Abstract
Clinical databases provide a rich source of data for answering clinical research questions. However, the variables recorded in clinical data systems are often identified by local, idiosyncratic, and sometimes redundant and/or ambiguous names (or codes) rather than unique, well-organized codes from standard code systems. This reality discourages research use of such databases, because researchers must invest considerable time in cleaning up the data before they can ask their first research question. Researchers at MIT developed MIMIC-II, a nearly complete collection of clinical data about intensive care patients. Because its data are drawn from existing clinical systems, it has many of the problems described above. In collaboration with the MIT researchers, we have begun a process of cleaning up the data and mapping the variable names and codes to LOINC codes. Our first step, which we describe here, was to map all of the laboratory test observations to LOINC codes. We were able to map 87% of the unique laboratory tests that cover 94% of the total number of laboratory tests results. Of the 13% of tests that we could not map, nearly 60% were due to test names whose real meaning could not be discerned and 29% represented tests that were not yet included in the LOINC table. These results suggest that LOINC codes cover most of laboratory tests used in critical care. We have delivered this work to the MIMIC-II researchers, who have included it in their standard MIMIC-II database release so that researchers who use this database in the future will not have to do this work.
Keywords: Vocabulary standards, laboratory tests, LOINC, MIMIC-II, secondary use, mapping guidelines
1. Introduction
Electronic health record (EHR) systems provide a rich source of clinical data for answering research questions. In an ideal world, the data from an EHR would have a simple internal structure and be coded according to nationally accepted vocabulary standards. However, most of the data in EHRs are not yet linked to standard codes, and a single EHR system may contain many different local names (codes) for a single clinical variable and conversely, may use the same name (or code) for different variables over time, especially when the EHR has a long history. Faced with such realities, researchers who want to analyze such databases must invest substantial time and effort to consolidate redundant codes and distinguish among different meanings for one code. We are collaborating with researchers at the Massachusetts Institute of Technology (MIT) to normalize the data contained in the publicly available Multiparameter Intelligent Monitoring in Intensive Care (MIMIC-II) database [1] to facilitate use by researchers. Our overall goal is to map all important MIMIC-II clinical variables (tests, clinical observations, drugs and problems) to nationally-endorsed vocabularies standards and along the way to remove duplicates and resolve ambiguity in the local names and codes we were standardizing. In this paper, we describe the process for mapping MIMIC-II laboratory test observations to Logical Observation Identifiers Names and Codes (LOINC®) [2] and our mapping guidelines that came out of this work.
2. Background
2.1. Secondary use databases
Large clinical databases are becoming increasingly available to researchers as more hospitals and practices adopt EHR systems. These databases cover a variety of clinical domains, including cancer, diabetes, gene therapy, and intensive care. MIMIC-II is an intensive care database with vast amounts (over 250 million rows) of information about critically ill patients along with physiologic tracings. MIMIC-II represents a source of information about the natural course of serious conditions, the effects of various treatments, and the predictive power of various symptoms, findings and physiologic measurements on outcomes [1].
Secondary use of the EHR data (i.e., using data collected during patient care for clinical research) is becoming increasingly important because EHRs contain clinical data collected over years to decades that would be time- and cost- prohibitive to collect prospectively. Many examples of the successful research use of clinical databases exist. For example, the first large study linking early use of erythromycin to infantile hypertrophic pyloric stenosis was based on a retrospective review of more than 14,000 infant records from a clinical EHR [3]. More recently, Kurreeman and colleagues published a study evaluating the relationship between patients’ specific genomic data and autoimmune antibody status and their risk of developing rheumatoid arthritis in a diverse ethnic population, also based on clinical EHR data [4].
Clinical databases are developed during the course of clinical care, not with a specific research focus, and therefore will not have the regularity or completeness of a database designed for a particular research protocol [5]. Observations in a clinical database are collected according to each patient’s clinical need, and data for a given variable will rarely be collected at regular intervals and will often be completely absent between episodes of care. Even though much of the data collected for clinical use are in a structured format, a care institution may assign multiple local names or codes to the same variable and a local code may change meaning over time. (Of course this also happens with research data bases collected over long periods.) Effort is needed to eliminate the redundancies and ambiguities and to regularize the content for research purposes. An even larger effort is necessary when the data comes from more than one institution that has not adopted standard vocabularies for their key content, because in that case not only is it necessary to clean up the data in each system, but the variables from the different care systems must be mapped to one another [6,7].
2.2. LOINC
The Health IT Standards Committee, which advises the National Coordinator for Health IT, recently recommended “a minimum necessary set of vocabulary standards that will enable…interoperable electronic health record data elements” [8]. These vocabulary standards specify LOINC for laboratory tests and diagnostic studies as well as many other categories of information. LOINC provides a universal code system for clinical observations and orders that: 1) provides a grouping code for redundant local codes (that may be necessary for local reasons); 2) enables EHRs and other data sources to send and receive computer understandable Health Level 7 (HL7) [9] messages and documents [10]; and 3) provides common codes for data captured from many different sources and/or time periods so that data points from many sources can be pooled and analyzed as a unit.
The LOINC database is maintained by the Regenstrief Institute and the latest release contains over 68,000 standard codes for identifying laboratory tests, clinical measures, survey instruments and narrative reports. Each LOINC term has six main parts: 1) component (the specific lab analyte or physiologic measure); 2) property (such as mass concentration or ratio); 3) timing (such as 24-hour collection or one point in time); 4) specimen type (e.g., blood or urine); 5) scale (such as quantitative or qualitative); and 6) method used (such as a specific lab technique or estimated versus measured) [10]. LOINC variables also include example units of measure when appropriate, short name, and frequency statistics for the most common tests [11].
