Abstract
Mass spectrometry imaging and profiling of individual cells and subcellular structures provide unique analytical capabilities for biological and biomedical research, including determination of the biochemical heterogeneity of cellular populations and intracellular localization of pharmaceuticals. Two mass spectrometry technologies—secondary ion mass spectrometry (SIMS) and matrix assisted laser desorption ionization mass spectrometry (MALDI MS)—are most often used in micro-bioanalytical investigations. Recent advances in ion probe technologies have increased the dynamic range and sensitivity of analyte detection by SIMS, allowing two- and three-dimensional localization of analytes in a variety of cells. SIMS operating in the mass spectrometry imaging (MSI) mode can routinely reach spatial resolutions at the submicron level; therefore, it is frequently used in studies of the chemical composition of subcellular structures. MALDI MS offers a large mass range and high sensitivity of analyte detection. It has been successfully applied in a variety of single-cell and organelle profiling studies. Innovative instrumentation such as scanning microprobe MALDI and mass microscope spectrometers enable new subcellular MSI measurements. Other approaches for MS-based chemical imaging and profiling include those based on near-field laser ablation and inductively-coupled plasma MS analysis, which offer complementary capabilities for subcellular chemical imaging and profiling.
Keywords: secondary ion mass spectrometry, matrix assisted laser desorption/ionization, subcellular profiling, elemental imaging, mass cytometry, tissue imaging
1. Introduction
Visualization is arguably the single most powerful capability we possess for analyzing the physical, chemical, and biological world around us. Though we as humans image the world primarily by observing interactions of matter with light in a narrow wavelength range, technology has increased our ability to use a greatly expanded portion of the electromagnetic spectrum, higher-order interactions of light with matter [1], and even to image by other phenomena such as compression waves through matter and the propensity of electrons to tunnel through it. While most imaging techniques provide chemical information about a subject, nearly all leave some ambiguity about its exact chemical composition. This is especially true for biological structures that are made up of a myriad of distinct compounds comprised largely of the same handful of elements, and where biomolecules of differing function may be similar in easily-visualized characteristics such as visible light absorbance profiles. Labeling biomolecules with a distinct and easily-detected aid, such as a radionuclide or fluorescent tag, is an effective and widely-used solution for selective imaging; however, this requires targeting of known molecules and also limits the output to preselected compounds. If one goal of visualization techniques is to detect and subsequently identify a broad range of chemical species that are present in a sample, including those heretofore unknown, chemical labeling is not the answer.
As an alternative to labeling for chemical characterization, one of the most successful analytical methods of the 20th century has been mass spectrometry (MS) with MS/MS capabilities. A half century ago, MS was shown to be useful as an imaging method [2]; now known as mass spectrometry imaging (MSI), it can be performed with secondary ion mass spectrometry (SIMS), matrix-assisted laser desorption ionization (MALDI), and other ionization sources [3–7]. Typically, the technique involves acquiring separate mass spectra in a spatially defined grid, then applying mass filters to the resulting spectra to generate ion images that reveal the distribution of specific chemical species. Acquisition can be accomplished in microprobe mode [8] by scanning a focused laser, ion beam, or sampling inlet across the sample surface, or in microscope mode [9] where the entire field of view is sampled at once and ions are conducted to a position-sensitive detector via a mass analyzer, which preserves spatial information. MSI is a powerful microscopic chemical imaging tool for several reasons: It offers the chemical specificity and versatility inherent to MS, analytes of interest need not be preselected or even known prior to analysis, and the number of co-registered ion images producible from a single MSI experiment is limited (in theory) only by the number of distinct ions detected and resolved in the spectra, which can number from tens to thousands. MSI has been applied extensively in a variety of fields including biomedical research [10], microbiology [11], plant biology [12], and in the pharmaceutical sciences [13, 14]. For procedural details, the reader is directed to the following resources that describe multiple protocols for a diverse array of MSI applications and techniques [15, 16].
Despite recent progress in MSI instrumentation, sample preparation, and data analysis methods, further advances are needed, especially as interest grows in the ability to reliably visualize the distributions of intact molecules at micron and submicron resolutions. Beyond the challenge of designing suitable micro- or nanoprobes for this purpose, sensitivity becomes a major issue. Specifically, the sampled surface area diminishes rapidly with a decrease in probe radius; a 50 nm circular probe can interrogate only 0.01% the surface area of a 5 μm spot. Therefore, a 10,000-fold increase in some combination of instrument sensitivity and volume sampled (e.g., as a deeper voxel) is required in order to detect a given analyte at the same mass fraction. The sensitivity issue is compounded by the matrix suppression effect (discussed in detail later), a general scarcity of larger molecules (e.g., proteins) relative to metabolites and endogenous inorganic ions, and the low sampling efficiency of many microprobe ionization sources, especially for thick insulating specimens [17, 18]. The combination of these issues effectively limits cell-scale MSI to a subset of abundant biomolecules, leaving much room for improvement.
In this review we focus on the reported cell and subcellular applications of MSI, innovative analytical instrumentation and methods that enable such studies, and current efforts to address the major challenges facing technological advances.
2. Secondary ion mass spectrometry (SIMS)
Developed in the 1960s, SIMS was the first mass spectrometric technique applied to chemical imaging [2] and has been used to image a broad range of biological specimens, including single cells [19]. Generally offering the highest spatial resolution (<50 nm) among the MSI approaches, it can provide relative [20] and absolute [21] quantitative chemical information. The scope of this discussion is confined to SIMS imaging applied at cellular length scales; a recent review by Boxer et al. [22] covers SIMS fundamentals and thoroughly examines broader biological applications.
SIMS is a surface analysis method that involves bombarding the sample with a beam of energetic primary ions (e.g., Cs+ or O−) in order to induce desorption of intact molecules, fragments, and atoms from the first few nanometers of the sample surface. This desorption event—termed sputtering—produces predominantly neutral species but also cations and anions. Secondary ions of a single polarity are extracted and subsequently transferred into a mass analyzer, usually a magnetic and/or electrostatic sector or a time-of-flight (TOF) analyzer, although quadrupole ion traps [23] and hybrid quadrupole-TOF analyzers [24] have also been effective in enabling MS/MS functionality. SIMS is a relatively hard ionization method compared with other ion sources such as ESI or MALDI. Detectable ions are typically limited to a narrow mass range of only a few hundred Daltons; however, cluster ion sources (e.g, C60+ and Bi3+) have effectively extended this limit to ~2 kDa [25]. Matrix enhancement has also been shown to extend the mass range considerably [26]. Fragments can frequently be characteristic of a particular biomolecular species or class, such as the choline “head group” (m/z 86), corresponding to any lipid of the phosphatidylcholine or sphingomyelin classes, or the C5H9+ fatty acid tail fragment, both observed in abundance from cell membranes [27]. CH− and CN− ions also generate ubiquitous biological background signals (and can in fact be produced by post-desorption recombination [28]), which is useful in pixel normalization procedures [29].
2.1. Dynamic SIMS
Dynamic SIMS is a well-developed technique for cellular MSI, and a recent article by Chandra [30] in the Encyclopedia of Mass Spectrometry provides a good overview of its history, preparation protocols, and current capabilities. The approach implements a constant, high fluence (>1013 primary ions/cm2 delivered to sample) bombardment of the sample by primary ions followed by analysis of resulting secondary ions, oftentimes by electric and or magnetic sector instruments. Although capable of providing excellent lateral resolution and imaging sensitivity on the order of parts-per-million [31], dynamic SIMS breaks many of the covalent bonds of larger biomolecule constituents. The result is a mass spectral output of mainly monatomic secondary ions along with some small organic fragments such as CN− and CH− [32]. Instrumentation usually allows collection of a small handful of selected ions simultaneously from a sample, e.g., the “new generation” nanoSIMS from CAMECA has up to seven detectors, each of which can be calibrated to a single m/z window [31]. Due to these limitations, dynamic SIMS ion images are frequently correlated with results from cell investigations using other high-resolution imaging techniques, including electron [33, 34], atomic force [35], and fluorescence microscopy [36, 37], in order to draw more meaningful conclusions.
Nonetheless, a large amount of information can be obtained from dynamic SIMS data alone. Elemental sulfur and phosphorous ion maps can be used to demarcate the general localization of proteins (with sulfur-containing cysteine and methionine residues) and nucleic acids (by phosphorous in the backbone) within cells, thereby providing a coarse molecular map upon which additional ion images can be superimposed [38]. Certain endogenous inorganic ions (e.g., Na+, K+, Mg2+, Ca2+, and Cl−) can reveal much about the physiological state of cells, such as membrane potential, membrane transport, and mitosis [30]. In SIMS, the K+:Na+ ratio within cells can also generally guide the selection of intact cultured specimens for imaging since this ratio will be approximately ten for healthy cells, whereas it will be lower for those that have been damaged or lysed [34].
In more specific applications, interactions of Ca2+ and Mg2+ with chromatin have been studied by dynamic SIMS in correlation with immunofluorescence in order to label scaffold proteins, determine a binding ratio for each ion, and show that the calcium exclusively localizes with the scaffold protein and also causes structural deformation when depleted [37]. This is particularly interesting when considered alongside a separate SIMS imaging study of dividing human glioblastoma tumor cells in culture showing that calcium is notably absent, specifically from the mitotic spindle region during metaphase [39], and also in comparison with normal dividing cells, which actually concentrate the calcium in the same region [34]. The depth-resolving capability of dynamic SIMS was crucial in these studies as it allowed separation of the spindle region from the outer cytoplasm, which if observed together, would not have shown the localization. Label-free dynamic SIMS has been informative in research on other disease states that involve characteristic endogenous elements. In Alzheimer’s disease for example, the subcellular localization of iron was mapped to specific subcellular compartments in the hippocampus of human patients, and also shown to colocalize with calcium in mineralized amyloid deposits in a mouse model [40, 41]. Soft tissue biomineralization (calcification) has been studied successfully with SIMS in correlation with transmission electron microscopy (TEM) and immunostaining to reveal organelle-level location of hydroxyapatite crystals (detected as a CaO− fragment) in epithelial cells [42].
Dynamic SIMS can be effective for high-resolution mapping of any unlabeled exogenous molecule that contains a unique element within the biological system under investigation. The element is used as a built-in tracer of the molecule, and in some cases, can elucidate related processes. This has become a powerful and well-established method for following the localization of drugs at size scales ranging from tissue sections [43] to single organelles [44]. Early work showed that halogens from steroids and pyrimidine analogs can be located within cells [45], and extensive SIMS investigations of boron neutron capture therapy (BNCT) cancer treatment pharmaceuticals have been successfully performed, largely by Chandra and colleagues [46, 47], for over two decades. In BNCT, glioblastoma tumor cells are loaded with boron atoms and then irradiated with low-energy neutrons; the neutrons cause no significant damage to the surrounding tissue but result in alpha decay of the boron nuclei, which are destructive within a ~10 μm radius, i.e., primarily to the cells containing them [44]. BNCT agents are a broad class of drugs designed to deliver the boron atoms preferentially to glioblastoma cells with a partition ratio (relative to healthy tissue) adequate to selectively destroy the tumor. Effectiveness, therefore, hinges critically on where the drug localizes within a tumor cell population and in what partition ratio, as well as where exactly the boron accumulates within the tumor cells. Because the cell nucleus is more sensitive to boron decay, it is the ideal target for the therapy. Since dynamic SIMS can quantitatively map the boron directly at subcellular resolution, it has been used successfully to evaluate the efficacy of a variety of BNCT pharmaceuticals [48, 49], compare responses to the treatment by multiple cell types within a cell co-culture simultaneously and quantitatively [50], and compare boron concentrations amongst subcellular regions [44].
Absolute quantification usually is not possible with MSI due to a variety of factors that affect the intensities of ion signals recorded across cells or tissues. In some cases, however, it has been successfully performed using dynamic SIMS. Inductively-coupled plasma atomic emission spectroscopy is used to create a set of “relative sensitivity factors” needed for quantitative MSI measurements of the elemental species of interest (e.g., B, Ca, K, Mg, Na) [47]. These factors permit absolute quantitation of the targeted species by comparing their signals (on each detector pixel) to that of carbon. The harsh nature of the high-current SIMS beam works advantageously because it ionizes the elements from various tissue or intracellular regions indiscriminately. This calibration approach has been used to determine the effect of a drug’s infusion time on treatment outcome [43], and for quantitative comparison of drug uptake by cells in different stages of the cell cycle [21].
