Surface enhanced Raman spectroscopy (SERS) is a rapid and highly sensitive spectroscopic technique that has the potential to measure chemical changes in bacterial cell surface properties in response to environmental changes. The objective of this study was to determine whether SERS had sufficient resolution to differentiate closely related bacteria within a genus grown on solid and liquid medium, and a single Arthrobacter strain grown in multiple chromate concentrations. Fourteen closely related Arthrobacter strains, based on their 16S rRNA gene sequences, were used in this study. After performing principal component analysis in conjunction with linear discriminate analysis, we used a novel, adapted cross-validation method, which more faithfully models the classification of spectra. All fourteen strains could be classified with up to 97% accuracy. The hierarchical trees comparing SERS spectra from the liquid and solid media data sets were different. Additionally, hierarchical trees created from the Raman data were different from those obtained using 16S rRNA gene sequences (a phylogenetic measure). A single bacterial strain grown on solid media culture with three different chromate levels also showed significant spectral distinction at discrete points identified by the new Elastic Net regularized regression method demonstrating the ability of SERS to detect environmentally induced changes in cell surface composition. This study demonstrates that SERS is effective in distinguishing between a large number of very closely related Arthrobacter strains and could be a valuable tool for rapid monitoring and characterization of phenotypic variations in a single population in response to environmental conditions.
Introduction
Techniques that allow for rapid identification of the physiological status of a single cell or microbial population is in demand because these could potentially be useful in exploring sample heterogeneity1, 2. Raman spectroscopy is a measure of the molecular composition of cellular surfaces via vibrations specific to molecular bonds and molecular symmetries on the surface of a sample3. The presence of certain bonds on cell surfaces relates to the diversity of carbohydrate, protein and lipid composition of the external cell surface. Many of the differences in the makeup of these molecules result from environmentally induced differential gene expression and as a result are indicative of how a cell responds to, and interacts with its environment. Surface enhanced Raman spectroscopy (SERS) has been shown to be an effective method for the detection and identification of many medically relevant microbes4. SERS has also been shown to be accurate in differentiating eight Bacillus species when multiple strains of each species were included to capture variations within a species5. However, investigations exploring the limit of discrimination based on small-scale physiology changes when closely related bacterial strains are grown under different culture conditions have been sparse.
Much of the research in microbial discrimination using SERS has been focused around phylogenetic classification6, but SERS has also been used to study the physiological status of bacterial cells7–10. The Raman spectra of single Clostridia cells in batch cultures was found to chemically vary between cells with clearly distinct cellular morphologies7. However, this study focused on single cell variation and did not statistically analyze these variations across the population. Other studies have also shown that SERS spectra are influenced by growth conditions such as temperature, media composition and incubation time (or growth stage)8–10. These studies, while still focused on phylogenetic classification, indicated that by changing culture conditions the hierarchical cluster analysis results also changed but each species could still be differentiated by a supervised statistical classification method, Support Vector Machines (SVM) or Linear Discriminant Analysis (LDA), with a high degree of accuracy.
The significant spectral differences between samples are often limited in number and can be difficult to identify among all the common spectra. Therefore, multivariate statistical methods are usually applied to extract biochemical information, statistically classify organisms and build relationship trees using the entirety of the spectrum4, 8, 11, 12. The method of statistical classification used is important because in practice, SERS could be used to identify the spectrum of an unknown sample by comparing it to a library of spectra from known bacteria. In the case where physiological status of an unknown is being assessed the library would be composed of spectra from controls grown under conditions relevant to the experiment. While a variety of statistical methods have been used that attempts to differentiate a bacterium based on its spectrum, they have yet to be done in a manner that emulates the process of validation of an “unknown sample.” We propose the use of an adapted cross validation approach to analyze the efficacy of using a set of known spectra to characterize an unknown spectrum.
This study was aimed at assessing the reliability and efficacy of SERS for discriminating very closely related microbial strains based on differences in surface composition when grown under different culture conditions. The two objectives are: 1) Determine whether SERS can differentiate between fourteen very closely related Arthrobacter strains grown on liquid versus solid medium, 2) Determine whether SERS can detect differences in surface composition of a single bacterial strain grown in different chromate concentrations. These two comparisons address the feasibility of using SERS to detect distinct surface compositional differences related to environmentally induced cellular responses.