LOINC includes a large spectrum of laboratory tests, including chemistry, pathology reports and mutation analyses that range from the common, e.g., blood hemoglobin and serum potassium, to the less common, e.g., CFTR gene p.3199 del6 (a specific cystic fibrosis gene mutation), to the rare, e.g., Ebola virus RNA. LOINC test codes vary in specificity from tests without a specimen type or method (“methodless”), e.g., “Glucose in Unspecified specimen” to very precise codes for tests run on a specific type of fluid by a specific method, e.g., “Beta globulin in CSF by electrophoresis.” For most chemistry analytes LOINC includes one code for “serum/plasma” (because the results are equivalent) and a different code for the same test run on whole blood because of differences in the results and/or handling of the two cases. When required by laboratory practice, LOINC defines codes specific to either plasma (e.g., ammonia concentration) or serum (e.g., rheumatoid arthritis nuclear antibodies). It also includes codes for clinical measures such as vital signs, body measurements, EKG results, and radiology study findings and many other clinical observations. In addition to variables gathered for clinical purposes, LOINC also includes those gathered principally for administrative purposes, for example, the Outcome and Assessment Information Set (OASIS)™ [12] and the Nursing Management Minimum Data Set (NMMDS) [13], which contain patient and administrative variables related to home health and nursing care.
LOINC has very good concept coverage, especially for common tests. Lin and colleagues found that after excluding the “narrative results” and “internal use only” variables, the LOINC December 2007 release had at least 90% concept coverage for laboratory tests from two large hospitals and 73% coverage for a reference laboratory which handles both common and specialized tests [14]. Many of the tests that could not be mapped, such as “NBC14:1_C16 NBS RATIO” and “MoM for Nuchal Translucency,” did not have the corresponding analyte in the 2007 LOINC database; however, those analytes have since been added along with thousands of other ones, so the coverage is even better.
Regenstrief provides two tools for browsing and mapping local codes to LOINC: RELMA® and the LOINC web search tool (http://search.loinc.org/). The RELMA auto-mapper takes all of the available local information for a given test, such as test name, units of measure, fluid, and method and returns a list of the most closely matched LOINC codes from which the user picks the best code. However, the auto-mapper is still under development and does not always find the best match. Using the LOINC websearch tool alone or RELMA without the auto-mapper, a user can quickly find matches, especially if he/she constrains the search to the most common terms that are consistent with the local terms’ units of measure.
2.3. Related work
The value of mapping local variables to LOINC for data aggregation and research has been recognized across a broad range of domains. The Agency for Healthcare Research and Quality (AHRQ) conducted a pilot study in Florida from 2007 to 2009 on predicting in-hospital mortality based on laboratory, administrative, and admission diagnosis data. In that study, the investigators mapped 55 laboratory tests from 22 different hospitals to LOINC so that the lab results could be aggregated across all of the participating hospitals and analyzed together with the administrative and diagnosis data. The mapping to LOINC was important to mortality predictions. Eleven of these 55 tests significantly improved the prediction of inpatient mortality compared to the use of the admission diagnosis data alone [15]. Kroth and colleagues used LOINC to unify the elements from ten different existing cephalometric measurement standards (used to define X-rays measurements for orthodontics) to create one common standard terminology for this domain [16].
Mapping every single variable in an EHR is not necessary to answer a specific research question; focusing on a limited set of variables may be more productive. For example, the eMERGE Network [6] mapped 157 phenotype variables that were created by extracting diagnoses symptoms and findings as phenotype data from five institutions’ EHRs to four standardized metadata and terminology resources: SNOMED CT, the Cancer Data Standards Registry and Repository (caDSR), the NCI thesaurus, and the Study Data Tabulation Model terminology. The researchers created an open-source web-based application, eleMAP, for searching caDSR and the National Center for Biomedical Ontology BioPortal. They mapped 95 data elements to existing terms in at least one vocabulary. The remaining 62 data elements required adding new elements to the caDSR and NCI thesaurus as well as post-coordination of the standard terms. Based on the lessons they learned mapping the initial set of 157 elements, the eMERGE researchers plan to refine and expand eleMAP for future mapping efforts [6].
Previous mapping efforts also offered important lessons to us. Nadkarni and Darer [17] mapped legacy ICD-9-CM codes used at their institution to SNOMED CT. They automatically mapped using available UMLS cross-mappings, and developed a manual process for mapping the remaining 35%. Despite the use of query expansion and other advanced information retrieval techniques, the authors had to use their knowledge of both the specific domain and the general English language to choose the right codes. They describe the process to be “especially onerous if a concept encountered in text does not exist in the terminology” [17].
2.4. MIMIC-II
2.4.1. Background and Scope
The MIMIC-II database was developed by MIT under a grant from the National Institute of Biomedical Imaging and Bioengineering. MIMIC-II contains data from the intensive care unit (ICU) at Beth Israel Deaconess Medical Center, a tertiary care hospital in Boston, Massachusetts from 2001 onward. The data were de-identified per HIPAA regulations [18] by: 1) removing the structured fields containing personal health information (PHI) (e.g., name, address, medical record number); 2) using MIT’s open-source algorithm [19] to remove PHI from all of the free text notes; and 3) randomly transforming dates into the future while maintaining the temporal sequence within single patients. Researchers who accept the data use agreement and have completed human subjects training can apply for permission to access the data [1].
MIMIC-II contains a broad range of both clinical and administrative information and encompasses approximately 240 million rows of data. It contains several different ICU types, including surgical, medical, cardiac, and neonatal. The database includes demographic data, ICD-9-CM discharge diagnoses codes, and detailed nursing data and other physiologic measures such as Glasgow Coma Score (GCS) and ventilator settings. Lab results include both ICU point-of-care (POC) tests as well as tests run in the hospital laboratory. Medications are recorded in a variety of formats including medication administration data as well as medication orders. MIMIC-II includes narrative notes such as daily progress notes, comprehensive discharge summaries, and radiology reports.
