CONSPECTUS
In DNA, bases pair in a molecular interaction that is both highly predictable and exquisitely specific. Therefore researchers have generally believed that the insertion of the matching nucleotide opposite a template base by DNA polymerases (pols) required Watson-Crick (W-C) hydrogen bond formation. However pioneering work by Kool and coworkers using hydrophobic base analogs such as the T isostere 2,4-difluorotoluene (F) showed that shape rather than H-bonding served as the primary source of specificity in DNA replication by certain pols. This steric hypothesis for DNA replication has gained popularity, perhaps discouraging further experimental studies to address potential limitations of this new idea.
The idea that shape trumps H-bonding in terms of pol selectivity largely hinges on the belief that fluorine is a poor H-bond acceptor. However, the shape complementarity model was embraced in the absence of any detailed structural data for match (F:A) and mismatch pairs (F:G, F:C, F:T) in DNA duplexes or at active sites of pols. Although the F and T nucleosides are roughly isosteric, it is unclear whether F:A and T:A pairs exhibit similar geometries. If the former pair is devoid of H-bonding, it will be notably wider than a T:A pair. Because shape/size and H bonding are intimately related, it may not be possible to separate these two properties. Thus the geometries of an isolated F:A pair in water may differ considerably from an F:A pair embedded in a stretch of duplex DNA, at the tight active site of an A-family replicative pol, or within the spacious active site of a Y-family translesion pol. The shape complementarity model may have more significance for pol accuracy than efficiency: this model appears to be most relevant for replicative pols that use specific residues to probe the identity of the nascent base pair from the minor groove side. However, compared with W-C H-bonds researchers have not fully considered the importance of such interactions that include H-bonds in terms of pol fidelity and the shape complementarity model.
This Account revisits the steric hypothesis for DNA replication in light of recent structural data and discusses the role of fluorine as an H-bond acceptor. Over the last five years, crystal structures have emerged for nucleic acid duplexes with F paired opposite to natural bases or located at the active sites of DNA pols. These data permit a more nuanced understanding of the role of shape in DNA replication and the capacity of fluorine to form H-bonds. These studies and additional research involving RNA or other fluorine-containing nucleoside analogs within duplexes indicate that fluorine engages in H-bonding in many cases. Although T and F are isosteric at the nucleoside level, replacement of a natural base by F in pairs often changes their shapes and sizes, and dF in DNA behaves differently from rF in RNA. Similarly, the pairing geometries observed for F and T opposite dATP, dGTP, dTTP or dCTP and their H-bonding patterns at the active site of a replicative pol differ considerably.
Introduction
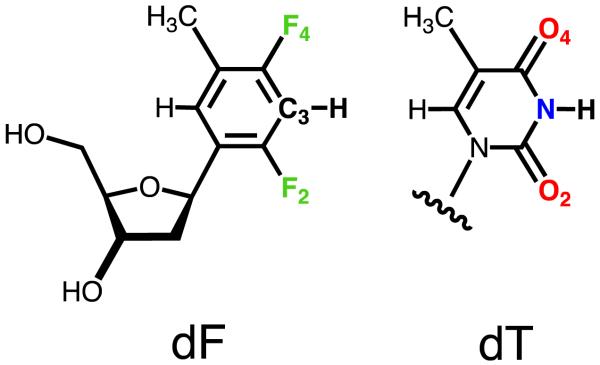
The finding that certain DNA polymerases (pols) insert dATP opposite a 2,4-difluorotoluene nucleoside analog (F, Figure 1) quite efficiently and accurately relative to template T is counterintuitive.1-3 Although we have known for a long time that enzymes rely to various degrees on shape and size differences to select their substrates,4,5 Watson-Crick hydrogen bonding (W-C H-bonding) is crucial for the pairing specificity and stability of DNA. That shape should ultimately be the major or sole determinant of replication fidelity as put forth in the so-called Steric Hypothesis for DNA Replication6-9 is therefore startling. Moreover, the shape complementarity model hinges largely on the assumption that despite its high electronegativity, organic fluorine is a poor H-bond acceptor.10,11 While this verdict is consistent with the environments of fluorine in small-molecule crystal structures, a host of physical-chemical data and the results of computational simulations, there are numerous reports of fluorine affecting crystal packing, protein-ligand and DNA-ligand interactions directly or indirectly despite the weakness of individual C-H…F, C-F…F, C-F…N or C-F…π contributions (refs. 12-15 and cited refs.). Although there is a priori no reason to believe that difluorotoluene violates the rule that fluorine hardly ever accepts H-bonds,11 it is obvious that without actual structural data for the analog in the relevant contexts - i.e. in a DNA duplex or at a pol active site - we cannot rule out that the supposedly hydrophobic T isostere might occasionally engage in H-bonding interactions. Shape, in spite of being a key player in enzyme function and activity, is by no means the only determinant of substrate specificity.
Figure 1.
Structures of dT and dF (or F).
Over the last five years my laboratory has determined crystal structures of DNA and RNA oligonucleotides that contain F or other fluorine-modified nucleoside analogs. We as well as others have also analyzed crystal structures of pol-DNA complexes with F in the template strand and at the active site. These data permit a more differentiated interpretation of the role of shape in DNA replication. In combination with experimental stability data the structures of duplexes containing fluorine-bearing residues support the view that fluorine can form energetically favorable H-bonds. Despite the growing database of structures containing F, predictions regarding the geometry of and contributions of fluorine to H-bonds in dF/rF match and mismatch pairs remain difficult. What is clear is that both, geometry and fluorine H-bonding are strongly influenced by the particular environment.
