Abstract
Genome-wide association studies have successfully identified hundreds of novel genetic variants associated with many complex human diseases. However, there is a lack of rigorous work on evaluating the statistical power for identifying these variants. In this paper, we consider sparse signal identification in genome-wide association studies and present two analytical frameworks for detailed analysis of the statistical power for detecting and identifying the disease-associated variants. We present an explicit sample size formula for achieving a given false non-discovery rate while controlling the false discovery rate based on an optimal procedure. Sparse genetic variant recovery is also considered and a boundary condition is established in terms of sparsity and signal strength for almost exact recovery of both disease-associated variants and nondisease-associated variants. A data-adaptive procedure is proposed to achieve this bound. The analytical results are illustrated with a genome-wide association study of neuroblastoma.
Keywords: False discovery rate, False non-discovery rate, High-dimensional data, Multiple testing, Oracle exact recovery
1. Introduction
Genome-wide association studies have emerged as an important tool for discovering regions of the genome that harbour genetic variants conferring risk for complex diseases. Such studies often involve scanning hundreds of thousands of single-nucleotide polymorphism markers in the genome. Many novel genetic variants have been identified from recent genome-wide association studies, including variants for age-related macular degenerative diseases (Klein et al., 2005), breast cancer (Hunter et al., 2007) and neuroblastoma (Maris et al., 2008). The Welcome Trust Case-Control Consortium (2007) published a study of seven diseases using 14 000 cases and 3000 shared controls and has identified several markers associated with each of these complex diseases. The success of these studies has provided solid evidence that the genome-wide association studies represent a powerful approach to the identification of genes involved in common human diseases.
Due to the genetic complexity of many common diseases, it is generally assumed that multiple genetic variants are associated with disease risk. The key question is to identify true disease-associated markers from the multitude of nondisease-associated markers. The most common approach to this is to perform a single marker score test derived from the logistic regression model and to control for multiplicity of testing using stringent criteria such as the Bonferroni correction (McCarthy et al., 2008). Similarly, the most commonly used approach for sample size analysis or power analysis for such large-scale association studies is based on detecting markers of given odds ratios using a conservative Bonferroni correction. However, this correction is often too conservative for large-scale multiple testing problems, leading to reduced power in detecting disease-associated markers. Analytical and simulation studies by Sabatti et al. (2003) have shown that the false discovery rate procedure of Benjamini & Hochberg (1995) can effectively control the false discovery rate for the dependent tests encountered in case–control association studies and increase power.
Despite the success of genome-wide association studies, questions remain as to whether current sample sizes are large enough to detect most or all of the disease markers. This is related to power and sample size analysis and is closely related to sparse signal detection and discovery. However, power analysis has not been addressed fully for simultaneous testing of hundreds of thousands of null hypotheses. Efron (2007) presented an important alternative for power analysis for large-scale multiple testing problems in the framework of local false discovery rate. Gail et al. (2008) investigated the probability of detecting the disease-associated markers in case–control genome-wide association studies in the top T largest chi-square values from the trend tests of association. The detection probability is related to false non-discovery and T is related to false discovery, although these terms are not explicitly used in that paper.
The goal of the present paper is to provide an analytical study of the power and sample size issues in genome-wide association studies. We treat the vector of the score statistics across all the markers as a sequence of Gaussian random variables. Since we expect only a small number of markers to be associated with disease, the true mean of the vector of score statistics should be very sparse, although the degree of sparsity is unknown. Our goal is to recover those sparse and relevant markers. This is deeply connected with hypothesis testing in the context of multiple comparisons and false discovery rate control. Both false discovery rate control and sparse signal recovery have been areas of intensive research in recent years. Sun & Cai (2007) showed that the large-scale multiple testing problem has an equivalent weighted classification formulation, in the sense that the optimal solution to the multiple testing problem is also the optimal decision rule for the weighted classification problem. They further proposed an optimal false discovery rate controlling procedure that minimizes the false non-discovery rate. Donoho & Jin (2004) studied the detection of sparse heterogeneous mixtures using higher criticism, focusing on testing the global null hypothesis against the alternative where only a fraction of the data comes from a normal distribution with a common nonnull mean. It is particularly important that the detectable region is identified on the amplitude or sparsity plane so that higher criticism can completely separate the two hypotheses asymptotically.
In this paper, we present analytical results on sparse signal recovery in case–control genome-wide association studies. We investigate two frameworks for detailed analysis of the statistical power for detecting and identifying the disease-associated markers. In a similar setting as in Gail et al. (2008), we first present an explicit sample-size formula for achieving a given false non-discovery rate while controlling the false discovery rate based on the optimal false discovery rate controlling procedure of Sun & Cai (2007). This provides results on how odds ratios and marker allele frequencies affect the power of identifying the disease-associated markers. We also consider sparse marker recovery, establishing the theoretical boundary for almost exact recovery of both the disease-associated markers and nondisease-associated markers. Our results further extend the amplitude or sparsity boundary of Donoho & Jin (2004) for almost exact recovery of the signals. Finally, we construct a data adaptive procedure to achieve this bound.