2.4.2. Database structure
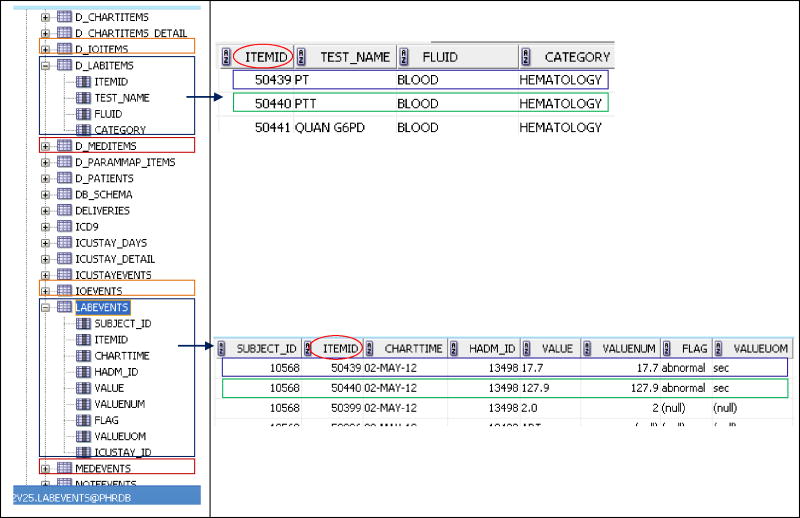
MIMIC-II contains a pair of related tables for each category of clinical information, a general strategy employed by most EHRs. One table in the pair carries patient observation data and includes one field that carries a code that identifies the specific observation, another field that identifies the patient, and several other fields that carry the date, value, and other information about the observation. The associated dictionary table provides descriptive information about each unique observation, e.g., the observation name, units of measure (when applicable), and distinct observation identifying code. MIMIC-II includes separate table pairs for: labs, medications, fluids and outputs, narrative notes, and data recorded during the course of routine clinical care such as vital signs and ventilator parameters. For the lab data, there is one table for patients’ lab results called “labevents” and a companion lab dictionary table called “d_labitems.” The lab results table includes a 5-digit local lab test code, test date and time, and value for the test observation; its corresponding lab dictionary table contains one row for each unique lab test that includes the corresponding 5-digit code, test name, specimen type, and lab category (see Figure 1). Another pair of tables with the same structure is “chartevents” and “d_chartitems.” Chartevents carries a wide variety of clinical information entered by nurses, including vital signs, lab results, medications, and other information such as ventilator parameters and physical exam findings. D_chartitems is its corresponding data dictionary. Some of the MIMIC-II patient data are duplicated across tables. For example, the hospital laboratory directly populated the lab results table, and nursing copied some of these same laboratory results into the chartevents table to facilitate their workflow. Similarly, the chartevents table was the primary repository of point of care (POC) tests performed in the ICU, and some of these results eventually populated the lab results table. As a result, some of the data in the lab results table was not in the chartevents table and vice versa. The database contains one table of orders (for medications only); complete documentation of the MIMIC-II table structure is available on the MIMIC-II website (http://physionet.org/mimic2/).
Figure 1.
Depiction of the MIMIC-II database structure. Note the pairs of patient result and associated data dictionary tables on the left. Details from the d_labitems and labevents tables are shown on the right. The d_chartitems and chartevents pair of tables have an analogous structure.
Each pair of tables contains sequential variable identifiers that the system automatically assigned as it created new records. Different pairs of tables have unique ranges of possible codes so there is no link between a given laboratory result in the lab results table and the same lab result in chartevents. Users could generate a new variable at will when they did not find what they were trying to enter. This led to creation of many different variable identifiers for the same observation concept. There are hundreds of examples of variables names that differ only in spaces between words, capitalization, and punctuation. For example, if a user intending to enter the result of a “Fingerstick Glucose” test (chartevents item # 807) typed in “fsbs” (fingerstick blood sugar), the system would not find the existing “Fingerstick Glucose” because it could not recognize synonyms or near matches for an entered term. So, it offered to make a new variable for “fsbs” and assigned it the next available local code, #1946. Spelling errors and variation in the abbreviations used by different providers led to many redundant local codes for the same concept, some of which were only used by a single provider for a few patients on a given day. Because the data in the lab results table came directly from the lab system, it contained fewer redundant codes. We describe these particulars to exemplify the kinds of problems researchers can face when they try to utilize clinical data systems with long histories.
3. Methods
In this report we describe the mapping and disambiguating of the laboratory tests in the MIMIC-II database. We tackled the laboratory test results first because laboratory data are so pertinent to most clinical research questions and are available in large quantities. The laboratory results file was the best organized and had the fewest codes per volume of data. We went through multiple cycles of mapping, preliminary review, and expert review. As we worked through the mapping process, we developed a set of basic mapping guidelines to help guide our own and others’ future mapping work.
3.1. Data extraction
Our first step was to extract the lab test information (test name, specimen fluid, and the lab domain) from the MIMIC-II lab data dictionary into a spreadsheet to make it easier to work with. Next, we pre-processed the test names to eliminate spelling errors, remove extra spacing and incorrect punctuation, and expand the abbreviations. Once the pre-processing was complete we were able to aggregate all of the MIMIC-II test names that represented a single concept.
3.2. Gathering supporting data for disambiguation
The information in the lab data dictionary was sometimes less specific than that required to directly map to LOINC codes. MIMIC-II, for example, did not distinguish among serum, plasma, or whole blood as the intravascular specimen for chemistry tests, and instead always listed “Blood” as the specimen type. This mismatch between the conceptualizations of such tests by MIMIC-II versus LOINC required extra investigation to clarify when to map to a LOINC term with specimen of whole blood and when to use one with serum/plasma, serum, or plasma. We were able to do this by distinguishing the MIMIC-II POC tests, which use whole blood as their specimen, from main laboratory tests that use serum or plasma (with a few exceptions).
For the tests performed in the hospital laboratory, we used detailed lab test information from three large reference laboratories [20,21,22] to determine the specimen type when only one specimen type was used. In modern laboratories plasma is the only acceptable specimen for ammonia, so we assigned the LOINC code for ammonia concentration in plasma. When more than one specimen type (e.g., serum or plasma) was acceptable, we mapped the MIMIC-II test to the LOINC code for serum/plasma. When MIMIC-II reported the specific specimen type in the lab data dictionary for specimens that were not an intravascular specimen, such as urine, cerebrospinal fluid or peritoneal fluid, mapping was easy. We also augmented the information about analyte and specimen type from the laboratory dictionary file with information about units of measure, result ranges, and average values of test results from the patient laboratory results table. When the test name or specimen type was ambiguous we narrowed the search for a LOINC code by finding tests with similar units and reference ranges within the three reference lab resources [20,21,22]. In a few cases, we examined the nursing notes and diagnosis codes for patients who had that test in order to expand the clinical context and pinpoint the exact test.