The Hypothesis
Looking back at a decade of debate regarding the hydrophobicity of the difluorotoluene analog, Sintim and Kool remained unconvinced of the existence of N-H…F or C-H…F H-bonds.8 A host of observations were offered in support of the strictly hydrophobic nature of F or dF. For example, dF partitions strongly to octanol in mixtures with water, unlike dT [logP(dF) = 0.78; logP(dT) = −1.1]. In the crystal structure of dF, F2, C3H and F4 appear not to form H-bonds, whereas in the structure of dT the corresponding O2, N3H and O4 do. The dipole moments of F and T differ strongly (1.84 vs. 4.19 Debye, resp.), as do the atomic charges of C3(F) and N3(T) (0.074 vs. 0.203, resp.) and F4(F) and O4(T) (−0.079 vs. −0.307, resp.). Incorporation of a single F in place of T into DNA opposite A destabilizes the duplex significantly; the Tm as assessed by UV melting drops by some 15°C.16 However, a 12mer DNA duplex with an F:A pair is more stable than duplexes with F:G, F:C or F:T mismatch pairs (by between 2 and 3°C). Perhaps this reflects a better shape correspondence and more optimal stacking with neighboring pairs as a result, or a residual electrostatic contribution that can be expected to be more favorable for a matching pair. Interestingly, when the relative stabilities of U, F and the toluyl (To) C-nucleoside analog paired with either A, G, U or C inside duplex RNA were compared, To pairs are clearly more destabilizing than F pairs (i.e. U:A, 37.8°C; F:A, 27.4°C; To:A, 23.0°C).17 This observation too leaves open the possibility that F pairing entails a residual H-bonding contribution, although it is also known that F furnishes improved stacking in DNA compared with both T and a benzyl C-nucleoside analog.18
At the level of the nucleoside, there is good evidence that dF is an isostere of dT. The C=O and C-F bond lengths are not too different. Both dF and dT favor the anti glycosidic conformation and the preferred sugar conformation with both lies in the Southern range (90% and 70% South for dF and dT, respectively). The NMR structure of a DNA duplex with an incorporated F:A pair appeared to be largely unchanged relative to B-from DNA,19 further bolstering the impression that dF and dT and perhaps the F:A and T:A pairs are of the same shape. However, the structure itself did not provide a detailed look at the F:A pair. Rather, the structural data simply support the notion that F:A does not significantly perturb the DNA duplex, i.e. by F or A or both adopting an extra-helical orientation. So the shapes may be similar, but the sizes (width of the base pair, i.e. glycosidic C1′…C1′ distance) could nevertheless differ substantially between the two. In fact, ab initio calculations and force field modeling of the F:A pair in duplex DNA were not suggestive of H-bonding and the resulting F4…H-N6 and C3-H…N1 distances of 3.32 Å and 3.52 Å, respectively,20 were around 0.5 Å longer than the corresponding distances in a T:A pair (Figure 2). Thus, the inability of fluorine in dF to accept H-bonds and the isosteric nature of the analog relative to T are two cornerstones of the steric hypothesis for DNA replication.
Figure 2.
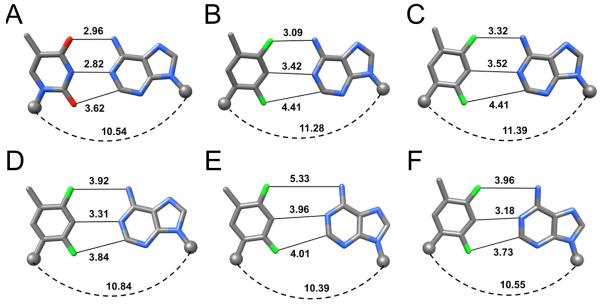
Geometries of F:A pairs. (A) Reference T:A pair (DDD, CGCGAATTCGCG; PDB ID 436D).32 (B) F:A in a modified DDD (CGCGAATFCGCG; PDB ID 3I8D).30 (C) Computational model of F:A.20 (D) rF:rA in duplex RNA (rCGCFAAUUAGCG; PDB ID 2G92).33 (E) F:A at the active site of the Y-family pol Dpo4 (PDB ID 2VA2).25 (F) F:A at the active site of the B-family pol RB69 (PDB ID 3QER).43 Distances are in Å, C1′ atoms are shown as spheres and the spacing between them is indicated by a dashed arc.
The central pillar of the hypothesis that electrostatic effects seem to play a minor role in accurate replication, ultimately triggering more extensive investigations into the importance of shape, is of course provided by the efficient and preferred (as judged by steady state kinetics; kcat/Km) incorporation by E. coli DNA polymerase I Klenow fragment (Kf) of dATP opposite template F relative to dATP insertion opposite T,1 as well as that of insertion of dFTP opposite A relative to dTTP opposite A.2 There is an asymmetry in the decrease of the efficiency in that F in the template is better tolerated than F as the incoming nucleotide triphosphate. And with Kf and other replicative pols, efficiency is more affected than fidelity.2,9,21 The asymmetry between dATP insertion opposite template F and dFTP insertion opposite template A becomes much more pronounced when pre-steady state kinetics are considered. Thus, the reduction in efficiency (kcat/Kd) is around 30-fold in the former case but much more pronounced (ca. 1000-fold) for dFTP vs. dTTP as the incoming nucleotide.22 Similar observations with respect to efficiency and accuracy were made with the human mitochondrial pol.23 Again, these data indicate the importance of H-bonding for efficient primer extension even with pols featuring tight active sites.
Compared to A- and B-family pols, the tolerance of Y-family translesion pols toward F in the template or dFTP as the incoming nucleotide is much more limited. The archaeal Dbh22 and Dpo424,25 pols as well as yeast pol η26 and human pol κ27 were all found to be dependent on H-bonding for efficient and accurate replication. Thus, it appears that only certain pols (i.e. those belonging to the A- and B-families) with tight active sites and amino acids probing the minor groove of the replicative and post-replicative base pairs22 are capable of accurate and relatively efficient replication, by selecting incoming and templating nucleotide pairs of optimal shape and size. Interestingly, Dpo4 (Y-family) and Kf (A-family) exhibit similar preferences vis-à-vis the gradually increasing sizes of base pairs between A and various thymine analogs (toluene, 2,4-difluorotoluene, 2,4-dichlorotoluene, 2,4-dibromotoluene, 2,4-diiodotoluene).24 For both pols the maximum efficiency is observed with the dichloro analog (either inside the template or as the incoming nucleotide triphosphate). However, given the important role attributed to shape and size, it is puzzling that Kf’s efficiencies of incorporating either dFTP or the 2′-deoxy diiodotoluene triphosphate analog (d[DIT]TP) opposite A are virtually identical. This indicates that steric effects alone cannot account for the overall selectivity of Kf.
