Abstract
The development of transgenic mouse lines that selectively label a subset of neurons provides unique opportunities to study detailed neuronal morphology and morphological changes under experimental conditions. In the present study, a mouse line in which a small number of retinal ganglion cells (RGCs) express yellow fluorescent protein (YFP) under control of the Thy-1 promoter was used (Feng et al., 2000). We characterized the number, distribution by retinal region and eccentricity of YFP-labeled RGCs using fluorescence microscopy and StereoInvestigator software (MicroBrightField, VT, USA). Then, we captured images of 4–6 YFP-expressing RGCs from each of 8 retinal regions by confocal microscopy, producing 3-dimensional and flattened data sets. A new semi-automated method to quantify the soma size, dendritic length and dendritic arbor complexity was developed using MetaMorph software (Molecular Devices, PA, USA). Our results show that YFP is expressed in 0.2% of all RGCs. Expression of YFP was not significantly different in central versus peripheral retina, but there were higher number of YFP expressing RGCs in the temporal quadrant than in the nasal. By confocal-based analysis, 58% of RGCs expressing YFP did so at a high level, with the remainder distributed in decreasing levels of brightness. Variability in detailed morphometric parameters was as great between two fellow retinas as in retinas from different mice. The analytic methods developed for this selective YFP expressing RGC model permit quantitative comparisons of parameters relevant to neuronal injury.
Keywords: mouse, retina, ganglion cell, glaucoma, optic nerve, neuropathy, yellow fluorescent protein
Introduction
Glaucoma is the second leading cause of blindness worldwide (Quigley and Broman, 2006) and its principal pathological feature is the death of RGCs (Quigley et al., 1981). Apoptosis is a final common pathway for RGC death both in human and experimental models of glaucoma (Quigley et al., 1995; Kerrigan et al., 1997). There is some evidence that, prior to cell death, RGCs undergo decreases in either soma size and dendritic arborization (Weber et al., 1998). In the spontaneous glaucoma exhibited by DBA/2J mice and experimental glaucoma in rats, there is temporary persistence of RGC bodies with an altered dendritic phenotype after axon loss (Jakobs et al., 2005; Buckingham et al., 2008; Son et al., 2010; Soto et al., 2011). Similarly, some neurons undergo change in structure prior to death in Alzheimer’s, Parkinson’s and Huntington’s diseases (Kisiswa et al., 2010).
Further support for alterations in RGCs prior to their final somal death process has been detected by functional testing after experimental injury. Pattern-evoked electroretinography (ERG) (Johnson et al., 1989) and detailed functional recordings (Weber and Harman, 2005) point to altered RGC responses in monkey models of glaucoma. ERG changes in waveforms thought to be derived from RGC layer cells are seen in rodent glaucoma (Holcombe et al., 2008; Kong et al., 2009; Porciatti and Ventura, 2009). If either structural or functional change in RGCs prior to cell death were measurable in human eyes, these parameters could serve as early indications of neuronal injury, potentially at a reversible stage. Such a biomarker would be useful in shortening the length of human clinical trials for neuroprotective agents.
The recent development of a transgenic mouse in which a small number of RGCs express yellow fluorescent protein (YFP) provides unique opportunities to study detailed structural change. In transgenic mice in which every RGC is labeled (Feng et al., 2000), individual RGC dendritic and axonal morphology cannot be resolved. By contrast, RGCs in the retina of selectively expressing YFP mice are sparsely labeled, so the individual cell characteristics of dendritic, somal, and axonal anatomy can be examined in detail and quantified. Our laboratory and others found it possible to image the same RGC over time in living mice of this strain (Walsh and Quigley, 2008; Leung et al., 2008). A recent report showed serial, in vivo observations of YFP-selective expressing RGCs in mice after optic nerve crush injury (Leung et al., 2011), suggesting dendritic shrinkage prior to death. By contrast, another in vivo study using optical coherence tomography suggested that the retina thickened prior to RGC death in mice after optic nerve crush (Gabriele et al., 2011).
To study changes in RGC morphology after experimental glaucoma or nerve injury in the YFP selective expression mice, we developed methods for detailed analysis of neuronal structure in whole mounted retina. These methods can specify the phenotypic alterations that seem likely to be occurring between the time of significant axonal injury and somal death. While in vivo imaging has the advantage of following individual RGCs over time, it can view only a small number of cells. The changes in corneal clarity in chronic experimental mouse glaucoma often reduce resolution of detailed measurement in vivo. Though the morphology of individual cells can be followed to some extent in the living eye, a detailed, 3-dimensional analysis cannot be performed as accurately in vivo as it can be with confocal imaging of fixed tissues or of retinal explants.
Past approaches to quantify neuronal morphology in neural tissues included the histological Golgi method, immunohistochemical delineation of the cytoskeleton (Jakobs et al., 2005), intracellular single-cell dye injections (Tauchi and Masland., 1985), or biolistic delivery of fluorescent dyes (Gan et al., 2000). In addition to mice expressing YFP in a select number of RGCs, Nathans and co-workers have produced transgenic mouse lines exhibiting sparse labeling of RGCs, amacrine cells and other retinal neurons (Badea et al., 2003; Badea et al., 2009). Quantitative evaluation of neuronal morphology has recently been improved by objective, semiautomated methods (Gensel et al., 2010) that provide quantitative data on axon length and dendrite branching complexity, also known as Sholl analysis (Sholl, 1953).
In vitro study of whole-mounted retinas allows detailed, quantitative observations on RGC dendritic, somal, and axonal components, and can be paired with immunolabeling of important additional biomarkers and non-RGC cells that may be specifically related to injury and disease. We and others have developed inducible models of chronic glaucoma in mice (Sappington et al., 2010; Cone et al., 2010) that produce RGC death and may permit study of initial changes in RGC morphology. By detailed analysis of the earliest phase of injury, it is possible that features can be detected that would be usable clinically to identify a reversible stage of RGC injury. This report details the characteristics of RGCs that selectively express YFP in mice, analysed by newly developed morphological methods and comparing data from different regions and eccentricities of the retina. A companion report will give the detailed changes from YFP mice after nerve crush and glaucoma.
1. Materials and methods
1.1 Animals
The mice from B6.Cg-Tg(Thy1-YFPH)2Jrs/J strain (Jackson Laboratory, Bar Harbor, Maine; cat. no. 3782) that express YFP in a small subset of RGCs were used. The study contains data from 6 eyes of three 6-month-old naïve mice and 14 control eyes of 4 to 10.5 month old mice that underwent an optic nerve crush procedure from an accompanying study (Kalesnykas et al., in preparation). All animals were treated in accordance with the ARVO Statement for the Use of Animals in Ophthalmic and Vision Research and the EC Directive 86/609/EEC for animal experiments, using protocols approved and monitored by the Johns Hopkins University School of Medicine Animal Care and Use Committee.
2.2 Tissue preparation
Animals were anesthetized with an intraperitoneal injection of ketamine, xylazine, and acepromazine (50, 10 and 2 mg/kg, respectively) and perfused transcardially for one minute with saline followed by 9 minutes with 4% paraformaldehyde in 0.1 M phosphate buffer solution (PB), pH 7.2. The superior pole of each eye was marked for orientation, the eyes were enucleated, and their retinas were detached from the sclera. Retinas were post-fixed as whole mounts for 24 hours in the same fixative solution. Next, retinas were washed with 0.1M PB, pH 7.2, flat mounted on a glass slide in glycerol with the RGC layer up and coverslipped.