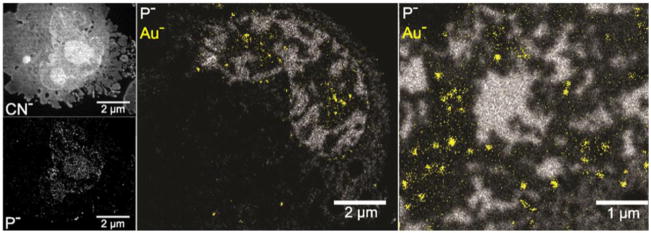
The SIMS approaches reported for BNCT research can be applied to the study of other drugs, provided the drug in question contains a unique element or can be labeled with one. In fact, SIMS imaging has been used in an assortment of other drug studies. Examples include investigating the anti-cancer drug Cisplatin’s cytotoxicity as it relates to cell calcium stores [51], evaluating the use of platinum-containing delivery agents for hadron therapy cancer treatment [52], and visualization of the copper-chelating drug ATN-224, used in prostate cancer treatment and located within cells by its characteristic MoS− and MoS2− fragments [36]. Dynamic SIMS imaging has proven useful for direct cancer detection, for example, by imaging the distribution of the melanoma marker iodobenzamide by iodine anion formation in mice, shown to be confined to submicron-sized melanosomes [53]. Another recent study by Wedlock et al. [38] focused on evaluating the distribution of a new potential gold-containing anti-cancer drug in order to elucidate its functional mechanism; dynamic SIMS was used in conjunction with energy-filtered electron microscopy in order to visualize the gold complex amongst cell contents. Remarkably clear, high-resolution SIMS images, such as those shown in Fig. 1, revealed not only that the gold penetrates the treated cells and aggregates within the nucleus and cytoplasm, but also that the cell morphology changes significantly after treatment. More importantly, the perfectly-registered ion images acquired by SIMS revealed that the gold complex was colocalized with sulfur rather than phosphate within the cell, suggesting that it functions through interference with thiol-containing proteins rather than direct interaction with DNA.
Figure 1. High resolution dynamic SIMS imaging reveals gold complex distribution within single human breast cancer cells.
The CN− ion (top left) reveals overall cellular structure while P− (bottom left) shows nucleic acid distribution. In other cells after treatment with a gold-containing anticancer complex, superimposed Au− and P− images (center and right) indicate that the Au accumulates as ~200 nm aggregates in and around the nucleus, segregated clearly from the DNA. Adapted from ref. [38]. Reproduced by permission of The Royal Society of Chemistry.
In cases where endogenous or exogenous species of interest do not include characteristic elements by which they may be easily detected, stable isotope labeling can used in dynamic SIMS experiments. Most commercial instruments provide sufficient mass resolution to discern nearly-isobaric ions; for example, 12C1H− and 13C−, which differ by 4.4 mDa [54]. If naturally-occurring but rare isotopes such as 13C and 15N can be incorporated into a drug, nutrient, or other molecule of interest, then they can be used to trace and quantify analyte distribution at a subcellular level. Lechene and colleagues have designed an impressive assortment of experiments utilizing isotopic labels with nanoSIMS to examine free fatty acid uptake by cultured adipocytes [29], molecular nitrogen fixation by bacteria in single cells of marine worms [20], protein synthesis in hair cells [55], cellular metabolism and DNA strand segregation [56]. In these studies, relative quantitation was performed by ratiometric measurements of enriched and naturally-abundant labeled elements, e.g., 13C:12C. Isotopic labeling has also been notably used by Kraft et al. [32, 57] to study phase separation of lipid membranes in supported bilayer models. Because isotopic-labeling does not significantly alter the shape of the lipids forming the membrane as a fluorescent tag might, SIMS can be used to study the unperturbed system. This work has been highly informative in the elucidation of nanoscale lipid layer organization, and we expect future studies will aid in understanding the function (and dysfunction) of membrane-bound proteins. Most recently this work has been extended to examine the effect of cholesterol incorporation on membrane behavior with SIMS and correlated atomic force microscopy (AFM) measurements [35].
2.2. Static SIMS (TOF-SIMS)
When SIMS is conducted with a primary ion fluence of <1013 ions/cm2 (the static limit), it is considered to be sampling an unperturbed surface, i.e., any particular sample area is probed by a primary ion only once. This is referred to as static SIMS or sometimes TOF-SIMS, since the pulsed primary ion beam is conveniently coupled directly to a TOF analyzer, which can acquire full mass spectra at each image pixel rather than monitoring a few preselected ions as with sector-based dynamic SIMS instruments [31]. Less material is ultimately ablated per unit surface area for ionization and detection (due to the static limit), which makes MSI of low-abundance analyte distributions more difficult and limited to a small dynamic range of concentrations as per-pixel ion counts are frequently in the single digits. However, static SIMS is well suited to the study of intact molecular ions and larger biomolecular fragments such as peptides [25], since it almost exclusively interrogates undamaged sample surfaces and measures across a broad, continuous mass range. With microprobes focusable to a submicron diameter, but not quite the <50 nm achievable with dynamic SIMS due to the surface availability of the analyte, static SIMS provides relatively more chemical information at lower but still “subcellular” lateral resolutions.
2.2.1 Static SIMS instrumentation advances
Polyatomic “cluster” primary ion beam sources are major recent innovations in static SIMS imaging, enabling new studies from a range of biological samples. In contrast with traditional monatomic sources (Cs+ and Ga+), cluster ions (SF5+, C60+, Bi3+ and Au3+) generally produce softer ionization and much greater secondary ion yields, especially at higher masses [58], thus extending the practical mass limit of static SIMS to approximately 2,000 Da [25] and permitting acquisition of more molecular information. While Bi3+ clusters have been shown to provide superior sensitivity and lateral resolution (>100 nm) [59, 60] with biomolecules, the C60+ cluster source [61] uniquely produces far less damage to organic samples upon impact in comparison with other cluster sources [62]. As a result, the C60+ primary ion beam can be applied at a fluence far exceeding the static limit in order to strip or “etch” material from a sample surface in fairly uniform layers. This enabled TOF-SIMS instruments to investigate subsurface cell contents and even generate three-dimensional (3-D) images [63].
Many state-of-the-art TOF-SIMS instruments are now equipped with such cluster sources, which enhance biological sample analysis [64]. Carado and colleagues [24, 65] added a C60+ source to a commercial MALDI quadrupole-TOF hybrid mass spectrometer (QSTAR XL, SCIEX), enabling molecular SIMS experiments to benefit from both the relatively high mass resolution (>12,000) and MS/MS (CID) capabilities of the spectrometer’s native configuration. These are both highly useful features for investigating complex biological samples such as cells that produce dense, information-rich spectra in the wider mass range now accessible, as many detected molecular ions could potentially be further identified by MS/MS. Another impressive instrumental design, called the “J-105 3D chemical imager”, has been introduced as a product of several groups in collaboration with Ionoptika, Ltd. (Manchester, UK) [66–68]; it includes several features known to be useful for successful cell-level SIMS imaging. The instrument utilizes a time-focusing ion buncher [67] to maximize sensitivity, in conjunction with a coaxial TOF analyzer to enable accurate (5 ppm), sensitive, and high-mass resolution (7000) measurements at a simultaneously-high (stated 200 nm beam focus limit) spatial resolution using an Au3+ cluster primary beam. A C60+ beam serves a dual purpose: as an alternative primary ion source and as an in-source etching and sputter-cleaning tool. The sophisticated sample handling system on the J-105 includes an Ar-filled glove box for air-free preparation and a cryo-cooled stage with a “cryoshield” apparatus to inhibit surface contamination by ice, as well as a semi-automatic freeze-fracture system to expose cell contents under vacuum [69]. Initial results indicate that the design indeed permits exceptional cellular chemical imaging to be performed in two and three dimensions with cluster SIMS [68]. More recently a C60+ source has been coupled with an FT-ICR analyzer for high (100,000) mass resolution, sub-ppm mass accuracy and CID MS/MS capability; this combination promises great subcellular MSI potential once its sensitivity is improved to permit a suitably small microprobe [70].
2.2.2. Static SIMS applications to cellular imaging
The application of static SIMS to cellular MSI has been mostly confined to a small assortment of detectable endogenous molecules comprised primarily of membrane phospholipids (reviewed in [71]), and a few other membrane-localized small molecules such as cholesterol [72, 73] and vitamin E [27]. These molecules are abundant on the surface of cells and in tissues of interest (e.g., brain [74] and spinal cord [75]), easily ionized, and often produce characteristic fragments that can be used to improve image quality [72]. The capability of static SIMS to image the distribution of membrane constituents, lipids in particular, is highly valuable in bioanalytical studies since few other methods can do so at all, and if a label is used, it may significantly perturb native chemistry. Microbes made a convenient initial static SIMS imaging biological model owing to their appropriate (~1 μm) cell size and tolerance to various chemical treatments during sample preparation [76, 77]. More recently, membrane composition analysis of cultured mammalian cells has also become possible. Ostrowski et al. [78] used static SIMS to visualize the distribution of membrane cholesterol in cultured macrophages and by correlation with fluorescence imaging, they relatively quantified the difference between control and cholesterol-enriched cellular populations in a single imaging experiment. Lipids have been relatively quantified between cell populations as well as between the membrane and cytoplasm of individual cultured cells [79]. Mammalian cardiac tissue and individual isolated cardiac cells have been imaged to visualize both tissue-level and subcellular localization of phosphocholine and other lipids such as cardiolipins, revealing complimentary distributions and correlating them with particular fatty acids [73]. Lanekoff et al. [80] used a stable-isotope labeling approach to determine membrane lipid exchange rates in PC12 cells by imaging the membrane to trace deuterated phospholipids that were incorporated into the cells from culture medium at various time points. Low rates of exchange were observed, leading the authors to conclude that small membrane compositional changes may result in large changes in cellular activity, e.g., exocytosis, as observed in their previous work with a comparable cell treatment [81]. Using a similar labeling approach, Kraft and coworkers [82] employed static SIMS to extensively study phase separation (a.k.a. “lipid rafts”) in supported model lipid membranes, applying principal component analysis (PCA) to enhance phospholipid distribution image contrast by utilizing subtle differences in species-specific fragmentation profiles.
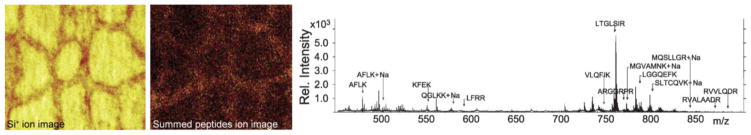
Prior successes with the visualization of membrane components notwithstanding, cellular MSI of the other macromolecule classes—carbohydrates, nucleic acids, and proteins (including peptides)—continues to be a main focus of development efforts for biological static SIMS. Observations of small, nonspecific amino acid- and nucleic acid-related fragment ions may be used for broad subcellular-mapping purposes [83]. When single characteristic ions are not available to define subcellular regions, statistical analysis of MSI data has been employed to find analyte signatures specific for particular image regions using methods that include PCA, [69] k-means clustering [84], and maximum autocorrelation factor [85], among others. Sulfur and phosphor stable isotope labels can also be used to distinguish different co-cultured cells by static SIMS [86]. However, imaging of the localization of intact large molecules such as proteins has been limited due to their in-source fragmentation, relatively low abundance, and difficulties in sputtering large molecules from highly intertwined cellular matrix and ion suppression effects. Nevertheless, instrumental and method developments have led to some significant and promising results. Altelaar et al. [26] were able to detect the neuropeptide APGWamide from Lymnaea stagnalis nervous tissue and image its distribution at a 3 μm spatial resolution with SIMS, and Komatsu et al. [25] demonstrated that human serum albumin deposited in a thin film could be digested in situ with acidified trypsin microdroplets to produce peptides in the 500–2000 Da mass range. These peptides were detectable with a Bi3+ cluster source. Nygren and colleagues [87] have shown that subcellular protein mapping is possible with a similar approach, as shown by their results in Fig. 2. Thyroglobulin, a 660 kDa protein produced in the thyroid gland, was digested on-tissue by trypsin and then over a dozen resulting tryptic peptides were detected simultaneously with <60 ppm mass accuracy. This allowed for protein identification by database search (MS-Digest, ProteinProspector) and also high-resolution (3 μm) mapping of the protein distribution, which was revealed to be localized heterogeneously around the epithelial border of pig thyroid follicle cells but not inside them. This serves as a valid proof-of-principle for the simultaneous analyte identification and visualization of the subcellular localization of large proteins by static SIMS. Piwowar and coworkers [88] have described a promising “top-down” approach to characterizing various subcellular regions by fractionating them via centrifugation and then profiling each fraction by static SIMS to obtain “reference spectra.” In theory, a library of these spectra can be used to identify subcellular features in chemical images as well as changes in analyte profiles corresponding to cellular activity.
Figure 2. Static SIMS imaging of a large protein at cell-scale resolution.
Thyroglobulin (660 kDa) in thyroid gland tissue is visualized using static matrix-enhanced SIMS after on-tissue trypsin digest. An Si+ ion image (left) of sectioned tissue reveals cell morphology since removal of colloid within the cells exposes the underlying silicon substrate. Summed signals of the detected tryptic peptides generates an ion image (middle) indicating protein localization along the epithelial cell borders. A mass spectrum (right) from the tissue on the right shows labeled tryptic peptides in the m/z 450–900 range. Images are represented in false-color scale ranging from black (low signal) through red to yellow (high signal); field of view is 500 × 500 μm. Adapted with permission from ref. [87], copyright 2010, John Wiley and Sons.