F Pairing in Duplex DNA
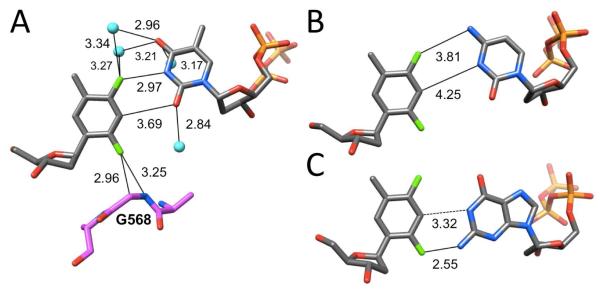
The B-form DNA duplex [d(CGCGAATTCGCG)]2, the so-called Dickerson-Drew Dodecamer (DDD), has long served as a crystallographic model system to study the sequence dependence of conformation, bending, hydration and metal ion binding (ref. 29 and cited refs.). We synthesized a modified DDD with F inserted in place of T8, d(CGCGAATFCGCG) and determined its structure bound to RNase H from Bacillus halodurans (BhRNase H) as crystals could not be grown for the duplex alone.30 The protein in the complex crystals merely serves as a scaffold and interacts with the G-tracts at both ends of the DDD.31 Thus the geometry of the central AATT (AATF) tract is not perturbed as a result of the presence of RNase H. At 1.6 Å, the structure is of sufficiently high resolution to allow accurate determination of inter-atomic distances [si file]. In the two independent F:A pairs observed in the structure, the F4(F)…N6(A) distances are 3.09 and 3.12 Å (Figure 2B). These are not too different from the corresponding O4(T)…N6(A) distances in structures of the native DDD or its complex with BhRNase H that vary between 2.96 and 3.11 Å (Figure 2A).30,32 At just slightly above 3 Å the experimentally determined distances involving fluorine are significantly shorter than those seen in computational models of F:A pairs in which the minimal separation reported amounts to 3.32 Å (Figure 2C).20 However, most of the modeled F:A pairs in DNA or RNA feature F4(F)…N6(A) separations that are more consistent with the sum of the van der Waals (vdW) radii of the interacting atoms (ca. 3.6 Å).17,30 Because the distance is around 3.1 Å in the crystal structure and thus about 15% shorter than the above sum of vdW radii, we can basically rule out a mere van der Waals contact between fluorine and the amine in the DDD F:A pairs. If we assume a bond length of around 1 Å for N-H, then the 3.1 Å F…N separation in the DDD F:A pairs matches the shortest F…H-N interactions in crystal structures of small molecules that were found to be around 2.1 Å.10 H-bond participation by organic fluorine may indeed be rare, but it is hard to avoid the impression that electrostatics have something to do with the observed configuration of F:A pairs in the DDD. Although incorporation of F in place of T into DNA destabilizes the duplex significantly, DNAs with F:A pairs melt at higher temperatures than those with F:G, F:C or F:T mismatch pairs,16 as pointed out in the introduction. Clearly, the stabilization afforded by a putative C-F…H-N H-bond is limited compared to the standard C=O…H-N H-bond. Either favorable stacking or residual H-bonding or both are likely at the origin of the higher stability of F:A relative to the mismatch pairs.
F Pairing in Duplex RNA
The effects of incorporation of the ribonucleotide analog of F, rF, into small interfering RNA molecules (siRNAs) on their activity have been studied in some detail.33-35 The modulation of the siRNA activity upon replacement of rU (U) by rF in the antisense (guide) strand was assayed in HeLa cells in vitro.33,34 When placed at the 5′-terminal end or opposite A in the interior of the guide strand, rF was quite well tolerated. Even when positioned adjacent to the Ago2 cleavage site, measurements of the luciferase luminescence revealed that the modified siRNA duplex maintains considerable activity as long as the pairing partner of rF is A. Thus, rF:A and C:A at the cleavage site affected the activity to a similar degree and these combinations were better tolerated than A:A or G:A mismatches that strongly impaired activity.33 A related study that assessed the interference of siRNAs with rF in place of U at 11 different positions of the guide strand demonstrated that activity and thermodynamic stability of the individual duplexes correlated quite well.34 However, an siRNA with rF at a central location of the guide strand retained high activity despite being strongly destabilizing. Interestingly, the crystal structure of the RNA duplex [r(CGCFAAUUAGCG)]2 at 1.6 Å resolution revealed a configuration of the rF:A pair with a separation between F4 and N6 that was considerably larger than that in the corresponding pair in the DDD (Figure 2D).33 At almost 4 Å, the distance exceeds the sum of van der Waals radii by some 10%, indicating a rather loose association of F and A in RNA and hinting at a potentially considerable range of the F:A geometry inside canonical duplexes. This finding is consistent with the results of molecular dynamics (MD) simulations for an RNA containing a central rF:A pair.36 Compared to a reference duplex with U:A the modified duplex exhibited significantly larger conformational fluctuations in the central portion, with frequent base pair openings and shearing.
Whereas single rF:A pairs in the central portion of the guide strand resulted in an only marginally reduced activity compared to native siRNA, three consecutive rF:A pairs covering the same region had a detrimental effect on the activity.35 Evidently the softer core of the latter construct is not tolerated by the RNA-induced silencing complex (RISC) and perhaps hampers cleavage catalyzed by Ago2. The activity of an siRNA duplex with a single U:G mismatch in the guide strand adjacent to the cleavage site was as low as that of the above duplex with three rF:A mismatches. And replacing the same U:G by rF:G or placing the rF:G mismatch two nucleotides upstream in the guide strand abolished the activity altogether.35 When we analyzed the rF:G mismatch in the crystal structure of the duplex [r(CGCFAAUUGGCG)]2 at 1.1 Å resolution, we found to our surprise that its geometry mimicked that of the native U:G wobble pair (Figure 3). Given the weak association between rF and A in the same sequence context this is rather puzzling. Not only does the F2 atom in rF form a close contact with the imino N1-H from G (3.03 Å) in the minor groove, but the 3.2 Å distance between C3-H (rF) and O6(G) is also suggestive of a C-H…O H-bond. By comparison, the C3-H…N1 distances in the dF:A (DDD; Figure 2B) and rF:A pairs (RNA; Figure 2D) are longer and not compatible with a significant role in pairing stability.
Figure 3.