2.3 YFP positive RGC quantification and distribution
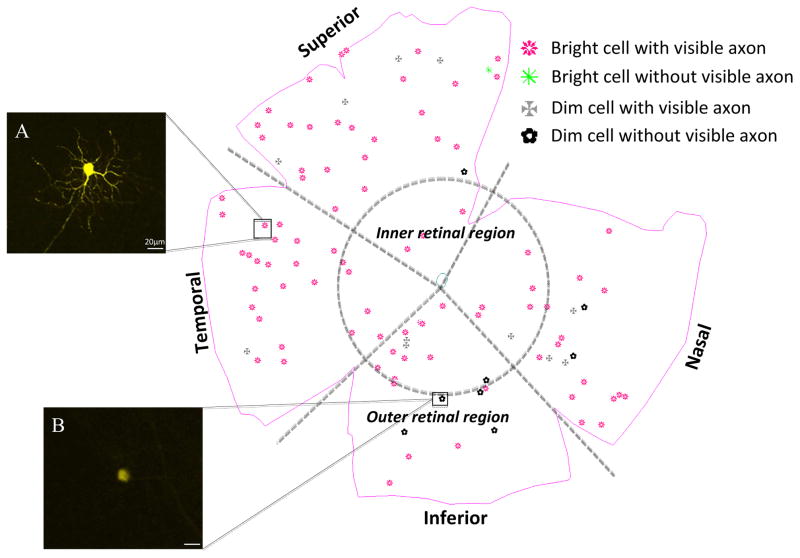
The number and distribution of RGCs expressing YFP was performed with Stereo Investigator software (MicroBrightField, USA), in an integrated hardware-software apparatus, consisting of an ECLIPSE E600 microscope (Nikon, Japan) with a 3-Chip CCD color video camera (HV-C20, Hitachi, Japan) and motorized stage (three-axis computer-controlled stepping motor system with a 0.1-m resolution) with a microcator (Heidenhain EXE 610C) attachment providing a 0.1-μm resolution in the z axis. RGCs were subjectively characterized based on whether their level of YFP expression was bright or dim and whether there was a visible axon. Four initial categories were thus established: 1) bright fluorescent cells with visible axon, 2) bright cells without visible axon; 3) dim cells with axon; 4) dim cells without axon. All RGCs, whether bright or dim, were marked as to their position in a retinal quadrant (superior, inferior, nasal and temporal), and by their central and peripheral position on a map of the whole mount for each retina. The line demarcating the central/peripheral division had a radius half the distance to the retinal whole mount periphery.
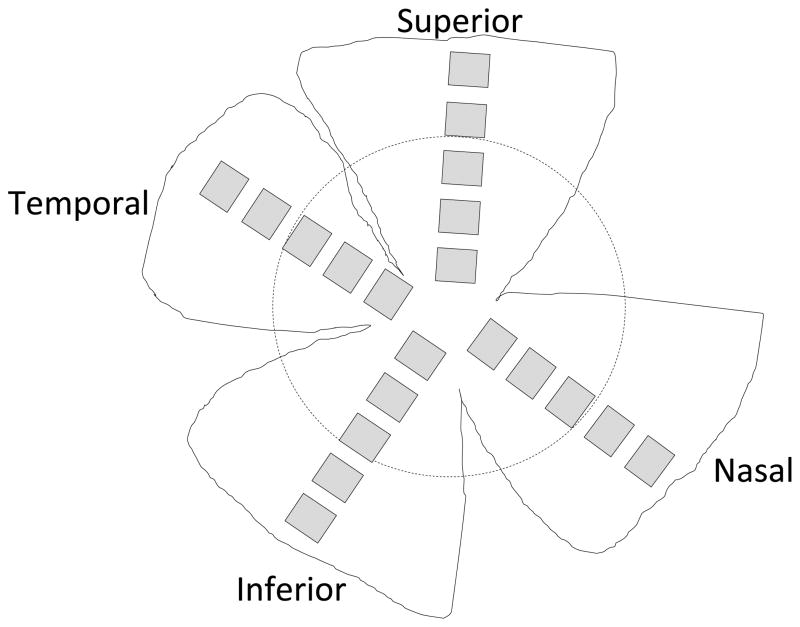
To provide an estimate of the proportion of RGCs that express YFP, we counted the total number of RGC layer cells in retinal flat mounts from normal, similar aged C57BL/6 mice, the base strain for the YFP mice. Briefly, perfusion-fixed retinal flat mounts were stained with 4′,6-diamidino-2-phenylindole (DAPI, 1:250, Invitrogen, Carlsbad, CA, USA). After buffer washes, retinas were cover-slipped with Fluorescent Mounting Medium (DakoCytomation, Carpinteria, CA, USA) and imaged using the same confocal microscope. Twenty 40x images were taken per retina, 5 fields from each of 4 quadrants (superior, nasal, inferior, temporal), equaling a 7% sample of total retinal area (Figure 1). Each image was taken at the level of the RGC layer and analyzed with MetaMorph software (version 7.7.0.0, Molecular Devices, Downington, PA, USA). DAPI counted neurons were subdivided into inner and outer retina and into quadrant-specific data. Because the confocal imaging was performed only at the level of the RGC layer, astrocytes of the nerve fiber layer were not included. All cells with a round or slightly oval nucleus and morphology compatible with that of neurons were identified manually by trained technical staff masked to the protocol details for each retina. Interobserver reproducibility of the method comparing 3 technicians showed a mean difference of only 0.5% between observer A and B, and a mean difference of 1.7% between observer A and observer C, counting a total of nearly 6,000 cells from 20 images in each test. Cells that were not counted included: 1) those with elongated nuclei more typical for endothelial cells or astrocytes and 2) polymorphonuclear cells assumed to be white blood cells.
Figure 1.
DAPI-positive nuclei were counted from 5 fields of 4 quadrants (superior, nasal, inferior, temporal), equaling a 7% sample of total retinal area.
Since the DAPI-labeled neurons included both amacrine cells and RGCs in the RGC layer, we have previously (Quigley et al., 2011) measured the proportion of DAPI-labeled neurons that are RGCs in mouse retina by immunolabeling for synuclein gamma (Sncg), a protein found in RGCs. As a proportion of all neurons labeled with DAPI in the RGC layer, Sncg positive cells represented 40.3 ± 5.9% of total neurons. There is no local difference in the number of amacrine cells in the mouse retina by quadrant or distance from the central retina (Jakobs et al., 2005).
2.4 Confocal imaging, Neurite outgrowth and Sholl Analysis
Four to six randomly selected RGCs from each of the 8 areas of the retina (e.g. peripheral/nasal, central/superior, etc) were imaged on the Zeiss LSM 510 Meta Confocal Microscope (Zeiss MicroImaging, Thornwood, NY, USA). Images were taken to include the entire dendritic field of each cell in a series along the z axis (perpendicular to the optic axis). The following channel settings for imaging were used: excitation at 514 nm = 17.9, pinhole =1 Airy unit, detector gain = 764, amplifier offset = 0.057, amplifier gain = 1. A measurement was made of the thickness of the dendritic fields in the z axis by noting the distance in microns from the axon hillock at the cell body to the last plane in which the dendrites were still visible. No cells were used in which the dendritic fields of two RGCs overlapped. The captured z stack images were converted into a 2-dimensional single projection image. This 2-dimensional image was then exported as 16-bit TIFF format grayscale image using LSM 510 software (Zeiss MicroImaging) for further analysis.
