charged nylon membrane (BrightStar-Plus; Ambion, Austin, TX). DNA probes were labeled with [α−32P]dCTP (Amersham Pharmacia Biotech) using High Prime (Roche Biochemicals), and hybridization was performed overnight for 42°C in ULTRAhyb hybridization buffer (Ambion). Washes were performed at 65°C, twice in 2 × SSC (1× SSC is 0.15 M NaCl and 0.015 M sodium citrate) and 0.1% SDS for 15 min, followed by two washes in 0.2 × SSC and 0.1% SDS for 15 min.
Measurement of Total Methyl-Cytosine Content
Total cytosine content was analyzed by HPCE. DNA extracted from leaf and endosperm of 12 individual plants of the W64A inbred line was subjected to HPCE as described by Fraga and coworkers (2000). Each sample was measured in triplicate.
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession numbers AY224046–AY224077.
Acknowledgments
We thank Marlis Nissen and Petra von Wiegen for endosperm isolation, Hans Hartings for advice on AFLP techniques, Epiphysage Group, Oviedo University for the HPCE analysis, and Vincenzo Rossi, Alexandra Mant, Silvio Zaina, Birger L. Møller, and Franceso Salamini for critical reading of the manuscript. This work was supported by a grant from the Danish National Research Foundation and partially by a grant from Ministero dell'Istruzione, dell'Università e della Ricerca–ll Fondo per gli Investimenti della Ricerca di Base (RBNE01TYZF) to A.V.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Gertrud Lund (gel@kvl.dk).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.017780.
References
- Adams, S., Vinkenoog, R., Spielman, M., Dickinson, H.G., and Scott, R.J. (2000). Parent-of-origin effects on seed development in Arabidopsis thaliana require DNA methylation. Development 127, 2493–2502. [DOI] [PubMed] [Google Scholar]
- Alleman, M., and Doctor, J. (2000). Genomic imprinting in plants: Observations and evolutionary implications. Plant Mol. Biol. 43, 147–161. [DOI] [PubMed] [Google Scholar]
- Baroux, C., Spillane, C., and Grossniklaus, U. (2002). Genomic imprinting during seed development. Adv. Genet. 46, 165–214. [DOI] [PubMed] [Google Scholar]
- Bianchi, M.W., and Viotti, A. (1988). DNA methylation and tissue-specific transcription of the storage protein gene of maize. Plant Mol. Biol. 11, 203–214. [DOI] [PubMed] [Google Scholar]
- Birchler, J.A. (1993). Dosage analysis of maize endosperm development. Annu. Rev. Genet. 27, 181–204. [DOI] [PubMed] [Google Scholar]
- Birchler, J.A., and Hart, J.R. (1987). Interaction of endosperm size factors in maize. Genetics 117, 309–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird, A.P., and Wolffe, A.P. (1999). Methylation-induced repression—Belts, braces, and chromatin. Cell 24, 451–454. [DOI] [PubMed] [Google Scholar]
- Bushell, C., Spielman, M., and Scott, R.J. (2003). The basis of natural and artificial postzygotic hybridization barriers in Arabidopsis species. Plant Cell 15, 1430–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman, V., Forrester, L., Sanford, J., Hastie, N., and Rossant, J. (1984). Cell lineage-specific undermethylation of mouse repetitive DNA. Nature 307, 284–286. [DOI] [PubMed] [Google Scholar]
- Chaudhuri, S., and Messing, J. (1994). Allele-specific parental imprinting of dzr1, a posttranscriptional regulator of zein accumulation. Proc. Natl. Acad. Sci. USA 91, 4867–4871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhury, A.M., Ming, L., Miller, C., Craig, S., Dennis, E.S., and Peacock, W.J. (1997). Fertilization-independent seed development in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 94, 4223–4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danilevskaya, O.N., Hermon, P., Hantke, S., Muszynski, M.G., Kollipara, K., and Ananiev, E.V. (2003). Duplicated fie genes in maize: Expression pattern and imprinting suggest distinct functions. Plant Cell 15, 425–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dilkes, B.P., Dante, R.A., Coelho, C., and Larkins, B.A. (2002). Genetic analyses of endoreduplication in Zea mays endosperm: Evidence of sporophytic and zygotic maternal control. Genetics 160, 1163–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich, K.C. (1993). Characterization of DBPm, a plant protein that binds to DNA containing 5-methylcytosine. Biochim. Biophys. Acta 1172, 108–116. [DOI] [PubMed] [Google Scholar]
- Finnegan, E.J., Peacock, W.J., and Dennis, E.S. (1996). Reduced DNA methylation in Arabidopsis thaliana results in abnormal plant development. Proc. Natl. Acad. Sci. USA 93, 8449–8454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraga, M.F., Rodriguez, R., and Canal, M.J. (2000). Rapid quantification of DNA methylation by high performance capillary electrophoresis. Electrophoresis 21, 2990–2994. [DOI] [PubMed] [Google Scholar]