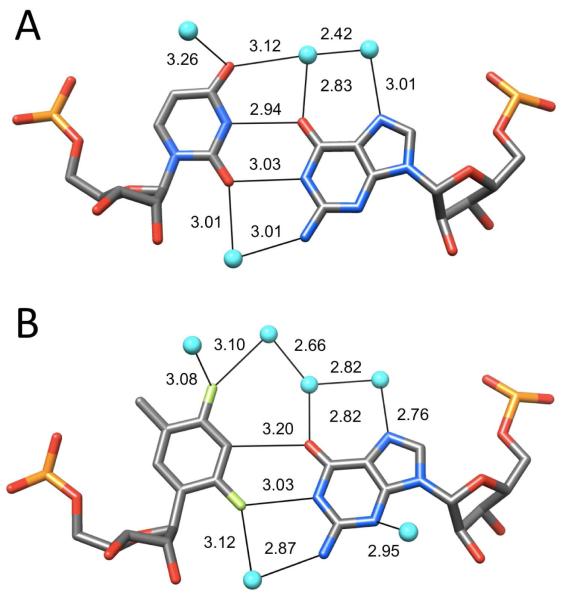
Geometry and hydration of RNA (A) U:G (rGGGGCUA:rUAGCUCC; PDB ID 434D)38 and (B) rF:G (rCGCFAAUUGGCG; PDB ID 2Q1O)35 pairs. Water molecules are cyan spheres and distances are in Å.
The mimicry of U:G by rF:G is not limited to the nucleotides but extends to the hydration of the pairs, with water molecules engaged in close contacts to F2 and F4 atoms (Figure 3B). Rather than focusing solely on the tight spacing between donor atoms and fluorine that are consistent with H-bond formation, the most convincing argument for electrostatics (not stacking) underlying the geometry of the rF:G pairs in our crystal structure is the displacement of G into the minor groove. Why, other than for favorable electrostatics, i.e. H-bonding, would rF and G slide along each other to assume a relative orientation that is basically identical with that seen for U and G (Figure 3)?
An MD simulation of F and G in B-DNA resulted in a loosely associated pair in a pseudo W-C configuration with F2(F) spaced at about 3.5 Å from N2(G) in the minor groove,37 inconsistent with our crystallographic results in the context of RNA. That rF in RNA behaves quite differently from F in DNA is not just supported by the tighter pairing between mismatched rF and G (Figure 3B) compared with the rF:A matching pair (Figure 2D), but by divergent effects of F incorporation into DNA and RNA on the thermodynamic stabilities of the respective duplexes. For example, we found that RNA with an rF:G mismatch melts at a slightly higher temperature than the same duplex with rF:A.35 This is noteworthy because the native duplex with U:A is of higher stability than the corresponding duplex with U:G. In addition, osmotic stress experiments demonstrated that significantly more water molecules are released upon melting of an RNA duplex with rF:G relative to the same duplex with rF:A, in line with our observation that the former base pair is surrounded by more water molecules in the crystal.33,35
Stabilizing H-bond Interactions by Fluorine
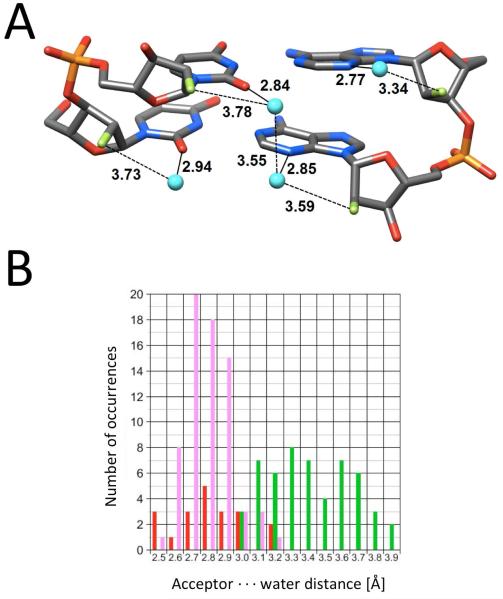
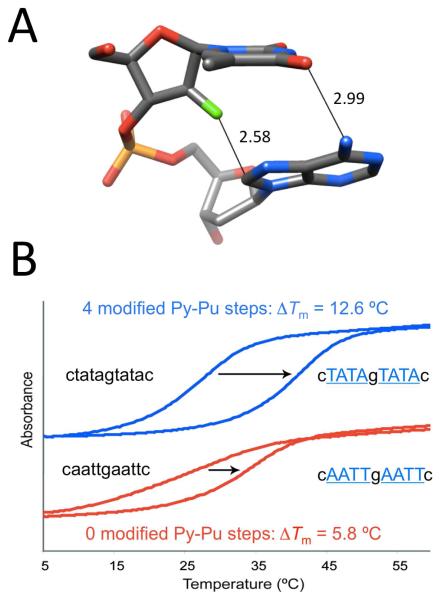
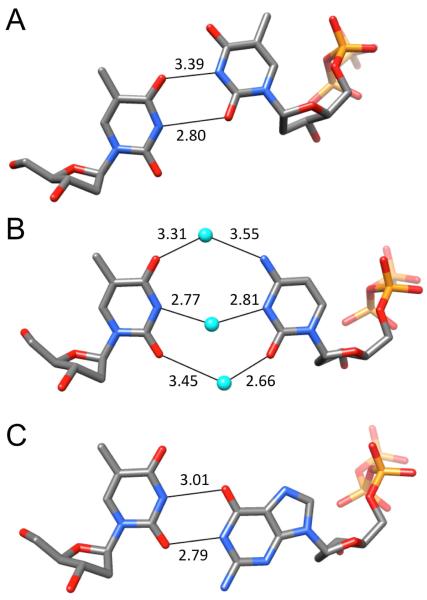
The crystallographic data for F incorporated into DNA and RNA, although still somewhat limited, demonstrate that fluorine doesn’t behave in a uniform manner. Rather, it is a sort of polar hydrophobe whose interactions appear to be influenced by the particular environment. Structural and thermodynamic studies relating to 2′-deoxy-2′-fluoro-ribonucleic acid (FRNA) and 2′-deoxy-2′-fluoro-arabinonucleic acid (FANA) provide further support for this dichotomy. Crystal structures of FRNA duplexes support the notion that fluorine replacing the ribose 2′-hydroxyl group is hydrophobic and does not engage in H-bond formation (Figure 4A).39,40 Fluorine polarizes the nucleobase and may strengthen W-C H-bonds but, unlike 2′-OH, is cannot act as a bridgehead for water molecules linking paired strands across the minor groove. This is evident from a comparison of the distances between water molecules and 2′-F, 2′-OH or base functions such as pyrimidine O2 and purine N3 (Figure 4B). Therefore, fluorine exposed to water is no match for H-bond acceptors such as hydroxyl, ketones or imines. Conversely, fluorine in the arabino configuration in FANA is sequestered from solvent inside the duplex and was found to engage in a tight contact with C8-H of the 3′-adjacent adenine in the crystal structure of an A-DNA duplex with incorporated FANA-Ts (Figure 5A).41 A short distance between non-bonded atoms alone, even if these constitute a potential donor-acceptor pair, cannot be taken as evidence for a stabilizing interaction. However, in the case of the F2′…H-C contact at FANA intra-strand pyrimidine-purine steps, there is experimental evidence from standard UV melts that such fluorine (pseudo) H-bonds are energetically favorable (Figure 5B).42 The FRNA and FANA examples provide a powerful demonstration of altered fluorine behavior as a consequence of an epimeric change and different environments.