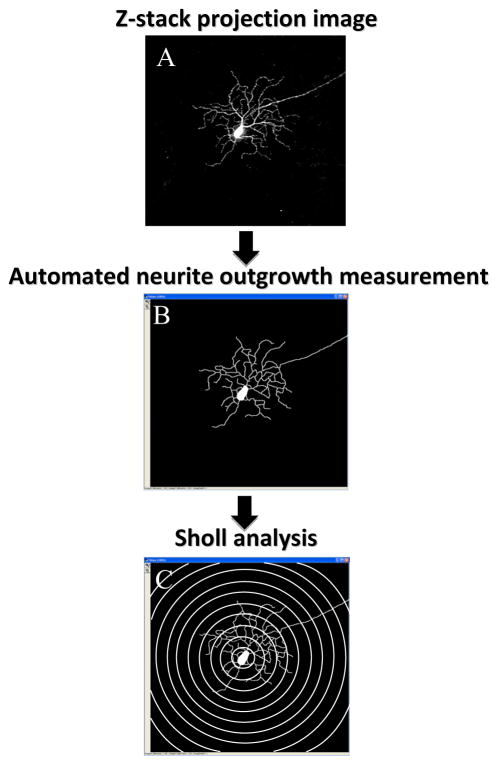
Confocal images that contained more than one cell of interest were modified to include only one cell at a time using Adobe Photoshop software (version 7.0, Adobe Systems, San Jose, CA, USA). Each cell was outlined, copied and pasted into a new image with a black background. Next, the image of each RGC was imported using an automated macro (see Supplement) to determine features of cell morphology and the mean intensity of YFP expression in its soma using MetaMorph Offline image analysis software (version 7.7.0.0, Molecular Devices). This included both a proprietary MetaMorph neurite outgrowth measurement program (Figure 2) and our specially-designed Sholl analysis based on a template of rings (Sholl, 1953), consisting of 10 concentric circles centered on the soma, from 50–500 μm increasing in 50 μm steps (Figure 2). Because of the diversity of YFP expression among RGCs, it was appropriate to analyse the bright and dim RGCs separately in the neurite outgrowth and Sholl analyses. For bright cells, the minimum intensity of neurite outgrowth was set to 3,500 grey levels. The latter intensity level was too high for dim cell neurite outgrowth to be recognized by the software. Therefore, the minimum intensity of neurite outgrowth for dim cells was set to 2,000 grey levels. However, in majority of cases only the primary processes of dim YFP cells could be visible by observer and detected using MetaMorph software (see Figure 3B). The neurite outgrowth analysis provided the cell soma area, total length of outgrowth, number of outgrowths that connected to cell soma, number of branches of all the processes, mean, median and maximum process length, and straightness of the neurite outgrowth.
Figure 2.
A graphical representation of neurite outgrowth and Sholl analysis measurements. A 2-dimensional 16-bit TIFF image (A) was generated from the z stack of images that were captured using confocal microscopy. The automated macro that contains both a proprietary neurite outgrowth measurement program (B) and a Sholl rings’ template (C) was applied for detailed morphological analysis.
Figure 3.
A map of YFP positive RGC distribution in the retina. Cells were classified according to YFP expression (bright or dim) and presence or absence of a visible axon. Classifications were: bright YFP cells with axon (A), bright YFP cells without axon, dim YFP cells with axon and dim YFP cells without axon(B). The selected cells were imaged and analyzed from the inner and outer parts of each quadrant. Scale bar = 20 μm.
The Sholl analysis is based on the number of dendritic processes that intersect each of the 10 concentric rings (Sholl, 1953). For this approach, the investigator exported the data for each ring into newly generated log images. Then, the automated Sholl macro determined the sum of intersections between the each ring and dendritic processes (Gensel et al., 2010).
2.5 Statistical analysis
Statistical analyses included the comparison of mean values with parametric statistical tests for data that were normally distributed and median values with non-parametric testing for those whose distributions failed normality testing. Nine parameters were used in cluster analysis in order to identify different subtypes of RGCs based on their morphological appearance: cell body size, number of primary processes, total neurite outgrowth, z depth of dendritic arbor stratification, maximum process length, mean process length, median process length, number of dendritic branches and straightness of the neurite outgrowth. Among the commonly used cluster analytic approaches (Day and Edelsbrunner, 1984) (single-link, complete-link and average-link), we chose the average-link method as the best compromise. Hierarchical clustering treats each data point as a singleton cluster, and then successively merges clusters until all points have been merged into a single remaining cluster. Single-link hierarchical clustering merges in each step the two clusters with the smallest minimum pairwise distance. In complete-link hierarchical clustering, each step merges the two clusters with the smallest maximum pairwise distance. Average-link clustering is a compromise between the sensitivity of complete-link clustering to outliers and the tendency of single-link clustering to form long chains that do not correspond to the intuitive notion of clusters as compact, spherical objects.
To compare the results of Sholl analysis between the bright and dim cells, we used as the count for Poisson regression the dendritic complexity (number of processes crossing the Sholl ring) every 50 μm from 50 to 500 μm. A mixed model was used to address the issue of repeated measurements from each group. Instead of different time points, we treated the dendritic complexity at each eccentricity as a point, allowing us to predict both the mean trajectory (fixed effect) and also predict the individual trajectory over time (random effect). Since our outcome is count, we used a non-linear mixed model to specify the Poisson distribution in the model. The model for mean count (fixed effect) is described by: Log Y= b0+b1*X+b2*group+b3*X*group, where Y is the count (dependent variable), X is the independent variable (dendritic complexity at 50 μm, 100 μm etc.), GROUP is the binary variable (0 = group 1; 1 = group 2); b0 is the intercept (the predicted count at 50 μm; b0 + b2 is the intercept for group 2 (predicted count at 50 μm); b1 is the slope for group 1 (how fast the count declines over X); b1 + b3 is the slope for group 2 (how fast the count declines over X). We put the intercept only in the random effect term. The random effect actually indicates how far the count over time of each individual is from predicted mean count (fixed effect). The data were analyzed using SPSS for Windows (v. 14.0, SPSS Inc., Chicago, IL, USA) or SAS software (v. 9.2, SAS Inc., Cary, NC, USA).
