Abstract
Optogenetics combines optical and genetic methods to rapidly and reversibly control neural activities or other cellular functions. Using genetic methods, specific cells or anatomical pathways can be sensitized to light through exogenous expression of microbial light activated opsin proteins. Using optical methods, opsin expressing cells can be rapidly and reversibly controlled by pulses of light of specific wavelength. With the high spatial temporal precision, optogenetic tools have enabled new ways to probe the causal role of specific cells in neural computation and behavior. Here, we overview the current state of the technology, and provide a brief introduction to the practical considerations in applying optogenetics in vivo to analyze neural circuit functions.
Keywords: Channelrhodopsin, archaerhodopsin, halorhodopsin, cell type specificity
Optogenetics is a new field being rapidly established upon the first demonstration of precise activation of neurons expressing a light-activated microbial opsin, channelrhodopsin-2, with pulses of blue light in 2005.1 Microbial (type I) opsins are classes of monolithic light activated proteins, encoded by small genes of under a kilobase long. Three major classes of microbial opsins have been adapted to optogenetically control cellular functions, channelrhodopsins, halorhodopsins, and archaerhodopsins (Figure 1). These opsin proteins can be easily expressed in specific cells through genetic modification, and have been successfully used in many model systems, from Caenorhabditis elegan, rodent to nonhuman primate, and human retina. Together with the ability achieved by optical methods to rapidly and reversibly illuminate genetically transduced cells, optogenetics has enabled the examination of the causal role of certain cellular functions in neural computation and diseases. A number of recent reviews and books have summarized various aspects of the current state of the field.2−8 Here, we hope to provide a brief overview of the technology and highlight the major considerations in applying it toward mapping neural circuits in vivo.
Figure 1.
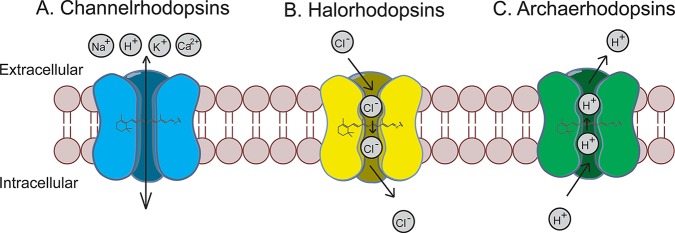
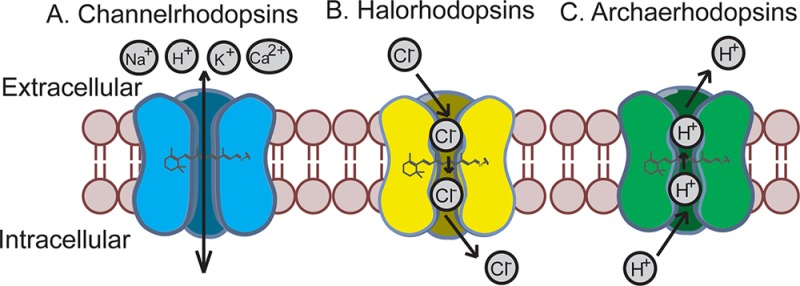
Optogenetic molecular sensors. Upon light illumination, channelrhodopsins passively transport Na+, K+, H+, Ca2+ down their electrochemical gradients to depolarize neurons (A); halorhodopsins actively pump Cl– into the cell to hyperpolarize neurons (B); archaerhodopsins actively pump H+ out of the cell to hyperpolarize neurons (C).
Optogenetic Molecular Sensors
Microbial (Type I) Opsins
Microbial (type I) opsins are photoactive proteins with seven transmembrane domains, widely spread in archaea, bacteria, algae, and fungi, where they are critical for light-sensing or photosynthetic functions. Microbial opsins have been studied since 1970s and were only recently adapted to control cellular functions as optogenetic sensors. Microbial opsins can be broadly categorized into transport rhodopsins and sensory rhodopsins.9 Transport rhodopsins include light-driven chloride pumps (halorhodopsins) and light-driven proton pumps (e.g., bacteriorhodopsins, archaerhodopsins, proteorhodopsins, etc.). Sensory rhodopsins include sensory rhodopsins I and II (phototaxis receptors in prokaryotes), channelrhodopsins (phototaxis receptors in algae), and others that mediate a variety of cellular functions (for reviews on opsin structure and function, see refs (9 and 10)). Most sensory rhodopsins function through recruiting intracellular signaling molecules without direct ion transport function. However, channelrhodopsins can mediate phototaxis as a light-gated cation channel at high light intensity and as a calcium channel at low light intensity.11
Microbial opsins share sequence homology and are phylogenetically clustered.12 Of these different classes, archaerhodopsins, halorhodopsins, and channelrhodopsins were found to directly convert photonic energy to ion transport across the membrane, and together they established the major classes of current molecular sensors for optogenetics. Type II animal rhodopsins found in higher eukaryotes are the mammalian homologues of type I microbial opsins. They share little sequence homology with microbial opsins, but exhibit similar 3-D structures. Type II animal opsins function through recruiting cellular kinases, instead of directly transporting ions. Thus, the small microbial opsins capable of directly coupling light to rapid ion transport represent a unique class of proteins as optogenetic sensors.