Experimental
Bacterial strains and growth conditions
Fourteen Arthrobacter strains (16-6, 16–22, 25–32, 31-31, 31–32, FB-24, J3-37, J3-40, J3-45, J3-46, J3-49, J3-51, J3-62, J3-73) were used in these experiments. All were previously isolated from soil contaminated with heavy metals and aromatic solvents, near Seymour (IN)13. These strains were chosen because of their high degree of genetic relatedness and characterized physiological differences14. The 16S rRNA genes sequences (DQ157984–DQ158006) were determined in a previous study15. All strains were plated, from original frozen stocks made from a single isolate, on to solid media of 0.1× nutrient broth plus Bacto Agar (15 g L−1) (DIFCO, BD Biosciences, Franklin Lakes, NJ) and incubated at 25°C. Liquid cultures were grown from single colonies after they reached a diameter of 1 mm then was inoculated into 0.1× nutrient broth and incubated at 25°C with shaking to an OD600 of 0.55; the end of the exponential growth phase to ensure consistent cellular morphology related to growth stage. One strain, Arthrobacter FB24 was chosen for analysis after growth on solid medium that included 0, 0.25, and 20 mM K2CrO4. This strain and chromate concentrations were chosen based on previous FB24 chromate resistance studies16, 17.
Sample preparation and Raman Imaging
Cultures grown in liquid media were collected by centrifugation at 14,000 g for 5 minutes. Media was removed and cells were washed with 1 mL of sterile MilliQ purified water. This process was repeated three times then cells were re-suspended in 200 μL of sterile MilliQ purified water. For cultures grown on solid medium, single colonies were picked from agar plates after they reached a diameter of 1 mm, and suspended in 1 mL of sterilized MilliQ purified water. Due to growth inhibition induced by the highest level of chromate (20 mM) it was necessary to use multiple colonies from the same plate to obtain sufficient biomass for analysis.
Cell suspensions were serially diluted with sterile MilliQ purified water to a 200-fold final dilution and each dilution was spotted on to roughened gold coated glass slides (EMP Corp, 239 Cherry Street, Ithaca, NY 14850). Gold coated slides were chosen over introduction of nanoparticles to cultures because the introduction of nanoparticles might affect gene expression and result in environmentally induced phenotypes that are undetermined18. Cells were dried on the slides in a 37°C incubator then stored at 4°C. All samples were analyzed using the SENTERRA confocal Raman system (Bruker Optics, Billerica, MA) fitted with a near-infrared 785 nm diode laser, and a 50× objective. An integration time of 140 seconds and a low laser power of 25 mW, at the source, were used for spectral acquisition to avoid damage to the samples. A spectral resolution of 9–15 cm−1 taken over a 90–3500 cm−1 spectral range with 25–35 spectra per strain was used for analysis.
Data preprocessing: outlier removal and normalization
All SERS data processing and statistical analysis was done using the R software19. Outliers were systematically removed by identifying spectra with large disparities because of technical errors including table vibration or failure to completely close the instrument doors (Fig. S1). All spectra were then cropped to the fingerprinting region of interest 500 – 1800 cm−1 20. Maximum intensity value was set to 1 to normalize spectra in each data set.
Statistical Classification
After normalization, both the liquid and solid media data sets were re-expressed using principal component analysis (PCA). PCA was performed with the prcomp function in the base R package19 in order to visualize data separation between strains. The PC plots were created with the mixOmics R package21, 22. The optimal number of principal components was used with linear discriminate analysis (LDA). The classification rates for each data set were assessed using an adapted cross-validation sampling method where a validation data set composed of three random spectra that were removed from each bacterial strain simultaneously. The validation set was classified after reanalyzing the remaining spectra in the training set using PCA/LDA classifier, which includes a separate cross-validation step for choosing the number of PCA components in the classifier. This process was repeated 100 times with the random removal of three spectra from each strain in every iteration. The number of spectra classified correctly was averaged in these repetitions.
Data for Arthrobacter strain FB24 grown in different chromate concentrations was preprocessed using the above method, then classified via Elastic net23. Elastic net is a version of penalized least squares that combines both Ridge and Lasso regression. Ridge regression shrinks (toward zero) the least square coefficients while Lasso both shrinks the coefficients and provides model selection. Analysis was performed using the glmnet.cv function in the glmnet package24. One must set the parameter α that controls the balance between the shrinkage and grouping of the wavenumbers. An alpha level of 1 is equivalent to Lasso regression while an alpha level of 0 is equivalent to Ridge regression. Classification was done at every level of α using 10-fold cross validation.