Ion signal suppression is a significant obstacle to detecting and quantifying analytes in chemically complex samples. This is clearly illustrated by the relatively small assortment of biomolecules that have been imaged by SIMS to date, compared with the large number of compounds known to be present within cells at concentrations that should be detectable using current SIMS instrumentation. Gunnarsson et al. [59] have shown that individual 300 nm artificial vesicles can be discerned by their characteristic lipid fragments using cluster static SIMS, suggesting that separate analysis of naturally-occurring intracellular organelles on the same size scale is entirely possible if signal suppression effects can be addressed. Fundamental studies on this subject have shown that the propensity for individual analytes to ionize over others in a desorbed mixture can be judged broadly by their relative gas-phase basicities [89]; consequently, the composition of the sputtered cloud of particles can greatly enhance or suppress individual ion yields. A single pulse of primary ions during the SIMS sputter event may generate only a few tens of unique gas-phase molecules; nevertheless, a staggeringly complex assortment of possible interactions and additional factors, such as local acidic proton availability and density in the region of desorption, add yet another layer of complexity [90]. Practical approaches to improving the coverage of cellular analytes, in part by addressing the signal suppression issue, have therefore been to increase ionization efficiency overall and/or bias ionization in favor of the analytes of interest. Metal-assisted SIMS and matrix-enhanced SIMS improve detection of a variety of bioanalytes by coating sample surfaces with either a thin layer of metal such as Ag or Au, or a typical MALDI matrix compound such as 2,5-dihydroxybenzoic acid [26, 91, 92]. Water can also act as an effective matrix for analytes in SIMS investigations. Controlling the sublimation-condensation equilibrium of water on the sample surface during analysis has been shown to enhance ion yields by up to two-fold during depth profiling experiments [93, 94]. Surprisingly, leaking water vapor directly over the sample has also produced significant enhancement of analyte detection [95]. Post-desorption photoionization is another way to generate more detectable ions from the neutral molecules that comprise the vast majority of desorption events [96]. None of these techniques has provided the cure-all to the signal suppression problem, but in combination with new instrumental configurations and sample preparation techniques, they may assist in the detection of additional species of interest.
2.2.3. Sample preparation for static SIMS imaging of cells
A successful SIMS imaging experiment hinges on a properly selected and executed sample preparation method, perhaps more than the data acquisition process itself. Much of the recent progress for static SIMS of biomolecules has been in the area of method development to improve the information yield of the technique. Unlike MALDI, laser ablation-inductively coupled plasma (LA-ICP) MS, or other ionization methods performed routinely at atmospheric pressure [17, 97, 98], SIMS is strictly a high-vacuum approach; therefore, cultured cell or tissue samples must be dry and/or frozen prior to analysis, leading to loss of cell viability. At the same time, several other general conditions must be met in order to produce high quality images: (i) the features of interest must be present at the sample surface; (ii) external contamination from culture medium, air, and other sources must be removed; (iii) chemical composition and cellular structure must be preserved, at least for the analyte of interest; (iv) cell morphology should be unperturbed; and (v) ion signal suppression should be minimized by removing suppressive constituents such as undesired salts and lipids, but without otherwise compromising the previously-stated conditions. This final condition is particularly difficult to meet given the inherent complexity of biological systems and the fact that wet chemical treatments, which are routine for non-imaging MS or tissue-level imaging, usually result in some delocalization of small molecules. This makes such approaches less suitable for high resolution imaging, especially when small and diffusible analytes are investigated [99].
One popular solution is cryogenic freeze-fracture preparation of cultured cells, introduced as a sample preparation approach for electron microscopy investigations in 1957 [100] and adapted for dynamic SIMS imaging by Chandra et al. in 1986 [99, 101]. This technique involves flash-freezing the cells while sandwiched between two substrates (e.g., Si wafers), then prying them apart in vacuum or inert gas in order to randomly expose various internal cell surfaces [102]. Freeze-fracture sample preparation has been frequently used in cell-level static SIMS imaging [103], including MSI of red blood cells [104] and tumor cells [48], and was also instrumental in the success of experiments investigating lysozyme lipid membrane permeability [105]. After flash-freezing, which reduces damage to cells by avoiding large ice crystal formation [106], samples may either be freeze-dried to remove water content or maintained at low temperature in a frozen-hydrated state in which water content remains present throughout the SIMS analysis. The frozen-hydrated approach requires careful temperature control at subsequent stages, but recent instrumentation advances such as the “mouse trap” spring-loaded in vacuo fracturing apparatus [107] streamline the procedure.
Comparison with other preparation techniques such as freeze drying and formalin fixation indicates that frozen-hydrated preparation provides the best physical and chemical cell preservation [66] and increases ion yields due to the water matrix enhancement effect [93, 108]. However, it was also recently reported that frozen-hydrated sample preparation may result in localized chemical image artifacts and inconsistent sputter rates and that freeze-drying samples avoids these drawbacks while adequately preserving cell morphology, chemical and elemental composition [109]. Simple non-cryogenic “wash and dry” and chemical substitution approaches for sample preparation have also been investigated and found to be successful by some groups, [110, 111], although it was noted by Berman et al. [110] that drying may lead to cell collapse and cell cytosol spreading, resulting in delocalization of diffusible species. Sjovall et al. [112] demonstrated subcellular “imprint-imaging” where cells are pressed onto an Ag surface, thereby transferring membrane components, which can then be imaged at sub-micron lateral resolution; nuclear and plasma membrane compositions of red blood cells were compared using this technique. Overall, these sample preparation approaches have varying procedural complexities and offer different levels of preservation for the physical and chemical integrity of specimens; therefore, the suitability of each must be evaluated on a per-case basis.
2.3. In-source milling, etching and three-dimensional imaging with SIMS
Aside from the traditional freeze-fracture technique, more recent work has demonstrated that clean internal cell planes can alternatively be exposed by in-SIMS milling or etching approaches, each of which have their own advantages. Grazing-incidence fast ion bombardment (FIB) milling has been used by Szakal et al. [84] to expose the contents of freeze-dried (but not fractured) HeLa cells, followed by imaging with static SIMS. Alternating mill and analysis cycles allowed depth-imaging of the cells in precise 20 nm increments. Weber et al. [33] took the same approach to expose and image a cross section of 1-μm diameter Bacillus bacterial spores by nanoSIMS, revealing elemental differences in the bacterial core, cortex, and coat layers. The latter work also demonstrated that the more involved “lift-out” FIB method often employed in TEM sample preparation allows consecutive removal of thin layers during 3-D sample analysis by MSI. In addition to these milling techniques, the low-damage impact of the C60+ ion primary beam on biomaterials has allowed its use as a high-angle rastered etching tool. Impressively, Kurczy and coworkers [113] used the approach to selectively remove a several nanometers-thick contamination overcoat to reveal nearly-unperturbed chemical features, such as patterned cholesterol film, without causing excessive signal loss. This surface-cleaning method was also shown to be highly effective with cultured and freeze-dried cells, removing culture medium-related surface contamination and improving cell:substrate contrast markedly [64]. The same study also compared various wash methods involved in cellular sample preparation and concluded that ammonium acetate provided the best overall results, in agreement with similar studies that used either this as a wash or ammonium formate [110, 111]. The etching technique has more recently been used to improve the identification of cells in co-culture by multivariate analysis of surface chemistry [114], and elsewhere to visualize the localization of antibiotics inside and outside of the bacteria Streptomyces coelicolor, demonstrating that one molecular species may be retained within the cell while another may not [115].
Milling and etching techniques can be combined with static SIMS analysis to produce 3-D ion images. Depth-resolved elemental ion image stacks have been reported for quite some time using dynamic SIMS instrumentation [39], but the ability of the C60+ beam to reliably and repeatedly etch an organic sample surface with a 12–30 nm depth resolution [116] now extends this approach to the detection of intact molecules and characteristic fragments. Fletcher and coworkers [117] reported early 3-D biomolecular imaging results using the C60+ beam in alternating etch/analysis modes, and the same group has now detected and visualized characteristic cytoplasmic and nucleus-abundant ions (phosphocholine at m/z 184 and adenine at m/z 136.1) in much smaller HeLa cells [69]. The C60+ beam can also be used to etch in conjunction with a second “analytical” beam, such as Au3+ or Bi3+, for higher imaging resolution and sensitivity, and has been applied for 3-D visualization of single thyroid carcinoma cells [118]. These initial proof-of-principle studies are encouraging, but reliable and informative 3-D MSI still faces a major challenge in the accurate 3-D localization of detected ions in some samples; for example, in cells that have significant topographical variations and heterogeneous ultrastructural consistency. Simple depth calibration has been performed by the detection of signal from a substrate (such as silicon) underlying a sample [83] or post-SIMS AFM measurements [64], and the effects of variables such as sample temperature, primary beam energy, and primary beam angle have been assessed [116, 119].
3. MALDI MS
Since its conception [120] and application to the analysis of large biomolecules [121], MALDI has rapidly grown to become one of the two most widely used ionization methods in biological MS alongside electrospray. MALDI incorporates analyte molecules into a matrix of organic substance crystals or liquid crystals and then irradiates the sample with a focused, pulsed or continuous laser beam. Absorption of the incident energy by the matrix leads to desorption of the analyte molecules and their ionization, often by gas-phase protonation or deprotonation reactions. The ions from the desorbed particle plume are then extracted from the source, analyzed (typically by TOF), and detected. MALDI MS has been effectively applied for analysis of proteins, peptides, lipids, DNA, and RNA, and is often the bioanalytical method of choice owing to its sensitivity, high impurity tolerance during analysis of complex mixtures, and ease of sample preparation.
3.1. Single-cell and subcellular MALDI MS profiling
In an MS profiling experiment, a single point (pixel) of a specimen is examined (MSI can be thought of as a set of MS profiling measurements performed at ordered array of locations). Several groups demonstrated in the mid-1990s that MALDI MS could be used effectively in profiling mode to interrogate the contents of individual cells [122–125]. In one such early experiment by Van Veelen and colleagues [122], large (<100 μm diameter soma) single neurons from L. stagnalis were either sampled in situ by micropipette or isolated and lysed into a small volume of matrix solution, dried on a conventional sample plate, and then investigated directly with MALDI MS. Both of these approaches produced remarkably high-quality spectra revealing both expected neuropeptides as well as unidentified species such as C-terminally elongated peptide variants. Jiménez and colleagues [126] took this work a step further by interrogating individual neurons in a simple neural circuit of L. stagnalis responsible for heartbeat modulation. They were able to show some variability in the peptidergic content of identified individual neurons, highlighting the value of MS profiling in revealing cell-to-cell heterogeneity. MS/MS analysis was used to sequence the peptides; the accuracy of this sequencing was supported by in situ hybridization, which showed that the transcripts responsible for the expression of the detected peptides were also present in the cells. This work established that beyond proof-of-principle, MS microanalysis techniques can yield important information regarding single-cell function. For example, single-cell measurements include the ability to relate peptides found in specific neurons to animal behaviors [127–130], as well as characterize rare post-translational modifications such as a D-amino acid in a peptide [131]. Peptide profiling experiments can be accomplished with smaller mammalian cells [132], and detailed protocols are now available to delineate the techniques [133]. MALDI MS profiling has also been used to profile metabolites in single HeLa cells and large microbes [134, 135]. Furthermore, recent work has shown that relative and even absolute quantitation is possible at the level of a few cells or even a single cell by various methods including isotopic labeling, succinic anhydride labeling and standard addition, with a 19 fmol limit of detection and a 64 fmol limit of quantitation demonstrated for the peptide cerebrin from small cell clusters via the standard addition approach [136].
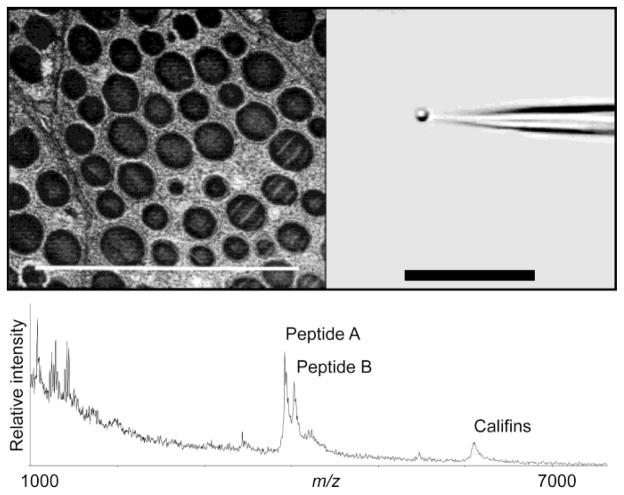
The contents of micron-sized organelles can be profiled using similar bioanalytical techniques. Individual secretory dense core vesicles from the atrial gland of Aplysia californica with 1–2 μm diameters and internal volumes of only a few hundred attoliters—known for containing high (mM) concentrations of peptide signaling molecules—were isolated by micropipette, deposited onto glass slides, rinsed quickly in NaCl-containing solution to remove externally-adsorbed material, then mixed with a picoliter amount of matrix and dried for analysis by MALDI MS, as shown in Fig. 3 [137]. Results revealed that many peptides from several genes were contained within individual vesicles; these peptide identities were confirmed by post-source decay fragmentation analysis of tissue-scale blots of the same gland from A. californica. Comparing the tissue blots to the single-vesicle profiles also revealed an informative difference: fully-processed califin peptides were detected from within individual vesicles while the blots showed yet-unassembled peptide subunits elsewhere. Clearly, with the proper collection techniques, MS profiling becomes a powerful tool for biological inquiry at the subcellular level.