3. Results
3.1 Number, distribution, and description of labeled RGC
The number of YFP-labeled RGCs per retina was 87.0 ± 29.2 cells (mean ± standard deviation (SD)). The number of labeled cells was counted in the peripheral retinal region and the inner retinal region of each retina (Figure 3). In the defined peripheral zone, the mean number of labeled RGCs was 58.1 ± 14.2 and in the inner retinal zone it was 28.9 ± 6.1 RGCs (t-test, p < 0.0001). Since the dividing circle between inner and outer retina had a radius that was one-half of the distance from the center to the periphery, the area of the peripheral zone was π(2r)2-πr2, or 3πr2, compared to an area of πr2 for the central zone. As described above, we previously measured the density of neurons in the RGC layer by DAPI labeling in 37 normal (non-YFP), C57BL/6 mouse retinas, dividing the retina into similar central and peripheral zones, as well as into the 4 quadrants, superior, inferior, nasal, and temporal. The density of RGC layer neurons was 6,934 ± 482 per mm2 in the central region and 5,960 ± 634 per mm2 in the peripheral region (n = 71 fields and 52 fields respectively; Mann-Whitney U test, p < 0.0001). This is a 16% higher neuronal density centrally. Thus, if we take into account the 3 times greater peripheral area measured and the higher central density, we would expect that a central region mean of 30 labeled cells would lead to an estimated mean of 77.4 cells in the periphery (30 * 3 * (5960/6934)), taking both area difference and regional density of all RGCs into account. Our observed mean number of 58.1 RGCs is less than this, suggesting that there is preferential YFP expression in central RGCs in this strain.
We also evaluated the number of YFP-labeled RGCs in each quadrant for a total of more than 1700 cells in the 20 eyes. The mean number of labeled cells for inferior, nasal, superior, and temporal quadrants was 19.2, 16.2, 24.3, and 27.4, respectively, indicating significantly higher YFP labeling in temporal quadrant as compared to nasal quadrant (ANOVA, p = 0.008). To be sure that the density of RGC layer cells was generally similar across quadrants, we summarized the mean density of RGC layer cells per mm2 in the same 4 quadrants of the 37 control mouse eyes from the prior study as above. There were small, but significant differences, in that the inferior and nasal quadrants had higher density than the superior and temporal quadrants (Table 1). However, these differences represent only 4% more RGC layer cells in the inferior and nasal quadrants compared to superiorly or temporally. Thus, higher proportion of YFP-labeled RGCs is distributed in superior and temporal quadrants.
Table 1.
Density of RGC layer cells by quadrant
| Quadrant data in RGC layer, cells/mm2 | ||||
|---|---|---|---|---|
| Inferior, n = 31 | Nasal, n = 27 | Superior, n = 33 | Temporal, n = 28 | |
| Mean ± SD | 6,729 ± 362 | 6,928 ± 533 | 6,162 ± 526 | 6,240 ± 492 |
| Median | 6,720 | 6,858 | 6,167 | 6,216 |
| One-way ANOVA, Bonferroni posthoc test, p values | ||||
| Inferior | X | n.s. | 0.001 | 0.01 |
| Nasal | X | 0.001 | 0.001 | |
| Superior | X | n.s. | ||
| Temporal | X | |||
n.s.-statistically non-significant difference.
The YFP-labeled cells could be subjectively distinguished as those that were bright (high YFP expression) and those that were dimmer in fluorescence (lower YFP expression) at the same confocal laser settings. With quantification of the soma body fluorescence, this division was made at particular settings of the image analytic software (see below). Further, the axon was visible in many RGCs, but not visible in others. We therefore totaled the mean number of the following types of RGCs: bright with axon, bright no axon, dimmer with axon, dimmer no axon. The brighter RGCs with axon represented 56% of labeled RGCs, while the dimmer with axon group was 30% and the dimmer without visible axon were 11%. Bright RGCs with no visible axon were 2% of the total.
Comparing central to peripheral retinal zones, we found no significant differences in the number of RGC classified by the bright versus dim ± axon segregation. The peripheral retina had 59.3% bright with axon, 27.8% dimmer with axon, and 10.9% dimmer without visible axon, while the central retina had 49.5%, 34.4% and 13.7% in these groups, respectively (chi-squared test, p = 0.069). Likewise, the distribution of labeled RGC classified in this way did not differ by quadrant of the retina (Table 2).
Table 2.
Appearance of labeled RGC by quadrant
| Bright with axon, % | Bright no axon, % | Dim with axon, % | Dim no axon, % | |
|---|---|---|---|---|
| Inferior | 58 | 1 | 29 | 12 |
| Nasal | 56 | 3 | 29 | 12 |
| Superior | 51 | 4 | 32 | 14 |
| Temporal | 59 | 2 | 29 | 10 |
3.2 Quantitative RGC soma intensity data
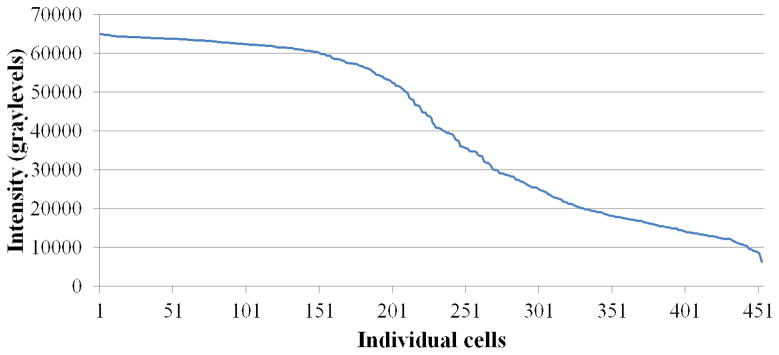
To represent the detailed anatomy of RGCs, we selected 4–6 YFP positive neurons from each of the 8 regions of each retina. At the same confocal microscopic setting for each RGC, we captured the images containing all of the dendrites in 454 cells and collapsed these images into a compressed z-stack showing the extent of all processes. The cell soma was then analysed for its intensity on a scale with up to 65,535 grey levels (the brighter cells would therefore have more grey levels). Many cells achieved the highest level of brightness, while others exhibited fewer total brightness levels. The cells that were subjectively classified as bright YFP cells during retinal mapping had an average intensity of 55,709 ± 12,082 (n = 271) grey levels (median = 61,017) when measured by MetaMorph software, whereas the cells that were classified as dim YFP cells had an average intensity of 19,409 ± 7,951 (n = 182) grey levels (median = 17,406; Mann-Whitney U test for difference between bright and dim groups, p ≤ 0.0001; data missing for one RGC). We inspected the data to find the most useful cutoff between brighter and dimmer cells by subjective analysis. Using the value of 30,000 levels, there were 21/272 (7.7%) of cells classified as bright with values below 30,000 and 20/182 (10.9%) of cells classified as dim with values greater than 30,000. Thus, the subjective grading placed more than 90% of cells in the appropriate grouping based on the quantitative measure. The graphical distribution of brightness levels for RGCs approximates an S-shaped curve (Figure 4).
Figure 4.
The graphical distribution of YFP brightness in grey levels of randomly selected and imaged RGCs approximates an S-shaped curve.
3.3 Depth of dendritic arbor stratification
The depth of dendritic arbor stratification of YFP positive RGCs was estimated only in cells classified as bright, because dim cells did not have enough YFP labeling in dendrites. For bright YFP cells, the mean distance from axon hillock at the soma to the final dendrites was 9.98 ± 3.64 μm (n = 271).
3.4 Quantitative RGC morphology
Among all YFP-positive RGCs, both bright and dim combined, the mean soma area was 586 ± 265 μm2, the total length of outgrowth was 1,564 ± 1,449 μm, the mean process length was 262 ± 236.8 μm, the median process length was 148 ± 178 μm, and the maximum process length was 816.8 ± 824. There were 5.1 ± 2.3 major outgrowths (dendrites plus the axon) connected to the cell soma and 81.6 ± 80.5 branches of all the outgrowths.