Figure 4.
Fluorine is a poor acceptor in the minor groove of FRNA. (A) Example of the water structure in [f(CGAAUUCG)]2 (PDB code 3P4A).39 (B) Comparison between distances to water molecules by O2′ (red), O2 (pyrimidine)/N3 (purine) (pink) and F2′ (green) in the minor grooves of FRNA or mixed FRNA/RNA duplexes.40
Figure 5.
Stabilizing effect of C-H…F interactions in FANA-modified DNAs. (A) Tight contact between FANA-T 2′-fluorine and C8-H of dA at a (faT)p(dA) Py-Pu step in an A-form duplex (PDB code 2FIL).41 (B) Incorporation of FANA residues (capital font in blue) into an oligo-2′-deoxynucleotide (small font) at Py-Pu steps leads to increased stability of the duplex with complementary RNA (UV melting assay) compared to a mixed FANA-DNA strand of identical composition but lacking a Py-Pu step.42
F Pairing at a Y-Family Pol Active Site
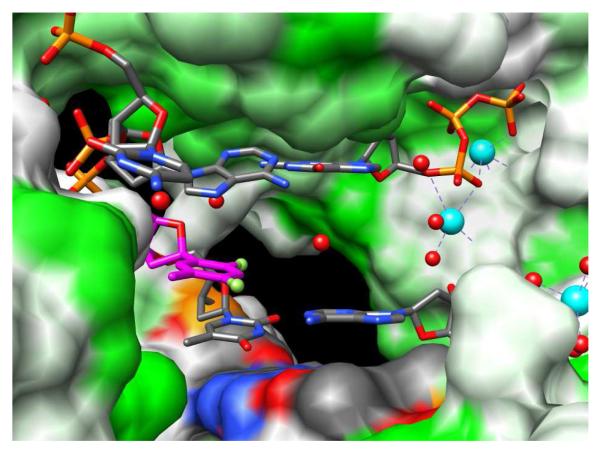
Y-family translesion pols possess spacious active sites (Figure 6), consistent with their roles of inserting dNTPs opposite and/or extending from adducted bases.22-27 With the Dpo4 pol from S. solfataricus insertion of dATP opposite F was ca. 5,000-fold less efficient than insertion opposite T, as measured by steady-state kinetics (kcat/Km).25 However, in the steady state Dpo4 exhibited a robust ability to discriminate between inserting either dATP or dGTP opposite F, as the efficiency in the latter case was reduced about 200-fold. Consistent with the >5,000-fold reduced efficiency in the steady state, no product (F:A) was formed in the pre-steady state, even at high dATP concentrations.25 Also in the pre-steady state, extension following an F:A pair was moderately inhibited relative to T:A, but no extension at all was seen following F:G.
Figure 6.
The roomy active site of the Dpo4 Y-family pol can accommodate two template bases (A and F in this so-called type II complex; PDB ID 2VA3).25 H-bonding is required between template base and incoming dNTP for efficient bypass. The pol surface is colored according to hydrophobicity (min=white; max=green), F carbons are magenta, and Ca2+ ions and water molecules are cyan and red spheres, respectively.
The crystal structure of Dpo4 with F:A at the active site revealed a loose association between the two, with F4(F) and N6(A) separated by >5 Å (Figure 2E).25 However, despite this large separation, the distance between C1′ atoms was basically identical to that in a native T:A pair (Figure 2). The geometry of the F:A pair seen here differs drastically from the tightly associated F and A in the core of the DDD (Figure 2B),30 indicating once again that the environment crucially affects shape and size of base pairs involving F. This is particularly evident in the structure of a ternary complex of Dpo4 with F lodged opposite G at the active site that allowed two independent observations of this pairing (Figure 7).25 Relative to F:A, the exocyclic N2 amino group of G pushes the difluorotoluene moiety further away (Figures 2E and 7A). Since neither G nor A form H-bonds to F at the Dpo4 active site, the pol ironically may rely solely on steric factors to discriminate between the two. In the second complex, G was completely ejected out of the active site (Figure 7B). It is clear that such a misalignment will prevent a proper line up between the 3′-terminal hydroxyl group of the primer and the α-phosphate of the incoming nucleotide (i.e. dGTP), required for efficient replication.
Figure 7.
Geometry of F:G at the active site of the Y-family pol Dpo4 (PDB ID 2V9W).25
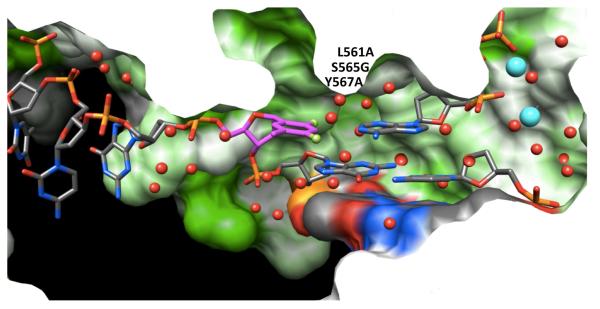
F Pairing at a B-Family Pol Active Site
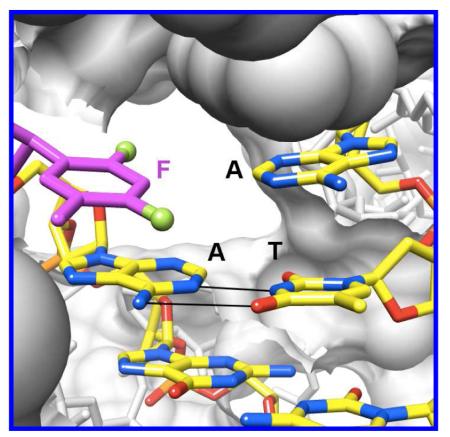
A- and B-family pols possess tight active sites, consistent with their ability to replicate DNA with high accuracy and efficiency (Figure 8). Thus, such pol active sites may select for standard size W-C pairs between natural purines and pyrimidines. Crystal structures for ternary complexes of the RB69 B-family pol (the L561A/S565G/Y567A triple mutant) with template F opposite dATP, dTTP, dGTP or dCTP were reported very recently.43 In the F:dATP complex, F and A were found to be more closely spaced (Figure 2F) relative to the situation in Dpo4 (Figure 2E). However, the distance of ca. 4 Å between F4(F) and N6(A) is inconsistent with H-bonding. The geometry of the F:A pair in the RB69 structure actually bears close resemblance to that of F:A in duplex RNA (Figure 2D), at once invalidating the authors’ suggestion that the geometries of pairs involving F at pol active sites and within isolated duplexes differ as a result of backbone constraints in the latter. As far as the steric hypothesis is concerned the F:A arrangement in RB69 is not a close mimic of T:A (i.e. Figure 2A). With regard to the ability of fluorine to participate in H-bonds, the structure of the complex with F:A at the RB69 active site stands in contrast to our observations for F:A in the DDD (Figure 2B).30
Figure 8.