Figure 3. Single-organelle mass profiling with MALDI.
Individual secretory granules from the A. californica atrial gland (left, TEM of tissue cross-section) are isolated manually using micropipette (right, video image) and prepared with matrix for MALDI-TOF analysis. Several peptides contained within the single granule are detected as shown in the mass spectrum (bottom). Scale bars are 10 μm. Adapted with permission from ref. [137], Macmillan Publishers Ltd: Nature Biotechnology, copyright 2000.
3.2. Cell-scale MALDI MSI
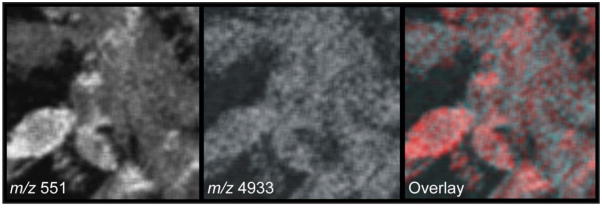
In a typical MALDI MSI experiment the laser probe is rastered across a sample, much like the primary ion beam during SIMS imaging. Caprioli et al. [3] were among the first to describe in detail the MALDI MSI approach and demonstrate its applicability to investigations of biological specimens. The low spatial resolution of subcellular MALDI MSI of larger cultured A. californica neurons was reported six years later [138]. The neurons were profiled at subcellular resolution with individually-deposited matrix microdroplets and also imaged at 50 μm lateral resolution to show that different relative peptide concentrations were observed in the cell soma as compared to its neurite outgrowths, which extended a few hundred microns from the soma. Recent MALDI instrumentation advances have resulted in a growing body of MSI studies performed at “cellular length scales” (usually defined as single micron-scale resolution or better), with true cell-per-pixel or subcellular resolutions achieved in a number of these cases. Instrumental features required to achieve subcellular MSI include the ability to focus the laser beam microprobe to submicron diameters and higher laser fluence in order to ionize adequate amounts of analytes from small sample areas [139]. One response to the probe focusing issue was presented by Altelaar and colleagues [140]; using a stigmatic SIMS instrument (TRIFT II, PHI Electronics) modified for MALDI MSI, a defocused laser was used to illuminate a large region of a sample for microscope-mode imaging. Shown to achieve 4 μm lateral resolution, the instrument was used to image contrasting localization patterns of several peptides in rat, mouse and human pituitary glands. An alternative to stigmatic imaging for high spatial resolution is to use sophisticated optics to focus the laser into a near-diffraction-limited microprobe directed at the sample. Several decades ago, Hillenkamp et al. [141] demonstrated this approach for elemental analysis with the laser microprobe mass analyzer (or LAMMA) in which the laser was introduced coaxially (with respect to the ion extraction path) from behind the sample via an immersion lens and a vacuum glass window, ablating material directly into a TOF analyzer. Spengler and Hubert [142] reported a similar system, delivering the laser coaxially from the other direction via a central aperture bored through the quartz optical lenses. In this configuration, the laser is focusable to a 260 nm diameter, enabling subcellular resolution with a scanning microprobe-type instrument. The effective lateral imaging resolution becomes limited by sampling efficiency and was found to fall between 0.6–1.5 μm, depending on sample consistency. The instrument has been used to image human renal carcinoma cells at a two micron resolution and detecting masses up to 5 kDa, as shown in Fig. 4 [143]. This work demonstrates that subcellular biological imaging with MALDI is possible and in fact, offers rich chemical information beyond the small molecules and fragments observed by SIMS and other methods.
Figure 4. MALDI MSI at subcellular spatial resolution.
MALDI analysis of cultured human renal cancer cells allows visualization of analytes across an extended m/z range relative to SIMS; vapor deposition of matrix permits 2 μm effective spatial resolution. Two ion images are overlaid (m/z 551 in red false color and m/z 4933 in greyscale) to reveal differences in the profiles of adjacent cells. Field of view shown is 100 × 100 μm. Adapted with permission from ref. [143], copyright 2010, John Wiley and Sons.
Spengler’s group [144] has also recently introduced another instrument employing a similar optical setup applied to an atmospheric pressure (AP) MALDI source combined with either an ion trap or an FT-ICR analyzer for high-sensitivity or high-mass resolution imaging, respectively. Initial demonstrations showed that patterned biomolecules can be detected at biologically-relevant sensitivities—tens of attomoles per pixel—using either mass analyzer. More recent results include detection and identification of ten neuropeptides in mouse pituitary gland, and 5 μm lateral resolution which reveals cell-scale heterogeneity [145]. Similarly, Setou and colleagues [146] have described results from their own high-resolution AP-MALDI instrument, or “mass microscope,” which is actually a microprobe-mode imaging instrument capable of 4 μm resolution. They have used their mass microscope to visualize varying relative concentrations of membrane phosphatidylcholines in the retinal layers of the Ambystoma mexicanum salamander eye at 7 μm lateral resolution. The high lateral resolution of the instrument allows single cell-per-pixel acquisition of 15 μm-diameter retinal cells without dissociation from the tissue where they are closely packed [97].
3.3 Sample preparation for MALDI-MSI
In initial MALDI single-cell imaging experiments [138], matrix was simply applied in a single drop directly over the cells and quickly dried with heat. This sufficed for large frozen A. californica cells imaged at a 50 μm raster size; however, with the goal of imaging much smaller mammalian cells, MALDI-MSI has been pushed toward ever higher resolutions. Unfortunately, the lack of appropriate matrix application techniques has become a limiting factor and thus, is a focus of ongoing method development efforts. Conventional tissue-level wet application techniques fall short, either due to analyte delocalization, leading to losses in spatial resolution, or insufficient sensitivity resulting from poor analyte extraction. MALDI matrix sublimation, originally proposed by Hankin et al. [147], is a promising method of matrix application for subcellular imaging since it produces uniform microcrystals on the sample surface and therefore provides good spatial resolution. With a subsequent reconstitution/recrystallization step, sensitivity is improved [148]. This effectively decouples the two functions of conventional matrix application—analyte extraction and matrix crystallization/analyte inclusion—thus allowing better control of each step. Also beneficial is the highly homogeneous, reproducible microcrystal formation, as well as the capability to extend the detectable masses to tens of kDa [149]. Another approach has been to modify the dimensions of the sample itself by thaw-mounting it on a stretchable material such as Parafilm-M and then physically enlarging its area prior to matrix application [150, 151]. This can be done with tissue sections and cell cultures and not only enables single-cell measurements and MS/MS peptide sequencing, but original sample dimensions can also be reconstructed using a software tool [152]. Finally, it is worth noting that while alternative (non-organic) matrices such as gold nanoparticles [153] and functionalized metal nanoparticles [154] have not been reported for the MSI of individual cells or subcellular components, these particles are on the single-nanometer size scale and support analyte detection without formation of larger crystals; we expect to see these used for single cell MSI in the near future.
4. Other mass spectrometric techniques applied to single cell-scale measurements
Traditional optical systems used for focused laser probes in MALDI have been impressively refined for lateral submicron resolution and therefore subcellular imaging, but ultimately they are still diffraction-limited to approximately half their emission wavelength, which is on the scale of hundreds of nanometers. Fiber optic laser interfaces have been utilized for some time to desorb [139, 155], ionize [156], and photodissociate [157] analytes in conjunction with a variety of mass analyzers and this method of laser probe delivery presents an alternative approach to improving MSI resolution via near-field focusing techniques. In scanning near-field optical microscopy (SNOM), laser light is directed at a finely-sharpened metal tip that is placed within nanometers of the sample surface. This significantly enhances laser fluence delivered in the vicinity of the tip, allowing localized ablation from craters <200 nm in diameter and ~20 nm deep [158]. SNOM-MS can be performed at atmospheric pressure and ablated material (ions and neutrals) is sampled into the MS vacuum chamber with a closely-positioned orifice, after which additional ions may be generated from the abundant neutrals by electron impact ionization. A TOF-MS system equipped with this desorption/ionization apparatus allowed researchers to analyze atoms and molecules including acetylcholine or 2,5-dihydroxybenzoic acid [159]. Zhu, Zenobi and colleagues [17] have recently shown that SNOM-MS transfers approximately 10% of all ablated material into the mass spectrometer compared to the ~1% ion sampling efficiency of AP-MALDI, a significant improvement.
When elemental information is needed, the inductively-coupled plasma (ICP) ionization source [160] provides quantitative and sensitive (pg/g LOD) [161] elemental and isotopic data. ICP-MS data is not influenced by the sample matrix or ion signal suppression effects, even in complex and heterogeneous biological specimens [162]. By directing laser-ablated sample material into the plasma with a carrier gas such as argon, LA-ICP-MS has been successfully applied to tissue-level imaging studies and has been reviewed by Becker [163]. Recent work by this same group [98] has also shown that combining ICP-MS with a modified laser microdissection apparatus allows for practical imaging spatial resolutions of 3–5 μm and SNOM-focusing methods produce submicron-scale ablation [164], which can be expected to improve resolution further, possibly to below 100 nm [161]. This research direction offers the exciting possibility of a highly sensitive, quantitative nanoscale elemental imaging capability at atmospheric pressure, conceivably even on live cells in tissue or culture.
Near-field sampling methods could also be advantageous when coupled with nanostructure-initiator mass spectrometry (NIMS), a matrix-free desorption/ionization method that relies on liquid “initiator”-filled nanopores in the substrate beneath the sample to desorb and ionize analytes. NIMS boasts impressive 700 ymol limits of detection in ideal conditions and a mass limit of 30 kDa [165, 166]. It has been applied to biomolecular MSI at the tissue level to visualize cancer tumors [167], and can be performed using either laser or ion microprobes, with the latter providing an ultimate lateral resolution of ~150 nm. Near-field laser focusing methods could bring laser probe-based NIMS down to a competitive size scale and also allow high-resolution NIMS MSI at atmospheric pressure.
For high throughput whole-cell measurements, mass cytometry is another clever microanalytical approach introduced recently by Bandura et al. [168]. In mass cytometry, cells are first labeled with assorted multiatom elemental (e.g., lanthanide isotope) antibody tags which can number in the dozens. Cells are then suspended in solution, nebulized and fed into an ICP source which disintegrates them completely; the resulting discrete ion packets are then analyzed by an orthogonal TOF and metal ions are detected at high-speed to generate 10+ spectra per individual cell. This promising new instrument allows absolute quantitation of tens of tagged target molecules with virtually no channel crosstalk and high-throughput rates of (theoretically) 3000 cells/s.
5. Conclusions
Among the broad array of single-cell, spatially-resolved analysis techniques currently available, mass spectrometry imaging offers distinct advantages. It is capable of obtaining rich chemical information at the cellular level, and offers non-targeted specificity and multiplexed detection [169]. Continued method development and instrumentation advances represent a burgeoning analytical area that promises to rapidly increase the applicability of MS in the fields of biological and biomedical research as sensitivity, dynamic range and consequently, the variety of detectable biomolecules are all increased. The microsampling and imaging approaches discussed here comprise a diverse toolbox, effective for probing the composition of cells with various levels of breadth in chemical sensitivity and spatial resolution.
Subcellular MSI approaches predominantly use a variety of SIMS techniques. Dynamic SIMS offers high sensitivity with the best lateral resolution and has been especially useful for mapping the localization of compounds labeled with rare elements. This approach most notably allows researchers to determine how anti-cancer drugs and other compounds having therapeutic potential are distributed between and within the cells of humans and other multicellular organisms. Static SIMS, restricted traditionally to visualizing the distribution of small molecules and molecular fragments, has recently been augmented by the introduction of softer-ionization, higher-yield cluster ion sources and accompanying instrumental advances. As a result, greater molecular information, molecular depth profiling, and MS/MS identification of detected species is possible with traditionally-static SIMS instrumentation. Accordingly, this approach is now a rapidly advancing MSI platform with great potential for direct subcellular mapping of the localization of unlabeled biomolecules of interest. Although new single-cell sample preparation methods allow direct access to cell contents for molecular MSI, ion signal suppression effects continue to pose a major obstacle to thorough detection and mapping of cell composition. This issue is a major focus of development efforts. Newer primary ion sources being explored—argon cluster [170], electrospray droplet impact [171], massive gold cluster [172], and high-brightness C60 plasma [173]—offer promising alternatives to the existing ion source repertoire and may provide advantages such as softer ionization, higher secondary ion yields, or smoother etching for 3-D analysis. Work by Matsuo and colleagues [174–176] with a high-energy (MeV) copper primary ion beam also presents the possibility of SIMS operation at atmospheric pressure and therefore, live-cell SIMS imaging, which would be an interesting and noteworthy development.