- Gierl, A., Lutticke, S., and Saedler, H. (1988). TnpA product encoded by the transposable element En-1 of Zea mays is a DNA binding protein. EMBO J. 20, 4045–4053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossniklaus, U., Vielle-Calzada, J.-P., Hoeppner, M.A., and Gagliano, W.B. (1998). Maternal control of embryogenesis by MEDEA, a polycomb-group gene in Arabidopsis. Science 280, 446–450. [DOI] [PubMed] [Google Scholar]
- Gruenbaum, Y., Naveh-Many, T., Cedar, H., and Razin, A. (1981). Sequence specificity of methylation in higher plant DNA. Nature 292, 860–862. [DOI] [PubMed] [Google Scholar]
- Guo, M., Rupe, M.A., Danilevskaya, O.N., Yang, X., and Hu, Z. (2003). Genome-wide mRNA profiling reveals heterochronic allelic variation and a new imprinted gene in hybrid maize endosperm. Plant J. 36, 30–44. [DOI] [PubMed] [Google Scholar]
- Haig, D., and Westoby, M. (1989). Parent-specific gene expression and the triploid endosperm. Am. Nat. 134, 147–155. [Google Scholar]
- Haig, D., and Westoby, M. (1991). Genomic imprinting in endosperm: Its effect on seed development in crosses between species, and between different ploidies of the same species, and its implications for the evolution of apomixis. Philos. Trans. Roy. Soc. London 333, 1–14. [Google Scholar]
- Inamdar, N.M., Ehrlich, K.C., and Ehrlich, M. (1991). CpG methylation inhibits binding of several sequence-specific DNA-binding proteins from pea, wheat, soybean and cauliflower. Plant Mol. Biol. 17, 111–123. [DOI] [PubMed] [Google Scholar]
- Jones, P.L., Veenstra, G.J., Wade, P.A., Vermaak, D., Kass, S.U., Landsberger, N., Strouboulis, J., and Wolffe, A.P. (1998). Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat. Genet. 19, 187–191. [DOI] [PubMed] [Google Scholar]
- Kass, S.U., Landsberger, N., and Wolffe, A.P. (1997). DNA methylation directs a time-dependent repression of transcription initiation. Curr. Biol. 7, 157–165. [DOI] [PubMed] [Google Scholar]
- Kermicle, J.L. (1970). Dependence of the R-mottled phenotype in maize on mode of sexual transmission. Genetics 66, 69–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita, T., Yadegari, R., Harada, J.J., Goldberg, R.B., and Fischer, R.L. (1999). Imprinting of the MEDEA Polycomb gene in the Arabidopsis endosperm. Plant Cell 11, 1945–1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowles, R.V., Yerk, G.L., Haas, K.M., and Phillips, R.L. (1997). Maternal effects influencing DNA endoreduplication in developing endosperm of Zea mays. Genome 40, 798–805. [DOI] [PubMed] [Google Scholar]
- Leblanc, O., Pointe, C., and Hernandez, M. (2002). Cell cycle progression during endosperm development in Zea mays depends on parental dosage effects. Plant J. 32, 1057–1066. [DOI] [PubMed] [Google Scholar]
- Lin, B.Y. (1982). Association of endosperm reduction with parental imprinting in maize. Genetics 100, 475–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, B.Y. (1984). Ploidy barrier to endosperm development. Genetics 107, 103–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund, G., Ciceri, P., and Viotti, A. (1995. a). Maternal-specific demethylation and expression of specific alleles of zein genes in the endosperm of Zea mays L. Plant J. 8, 571–581. [DOI] [PubMed] [Google Scholar]
- Lund, G., Messing, J., and Viotti, A. (1995. b). Endosperm-specific demethylation and activation of specific alleles of α-tubulin genes of Zea mays L. Mol. Gen. Genet. 246, 716–722. [DOI] [PubMed] [Google Scholar]
- Luo, M., Bilodeau, P., Dennis, E.S., Peacock, W.J., and Chaudhury, A. (2000). Expression and parent-of-origin effects for FIS2, MEA, and FIE in the endosperm and embryo of developing Arabidopsis seeds. Proc. Natl. Acad. Sci. USA 97, 10637–10642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer, P., Niedenhof, I., and ten Lohuis, M. (1994). Evidence for cytosine methylation of non-symmetrical sequences in transgenic Petunia hybrida. EMBO J. 1, 32084–2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nan, X., Ng, H.H., Johnson, C.A., Laherty, C.D., Turner, B.M., Eisenman, R.N., and Bird, A. (1998). Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature 393, 386–389. [DOI] [PubMed] [Google Scholar]
- Oakeley, E.J., Podesta, A., and Jost, J.P. (1997). Developmental changes in DNA methylation of the two tobacco pollen nuclei during maturation. Proc. Natl. Acad. Sci. USA 94, 11721–11725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohad, N., Margossian, L., Hsu, Y.C., Williams, C., Repetti, P., and Fischer, R.L. (1996). A mutation that allows endosperm development without fertilization. Proc. Natl. Acad. Sci. USA 93, 5319–5324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prendergast, G.C., and Ziff, E.B. (1991). Methylation-sensitive sequence-specific DNA binding by the Myc basic region. Science 251, 186–189. [DOI] [PubMed] [Google Scholar]