3.3. Assigning codes
We attempted to map each MIMIC-II lab code to the most precise LOINC code possible. We depended on reference laboratory information to find the right intravascular specimen for MIMIC-II tests done in the central lab, and otherwise mapped to the LOINC codes whose specimens were at the same level of specificity as the specimen type named in the MIMIC-II test dictionary. For example, if the test in MIMIC-II had a specimen type of “Other body fluid,” we mapped to a LOINC code with specimen type “Body fluid” even if we were able to discern that the test was done on a more specific fluid such as pericardial fluid based on other data, because the test could potentially be used for other specimen types in the future.
We classified our mappings as: 1) an exact mapping when it had the same analyte name, unit of measure and given or inferred specimen type; 2) an ambiguous but likely map when the tests in the MIMIC-II database were mapped to the exact analyte but there was some question about the likely specimen type and/or unit of measure; and 3) no match. Tests defined as no match fell into three categories: 1) the MIMIC-II test name was too vague or incomplete to know what was meant; 2) the measurement in question was in the lab result table but was not literally a laboratory test; and 3) we understood what they meant but there was not yet a LOINC code that matched precisely to their concept.
3.4. Annotators and tools
The initial mappings of the entire lab variable set were done together by a fourth year medical student and a clinical informatics fellow (SA) using the RELMA auto-mapper version 4.3 (see Figure 2). The same two annotators then individually reviewed the initial results and flagged questionable mappings. Subsequently, a LOINC expert (CJM) reviewed all of the mapping results on paper, resolved some of the questionable mappings, and marked additional mappings that needed investigation. After our first run using the auto-mapper, we manually refined the mappings using RELMA and the LOINC websearch tool. Most of the tests required only one pass by the expert reviewer to establish the correct mapping. A small subset of tests required multiple iterations of mapping and expert review to establish the correct mapping. The initial expert review and feedback about the entire set of mappings improved the accuracy of the junior and more experienced annotator’s mappings so that only a few targeted tests required the expert’s attention during subsequent cycles.
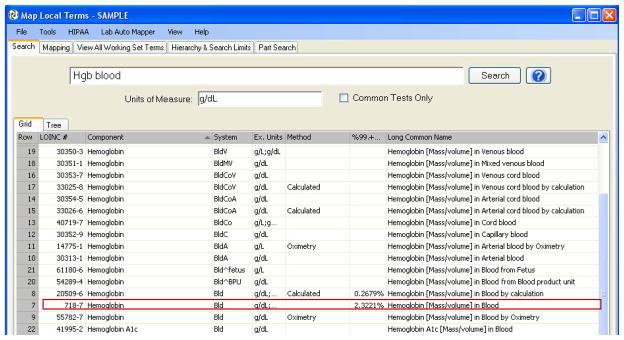
Figure 2.
The RELMA auto-mapper takes the available local test information and returns the best matches in LOINC. In this example, the local data was “Hgb” (test name), “blood” (the specimen), and “g/dL” (the units of measure). The table of likely LOINC matches illustrates the specificity of different parameters including units, method, specimen, and statistical rank. The red box indicates the best match in this example.
Results
4.1. Mapping results
MIMIC-II version 2.1 contains 661 unique local items in the lab dictionary table, a few of which are redundant local test names that represent the same test concept. The initial mapping run for these 661 tests took approximately 48 working hours or approximately 4 minutes per test. Subsequent review cycles likely tripled or quadrupled that time. Of the 661 items, we were able to map 483 tests (73.1%) to LOINC as exact matches, and an additional 92 (13.9%) as ambiguous but likely matches, giving a total of 575 out of 661 tests (87%) that we were able to map (see Table 1 for the classification of overall mapping results). The ambiguity regarding those 92 tests can be further classified as shown in Table 1, with over 80% due to missing information about the unit of measure. The majority of these 92 tests were CD cell marker tests counts which can be reported as percentages or as absolute counts. We mapped these tests to the LOINC code that would have been reported as a percent count because the numeric values of these tests were 2-digit integers consistent with percent measurements. We classified the CD mappings as ambiguous, which was a very conservative position because the numeric values spoke so loudly in favor of our classification. Detailed examples of tests with other types of ambiguities are given in Table 2, including analyte names that were not clear, such as “ANTI-MC” and “AM BIUR,” and vague specimen types, e.g., “OTHER BODY FLUID.”
Table 1.
LOINC mapping classification for MIMIC-II lab tests. The indented numbers and percentages are the values for the subcategories within the major categories.
| Number of tests in each category (N = 661) | % of total | |
|---|---|---|
| Exact match | 483 | 73.1% |
|
| ||
| Ambiguous but likely match | 92 | 13.9% |
| - No unit of measure | 77 | 11.6% |
| - Multiple units of measure | 4 | 0.6% |
| - Ambiguous name | 8 | 1.2% |
| - Uncertain specimen type | 2 | 0.3% |
| - No enough information about method | 1 | 0.2% |
|
| ||
| Unable to map | 86 | 13.0% |
| - Ambiguous name | 51 | 7.7% |
| - Not a lab test | 10 | 1.5% |
| - No LOINC code available | 25 | 3.8% |
Table 2.