The tight active site of the RB69 B-family pol with template F opposite dATP (PDB ID 3QER).43 Note that portions of the pol surface are sliced away to allow a view into the active site from the major groove side and that access to the minor groove is blocked compared with Dpo4 (Figure 6; please see that figure for color codes).
That the tight active site of the B-family pol can be conducive to fluorine H-bonding was demonstrated by the crystal structure of RB69 with mismatched F:dTTP (Figure 9A). Not only did F4(F) engage in a contact <3 Å with N3(T), but both Cα and amide N-H of glycine 568 that seals the minor groove were located quite close to the F2 atom (similar in the other structures). However, these latter contacts involving F2 as well as those between F4 and water molecules wedged into the active site do not by themselves provide strong support for fluorine H-bonding (Figure 9A). Moreover, the mutations in the RB69 pol studied affect amino acids L561, S565 and Y567 that line the minor groove (see Figure 8 for orientation). Potential effects of the latter on the F pairing mode can therefore not be fully evaluated. By comparison, F and dCTP lie far apart in the active site (Figure 9B), providing a rationale for the slightly more efficient incorporation of T (kpol/Kd = 2.8 × 10−2 μM−1s−1) over C (kpol/Kd = 1.3 × 10−2 μM−1 s−1) opposite F (kpol/Kd,F:dATP = 1.6 μM−1s−1). The preferred incorporation of dATP opposite template F is likely a consequence of the overall fit within the active site and the enhanced stacking of the incoming purine base relative to T and C. Insertion of dGTP opposite F proceeds with the lowest efficiency (kpol/Kd = 9 × 10−3 μM−1s−1) and the structure reveals a buckled orientation of G (Figure 9C) with diminished stacking on the penultimate base pair. However, the (F)F2…H-N2(G) and (F)C3-H…H-N1(G) distances are unrealistically short in the structure (Figure 9C) and are well below what can be expected for a fluorine H-bond and a C-H…H-N van der Waals interaction, respectively.
Figure 9.
Geometry of (A) F:dTTP (PDB ID 3QEP), (B) F:dCTP (PDB ID 3QEI), and (C) F:dGTP (PDB ID 3QES) at the active site of the B-family pol RB69.43 Pol residues A567, G568 and A569 probing the nascent base pair from the minor groove side are depicted with carbon atoms colored in magenta (similar for F:dCTP and F:dGTP) and selected water molecules are cyan (F:dTTP).
Looking at these four base pairs involving F and incoming dNTPs at the RB69 active site (Figures 2F, 9) and comparing them to the corresponding pairs with T (Figures 2A, 10), it becomes clear that template F and T behave quite differently. Thus, it is hard to avoid the impression that one may have compared apples and oranges by simply invoking sterics as the source of the ability of replicative pols to preferentially incorporate dATP opposite F.
Figure 10.
Geometry of (A) T:dTTP (PDB ID 3QET), (B) T:dCTP (PDB ID 3QEV), and (C) T:dGTP (PDB ID 3QEX) at the RB69 active site.43
Conclusions
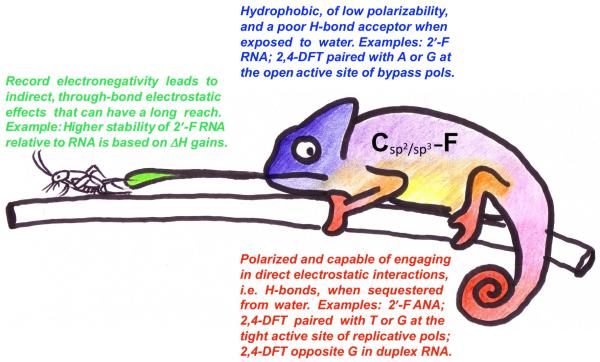
Crystal structures of DNAs, RNAs and pol-DNA complexes containing dF (rF in RNA) or other fluorine-modified nucleosides (FRNA, FANA) determined in recent years demonstrate that the standard view of fluorine as essentially hydrophobic and unable to participate in H-bonds belies a more complex behavior that appears to be influenced by the molecular environment and involves participation in H-bonds in many cases (Figure 11). Fluorine in a polar environment, i.e. exposed to water as in FRNA (Figure 4) does not engage in H-bonding. By contrast, when sequestered from solvent, i.e. in the core of duplexes (Figures 2B, 3B, 5A) or at active sites of replicative pols (Figure 9A), fluorine can participate in H-bonds. However, such H-bonds provide only weak constraints as can be seen from the wide range of geometries exhibited by F:A pairs (Figure 2) or the divergent geometries of F:G in duplex RNA (Figure 3B), at the active site of the Dpo4 pol (Figure 7), or at the active site of the RB69 pol (Figure 9C). Based on the currently available data, F behaves quite differently in DNA and RNA. Clearly, a key assumption of the steric hypothesis for DNA replication, namely that the F analog is unable to engage in H-bonds is no longer tenable. Further, the pairing geometries of F (Figure 9) and T (Figure 10) at a pol active site are quite different, indicating that F is a rather poor shape mimic of T even when it does engage in H-bonding.
Figure 11.
Organic mfluorine is a chemical chameleon, its ‘color’ (electronic preference) depending on the local environment.
Supplementary Material
Acknowledgments
I would like to thank the US National Institutes of Health for continuous support (R01 GM055237, P01 ES005355) and past and present members of the laboratory for their contributions.
BIOGRAPHICAL INFORMATION
Martin Egli is Professor in the Department of Biochemistry at the School of Medicine, Vanderbilt University, Nashville, Tennessee, USA.