However, there were significant differences between bright and dim RGCs in all of these parameters (Table 3).
Table 3.
Neurite outgrowth estimates from bright and dim YFP cells
| Parameter | Brighter Cells | Dimmer Cells | p value* |
|---|---|---|---|
| Soma size, μm2 | 668 ± 268.8 | 464 ± 205.8 | ≤0.001 |
| Primary processes | 6.1 ± 2.0 | 3.6 ± 1.8 | ≤0.001 |
| Total neurite outgrowth, μm | 2,392 ± 1245 | 327.2 ± 606 | ≤0.001 |
| Mean process length, μm | 392.5 ± 211 | 67.1 ± 100 | ≤0.001 |
| Median process length, μm | 215.6 ± 196.4 | 46.7 ± 67.5 | ≤0.001 |
| Maximum process length, μm | 1,242 ± 774.5 | 180.5 ± 350 | ≤0.001 |
| Branches | 126.4 ± 71 | 14.6 ± 33.5 | ≤0.001 |
t-test, not corrected for multiple comparisons. Values are presented as mean ± SD
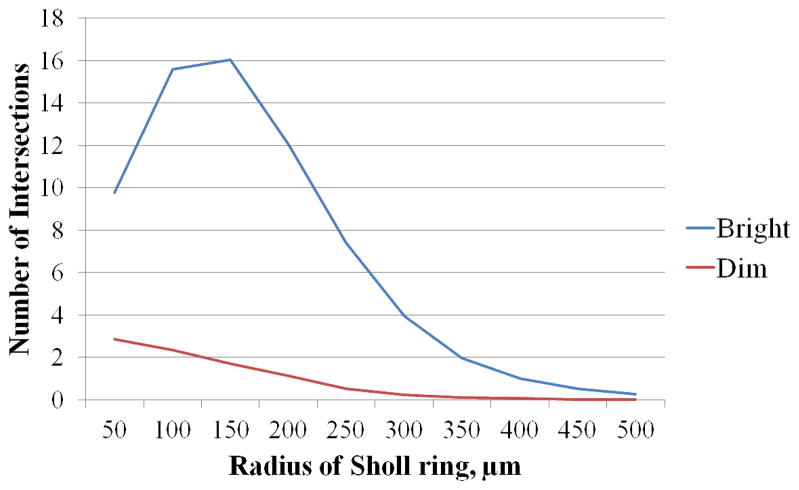
The Sholl analysis indicated the number of processes intersecting each sequentially wider circle away from the RGC soma. This measure, called dendritic complexity, was highest among bright RGCs at the radii 100–150 μm and among dim RGCs at the radii 50 μm from the RGC soma (Figure 5). Statistical modeling was applied to compare the curves of dendritic complexity between bright and dim RGCs. The dendritic complexity of bright and dim cells differed significantly both at each intercept (50 μm, 100 μm etc.) and in a slope produced by all intercepts (Poisson regression in a mixed model, p < 0.0001).
Figure 5.
The Sholl analysis of processes intersecting each concentric circle at a radius of 50 μm to 500 μm from the RGC soma. The dendritic complexity was highest among bright RGCs at the radii 100–150 μm from the RGC soma and highest among dim RGCs at the radii of 50 μm. The dendritic complexity of bright and dim cells differed significantly both at each intercept (50 μm, 100 μm etc.) and in a slope produced by all intercepts (poisson regression in a mixed model, p < 0.0001).
3.5 Eccentricity differences
We compared the analysis for 279 YFP-positive RGCs from the peripheral retina and 175 RGCs from the central retina, including all cells that were imaged, both bright and dim. The mean intensity of the analyzed cells did not differ between the groups and was 39,932 ± 20,637 and 42,043 ± 20,191 grey levels, respectively (Mann-Whitney U test, p = 0.40). Cell body size was significantly larger in the peripheral compared to central RGCs. None of the other neurite outgrowth parameters or dendritic complexity differed significantly between the central and peripheral retinal zones (Table 4; all p values ≥ 0.123, except for the body size (p ≤ 0.001).
Table 4.
The neurite outgrowth data comparing central and peripheral retina
| Parameter | Central retina | Peripheral retina |
|---|---|---|
| Soma size, μm2 | 530.5 ± 249* | 621 ± 269* |
| Number of processes | 5.05 ± 2.1 | 5.2 ± 2.4 |
| Total neurite outgrowth, μm | 1,642 ± 1,484 | 1,514 ± 1,427 |
| Mean process length, μm | 282.5 ± 243.8 | 249 ± 231.4 |
| Median process length, μm | 164.6 ± 196.6 | 137.3 ± 164.6 |
| Maximum process length, μm | 857.8 ± 843.4 | 791 ± 812 |
| Number of branches | 88.6 ± 85 | 77.2 ± 77.3 |
t-test, p ≤ 0.001. Values are presented as mean ± SD
3.6 Fellow eye comparison
We sought to determine if the expression of YFP was more similar in fellow eyes of the same mouse than in two unrelated eyes. To do this, we compared retinal data on neurite outgrowth and Sholl analysis in ANOVA models in which one independent variable was whether eyes were from the same animal. Among the 8 parameters that were compared, only mean process length was more closely related in fellow eyes than in eyes from different mice. Other parameters did not differ significantly.
3.7 Cluster analysis
The cluster analysis was done only on bright YFP cells. Six different clusters of YFP positive RGCs were identified based on morphological criteria with the statistical approach described. The main details of each cluster are presented in Table 5.
Table 5.