Examples of MIMIC-II tests that were mapped as ambiguous but likely matches.
| MIMIC-II test name/specimen type | Mapping issue | Supplemental data | Mapped to LOINC code | LOINC test name | Comments |
|---|---|---|---|---|---|
| ANTI-MC/BLOOD | Ambiguous name | Discharge summary text: “She had antibodies sent for acetylcholine receptors and thyroid antibodies which were still pending at the time of discharge” Mayo website[22]: Microsomal antibody = thyroperoxidase antibody | 32042-4 | Thyroperoxidase Ab [Presence] in Serum | |
| AM BIUR/URINE | Ambiguous name | ARUP website [21]: urate crystals are birefringent Sample values: none, few, mod, many |
12454-5 | Urate crystals amorphous [Presence] in Urine sediment by Light microscopy | Information from ARUP helped define am bi |
| PG/OTHER BODY FLUID | Uncertain body fluid | Patient notes: specimen was from amniotic fluid | 48785-0 | Prostaglandin D2 [Mass/volume] in Body fluid | We did not map to a specific amniotic fluid code because the local MIMIC-II code could potentially be used for different body fluids for different patients. |
| FREE TEST/BLOOD | Multiple units of measure | Units of measure: ng/dL and pg/mL | 2991-8 | Testosterone Free [Mass/volume] in Serum or Plasma | This LOINC code has both MIMIC-II units of measure specified as potential units. |
We could not map 86 out of the original 661 tests (13%). Nearly 60% of these 86 had MIMIC-II test names that were not clear (e.g., “Other”), 11% were not laboratory tests (e.g., “Intubated”), and almost 30% were tests that had not yet been included in the LOINC database (e.g., “Nucleated red blood cells” in the specimen pleural fluid). Table 3 gives detailed explanations of these and other tests that we were unable to map in each category. We have submitted the tests without available LOINC codes to Regenstrief Institute using the formal submission process (http://loinc.org/submissions).
Table 3.
Examples of MIMIC-II tests that we were not able to map.
| MIMIC-II test name/specimen type | Mapping issue | Comments |
|---|---|---|
| KAPPA/BLOOD | Ambiguous name | “Kappa” could refer to either free kappa immunoglobulin light chains or the kappa B-lymphocyte cell marker. There was not enough supplemental information in the database to disambiguate. |
| OTHER/JOINT FLUID | Ambiguous name | The results for this test were numeric with a range of 0–51 and % for the unit of measure. There was no other information to help disambiguate the term. |
| RED HOLD/BLOOD | Not a lab test | This represents an extra red-top tube of blood available for the patient in case further tests are needed. |
| INTUBATED/BLOOD | Not a lab test | This conveys the clinical information to the laboratory that the patient is on mechanical ventilation at the time the sample was taken. |
| NUCLEATED RED BLOOD CELLS/PLEURAL FLUID | No LOINC code available | LOINC has nucleated red blood cell codes for other specimen types including blood, CSF, and synovial fluid, but not for pleural fluid. |
| HEMATOCRIT/JOINT FLUID | No LOINC code available | LOINC has hematocrit codes for other specimen types including blood, urine, bone marrow, and dialysis fluid, but not for joint fluid. |
Overall, we mapped 575 MIMIC-II lab tests to 559 LOINC codes. The majority (544 tests) were mapped to LOINC with a 1 to 1 relationship; however, there was some redundancy of local lab tests. One set of three MIMIC-II tests represented the same test and we mapped them to one LOINC code. There were 14 pairs of MIMIC-II test codes (a total of 28) that represented one test concept so we mapped those 28 MIMIC-II tests to 14 LOINC codes with a 2 to 1 relationship (Table 4). We have delivered these mappings to the MIT research group, who have included them the latest release (2.6) of the database.
Table 4.
Examples of multiple MIMIC-II tests mapped to a single LOINC code
| LOINC code | LOINC description | MIMIC-II itemid | MIMIC-II test name | MIMIC-II fluid | MIMIC-II category |
|---|---|---|---|---|---|
| 718-7 | Hemoglobin [Mass/volume] in Blood | 50007 | HGB | BLOOD | BLOOD GAS |
| 50386 | HGB | BLOOD | HEMATOLOGY | ||
| 50184 | [Hgb] | BLOOD | CHEMISTRY | ||
| 13362-9 | Collection duration of Urine | 50256 | <COLLECT> | URINE | CHEMISTRY |
| 50268 | HOURS | URINE | CHEMISTRY | ||
| 1959-6 | Bicarbonate [Moles/volume] in Blood | 50022 | TCO2 | BLOOD | BLOOD GAS |
| 50025 | TOTAL CO2 | BLOOD | BLOOD GAS | ||
| 2350-7 | Glucose [Mass/volume] in Urine | 50641 | GLUCOSE | URINE | HEMATOLOGY |
| 50266 | GLUCOSE | URINE | CHEMISTRY | ||
| 26464-8 | Leukocytes [#/volume] in Blood | 50316 | WBC | BLOOD | HEMATOLOGY |
| 50468 | WBC | BLOOD | HEMATOLOGY | ||
| 26478-8 | Lymphocytes/100 leukocytes in Blood | 50315 | LYMPH | BLOOD | HEMATOLOGY |
| 50408 | LYMPHS | BLOOD | HEMATOLOGY | ||
| 2756-5 | pH of Urine | 50653 | PH | URINE | HEMATOLOGY |
| 50297 | pH | URINE | CHEMISTRY | ||
| 28009-9 | Volume of Urine | 50285 | VOLUME | URINE | CHEMISTRY |
| 50259 | <VOL-U> | URINE | CHEMISTRY | ||
| 41284-1 | Epithelial cells.non-squamous [#/area] in Urine sediment by Microscopy high power field | 50651 | NONSQ EPI | URINE | HEMATOLOGY |
| 50652 | NSQ EPI | URINE | HEMATOLOGY |
4.2. Mapping guidelines
During the course of our mapping efforts, we developed the following mapping procedure with some specific recommendations for certain steps. We have included some resources that were not available at the time we did our lab mappings (marked with a *) but which will be useful in our future work.
-
Obtain LOINC and RELMA
-
1
Go to the LOINC website (www.loinc.org) and download the LOINC database, RELMA, and the LOINC and RELMA user guides (all four of these are available as a single download package).
-
2
The website also has both on-line and downloadable tutorials and other training materials, as well as a table of the top 2000+ lab tests*[23], which includes helpful advice about how to map tests that are subject to mapping errors.
-
1
-
Data extraction
-
3
Determine the most important code group to map
Recommendation: Focus on the code group directly relevant to your research or project goals first.
-
4
Focus first on variables that are most common
-
5
Find and aggregate the local terms that represent a single concept
Recommendation: Pre-process the local terms in order to eliminate difference due to extra spaces, capitalization and punctuation. Edit the terms that are abbreviations, truncations or typographical errors to a standard name form. You can do this manually, but RELMA also has a “Spell Check” program that will find words and units strings in your source file that it does not recognize*. Most of these will be typos and unconventional abbreviations of common laboratory name and units of measure, and this same program will help you to convert them to the correct spellings or acronyms.
