Abstract
TGFβ3 is essential for palate development, particularly during the late phase of palatogenesis when the disintegration of the palatal medial edge seam (MES) occurs resulting in mesenchymal confluence. The MES is composed of medial-edge epithelium (MEE) of opposite palatal shelves; its complete disintegration is essential for mediating correct craniofacial morphogenesis. This phenomenon is initiated by TGFβ3 upon adherence of opposing palatal shelves, and subsequently epithelial-mesenchymal transition (EMT) instigates the loss of E-Cadherin, causing the MES to break into small epithelial islands forming confluent palatal mesenchyme, however, apoptosis and cell migration or in combination of all are other established mechanisms of seam disintegration. To investigate the molecular mechanisms that cause this E-Cadherin loss, we isolated and cultured murine embryonic primary MES cells from adhered palates and employed several biological approaches to explore the mechanism by which TGFβ3 facilitates palatal seam disintegration. Here, we demonstrate that TGFβ3 signals by activating both Smad-dependent and Smad-independent pathways. However, activation of the two most common EMT related transcription factors, Snail and SIP, was facilitated by Smad-independent pathways, contrary to the commonly accepted Smad-dependent pathway. Finally, we provide the first evidence that TGFβ3-activated Snail and SIP1, combined with Smad4, bind to the E-Cadherin promoter to repress its transcription in response to TGFβ3 signaling. These results suggest that TGFβ3 uses multiple pathways to activate Snail and SIP1 and these transcription factors repress the cell-cell adhesion protein, E-Cadherin, to induce palatal epithelial seam EMT. Manipulation and intervention of the pathways stimulated by TGFβ3 during palate development may have a significant therapeutic potential.
Keywords: Epithelial-Mesenchymal Transition, EMT, Transforming Growth Factor β3, TGFβ3, E-Cadherin, Medial Epithelial Seam, MES
Introduction
Cleft palate, the most common craniofacial deformity, can arise due to a lack of palatal shelf adhesion (Gritli-Linde, 2007). Palatal shelves grow out bilaterally from the internal surfaces of the maxillary processes, elongate on each side of the tongue and become horizontal above the tongue as it descends. As soon as the opposing shelves reach each other, the lateral surfaces of the underlying basal medial edge epithelia (MEE) cells contact with the opposite palatal shelves and subsequently form a medial epithelial seam (MES) (Ferguson, 1988). Complete disintegration of the MES is essential to form a single confluent palate craniofacial structure (Ferguson, 1988). Along with apoptosis and cellular migration, epithelial mesenchymal transition (EMT) has been proposed as a major palatal seam disintegration mechanism (Nawshad, 2008). EMT is an alteration in phenotype of an epithelial cell that is unified as an articulate sheet with apical-basal polarity to a combination of mesenchymal cells with less articulate, more migratory capacity, that also lacks the apical-basal polarity of epithelial cells (Thiery, 2003b). One of the most prominent features of EMT is the loss of the cell-cell adhesion protein E-Cadherin. Many transcription factor proteins, such as Snail, Slug, E12/E47, SIP-1, ZEB-1, and Twist directly bind to the E-Cadherin gene promoter to inhibit its transcription (Peinado et al., 2004a, Peinado et al., 2004b)
The stages of palatal development are coordinated and controlled by various proteins such as growth factors like transforming growth factor (TGFβ). The MES disintegration phase is regulated by TGFβ3 (Shuler et al., 1992). Following adherence of opposing palatal shelves and formation of the seam, the basal MES cells show increased expression of TGFβ3 (Nawshad et al., 2004) which occurs until EMT is complete (Tudela et al., 2002). Several lines of evidence suggest the importance of TGFβ3-mediated MES disintegration. TGFβ3-knockout mice (Proetzel et al., 1995, Taya et al., 1999), as well as some naturally TGFβ3-null avian systems (Sun et al., 1998) have complete penetrance for cleft palate. Furthermore, treatment of palates from TGFβ3-knockout mice with exogenous TGFβ3 is sufficient to rescue palatal fusion (Taya et al., 1999). While all isoforms (i.e., 1, 2, and 3) of TGFβ are expressed during different stages of palate development between 11–16.5 days post coitum (dpc), it is TGFβ3 that is expressed highly in the palatal seam region at 14–15 dpc when disintegration is about to begin, making it a likely candidate to induce palatal EMT (Tudela et al., 2002, Xia & Cheng, 2005, Dudas et al., 2004, Blavier et al., 2001). TGFβ3 signaling causes the transformation of epithelial seam cells into mesenchymal cells, as well as apoptosis and cell migration, which results in a confluent palate (Carette & Ferguson, 1992). Following palatal confluency, the expression of TGFβ3 ceases at 16 dpc, assuming that seam disintegration by EMT processes is achieved (Proetzel et al., 1995).
The transformation of palate medial-edge epithelium to mesenchyme has been well documented (Fitchett & Hay, 1989, Kaartinen et al., 1997, Nawshad et al., 2004). However, signaling mechanisms that promote EMT during palatogenesis have only recently been investigated in detail. TGFβ3 is known to have an essential role in palate development, including the transformation of MES cells into the mesenchymal morphology (Brunet et al., 1995, Nawshad & Hay, 2003b, Nawshad et al., 2005).
Herein, we provide evidence that during palatal EMT, TGFβ3 induces both Smad-dependent and Smad-independent pathways to activate several EMT-related transcription factors, such as Snail and SIP1 to repress the cell-cell adhesion molecule E-Cadherin, thereby inducing EMT.
Materials and Methods
Culture of embryonic palate and isolation of primary MES cells
Timed-pregnant CF1 mice (Charles Rivers Laboratories, Inc, MA) were obtained for this study. Primary Medial edge epithelial seam (MES) cell culture: The primary method used for palatal medial epithelial seam (MES) cell culture from 14 dpc mouse embryos has been explained in detail in (Ahmed et al., 2007, Nawshad et al., 2007). In short, isolated palates were cultured at 37°C in a humidified gas mixture incubator (5% CO2 and 95% air), palatal MES cells are cultured in flasks and collected when they reach approximately 80% confluency prior to any exogenous treatment. From one pair of palates, enough cells may be isolated to seed in one T-25 flask. These cells are divided 1:2 every three days for as many as five times to yield enough MES cells to preserve healthy growth and morphology for experiments and storage. As described in (Ahmed et al., 2007, Nawshad et al., 2007), these primary cells undergo clonal analysis for homogeneity of the MES cells to confirm 100% homogeneity; it is necessary to ensure the MES cells are not contaminated by surrounding mesenchymal cells using cobblestone epithelial morphology and positive staining with E-Cadherin, Desmoplakin, and Desmoglein for the classification of epithelial clone type. In vivo, MES cells produce optimum endogenous TGFβ3; however, when MES cells are removed from primary tissue, TGFβ3 significantly reduces its expression to levels that are not sufficient to stimulate cellular changes. To mimic the in vivo model system, exogenous TGFβ3 was added at different doses at different stages, as shown by us earlier (LaGamba et al., 2005). To demonstrate protein expression (such as TGFβ3 as shown in Fig. 1), we collected the embryonic palates at 14, 14.5, 15, 15.5, 16, 16.5 and 17 dpc, followed by Buin’s fixation and paraffin embedding of the embryonic palates.
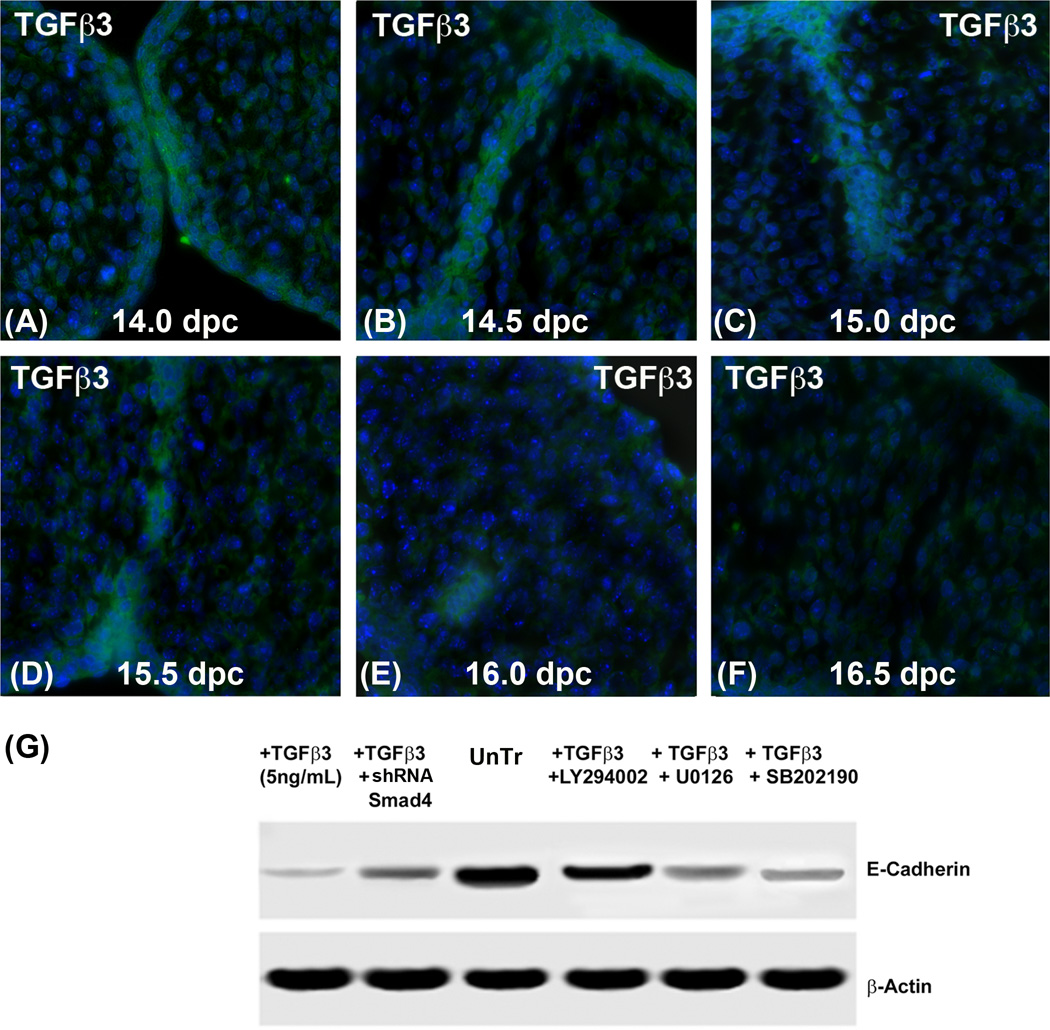
Fig. 1. Expression of TGFβ3 and E-Cadherin as palatal seam disintegrates.
Immunofluorescence protein expression assay was performed at different stages of palate development in vivo. Palates were collected at different stages (14.0, 14.5, 15, 15.5, 16 and 16.5 dpc) from murine CF1 pregnant mice. (A) At 14.0 dpc, TGFβ3 was highly expressed in the approaching opposite palates (A) as well as newly formed full length seam epithelial cells at 14.5 dpc (B). Fifteen and 15.5 dpc (C & D), the expression of TGFβ3 was reduced as seam epithelial cells broke into small islands. (E) By 16.0 dpc, the seam was significantly disintegrated; only one or two epithelial islands show TGFβ3 expression. (F) By 16.5 dpc, when the epithelial seam is completely disintegrated, no epithelial seam remained to express TGFβ3. (G) MES cells were treated with a Smad inhibitor (pRetrosuper-shRNA Smad4) for 48 hours, as well as small synthetic chemical inhibitors – SB202190 (20µM), an inhibitor of p38MAPK; U0126 (20µM), an inhibitor of MEK1/2 and LY294002 (20µM), an inhibitor of PI3 Kinase for 60 min. Following these treatments, exogenous recombinant TGFβ3 (5ng/mL) was added for another 24 h in the presence of these inhibitors. The results demonstrate that MES cells treated with exogenous TGFβ3 (5ng/mL) for 24 h had low levels of E-Cadherin expression. Inhibition of Smad-dependent and Smad-independent signalling reversed TGFβ3 mediated E-cadherin loss, except PI3 kinase pathway, as seen by the treatment with LY294002. Inhibiting TGFβ3 signaling with a blocking antibody resulted in higher expression of E-Cadherin compared to the MES cells treated with only TGFβ3.