2. Problem setup and score statistics
We consider a case-control genome-wide association study with m markers genotyped, where the minor allele frequency for marker i is pi. Let Gi = 0, 1, 2 be the number of minor alleles at marker i (i = 1, …, m), where Gi ∼ Bin(2, pi). Assume that the total sample size is n and n1 = rn and n2 = (1 − r)n are the sample sizes for cases and controls. Let Y = 1 for diseased and Y = 0 for nondiseased individuals. Suppose that in the source population, the probability of disease is given by
where only a very small fraction of the bis are nonzero. Gail et al. (2008) showed that for rare diseases or for more common diseases over a confined age range such as 10 years, for case-control population, if the markers are independent of each other, it follows that
| (1) |
approximately. This implies that a logistic regression model can be fitted for each single marker i separately.
Based on the results of Prentice & Pyke (1979), the maximum likelihood estimation for a cohort study applied to case-control data with model (1) yields a fully efficient estimate of bi and a consistent variance estimate. For a given case-control study, let j be the index for individuals in the samples, Yj be the disease status of the j th individual and Gij be the genotype score for the jth individual at the ith marker. The profile score statistic to test can be written as
where . In the following, we let pri0, Ei0, vari0 denote the probability, expectation and variance calculated under the null hypothesis , and pri1, Ei1, vari1 those calculated under the alternative hypothesis . Under , pri0(Yj = y | Gij = g) = K I (y = 1) + (1 − K) I (y = 0), where I (·) is the indicator function. We have Ei0{U(bi) | Y} = 0, vari0{U(bi) | Y} = 2pi (1 − pi)r(1 − r)n.
Under the alternative, if bi is known,
where ai can be determined by , with D being the disease prevalence and . Then
and
where , , and
Let Xi = U(bi)/vari0{U(bi)|Y}1/2 be the score statistic for testing the association between the marker i and the disease. If bi is known, then under , Xi ∼ N(0, 1) and under , Xi ∼ N(μn,i, σi) asymptotically, where
| (2) |
| (3) |
Given bi, pi, r and D, μn,i and σi can be determined. The alternative mean μn,i increases with the sample size n at the order √n. Intuitively, as the sample size increases, it is easier to distinguish the signals and zeros. So, the problem of power analysis and sample size analysis can be formulated as the sparse normal mean problem: we have score statistics Xi, (i = 1, …, m) and only a very small proportion ∊m of them have nonzero means, and the goal is to identify those markers whose score statistics have nonzero means.
3. Marker recovery
3.1. Effect size and false discovery rate control
As in Genovese & Wasserman (2002) and Sun & Cai (2007), we use the marginal false discovery rate, defined as mfdr = E(N01)/E(R), and marginal false non-discovery rate, defined as mfnr = E(N10)/E(S), as our criteria for multiple testing, where R is the number of rejections, N01 is the number of nulls among these rejections, S is the number of non-rejections and N10 is the number of nonnulls among these nonrejections. Genovese & Wasserman (2002) and Sun & Cai (2007) showed that under weak conditions, the marginal false discovery rate (mfdr) and the false discovery rate (fdr), the marginal false non-discovery rate (mfnr) and the false non-discovery rate (fnr) are asymptotically the same in the sense that mfdr = fdr + O(m−1/2) and mfnr = fnr + O(m−1/2). In the following, we use mfdr and mfnr for our analytical analysis, but we use the notation, fdr and fnr.
Consider the sequence of score statistics Xi (i = 1, …, m), as defined in the previous section, where under , Xi ∼ N(0, 1) and under , Xi ∼ N(μn,i, ), μn,i ≠ 0, σi ⩾ 1. Usually, μn,i and σi are different across different disease-associated markers due to different minor allele frequencies and different effect sizes as measured by the odds ratios; see (2) and (3). However, since most of the markers in genome-wide association studies are not rare and the observed odds ratios range from 1.2 to 1.5, we can reasonably assume that they are on a comparable scale. In addition, for the purpose of sample size calculation, one should always consider the worst case scenarios for all the markers, which leads to a conservative estimate of the required sample sizes. To simplify the notation and analysis and to obtain closed-form analytical results, we thus assume that all the markers considered have the same minor allele frequency and all the relevant markers have the same effect size. Specifically, we assume that under , Xi ∼ N(μ, σ2), σ ⩾ 1. Suppose that the proportion of nonnull effects is ∊m. Defining a sequence of binary latent variables, θ = (θ1, … ,θm), the model under consideration can be stated as follows:
| (4) |
Assume that σ is known. Our goal is to find the minimum μ that allows us to identify θ with the false discovery rate asymptotically controlled at α1 and the false non-discovery rate asymptotically controlled at α2. We particularly consider the optimal false discovery rate controlling procedure of Sun & Cai (2007), which simultaneously controls the false discovery rate at α1 and minimizes the false non-discovery rate asymptotically. Different from the p-value-based false discovery rate procedures, this approach considers the distribution of the test statistics and involves the following.
Algorithm 1. Calculating the optimal false discovery rate.
Step 1. Given the observation X = (X1, …, Xm), estimate the nonnull proportion ∊̂m using the method in Cai & Jin (2010). This estimator is based on Fourier transformation and the empirical characteristic function, which can be written as , where η ∈ (0, 1/2) is a tuning parameter, which needs to be small for the sparse case considered in this paper. Based on our simulations, η = 10−4 works well.
- Step 2. Use a kernel estimate f̂ to estimate the mixture density f of the Xi,
where K is a kernel function and ρ determines the bandwidth. We use ρ = 1.34 m−1/5 as recommended by Silverman (1986).(5) Step 3. Compute T̂i = (1 − ∊̂m) φ (Xi)/f̂(Xi), where φ(.) is the standard normal density function.