-
3
-
Gather supporting data for disambiguation
-
6
Collect descriptive statistics for the code values in the database if available
Recommendation: Collect information on the unit of measure, specimen type, and result statistics such as mean and range of result values across patients.
Alternate recommendation: Though you will usually be starting with the database designed to collect the data, you may have access to the HL7 message stream that populated such a database. If so, we strongly recommend using the RELMA HL7 import function that will convert a large set (use 1–2 million) of HL7 messages to a database that will carry one record per unique order name and test name, and each such record will include units of measure, sample results and reference ranges, and sample values taken from the HL7 message stream*.
-
7
Use information from trusted external laboratory websites and domain experts to help clarify details about the local terms, including the appropriate specimen type and resolving non-standard abbreviations
Recommendation: Cross-reference the information obtained in step 6 with information from multiple reference websites (preferably in a similar clinical setting) to find the best relevant reference, but do not force the local data to match the reference site information. This step will not usually be necessary if your laboratory test file (or other local lab data source) always includes the analyte and specimen, as well as units of measure, reference ranges, and sample data for each test you are trying to map.
-
6
-
Assigning codes
-
8
Assign the most specific codes supported by the data
Recommendation: Use all of the supplementary information gathered in step 6 to pick the most specific code possible, but do not invent distinctions that are not present in the local name. For quantitative variables, if the local unit of measure is not available, look to sample data to help pick the right LOINC codes.
Please note: there will likely be multiple iterations of Steps 9–11 for some variables.
-
8
-
Annotators and tools
-
9
Use the RELMA auto-mapper for the initial mappings by the junior annotator
Recommendations: 1) Use the RELMA spell-checker to evaluate both the list of local test names and units of measure*; 2) Constrain RELMA to consider only the top 2000+ tests first* and set the system to require the LOINC property to be consistent with the reported units of measures; 3) Relax the top 2000+ constraint when you have finished mapping all of the common tests and have to find more unusual test that will not be in the top 2000+.
-
10
Let a more experienced annotator review the mappings using RELMA or LOINC websearch
Recommendation: The more experienced annotator should flag questionable mappings specifically for expert review.
-
11
Let a domain expert annotator review the final mappings
Recommendation: The domain expert annotator should review all of the initial mappings and provide feedback on specific terms as well as general mapping techniques. This will increase the accuracy of the junior and more experienced annotators’ mappings and improve efficiency by allowing the expert annotator to focus on specific mapping questions in subsequent iterations.
-
12
Include metadata with final mappings: the version of the standard, the date for the final mapping, and the annotators’ contact information
-
9
5. Discussion
We found that although normalizing data is labor-intensive, it only has to be done once, saving each researcher the time of doing it themselves. By mapping redundant local codes to unique LOINC codes, data that were previously scattered among different tables and local variables can be consolidated into a single variable based on the standard code, thus assuring complete data capture for that variable. We were able to map distinct MIMIC-II lab test codes that cover 94% of the recorded patient results, demonstrating that LOINC has good coverage for the laboratory tests within a clinical ICU database. The tests that we were unable to map fell into many of the same categories that were described by Lin and colleagues [14]: 1) test names that were completely ambiguous; 2) variables that were not lab tests but that contained information related to the order and were meant only for local use; and 3) tests without a corresponding LOINC code. We did not encounter any terms in the other two categories identified by Lin et al (tests with narrative results and tests with overly specified methods).
5.1. Time/personnel needed to map
Mapping local terms from a secondary use database is time-consuming. Our first pass mapping took on average 4 minutes per term, and including the subsequent review and mapping iterations, we likely spent an average of at least 15 minutes mapping each term. These numbers are worst case estimates because our primary clinically-trained mappers (a medical student and clinical informatics fellow) had no previous experience with LOINC, laboratory dictionaries and term mapping, so they do not represent the time that experienced LOINC mappers would take. Some organizations that provide mapping consulting services are listed in the voluntary directory of LOINC adopters on the main LOINC website (http://loinc.org/adopters). The complete process, including data extraction, mapping and project coordination can take longer. For example the hospitals in the AHRQ pilot study report anywhere from 34 (for experienced LOINC mappers) to 145 minutes total time per term [15].
5.2. Limitations
While we have been fairly successful to date in mapping the MIMIC-II lab tests to standard vocabularies, in certain cases it was impossible to map a specific item. Given that the data are de-identified, we could not go back to the original hospital database to resolve questions such as missing data or implausible values. The fact that the MIMIC-II data included randomized dates confounded our attempts to resolve temporal questions such as whether the reason for different units of measure for one test was due to a change in methodology, instrument, or change in unit reporting practice because we could not put the results across all patients in true date order to find a point in time when the units changed.
In addition, as clinical practices changed, new variables were appropriately added to the database to capture that data. For example, version 2.6 of the MIMIC-II database contains 713 unique lab items (compared to 661 in versions 2.1 and 2.5), so there are 52 new items that need mapping. This reality highlights the need for ongoing maintenance that is necessary as long as health systems continue to use local codes instead of standard vocabularies.
5.3. Comparison to previous work and AHIMA guidelines
Our efforts in mapping the clinical variables used in the MIMIC-II database differed from the eMERGE mapping effort not only in the standard terminologies that we mapped to, but also in that each of the five eMERGE sites created its own well-defined, limited data dictionary containing 26 to 44 local terms [6], while we worked with a large set of terms, many of which were difficult to disambiguate since we did not have access to the full medical record or the source system’s schema. However, a common theme we found was that mapping is an iterative process and that the lessons learned in one cycle are valuable not only for the next iteration but also for other researchers. Our work was also different from Nadkarni and Darer’s work [17] because the target of our mapping effort was the idiosyncratic names and codes from an EMR database, not another standard terminology designed for a similar purpose. Nevertheless, our work was similar in that we also found that significant manual effort was needed for obtaining reliable mappings, and that relevant external resources were necessary to achieve complete and accurate mappings.