Cell culture/Treatment conditions/Antibodies
MES cells were cultured in DMEM media that contained different concentrations of FBS (10% to induce MES growth and confluency and 1% serum starvation to abolish the superfluous effect on MES cellular biofunction) with 1% penicillin/streptomycin. TGFβ3 (R&D Systems, CA) was used at concentrations of 2, 5, and 10ng/mL. Untreated MEE (no TGFβ3) cells were used as a control. TGFβ3 was changed every 24 hours (h). Blocking TGFβ3 antibody was purchased from R&D Systems, CA, and was used at 10ng/mL for 24 h to abolish the effect of endogenous TGFβ3. To inhibit the Smad-dependent pathway, small hairpin RNA (shRNA) Smad4, pRetrosuper-shSmad4 (Addgene, MA), was used which specifically targets the coding region of murine Smad4. A −800 bp E-Cadherin gene promoter construct (pGL3-E075, Wild type from −650- to +95 generously provided by J. Behrens, University Erlangen-Nürnberg, Erlangen, Germany) was cloned into the pGL3-Lux reporter plasmid. The pGL3-E075-Lux, its mutant constructs (see “Site specific mutagenesis” below) and pRetrosuper-shSmad4 were transfected into the MES cells by gently adding solution containing 10µl of Lipofectamine 2000 in 300µl of Opti-MEM medium for 24 h, as described previously by us (Medici & Nawshad, 2010). The Smad-independent pathways were blocked using commercially available small synthetic chemical inhibitors, such as SB202190 (20µM), an inhibitor of p38MAPK (Sigma-Aldrich, MO); U0126 (20µM), an inhibitor of MEK1/2 (Cell Signaling, MA); and LY294002 (20µM), an inhibitor of PI3 Kinase (Cell Signaling, MA) for 60 min. Following all inhibitions (pRetrosuper-shSmad4 and chemical inhibitors), TGFβ3 (5ng/mL) was added in the presence of these molecules up to 48 h prior to mRNA extraction at 0, 12, 24, 36 and 48 h (see “Reverse Transcriptase PCR” below). All antibodies and inhibitors were used at the concentration and time point recommended by the respective manufacturer/provider as well as confirmed by us previously (Nawshad & Hay, 2003a, Ahmed et al., 2007, Iordanskaia & Nawshad, 2011, Nawshad et al., 2007). For Immunofluorescence, primary antibodies used (and their source) included the following: TGFβ3 (R&D systems, CA), E-Cadherin (kindly provided by Dr. James Wahl, University of Nebraska Medical Center), Vimentin (Sigma-Aldrich, MO), Fibronectin (Abcam, MA), N-Cadherin (kindly provided by Dr. Keith R. Johnson, University of Nebraska Medical Center), ZO-1 (Invitrogen, CA), Keratin 14 (Abcam, MA), pSmad2, Smad2 and Smad4 (Cells Signaling, MA). For protein expression of MEE cells by western blot, the cells were serum starved for 24 h, followed by treatment with TGFβ3 (5 and 10ng/mL) in 1.0% FBS DMEM for 15 minutes up to 48 h and total, nuclear and cytoplasmic proteins were extracted. Immunoblotting primary antibodies used (and their source) included the following: E-Cadherin (Santa Cruz, CA), Snail-ChIP grade (Abcam, MA), SIP1 (Novus Biologicals, CO), β-Actin (Santa Cruz, CA), ERK 1/2 (Santa Cruz, CA), pERK 1/2 (Santa Cruz, CA), p38MAPK (Santa Cruz, CA), p-p38MAPK (Santa Cruz, CA), AKT 1/2 (Santa Cruz, CA), pAKT 1/2 (Santa Cruz, CA), MEK 1/2 (Santa Cruz, CA), and pMEK 1/2 (Santa Cruz, CA). Immunoblotting secondary antibody used was Alexa Fluor 680 (Molecular probes, OR). All antibodies and inhibitors were used at the concentration and time point recommended by the respective manufacturer/provider. Supplementary Figure 1 (A–G) shows the levels of protein repression by westernblot of all inhibitory treatment conditions.
Reverse Transcriptase (RT) PCR
As the methods mentioned before by us (LaGamba et al., 2005), 24 h serum starved (1% FBS) MES cells were subjected to TGFβ3 (5ng/ml) treatment for 0 to 48 h with or without inhibitors (see “Treatment conditions” above). RNA was harvested using the RNeasy Mini Kit (Qiagen, CA) according to the manufacturer’s instructions. RNA integrity was assessed using formaldehyde gels in 1×TAE buffer and RNA purity and concentration were determined by the 260/280 ratio on a Nanodrop 2000C (Thermoscientific, MA). RT-PCR was performed using the SuperScriptIII RT-PCR kit (Invitrogen, CA) according to the manufacturer’s protocol. The sequences of primers were obtained from the Invitrogen online PCR primer design site, and were synthesized at the Molecular Biology Core Facility, UNMC. Reactions were stimulated using a PTC-100 thermal cycler (MJ Research, Inc. MA), with 30 cycles per sample. Samples were run on 1.5% agarose gels that contained ethidium bromide (1.5µL) and were visualized using a ChemiDoc Imager (Bio-Rad, CA). Sequences of primers used are detailed in the Supplementary Table 1A.
Immunofluorescence and Immunobloting
Both MES cells and embryonic palates from 14.0–16.5 dpc underwent Immunofluorescence and Immunoblotting techniques as described by us previously (Ahmed et al., 2007, Nawshad & Hay, 2003a, Nawshad et al., 2007, Iordanskaia & Nawshad, 2010). For protein expression of MES cells by Western blot, the cells were grown to confluency in 10% FBS and serum starved in 1% FBS for 24 h, followed by treatment with TGFβ3 (5 and 10ng/mL) in 1.0% FBS DMEM for 0 to 24 h for total protein extraction. For nuclear and cytoplasmic proteins, we used the nuclear extraction kit from Chemicon Nuclear Extraction Kit (Millipore, MA) as done by us previously (Ahmed et al., 2007, Iordanskaia & Nawshad, 2011). The concentration of the nuclear, cytoplasmic and total proteins was obtained with the Genesys 10 UV scanner (Thermoscientific, MA) at 595 nm. 25µg of protein extract was electrophoresed on a 10% denaturing gel and transferred onto a nitrocellulose membrane. The membranes were blocked with gelatin, washed with PBS-Tween, incubated with the respective antibodies and reacted with anti-goat (1:1000) and anti-rabbit (1:2000) secondary antibodies (Cell Signaling, MA). The bands were then visualized by using an odyssey scanner (Li-Cor, NE). Intensity of the band was measured using the Carestream Molecular Imaging Software version 5.3.1 (Rochester, NY). To perform a t-test analysis of mean intensity measurements, a ROI analysis was done from the data to Microsoft Excel software from the exported “.txt” files. Data points for all samples are paired by spatial arrangement on gel and compared pairwise to minimize the impact of subtle background artifacts on image analysis.
For protein expression on embryonic palates and MES cells by Immunofluorescence, 8µm sections of 14.0 to 16.5 dpc palates and MES cells underwent Immunofluorescence as described by us previously (Ahmed et al., 2007, Nawshad & Hay, 2003a, Nawshad et al., 2007). Immunofluorescence secondary antibodies were obtained from: Rhodamine, 1:100 (Invitrogen, CA) and FITC, 1:200 (Jackson ImmunoResearch, PA).
Scratch Wound Assay
The Scratch Wound Assay was conducted as described in (Nawshad et al., 2007) but with minor modification. MES cells were grown to 80% confluency in 6-well culture plates, and a uniform straight line scratch was made with a sterile pipette tip. Scratches in TGFβ3 (2, 5 and 10ng/mL) treated and untreated wells were examined for 48 hours. The migration of cells (or gap filling) was monitored every 12 hours with phase-contrast microscopy where cells were morphologically assessed for the migratory phenotype.
Cell Motility Assay
The Cell Motility Assay was conducted as detailed in Nawshad et al. (2007). 8 µm pore size Transwell migration chambers of a 6-well plate (BD BioCoat, MA) were used for migration analyses. 5×105 MES cells were seeded in the presence of 5ng/mL TGFβ3 in 8µm pore size Transwell migration upper chambers of a 6-well plate. Treated and untreated TGFβ3 cells were allowed to migrate through the filter toward media containing serum (10%) for 24 to 48 hours at 37 °C. Cells that did not migrate through the filter were removed with a cotton swab from inside the upper chamber. Each filter was fixed in 4% Paraformaldehyde for 10 minutes, washed three times, each time for five minutes with 1x PBS, placed in Hematoxylin stain (Dako, Mayer’s hematoxylin) for 20 minutes, rinsed with water, and placed in bluing reagent (alkaline solution such as a weak ammonia solution, 0.08% in water) until the stain turned blue. Subsequently, the filters were washed again using deionized water. Migrating MES cells on the lower side of the filter were randomly counted at 10 areas per field by phase-contrast microscopy. The mean of the 10 areas was determined and is represented in the bar graph in Fig. 4C.
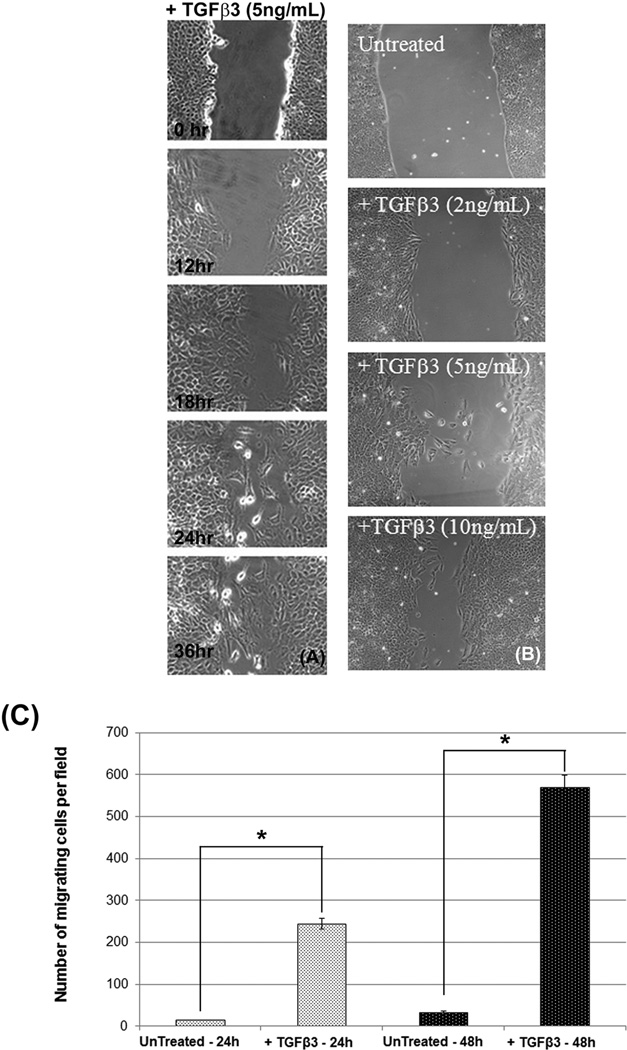
Fig. 4. TGFβ3 facilitates MES cell migration.