- Step 4. Let
then reject all H(i) (i = 1, …, k). This adaptive step-up procedure considers the average posterior probability of being null and is adaptive to both the global feature ∊m and local feature of φ(Xi)/f(Xi).
The procedure has a close connection with a weighted classification problem. Consider the loss function
| (6) |
The Bayes rule under the loss function (6) is
| (7) |
where f0(x) = φ (x; 0, 1) and f1(x) = φ (x; μ, σ2). Sun & Cai (2007) show that the threshold λ is a decreasing function of false discovery rate, and for a given level α1, one can determine a unique thresholding λ to ensure the false discovery rate level to be controlled under α1. The following theorem gives the minimal signal μ required in order to obtain the pre-specified false discovery rate and false non-discovery rate using this optimal procedure.
Theorem 1. Consider the model in (4) and the optimal oracle procedure of Sun & Cai (2007). In order to control the false discovery rate under α1 and the false non-discovery rate under α2, the minimum μ has to be no less than (σ2 − 1)d̂, where (ĉ, d̂) are the roots of the equations
| (8) |
where Φ is the cumulative density function of the standard normal distribution, and
As discussed in § 2, in genome-wide association studies, μ = √nμ1, where μ1 as defined in (2) can be determined by the effect of the marker, log odds ratio b, the minor allele frequency p and the case ratio or control ratio r. The following corollary provides the minimum sample size needed in case-control genome-wide association studies in order to control false discovery rate and false non-discovery rate.
Corollary 1. Consider a case-control genome-wide association study, with m markers, a total sample size of n, with n1 = nr cases and n2 = (1 − r)n controls. Assume that all the markers have the same minor allele frequency of pi = p and all the relevant markers have the same log odds ratio bi = b. In order to control false discovery rate and false non-discovery rate under α1 and α2 asymptotically, the minimum total sample size must be at least n = {d̂(σ2 − 1)/μ1}2, where d̂ is the root of (8), and μ1 and σ1 are defined in (2) and (3).
Corollary 1 gives the minimum sample size required in order to identify the markers with minor allele frequency p and effective odds ratio of b with false discovery rate less than α1 and false non-discovery rate less than α2 using the optimal false discovery rate controlling procedure of Sun & Cai (2007). Since this procedure asymptotically controls false discovery rate and minimizes false non-discovery rate, the sample size obtained here is a lower bound. This implies that asymptotically, one cannot control both false discovery rate and false non-discovery rate using a smaller sample size with any other procedure. In practice, α2 should be set to less than ∊m, which can be achieved by not rejecting any nulls.
3.2. Illustration
We present the minimum sample sizes needed for different levels of false discovery rate and false non-discovery rate in Fig. 1 for ∊m = 2 × 10−4, which corresponds to assuming 100 disease-associated markers. We assume a disease prevalence of D = 0.10. As expected, the sample size needed increases as the false discovery rate level or the target false non-discovery rate level decreases. It also decreases with higher signal strength. The false non-discovery rate levels presented in the plot are very small since it cannot exceed the proportion of the relevant markers ∊m. When m is extremely large, ∊m is usually very small. In Fig. 1, ∊m = 2 × 10−4 and false non-discovery rate we choose should be less than ∊m, since fnr = ∊m can be achieved by not rejecting any of the markers. Among the 100 relevant markers, fnr = 10−4, 2 × 10−5 and 10−6 corresponds to an expected number of nondiscovered relevant markers of fewer than 50, 10 and 0.5, respectively. As a comparison, with the same sample size for fdr = 0.05 and fnr = 10−4, 2 × 10−5 and 10−6, the power of the one-sided score test using the Bonferonni correction to control the genome-wide error rate at 0.05 is 0.22, 0.64 and 0.95, respectively. We observe that the sample sizes needed strongly depend on the false non-discovery rate and the effect size of the markers, but are less dependent on false discovery rate. Similar trends were observed for ∊m = 2 × 10−5, which corresponds to assuming 10 disease-associated markers.
Fig. 1.
Minimum sample size needed for a given false discovery rate and false non-discovery rate for a genome-wide association study with m = 500 000 markers and the relevant markers proportion ∊m = 2 × 10−4. Assume that the case proportion r = 0.40 and the minor allele frequency p = 0.40. (a) fdr = 0.05 and different values of odds ratio; (b) fdr = 0.20 and different values of odds ratio; (c) odds ratio of 1.20 and different values of false discovery rate; (d) odds ratio of 1.50 and different values of false discovery rate. For each plot, the solid, dash and dot lines correspond to fnr = 10−4, 2 × 10−5 and 10−6, respectively.
We performed simulation studies to evaluate the sample size formula presented in Corollary 1. We assume that m = 500 000 markers are tested with 100 being associated with disease, which corresponds to ∊m = 2 × 10−4. We first considered the setting that all the 100 relevant markers have the same odds ratio on disease risk with a minor allele frequency of 0.40. For a pre-specified fdr = 0.05 and fnr = 10−4, we determined the sample size based on Corollary 1. We then applied the false discovery rate controlling procedure of Sun & Cai (2007) to the simulated data and examined the values of the observed false discovery rates and false non-discovery rates. Table 1 shows the empirical false discovery rates and false non-discovery rates over 100 simulations for different values of the odds ratios. The empirical false discovery rates and false non-discovery rates are very close to the pre-specified values, indicating that the sample size formula can indeed result in the specified false discovery rate and false non-discovery rate.