REFERENCES
- 1.Moran S, Ren RX-F, Rumney S, Kool ET. Difluorotoluene, a Nonpolar Isostere of Thymine, Codes Specifically and Efficiently for Adenine in DNA Replication. J. Am. Chem. Soc. 1997;119:2056–2057. doi: 10.1021/ja963718g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moran S, Ren RX, Kool ET. A Thymidine Triphosphate Shape Analog Lacking Watson-Crick Pairing Ability is Replicated With High Sequence Selectivity. Proc. Natl. Acad. Sci. USA. 1997;94:10506–10511. doi: 10.1073/pnas.94.20.10506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morales JC, Kool ET. Efficient Replication Between Non-Hydrogen-Bonded Nucleoside Shape Analogs. Nat. Struct. Biol. 1998;5:950–954. doi: 10.1038/2925. [DOI] [PubMed] [Google Scholar]
- 4.Fischer E. Einfluss der Configuration auf die Wirkung der Enzyme. Ber. Dtsch. Chem. Ges. 1894;27:2985–2993. [Google Scholar]
- 5.Koshland DE., Jr. The Key-Lock Theory and the Induced Fit Theory. Angew. Chem. Int. Ed. 1994;33:2375–2378. [Google Scholar]
- 6.Kool ET. Hydrogen Bonding, Base Stacking, and Steric Effects in DNA Replication. Ann. Rev. Biophys. Biomol. Struct. 2001;30:1–22. doi: 10.1146/annurev.biophys.30.1.1. [DOI] [PubMed] [Google Scholar]
- 7.Kool ET. Active Site Tightness and Substrate Fit in DNA Replication. Ann. Rev. Biochem. 2002;71:191–219. doi: 10.1146/annurev.biochem.71.110601.135453. [DOI] [PubMed] [Google Scholar]
- 8.Sintim HO, Kool ET. The Difluorotoluene Debate - A Decade Later. Chem. Comm. 2006;35:3665–3675. doi: 10.1039/b605414e. [DOI] [PubMed] [Google Scholar]
- 9.Lee I, Berdis AJ. Non-Natural Nucleotides as Probes for the Mechanism and Fdelity of DNA Polymerases. Biochim. Biophys. Acta. 2010;1804:1064–1080. doi: 10.1016/j.bbapap.2009.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Howard JAK, Hoy VJ, O’Hagan D, Smith GT. How Good is Fluorine as a Hydrogen Bond Acceptor? Tetrahedron. 1996;52:12613–12622. [Google Scholar]
- 11.Dunitz JD. Organic Fluorine: Odd Man Out. ChemBioChem. 2004;5:614–621. doi: 10.1002/cbic.200300801. [DOI] [PubMed] [Google Scholar]
- 12.Rybalova TV, Bagryanskaya IY. C-F…π, F…H, and F…F Intermolecular Interactions and F-Aggregation: Role in Crystal Engineering of Fluoroorganic Compounds. J. Struct. Chem. 2009;50:741–753. [Google Scholar]
- 13.Zhou P, Zou J, Tian F, Shang Z. Fluorine Bonding - How Does It Work in Protein-Ligand Interactions? J. Chem. Inf. Model. 2009;49:2344–2355. doi: 10.1021/ci9002393. [DOI] [PubMed] [Google Scholar]
- 14.Chopra D, Guru Row TN. Role of Organic Fluorine in Crystal Engineering. Cryst. Eng. Comm. 2011;13:2175–2186. [Google Scholar]
- 15.Sun Z, McLaughlin LW. Probing the Nature of Three-Centered Hydrogen Bonds in Minor-Groove Ligand-DNA Interactions: The Contribution of Fluorine Hydrogen Bonds to Complex Stability. J. Am. Chem. Soc. 2007;129:12531–12536. doi: 10.1021/ja073981p. [DOI] [PubMed] [Google Scholar]
- 16.Schweitzer BA, Kool ET. Hydrophobic, Non-Hydrogen-Bonding Bases and Base Pairs in DNA. J. Am. Chem. Soc. 1995;117:1863–1872. doi: 10.1021/ja00112a001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koller AN, Bozilovic J, Engels JW, Gohlke H. Aromatic N Versus Aromatic F: Bioisosterism Discovered in RNA Base Pairing Interactions Leads to a Novel Class of Universal Base Analogs. Nucleic Acids. Res. 2010;38:3133–3146. doi: 10.1093/nar/gkp1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guckian K, Schweitzer BA, Ren RX-F, Sheils CJ, Paris PL, Tahmassebi DC, Kool ET. Experimental Measurement of Aromatic Stacking in the Context of Duplex DNA. J. Am. Chem. Soc. 1996;118:8182–8183. doi: 10.1021/ja961733f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guckian KM, Krugh TR, Kool ET. Solution Structure of a Duplex DNA Containing a Replicable Difluorotoluene-Adenine Pair. Nat. Struct. Biol. 1998;5:954–959. doi: 10.1038/2930. [DOI] [PubMed] [Google Scholar]
- 20.Wang X, Houk KN. Difluorotoluene, a thymine isostere, does not hydrogen bond at all. Chem. Commun. 1998:2631–2632. [Google Scholar]
- 21.Kim T, Brieba LG, Ellenberger T, Kool ET. Functional Evidence for a Small and Rigid Active Site in a High Fdelity DNA Polymerase: Probing T7 DNA Polymerase with Variably Sized Base Pairs. J. Biol. Chem. 2006;281:2289–2295. doi: 10.1074/jbc.M510744200. [DOI] [PubMed] [Google Scholar]
- 22.Potapova O, Chan C, DeLucia AM, Helquist SA, Kool ET, Grindley NDF, Joyce CM. DNA Polymerase Catalysis in the Absence of Watson-Crick Hydrogen Bonds: Analysis by Single-Turnover Kinetics. Biochemistry. 2006;45:890–898. doi: 10.1021/bi051792i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee HR, Helquist SA, Kool ET, Johnson KA. Importance of Hydrogen Bonding for Efficiency and Specificity of the Human Mitochondrial DNA Polymerase. J. Biol. Chem. 2008;283:14402–14410. doi: 10.1074/jbc.M705007200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mizukami S, Kim T, Helquist SA, Kool ET. Varying DNA Base-Pair Size in Subangstrom Increments: Evidence for a Loose, Not Large, Active Site in Low-Fidelity Dpo4 Polymerase. Biochemistry. 2006;45:2772–2778. doi: 10.1021/bi051961z. [DOI] [PubMed] [Google Scholar]