MALDI-MS is well-established for tissue-level imaging and has become almost routine for sensitive subcellular profiling experiments. For biological MSI, MALDI offers the advantages of a wide molecular mass range and soft ionization for high intact molecular ion yields. Recent advances in both sample preparation methodologies and high-resolution MSI instrumentation have enabled its application at single-cell and subcellular spatial resolutions. MALDI-MSI has progressed to the point where visualization of bioanalyte distributions can be made more routine, accessible, and informative at these length scales. Likewise, laser ablation-based techniques such as SNOM-MS, LA-ICP-MS and NIMS hold realistic potential to become highly sensitive and quantitative subcellular sampling platforms. These nontraditional approaches can operate under atmospheric conditions and therefore hold real promise for achieving the exciting ability to profile live cells.
Highlights.
Advances in MSI technologies allow imaging at cellular resolution in tissues
SIMS enables imaging of elements/small molecules at subcellular spatial resolution
New SIMS cluster ion sources greatly enhance the ability to probe cell contents
MALDI allows sensitive single-cell and organelle profiling over a broad mass range
Acknowledgments
This work was supported by Award No. P30 DA018310 from the National Institute on Drug Abuse (NIDA) and the Department of Energy by Award No. DE-SC0006642. The content is solely the responsibility of the authors and does not necessarily represent the official views of the funding agencies. We also thank Stephanie Baker for assistance with manuscript preparation and reviewers for helpful feedback.
Footnotes
The authors declare they have no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Scarcelli G, Yun SH. Confocal brillouin microscopy for three-dimensional mechanical imaging. Nat Photon. 2008;2:39–43. doi: 10.1038/nphoton.2007.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Castaing R, Slodzian G. Microanalyse par emission ionique secondaire. J Microscopie. 1962;1:395–410. [Google Scholar]
- 3.Caprioli RM, Farmer TB, Gile J. Molecular imaging of biological samples: Localization of peptides and proteins using MALDI-TOF MS. Anal Chem. 1997;69:4751–60. doi: 10.1021/ac970888i. [DOI] [PubMed] [Google Scholar]
- 4.Wiseman JM, Ifa DR, Song Q, Cooks RG. Tissue imaging at atmospheric pressure using desorption electrospray ionization (DESI) mass spectrometry. Angew Chem Int Ed. 2006;45:7188–92. doi: 10.1002/anie.200602449. [DOI] [PubMed] [Google Scholar]
- 5.Kertesz V, Ford MJ, Van Berkel GJ. Automation of a surface sampling probe/electrospray mass spectrometry system. Anal Chem. 2005;77:7183–9. doi: 10.1021/ac0510742. [DOI] [PubMed] [Google Scholar]
- 6.Nemes P, Vertes A. Atmospheric-pressure molecular imaging of biological tissues and biofilms by laesi mass spectrometry. J Vis Exp. 2010:1–4. doi: 10.3791/2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Laskin J, Heath BS, Roach PJ, Cazares L, Semmes OJ. Tissue imaging using nanospray desorption electrospray ionization mass spectrometry. Anal Chem. 2011;84:141–8. doi: 10.1021/ac2021322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liebl H. Ion microprobe mass analyzer. J Appl Phys. 1967;38:5277–83. [Google Scholar]
- 9.Schueler B, Sander P, Reed DA. A time-of-flight secondary ion microscope. Vacuum. 1990;41:1661–4. [Google Scholar]
- 10.Chughtai K, Heeren RMA. Mass spectrometric imaging for biomedical tissue analysis. Chem Rev. 2010;110:3237–77. doi: 10.1021/cr100012c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Watrous JD, Dorrestein PC. Imaging mass spectrometry in microbiology. Nat Rev Micro. 2011;9:683–94. doi: 10.1038/nrmicro2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaspar S, Peukert M, Svatos A, Matros A, Mock H-P. MALDI-imaging mass spectrometry – an emerging technique in plant biology. Proteomics. 2011;11:1840–50. doi: 10.1002/pmic.201000756. [DOI] [PubMed] [Google Scholar]
- 13.Sugiura Y, Setou M. Imaging mass spectrometry for visualization of drug and endogenous metabolite distribution: Toward in situ pharmacometabolomes. J Neuroimmune Pharmacol. 2010;5:31–43. doi: 10.1007/s11481-009-9162-6. [DOI] [PubMed] [Google Scholar]
- 14.Solon EG, Schweitzer A, Stoeckli M, Prideaux B. Autoradiography, MALDI-MS, and SIMS-MS imaging in pharmaceutical discovery and development. AAPS J. 2010;12:11–26. doi: 10.1208/s12248-009-9158-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rubakhin S, Sweedler J. Mass spectrometry imaging: Principles and protocols. Humana Press; New York: 2010. [Google Scholar]
- 16.Setou M. Imaging mass spectrometry : Protocols for mass microscopy. Tokyo: Springer; 2010. [Google Scholar]
- 17.Zhu L, Stadler J, Schmitz TA, Krumeich F, Zenobi R. Atmospheric pressure sampling for laser ablation based nanoscale imaging mass spectrometry: Ions or neutrals? J Phys Chem C. 2011;115:1006–13. [Google Scholar]
- 18.McCombie G, Knochenmuss R. Enhanced MALDI ionization efficiency at the metal-matrix interface: Practical and mechanistic consequences of sample thickness and preparation method. J Am Soc Mass Spectrom. 2006;17:737–45. doi: 10.1016/j.jasms.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 19.Galle P. Sur une nouvelle methode d’analyse cellulaire utilisant le phenomene d’emission ionique secondaire. Ann Phys Biol Med. 1970;42:83–94. [Google Scholar]
- 20.Lechene CP, Luyten Y, McMahon G, Distel DL. Quantitative imaging of nitrogen fixation by individual bacteria within animal cells. Science. 2007;317:1563–6. doi: 10.1126/science.1145557. [DOI] [PubMed] [Google Scholar]
- 21.Chandra S, Tjarks W, Lorey DR, Ii, Barth RF. Quantitative subcellular imaging of boron compounds in individual mitotic and interphase human glioblastoma cells with imaging secondary ion mass spectrometry (SIMS) J Microsc. 2008;229:92–103. doi: 10.1111/j.1365-2818.2007.01869.x. [DOI] [PubMed] [Google Scholar]
- 22.Boxer SG, Kraft ML, Weber PK. Advances in imaging secondary ion mass spectrometry for biological samples. Ann Rev Biophys. 2009;38:53–74. doi: 10.1146/annurev.biophys.050708.133634. [DOI] [PubMed] [Google Scholar]
- 23.Setou M, Kurabe N. Mass microscopy: High-resolution imaging mass spectrometry. J Electron Microsc. 2011;60:47–56. doi: 10.1093/jmicro/dfq079. [DOI] [PubMed] [Google Scholar]
- 24.Carado A, Passarelli MK, Kozole J, Wingate JE, Winograd N, Loboda AV. C60 secondary ion mass spectrometry with a hybrid-quadrupole orthogonal time-of-flight mass spectrometer. Anal Chem. 2008;80:7921–9. doi: 10.1021/ac801712s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Komatsu M, Murayama Y, Hashimoto H. Protein fragment imaging using ink jet printing digestion technique. Appl Surf Sci. 2008;255:1162–4. [Google Scholar]
- 26.Altelaar AFM, Minnen Jv, Jimenez CR, Heeren RMA, Piersma SR. Direct molecular imaging of Lymnaea stagnalis nervous tissue at subcellular spatial resolution by mass spectrometry. Anal Chem. 2005;77:735–41. doi: 10.1021/ac048329g. [DOI] [PubMed] [Google Scholar]
- 27.Monroe EB, Jurchen JC, Lee J, Rubakhin SS, Sweedler JV. Vitamin E imaging and localization in the neuronal membrane. J Am Chem Soc. 2005;127:12152–3. doi: 10.1021/ja051223y. [DOI] [PubMed] [Google Scholar]
- 28.McMahon G, Saint-Cyr HF, Lechene C, Unkefer CJ. Cn secondary ions form by recombination as demonstrated using multi-isotope mass spectrometry of 13C- and 15N-labeled polyglycine. J Am Soc Mass Spectrom. 2006;17:1181–7. doi: 10.1016/j.jasms.2006.04.031. [DOI] [PubMed] [Google Scholar]
- 29.Kleinfeld AM, Kampf JP, Lechene C. Transport of 13C-oleate in adipocytes measured using multi imaging mass spectrometry. J Am Soc Mass Spectrom. 2004;15:1572–80. doi: 10.1016/j.jasms.2004.07.010. [DOI] [PubMed] [Google Scholar]
- 30.Chandra S. Subcellular imaging of cells and tissues with dynamic secondary ion mass spectrometry. In: Gross ML, Caprioli RM, editors. The Encyclopedia of Mass Spectrometry. Elsevier; 2010. pp. 469–80. [Google Scholar]
- 31.CAMECA NanoSIMS 50/50l. SIMS microprobe for ultra fine feature analysis. 2011 Oct 23; http://www.cameca.fr/instruments-for-research/nanosims.aspx.