Cell migration was assessed by scratch wound assays of MES cells and observed by phase contrast microscopy every 12 hours up to 36 hours after they were scratched. MES cells became more migratory upon treatment with TGFβ3 (5ng/mL) in a time-dependent manner. Wounds begin to fill with migratory mesenchymal cells following TGFβ3 treatment for 12 h, and the number of migratory cells increased significantly as they continued to receive TGFβ3 for up to 36 hours. No migration was observed at 0 h (A). MES cells were also treated with different doses (2, 5, and 10ng/mL) of TGFβ3 for 24 hours. The results suggest that the higher doses cause increased cell migration (B).. Cell Motility Assay results show that only a few cells migrated in control MES cells at 24 h and 48 h, but at the same time points a significant number of MES cells migrated to the lower side of the upper chambers following TGFβ3 treatment, with the most migration observed at 48 h (C). Both TGFβ treatment (24 and 48 h) groups differed significantly from their respective negative control (untreated) groups, (p ≤ 0001, as indicated by *).
Chromatin immunoprecipitation (ChIP) assay
Equal quantities (4×107 cells) of the control (5% FBS), TGFβ3 (5ng/ml) stimulated MEE cells in 80%–90% monolayers were fixed with 1% formaldehyde for 10 min at RT for chromatin cross-linking. The reaction was stopped by adding glycine to a final concentration of 0.125M and the cells were immediately washed twice with ice-cold PBS and harvested by scraping cells in ice-cold 1×PBS+PMSF. The Simple ChIPTM Enzymatic Chromatin IP kit (Magnetic beads) (Cell Signaling, MA) was used according to the method successfully used by us previously (Iordanskaia & Nawshad, 2011). Briefly, cross-linked chromatin was isolated from the lysates by sonication (3 × 6 sec each) using a Microson XL 2000 sonicator. Immune precipitation of the cross-linked chromatin with rabbit anti-Smad4 antibody (Cell Signaling, MA), anti-Snail-ChIP grade (Abcam, MA) and anti-SIP1 (Novus Biologicals, CO), were subsequently performed. For the negative control, rabbit IgG and anti-actin antibody were used (Cell Signaling, MA) and for the positive control, Histone H3 antibody was used. The 100 ml of binding buffer from the Chromatin IP kit (Cell Signaling, MA), containing 10 to 20µg chromatin DNA were incubated overnight with antibodies at 4°C. The subsequent binding with magnetic beads, washing of immunocomplexes and DNA extraction were carried out following the manufacturer’s protocol. The mean ±SD obtained from three independent experiments were compared. Purified DNA was analyzed by PCR with primers specific for E-Pal and E-Box in the E-cadherin promoter. Reactions were performed in a 20µl volume according to the manufacturer’s recommendations. The sequences of primers used are detailed in the Supplementary Table 1B. Resulting DNA fragments were detected by electrophoresis in 1.5% agarose gel.
Site-directed mutagenesis
A −800 bp E-Cadherin gene promoter construct (pGL3-E075, Wild type from -−650- to +95 generously provided by J. Behrens, University Erlangen-Nürnberg, Erlangen, Germany) was cloned into the pGL3-Lux reporter plasmid. Site-directed mutagenesis (Mutant-Max) was used to create mutant −800bp promoter constructs. The mutant sites were confirmed by sequencing and then transfected into cells for luciferase assays as described above. The following regions were mutated: E-pal mut : plasmid pGL3-E-paL, GC was mutated to TT; E-Box Mut plasmid pGL3-E-box, CAC were mutated to TGT; and E-pal & E-box Mut plasmid pGL3-E-pal & E-box, GC were mutated to TT and CAC were mutated to TGT. These mutant constructs, pGL3-E-paL and pGL3-E-box, were created using pGL3-E075 as a template in the mutation PCR reaction (QuickChange, Stratagen, CA). QuickChange PCR mutation primer pairs:
Transfection and luciferase assay
To measure the TGFβ-mediated effect on E-Cadherin promoter activity, we measured expression of the firefly luciferase in MES cells transiently transfected with the plasmid harbouring the murine E-Cadherin promoter (-800bp; GL3-E075-WT plasmid) or derivative plasmids containing mutations in the E-pal site: plasmid pGL3-E-paL, E-Box site: plasmid pGL3-E-box, and both E-pal and E-Box sites: plasmid pGL3-E-pal & E-box. The detail of the luciferase protocol has been mentioned previously by us (Ahmed et al., 2007, Iordanskaia & Nawshad, 2011). The Lipofectamine 2000 transfection reagent (Invitrogen, CA) was used following the manufacturer’s protocol. Transfection of MES cells with pGL3-Lux vectors without the E-Cadherin promoter region (empty vector) was performed as a negative control. Recombinant TGFβ3 (5ng/ml) was added to the transfected cells. After 24h of TGFβ treatment, luciferase activity was detected using the Sirius luminometer (Berthold Detection Systems, NC). The results are reported as the mean ± SD of at least three independent experiments.
Statistical Analysis
Data from at least three replicates for each parameter were evaluated and analyzed for significance by SPSS 14.0. The treatment groups included TGFβ3 (5ng/mL and 10ng/mL) for 24 and 48 h and the control groups were empty vector treated and untreated cells (-ve control). The observation times were collapsed due to the convenience of the study, and one-way ANOVA was conducted. The significance level was set as 0.05. A p-value of ≤ 0.05 was considered significant. The one-way ANOVA indicated that the values differ significantly across the treatment groups. Bonferroni post-hoc comparisons of the treatment groups indicated that the two negative control groups (empty vector and untreated cells) and treated groups significantly differ from each other as expected (p ≤ 0001). All TGFβ treatment groups also differed significantly from the negative control groups, (p ≤ 0001).
Results
TGFβ3 expression in different stages of murine MES cells (in vivo)
To demonstrate the expression of TGFβ3, which is essential for palatal MES disintegration, we performed an immunofluorescence protein expression assay. TGFβ3 was strongly expressed at 14.0 dpc, when palatal shelves are about to make contact (Fig. 1A) and at 14.5 dpc, when the seam is formed (Fig. 1B). However, as the seam started to disintegrate at 15.0 dpc, the number of remaining epithelial cells decreased and the expression of TGFβ3 was restricted on the remaining epithelial seam (Fig. 1C). By 15.5 dpc, TGFβ3 was expressed in the epithelial islands only (Fig. 1D). During the later stages of palate development at 16.0 dpc, the number of TGFβ3 expressing cells was limited (Fig. 1E), and by the time the seam was completely disintegrated at 16.5 dpc (Fig. 1F), no epithelial cells remained as mesenchymal confluency occurred, resulting in the absence of any TGFβ3 positive cells.
In order to determine the pathways TGFβ3 uses during palatal EMT, we selectively blocked downstream molecules of the Smad-dependent and Smad-independent pathways utilized by TGFβ3. We performed protein expression analysis by immunoblotting protein extracts derived from MES cells that were treated with Smad-independent pathway inhibitors (such as SB202190, an inhibitor of p38MAPK; U0126, an inhibitor of ERK1/2; and LY294002, an inhibitor of phosphatidylinositol 3-kinase [PI3K]) in the presence of 5ng/mL exogenous TGFβ3. To block the Smad-dependent pathway, we treated MES cells with shRNA Smad4 and 5ng/mL exogenous TGFβ3. The results showed that MES cells treated with exogenous TGFβ3 (5ng/mL) had low expression of E-Cadherin. However, the expression of E-Cadherin protein in the MES cells treated with TGFβ3 blocking antibody showed increased E-Cadherin expression when compared to MES cells treated with only TGFβ3. Interestingly, when the Smad-dependent and Smad-independent pathways were blocked, E-Cadherin expression remained moderate but not as low as TGFβ3 treatment alone, suggesting that TGFβ3 uses both Smad-dependent and Smad-independent pathways to regulate E-Cadherin expression, except for PI3 kinase pathway with LY294002) (Fig. 1G).
Effect of TGFβ3 on epithelial and mesenchymal markers
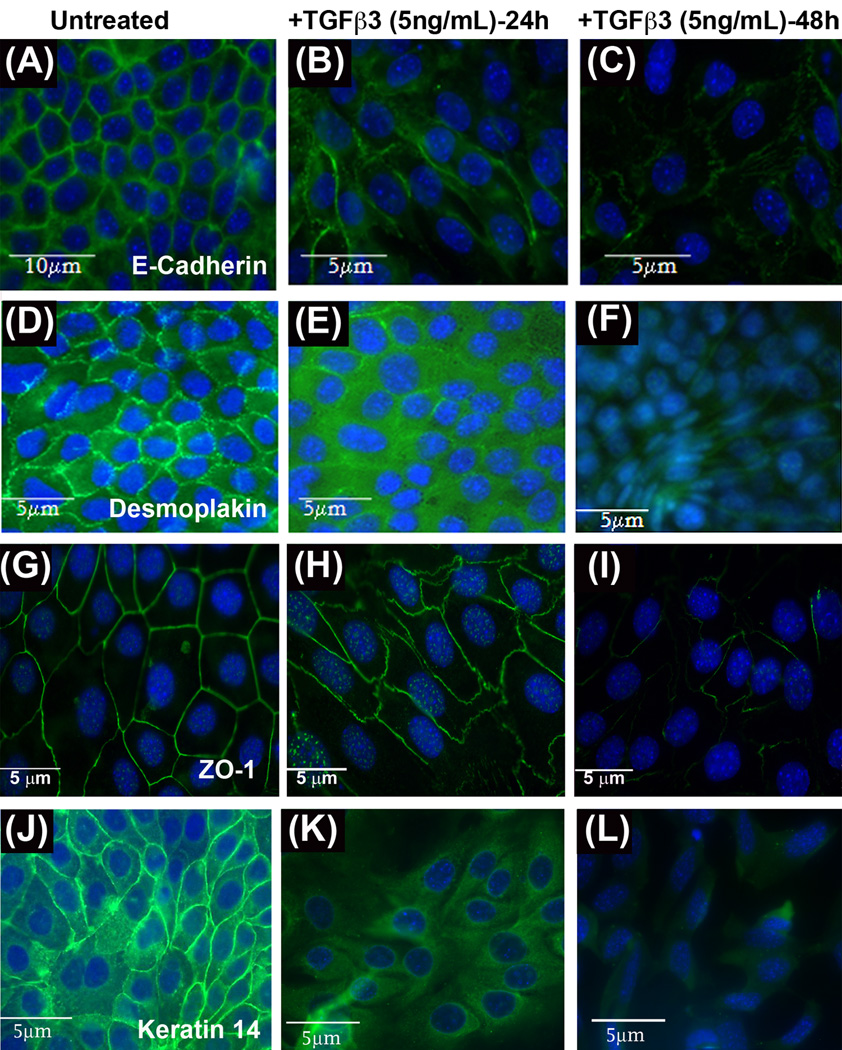
As shown by our previous results (Ahmed et al., 2007), TGFβ3 pathways activate the EMT process in palatal development by downregulating epithelial markers like E-Cadherin. We sought to determine the effect of TGFβ3 on other cell-cell adhesion proteins, such as Desmoplakin, Keratin 14 and ZO1. To show the changes that occur in epithelial markers due to TGFβ3 pathway activation, we performed immunofluorescence on MES cells treated with exogenous recombinant TGFβ3 (5ng/mL) for 24 and 48 h. Single staining immunofluorescence results of E-Cadherin showed a higher expression of E-Cadherin at the membrane in untreated (control) MES cells (Fig. 2A). However, within 24 h of TGFβ3 treatment, E-Cadherin expression was reduced and MES cells began to elongate (Fig. 2B). E-Cadherin expression was barely detectable after 48 h of TGFβ3 treatment in MES cells (Fig. 2C). Single staining immunofluorescence results of Desmoplakin demonstrated high expression of Desmoplakin in punctate form in untreated MES cell borders (Fig. 2D). After MES cells were treated with TGFβ3 for 24 h, Desmoplakin became diffusely expressed in the cytoplasm (Fig. 2E), and by 48 hours following treatment, no expression of Desmoplakin was observed (Fig. 2F). As we expected, ZO1 was strongly expressed in a uniform homogenous manner during single staining immunofluorescence in the control MES cell membranes (Fig. 2G), but the expression was significantly disorganized and reduced within 24 and 48 h (Figs. 2H and I) as MES cells lost their epithelial architecture.