- Rabinowicz, P.D., Schutz, K., Dedhia, N., Yordan, C., Parnell, L.D., Stein, L., McCombie, W.R., and Martienssen, R.A. (1999). Differential methylation of genes and retrotransposons facilitates shotgun sequencing of the maize genome. Nat. Genet. 23, 305–308. [DOI] [PubMed] [Google Scholar]
- Razin, A., Webb, C., Szyf, M., Yisraeli, J., Rosenthal, A., Naveh-Many, T., Sciaky-Gallili, N., and Cedar, H. (1984). Variations in DNA methylation during mouse cell differentiation in vivo and in vitro. Proc. Natl. Acad. Sci. USA 81, 2275–2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reik, W., Dean, W., and Walter, J. (2001). Epigenetic reprogramming in mammalian development. Science 293, 1089–1093. [DOI] [PubMed] [Google Scholar]
- Reina-Lopez, G.E., Simpson, J., and Ruiz-Herrera, J. (1997). Differences in DNA methylation patters are detectable during the dimorphic transition of fungi by amplification of restriction polymorphisms. Mol. Gen. Genet. 253, 703–710. [DOI] [PubMed] [Google Scholar]
- Rossant, J., Sanford, J.P., Chapman, V.M., and Andrews, G.K. (1986). Undermethylation of structural gene sequences in extraembryonic lineages of the mouse. Dev. Biol. 7, 567–573. [DOI] [PubMed] [Google Scholar]
- Scholten, S., Lorz, H., and Kranz, E. (2002). Paternal mRNA and protein synthesis coincides with male chromatin decondensation in maize zygotes. Plant J. 32, 221–231. [DOI] [PubMed] [Google Scholar]
- Scott, R.J., Spielman, M., Bailey, J., and Dickinson, H.G. (1998). Parent-of-origin effects on seed development in Arabidopsis thaliana. Development 125, 3329–3341. [DOI] [PubMed] [Google Scholar]
- Sleutels, F., and Barlow, D.P. (2002). The origins of genomic imprinting in mammals. Adv. Genet. 46, 119–163. [DOI] [PubMed] [Google Scholar]
- Springer, P.S., Holding, D.R., Groover, A., Yordan, C., and Martienssen, R.A. (2000). The essential Mcm7 protein PROLIFERA is localized to the nucleus of dividing cells during the G1 phase and is required maternally for early Arabidopsis development. Development 127, 1815–1822. [DOI] [PubMed] [Google Scholar]
- Stäiger, D., Kaulen, H., and Schell, J. (1989). A CACGTG motif of the Antirrhinum majus chalcone synthase promoter is recognized by an evolutionary conserved nuclear protein. Proc. Natl. Acad. Sci. USA 86, 6930–6934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturaro, M., and Viotti, A. (2001). Methylation of the Opaque2 box in zein genes is parent-dependent and affects O2 DNA binding activity in vitro. Plant Mol. Biol. 46, 549–560. [DOI] [PubMed] [Google Scholar]
- Takagi, N., and Sasaki, M. (1975). Preferential inactivation of the paternally derived X chromosome in the extraembryonic membranes of the mouse. Nature 256, 640–642. [DOI] [PubMed] [Google Scholar]
- Vielle-Calzada, J.P., Baskar, R., and Grossniklaus, U. (2000). Delayed activation of the paternal genome during seed development. Nature 404, 91–94. [DOI] [PubMed] [Google Scholar]
- Vielle-Calzada, J.P., Thomas, J., Spillane, C., Coluccio, A., Hoeppner, M.A., and Grossniklaus, U. (1999). Maintenance of genomic imprinting at the Arabidopsis medea locus requires zygotic DDM1 activity. Genes Dev. 13, 2971–2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinkenoog, R., Spielman, M., Adams, S., Fischer, R.L., Dickinson, H.G., and Scott, R.J. (2000). Hypomethylation promotes autonomous endosperm development and rescues postfertilization lethality in fie mutants. Plant Cell 12, 2271–2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos, P., et al. (1995). AFLP: A new technique for DNA fingerprinting. Nucleic Acids Res. 23, 4407–4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weijers, D., Geldner, N., Offringa, R., and Jurgens, G. (2001). Seed development: Early paternal gene activity in Arabidopsis. Nature 414, 709–710. [DOI] [PubMed] [Google Scholar]
- Yadegari, R., Kinoshita, T., Lotan, O., Cohen, G., Katz, A., Nakashima, K., Harada, J.J., Goldberg, R.B., Fischer, R.L., and Ohad, N. (2000). Mutations in the FIE and MEA genes that encode interacting polycomb proteins cause parent-of-origin effects on seed development by distinct mechanisms. Plant Cell 12, 2367–2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, D.L., Ehrlich, K.C., Supakar, P.C., and Ehrlich, M. (1989). A plant DNA-binding protein that recognizes 5-methylcytosine residues. Mol. Cell. Biol. 9, 1351–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]