- 32.Kraft ML, Weber PK, Longo ML, Hutcheon ID, Boxer SG. Phase separation of lipid membranes analyzed with high-resolution secondary ion mass spectrometry. Science. 2006;313:1948–51. doi: 10.1126/science.1130279. [DOI] [PubMed] [Google Scholar]
- 33.Weber PK, Graham GA, Teslich NE, Chan WM, Ghosal S, Leighton TJ, et al. NanoSIMS imaging of bacillus spores sectioned by focused ion beam. J Microsc. 2010;238:189–99. doi: 10.1111/j.1365-2818.2009.03336.x. [DOI] [PubMed] [Google Scholar]
- 34.Chandra S. Studies of cell division (mitosis and cytokinesis) by dynamic secondary ion mass spectrometry ion microscopy: LLC-PK1 epithelial cells as a model for subcellular isotopic imaging. J Microsc. 2001;204:150–65. doi: 10.1046/j.1365-2818.2001.00944.x. [DOI] [PubMed] [Google Scholar]
- 35.Anderton CR, Lou K, Weber PK, Hutcheon ID, Kraft ML. Correlated AFM and nanoSIMS imaging to probe cholesterol-induced changes in phase behavior and non-ideal mixing in ternary lipid membranes. Biochim Biophys Acta. 2011;1808:307–15. doi: 10.1016/j.bbamem.2010.09.016. [DOI] [PubMed] [Google Scholar]
- 36.Lau KH, Christlieb M, SchrÖder M, Sheldon H, Harris AL, Grovenor CRM. Development of a new bimodal imaging methodology: A combination of fluorescence microscopy and high-resolution secondary ion mass spectrometry. J Microsc. 2010;240:21–31. doi: 10.1111/j.1365-2818.2010.03380.x. [DOI] [PubMed] [Google Scholar]
- 37.Strick R, Strissel PL, Gavrilov K, Levi-Setti R. Cation–chromatin binding as shown by ion microscopy is essential for the structural integrity of chromosomes. J Cell Biol. 2001;155:899–910. doi: 10.1083/jcb.200105026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wedlock LE, Wedlock MRK, Cliff JB, Filgueira L, Saunders M, Berners-Price SJ. Visualising gold inside tumour cells following treatment with an antitumour gold(i) complex. Metallomics. 2011;3:917–25. doi: 10.1039/c1mt00053e. [DOI] [PubMed] [Google Scholar]
- 39.Chandra S. 3D subcellular SIMS imaging in cryogenically prepared single cells. Appl Surf Sci. 2004;231–232:467–9. [Google Scholar]
- 40.Quintana C, Wu TD, Delatour B, Dhenain M, Guerquin-Kern JL, Croisy A. Morphological and chemical studies of pathological human and mice brain at the subcellular level: Correlation between light, electron, and nanoSIMS microscopies. Microsc Res Tech. 2007;70:281–95. doi: 10.1002/jemt.20403. [DOI] [PubMed] [Google Scholar]
- 41.Quintana C, Bellefqih S, Laval JY, Guerquin-Kern JL, Wu TD, Avila J, et al. Study of the localization of iron, ferritin, and hemosiderin in Alzheimer’s disease hippocampus by analytical microscopy at the subcellular level. J Struct Biol. 2006;153:42–54. doi: 10.1016/j.jsb.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 42.Azari F, Vali H, Guerquin-Kern JL, Wu TD, Croisy A, Sears SK, et al. Intracellular precipitation of hydroxyapatite mineral and implications for pathologic calcification. J Struct Biol. 2008;162:468–79. doi: 10.1016/j.jsb.2008.03.003. [DOI] [PubMed] [Google Scholar]
- 43.Smith DR, Chandra S, Barth RF, Yang W, Joel DD, Coderre JA. Quantitative imaging and microlocalization of boron-10 in brain tumors and infiltrating tumor cells by SIMS ion microscopy: Relevance to neutron capture therapy. Cancer Res. 2001;61:8179–87. [PubMed] [Google Scholar]
- 44.Chandra S, Kabalka GW, Lorey DR, Ii, Smith DR, Coderre JA. Imaging of fluorine and boron from fluorinated boronophenylalanine in the same cell at organelle resolution by correlative ion microscopy and confocal laser scanning microscopy. Clin Cancer Res. 2002;8:2675–83. [PubMed] [Google Scholar]
- 45.Berry JP, Galle P, Chassoux D, Escaig F, Linarez-Cruz LG, Lespinats G. Mapping of intracellular halogenous molecules by low and high resolution SIMS microscopy. Biol Cell. 1992;74:93–8. doi: 10.1016/0248-4900(92)90013-q. [DOI] [PubMed] [Google Scholar]
- 46.Chandra S, Morrison GH. Imaging ion and molecular transport at subcellular resolution by secondary ion mass spectrometry. Int J Mass Spectrom Ion Processes. 1995;143:161–76. [Google Scholar]
- 47.Ausserer WA, Ling YC, Chandra S, Morrison GH. Quantitative imaging of boron, calcium, magnesium, potassium, and sodium distributions in cultured cells with ion microscopy. Anal Chem. 1989;61:2690–5. doi: 10.1021/ac00199a002. [DOI] [PubMed] [Google Scholar]
- 48.Kabalka GW, Shaikh AL, Barth RF, Huo T, Yang W, Gordnier PM, et al. Boronated unnatural cyclic amino acids as potential delivery agents for neutron capture therapy. Appl Radiat Isot. 2011;69:1778–81. doi: 10.1016/j.apradiso.2011.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kabalka GW, Yao ML, Marepally SR, Chandra S. Biological evaluation of boronated unnatural amino acids as new boron carriers. Appl Radiat Isot. 2009;67:S374–S9. doi: 10.1016/j.apradiso.2009.03.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lorey DR, Ii, Morrison GH, Chandra S. Dynamic secondary ion mass spectrometry analysis of boron from boron neutron capture therapy drugs in co-cultures: Single-cell imaging of two different cell types within the same ion microscopy field of imaging. Anal Chem. 2001;73:3947–53. doi: 10.1021/ac0103266. [DOI] [PubMed] [Google Scholar]
- 51.Chandra S. Quantitative imaging of chemical composition in single cells by secondary ion mass spectrometry: Cisplatin affects calcium stores in renal epithelial cells. In: Rubakhin SS, Sweedler JV, editors. Mass spectrometry imaging: Principles and protocols. 1. New York: Humana Press; 2010. pp. 113–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Usami N, Furusawa Y, Kobayashi K, Lacombe S, Reynaud-Angelin A, Sage E, et al. Mammalian cells loaded with platinum-containing molecules are sensitized to fast atomic ions. Int J Radiat Biol. 2008;84:603–11. doi: 10.1080/09553000802199846. [DOI] [PubMed] [Google Scholar]
- 53.Guerquin-Kern JL, Hillion F, Madelmont JC, Labarre P, Papon J, Croisy A. Ultra-structural cell distribution of the melanoma marker iodobenzamide: Improved potentially of SIMS imaging in life sciences. BioMed Eng Online. 2004:3. doi: 10.1186/1475-925X-3-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Clode PL, Stern RA, Marshall AT. Subcellular imaging of isotopically labeled carbon compounds in a biological sample by ion microprobe (nanoSIMS) Microsc Res Tech. 2007;70:220–9. doi: 10.1002/jemt.20409. [DOI] [PubMed] [Google Scholar]
- 55.Zhang D-S, Piazza V, Perrin BJ, Rzadzinska AK, Poczatek JC, Wang M, et al. Multi-isotope imaging mass spectrometry reveals slow protein turnover in hair-cell stereocilia. Nature. 2012;481:520–4. doi: 10.1038/nature10745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Steinhauser ML, Bailey AP, Senyo SE, Guillermier C, Perlstein TS, Gould AP, et al. Multi-isotope imaging mass spectrometry quantifies stem cell division and metabolism. Nature. 2012;481:516–9. doi: 10.1038/nature10734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kraft ML, Fishel SF, Marxer CG, Weber PK, Hutcheon ID, Boxer SG. Quantitative analysis of supported membrane composition using the nanoSIMS. Appl Surf Sci. 2006;252:6950–6. [Google Scholar]
- 58.Kollmer F. Cluster primary ion bombardment of organic materials. Appl Surf Sci. 2004;231–232:153–8. [Google Scholar]
- 59.Gunnarsson A, Kollmer F, Sohn S, Höök F, Sjövall P. Spatial-resolution limits in mass spectrometry imaging of supported lipid bilayers and individual lipid vesicles. Anal Chem. 2010;82:2426–33. doi: 10.1021/ac902744u. [DOI] [PubMed] [Google Scholar]
- 60.Dubey M, Brison J, Grainger DW, Castner DG. Comparison of Bi1+, Bi3+ and C60+ primary ion sources for ToF-SIMS imaging of patterned protein samples. Surf Interface Anal. 2011;43:261–4. doi: 10.1002/sia.3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Weibel D, Wong S, Lockyer N, Blenkinsopp P, Hill R, Vickerman JC. A C60 primary ion beam system for time of flight secondary ion mass spectrometry: Its development and secondary ion yield characteristics. Anal Chem. 2003;75:1754–64. doi: 10.1021/ac026338o. [DOI] [PubMed] [Google Scholar]
- 62.Nygren H, Malmberg P, Nilsson M, Kriegeskotte C, Arlinghaus HF. The cytochemistry of anaplastic thyroid tumour cells and differentiated thyrocytes analyzed by TOF-SIMS and depth profiling. Appl Surf Sci. 2008;255:1285–8. [Google Scholar]
- 63.Breitenstein D, Rommel CE, Möllers R, Wegener J, Hagenhoff B. The chemical composition of animal cells and their intracellular compartments reconstructed from 3D mass spectrometry. Angew Chem Int Ed. 2007;46:5332–5. doi: 10.1002/anie.200604468. [DOI] [PubMed] [Google Scholar]
- 64.Brison J, Benoit DSW, Muramoto S, Robinson M, Stayton PS, Castner DG. ToF-SIMS imaging and depth profiling of hela cells treated with bromodeoxyuridine. Surf Interface Anal. 2011;43:354–7. doi: 10.1002/sia.3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Carado A, Kozole J, Passarelli M, Winograd N, Loboda A, Wingate J. Cluster SIMS with a hybrid quadrupole time-of-flight mass spectrometer. Appl Surf Sci. 2008;255:1610–3. doi: 10.1021/ac801712s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rabbani S, Fletcher JS, Lockyer NP, Vickerman JC. Exploring subcellular imaging on the buncher-ToF j105 3D chemical imager. Surf Interface Anal. 2011;43:380–4. [Google Scholar]
- 67.Hill R, Blenkinsopp P, Thompson S, Vickerman J, Fletcher JS. A new time-of-flight SIMS instrument for 3D imaging and analysis. Surf Interface Anal. 2011;43:506–9. [Google Scholar]
- 68.Fletcher JS, Rabbani S, Henderson A, Blenkinsopp P, Thompson SP, Lockyer NP, et al. A new dynamic in mass spectral imaging of single biological cells. Anal Chem. 2008;80:9058–64. doi: 10.1021/ac8015278. [DOI] [PubMed] [Google Scholar]
- 69.Fletcher JS, Rabbani S, Henderson A, Lockyer NP, Vickerman JC. Three-dimensional mass spectral imaging of HeLa-M cells – sample preparation, data interpretation and visualisation. Rapid Commun Mass Spectrom. 2011;25:925–32. doi: 10.1002/rcm.4944. [DOI] [PubMed] [Google Scholar]
- 70.Smith DF, Robinson EW, Tolmachev AV, Heeren RMA, Paša-Tolić L. C60 secondary ion Fourier transform ion cyclotron resonance mass spectrometry. Anal Chem. 2011;83:9552–6. doi: 10.1021/ac2023348. [DOI] [PubMed] [Google Scholar]
- 71.Passarelli MK, Winograd N. Lipid imaging with time-of-flight secondary ion mass spectrometry (ToF-SIMS) Biochim Biophys Acta. 2011;1811:976–90. doi: 10.1016/j.bbalip.2011.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Piehowski PD, Carado AJ, Kurczy ME, Ostrowski SG, Heien ML, Winograd N, et al. MS/MS methodology to improve subcellular mapping of cholesterol using TOF-SIMS. Anal Chem. 2008;80:8662–7. doi: 10.1021/ac801591r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jerigova M, Biro C, Kirchnerova J, Chorvatova A, Chorvat D, Lorenc D, et al. Chemical imaging of cardiac cell and tissue by using secondary ion mass spectrometry. Mol Imag Biol. 2010:1–10. doi: 10.1007/s11307-010-0460-4. [DOI] [PubMed] [Google Scholar]
- 74.Benabdellah F, Seyer A, Quinton L, Touboul D, Brunelle A, Laprévote O. Mass spectrometry imaging of rat brain sections: Nanomolar sensitivity with MALDI versus nanometer resolution by TOF-SIMS. Anal Bioanal Chem. 2010;396:151–62. doi: 10.1007/s00216-009-3031-2. [DOI] [PubMed] [Google Scholar]
- 75.Monroe EB, Annangudi SP, Hatcher NG, Gutstein HB, Rubakhin SS, Sweedler JV. SIMS and MALDI MS imaging of the spinal cord. Proteomics. 2008;8:3746–54. doi: 10.1002/pmic.200800127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ostrowski SG, Van Bell CT, Winograd N, Ewing AG. Mass spectrometric imaging of highly curved membranes during Tetrahymena mating. Science. 2004;305:71–3. doi: 10.1126/science.1099791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kurczy ME, Piehowski PD, Van Bell CT, Heien ML, Winograd N, Ewing AG. Mass spectrometry imaging of mating Tetrahymena show that changes in cell morphology regulate lipid domain formation. Proc Natl Acad Sci USA. 2010;107:2751–6. doi: 10.1073/pnas.0908101107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ostrowski SG, Kurczy ME, Roddy TP, Winograd N, Ewing AG. Secondary ion MS imaging to relatively quantify cholesterol in the membranes of individual cells from differentially treated populations. Anal Chem. 2007;79:3554–60. doi: 10.1021/ac061825f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kurczy ME, Kozole J, Parry SA, Piehowski PD, Winograd N, Ewing AG. Relative quantification of cellular sections with molecular depth profiling ToF-SIMS imaging. Appl Surf Sci. 2008;255:1158–61. doi: 10.1016/j.apsusc.2008.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lanekoff I, Sjövall P, Ewing AG. Relative quantification of phospholipid accumulation in the PC12 cell plasma membrane following phospholipid incubation using TOF-SIMS imaging. Anal Chem. 2011;83:5337–43. doi: 10.1021/ac200771g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Uchiyama Y, Maxson MM, Sawada T, Nakano A, Ewing AG. Phospholipid mediated plasticity in exocytosis observed in PC12 cells. Brain Res. 2007;1151:46–54. doi: 10.1016/j.brainres.2007.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Anderton CR, Vaezian B, Lou K, Frisz JF, Kraft ML. Identification of a lipid-related peak set to enhance the interpretation of TOF-SIMS data from model and cellular membranes. Surf Interface Anal. 2011 Early View. [Google Scholar]