Hierarchical trees
Agglomerative, neighbor-joining, hierarchical trees were created from the two Raman spectral data sets (cells grown in liquid or solid culture), and the 16S rRNA gene sequences from each strain. Raman data trees were constructed from the mean spectra of each strain, and a Euclidean distance measure was calculated using the hc function in the mclust package25. Bootstrap values were calculated from 100 replications. The number of times a certain `leaf' was maintained divided by 100 equals the bootstrap p-value. The 16S rRNA gene sequence (DQ157984–DQ158006) hierarchical tree was constructed within MacVector26, using a Jukes-Cantor distance measure. Two strains, J345 and J346, not included in the previously published tree15 are known to have identical 16S rRNA gene sequences to strains to J336 and J340, respectively, in that publication.
Results and Discussion
Data preprocessing and visualization
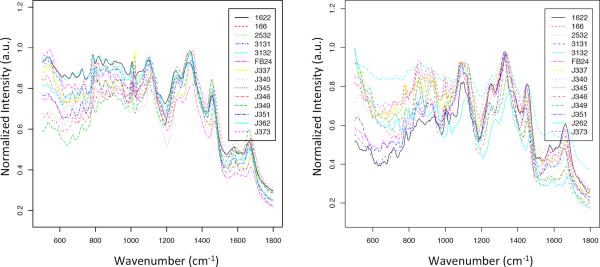
After the elimination of 10 outliers from the solid media data set and zero from the liquid data set, each were composed of 399 spectra and 470 spectra, respectively. Other more involved methods of normalization were forgone in favor of maintaining spectral decay, small spectral variations and mass flux information. These would have otherwise been removed or diluted with other previously reported methods of smoothing and baseline correction27. Failure to preserve this information might compromise the detection of differences between strains that occur in narrow regions of the spectra. The spectra of cells grown on solid media were on an average noisier, had more background signal, than those from liquid media cultures.
The differences between spectra belonging to the same bacterial strain grown under different culture conditions indicated differences in surface composition of cells between the two growth forms (Fig. S2). These differences in spectra were likely due to well established physiological differences between bacterial cells grown on solid surfaces versus liquid suspension28. Many pathogenic bacteria species have been shown to increase their expression of surface antigens when grown on solid media, as opposed to liquid29–31. Similar results were found in the cell surface macromolecules of common freshwater bacteria32. Colonies growing on solid media are not called biofilms but their physiology is probably more similar to a biofilm than to planktonic cells grown in liquid media. The distinct differences in the spectra of Arthrobacter strains grown in solid or liquid media suggests that SERS could detect variability in cell surface composition due to differences in growth environment. The recognition of dissimilarity between liquid and solid culture conditions are important when addressing potential future applications of Raman spectroscopy for use in bacterial discrimination.
Principal component analysis (PCA) showed that the first two principal components (PCs) accounted for a total of 77.7% and 77.2% of the total variance in the data obtained from the liquid media and solid culture data sets, respectively (Fig. S3). PCA is a linear dimensionality reduction technique that is often used with Raman data4, 8, 23, 33 for visualization and dimension reduction prior to linear discriminate analysis (LDA). The PC plots of the solid and liquid culture data sets showed separation between some but not all clusters, and clustering patterns were not similar between culturing conditions. If the spectra from each strain have a large percentage of their variation explained by the first two PCs, and definite repeatable similarities they should cluster together in the coordinate space. Other studies that have used PC plots to show the differentiation of SERS spectra from different bacterial strains have shown greater distinction, but have included fewer strains in their analysis4, 34, 35.
Statistical Classification
The goal of statistical classification of this spectral data was to determine the effectiveness of SERS in distinguishing between all fourteen Arthrobacter strains, and investigate differences in clustering between solid and liquid media data sets. The adapted cross-validation method of PCA+LDA, in 100 trials of leave-three-out, showed that the mean number of correct classifications for all liquid and solid media cultures of each individual strain were very similar (ranging from 85–100%) (Table 1). This indicated that one strain did not disproportionately contribute to the overall classification error rate. These results also indicated that error rates were more likely due to spectral noise than underlying similarity of surface molecule composition between strains. The mean classification rate for the liquid and solid media data sets was 0.971 (97.1%) and 0.944 (94.4%), respectively. The mean classification rates were similar to other bacterial SERS studies using LDA5, but the classification rates are noteworthy given that fourteen strains of highly related strains were included.