Table 1.
Empirical false discovery rate and false non-discovery rate (10−4 unit) and their standard errors for the optimal false discovery rate procedure based on 100 simulations where the sample sizes are determined by Corollary 1 for the independent sequences with the same alternative odds ratio for fdr = 0.05 and fnr = 10−4.
| Independent sequence with the same alternative odds ratios | Short-ranged dependent sequence with different alternative odds ratios | |||
|---|---|---|---|---|
| Odds ratio | Empirical fdr | Empirical fnr | Empirical fdr | Empirical fnr |
| 1.20 | 0.050 (0.028) | 1.04 (0.10) | 0.051 (0.030) | 1.04 (0.10) |
| 1.23 | 0.048 (0.028) | 1.03 (0.12) | 0.051 (0.032) | 1.04 (0.11) |
| 1.26 | 0.045 (0.030) | 1.04 (0.11) | 0.055 (0.033) | 1.04 (0.11) |
| 1.29 | 0.046 (0.030) | 1.15 (0.13) | 0.053 (0.031) | 1.03 (0.11) |
| 1.33 | 0.053 (0.033) | 1.04 (0.11) | 0.054 (0.032) | 1.05 (0.10) |
| 1.36 | 0.047 (0.029) | 1.01 (0.13) | 0.049 (0.029) | 1.00 (0.12) |
| 1.39 | 0.051 (0.030) | 1.02 (0.11) | 0.051 (0.033) | 1.01 (0.11) |
| 1.43 | 0.046 (0.033) | 1.05 (0.12) | 0.044 (0.029) | 1.04 (0.11) |
| 1.46 | 0.046 (0.027) | 1.04 (0.10) | 0.046 (0.025) | 1.02 (0.11) |
| 1.50 | 0.051 (0.027) | 1.03 (0.13) | 0.053 (0.033) | 1.03 (0.12) |
fdr, false discovery rate; fnr, false non-discovery rate.
Since our power and sample size calculation is derived under the assumption of common effect sizes for all associated markers and independent test statistics, we also evaluated our results under the assumption of short-range dependency and unequal effect sizes. To approximately mimic the dependency structure among markers due to linkage disequilibrium, we assumed a block-diagonal covariance structure for the score statistics with exchangeable correlation of 0.20 for block sizes of 50. We simulated marker effects with a mean odds ratio ranging from 1.20 to 1.50. For a given mean odds ratio, we obtained the sample size and simulated the corresponding marker effects by generating μn,i from N (4.40, 1.00). We then applied the false discovery rate controlling procedure and calculated the empirical false discovery rates and false non-discovery rates for different mean odds ratios. The results are presented in Table 1, indicating that the false discovery rate controlling procedure can indeed control false discovery rates and obtain the pre-specified false non-discovery rates.
4. A procedure for sparse marker recovery
4.1. Oracle sparse marker discovery
Besides controlling false discovery rate and false non-discovery rate, an important alternative in practice is to control the numbers of false discoveries and false non-discoveries. One limitation with the use of false discovery rate and false non-discovery rate in power calculations is that they do not provide a very clear idea about roughly how many markers are falsely identified and how many are missed until the analysis is conducted. In genome-wide association studies, the goal is to make the number of misidentified markers, including both the false discoveries and false non-discoveries, converge to zero when the number of the markers m and the sample sizes are sufficiently large. In this section, we investigate the conditions on the parameters μ = μm and ∊m in (4) that can lead to almost exact recovery of the null and nonnull markers. This in turn provides a useful formula for the minimum sample size required for an almost exact recovery of disease-associated markers.
Consider (4) and a decision rule δi (i = 1, …, m). We define false discoveries as the expected number of null scores that are misclassified to the alternative, and false non-discoveries as the expected number of nonnull scores that are misclassified to null,
Define the loss function as
which is simply the Hamming distance between the true θ and its estimate δ. Note that E{L(θ, δ)} = fd + fn.
Under (4), the Bayes oracle rule (7) with λ = 1, defined as
| (9) |
minimizes the risk E{L(θ, δ)}. We consider the condition on μm and ∊m such that this risk converges to zero as m goes to infinity, where the nonnull markers are assumed to be sparse. We calibrate ∊m and μm as follows. Define
| (10) |
which measures the sparsity of the disease-associated markers, and
| (11) |
which measures the strength of the disease-associated markers. Here we assume that the disease markers are sparse. We first study the relationship between τ and β so that both false discoveries and false non-discoveries converge to zero as m goes to infinity.
Theorem 2. Consider (4) and reparameterize ∊m and μ = μm in terms of β and τ as in (10) and (11). For any γ ⩾ 0, under the condition
| (12) |
the Bayes rule (9) guarantees that both false discoveries and false non-discoveries converge to zero with convergence rate 1/{mγ (log m)1/2}.
Theorem 2 shows the relationship between the strength of the signal and the convergence rate of the expected false discoveries and false non-discoveries. As expected, a stronger nonzero signal leads to faster convergence of the expected number of false identifications to zero and easier separation of the alternative from the null.