Subtypes of RGCs that were grouped using cluster analysis
| Parameter | Cluster 1 | Cluster 2 | Cluster 3 | Cluster 4 | Cluster 5 | Cluster 6 |
|---|---|---|---|---|---|---|
| Depth of dendritic stratification, μm | 14.9 ± 4.4 | 8.9 ± 2.5 | 9.2 ± 2.6 | 11.8 ± 2.8 | 8.7 ± 2.1 | 8.2 ± 3.1 |
| Total outgrowth, μm | 2,689 ± 752 | 1,890 ± 518 | 3,422 ± 809 | 1,022 ± 581 | 3,159 ± 709 | 837 ± 578 |
| Number of processes | 5.5 ± 1.2 | 7 ± 1 | 5.6 ± 1.2 | 4.9 ± 1.1 | 7.8 ± 1.3 | 4.3 ± 1.1 |
| Mean process length, μm | 497 ± 180 | 273 ± 76 | 613 ± 116 | 209 ± 122 | 413 ± 99 | 190 ± 120 |
| Number of branches | 165 ± 43 | 97 ± 33 | 169 ± 47 | 50 ± 35 | 172 ± 40 | 37 ± 29 |
| Cell body area, μm2 | 518 ± 158 | 713 ± 218 | 626 ± 193 | 448 ± 175 | 896 ± 192 | 486 ± 154 |
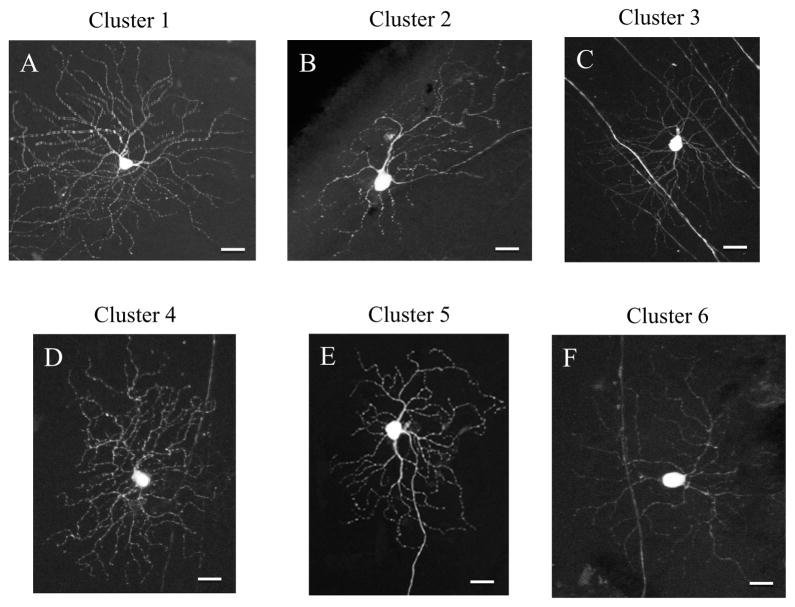
RGCs from cluster 1 had the greatest depth of dendritic stratification and were among the highest in mean process length (Figure 6A). However, they did not have the largest average cell body size. Cluster 2 RGCs had bigger cell soma than cells in cluster 1, but fewer branches, smaller total outgrowth and stratified their dendritic arbor closer to the RGC layer (Figure 6B). Cells from cluster 3 had the highest total outgrowth of the dendritic arbor and the longest mean process length among all RGC subtypes (Figure 6C). Clusters 4 and 6 were similar in neurite outgrowth parameters, but stratified their dendritic arbors at different depths; cells from cluster 4 stratified deeper in the inner nuclear layer, whereas cluster 6 cells stratified closer to the RGC layer (Figure 6D and F). Cluster 5 cells had the largest soma size and one of the longest total outgrowth values, but the depth of dendritic arbor stratification was one of those close to the RGC layer (Figure 6E).
Figure 6.
YFP expressing RGCs were grouped into 6 different clusters of cells according to their neurite outgrowth parameters, soma size and depth of stratification in the inner plexiform layer. Cells from cluster 1 (A) had the highest depth of dendritic stratification. Cluster 2 cells (B) had bigger cell soma than cells in cluster 1, whereas cells from cluster 3 (C) had the highest total outgrowth of the dendritic arbor. Cells that were identified as cluster 4 (D) had low number of primary processes and dendritic branches, whereas cluster 5 cells (E) were the biggest in soma size and have one of the longest total outgrowth. Cells from cluster 6 (F) had the lowest number of branches and primary processes. Scale bar = 20 μm.
4. Discussion
Twenty-five transgenic mouse lines that express fluorescent proteins in motor neurons and RGCs were reported previously (Feng et al., 2000). Some of these strains express the fluorescent marker only in a small number of neurons, allowing study of individual cell details, such as dendrites, soma and axon. Our study described semi-automated analysis of the distribution and morphology of RGCs of one of these strains that selectively express YFP under the control of the Thy-1 promoter. We found that approximately one RGC out of five hundred express YFP. The distribution of YFP-expressing RGCs varied by quadrant with the highest number in the temporal quadrant and the lowest in the nasal quadrant. Taking account of the density of RGCs by eccentricity, we found that there was a tendency for proportionately more YFP-expressing RGC in the central retina. The detailed neurite outgrowth parameters did not differ by region, except for RGC body size. RGCs located in the central retina had significantly smaller cell body area than RGCs located in the periphery.
The degree of YFP expression in RGCs in this strain varied substantially, such that one could subjectively separate fluorescent cells into a majority that were bright and a smaller proportion that were dim. We quantitatively corroborated the distinction between bright and dim cells by intensity measurements of their cell soma, and future investigations might choose to utilize a quantitative cutoff value for cell soma brightness as the separation method when RGCs expressing YFP are grouped. In the bright cells, the details of soma, dendrites and axon were similar to that in Golgi-type staining. In the dim cells, the low intensity of YFP expression showed the soma, but limited the fluorescence visibility to the proximal processes. During experimental conditions in which the normal morphology may change, the proportion of dim cells might increase due to downregulation of Thy-1 gene expression and/or actual contraction of dendrites. If Thy-1 expression is downregulated, the content of YFP in RGCs may decrease at a rate proportional to the turnover rate of YFP, which to our knowledge is unknown. If cell soma brightness decreases and dendritic visibility falls due to these factors, the resulting RGCs may be indistinguishable from the dim cell population of control eyes. Therefore, evaluation of the total YFP-expressing cell numbers as well as proportion of bright and dim cells (or the distribution by brightness using quantitative soma brightness values) will need to be used to draw conclusions about morphological alterations in experimental eyes.
Our cluster analysis separated RGCs into 6 subtypes according to their morphologic criteria and the depth of dendritic arbor stratification. Previous studies on mouse retinas classified RGCs into different subtypes based on their morphology, including the depth of dendritic arbor stratification (Kong et al., 2005; Sun et al., 2002; Badea and Nathans, 2004). Sun et al. (2002) introduced a classification of RGC morphology based on soma size, dendritic field diameter and branching in the inner plexiform layer (IPL), identifying 4 major subtypes with one unclassified group (Sun et al., 2002). Badea and Nathans (2004) classified RGCs as monostratified and bistratified with subsequent subgrouping into 12 clusters based on dendritic arbor area and normalized branching distance in the inner and outer IPL (Badea and Nathans, 2004). Kong and co-workers, using dendritic field area, mean stratification depth and density of dendritic arbor, differentiated 11 RGC subtypes, excluding bistratified and melanopsin-containing cells (Kong et al., 2005). Leung et al. (2011) found 6 morphological classes of RGCs in selective YFP-expressing mice, based on cell body size, dendritic field, total branching length, and distance from the optic disc. We found that RGCs could be grouped into 6 clusters, as well. For example, our cluster 1 of RGCs had the highest depth of dendritic stratification and one of the highest mean process length and was comparable to cluster 1 from Leung et al. (2011). Similarly, our clusters 4, 5, and 6 had similar cell body size, total outgrowth and number of branches to their clusters 5, 4 and 6, respectively (Leung et al., 2011). Two of our clusters did not have apparent correlates to those seen by Leung. One was cluster 2, with large soma, but relatively few branches and low total outgrowth. The other was cluster 3, with the highest total outgrowth and the longest processes. The selection and visibility of various RGC types may differ depending on the observation of neurons in vivo compared to in vitro. In general, the diversity of morphology in mouse RGCs seems much less than that of the cat, rabbit or monkey retina (Roska and Werblin, 2001; O’Brien et al., 2002; Rockhill et al., 2002; Dacey et al., 2003).