Table 1.
Mean classification rate of the adapted cross-validation method of PCA+LDA
| Mean classification rate* for each strain | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Strain | 1622 | 166 | 2532 | 3131 | 3132 | FB24 | J337 | J340 | J345 | J346 | J349 | J351 | J362 | J373 |
| Liquid | 1.00 | 1.00 | 0.95 | 1.00 | 0.97 | 1.00 | 0.98 | 0.97 | 0.99 | 0.89 | 0.96 | 1.00 | 0.89 | 0.96 |
| Solid | 0.90 | 0.88 | 0.96 | 0.88 | 1.00 | 0.85 | 1.00 | 0.87 | 1.00 | 1.00 | 1.00 | 0.90 | 1.00 | 1.00 |
Each rate was assessed using an adapted cross-validation sampling method in which three random spectra from each bacterial strain were removed simultaneously and classified against the remaining data set after it was reanalyzed using PCA/LDA. Total of 100 trials of leave-three-out was performed. A rate of 1.00 indicates 100% correct classification for all trials.
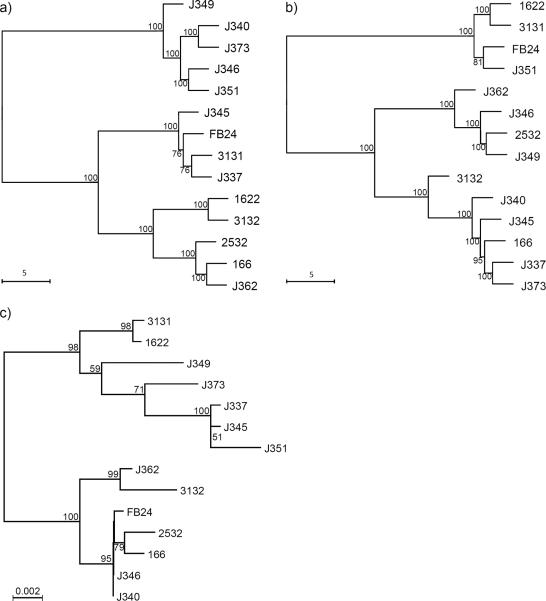
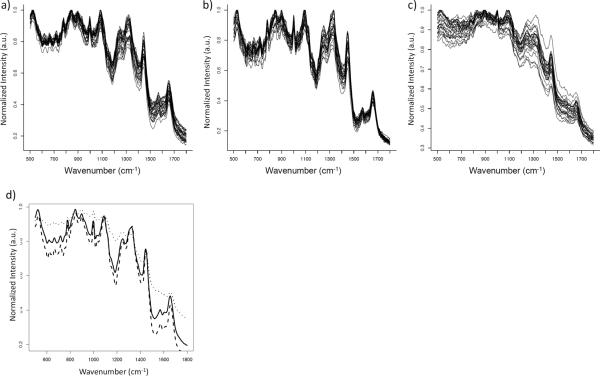
Given that LDA demonstrated the ability to reliably discriminate between the spectra of each strain, the average spectra of each strain were used to create the hierarchical tress. These spectra had notable differences in the regions 500–1000 cm−1 and 1500–1700 cm−1 (Fig. 1). Comparison of the hierarchical trees of SERS data from the two culture conditions showed the relative position of each strain was very different (Fig. 2a and b). Surprisingly, there was not a single relationship conserved between trees, but bootstrap values >70 at every branch indicated a high degree of confidence.
Fig. 1.
Mean SERS spectra for all fourteen Arthrobacter strains grown in a) liquid and b) on solid media. Cells were dried on gold coated glass slides and spectra recorded with an excitation wavelength of 785 nm.
Fig. 2.
Neighbor-joining hierarchical trees of the Euclidean distance of mean SERS spectrum from each strain grown in a) liquid, b) solid media, and Jukes-Cantor distance measure of c) 16S rRNA gene sequences (modified from13). Bootstrap values are indicated at nodes when >50.