In April 2010, at the time we started our mapping work, guidelines for this process were scarce, and therefore we developed our own mapping rules. In April 2011, AHIMA published a consensus opinion on mapping best practices, which includes establishing the reason for mapping, defining a use case, developing mapping rules, and testing and validating the rules [24]. However, AHIMA’s work was not based on a direct mapping effort and did not deal with many of the very specific issues we encountered. Nevertheless, we are pleased that our mapping guidelines generally conform to the AHIMA practice guidelines.
AHIMA specifies four essential steps: 1) establishing a business case; 2) defining a specific use case; 3) developing rules for implementation; and 4) testing the rules with a pilot phase [24]. We began this mapping project to facilitate data reuse for retrospective clinical studies (the business case). Second, keeping in mind that the MIMIC dataset is publicly available and most researchers who will use the dataset subsequent to our efforts will have knowledge of standard terminologies, we selected LOINC as the target (the use case). Third, we developed the mapping rules and identified software to assist our efforts (rules for implementation). Fourth, we went through several reviews until satisfied with the quality of our mappings, and we then used the coded data for a clinical research study that has been submitted for publication (the pilot phase for testing the rules). Finally, we hope that the LOINC codes included in the current MIMIC-II release will be used and therefore further validated by the data owners and licensees.
5.4. Future directions
We are currently working on mapping other patient data such as vital signs, ventilator parameters, and lab tests from chartevents to LOINC and medications to RxNorm. This recent work has been slower than the lab results mapping due to greater local code redundancy and naming idiosyncrasies in the chartevents table (compared to the lab variables), as well as the inherent complexity in medication mapping due to the wide range of specificity in both MIMIC-II drug data as well as RxNorm codes. For example, some MIMIC-II tables have brand and generic drug names while others have one or the other. In addition, some tables have specific dose and drug form information while others do not specify this level of detail. RxNorm has unique codes for various combinations of drug name, dose, and drug form. Although in our mapping procedure we stated that in most cases, it is best to map to the most specific code possible, for medications it may make sense to first map all MIMIC-II drugs to ingredient alone to create a set of drug mappings with the same level of specificity, and then map drugs that have more available information to more specific drug codes.
6. Conclusion
The goal of this work is to facilitate the secondary use of a large, third-party, publicly available collection of clinical data. Although some aspects of our work are similar to previous efforts in mapping proprietary coded data to standards, mapping codes that are noisy, incomplete and unfamiliar presents unique challenges. Overcoming these challenges resulted in standardization of 87% of local laboratory codes to LOINC and inclusion of the standard codes in the distribution of the MIMIC-II database. We have seen the value of normalizing data firsthand as we have used the MIMIC-II database for research purposes. As we progressed in our mapping efforts, we discovered new redundant local variables that we then incorporated into our research datasets, which made the data more complete and strengthened the conclusions we were able to draw from the analysis. Using the mapped data, we have submitted a study for publication on obesity and relationship of body mass index to outcomes during and after critical illness. By delivering our mapping work back to MIT to include in future releases, others will have the benefits of standardization. Equally important to providing the standard codes for the data, or perhaps even more important, are our mapping guidelines, which will become very useful as hospitals are incented to adopt EHRs [25,26] and both hospitals and vendors begin mapping their local codes to the standard terminologies. Finally, with the understanding that not all EHR data can immediately be coded to standards, we sincerely hope that the new generations of EHR systems will maximize the use of standards. Our study adds to the growing body of evidence that existing terminology standards already capture most of the useful concepts in clinical data.
Highlights.
Current publicly available clinical databases are not coded using national vocabulary standards
We are mapping the MIMIC-II ICU database to standard terminologies such as LOINC
Our mappings are incorporated into successive releases of the database
We have developed mapping guidelines based on our experience
Acknowledgments
Many thanks to all our collaborators at MIT, including Drs. Roger Mark and Daniel Scott. This work would have not been possible without their dedication to collecting and making the data publicly available and without their continuous support in providing (where possible) additional information for our mapping efforts. Thanks also to our student, Dr. Howard Ching, who helped with the initial mapping effort. Finally, thank you to our anonymous reviewers for their insightful comments and suggestions.
This work was supported by the Intramural Research Program of the National Library of Medicine, National Institutes of Health.
Footnotes
Potential reviewers:
Daniel J Vreeman PT, DPT; Indiana University School of Medicine; dvreeman@regenstrief.org
Erik van Mulligen PhD; Erasmus University; e.vanmulligen@erasmusmc.nl
Jonathan Darer MD, MPH; Geisinger Health System; jddarer@geisinger.edu
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errorsmaybe discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Swapna Abhyankar, Email: swapna.abhyankar@nih.gov.
Dina Demner-Fushman, Email: dina.demner@nih.gov.
Clement J. McDonald, Email: clement.mcdonald@nih.gov.
References
- 1.Saeed M, Villarroel M, Reisner AT, Clifford G, Lehman LW, Moody G, Heldt T, Kyaw TH, Moody B, Mark R. Multiparameter Intelligent Monitoring in Intensive Care II (MIMIC –II): A public-access intensive care unit database. Crit Care Med. 2011 Jan 28;39(5) doi: 10.1097/CCM.0b013e31820a92c6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Regenstrief Institute. [Accessed October 3, 2011];Logical Observation Identifiers Names and Codes. Web site. http://loinc.org/. Updated September 28, 2011.