To date, the majority of methods that are used to reconstruct and evaluate neuronal morphology are manual (for review see Donohue and Ascoli, 2011). The process is usually time consuming and prone to inter-individual variability. The most common approach to quantitatively evaluate number of dendritic branches in modern neurobiology is applying the Sholl analysis (Sholl, 1953). The method is relatively fast, easy to use and meaningful. Recently, Gensel et al. (2010) introduced semi-automated method for Sholl analysis (Gensel et al., 2010). Our neurite outgrowth approach efficiently and in an objective manner quantified neuronal morphology by assessing cell body size, length of the dendritic arbor and number of branches. The advantage of adding Sholl analysis for studies of neurodegeneration is to provide the number of dendritic branches at various distances from the cell body. In this way, we can not only describe the overall dendritic values, but we can dissect whether changes in dendrites are greater close to or further from the cell body.
In our hands, the imaging of one cell took about four minutes, including the capture of serial images of the entire dendritic arbor into a z-stack projection and flattening into a 2-dimensional 16-bit TIFF image. The subsequent analysis to complete the neurite outgrowth and Sholl steps took less than a minute per neuron, including importing the image into software, placing the Sholl template, applying the neurite outgrowth application module and counting the number of intersections between each Sholl ring and neurite outgrowth. Importantly, the neurite outgrowth analysis is free from inter-individual variations once the parameters for the procedure are set. The software identifies the structural appearance of the neuron and quantifies its morphology in an automated manner. Thus, the method is time-efficient and free from obvious bias in quantitative estimation of neuronal morphology.
RGCs from YFP and cyan fluorescent protein selectively expressing mouse strains have been studied in vivo by confocal and scanning laser ophthalmoscopy (Walsh and Quigley, 2008; Leung et al., 2008). Sequential in vivo imaging of these YFP mice was used to study RGC morphology after optic nerve crush (Leung et al., 2011). The authors reported progressive dendritic shrinkage, followed by loss of the axon and cell body over a 6 month period. In vivo imaging offers valuable information about the prospective changes of single cell retinal morphology over time. This circumvents some weaknesses of cross-sectional analysis of RGC morphology when using histological tissues. However, in vivo imaging is somewhat limited in the assessment of detailed changes in number of RGCs across different retinal sectors. Furthermore, glaucoma models in rodents frequently lead to optical defects in the cornea that may preclude in vivo detection of early phenotypical changes in RGC morphology.
Some limitations of in vivo imaging of YFP RGCs can be successfully solved in studies on fixed, post-mortem tissue. Quantification of all YFP positive RGCs permits the study of the greatest possible number of neurons from each retina, increasing statistical power. Furthermore, fixed, retinal whole mount evaluation offers better resolution of cellular processes, the ability to analyze cells that are located in the periphery, and a more detailed 3-dimensional visualization of neuronal morphology. Furthermore, it is possible to perform double labeling for other markers in fixed tissues, but not with in vivo study. Yet, the studies of this strain in fixed tissue lack the power of study of individual RGCs over time, which avoids the need to perform cross-sectional comparisons over time. We have also documented here the need to account for downregulation of YFP expression as a factor in fixed tissue studies.
In summary, the present study introduces semi-automated methods to evaluate neuronal morphology. In the selectively YFP-expressing mouse, we describe mean RGC number, the distribution by retinal quadrants and eccentricity, and detailed RGC morphology. YFP expressing RGCs were homogeneously distributed across the retina and could be statistically grouped into 6 subtypes by cluster analysis. These methods will be utilized in a companion study of the effects of optic nerve crush and experimental glaucoma in these mice.
Supplementary Material
Highlights.
YFP expressing retinal ganglion cells were studied.
A semi-automated method was used to analyze the morphology of these cells.
Quantitative measures of morphometric parameters were obtained.
This method can benefit in studies of cell changes in eye disease models.
Acknowledgments
Supported in part by PHS Research Grants EY 02120, EY 01765, Academy of Finland, Evald and Hilda Nissi Foundation, and by unrestricted gifts from William T. Forrester, Saranne and Livingston Kosberg, and the Leonard Wagner Trust, New York.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Badea TC, Nathans J. Quantitative analysis of neuronal morphologies in the mouse retina visualized by using a genetically directed reporter. J Comp Neurol. 2004;480:331–351. doi: 10.1002/cne.20304. [DOI] [PubMed] [Google Scholar]
- Badea TC, Wang Y, Nathans J. A noninvasive genetic/pharmacologic strategy for visualizing cell morphology and clonal relationships in the mouse. J Neurosci. 2003;23:2314–2322. doi: 10.1523/JNEUROSCI.23-06-02314.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badea TC, Hua ZL, Smallwood PM, Williams J, Rotolo T, Ye X, Nathans J. New mouse lines for the analysis of neuronal morphology using CreER(T)/loxP-directed sparse labeling. PLoS One. 2009;4:e7859. doi: 10.1371/journal.pone.0007859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckingham BP, Inman DM, Lambert W, Oglesby E, Calkins DJ, Steele MR, Vetter ML, Marsh-Armstrong N, Horner PJ. Progressive ganglion cell degeneration precedes neuronal loss in a mouse model of glaucoma. J Neurosci. 2008;28:2735–2744. doi: 10.1523/JNEUROSCI.4443-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cone FE, Gelman SE, Son JL, Pease ME, Quigley HA. Differential susceptibility to experimental glaucoma among 3 mouse strains using bead and viscoelastic injection. Exp Eye Res. 2010;91:415–424. doi: 10.1016/j.exer.2010.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dacey DM, Peterson BB, Robinson FR, Gamlin PD. Fireworks in the primate retina: in vitro photodynamics reveals diverse LGN-projecting ganglion cell types. Neuron. 2003;37:15–27. doi: 10.1016/s0896-6273(02)01143-1. [DOI] [PubMed] [Google Scholar]
- Day WH, Edelsbrunner H. Efficient algorithms for agglomerative hierarchical clustering methods. J Classification. 1984;1:1–24. [Google Scholar]
- Donohue DE, Ascoli GA. Automated reconstruction of neuronal morphology: An overview. Brain Res Rev. 2011;67:94–102. doi: 10.1016/j.brainresrev.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng G, Mellor RH, Bernstein M, Keller-Peck C, Nguyen QT, Wallace M, Nerbonne JM, Lichtman JW, Sanes JR. Imaging neuronal subsets in transgenic mice expressing multiple spectral variants of GFP. Neuron. 2000;28:41–51. doi: 10.1016/s0896-6273(00)00084-2. [DOI] [PubMed] [Google Scholar]