LDA and support vector machines (SVM) have been the supervised statistical classification methods of choice used in conjunction with the Raman data, and have proven to be very effective for the analysis of spectra when paired with a variety of cross validation methods such as 10-fold or leave one out cross validation36. However, these validation methods do not adequately address the manner in which SERS would be used to identify samples with an unknown phenotypic characteristic. Therefore, an adapted cross-validation approach was used that more faithfully models the research problem that would arise when attempting to classify unknown spectra against a library of known spectra. This method is proposed as the most appropriate method for cross-validation because it adequately addresses the functionality of using more than one spectrum for bacterial classification. It also limits the reduction of power that occurs when a higher fold cross-validation is used with a limited data set by uniformly reducing the power across all strain groups before prediction.
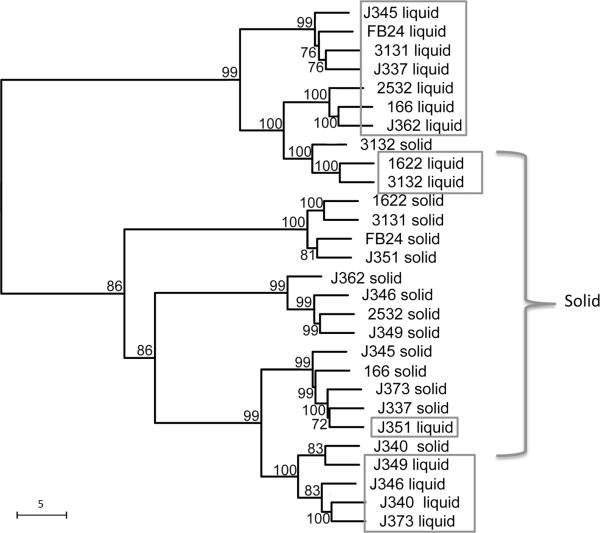
To investigate how statistical classification is affected by environmentally induced phenotypes compared to strain ID, two data sets were combined into a single tree (Fig. 3). This tree indicated that culture condition, not strain ID is the dominant determinant of hierarchical relationship between branches. While not all liquid or solid media culture grown strains clustered together, two of the three clusters were composed solely of strains from a single culture condition, and all clusters had a majority of branches belonging to single media type. Only two preparations of the same strain clustered together: 31–32 and J3–40. This means that the molecular composition of the surface of these fourteen Arthrobacter strains not only vary considerably with growth conditions, but vary less between strains sharing the same growth condition than between the same strain grown in different conditions. This result is expected due to the extensive phenotypic differences documented in strains grown in various culture forms15, 29 as discussed earlier and the studies showing that culture conditions can have a great effect on the resulting Raman spectra8, 10.
Fig. 3.
Neighbor-joining tree of Euclidean distances from combined SERS data of both the liquid and solid media data sets. Bootstrap values are indicated at nodes when >50.
The SERS hierarchical cluster results when compared with those obtained from 16S rRNA gene sequences showed minimum correspondence between relative positions of strains in the two trees (Fig. 2). There were two strains, J3–40 and J3–46, which have identical 16S rRNA gene sequences (Fig. 2c), but have distinct surface compositions according to SERS measurements (Fig. 2a and b). These results correspond to documented physiological differences between these two strains in growth rates and heavy metal tolerance14. There are only two strains whose relationship is conserved between the 16S rRNA sequence tree and the solid media SERS tree: strains 16–22 and 31-31 (Fig 2a and c). The clusters shown in the Raman trees were, for the most part, not similar to those in the 16S rRNA gene sequence tree. This was anticipated because the 16S rRNA gene sequence is a phylogenetic measure of relatedness, and is not expected to correlate to bacterial phenotype differentiating very closely related strains. However, other studies in bacterial spectral clustering via Raman spectroscopy have found correspondence between the hierarchical clustering from Raman data and 16S rRNA gene sequence8, 37. The difference between the results of this study and previous published results is possibly rooted in both the number of strains analyzed and their degree of phylogenetic similarity. However, other literature has expressed skepticism over the idea of using SERS for phylogenetic classification38.