Fig. 2. TGFβ3 regulates E-Cadherin, Fibronectin, Vimentin and N-Cadherin.
Single staining immunofluorescence (FITC) protein expression assays were performed on homogenous MES cells treated with TGFβ3 (5ng/mL) for 24h and 48 h. E-Cadherin (A, B & C), Desmoplakin (D, E & F), ZO1 (G, H & I) and Keratin 14 (J, K & L) were highly expressed at the cell membrane of untreated (control, A, D, G & J) MES cells, reduced by 24 h (B, E, H and K), and lost by 48 h of TGFβ3 treatment (C, F, I and L). Unlike other three epithelial markers, Keratin 14, however, is expressed in the cell membrane and in the cytoplasm in untreated cells. And is relocated mostly in the cytoplasm in response to TGFβ3 (24h) before completely repressed by 48h
Unlike rest of the EMT markers, Keratin 14, which is not only expressed in the cell membrane but also in the cytoplasm (Fig. 2J), also showed a significant loss of its expression in the cell membrane in response to TGFβ3 treatment. Its expression was translocated and re arranged within the cytoplasm (Fig 2K) and eventually was lost as the cells acquire full mesenchymal phenotype by 48 h (Fig. 2L). Taken together, these combined results suggest that in response to TGFβ3, expression of epithelial proteins is significantly reduced while mesenchymal protein expression is increased, as MES cells undergo EMT.
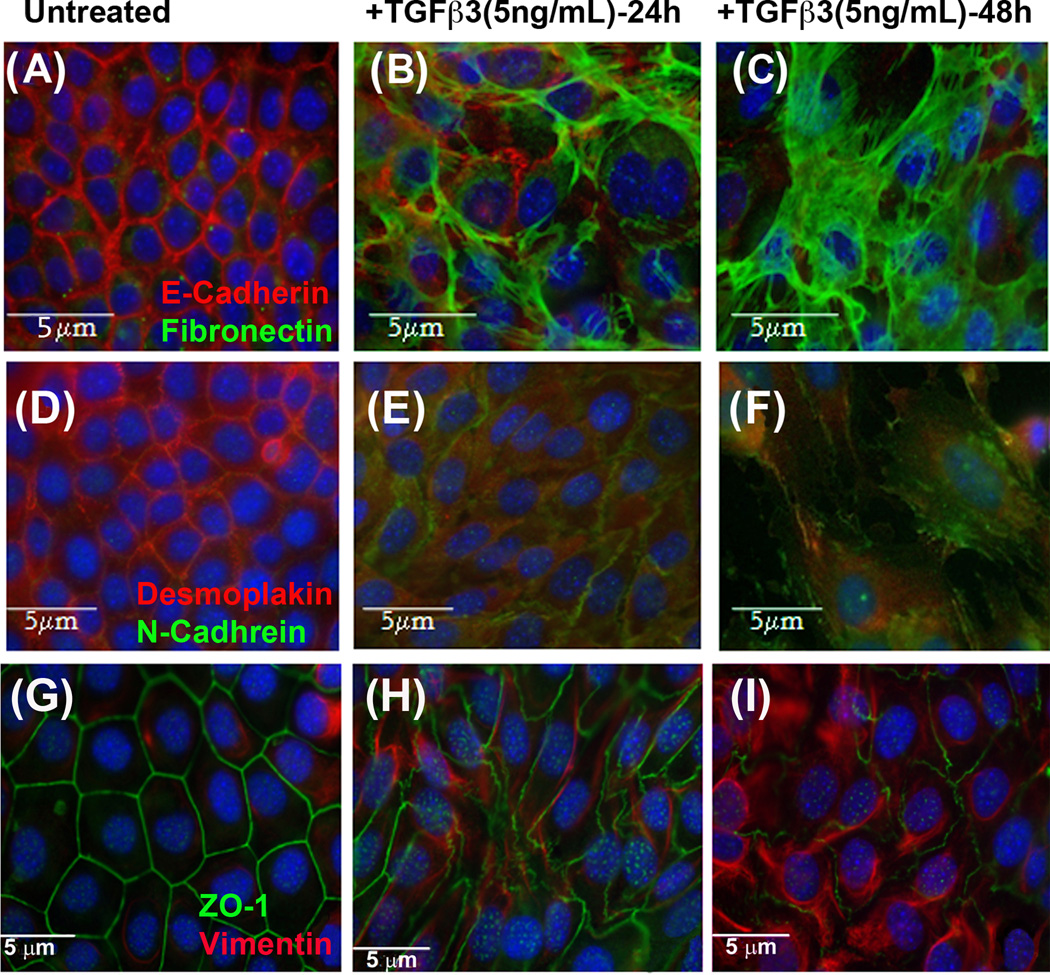
To demonstrate the role of TGFβ3 in regulating mesenchymal markers such as Vimentin, Fibronectin, and N-Cadherin in parallel to epithelial markers, double staining immunofluorescence results of E-Cadherin and Fibronectin showed high expression of E-Cadherin and no expression of Fibronectin in untreated MES cells (Fig. 3A). However, following 24 hours of TGFβ3 treatment, Fibronectin was beginning to be detected in MES cells while expression of E-Cadherin was decreasing (Fig. 3B). High expression of Fibronectin and almost no expression of E-Cadherin were observed in 48 h TGFβ3 treated MES cells (Fig. 3C). After double staining immunofluorescence of Desmoplakin with N-Cadherin, we observed no expression of N-Cadherin and increased expression of Desmoplakin in untreated MES cells (Fig. 3D). However, N-Cadherin began to be expressed at 24 h (Fig. 3E) and increased in intensity by 48 h following TGFβ3 treatment in MES cells (Fig. 3F). Double staining immunofluorescence of ZO1 and Vimentin revealed a high expression of ZO1 and no expression of Vimentin in control MES cells (Fig. 4G). Once MES cells were treated with TGFβ3 for 24 h, Vimentin begin to be expressed, and ZO1 expression became disorganized and reduced (Fig. 3H). Forty-eight hours following TGFβ3 treatment, MES cells showed increased expression of Vimentin and reduced expression of ZO1 compared to control MES cells (Fig. 3I).
Fig. 3. TGFβ3 regulates Desmoplakin, Fibronectin and N-Cadherin.
Double staining immunofluorescence protein expression assays were performed on homogenous MES cells that were treated with TGFβ3 (5ng/mL) for 24 and 48 h. In double staining, E-Cadherin (A, Rhodamine), Desmoplakin (D, Rhodamine) and ZO1 (G, FITC) were highly expressed while Fibronectin (A, FITC), N-Cadherin (D, FITC) and Vimentin (G, Rhodamine) expression were not seen in untreated control cells. As expression of Fibronectin, Vimentin, and N-Cadherin began to increase, in response TGFβ3 for 24 h (B, E & H), E-Cadherin, Desmoplakin and ZO1 expressions were reduced and significantly lost by 48 h (C, F & I), at which stage, Fibronectin, Vimentin, and N-Cadherin were highly expressed.
Similar results were observed in double staining immunofluorescence of E-Cadherin with Vimentin and N-Cadherin. Vimentin and N-Cadherin were highly expressed, compared to untreated cells (Suppl. Figs. A & D), by 48 hours following treatment with TGFβ3 (5ng/mL) (Suppl. Figs. 2C and F), while E-Cadherin expression was dramatically reduced by 24 h (Suppl. Figs. 2B and E) and lost by 48 h following TGFβ3 treatment (Suppl. Figs. 2C & F). As expected with regard to untreated MES cells, after double staining immunofluorescence of Desmoplakin with Fibronectin, we observed no expression of Fibronectin and increased expression of Desmoplakin in untreated MES cells (Suppl. Figs. 2G). However, Fibronectin began to be expressed at 24 h (Suppl. Fig. 2H) and increased in intensity by 48 hours following TGFβ3 treatment in MES cells (Suppl. Fig. 2I). Desmoplakin was highly expressed and no expression of Fibronectin was observed (Suppl. Fig. 2G)
Role of TGFβ3 in cell migration
To demonstrate the functional characteristics of EMT in response to TGFβ3, we undertook scratch wound and cell motility assays. In this experiment, cultured MES cells were treated with TGFβ3 (5ng/mL), and the cells were observed and photographed by phase-contrast microscopy at 0, 12, 24, 36 and 48 h following wounding (Fig. 4A). Scratch wound assay results indicated an increased level of migration as MES cells were treated for longer periods of time with TGFβ3. Migration of MES cells was increased in a time dependent fashion from 0, 12, 24, 36 and 48 h following treatment. We also showed that the migration of MES cells is dose dependent. As doses of TGFβ3 treatment were increased from 0 (untreated) to 2, 5, and 10ng/mL of TGFβ3 (Fig. 4B), MES cells became more migratory. A cell motility assay was performed on MES cells to show the quantitative results of cell migration in response to 5ng/mL TGFβ3 treatment in 8 µm pore size Transwell migration chambers of 6-well plates (BD BioCoat, MA) for 24 and 48 h. Results indicated a significantly increased number of migratory cells in response to 24 h of TGFβ3 treatment, with significantly higher migration observed at 48 h, while very few cells migrated in control chambers at both time points (Fig. 4C).
Evidence of TGFβ3 Smad-dependent and Smad-independent pathways being activated
Previously, it was shown that activation of TGFβ3 Smad-dependent pathways cause Smad phosphorylation and translocation from the cytoplasm to the nucleus. This translocation event is accepted evidence of Smad pathway activation (Derynck & Zhang, 2003, Itoh et al., 2003, Massague et al., 2005). To investigate the activity of Smad proteins in response to TGFβ3, an immunofluorescence protein expression assay was performed with MES cells treated with 5ng/mL exogenous TGFβ3 for 3 h. Following TGFβ3 treatment, pSmad2 and Smad4 were observed in both the cytoplasm and the nucleus in MES cells within 3 h (Figs. 5A and B). In the absence of TGFβ3, Smad2 remained only in the cytoplasm and there was no expression of pSmad2 either in the cytoplasm or the nucleus (DAPI, Figs. 5C and D). After cells were treated with blocking TGFβ3 antibody (10ng/mL) for 24 h, Smad4 remained solely in the cytoplasm (Figs. 5E and G), while the nucleus (DAPI) was complete devoid of any Smad4 expression (Fig. 2F). Moreover, there was no change in morphology of the MES cells (Fig. 5G). Once we increased the dose of exogenous TGFβ3 to 10ng/mL, Smad4 completely moved to the nucleus (Figs. 5H and I), with no expression in the cytoplasm.