- 83.Breitenstein D, Rommel CE, Stolwijk J, Wegener J, Hagenhoff B. The chemical composition of animal cells reconstructed from 2D and 3D ToF-SIMS analysis. Appl Surf Sci. 2008;255:1249–56. [Google Scholar]
- 84.Szakal C, Narayan K, Fu J, Lefman J, Subramaniam S. Compositional mapping of the surface and interior of mammalian cells at submicrometer resolution. Anal Chem. 2011;83:1207–13. doi: 10.1021/ac1030607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Henderson A, Fletcher JS, Vickerman JC. A comparison of PCA and MAF for ToF-SIMS image interpretation. Surf Interface Anal. 2009;41:666–74. [Google Scholar]
- 86.Tyler BJ, Takeno MM, Hauch KD. Identification and imaging of 15N labeled cells with ToF-SIMS. Surf Interface Anal. 2011;43:336–9. doi: 10.1002/sia.3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Nygren H, Malmberg P. High-resolution imaging and proteomics of peptide fragments by TOF-SIMS. Proteomics. 2010;10:1694–8. doi: 10.1002/pmic.200900782. [DOI] [PubMed] [Google Scholar]
- 88.Piwowar A, Fletcher J, Lockyer N, Vickerman J. Top-down approach to studying biological components using ToF-SIMS. Surf Interface Anal. 2011;43:265–8. [Google Scholar]
- 89.Jones E, Lockyer N, Kordys J, Vickerman J. Suppression and enhancement of secondary ion formation due to the chemical environment in static-secondary ion mass spectrometry. J Am Soc Mass Spectrom. 2007;18:1559–67. doi: 10.1016/j.jasms.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 90.Sostarecz AG, Cannon DM, Jr, McQuaw CM, Sun S, Ewing AG, Winograd N. Influence of molecular environment on the analysis of phospholipids by time-of-flight secondary ion mass spectrometry. Langmuir. 2004;20:4926–32. doi: 10.1021/la0496892. [DOI] [PubMed] [Google Scholar]
- 91.Altelaar AFM, Klinkert I, Jalink K, De Lange RPJ, Adan RAH, Heeren RMA, et al. Gold-enhanced biomolecular surface imaging of cells and tissue by SIMS and MALDI mass spectrometry. Anal Chem. 2006;78:734–42. doi: 10.1021/ac0513111. [DOI] [PubMed] [Google Scholar]
- 92.McDonnell LA, Piersma SR, Altelaar AFM, Mize TH, Luxembourg SL, Verhaert PDEM, et al. Subcellular imaging mass spectrometry of brain tissue. J Mass Spectrom. 2005;40:160–8. doi: 10.1002/jms.735. [DOI] [PubMed] [Google Scholar]
- 93.Piwowar AM, Fletcher JS, Kordys J, Lockyer NP, Winograd N, Vickerman JC. Effects of cryogenic sample analysis on molecular depth profiles with TOF-secondary ion mass spectrometry. Anal Chem. 2010;82:8291–9. doi: 10.1021/ac101746h. [DOI] [PubMed] [Google Scholar]
- 94.Piehowski PD, Kurczy ME, Willingham D, Parry S, Heien ML, Winograd N, et al. Freeze-etching and vapor matrix deposition for ToF-SIMS imaging of single cells. Langmuir. 2008;24:7906–11. doi: 10.1021/la800292e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Mouhib T, Delcorte A, Poleunis C, Bertrand P. Organic secondary ion mass spectrometry: Signal enhancement by water vapor injection. J Am Soc Mass Spectrom. 2010;21:2005–10. doi: 10.1016/j.jasms.2010.08.013. [DOI] [PubMed] [Google Scholar]
- 96.Brummel CL, Willey KF, Vickerman JC, Winograd N. Ion beam induced desorption with postionization using high repetition femtosecond lasers. Int J Mass Spectrom Ion Processes. 1995;143:257–70. [Google Scholar]
- 97.Roy MC, Nakanishi H, Takahashi K, Nakanishi S, Kajihara S, Hayasaka T, et al. Salamander retina phospholipids and their localization by MALDI imaging mass spectrometry at cellular size resolution. J Lipid Res. 2011;52:463–70. doi: 10.1194/jlr.M010546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Becker JS, Niehren S, Matusch A, Wu B, Hsieh HF, Kumtabtim U, et al. Scaling down the bioimaging of metals by laser microdissection inductively coupled plasma mass spectrometry (LMD-ICP-MS) Int J Mass spectrom. 2010;294:1–6. [Google Scholar]
- 99.Chandra S, Bernius MT, Morrison GH. Intracellular localization of diffusible elements in frozen-hydrated biological specimens with ion microscopy. Anal Chem. 1986;58:493–6. doi: 10.1021/ac00293a053. [DOI] [PubMed] [Google Scholar]
- 100.Steere RL. Electron microscopy of structural detail in frozen biological specimens. J Biophys Biochem Cytol. 1957;3:45–60. doi: 10.1083/jcb.3.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Chandra S, Morrison GH, Wolcott CC. Imaging intracellular elemental distribution and ion fluxes in cultured cells using ion microscopy: A freeze-fracture methodology. J Microsc. 1986;144:15–37. doi: 10.1111/j.1365-2818.1986.tb04670.x. [DOI] [PubMed] [Google Scholar]
- 102.Roddy TP, Cannon DM, Ostrowski SG, Winograd N, Ewing AG. Identification of cellular sections with imaging mass spectrometry following freeze fracture. Anal Chem. 2002;74:4020–6. doi: 10.1021/ac025574w. [DOI] [PubMed] [Google Scholar]
- 103.Colliver TL, Brummel CL, Pacholski ML, Swanek FD, Ewing AG, Winograd N. Atomic and molecular imaging at the single-cell level with TOF-SIMS. Anal Chem. 1997;69:2225–31. doi: 10.1021/ac9701748. [DOI] [PubMed] [Google Scholar]
- 104.Pacholski ML, Cannon DM, Ewing AG, Winograd N. Static time-of-flight secondary ion mass spectrometry imaging of freeze-fractured, frozen-hydrated biological membranes. Rapid Commun Mass Spectrom. 1998;12:1232–5. doi: 10.1002/(SICI)1097-0231(19980930)12:18<1232::AID-RCM319>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 105.Cliff B, Lockyer N, Jungnickel H, Stephens G, Vickerman JC. Probing cell chemistry with time-of-flight secondary ion mass spectrometry: Development and exploitation of instrumentation for studies of frozen-hydrated biological material. Rapid Commun Mass Spectrom. 2003;17:2163–7. doi: 10.1002/rcm.1169. [DOI] [PubMed] [Google Scholar]
- 106.Pegg DE. Principles of cryopreservation. In: DJ G, SG N, editors. Cryopreservation and freeze-drying protocols. 2. Totowa, NJ: Humana Press; 2007. pp. 39–57. [Google Scholar]
- 107.Lanekoff I, Kurczy ME, Hill R, Fletcher JS, Vickerman JC, Winograd N, et al. Time of flight mass spectrometry imaging of samples fractured in situ with a spring-loaded trap system. Anal Chem. 2010;82:6652–9. doi: 10.1021/ac101243b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Conlan XA, Lockyer NP, Vickerman JC. Is proton cationization promoted by polyatomic primary ion bombardment during time-of-flight secondary ion mass spectrometry analysis of frozen aqueous solutions? Rapid Commun Mass Spectrom. 2006;20:1327–34. doi: 10.1002/rcm.2446. [DOI] [PubMed] [Google Scholar]
- 109.Chandra S. Challenges of biological sample preparation for SIMS imaging of elements and molecules at subcellular resolution. Appl Surf Sci. 2008;255:1273–84. [Google Scholar]
- 110.Berman E, Fortson S, Checchi K, Wu L, Felton J, Wu K, et al. Preparation of single cells for imaging/profiling mass spectrometry. J Am Soc Mass Spectrom. 2008;19:1230–6. doi: 10.1016/j.jasms.2008.05.006. [DOI] [PubMed] [Google Scholar]
- 111.Malm J, Giannaras D, Riehle M, Gadegaard, Sjovall P. Fixation and drying protocols for the preparation of cell samples for time-of-flight secondary ion mass spectrometry analysis. Anal Chem. 2009;81:7197–205. doi: 10.1021/ac900636v. [DOI] [PubMed] [Google Scholar]
- 112.Sjövall P, Lausmaa J, Nygren H, Carlsson L, Malmberg P. Imaging of membrane lipids in single cells by imprint-imaging time-of-flight secondary ion mass spectrometry. Anal Chem. 2003;75:3429–34. doi: 10.1021/ac0207675. [DOI] [PubMed] [Google Scholar]
- 113.Kurczy ME, Piehowsky PD, Willingham D, Molyneaux KA, Heien ML, Winograd N, et al. Nanotome cluster bombardment to recover spatial chemistry after preparation of biological samples for SIMS imaging. J Am Soc Mass Spectrom. 2010;21:833–6. doi: 10.1016/j.jasms.2010.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Barnes CA, Brison J, Robinson M, Graham DJ, Castner DG, Ratner BD. Identifying individual cell types in heterogeneous cultures using secondary ion mass spectrometry imaging with C60 etching and multivariate analysis. Anal Chem. 2011;84:893–900. doi: 10.1021/ac201179t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Vaidyanathan S, Fletcher JS, Goodacre R, Lockyer NP, Micklefield J, Vickerman JC. Subsurface biomolecular imaging of streptomyces coelicolor using secondary ion mass spectrometry. Anal Chem. 2008;80:1942–51. doi: 10.1021/ac701921e. [DOI] [PubMed] [Google Scholar]
- 116.Lu C, Wucher A, Winograd N. Molecular depth profiling of buried lipid bilayers using C60-secondary ion mass spectrometry. Anal Chem. 2011;83:351–8. doi: 10.1021/ac102525v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Fletcher JS, Lockyer NP, Vaidyanathan S, Vickerman JC. TOF-SIMS 3D biomolecular imaging of xenopus laevis oocytes using buckminsterfullerene (C60) primary ions. Anal Chem. 2007;29:2199–206. doi: 10.1021/ac061370u. [DOI] [PubMed] [Google Scholar]
- 118.Malmberg P, Kriegeskotte C, Arlinghaus HF, Hagenhoff B, Holmgren J, Nilsson M, et al. Depth profiling of cells and tissues by using C60+ and SF5+ as sputter ions. Appl Surf Sci. 2008;255:926–8. [Google Scholar]
- 119.Sjövall P, Rading D, Ray S, Yang L, Shard AG. Sample cooling or rotation improves C60 organic depth profiles of multilayered reference samples: Results from a vamas interlaboratory study. J Phys Chem B. 2009;114:769–74. doi: 10.1021/jp9095216. [DOI] [PubMed] [Google Scholar]
- 120.Karas M, Bachmann D, Hillenkamp F. Influence of the wavelength in high-irradiance ultraviolet laser desorption mass spectrometry of organic molecules. Anal Chem. 1985;57:2935–9. [Google Scholar]
- 121.Tanaka K, Waki H, Ido Y, Akita S, Yoshida Y, Yoshida T, et al. Protein and polymer analyses up to m/z 100 000 by laser ionization time-of-flight mass spectrometry. Rapid Commun Mass Spectrom. 1988;2:151–3. [Google Scholar]
- 122.Van Veelen PA, Jiménez CR, Li KW, Wildering WC, Geraerts WPM, Tjaden UR, et al. Direct peptide profiling of single neurons by matrix-assisted laser desorption-ionization mass spectrometry. Org Mass Spectrom. 1993;28:1542–6. [Google Scholar]
- 123.Jiménez CR, Van Veelen PA, Li KW, Wildering WC, Geraerts WPM, Tjaden UR, et al. Neuropeptide expression and processing as revealed by direct matrix- assisted laser desorption ionization mass spectrometry of single neurons. J Neurochem. 1994;62:404–7. doi: 10.1046/j.1471-4159.1994.62010404.x. [DOI] [PubMed] [Google Scholar]
- 124.Li KW, Hoek RM, Smith F, Jiménez CR, Van Der Schors RC, Van Veelen PA, et al. Direct peptide profiling by mass spectrometry of single identified neurons reveals complex neuropeptide-processing pattern. J Biol Chem. 1994;269:30288–92. [PubMed] [Google Scholar]
- 125.Garden RW, Moroz LL, Moroz TP, Shippy SA, Sweedler JV. Excess salt removal with matrix rinsing: Direct peptide profiling of neurons from marine invertebrates using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. J Mass Spectrom. 1996;31:1126–30. doi: 10.1002/(SICI)1096-9888(199610)31:10<1126::AID-JMS403>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 126.Jiménez CR, Li KW, Dreisewerd K, Spijker S, Kingston R, Bateman RH, et al. Direct mass spectrometric peptide profiling and sequencing of single neurons reveals differential peptide patterns in a small neuronal network. Biochemistry (Mosc) 1998;37:2070–6. doi: 10.1021/bi971848b. [DOI] [PubMed] [Google Scholar]