Light Mediated Ion Transport
Light mediated ion transport by archaerhodopsin, halorhodopsin, and channelrhodopsin proteins starts with the photoisomerization of the chromophore retinal (vitamin A-aldehyde). In the dark state, all-trans retinal binds to a conserved lysine residue within the seventh transmembrane domain, forming a protonated Schiff base.9 Upon absorption of a photon, photoisomerization of all-trans retinal to 13-cis retinal causes a sequence of protein conformational changes, which results in the transport of ions across the membrane. Different intermediate states during each photochemical cycle (photocycle) have been well studied for many microbial opsins, and each intermediate state can be distinguished based on the absorption spectrum determined by the overall structure of the protein. In general, opsin proteins cycle through a series of nonconducting and conducting states during each photocycle. The kinetic transitions between these different states determine the efficiency and the speed of light evoked ion transport, which ultimately determines an opsin’s ability to optogenetically control cells.
Channelrhodopsins for Neural Activation
Channelrhodopsins are light-gated ion channels found in green algae, and function as sensory rhodopsins homologous to phototaxis receptors and light-driven ion transporters in prokaryotes. One member of this class, channelrhodopsin-2 (ChR-2) from the green alga Chlamydomonas reinhardtii, was the first opsin used to control neural activities demonstrated by Boyden et al.1 and a few other groups around the same time.13 The widely used optogenetic sensor ChR2 is the N-terminal domain (315 amino acids) of the native algae channelrhodopsin-2, containing a channel with light-gated passive conductance to a number of monovalent and divalent cations, such as, Na+, K+, H+, and Ca2+.14 It was recently suggested that ChR2 is a proton pump with a leak that shows ion channel properties.15 Since the active proton pumping process would be much slower than the passive ion flow process, the overall effect of ChR2 would be mainly dominated by its channel properties.
When expressed in the plasma membrane, light illumination leads to ChR2 protein conformational changes, opening up a channel that lets in positively charged ions to depolarize the cell. As a channel, the photonic energy upon absorption of a photon is consumed to open the channel. Once the channel is open, the subsequent ionic current through the channel is passive and does not need additional photonic energy. As a result, channelrhodopsins are in general more efficient in transporting ions than light-gated ion pumps (such as 'halorhodopsins or archaerhodopsins). However, ChR2 has small single channel conductance comparing to endogenous ion channels,14−16 and thus, sufficient ChR2 proteins are needed to effectively activate a cell.
The success of using ChR2 to rapidly and reversibly drive neural activation on the millisecond time scale has motivated the development of a number of channelrhodopsin variants with improved photocurrent, altered kinetics, or shifted color spectrum. For example, ChR2 point mutants ChR2(H134R), ChR2(T159C), and several chimeras of ChR1 and ChR2 (ChIEF, ChRGR) exhibit higher amplitude photocurrent capable of supporting more efficient and precise control.16−20 CheTA showed faster kinetics, synergistic with higher amplitude photocurrent to support precise control of neural firing.21 Point mutants ChR2 (D156A) and ChR2 (C128A/S) showed slower responses upon light illumination, and are capable of sustaining long-term depolarization.22,23 VChR1 from Volvox carteri, MChR1 from M. viride, and chimeras C1 V1 are red-shifted,24−26 whereas ChR2 (L132C) exhibited enhanced calcium permeability.27 These ChR2 variants provided a variety of options to activate specific cells at different time scales to analyze their functions in neural circuit computation.