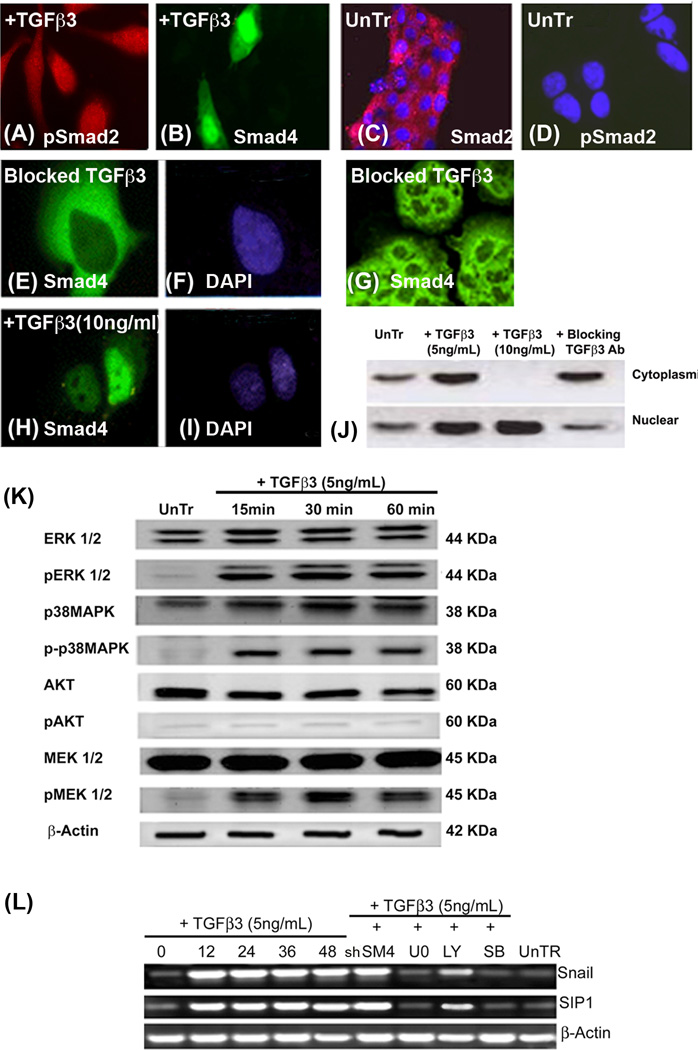
Fig. 5. TGFβ3 induces Smad-dependent and Smad-independent pathways in the palatal seam cells to activate the EMT related transcription factors, Snail and SIP1.
Immunofluorescence was performed on homogenous palatal MES cells that were treated TGFβ3 (5 and 10ng/mL) for 3 h to demonstrate the Smad translocation in response to TGFβ3. In the presence of 5ng/mL of TGFβ3, pSmad2 and Smad4 are seen in the cytoplasm and nucleus (A and B respectively). In the absence of TGFβ3, Smad2 (C) remains in the cytoplasm while pSmad2 (D) is not seen in either nuclei (DAPI) or cytoplasm. MES cells treated with blocking TGFβ3 antibody for 24 h showed no expression of Smad4 in the nucleus (E), but only in the cytoplasm (F, DAPI), and there was no change of morphology in MES cells as they remained epithelial (G). Once the dose of exogenous TGFβ3 was increased to 10ng/mL for 3 h, Smad4 completely moved to nucleus (I, DAPI), and no cytoplasmic expression was observed (H). Immunoblotting analysis of nuclear and cytoplasmic proteins from untreated MES cells (control) and those treated with TGFβ3 (5ng/mL), demonstrated that Smad4 protein expression was higher in both the cytoplasm and nucleus compared to its low expression in control cells. However, cells treated with 10ng/mL TGFβ3 showed only nuclear expression of Smad4. MES cells treated with blocking TGFβ3 antibody showed much higher expression of cytoplasmic Smad4 protein than nuclear (Fig. J). (K) MES cells were treated with TGFβ3 (5ng/mL) for 15, 30, and 60 min., and protein from these cells was extracted for immunoblotting – control cells were untreated (UnTr) at 0 (not shown) and 60 min (shown). Results showed phosphorylation of ERK, p38MAPK and MEK1/2, but not AKT in MES cells treated with 5ng/mL exogenous TGFβ3. These proteins were not phosphorylated in control, untreated cells harvested at either 0 (not shown) or 60 min. (L) mRNA was collected from the MES cells treated with TGFβ3 (5ng/mL) for 0, 12, 24, 36, and 48 h. Some groups of MES cells were treated with a Smad inhibitor (Smad4 shRNA; shSM4) for 48 h and Smad-independent pathways were blocked using small synthetic chemical inhibitors – LY294002 to block PI3 Kinase (20µM), SB202190 for p38MAPK (20µM) and U0126 for MEK1/2 (20µM) – for 60 min. mRNA expression of Snail and SIP1 transcription factors increased dramatically within 12 h of TGFβ3 treatment and remained high until 48 hours. When Smad4 was inhibited, mRNA expression of these transcription factors stayed unaffected compared to 48 h of TGFβ3 treatment. However, Snail and SIP1 mRNA expression was drastically reduced when cells were treated with inhibitors of the Smad-independent pathways, except for PI3 kinase.
Immunoblotting results from nuclear and cytoplasmic protein fractions demonstrated a low expression of Smad4 in control MES cells (no TGFβ3) in both the cytoplasm and the nucleus compared to an increase in the steady state levels of Smad4 protein expression in the cytoplasm and the nucleus of MES cells that were treated with 5ng/mL of TGFβ3 for 3 h. However, when MES cells were treated with 10ng/mL TGFβ3, Smad4 was localized only in the nucleus. Inhibition of TGFβ3 signaling by treatment with blocking TGFβ3 antibody resulted in higher Smad4 expression in the cytoplasm compared to that found in the nucleus (Fig. 5J).
In order to provide evidence in support of the activation of Smad-independent pathways by TGFβ3 during palate development, an immunoblotting experiment was performed with MES cell proteins treated with exogenous TGFβ3 (5ng/mL) for 0 (control), 15, 30 and 60 minutes, since the phosphorylation of these proteins happens fairly quickly. Our results demonstrate the phosphorylation of ERK, p38MAPK, and MEK1/2, but not AKT in MES cells and, as we expected, no phosphorylation of any of these proteins was observed in control cells. These results support the notion that in response to TGFβ3, Smad-independent pathways are activated, as noted by phosphorylation of ERK, p38MAPK, and MEK1/2, but not PI3K (Fig. 5K).
MES cells were treated with exogenous TGFβ3 (5ng/mL) and mRNA was collected at 0, 12, 24, 36 and 48 hours. To establish the exact pathways that are activated, we blocked the Smad-dependent pathway with shRNA Smad4 and its independent pathways with inhibitors, SB202190 (for p38MAPK), U0126 (for MEK1/2), and LY294002 (for PI3 Kinase). Following reverse transcriptase PCR, our results (Fig. 5L) showed that in response to TGFβ3 (5ng/mL), the mRNA expression of Snail and SIP1 transcription factors was significantly increased in a time dependent fashion, from 0 to 48 hours. However, inhibition of the Smad-independent pathways blocked TGFβ3 induced Snail and SIP1 mRNA expression, except for PI3 kinase pathway with LY294002. Interestingly, we observed that the TGFβ3 stimulated expression of these transcription factors was unaffected when the Smad-dependent pathway was blocked. Here we show that TGFβ3 pathways induce several factors such as Snail and SIP1, common EMT related transcription factors are activated in response to TGFβ3, which trigger the EMT process.
Binding of Snail, SIP1 and Smad4 to the E-Cadherin promoter
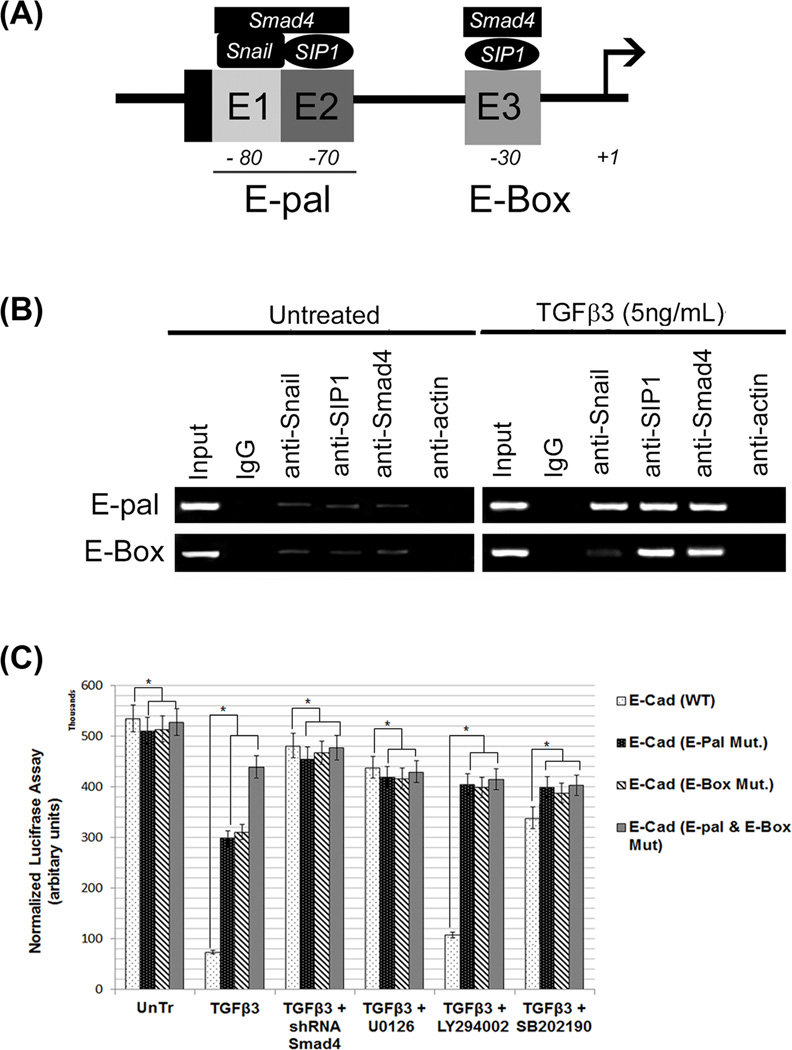
Since the most common molecular change which promotes and is often used to characterize EMT is the loss of E-cadherin expression, we performed sequence analysis of the promoter region of the murine E-cadherin gene. Binding of the Snail, SIP1 and Smad4 proteins to the endogenous loci of these regions was confirmed by chromatin immunoprecipitation (ChIP; Fig. 6A). ChIP was performed using antibodies against IgG (negative control), Snail, SIP1, Smad4 and actin, and PCR was then performed with primers specific for the E-pal and other E-box sites. Untreated MES cells showed no signal with any antibodies, whereas TGFβ3 treated cells showed a significantly increased signal with antibodies against Snail, SIP1 and, Smad4 (Fig. 6B). Actin was not found to be associated with these binding regions. Snail, SIP1 and Smad4 bind to the E-pal region, whereas only SIP1 and Smad4 bind to the proximal E-box. To avoid the possibility of the observed PCR signals being the result of over-amplification, we conducted real-time PCR for a quantitative assessment of ChIP signals (results not shown).
Fig. 6. Smad4-Snail-SIP1 binds to the promoter of the E-cadherin gene.
(A) A schematic diagram of the proposed interaction between the Snail-SIP1-Smad4 transcription complex and the E-Cadherin gene promoter is shown to demonstrate our hypothesis of transcriptional repression. The transcription initiation site (+1) is indicated by arrows. Positions of regulatory E-boxes in the E-cadherin promoter (E1–E3) (bound by E-cadherin repressors) are indicated by grey boxes. E1 and E2 boxes are adjacent in the mouse promoter, forming the E-pal element (Moreno-Bueno et al., 2009) (B) Binding of the Snail-SIP1-Smad4 complex to the endogenous loci was confirmed by chromatin immunoprecipitation. Chromatin was immunoprecipitated with antibodies against IgG (negative control), Smad4, Snail, SIP1 and β-actin (non-specific control). PCR analysis using precipitated DNA showed binding of Snail-SIP1-Smad4 and SIP1-Smad4 to E-pal and E-box regions respectively, of the E-Cadherin promoter, in response to TGFβ3 (5ng/mL) treatment for 24 h. No signal was observed from untreated cells and β-Actin was not detected to interact with these binding sites. (C) Using a wild type (−650- to +95) pGL3-Lux-E-Cadherin reporter plasmid and three mutant plasmids (mutations on E-Box, E-pal and both E-box and E-pal), we were able to demonstrate that these binding sites are critical for E-Cadherin promoter activity. Luciferase assays, performed in MES cells treated with 5 and 10ng/mL TGFβ3 with wild type and mutant plasmids, showed a high level of E-Cadherin promoter activity in all groups of untreated cells. TGFβ3 (5ng/mL) treatment significantly reduced E-Cadherin activity in the wild type transfected cells. E-pal and E-box mutations reversed TGFβ3’s inhibitory effect on promoter activity, with the double mutant demonstrating the most significant effect. Repression of E-Cadherin gene activity by TGFβ3 can be restored and reversed by blocking both Smad dependent and Smad independent pathways (MEK1/2 blocked with U0126, p38MAPK blocked with SB202190), except for the PI3 kinase pathway (blocked with LY294002). Both TGFβ treatment groups differed significantly from the negative control (untreated and empty vector treated cells) groups, (p ≤ 0001, as indicated by *).