- 127.Jing J, Sweedler JV, Cropper EC, Alexeeva V, Park JH, Romanova EV, et al. Feedforward compensation mediated by the central and peripheral actions of a single neuropeptide discovered using representational difference analysis. J Neurosci. 2010;30:16545–58. doi: 10.1523/JNEUROSCI.4264-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Proekt A, Vilim FS, Alexeeva V, Brezina V, Friedman A, Jing J, et al. Identification of a new neuropeptide precursor reveals a novel source of extrinsic modulation in the feeding system of Aplysia. J Neurosci. 2005;25:9637–48. doi: 10.1523/JNEUROSCI.2932-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Sweedler JV, Li L, Rubakhin SS, Alexeeva V, Dembrow NC, Dowling O, et al. Identification and characterization of the feeding circuit-activating peptides, a novel neuropeptide family of Aplysia. J Neurosci. 2002;22:7797–808. doi: 10.1523/JNEUROSCI.22-17-07797.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Floyd PD, Li L, Rubakhin SS, Sweedler JV, Horn CC, Kupfermann I, et al. Insulin prohormone processing, distribution, and relation to metabolism in Aplysia californica. J Neurosci. 1999;19:7732–41. doi: 10.1523/JNEUROSCI.19-18-07732.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Bai L, Romanova EV, Sweedler JV. Distinguishing endogenous d-amino acid-containing neuropeptides in individual neurons using tandem mass spectrometry. Anal Chem. 2011;83:2794–800. doi: 10.1021/ac200142m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Rubakhin SS, Churchill JD, Greenough WT, Sweedler JV. Profiling signaling peptides in single mammalian cells using mass spectrometry. Anal Chem. 2006;78:7267–72. doi: 10.1021/ac0607010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Rubakhin SS, Sweedler JV. Characterizing peptides in individual mammalian cells using mass spectrometry. Nat Protoc. 2007;2:1987–97. doi: 10.1038/nprot.2007.277. [DOI] [PubMed] [Google Scholar]
- 134.Amantonico A, Urban PL, Fagerer SR, Balabin RM, Zenobi R. Single-cell MALDI-MS as an analytical tool for studying intrapopulation metabolic heterogeneity of unicellular organisms. Anal Chem. 2010;82:7394–400. doi: 10.1021/ac1015326. [DOI] [PubMed] [Google Scholar]
- 135.Miura D, Fujimura Y, Yamato M, Hyodo F, Utsumi H, Tachibana H, et al. Ultrahighly sensitive in situ metabolomic imaging for visualizing spatiotemporal metabolic behaviors. Anal Chem. 2010;82:9789–96. doi: 10.1021/ac101998z. [DOI] [PubMed] [Google Scholar]
- 136.Rubakhin SS, Sweedler JV. Quantitative measurements of cell cell signaling peptides with single-cell MALDI MS. Anal Chem. 2008;80:7128–36. doi: 10.1021/ac8010389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Rubakhin SS, Garden RW, Fuller RR, Sweedler JV. Measuring the peptides in individual organelles with mass spectrometry. Nat Biotech. 2000;18:172–5. doi: 10.1038/72622. [DOI] [PubMed] [Google Scholar]
- 138.Rubakhin SS, Greenough WT, Sweedler JV. Spatial profiling with MALDI MS: Distribution of neuropeptides within single neurons. Anal Chem. 2003;75:5374–80. doi: 10.1021/ac034498+. [DOI] [PubMed] [Google Scholar]
- 139.Dreisewerd K, Schürenberg M, Karas M, Hillenkamp F. Influence of the laser intensity and spot size on the desorption of molecules and ions in matrix-assisted laser desorption/ionization with a uniform beam profile. Int J Mass Spectrom Ion Processes. 1995;141:127–48. [Google Scholar]
- 140.Altelaar AFM, Taban IM, McDonnell LA, Verhaert PDEM, de Lange RPJ, Adan RAH, et al. High-resolution MALDI imaging mass spectrometry allows localization of peptide distributions at cellular length scales in pituitary tissue sections. Int J Mass spectrom. 2007;260:203–11. [Google Scholar]
- 141.Hillenkamp F, Unsöld E, Kaufmann R, Nitsche R. Laser microprobe mass analysis of organic materials. Nature. 1975;256:119–20. doi: 10.1038/256119a0. [DOI] [PubMed] [Google Scholar]
- 142.Spengler B, Hubert M. Scanning microprobe matrix-assisted laser desorption ionization (SMALDI) mass spectrometry: Instrumentation for sub-micrometer resolved LDI and MALDI surface analysis. J Am Soc Mass Spectrom. 2002;13:735–48. doi: 10.1016/S1044-0305(02)00376-8. [DOI] [PubMed] [Google Scholar]
- 143.Bouschen W, Schulz O, Eikel D, Spengler B. Matrix vapor deposition/recrystallization and dedicated spray preparation for high-resolution scanning microprobe matrix-assisted laser desorption/ionization imaging mass spectrometry (SMALDI-MS) of tissue and single cells. Rapid Commun Mass Spectrom. 2010;24:355–64. doi: 10.1002/rcm.4401. [DOI] [PubMed] [Google Scholar]
- 144.Koestler M, Kirsch D, Hester A, Leisner A, Guenther S, Spengler B. A high-resolution scanning microprobe matrix-assisted laser desorption/ionization ion source for imaging analysis on an ion trap/Fourier transform ion cyclotron resonance mass spectrometer. Rapid Commun Mass Spectrom. 2008;22:3275–85. doi: 10.1002/rcm.3733. [DOI] [PubMed] [Google Scholar]
- 145.Guenther S, Römpp A, Kummer W, Spengler B. AP-MALDI imaging of neuropeptides in mouse pituitary gland with 5 μm spatial resolution and high mass accuracy. Int J Mass spectrom. 2011;305:228–37. [Google Scholar]
- 146.Harada T, Yuba-Kubo A, Sugiura Y, Zaima N, Hayasaka T, Goto-Inoue N, et al. Visualization of volatile substances in different organelles with an atmospheric-pressure mass microscope. Anal Chem. 2009;81:9153–7. doi: 10.1021/ac901872n. [DOI] [PubMed] [Google Scholar]
- 147.Hankin J, Barkley R, Murphy R. Sublimation as a method of matrix application for mass spectrometric imaging. J Am Soc Mass Spectrom. 2007;18:1646–52. doi: 10.1016/j.jasms.2007.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Monroe EB, Koszczuk BA, Losh JL, Jurchen JC, Sweedler JV. Measuring salty samples without adducts with MALDI MS. Int J Mass spectrom. 2007;260:237–42. [Google Scholar]
- 149.Yang J, Caprioli RM. Matrix sublimation/recrystallization for imaging proteins by mass spectrometry at high spatial resolution. Anal Chem. 2011;83:5728–34. doi: 10.1021/ac200998a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Monroe EB, Jurchen JC, Koszczuk BA, Losh JL, Rubakhin SS, Sweedler JV. Massively parallel sample preparation for the MALDI MS analyses of tissues. Anal Chem. 2006;78:6826–32. doi: 10.1021/ac060652r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Tucker KR, Lanni EJ, Serebryannyy LA, Rubakhin SS, Sweedler JV. Stretched tissue mounting for MALDI mass spectrometry imaging. Anal Chem. 2011;83:9181–5. doi: 10.1021/ac201857k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Zimmerman T, Rubakhin S, Sweedler J. MALDI mass spectrometry imaging of neuronal cell cultures. J Am Soc Mass Spectrom. 2011;22:828–36. 36. doi: 10.1007/s13361-011-0111-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Goto-Inoue N, Hayasaka T, Zaima N, Kashiwagi Y, Yamamoto M, Nakamoto M, et al. The detection of glycosphingolipids in brain tissue sections by imaging mass spectrometry using gold nanoparticles. J Am Soc Mass Spectrom. 2010;21:1940–3. doi: 10.1016/j.jasms.2010.08.002. [DOI] [PubMed] [Google Scholar]
- 154.Taira S, Sugiura Y, Moritake S, Shimma S, Ichiyanagi Y, Setou M. Nanoparticle-assisted laser desorption/ionization based mass imaging with cellular resolution. Anal Chem. 2008;80:4761–6. doi: 10.1021/ac800081z. [DOI] [PubMed] [Google Scholar]
- 155.Cecchetti W, Polloni R, Maccioni AM, Traldi P. A simple instrumental approach for laser induced ‘hard’ evaporation of samples. Org Mass Spectrom. 1986;21:517–8. [Google Scholar]
- 156.Hand OW, Emary WB, Winger BE, Cooks RG. Depth profiling of multilayered samples and comparisons of secondary ion and laser desorption mass spectra in the same instrument. Int J Mass Spectrom Ion Processes. 1989;90:97–118. [Google Scholar]
- 157.Louris JN, Brodbelt JS, Cooks RG. Photodissociation in a quadrupole ion trap mass spectrometer using a fiber optic interface. Int J Mass Spectrom Ion Processes. 1987;75:345–52. [Google Scholar]
- 158.Stöckle R, Setz P, Deckert V, Lippert T, Wokaun A, Zenobi R. Nanoscale atmospheric pressure laser ablation-mass spectrometry. Anal Chem. 2001;73:1399–402. doi: 10.1021/ac001440b. [DOI] [PubMed] [Google Scholar]
- 159.Kossakovski DA, O’Connor SD, Widmer M, Baldeschwieler JD, Beauchamp JL. Spatially resolved chemical analysis with an nsom-based laser desorption microprobe. Ultramicroscopy. 1998;71:111–5. [Google Scholar]
- 160.Zoriy MV, Becker JS. Near-field laser ablation inductively coupled plasma mass spectrometry: A novel elemental analytical technique at the nanometer scale. Rapid Commun Mass Spectrom. 2009;23:23–30. doi: 10.1002/rcm.3839. [DOI] [PubMed] [Google Scholar]
- 161.Becker JS, Gorbunoff A, Zoriy M, Izmer A, Kayser M. Evidence of near-field laser ablation inductively coupled plasma mass spectrometry (NF-LA-ICP-MS) at nanometre scale for elemental and isotopic analysis on gels and biological samples. J Anal At Spectrom. 2006;21:19–25. [Google Scholar]
- 162.Zayed A, Shoeib T, Taylor SE, Jones GDD, Thomas AL, Wood JP, et al. Determination of Pt-DNA adducts and the sub-cellular distribution of Pt in human cancer cell lines and the leukocytes of cancer patients, following mono- or combination treatments, by inductively-coupled plasma mass spectrometry. Int J Mass spectrom. 2011;307:70–8. [Google Scholar]
- 163.Becker JS. Bioimaging of metals in brain tissue from micrometre to nanometre scale by laser ablation inductively coupled plasma mass spectrometry: State of the art and perspectives. Int J Mass spectrom. 2010;289:65–75. [Google Scholar]
- 164.Zoriy MV, Kayser M, Becker JS. Possibility of nano-local element analysis by near-field laser ablation inductively coupled plasma mass spectrometry (LA-ICP-MS): New experimental arrangement and first application. Int J Mass spectrom. 2008;273:151–5. [Google Scholar]
- 165.Greving MP, Patti GJ, Siuzdak G. Nanostructure-initiator mass spectrometry metabolite analysis and imaging. Anal Chem. 2010;83:2–7. doi: 10.1021/ac101565f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Northen TR, Yanes O, Northen MT, Marrinucci D, Uritboonthai W, Apon J, et al. Clathrate nanostructures for mass spectrometry. Nature. 2007;449:1033–6. doi: 10.1038/nature06195. [DOI] [PubMed] [Google Scholar]
- 167.Reindl W, Bowen BP, Balamotis MA, Green JE, Northen TR. Multivariate analysis of a 3D mass spectral image for examining tissue heterogeneity. Integr Biol. 2011;3:460–7. doi: 10.1039/c0ib00091d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Bandura DR, Baranov VI, Ornatsky OI, Antonov A, Kinach R, Lou X, et al. Mass cytometry: Technique for real time single cell multitarget immunoassay based on inductively coupled plasma time-of-flight mass spectrometry. Anal Chem. 2009;81:6813–22. doi: 10.1021/ac901049w. [DOI] [PubMed] [Google Scholar]
- 169.Lin Y, Trouillon R, Safina G, Ewing AG. Chemical analysis of single cells. Anal Chem. 2011;83:4369–92. doi: 10.1021/ac2009838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Yamada I, Matsuo J, Toyoda N, Kirkpatrick A. Materials processing by gas cluster ion beams. Mater Sci Eng R Rep. 2001;34:231–95. [Google Scholar]
- 171.Sakai Y, Iijima Y, Mukou S, Hiraoka K. Molecular depth profiling of polystyrene by electrospray droplet impact. Surf Interface Anal. 2011;43:167–70. [Google Scholar]
- 172.Fernandez-Lima FA, Post J, DeBord JD, Eller MJ, Verkhoturov SV, Della-Negra S, et al. Analysis of native biological surfaces using a 100 kv massive gold cluster source. Anal Chem. 2011;83:8448–53. doi: 10.1021/ac201481r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Ji Q, Chen Y, Ji L, Hahto S, Leung K-N, Lee TG, et al. Development of C60 plasma ion source for time-of-flight secondary ion mass spectrometry applications. Rev Sci Instrum. 2008;79:02B309-02B-4. doi: 10.1063/1.2804912. [DOI] [PubMed] [Google Scholar]
- 174.Yamada H, Nakata Y, Ninomiya S, Seki T, Aoki T, Tamura J, et al. MeV-energy probe SIMS imaging of major components in washed and fractured animal cells. Surf Interface Anal. 2011;43:363–6. [Google Scholar]
- 175.Yamada H, Ichiki K, Nakata Y, Ninomiya S, Seki T, Aoki T, et al. MeV-energy probe SIMS imaging of major components in animal cells etched using large gas cluster ions. Nucl Instrum Methods Phys Res B. 2010;268:1736–40. [Google Scholar]
- 176.Nakata Y, Honda Y, Ninomiya S, Seki T, Aokic T, Matsuo J. Matrix-free high-resolution imaging mass spectrometry with high-energy ion projectiles. J Mass Spectrom. 2009;44:128–36. doi: 10.1002/jms.1482. [DOI] [PubMed] [Google Scholar]