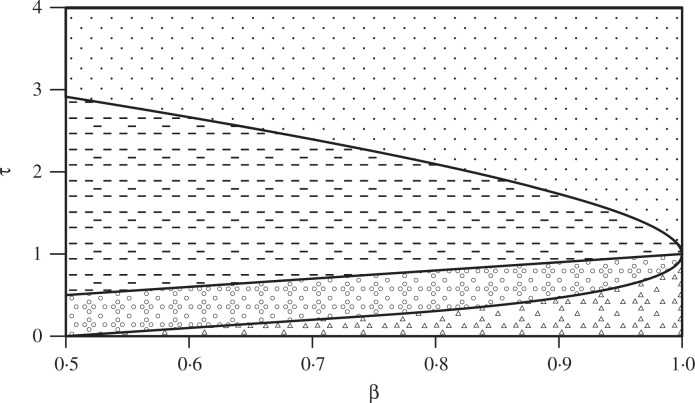
In (12), when γ = 0, the convergence rate of false discoveries and false non-discoveries is the slowest, 1/(log m)1/2. Letting σ → 1 yields the phase diagram in Fig. 2, which is an extension to the phase diagram in Donoho & Jin (2004). Three lines separate the τ − β plane into four regions. The detection boundary
separates the detectable region and the undetectable region, when μ exceeds the detection boundary, the null hypothesis H0 : Xi ∼ N(0, 1), (i = 1, …, m) and the alternative : Xi ∼ (1 − ∊m)N(0, 1) + ∊m N (μ, 1), (i = 1, …, m) separate asymptotically. Above the estimation boundary τ = β, it is possible not only to detect the presence of nonzero means, but also to estimate these means. These two regions were identified by Donoho & Jin (2004). Based on Theorem 2, we can obtain the almost exact recovery boundary
which provides the region that we can recover the whole sequence of θ with a very high probability converging to one when m → ∞, since
where ≍ represents asymptotic equivalence. In other words, in this region, we can almost fully classify the θi into the nulls and the non-nulls.
Fig. 2.
Four regions of the τ − β plane, where ∊m = m−β represents the percentage of the true signals and μm = (2τ log m)1/2 measures the strength of the signals: almost exact recovery (dots); estimable (dashes); detectable (circles); undetectable (triangles).
In large-scale genetic association studies, μ = √nμ1. Based on Theorem 2, we have the following corollary.
Corollary 2. Consider the same model as in Corollary 1. If the sample size
then both false discoveries and false non-discoveries of the oracle rule converge to zero at rate 1/{mγ (log m)1/2}.
4.2. An adaptive sparse recovery procedure
Theorem 2 shows that the convergence rate 1/{mγ (log m)1/2} can be achieved using the oracle rule (9). However, this rule involves unknown parameters, ∊m, f1(x) and f0(x), which need to be estimated. Estimation errors can therefore affect the convergence rate. We instead propose an adaptive estimation procedure for model (4) that can achieve the same convergence rate for false discoveries and false non-discoveries under a slightly stronger condition than that in Theorem 2.
The Bayes oracle rule given in (9) can be rewritten as
| (13) |
where f (x) = (1 − ∊m) f0(x) + ∊m f1(x) is the mixture density of the Xi. The density f is typically unknown, but is easily estimable. Let f̂(x) be a kernel density estimate based on the observations X1, X2, …, Xm, defined by (5). For i = 1, …, m, if |Xi| > {2(1 + γ) log m}1/2, then reject the null ; otherwise, calculate Ŝ(Xi) = f̂(Xi)/φ(Xi) and reject the null if and only if Ŝ(Xi) > 2. The threshold value 2 is based on (13) since the proportion ∊m is vanishingly small in our setting.
The next theorem shows that this adaptive procedure achieves the same convergence rate as the optimal Bayesian rule under a slightly stronger condition.
Theorem 3. Consider (4) and reparameterize ∊m = n−β and μ = μm = (2τ log m)1/2 as in (10) and (11). Suppose that the Gaussian kernel density estimate f̂, defined in (5), with ρ = 1.34 × m−1/5 is used in the adaptive procedure. For any γ ⩾ 0, if
then the adaptive procedure guarantees that both false discoveries and false non-discoveries converge to zero with convergence rate 1/{mγ (log m)1/2}.
4.3. Illustration of marker recovery
The sample sizes needed to obtain an almost exact recovery of the true disease-associated markers are presented in Fig. 3 for ∊m = 2 × 10−4 and ∊m = 2 × 10−5, which correspond to 100 and 10 disease-associated markers. We assume a disease prevalence of D = 0.10. We observe that a larger γ implies a faster rate of convergence, and therefore also larger sample size required to the achieve the rate. Generally speaking, the sample sizes in Fig. 3 are much larger than those in Fig. 1, simply because almost exact recovery of the disease-associated markers is a much stronger goal than controlling the false discovery rate and false non-discovery rate at given levels. In addition, for markers with the same effect size, it is easier to achieve full recovery of the relevant markers for the sparser cases, see of Fig. 3(b).
Fig. 3.

Minimum sample size required for almost exact recovery of the disease-associated markers for a case-control genome-wide association study with the number of markers m = 500 000, case proportion r = 0.40, minor allele frequency p = 0.40 for all the markers. (a) The relevant markers proportion is ∊m = 2 × 10−4; (b) The relevant markers proportion is ∊m = 2 × 10−5. For each plot, the solid, dash and dot lines correspond to γ values of 0.00, 0.20 and 0.40.