Arthrobacter FB24 grown in different chromate conditions
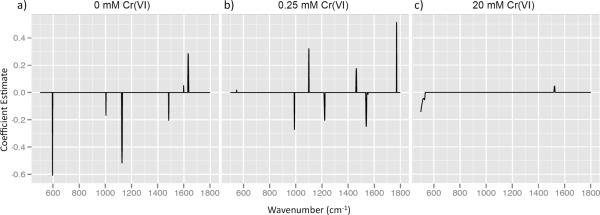
To investigate the applicability of SERS to identify induced changes in the cell surface in response to an environmentally relevant factor, a single Arthrobacter strain was grown in different chromate conditions. Perfect statistical classification of FB24 SERS spectra grown in three chromate concentrations (0, 0.25, and 20 mM) was achieved at every level of α from 0 to 1. This indicates that classification can be achieved from relatively small regions of the spectrum (α close to 1) or when almost the entirety of the spectrum is used (α close to 0). Elastic Net was chosen as the method of statistical classification because, unlike LDA, it allows for the visualization of discriminating wavenumbers for each strain. Plotting the coefficient estimates provided visualization of the α set at 0.489 (Fig. 4). Strain FB24 grown in 0 mM and 0.25 mM Cr(VI) was associated with five or six spectral regions, but when grown in the presence of 20 mM Cr(VI) was not strongly associated with any spectral region.
Fig. 4.
Elastic Net coefficient estimates for alpha = 0.489 for Arthrobacter strain FB24 grown in a) 0, b) 0.25 and c) 20 mM Cr(VI). A peak indicates that correct classification of spectra is associated with the corresponding spectral region. A positive peak indicates higher intensity in that spectral region than other chromate levels and a negative peak indicates lower intensity.
The three chromate concentrations were chosen based on FB24 chromate resistance studies that found a chromate efflux transport protein16 and many other membrane associated proteins up-regulated when chromate levels were greater than 5 mM17. We expected and found that Raman could also detect differences in surface proteins. Unlike proteomic analysis that requires a massive quantity of cells for analysis, SERS requires much fewer cells and should be able to assess cell-to-cell variability.
Spectra of FB24 grown in 0.25 mM Cr(VI) showed an overall decrease in spectral intensity (Fig. 5d). This result was similar to another study in which microbes were grown in the presence of toxic metals at similar concentrations35. The large regions of the spectrum that corresponded to molecules known to be involved in biosorption of metals (approximately 800–1150, 1250–1450, and 1550–1750 wavenumbers) decreased in intensity when Lactobacillus kefir was grown in the presence of low concentrations of heavy metals. Spectra of FB24 grown in 20 mM Cr(VI) were much more variable compared to the other two growth conditions (Fig. 5a,b,c). A new hypothesis can be generated from this result; cell-to-cell variation in phenotype of FB24 grown in a high chromate environment causes high spectral variation due to the high variation in expressed surface molecules between cells. This hypothesis warrants further investigation in the future as a means of exploring the potential of SERS to determine phenotypic differences between single cells within a particular population.
Fig. 5.
Replicate SERS spectra after removal of outliers and normalization of Arthrobacter strain FB24 grown under three chromate conditions, a) 0 mM Cr(VI), b) 0.25 mM Cr(VI) and c) 20 mM Cr(VI) and d) comparison of average SERS spectra from each chromate concentration depicted in a–c.
The quality and consistency of Raman spectra is highly dependent on sample preparation and instrument parameters. A single Raman spectrum has many complicated features that are distorted or masked by perturbation, or “noise” from various sources of measurement error, such as, sample preparation, environmental instabilities and fluorescence. Another observed trend was an overall increase in spectral intensity with a high chromate environment (Fig. 5d); this may be due to noise-causing factors such as sample fluorescence. The large variation in spectra from FB24 grown in 20 mM chromate does show a potential for SERS to be used to detect changes in the biochemical composition of a sample as it relates to its function in the environment. It is possible that SERS signatures of bacterial cultures may serve as a fingerprint and allow for the study of specific surface phenotypic changes in cells performing a desired function such as metal reduction39.