- Gabriele ML, Ishikawa H, Schuman JS, Ling Y, Bilonick RA, Kim JS, Kagemann L, Wollstein G. Optic nerve crush mice followed longitudinally with spectral domain optical coherence tomography. Invest Ophthalmol Vis Sci. 2011;52:2250–2254. doi: 10.1167/iovs.10-6311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan WB, Grutzendler J, Wong WT, Wong RO, Lichtman JW. Multicolor “DiOlistic” labeling of the nervous system using lipophilic dye combinations. Neuron. 2000;27:219–225. doi: 10.1016/s0896-6273(00)00031-3. [DOI] [PubMed] [Google Scholar]
- Gensel JC, Schonberg DL, Alexander JK, McTigue DM, Popovich PG. Semiautomated Sholl analysis for quantifying changes in growth and differentiation of neurons and glia. J Neurosci Methods. 2010;190:71–79. doi: 10.1016/j.jneumeth.2010.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holcombe DJ, Lengefeld N, Gole GA, Barnett NL. Selective inner retinal dysfunction precedes ganglion cell loss in a mouse glaucoma model. Br J Ophthalmol. 2008;92:683–688. doi: 10.1136/bjo.2007.133223. [DOI] [PubMed] [Google Scholar]
- Jakobs TC, Libby RT, Ben Y, John SW, Masland RH. Retinal ganglion cell degeneration is topological but not cell type specific in DBA/2J mice. J Cell Biol. 2005;171:313–325. doi: 10.1083/jcb.200506099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MA, Drum BA, Quigley HA, Sanchez RM, Dunkelberger GR. Pattern-evoked potentials and optic nerve fiber loss in monocular laser-induced glaucoma. Invest Ophthalmol Vis Sci. 1989;30:897–907. [PubMed] [Google Scholar]
- Kerrigan LA, Zack DJ, Quigley HA, Smith SD, Pease ME. TUNEL-positive ganglion cells in human primary open-angle glaucoma. Arch Ophthalmol. 1997;115:1031–1035. doi: 10.1001/archopht.1997.01100160201010. [DOI] [PubMed] [Google Scholar]
- Kisiswa L, Dervan AG, Albon J, Morgan JE, Wride MA. Retinal ganglion cell death postponed: giving apoptosis a break? Ophthalmic Res. 2010;43:61–78. doi: 10.1159/000247591. [DOI] [PubMed] [Google Scholar]
- Kong JH, Fish DR, Rockhill RL, Masland RH. Diversity of ganglion cells in the mouse retina: unsupervised morphological classification and its limits. J Comp Neurol. 2005;489:293–310. doi: 10.1002/cne.20631. [DOI] [PubMed] [Google Scholar]
- Kong YX, Crowston JG, Vingrys AJ, Trounce IA, Bui VB. Functional changes in the retina during and after acute intraocular pressure elevation in mice. Invest Ophthalmol Vis Sci. 2009;50:5732–5740. doi: 10.1167/iovs.09-3814. [DOI] [PubMed] [Google Scholar]
- Leung CK, Lindsey JD, Crowston JG, Ju WK, Liu Q, Bartsch DU, Weinreb RN. In vivo imaging of murine retinal ganglion cells. J Neurosci Methods. 2008;168:475–478. doi: 10.1016/j.jneumeth.2007.10.018. [DOI] [PubMed] [Google Scholar]
- Leung CK, Weinreb RN, Li ZW, Liu S, Lindsey JD, Choi N, Liu L, Cheung CY, Ye C, Qiu K, Chen LJ, Yung WH, Crowston JG, Pu M, So KF, Pang CP, Lam DS. Long-term in vivo imaging and measurement of dendritic shrinkage of retinal ganglion cells. Invest Ophthalmol Vis Sci. 2011;52:1539–1547. doi: 10.1167/iovs.10-6012. [DOI] [PubMed] [Google Scholar]
- O’Brien BJ, Isayama T, Richardson R, Berson DM. Intrinsic physiological properties of cat retinal ganglion cells. J Physiol. 2002;538:787–802. doi: 10.1113/jphysiol.2001.013009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porciatti V, Ventura LM. Physiologic significance of steady-state pattern electroretinogram losses in glaucoma: clues from simulation of abnormalities in normal subjects. J Glaucoma. 2009;18:535–542. doi: 10.1097/IJG.0b013e318193c2e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quigley HA, Addicks EM, Green WR, Maumenee AE. Optic nerve damage in human glaucoma. II. The site of injury and susceptibility to damage. Arch Ophthalmol. 1981;99:635–649. doi: 10.1001/archopht.1981.03930010635009. [DOI] [PubMed] [Google Scholar]
- Quigley HA, Broman AT. The number of people with glaucoma worldwide in 2010 and 2020. Br J Ophthalmol. 2006;90:262–267. doi: 10.1136/bjo.2005.081224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quigley HA, Cone FE, Gelman SE, Yang Z, Son JL, Oglesby EN, Pease ME, Zack DJ. Lack of neuroprotection against experimental glaucoma in c-Jun N-terminal kinase 3 knockout mice. Exp Eye Res. 2011;92:299–305. doi: 10.1016/j.exer.2011.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quigley HA, Nickells RW, Kerrigan LA, Pease ME, Thibault DJ, Zack DJ. Retinal ganglion cell death in experimental glaucoma and after axotomy occurs by apoptosis. Invest Ophthalmol Vis Sci. 1995;36:774–786. [PubMed] [Google Scholar]
- Rockhill RL, Daly FJ, MacNeil MA, Brown SP, Masland RH. The diversity of ganglion cells in a mammalian retina. J Neurosci. 2002;22:3831–3843. doi: 10.1523/JNEUROSCI.22-09-03831.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roska B, Werblin F. Vertical interactions across ten parallel, stacked representations in the mammalian retina. Nature. 2001;410:583–587. doi: 10.1038/35069068. [DOI] [PubMed] [Google Scholar]
- Sappington RM, Carlson BJ, Crish SD, Calkins DJ. The microbead occlusion model: a paradigm for induced ocular hypertension in rats and mice. Invest Ophthalmol Vis Sci. 2010;51:207–216. doi: 10.1167/iovs.09-3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sholl DA. Dendritic organization in the neurons of the visual and motor cortices of the cat. J Anat. 1953;87:387–406. [PMC free article] [PubMed] [Google Scholar]
- Son JL, Soto I, Oglesby E, Lopez-Roca T, Pease ME, Quigley HA, Marsh-Armstrong N. Glaucomatous optic nerve injury involves early astrocyte reactivity and late oligodendrocyte loss. Glia. 2010;58:780–789. doi: 10.1002/glia.20962. [DOI] [PubMed] [Google Scholar]
- Soto I, Pease ME, Son JL, Shi X, Quigley HA, Marsh-Armstrong N. Retinal ganglion cell loss in a rat ocular hypertension model is sectorial and involves early optic nerve axon loss. Invest Ophthalmol Vis Sci. 2011;52:434–441. doi: 10.1167/iovs.10-5856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W, Li N, He S. Large-scale morphological survey of mouse retinal ganglion cells. J Comp Neurol. 2002;451:115–126. doi: 10.1002/cne.10323. [DOI] [PubMed] [Google Scholar]
- Tauchi M, Masland RH. Local order among the dendrites of an amacrine cell population. J Neurosci. 1985;5:2494–2501. doi: 10.1523/JNEUROSCI.05-09-02494.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh MK, Quigley HA. In vivo time-lapse fluorescence imaging of individual retinal ganglion cells in mice. J Neurosci Methods. 2008;169:214–221. doi: 10.1016/j.jneumeth.2007.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber AJ, Harman CD. Structure-function relations of parasol cells in the normal and glaucomatous primate retina. Invest Ophthalmol Vis Sci. 2005;46:3197–3207. doi: 10.1167/iovs.04-0834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber AJ, Kaufman PL, Hubbard WC. Morphology of single ganglion cells in the glaucomatous primate retina. Invest Ophthalmol Vis Sci. 1998;39:2304–2320. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.