- 3.Mahon BE, Rosenman MB, Kleiman MB. Maternal and infant use of erythromycin and other macrolide antibiotics as risk factors for infantile hypertrophic pyloric stenosis. J Pediatr. 2001 Sep;139(3):380–4. doi: 10.1067/mpd.2001.117577. [DOI] [PubMed] [Google Scholar]
- 4.Kurreeman F, Liao K, Chibnik L, et al. Genetic basis of autoantibody positive and negative rheumatoid arthritis risk in a multi-ethnic cohort derived from electronic health records. Am J Hum Genet. 2011 Jan 7;88(1):57–69. doi: 10.1016/j.ajhg.2010.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gliklich RE, Dreyer NA, editors. (Prepared by Outcome DEcIDE Center [Outcome Sciences, Inc. d/b/a Outcome] under Contract No. HHSA29020050035I TO3.) AHRQ Publication No.10-EHC049. 2. Rockville, MD: Agency for Healthcare Research and Quality; Sep, 2010. Registries for Evaluating Patient Outcomes: A User’s Guide. [Google Scholar]
- 6.Pathak J, Wang J, Kashyap S, Basford M, Li R, Masys DR, Chute CG. Mapping clinical phenotype data elements to standardized metadata repositories and controlled terminologies: the eMERGE Network experience. J Am Med Inform Assoc. 2011 Jul 1;18(4):376–86. doi: 10.1136/amiajnl-2010-000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen ES, Melton GB, Engelstad ME, Sarkar IN. Standardizing Clinical Document Names Using the HL7/LOINC Document Ontology and LOINC Codes. AMIA Annu Symp Proc. 2010 Nov 13;2010:101–5. [PMC free article] [PubMed] [Google Scholar]
- 8.Clinical Quality Measures Workgroup and Vocabulary Task Force. Letter to: Dr Farzad Mostashari, National Coordinator for Health Information. Health Information Technology Standards Committee; Washington, DC: 2011. Aug 7, Available from: http://healthit.hhs.gov/portal/server.pt/gateway/PTARGS_0_12811_955546_0_0_18/HITSC_CQMWG_VTF_Transmit_090911.pdf. [Google Scholar]
- 9.Health Level Seven International [Internet] Ann Arbor, MI: Health Level Seven International; [Accessed February 12, 2012]. ©2007–2012. http://www.hl7.org/index.cfm. [Google Scholar]
- 10.McDonald CJ, Huff SM, Mercer K, Hernandez JA, Vreeman DJ, editors. Logical Observations Identifiers Names and Codes (LOINC) Users’ Guide. Indianapolis (IN): Regenstrief Institute; 2011. Dec, Available from: http://loinc.org/downloads/files/LOINCManual.pdf. [Google Scholar]
- 11.LOINC Mapper’s Guide to the Top 2000+ Lab Observations - Introduction to Version 1.0. Indianapolis (IN): Regenstrief Institute; 2011. May, Available from: http://loinc.org/usage/obs/introduction-to-the-mappers-guide-for-the-top-2000-plus-loinc-laboratory-observations.pdf/view. [Google Scholar]
- 12.Centers for Medicare and Medicaid Services. OASIS Overview. Washington (DC): Department of Health and Human Services; [Accessed February 11, 2012]. https://www.cms.gov/OASIS/ [Google Scholar]
- 13.Huber D, Schumacher L, Delaney C. Nursing Management Minimum Data Set (NMMDS) J Nurs Adm. 1997;27:42–48. doi: 10.1097/00005110-199704000-00011. [DOI] [PubMed] [Google Scholar]
- 14.Lin MC, Vreeman DJ, McDonald CJ, Huff SM. A Characterization of Local LOINC Mapping for Laboratory Tests in Three Large Institutions. Methods Inf Med. 2011 Feb;50:105–114. doi: 10.3414/ME09-01-0072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Adding Clinical Data to Statewide Administrative Data. Final Report Submitted to the Agency for Healthcare Research and Quality. Tallahassee (FL): Agency for Health Care Administration (AHCA), Florida Center for Health Information and Policy Analysis; 2010. Mar, AHRQ Contract #07-10042. [Google Scholar]
- 16.Kroth PJ, Daneshvari S, Harris EF, Vreeman DJ, Edgar HJ. Using LOINC to link 10 terminology standards to one unified standard in a specialized domain. J Biomed Inform. 2011 Oct 19; doi: 10.1016/j.jbi.2011.10.003. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nadkarni PM, Darer JA. Migrating existing clinical content from ICD-9 to SNOMED. JAMIA. 2010;17:602–607. doi: 10.1136/jamia.2009.001057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Standards for Privacy of Individually Identifiable Health Information Final Rule. Entry Type: Rule. Dates: This final rule is effective on October 15, 2002; pp. 53182–53273. (92 pages); Document Citation: 67 FR 53182. [PubMed] [Google Scholar]
- 19.Neamatullah I, Douglass M, Lehman LH, Reisner A, Villarroel M, Long WJ, Szolovits P, Moody GB, Mark RG, Clifford GD. Automated De-Identification of Free-Text Medical Records. BMC Medical Informatics and Decision Making. 2008;8:32. doi: 10.1186/1472-6947-8-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pathology Associates Medical Laboratories. [Accessed August 1, 2011];PAML Reference Ranges Report. Web site. http://etd.paml.com/etd/refrangereport.php.
- 21.ARUP Laboratories. [Accessed August 1, 2011];ARUP Laboratory Test Directory. Web site. http://www.aruplab.com/
- 22.Mayo Medical Laboratories. Test Catalog. Mayo Medical Laboratories; [Accessed August 1, 2011]. Web site. http://www.mayomedicallaboratories.com/test-catalog/index.html. [Google Scholar]
- 23.LOINC Mapper’s Guide to the Top 2000+ Lab Observations - Introduction to Version 1.0. Indianapolis (IN): Regenstrief Institute; 2011. May, Available from: http://loinc.org/usage/obs/introduction-to-the-mappers-guide-for-the-top-2000-plus-loinc-laboratory-observations.pdf/view. [Google Scholar]
- 24.AHIMA. Data Mapping Best Practices. Journal of AHIMA. 2011 Apr;82(4):46–52. Available from: http://healthdataanalysisupdate.org/?p=97. [PubMed] [Google Scholar]
- 25.American Recovery and Reinvestment Act (ARRA) of 2009, Pub. L. No. 111–5, 123 Stat. 115, 516. [Accessed October 3, 2011]; http://www.gpo.gov/fdsys/pkg/PLAW-111publ5/html/PLAW-111publ5.htm. Approved February 17, 2009. Effective February 17, 2009.
- 26.Centers for Medicare and Medicaid Services. Medicare and Medicaid programs; Electronic health record incentive program; Final Rule, 42 C.F.R. Sect. 412, 413, 422 et al. [Accessed October 3, 2011]; http://edocket.access.gpo.gov/2010/pdf/2010-17207.pdf. Revised July 28, 2010. Effective September 27, 2010.