- 25.Irimia A, Eoff RL, Pallan PS, Guengerich FP, Egli M. Structure and Activity of Y-class DNA Polymerase Dpo4 from Sulfolobus solfataricus with Templates Containing the Hydrophobic Thymine Analog 2,4-Difluorotoluene. J. Biol. Chem. 2007;282:36421–36433. doi: 10.1074/jbc.M707267200. [DOI] [PubMed] [Google Scholar]
- 26.Washington MT, Helquist SA, Kool ET, Prakash L, Prakash S. Requirement of Watson-Crick Hydrogen Bonding for DNA Synthesis by Yeast DNA Polymerase η. Mol. Cell. Biol. 2003;23:5107–5112. doi: 10.1128/MCB.23.14.5107-5112.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wolfle WT, Washington MT, Kool ET, Spratt TE, Helquist SA, Prakash L, Prakash S. Evidence for a Watson-Crick Hydrogen Bonding Requirement in DNA Synthesis by Human DNA Polymerase κ. Mol. Cell. Biol. 2005;25:7137–7143. doi: 10.1128/MCB.25.16.7137-7143.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hogg M, Wallace SS, Doublié S. Bumps in the Road: How Replicative DNA Polymerases See DNA Damage. Curr. Opin. Struct. Biol. 2005;15:86–93. doi: 10.1016/j.sbi.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 29.Egli M. DNA-Cation Interactions: Quo Vadis? Chem. Biol. 2002;9:277–286. doi: 10.1016/s1074-5521(02)00116-3. [DOI] [PubMed] [Google Scholar]
- 30.Pallan PS, Egli M. The Pairing Geometry of the Hydrophobic Thymine Analog 2,4-Difluorotoluene in Duplex DNA as Analyzed by X-ray Crystallography. J. Am. Chem. Soc. 2009;131:12548–12549. doi: 10.1021/ja905739j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pallan PS, Egli M. Insights into RNA/DNA Hybrid Recognition and Processing by RNase H from the Crystal Structure of a Non-specific Enzyme-dsDNA Complex. Cell Cycle. 2008;7:2562–2569. doi: 10.4161/cc.7.16.6461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tereshko V, Minasov G, Egli M. The Dickerson-Drew B-DNA Dodecamer Revisited - At Atomic Resolution. J. Am. Chem. Soc. 1999;121:470–471. [Google Scholar]
- 33.Xia J, Noronha A, Toudjarska I, Li F, Akinc A, Braich R, Rajeev KG, Egli M, Manoharan M. siRNAs with a Ribo-difluorotoluyl Nucleotide: Structure, RISC-mediated Recognition, and Silencing. ACS Chem. Biol. 2006;1:176–183. doi: 10.1021/cb600063p. [DOI] [PubMed] [Google Scholar]
- 34.Somoza A, Chelliserrykattil J, Kool ET. The Roles of Hydrogen Bonding and Sterics in RNA Interference. Angew. Chem. Int. Ed. 2006;45:4994–4997. doi: 10.1002/anie.200601311. [DOI] [PubMed] [Google Scholar]
- 35.Li F, Pallan PS, Maier MA, Rajeev KG, Mathieu SL, Kreutz C, Fan Y, Sanghvi J, Micura R, Rozners E, Manoharan M, Egli M. Crystal Structure, Stability and in vitro RNAi Activity of Oligoribonucleotides Containing the Ribo-difluorotoluyl Nucleotide: Insights into Substrate Requirements by the Human RISC Ago2 Enzyme. Nucleic Acids Res. 2007;35:6424–6438. doi: 10.1093/nar/gkm664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zacharias M, Engels JW. Influence of a Fluorobenzene Nucleobase Analogue on the Conformational Flexibility of RNA Studied by Molecular Dynamics Simulations. Nucleic Acids Res. 2004;32:6304–6311. doi: 10.1093/nar/gkh971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pfaff DA, Clarke KM, Parr TA, Cole JM, Geierstanger BH, Tahmassebi DC, Dwyer TJ. Solution Structure of a DNA Duplex Containing a Guanine-Difluorotoluene Pair: A Wobble Pair without Hydrogen Bonding? J. Am. Chem. Soc. 2008;130:4869–4878. doi: 10.1021/ja7103608. [DOI] [PubMed] [Google Scholar]
- 38.Mueller U, Schubel H, Sprinzl M, Heinemann U. Crystal Structure of Acceptor Stem of tRNAAla from Escherichia coli Shows Unique G.U Wobble Base Pair at 1.16 Å Resolution. RNA. 1999;5:670–677. doi: 10.1017/s1355838299982304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Manoharan M, Akinc A, Pandey RK, Qin J, Hadwiger P, John M, Mills K, Charisse K, Maier MA, Nechev L, Greene EM, Pallan PS, Rozners E, Rajeev KG, Egli M. Unique Gene-silencing and Structural Properties of 2'-F Modified siRNAs. Angew. Chem. Int. Ed. 2011;50:2284–2288. doi: 10.1002/anie.201006519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pallan PS, Greene E, Jicman P, Pandey R, Manoharan M, Rozners E, Egli M. Unexpected Origins of the Enhanced Pairing Affinity of 2'-Fluoro-modified. RNA. Nucleic Acids Res. 2011;39:3482–3495. doi: 10.1093/nar/gkq1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li F, Sarkhel S, Wilds CJ, Wawrzak Z, Prakash TP, Manoharan M, Egli M. 2′-Fluoroarabino- and Arabinonucleic Acid Show Different Conformations, Resulting in Deviating RNA Affinities and Processing of Their Heteroduplexes with RNA by RNase H. Biochemistry. 2006;45:4141–4152. doi: 10.1021/bi052322r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yahyaee Anzahaee M, Watts JK, Alla NR, Nicholson AW, Damha MJ. Energetically Important C-H…F-C Pseudohydrogen Bonding in Water: Evidence and Application to Rational Design of Oligonucleotides with High Binding Affinity. J. Am. Chem. Soc. 2011;133:728–731. doi: 10.1021/ja109817p. [DOI] [PubMed] [Google Scholar]
- 43.Xia S, Eom SH, Konigsberg WH, Wang J. Structural Basis for Differential Insertion Kinetics of dNMPs Opposite a Difluorotoluene Nucleotide Residue. Bochemistry. 2012;51:1476–1485. doi: 10.1021/bi2016487. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.