We performed simulation studies to investigate how sample size affects the convergence rate of false discoveries and false non-discoveries. We considered the same setting as above, where the number of markers m = 500 000, case proportion r = 0.40, minor allele frequency p = 0.40 for all the markers and the relevant markers proportion is ∊m = 2 × 10−4. For a given odds ratio and a given γ parameter, we obtained the minimum sample size n for almost exact recovery and evaluated the convergence rate of the oracle and the adaptive procedures under different sample sizes. Table 2 shows the average number of fd+fn over 100 simulations. We observe a clear decrease of fd+fn as we increase the sample sizes for both the oracle and the adaptive procedures. For the adaptive procedure, when the sample size is larger than the minimum sample size, the decrease in fd+fn is small since the minimum sample size has already achieved almost exact recovery.
Table 2.
The performances of the oracle and adaptive false discovery and false non-discovery controlling procedures for varying sample sizes, where n is the theoretical sample size. For a given γ, the average number of fd+fn and its standard error over 100 simulations are shown for each sample size and odds ratio combination.
| γ = 0 | γ = 0.2 | ||||||
|---|---|---|---|---|---|---|---|
| or | 0.8n | n | 1.2n | 0.8n | n | 1.2n | |
| o | 0.77 (0.89) | 0.14 (0.35) | 0.02 (0.14) | 0.08 (0.27) | 0.00 (0.00) | 0.00 (0.00) | |
| 1.20 | a | 3.08 (3.11) | 3.10 (3.23) | 2.51 (3.04) | 2.57 (3.02) | 3.12 (3.24) | 2.59 (3.12) |
| o | 0.57 (0.79) | 0.13 (0.37) | 0.01 (0.10) | 0.03 (0.17) | 0.00 (0.00) | 0.00 (0.00) | |
| 1.30 | a | 3.93 (4.22) | 2.61 (2.97) | 2.92 (3.36) | 3.41 (3.99) | 2.66 (3.00) | 2.92 (3.34) |
| o | 0.50 (0.72) | 0.1 (0.30) | 0.04 (0.20) | 0.02 (0.14) | 0.00 (0.00) | 0.01 (0.10) | |
| 1.40 | a | 3.31 (3.30) | 2.57 (3.11) | 2.89 (3.45) | 2.97 (3.39) | 2.55 (3.11) | 2.84 (3.42) |
or, odds ratio; o, oracle; a, adaptive.
5. Genome-wide association study of neuroblastoma
Neuroblastoma is a paediatric cancer of the developing sympathetic nervous system and is the most common form of solid tumour outside the central nervous system. It is a complex disease, with rare familial forms occurring due to mutations in paired-like homeobox 2b or anaplastic lymphoma kinase genes (Mosse et al., 2008), and several common variations being enriched in sporadic neuroblastoma cases (Maris et al., 2008). The latter genetic associations were discovered in a genome-wide association study of sporadic cases, compared with children without cancer, conducted at The Children’s Hospital of Philadelphia. After initial quality controls on samples and marker genotypes, our discoveries dataset contained 1627 neuroblastoma case subjects of European ancestry, each of which contained 479 804 markers. To correct the potential effects of population structure, 2575 matching control subjects of European ancestry were selected based on their low identity-by-state estimates with case subjects. Analysis of this dataset has led to identification of several markers associated with neuroblastoma, including three markers on 6p22 containing the predicted genes FLJ22536 and FLJ44180 with allelic odds ratio of about 1.4 (Maris et al., 2008), and markers in BARD1 genes on chromosome 2 (Capasso et al., 2009) with odds ratio of about 1.68 in high-risk cases. The question that remains to be answered is whether the current sample size is large enough to identify all the neuroblastoma-associated markers.
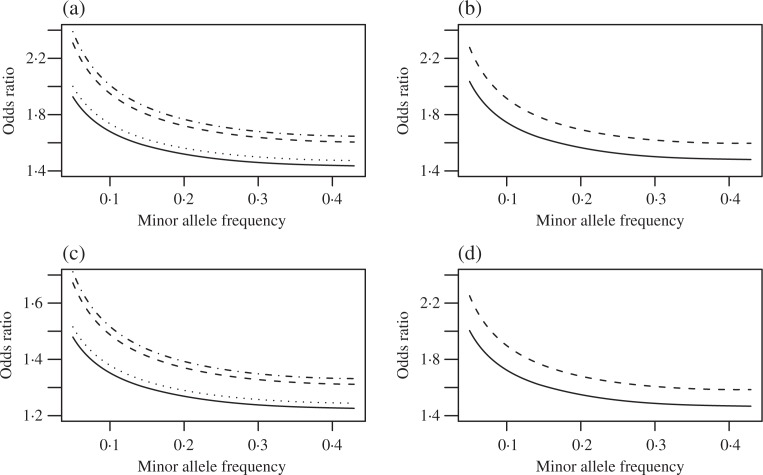
We estimated ∊m = 2 × 10−4 using the procedure of Cai and Jin (2010) with tuning parameter η = 10−4. A small value of η was chosen to reflect that we expect a very small number of disease-associated markers. With this estimate, we assume that there are 96 markers associated with neuroblastoma. For the given sample size and number of markers to be tested, Fig. 4(a) shows the minimum detectable odds ratios for markers with different minor allele frequencies for various false discovery rate and false non-discovery rate levels. The false non-discovery rates of 10−4, 5 × 10−5 and 10−5 correspond to an expected number of nondiscovered markers of fewer than 48, 24 and 4.8, respectively. This plot indicates that with the current sample size, it is possible to recover about half of the disease-associated markers with an odds ratio around 1.4 and a minor allele frequency of around 20%.