EMT related transcription factors, Snail and SIP1 bind to specific binding sites (E-pal and E-box) on the E-Cadherin promoter gene. However, to demonstrate whether these transcription factors are functional and that they bind to the specific sites during palate development, we tested E-Cadherin promoter activity while mutating these binding sites. We have already shown that TGFβ3 represses the expression of E-Cadherin protein levels during palate development (Figs. 1, 2, 3 and 6). However, in this experiment involving a Luciferase reporter gene assay, we demonstrated the crucial roles that these binding sites play in response to TGFβ3. Four plasmids were used: Wild type (WT); E-pal with GC region mutated to TT; E-box with CAC region mutated to TGT; and a plasmid with both E-pal and E-box mutated, GC to TT and CAC to TGT. According to the Luciferase reporter gene assay results (Fig. 6C), in WT transfected cells, E-Cadherin has the highest promoter activity in untreated cells and the lowest activity in cells that have been treated with TGFβ3 (5ng/mL). In the cells transfected with the E-pal mutated reporter, the promoter activity in TGFβ3 treated MES cells was higher compared to wild type transfected cells treated with TGFβ3. In cells transfected with the E-box mutated plasmid, the promoter activity following TGFβ3 treatment was also higher compared to wild type transfected MES cells, similar to the E-pal reporter results. The promoter activity of the E-Cadherin reporter containing the double mutation demonstrated the highest reporter activity amongst all the mutated plasmids in response to TGFβ3 treatment compared to wild type transfection in MES cells (Fig. 6C). And when the Smad-dependent and Smad-independent pathways were blocked, E-Cadherin expression remained moderate but not as low as TGFβ3 treatment alone, suggesting that TGFβ3 uses both Smad-dependent and Smad-independent pathways to regulate E-Cadherin gene activity, except for PI3 kinase pathway with LY294002) (Fig. 6C).
Discussion
Palatogenesis is a crucial stage in craniofacial development in higher vertebrates (amniotes), such as rodents and humans (Hay, 2005). All stages of palate development, such as elongation, elevation and adhesion, are important; however, the last step during which the epithelial seam forms due to palatal shelf contact with an opposing shelf (and subsequently disintegrates to achieve a confluent palate) is of particular interest in this study. The medial edge epithelia (MEE) of the palatal shelves that arise from the maxillary processes make contact to form a bilayered midline seam composed of epithelial cells (Ferguson, 1988). This seam is a hindrance for successful palatal confluency; therefore, to achieve complete palatal confluency, the seam must disintegrate to form a single confluent palate craniofacial structure. In the absence of palatal seam disintegration, cleft palate, the second most common birth defect and most common craniofacial deformity, may develop (Nawshad, 2008). Apoptosis, epithelial mesenchymal transition (EMT) and migration are common mechanisms of seam disintegration (Nawshad, 2008). In this study, we explored the mechanisms of TGFβ3 signaling, which is essential for seam disintegration and activates EMT.
EMT consists of the entire series of events involved in the transition of epithelial to mesenchymal cell characteristics (Hay, 1995, Thiery & Sleeman, 2006). During EMT, epithelial proteins and characteristics are compromised, which allows the proteins and properties of mesenchymal cells to be activated (Hay, 1995, Thiery & Sleeman, 2006). Epithelial and mesenchymal cells have their own unique characteristics and morphology (Schock & Perrimon, 2002). Epithelial cells can be arranged in a single layer, or they can form layers of cells that are in close contact with neighboring cells connected by various junctions, such as: tight junctions, adherent junctions, and desmosomes (Horwitz & Webb, 2003). E-Cadherin is the most common adherent protein, while Occludin, Claudins, and Zonula Occludens (ZO 1, 2, 3) are important components of tight junction strands (Shimoyama et al., 1989, Takeichi, 1993, Takeichi, 1988a, Takeichi, 1988b). Desmosomes provide additional strength for intercellular adhesion and are structurally similar to adherent junctions. Desmoplakin, Plakophilin, and Plakoglobin are the common desmosomal proteins (Thiery, 2003a). All these junctional proteins help epithelial cells to remain in a regimented structure (Shook & Keller, 2003) and during EMT, these proteins are lost to promote significant changes in epithelial cell structure and behaviour by giving up their functional and phenotypic characteristics and acquiring mesenchymal cells’ characteristics (Takeichi, 1993, Takeichi, 1995). Mesenchymal cells are irregular in shape and have weaker adhesions, which facilitates migration. In comparison with epithelial sheets, mesenchymal cells have more elongated shapes and front-to-back polarity; they also move individually and are more dynamic. Interestingly, TGFβ is known to be a potent inducer of EMT in both development and tumorigenesis (Zavadil & Bottinger, 2005, Valcourt et al., 2005, Tian & Schiemann, 2009, Derynck & Feng, 1997). TGFβ signals through Smad-dependent and Smad-independent pathways (Akhurst & Derynck, 2001, Derynck, 1994). In the Smad-dependent pathway, Smad phosphorylation and translocation to the nucleus is regulated by TGF-β receptors, early endosomes, phosphatidylinositol 3-kinase (PI3K) and the Smad anchor for receptor activation (SARA) (Bakin et al., 2000, Itoh et al., 2002, Itoh et al., 2003, Panopoulou et al., 2002, Tsukazaki et al., 1998). Several signaling pathways can trigger the activation of EMT related transcription factors, such as Snail and SIP1, which induces loss of cell-cell adhesion genes, such as E-Cadherin (Cano et al., 2000, Thiery & Sleeman, 2006). It was elegantly shown in the transgenic mice model system that palatal epithelium to employs both pathways that demonstrate the complicated nature of TGF-β signaling mechanisms in palate development (Xu et al., 2008). Interestingly, they have shown that palatal epithelial tissue specific disruption of Smad4 did not adversely affect palatal fusion, but when both Smad4 and p38 functions are conceded, it resulted in cleft palate. They propose that p38 and Smad4 are functionally redundant in facilitating TGFβ signaling in various circumstances during palatogenesis and concluded that the fact that both pathways are functional in tandem elucidates the intricate nature of TGFβ signaling mechanisms in palatogenesis. In this study, we explore the mechanisms behind and show that during palatal EMT, TGFβ3 activates both Smad-dependent and Smad-independent pathways to induce Snail and SIP1 to promote MES EMT, resulting in complete disintegration of the palatal seam. Although it has been shown previously that during palatogenesis, PI3 kinase pathway active (Kang & Svoboda, 2002, Yu et al., 2008), we, however, were not able to see any activation of PI3 kinase, as demonstrated by no phospho AKT protein expression (Fig. 5K) or by limited or no effect on E-Cadherin protein (Fig. 1G) expression or gene activity (Fig. 6C) when PI3 kinase was blocked with LY294402.
First, we established that TGFβ3 is expressed in the palatal seam as it is formed at 14.5 dpc and is highly expressed throughout the full length, unbroken palatal seam (Fig. 1A). We observed that as the seam began to diminish (Fig. 1B–F), expression of TGFβ3 remained limited only to the smaller islands of epithelial cells until the seam completely disintegrated (Fig. 1E) and no TGFβ3 expressing cells were present. Therefore, as the palatal seam disintegrates and cells undergo EMT, the expression of TGFβ in the seam region also decreases until it is no longer present (Fig. 1F), indicating that TGFβ3 may play a role in seam disintegration and the acquisition of a mesenchymal phenotype. We were also able to demonstrate an association between the expression of E-Cadherin and TGFβ3 signaling pathways. MES cells treated with TGFβ3 showed low E-Cadherin protein levels, but when both the Smad-dependent and Smad-independent pathways were blocked, expression of E-Cadherin was restored even in the presence of TGFβ3 (Fig. 1G). These results support the notion that during palatal EMT, TGFβ3 uses Smad-dependent and Smad-independent pathways (sans PI3K) to reduce E-Cadherin protein levels.
We also observed that the E-Cadherin expression level was reduced by TGFβ3 during palatal development in addition to other epithelial markers such as Desmoplakin and ZO1. At the same time, key mesenchymal proteins, such as Vimentin, Fibronectin, and N-Cadherin were induced following TGFβ3 treatments (Figs. 2, 3 and Suppl. Fig. 2). These results support that premise of functional EMT, whereby palatal cells lose expression of epithelial adhesion proteins and become isolated. At that stage, increased expression of mesenchymal proteins cause these isolated cells to become motile and migratory (Figs. 4A, B, and C). Subsequently, this phenotypic transition initiates the EMT processes involved in palate development.
We established that in response to TGFβ3, both Smad-dependent and Smad-independent pathways are active in palatal seam cells (Fig. 5). Translocation of Smad4, from the cytoplasm to the nucleus, is an indication of an activated Smad pathway (Abdollah et al., 1997). In our study, we showed the presence of phosphorylated Smad2 and Smad4 in the cytoplasm and nucleus following treatment with TGFβ3 as evidence of the Smad-dependent pathway being activated (Figs. 5A and B). Whereas in the absence of TGFβ3, Smad2 remains in the cytoplasm, is not phosphorylated and cellular morphology remains epithelial (Fig. 5C and D). Interestingly, TGFβ3 also activated Smad-independent pathways by phosphorylating p38MAPK, MEK1/2, and ERK1/2 (but not AKT). These results indicate that both Smad-dependent and Smad-independent pathways are functional during palatal EMT. TGFβ3 is highly expressed in the palatal seam region immediately upon seam formation and its expression is sustained until the seam is completely disintegrated. TGFβ3 expression coincides with expression of Snail, a gene crucial to the process of EMT. This suggests that TGFβ3 and Snail are expressed simultaneously during seam formation, thus making Snail a potential TGFβ3-influenced target, thereby regulating EMT (Martinez-Alvarez et al., 2004, Pungchanchaikul et al., 2005). In this study, we intended to explore the multiple pathways that TGFβ3 activates to induce EMT related transcription factors (e.g., Snail and SIP1) to promote cell migration. These transcription factors have been shown to be regulators of EMT during palatal development. Our results are in accord with previous findings, as TGFβ3 has been shown to be expressed only in MES at the time of fusion and completely absent in MES cells before adhesion (Kaartinen et al., 1997, Martinez-Alvarez et al., 2004, Martinez-Alvarez et al., 2000). TGFβ3 expression in MES also results in downregulation of E-Cadherin expression, which is a major feature of EMT (Batlle et al., 2000, Cano et al., 2000). While the expression of these EMT related transcription factors have been associated with palatal EMT, in this study we demonstrate that the inhibition of TGFβ3 Smad-independent pathways resulted in deactivation of these transcription factors while blocking the Smad pathway had no effect. Therefore, we conclude that the Snail and SIP1 transcription factors are activated through TGFβ3 Smad-independent pathways during palatal development, but not via PI3 Kinase (Fig. 5L).
Downregulation of E-Cadherin is one of the most common molecular changes associated with EMT during palatal development. In our study, we performed experiments to investigate the mechanisms of activation of transcription factors through multiple TGFβ pathways and how these transcription factors bind to the E-cadherin promoter (Fig. 6A). We found that Smad4 (activated by the Smad-dependent pathway) and Snail and SIP1 (activated by Smad-independent pathways) interact with the E-pal and E-box sites of the E-Cadherin promoter (Fig. 6B). Interestingly, upon TGFβ3 treatment, the E-pal site harbors Snail-SIP1 and Smad4 whereas the E-box site harbors SIP1 and Smad4, but not Snail. It is therefore likely that these transcription factors would affect E-Cadherin gene activity.