Conclusions
Raman spectroscopy is an attractive technique for bacterial discrimination due to its capacity for fast sample preparation and small-scale measurement of molecular surface composition. The results of the adapted cross-validation method showed that SERS was able to measure distinct molecular differences on the surface of all fourteen closely related Arthrobacter strains with very low error rates. The hierarchical relationships between strains from the two datasets (i.e. liquid and solid cultures) were dissimilar. Elastic Net analysis was effective in identifying discrete spectral differences in a single strain grown under three different chromate concentrations. The approaches used in our study for spectral analysis indicated that SERS has the potential to identify molecular differences between cells due to exposure to different environmental factors. Our results demonstrate that SERS could be an effective technique for monitoring environmentally induced biochemical changes within environments of interest such as chromate (or other heavy metal) exposure. These biochemical changes may lead to the identification of differences in physiological responses of bacteria to variations in environmental conditions at a single cell as well as community level. Further investigation is needed to determine the effectiveness of using SERS to reproducibly identify discriminating spectral regions in response to specific environmental conditions when cell-to-cell variation is high. Additionally, the conservation of discriminating spectra between different bacterial strains still needs to be tested.
Supplementary Material
Acknowledgements
This work was supported by funding from the NIH-NIEHS R01 ES017066-3 award. The authors thank Dr. Sandeep Ravindranath for assistance with the Confocal Raman measurements.
References
- 1.Smits WK, Kuipers OP, Veening JW. Nat Rev Microbiol. 2006;4:259–271. doi: 10.1038/nrmicro1381. [DOI] [PubMed] [Google Scholar]
- 2.Stewart PS, Franklin MJ. Nat Rev Microbiol. 2008;6:199–210. doi: 10.1038/nrmicro1838. [DOI] [PubMed] [Google Scholar]
- 3.Le Ru E, Etchegoin PG. Principles of surface enhanced Raman spectroscopy, and related plasmonic effects. Elsevier Science; Oxford: 2009. [Google Scholar]
- 4.Jarvis RM, Goodacre R. Anal Chem. 2004;76:40–47. doi: 10.1021/ac034689c. [DOI] [PubMed] [Google Scholar]
- 5.Hutsebaut D, Vandroemme J, Heyrman J, Dawyndt P, Vandenabeele P, Moens L, de Vos P. Syst Appl Microbiol. 2006;29:650–660. doi: 10.1016/j.syapm.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 6.Jarvis RM, Goodacre R. Chem Soc Rev. 2008;37:931–936. doi: 10.1039/b705973f. [DOI] [PubMed] [Google Scholar]
- 7.Schuster KC, Reese I, Urlaub E, Gapes JR, Lendl B. Anal Chem. 2000;72:5529–5534. doi: 10.1021/ac000718x. [DOI] [PubMed] [Google Scholar]
- 8.Hutsebaut D, Maquelin K, De Vos P, Vandenabeele P, Moens L, Puppels GJ. Anal Chem. 2004;76:6274–6281. doi: 10.1021/ac049228l. [DOI] [PubMed] [Google Scholar]
- 9.Schuster KC, Urlaub E, Gapes JR. J Microbiol Methods. 2000;42:29–38. doi: 10.1016/s0167-7012(00)00169-x. [DOI] [PubMed] [Google Scholar]
- 10.Harz M, Rosch P, Peschke KD, Ronneberger O, Burkhardt H, Popp J. Analyst. 2005;130:1543–1550. doi: 10.1039/b507715j. [DOI] [PubMed] [Google Scholar]
- 11.Goeller LJ, Riley MR. Appl Spectrosc. 2007;61:679–685. doi: 10.1366/000370207781393217. [DOI] [PubMed] [Google Scholar]
- 12.Maquelin K, Kirshner C, Choo-Smith LP, van den Braak N, Endtz HP, Naumann D, Puppels GJ. J Microbiol Methods. 2002;51:255–271. doi: 10.1016/s0167-7012(02)00127-6. [DOI] [PubMed] [Google Scholar]