Fig. 4.
Power analysis for neuroblastoma genome-wide association study. Plots (a) and (b) assume that ∊ = 2 × 10−4, which corresponds to 98 relevant markers and plots (c) and (d) assume that ∊ = 10−4, which corresponds to 49 relevant markers. (a) and (c): detectable odds ratios for markers with different minor allele frequencies. For plot (a), the solid, dot, dash and dot-dash lines correspond to (fdr = 5%, fnr = 10−4), (fdr = 1%, fnr = 10−4), (fdr = 5%, fnr = 10−5), and (fdr = 1% and fnr = 10−5), respectively. For plot (c), the solid, dot, dash and dot-dash lines correspond to (fdr = 5%, fnr = 5 × 10−5), (fdr = 1%, fnr = 5 × 10−5), (fdr = 5%, fnr = 5 × 10−6), and (fdr = 1% and fnr = 5 × 10−6), respectively. (b) and (d): detectable odds ratios for markers with different minor allele frequencies for almost exact recovery with different convergence rates determined by γ = 0.10 (solid) and 0.40 (dash).
Figure 4(b) shows the minimum detectable odds ratios for markers with different minor allele frequencies for almost exact recovery at different rates of convergence as specified by the parameter γ, indicating that the current sample size is not large enough to obtain almost exact recovery of all the disease-associated markers with odds ratio around 1.4.
Figure 4(c) and (d) shows similar plots if we assume that ∊m = 10−4, which implies that there are 48 associated markers. This plot indicates that with the current sample size, it is possible to recover most of the disease-associated markers with odds ratio around 1.4 and minor allele frequency around 20% for false discovery rates of 1 or 5%. However, the sample size is still not large enough to obtain almost exact recovery of all disease-associated markers with odds ratio around 1.4.
6. Discussion
Our work complements and differs from that of Zaykin & Zhivotovsky (2005) and Gail et al. (2008), who based their analyses on the individual marker P-values, aiming to evaluate how likely the markers selected based on the P-value ranking are disease-associated. Although both papers considered the issues of false positives and false negatives, neither links these to false discovery and false non-discovery rates in a formal way. Our analysis is mainly based on the distribution of the score statistics derived from the logistic regressions for case-control data and is set in the framework of multiple testing and sparse signal recovery. In the setting of thousands of hypotheses, it is natural to consider the power and sample size in terms of false discovery and false non-discovery rates, which correspond to Type I and Type II errors in single hypothesis testing. In addition, we have derived an explicit sample size formula under the assumption of fixed allele frequency and fixed common marker effect.
In presenting the results for genome-wide association studies, we made several simplifications. First, we assumed that the score statistics were independent across all the m markers. This obviously does not hold due to linkage disequilibrium among the markers. However, we expect such dependency to be short-range and our recent unpublished results show that the optimal false discovery rate controlling procedure of Sun & Cai (2007) is still valid and remains optimal. The results and the sample size formulae remain valid for such short-range dependent score statistics. Zaykin & Zhivotovsky (2005) and Gail et al. (2008) showed that correlations of P-values within linkage disequilibrium blocks of markers or among such blocks have little effect on selection or detection probabilities because such correlations do not extend beyond a small portion of the genome. It is likely, therefore, that our results were also little affected by such correlations. This was further verified in our simulation studies shown in Table 1. Second, to simplify the presentation and the derivation and to obtain a clean sample size formula as we presented in Corollaries 1 and 2, we assumed that all the markers had the same minor allele frequency and the effect sizes of the relevant markers were the same. One can only employ simulation for power analysis if the minor allele frequencies and the marker effects are all different. In fact, the analytical results that were used to validate their simulations in Gail et al. (2008) were also derived under the assumption of fixed allele frequency and fixed common effect. As in most of the power calculations, in practice, one should consider different scenarios in terms of minor allele frequencies and odds ratios.
Acknowledgments
This research was supported by grants from the National Institutes of Health and the National Science Foundation. We would like to thank the reviewers and associate editor for helpful comments.
Appendix
We present the proofs of Theorem 1, Theorem 2 and Theorem 3. We use the symbol ≍ to represent asymptotic equivalence. If a ≍ b, then a = O(b) and b = O(a).
Proof of Theorem 1. When δi = 1, we have
which is equivalent to (Xi − μ)/σ ∈ (−∞, −c − dσ) ∪ (c − dσ, +∞), where
| (A1) |
Since
we have
| (A2) |
where c1 = α1∊m /{(1 − α1)(1 − ∊m)}. Similarly, since
we have
| (A3) |
where c2 = (1 − α2)∊m /{α2(1 − ∊m)}. Setting (A2) and (A3) as equalities and simplifying, we obtain (8). Let (ĉ, d̂) be the solution of this equation, then (A1) leads to μ̂ = d̂(σ2 − 1).
Proof of Theorem 2. Define c and d as in (A1). Since
in order for false discoveries to converge to zero, −cσ + d should be negative and small enough. In addition, since
as m → ∞, −cσ + d < 0. Because Φ(−cσ − d) ⩽ Φ(−cσ + d), we have
where
In order for false discoveries to converge to zero with the rate 1/{mγ (log m)1/2}, we require 1 − C1 ⩽ − γ, which leads to τ ⩾ {(1 + γ)1/2 + σ (1 + γ − β)1/2}2. Similarly,
and
As m → ∞, in order for c − dσ < 0, we require {β(σ2 − 1) + τ}1/2 < σ/τ1/2, which implies τ > β. Then,
where
It is easy to show that
is equivalent to τ ⩾ {(1 + γ)1/2 + σ (1 + γ − β)1/2}2.