We showed, by luciferase assay, that E-Cadherin promoter activity is significantly repressed in response to TGFβ3, in a dose dependent manner (Fig. 6C); similar findings with other TGFβ isoforms were shown in cancer and developmental systems (Lee et al., 2006). E-Cadherin is known to be regulated by many EMT related transcription factors, and several of these transcription factors are activated by TGFβ signaling (Peinado et al., 2007). Accordingly, our findings show that TGFβ3 pathways repress the expression of E-Cadherin and also activate the Snail and SIP1 transcription factors. We also discovered these transcription factors bind to specific binding sites on E-Cadherin to repress its expression, resulting in subsequent execution of the EMT program. There have been several studies analyzing the sequences of these binding sites (Snail and SIP1) of the E-Cadherin promoter, which led to the identification of E-box and E-pal binding sites (Nawshad et al., 2007, Peinado et al., 2007). In this study, we investigated the importance of the E-pal and E-box binding sites on the E-Cadherin promoter in relation to their respective transcription factors in response to TGFβ3 signaling. We were able to establish that E-Cadherin promoter activity remained de-repressed in response to TGFβ3 when both E-pal and E-box sites were mutated. And we also established that such repression of E-Cadhetin gene activity is facilitated by using both Smad dependent and Smad independent pathways (except PI3 kinase) Therefore, the E-pal and E-box binding sites of the E-Cadherin promoter play an essential role in the downregulation of E-Cadherin in response to TGFβ3 signaling during EMT palatal development (Fig. 6C).
Conclusions
Disappearance of palate medial-edge epithelium during craniofacial development is a topic of much controversy. Whereas most of the early work in this field has provided evidence for EMT, others have suggested that apoptosis or cell migration may be the major fate of the MEE with valid and convincing data (Nawshad, 2008). Recently, we have proposed that a combination of both EMT and apoptosis function chronologically to ensure complete disintegration of the palatal seam (Ahmed et al., 2007). The purpose of our work was not to provide further support for in vivo EMT, but rather, to establish the importance of Smad-dependent and -independent pathways that result in Snail and SIP1 activation. Surprisingly, these EMT related transcription factors (Snail and SIP1) were regulated by Smad-independent pathways, rather than the commonly believed Smad pathways. This work provides a foundation to isolate differences between Smad-mediated Smad4 activation and Smad-independent Snail and SIP1 activation, by pathways that eventually cooperate forming complexes to bind to the E-Cadherin promoter to repress its expression and activity. The unique mechanism of cell adhesion loss described here will provide new insight into EMT in other systems of embryonic development and pathology.
Supplementary Material
Supplementary Figure 1. (A) Westernblot was performed on homogenous palatal MES cells that were treated with small hairpin RNA (shRNA) Smad4 (pRetrosuper-shSmad4 (Addgene, MA) for 48 h to inhibit the Smad-dependent pathway), was used which specifically targets the coding region of murine Smad4. Empty vector and scrambled shRNA vector (scr) were also used as controls. (B) To abolish the effect of endogenous TGFβ3, cells were treated with blocking TGFβ3 antibody was used at 10ng/mL for 24 h. MES cells were treated with a Smad-independent pathways were blocked using small synthetic chemical inhibitors for 60 min – (C) U0126 for MEK1/2 (20µM), (D) SB202190 for p38MAPK (20µM) and (E) LY294002 to block PI3 Kinase (20µM). (F) All treatment showed significant loss of the targeted protein. Intensity of the band was measured using the Carestream Molecular Imaging Software version 5.3.1 (Rochester, NY). To perform a t-test analysis of mean intensity measurements, a ROI analysis was done from the data to Microsoft Excel software from the exported “.txt” files. (G) In double staining, Keratin 14 (K14) (Rhodamine) was expressed in untreated (i) as well as 24 (ii) and 48 h (iii) of TGFβ3 (5ng/mL) treatments while Vimentin (FITC; iv), confirm its mesenchynal transformation at 48 h.
Supplementary Figure 2. Double staining immunofluorescence protein expression assays were performed on homogenous MES cells that were treated with TGFβ3 (5ng/mL) for 24 and 48 h. In double staining, E-Cadherin (A, FITC and D, Rhodamine) and Desmoplakin (G, Rhodamine) were highly expressed while Vimentin (A, Rhodamine), N-Cadherin (D, FITC) and Fibronectin (G, FITC) expression were not seen in untreated control cells. As expression of Vimentin, Fibronectin and N-Cadherin began to increase, in response TGFβ3 for 24 h (B, E & H), E-Cadherin and Desmoplakin expressions were reduced and significantly lost by 48 h (C, F & I), at which stage, Vimentin, Fibronectin and N-Cadherin were highly expressed.
Acknowledgements
We thank Dr. Shaukat Dedhar (University of British Columbia, Vancouver, Canada) for generously providing us with the pGL3-E-cad-Lux luciferase construct. We also thank Hector Peinado and Amparo Cano (Instituto de Investigaciones Biomedicas, Madrid) for their helpful advice regarding this study. This research was supported by NIDCR, NIH grant, R01DE017986 to Dr. Ali Nawshad.
References
- Abdollah S, Macias-Silva M, Tsukazaki T, Hayashi H, Attisano L, Wrana JL. TbetaRI phosphorylation of Smad2 on Ser465 and Ser467 is required for Smad2-Smad4 complex formation and signaling. J Biol Chem. 1997;272:27678–27685. doi: 10.1074/jbc.272.44.27678. [DOI] [PubMed] [Google Scholar]
- Ahmed S, Liu CC, Nawshad A. Mechanisms of palatal epithelial seam disintegration by transforming growth factor (TGF) beta3. Dev Biol. 2007;309:193–207. doi: 10.1016/j.ydbio.2007.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhurst RJ, Derynck R. TGF-beta signaling in cancer--a double-edged sword. Trends Cell Biol. 2001;11:S44–S51. doi: 10.1016/s0962-8924(01)02130-4. [DOI] [PubMed] [Google Scholar]
- Bakin AV, Tomlinson AK, Bhowmick NA, Moses HL, Arteaga CL. Phosphatidylinositol 3-kinase function is required for transforming growth factor beta-mediated epithelial to mesenchymal transition and cell migration. J Biol Chem. 2000;275:36803–36810. doi: 10.1074/jbc.M005912200. [DOI] [PubMed] [Google Scholar]
- Batlle E, Sancho E, Franci C, et al. The transcription factor snail is a repressor of E-cadherin gene expression in epithelial tumour cells. Nat Cell Biol. 2000;2:84–89. doi: 10.1038/35000034. [DOI] [PubMed] [Google Scholar]
- Blavier L, Lazaryev A, Groffen J, Heisterkamp N, Declerck YA, Kaartinen V. TGF-beta3-induced palatogenesis requires matrix metalloproteinases. Mol Biol Cell. 2001;12:1457–1466. doi: 10.1091/mbc.12.5.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet CL, Sharpe PM, Ferguson MW. Inhibition of TGF-beta 3 (but not TGF-beta 1 or TGF-beta 2) activity prevents normal mouse embryonic palate fusion. Int J Dev Biol. 1995;39:345–355. [PubMed] [Google Scholar]
- Cano A, Perez-Moreno MA, Rodrigo I, et al. The transcription factor snail controls epithelial-mesenchymal transitions by repressing E-cadherin expression. Nat Cell Biol. 2000;2:76–83. doi: 10.1038/35000025. [DOI] [PubMed] [Google Scholar]
- Carette MJ, Ferguson MW. The fate of medial edge epithelial cells during palatal fusion in vitro: an analysis by DiI labelling and confocal microscopy. Development. 1992;114:379–388. doi: 10.1242/dev.114.2.379. [DOI] [PubMed] [Google Scholar]
- Derynck R. TGF-beta-receptor-mediated signaling. Trends Biochem Sci. 1994;19:548–553. doi: 10.1016/0968-0004(94)90059-0. [DOI] [PubMed] [Google Scholar]
- Derynck R, Feng XH. TGF-beta receptor signaling. Biochim Biophys Acta. 1997;1333:F105–F150. doi: 10.1016/s0304-419x(97)00017-6. [DOI] [PubMed] [Google Scholar]
- Derynck R, Zhang YE. Smad-dependent and Smad-independent pathways in TGF-beta family signalling. Nature. 2003;425:577–584. doi: 10.1038/nature02006. [DOI] [PubMed] [Google Scholar]
- Dudas M, Nagy A, Laping NJ, Moustakas A, Kaartinen V. Tgf-beta3-induced palatal fusion is mediated by Alk-5/Smad pathway. Dev Biol. 2004;266:96–108. doi: 10.1016/j.ydbio.2003.10.007. [DOI] [PubMed] [Google Scholar]
- Ferguson MW. Palate development. Development. 1988;(103 Suppl):41–60. doi: 10.1242/dev.103.Supplement.41. [DOI] [PubMed] [Google Scholar]
- Fitchett JE, Hay ED. Medial edge epithelium transforms to mesenchyme after embryonic palatal shelves fuse. Dev Biol. 1989;131:455–474. doi: 10.1016/s0012-1606(89)80017-x. [DOI] [PubMed] [Google Scholar]
- Gritli-Linde A. Molecular control of secondary palate development. Dev Biol. 2007;301:309–326. doi: 10.1016/j.ydbio.2006.07.042. [DOI] [PubMed] [Google Scholar]
- Hay ED. An overview of epithelio-mesenchymal transformation. Acta Anat. 1995;154:8–20. doi: 10.1159/000147748. [DOI] [PubMed] [Google Scholar]
- Hay ED. The mesenchymal cell, its role in the embryo, and the remarkable signaling mechanisms that create it. Dev Dyn. 2005;233:706–720. doi: 10.1002/dvdy.20345. [DOI] [PubMed] [Google Scholar]
- Horwitz R, Webb D. Cell migration. Curr Biol. 2003;13:R756–R759. doi: 10.1016/j.cub.2003.09.014. [DOI] [PubMed] [Google Scholar]
- Iordanskaia T, Nawshad A. Mechanisms of transforming growth factor b induced cell cycle arrest in palate development. J Cell Physiol. 2010 doi: 10.1002/jcp.22477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iordanskaia T, Nawshad A. Mechanisms of transforming growth factor beta induced cell cycle arrest in palate development. J Cell Physiol. 2011;226:1415–1424. doi: 10.1002/jcp.22477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh F, Divecha N, Brocks L, et al. The FYVE domain in Smad anchor for receptor activation (SARA) is sufficient for localization of SARA in early endosomes and regulates TGF-beta/Smad signalling. Genes Cells. 2002;7:321–331. doi: 10.1046/j.1365-2443.2002.00519.x. [DOI] [PubMed] [Google Scholar]
- Itoh S, Thorikay M, Kowanetz M, et al. Elucidation of Smad requirement in transforming growth factor-beta type I receptor-induced responses. J Biol Chem. 2003;278:3751–3761. doi: 10.1074/jbc.M208258200. [DOI] [PubMed] [Google Scholar]
- Kaartinen V, Cui XM, Heisterkamp N, Groffen J, Shuler CF. Transforming growth factor-beta3 regulates transdifferentiation of medial edge epithelium during palatal fusion and associated degradation of the basement membrane. Dev Dyn. 1997;209:255–260. doi: 10.1002/(SICI)1097-0177(199707)209:3<255::AID-AJA1>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- Kang P, Svoboda KK. PI-3 kinase activity is required for epithelial-mesenchymal transformation during palate fusion. Dev Dyn. 2002;225:316–321. doi: 10.1002/dvdy.10161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagamba D, Nawshad A, Hay ED. Microarray analysis of gene expression during epithelial-mesenchymal transformation. Dev Dyn. 2005;234:132–142. doi: 10.1002/dvdy.20489. [DOI] [PubMed] [Google Scholar]