- 13.Nakatsu CH, Carmosini N, Baldwin B, Beasley F, Kourtev P, Konopka A. Appl Environ Microbiol. 2005;71:7679–7689. doi: 10.1128/AEM.71.12.7679-7689.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beasley F. MS thesis. Purdue University; 2004. [Google Scholar]
- 15.Vargha M, Takáts Z, Konopka A, Nakatsu CH. J Microbiol Methods. 2006;66:399–409. doi: 10.1016/j.mimet.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 16.Henne KL, Nakatsu CH, Thompson DK, Konopka AE. BMC Microbiol. 2009;9:199. doi: 10.1186/1471-2180-9-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Henne KL, Turse JE, Nicora CD, Lipton MS, Tollaksen SL, Lindberg C, Babnigg G, Giometti CS, Nakatsu CH, Thompson DK, Konopka AE. J Proteome Res. 2009;8:1704–1716. doi: 10.1021/pr800705f. [DOI] [PubMed] [Google Scholar]
- 18.Kahraman M, Yazici MM, Sahin F, Culha M. Langmuir. 2008;24:894–901. doi: 10.1021/la702240q. [DOI] [PubMed] [Google Scholar]
- 19.R Development Core Team . R: A langauge, and environment for statistical computing. Version 2.13.0 2012. [Google Scholar]
- 20.de Groot PJ, Postma GJ, Melssen WJ, Buydens LMC, Deckert V, Zenobi R. Anal Chim Acta. 2001;446:71–83. [Google Scholar]
- 21.Gonzalez I, Le Cao K, Dejean S. mixOmics: Omics Data Integration Project. R package version 3.0 2011. [Google Scholar]
- 22.Le Cao K, Gonzalez I, Dejean S. Bioinformatics. 2009;25:2855–2856. doi: 10.1093/bioinformatics/btp515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou H, Hastie T. J R Stat Soc B. 2005;67:301–320. [Google Scholar]
- 24.Friedman J, Hastie T, Tibshirani R. J Stat Softw. 2010;33:1–22. [PMC free article] [PubMed] [Google Scholar]
- 25.Fraley C, Raftery AE. Model-based clustering/normal mixture modeling. R package verison 3.4.11 2012. [Google Scholar]
- 26.Olson SA. Methods Mol Biol. 1994:195–201. doi: 10.1385/0-89603-276-0:195. [DOI] [PubMed] [Google Scholar]
- 27.Beleites C, Sergo V. Chemometric Analysis of Spectroscopic Data in R: hyperSpec. R package version 0.95 2012. [Google Scholar]
- 28.Zobell CE. J Bacteriol. 1943;46:39–56. doi: 10.1128/jb.46.1.39-56.1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cheung AL, Fischetti VA. Infect Immun. 1988;56:1061–1065. doi: 10.1128/iai.56.5.1061-1065.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Evans DG, Silver RP, Evans DJ, Jr., Chase DG, Gorbach SL. Infect Immun. 1975;12:656–667. doi: 10.1128/iai.12.3.656-667.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.DeVoe IW, Gilchrist JE. J Exp Med. 1975;141:297–305. doi: 10.1084/jem.141.2.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McEldowney S, Fletcher M. J Gen Microbiol. 1986;132:513–523. [Google Scholar]
- 33.Maquelin K, Choo-Smith LP, van Vreeswijk T, Endtz HP, Smith B, Bennett R, Bruining HA, Puppels GJ. Anal Chem. 2000;72:12–19. doi: 10.1021/ac991011h. [DOI] [PubMed] [Google Scholar]
- 34.Guicheteau J, Argue L, Emge D, Hyre A, Jacobson M, Christesen S. Appl Spectrosc. 2008;62:267–272. doi: 10.1366/000370208783759623. [DOI] [PubMed] [Google Scholar]
- 35.Gerbino E, Mobili P, Tymczyszyn EE, Frausto-Reyes C, Araujno-Andrade C, Gomez-Zavaglia A. J Appl Microbiol. 2011;112:363–371. doi: 10.1111/j.1365-2672.2011.05210.x. [DOI] [PubMed] [Google Scholar]
- 36.Gaus K, Rosch P, Petry R, Peschke KD, Ronneberger O, Burkhardt H, Baumann K, Popp J. Biopolymers. 2006;82:286–290. doi: 10.1002/bip.20448. [DOI] [PubMed] [Google Scholar]
- 37.Lopez-Diez EC, Goodacre R. Anal Chem. 2004;76:585–591. doi: 10.1021/ac035110d. [DOI] [PubMed] [Google Scholar]
- 38.Efrima S, Zeiri L. J Raman Spectrosc. 2009;40:277–288. [Google Scholar]
- 39.Walter A, Kuhuri S, Reinicke M, Bocklitz T, Schumacher W, Roesch P, Merten D, Buchel G, Kothe E, Popp J. J Raman Spectrosc. 2011 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.