Proof of Theorem 3. Define S(x) = f (x)/φ(Xi), S̃(x) = S(x)/(1 − ∊m). We have
say. We control (A), (B) and (C), respectively. First, we have
Using the same technique as the proof of Theorem 2, if τ ⩾ {(1 + γ)1/2 + σ (1 + γ − β)1/2}2, then
Consider the set 𝒳i = {Xi : |Xi| ⩽ {2(1 + γ) log m}1/2}, for all Xi ∈ 𝒳i,
and
Since the alternative density can be modelled as a Gaussian location-scale mixture, we have
where ∊m,l is the mixture proportion, representing the probability that Xj is from N (μl, σl)). Under the condition
and σl > 1 for all l = 1, …, L, we have f (Xi)/φ(Xi) < mβ′, for all Xi. Then,
Therefore, for m sufficiently large,
Let h = m−ρ, we have
Further, we take K (y) = φ(y) or any other symmetric kernel with a smaller tail than φ when |y| > {2(1 + γ) log m}1/2. Using the fact that ∫ K (y) dy = 1 and ∫ yK (y) dy = 0,
Let Hl(x) be the lth Hermite polynomial. For Xi ∈ 𝒳i,
Therefore, for m sufficiently large,
Let , l ⩾ k. For example, . Given Xi ∈ 𝒳i, define
then
For k ≠ i,
By McDiarmid’s inequality (McDiarmid, 1989),
Therefore,
Thus,
Similarly,
Using the same technique as the proof of Theorem 2, if
then (D) ⩾ Cm−1+β−γ /(log m)1/2. Finally,
Thus,
References
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Statist. Soc. B. 1995;57:289–300. [Google Scholar]
- Cai T, Jin J. Optimal rates of convergence for estimating the null and proportion of non-null effects in large-scale multiple testing. The Ann Statist. 2010;38:100–45. [Google Scholar]
- Capasso M, Hou C, Asgharzadeh S, et al. Common variations in the bard1 tumor suppressor gene influence susceptibility to high-risk neuroblastoma. Nature Genet. 2009;41:718–23. doi: 10.1038/ng.374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donoho D, Jin J. Higher criticism for detecting sparse heterogeneous mixtures. Ann Statist. 2004;32:962–94. [Google Scholar]
- Efron B. Size, power and false discovery rates. Ann Statist. 2007;35:1351–77. [Google Scholar]
- Gail M, Pfeiffer R, Wheeler W, Pee D. Probability of detecting disease-associated single nucleotide polymorphisms in case-control genome-wide association studies. Biostatistics. 2008;9:201–15. doi: 10.1093/biostatistics/kxm032. [DOI] [PubMed] [Google Scholar]
- Genovese C, Wasserman L. Operating characteristic and extensions of the false discovery rate procedure. J. R. Statist. Soc. B. 2002;64:499–517. [Google Scholar]
- Hunter D, Kraft P, Jacobs K, et al. A genome-wide association study identifies alleles in fgfr2 associated with risk of sporadic postmenopausal breast cancer. Nature Genet. 2007;39:870–4. doi: 10.1038/ng2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein R, Zeis C, Chew E, Tsai J, Sackler R, Haynes C, Henning A, SanGiovanni J, Mane S, Mayne S, Bracken M, Ferris F, Ott J, Barnstable C, Hoh J. Complement factor h polymorphism in age-related macular degeneration. Science. 2005;3085:385–9. doi: 10.1126/science.1109557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maris J, Yael P, Bradfield J, Hou C, Monni S, Scott R, Asgharzadeh S, Attiveh E, Diskin S, Laudenslager M, et al. A genome-wide association study identifies a susceptibility locus to clinically aggressive neuroblastoma at 6p22. New Engl J Med. 2008;358:2585–93. doi: 10.1056/NEJMoa0708698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy M, Abecasis G, Cardon L, Goldstein D, Little J, Ioannidis J, Hirschhorn J. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nature Rev Genet. 2008;9:356–69. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- McDiarmid C. On the method of bounded differences. Surveys in Combinatorics. 1989;141:148–88. [Google Scholar]
- Mosse Y, Laudenslager M, Longo L, et al. Identification of alk as a major familial neuroblastoma predisposition gene. Nature. 2008;455:930–35. doi: 10.1038/nature07261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prentice R, Pyke R. Logistic disease incidence models and case-control studies. Biometrika. 1979;66:403–11. [Google Scholar]
- Sabatti C, Service S, Freimer N. False discovery rates in linkage and association linkage genome screens for complex disorders. Genetics. 2003;164:829–33. doi: 10.1093/genetics/164.2.829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman BW. Density Estimation for Statistics and Data Analysis. London: Chapman and Hall; 1986. [Google Scholar]
- Sun W, Cai T. Oracle and adaptive compound decision rules for false discovery rate control. J Am Statist Assoc. 2007;102:901–12. [Google Scholar]
- Welcome Trust Case-Control Consortium Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–78. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaykin D, Zhivotovsky L. Ranks of genuine associations in whole-genome scans. Genetics. 2005;171:813–23. doi: 10.1534/genetics.105.044206. [DOI] [PMC free article] [PubMed] [Google Scholar]