- Lee JM, Dedhar S, Kalluri R, Thompson EW. The epithelial-mesenchymal transition: new insights in signaling, development, and disease. J Cell Biol. 2006;172:973–981. doi: 10.1083/jcb.200601018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Alvarez C, Blanco MJ, Perez R, et al. Snail family members and cell survival in physiological and pathological cleft palates. Dev Biol. 2004;265:207–218. doi: 10.1016/j.ydbio.2003.09.022. [DOI] [PubMed] [Google Scholar]
- Martinez-Alvarez C, Tudela C, Perez-Miguelsanz J, O'kane S, Puerta J, Ferguson MW. Medial edge epithelial cell fate during palatal fusion. Dev Biol. 2000;220:343–357. doi: 10.1006/dbio.2000.9644. [DOI] [PubMed] [Google Scholar]
- Massague J, Seoane J, Wotton D. Smad transcription factors. Genes Dev. 2005;19:2783–2810. doi: 10.1101/gad.1350705. [DOI] [PubMed] [Google Scholar]
- Medici D, Nawshad A. Type I collagen promotes epithelial-mesenchymal transition through ILK-dependent activation of NF-kappaB and LEF-1. Matrix Biol. 2010;29:161–165. doi: 10.1016/j.matbio.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Bueno G, Peinado H, Molina P, et al. The morphological and molecular features of the epithelial-to-mesenchymal transition. Nat Protoc. 2009;4:1591–1613. doi: 10.1038/nprot.2009.152. [DOI] [PubMed] [Google Scholar]
- Nawshad A. Palatal seam disintegration: to die or not to die? that is no longer the question. Dev Dyn. 2008;237:2643–2656. doi: 10.1002/dvdy.21599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawshad A, Hay ED. TGF{beta}3 signaling activates transcription of the LEF1 gene to induce epithelial mesenchymal transformation during mouse palate development. J Cell Biol. 2003a;163:1291–1301. doi: 10.1083/jcb.200306024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawshad A, Hay ED. TGFbeta3 signaling activates transcription of the LEF1 gene to induce epithelial mesenchymal transformation during mouse palate development. J Cell Biol. 2003b;163:1291–1301. doi: 10.1083/jcb.200306024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawshad A, Lagamba D, Hay ED. Transforming growth factor beta (TGFbeta) signalling in palatal growth, apoptosis and epithelial mesenchymal transformation (EMT) Arch Oral Biol. 2004;49:675–689. doi: 10.1016/j.archoralbio.2004.05.007. [DOI] [PubMed] [Google Scholar]
- Nawshad A, Lagamba D, Polad A, Hay ED. Transforming growth factor-beta signaling during epithelial-mesenchymal transformation: implications for embryogenesis and tumor metastasis. Cells Tissues Organs. 2005;179:11–23. doi: 10.1159/000084505. [DOI] [PubMed] [Google Scholar]
- Nawshad A, Medici D, Liu CC, Hay ED. TGFbeta3 inhibits E-cadherin gene expression in palate medial-edge epithelial cells through a Smad2-Smad4-LEF1 transcription complex. J Cell Sci. 2007;120:1646–1653. doi: 10.1242/jcs.003129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panopoulou E, Gillooly DJ, Wrana JL, et al. Early endosomal regulation of Smad-dependent signaling in endothelial cells. J Biol Chem. 2002;277:18046–18052. doi: 10.1074/jbc.M107983200. [DOI] [PubMed] [Google Scholar]
- Peinado H, Ballestar E, Esteller M, Cano A. Snail mediates E-cadherin repression by the recruitment of the Sin3A/histone deacetylase 1 (HDAC1)/HDAC2 complex. Mol Cell Biol. 2004a;24:306–319. doi: 10.1128/MCB.24.1.306-319.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peinado H, Olmeda D, Cano A. Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nature reviews. 2007;7:415–428. doi: 10.1038/nrc2131. [DOI] [PubMed] [Google Scholar]
- Peinado H, Portillo F, Cano A. Transcriptional regulation of cadherins during development and carcinogenesis. Int J Dev Biol. 2004b;48:365–375. doi: 10.1387/ijdb.041794hp. [DOI] [PubMed] [Google Scholar]
- Proetzel G, Pawlowski SA, Wiles MV, et al. Transforming growth factor-beta 3 is required for secondary palate fusion. Nat Genet. 1995;11:409–414. doi: 10.1038/ng1295-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pungchanchaikul P, Gelbier M, Ferretti P, Bloch-Zupan A. Gene expression during palate fusion in vivo and in vitro. J Dent Res. 2005;84:526–531. doi: 10.1177/154405910508400608. [DOI] [PubMed] [Google Scholar]
- Schock F, Perrimon N. Molecular mechanisms of epithelial morphogenesis. Annu Rev Cell Dev Biol. 2002;18:463–493. doi: 10.1146/annurev.cellbio.18.022602.131838. [DOI] [PubMed] [Google Scholar]
- Shimoyama Y, Hirohashi S, Hirano S, et al. Cadherin cell-adhesion molecules in human epithelial tissues and carcinomas. Cancer Res. 1989;49:2128–2133. [PubMed] [Google Scholar]
- Shook D, Keller R. Mechanisms, mechanics and function of epithelial-mesenchymal transitions in early development. Mech Dev. 2003;120:1351–1383. doi: 10.1016/j.mod.2003.06.005. [DOI] [PubMed] [Google Scholar]
- Shuler CF, Halpern DE, Guo Y, Sank AC. Medial edge epithelium fate traced by cell lineage analysis during epithelial-mesenchymal transformation in vivo. Dev Biol. 1992;154:318–330. doi: 10.1016/0012-1606(92)90071-n. [DOI] [PubMed] [Google Scholar]
- Sun D, Vanderburg CR, Odierna GS, Hay ED. TGFbeta3 promotes transformation of chicken palate medial edge epithelium to mesenchyme in vitro. Development. 1998;125:95–105. doi: 10.1242/dev.125.1.95. [DOI] [PubMed] [Google Scholar]
- Takeichi M. The cadherins: cell-cell adhesion molecules controlling animal morphogenesis. Development. 1988a;102:639–655. doi: 10.1242/dev.102.4.639. [DOI] [PubMed] [Google Scholar]
- Takeichi M. Cadherins: key molecules for selective cell-cell adhesion. IARC Sci Publ. 1988b:76–79. [PubMed] [Google Scholar]
- Takeichi M. Cadherins in cancer: implications for invasion and metastasis. Curr Opin Cell Biol. 1993;5:806–811. doi: 10.1016/0955-0674(93)90029-p. [DOI] [PubMed] [Google Scholar]
- Takeichi M. Morphogenetic roles of classic cadherins. Curr Opin Cell Biol. 1995;7:619–627. doi: 10.1016/0955-0674(95)80102-2. [DOI] [PubMed] [Google Scholar]
- Taya Y, O'kane S, Ferguson MW. Pathogenesis of cleft palate in TGF-beta3 knockout mice. Development. 1999;126:3869–3879. doi: 10.1242/dev.126.17.3869. [DOI] [PubMed] [Google Scholar]
- Thiery JP. Cell adhesion in development: a complex signaling network. Curr Opin Genet Dev. 2003a;13:365–371. doi: 10.1016/s0959-437x(03)00088-1. [DOI] [PubMed] [Google Scholar]
- Thiery JP. Epithelial-mesenchymal transitions in development and pathologies. Curr Opin Cell Biol. 2003b;15:740–746. doi: 10.1016/j.ceb.2003.10.006. [DOI] [PubMed] [Google Scholar]
- Thiery JP, Sleeman JP. Complex networks orchestrate epithelial-mesenchymal transitions. Nat Rev Mol Cell Biol. 2006;7:131–142. doi: 10.1038/nrm1835. [DOI] [PubMed] [Google Scholar]
- Tian M, Schiemann WP. The TGF-beta paradox in human cancer: an update. Future Oncol. 2009;5:259–271. doi: 10.2217/14796694.5.2.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukazaki T, Chiang TA, Davison AF, Attisano L, Wrana JL. SARA, a FYVE domain protein that recruits Smad2 to the TGFbeta receptor. Cell. 1998;95:779–791. doi: 10.1016/s0092-8674(00)81701-8. [DOI] [PubMed] [Google Scholar]
- Tudela C, Formoso MA, Martinez T, et al. TGF-beta3 is required for the adhesion and intercalation of medial edge epithelial cells during palate fusion. Int J Dev Biol. 2002;46:333–336. [PubMed] [Google Scholar]
- Valcourt U, Kowanetz M, Niimi H, Heldin CH, Moustakas A. TGF-beta and the Smad signaling pathway support transcriptomic reprogramming during epithelial-mesenchymal cell transition. Mol Biol Cell. 2005;16:1987–2002. doi: 10.1091/mbc.E04-08-0658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia W, Cheng CY. TGF-beta3 regulates anchoring junction dynamics in the seminiferous epithelium of the rat testis via the Ras/ERK signaling pathway: An in vivo study. Dev Biol. 2005;280:321–343. doi: 10.1016/j.ydbio.2004.12.036. [DOI] [PubMed] [Google Scholar]
- Xu X, Han J, Ito Y, Bringas P, Jr, Deng C, Chai Y. Ectodermal Smad4 and p38 MAPK are functionally redundant in mediating TGF-beta/BMP signaling during tooth and palate development. Dev Cell. 2008;15:322–329. doi: 10.1016/j.devcel.2008.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W, Kamara H, Svoboda KK. The role of twist during palate development. Dev Dyn. 2008;237:2716–2725. doi: 10.1002/dvdy.21627. [DOI] [PubMed] [Google Scholar]
- Zavadil J, Bottinger EP. TGF-beta and epithelial-to-mesenchymal transitions. Oncogene. 2005;24:5764–5774. doi: 10.1038/sj.onc.1208927. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. (A) Westernblot was performed on homogenous palatal MES cells that were treated with small hairpin RNA (shRNA) Smad4 (pRetrosuper-shSmad4 (Addgene, MA) for 48 h to inhibit the Smad-dependent pathway), was used which specifically targets the coding region of murine Smad4. Empty vector and scrambled shRNA vector (scr) were also used as controls. (B) To abolish the effect of endogenous TGFβ3, cells were treated with blocking TGFβ3 antibody was used at 10ng/mL for 24 h. MES cells were treated with a Smad-independent pathways were blocked using small synthetic chemical inhibitors for 60 min – (C) U0126 for MEK1/2 (20µM), (D) SB202190 for p38MAPK (20µM) and (E) LY294002 to block PI3 Kinase (20µM). (F) All treatment showed significant loss of the targeted protein. Intensity of the band was measured using the Carestream Molecular Imaging Software version 5.3.1 (Rochester, NY). To perform a t-test analysis of mean intensity measurements, a ROI analysis was done from the data to Microsoft Excel software from the exported “.txt” files. (G) In double staining, Keratin 14 (K14) (Rhodamine) was expressed in untreated (i) as well as 24 (ii) and 48 h (iii) of TGFβ3 (5ng/mL) treatments while Vimentin (FITC; iv), confirm its mesenchynal transformation at 48 h.
Supplementary Figure 2. Double staining immunofluorescence protein expression assays were performed on homogenous MES cells that were treated with TGFβ3 (5ng/mL) for 24 and 48 h. In double staining, E-Cadherin (A, FITC and D, Rhodamine) and Desmoplakin (G, Rhodamine) were highly expressed while Vimentin (A, Rhodamine), N-Cadherin (D, FITC) and Fibronectin (G, FITC) expression were not seen in untreated control cells. As expression of Vimentin, Fibronectin and N-Cadherin began to increase, in response TGFβ3 for 24 h (B, E & H), E-Cadherin and Desmoplakin expressions were reduced and significantly lost by 48 h (C, F & I), at which stage, Vimentin, Fibronectin and N-Cadherin were highly expressed.