Halorhodopsins for Neural Silencing
Halorhodopsins, a class of light-driven inward chloride pump, were applied to hyperpolarize neurons in 2007.28,29 The first molecule of this class, halorhodopsin from Natronomonas pharaonis (Halo/NpHR), a yellow-light activated chloride pump, when expressed in neurons can mediate rapid and reversible silencing of neural activities by pumping chloride into a cell. However, Halo exhibited poor membrane targeting in mammalian neurons, which limited its use in vivo. Tagging Halo/NpHR with membrane targeting sequences, such as the trafficking sequence of a potassium channel, has been successful, and Halo2.0/eNpHR, Halo3.0/eNpHR3.0 exhibited improved photocurrent when expressed in mammalian neurons.30−33
In general, halorhodopsins, including many naturally occurring members of this class and engineered mutants aiming at improving kinetics, have a slow recovery rate after extended illumination, because of nonconducting intermediate states within each photocycle.34 However, blue light could accelerate Halo recovery from minutes to milliseconds.28,35 This slow recovery kinetics should be considered for experiments requiring long duration silencing. In addition, constant transport of Cl– into Halo expressing cells may change intracellular or extracellular Cl– concentration, and thus alter the reversal potential of GABA receptors, leading to neural modulation effects secondary to that produced by Halo. For example, Halo3.0/eNpHR3.0 mediated Cl– influx was shown to drastically alter GABAA receptor reversal potential in hippocampal neurons, which in turn altered inhibitory synaptic transmission and led to increased excitability of Halo expressing neurons for seconds after light mediated optogenetic silencing effects.36
Archaerhodopsins for Neural Silencing
Archaerhodopsins, a class of light-driven outward proton pumps, have been applied to optogenetically silence neural activities in 2010.35 For example, Archearhodopsin-3 from Halorubrum sodomense (Arch), a green-yellow light activated proton pump, when expressed in neurons mediated rapid and reversible silencing of neural activities by pumping protons out of a cell to hyperpolarize the cell. Arch traffics well to the plasma membrane in mammalian neurons and is capable of exerting powerful inhibition in vivo. Several variants of archaerhodopsins have since been developed, including ArchT from Halorubrum strain TP009 with improved photocurrent, capable of silencing larger brain volumes,37 and Mac from Leptosphaeria maculans with blue-shifted action spectrum.35 We have demonstrated that the outward proton flow mediated by Arch did not alter cellular pH, due to the intrinsic compensatory mechanisms. However, it remains to be established whether constant outward proton flow would acidify the extracellular environment, which may influence the excitability of surrounding neurons that do not express Arch, leading to neural modulation effects secondary to that produced by Arch.
Molecular Considerations of Optogenetic Efficacy and Precision
The efficacy and precision of optogenetic control relies on the molecular properties of opsin proteins. For example, the kinetics of the opsin photocycle determines the kinetics of single opsin photocurrent, and the number of opsin proteins presented on the plasma membrane determines the bulk photocurrent that is directly linked to the efficiency of optogenetic control. These aspects are being improved, with new opsin variants arising at a rapid rate. Key considerations for future development include continued improvement on the magnitude and the kinetics of photocurrent for more efficient and rapid control, narrower action spectrum for true multicolor control, and near-infrared sensing ability for larger tissue volume control.
The magnitude of the photocurrent depends upon the speed of ion transport by single opsin proteins and the total number of opsin proteins in the membrane. In general, channelrhodopsins can mediate larger photocurrent, since the photonic energy is used to gate the opening of the channel instead of directing ion transport, whereas for halorhodopsins and archaerhodopsins ion transport is directly linked to the absorption of photons. Nonetheless, all three classes of microbial opsins have been engineered to provide sufficient control of neuronal excitability in vivo to influence behavior across a number of species.
The response time to light illumination, the rate of turning ion transport on or off, is directly determined by the kinetics of the intermediate state transitions within each opsin photocycle. Since ion transport by halorhodopsins and archaerhodopsins is directly linked to photon absorption, these two classes of optogenetic sensors have rapid on/off kinetics, on the order of a few milliseconds in general. In contrast, ion transport by channelrhodopsins does not rely on the continued absorption of photons, and thus, the kinetics could vary among different variants with the off rates ranging from a few milliseconds to minutes.4 The kinetics of the opsins determines the perturbation and control that each opsin is capable of in perturbing neural circuit dynamics. Prolonged activation of opsins, however, may lead to an overall increase in membrane conductance, making it harder to depolarize or hyperpolarize a neuron.
The action spectrum for most opsins is broad. For example, the blue light activated ChR2 has a peak activation wavelength at 470 nm, but can be activated with >50% efficacy by light of 430–520 nm,14 the yellow light activated Halo/NpHR, and the green light activated Arch can be activated by light of 500–600 nm with >50% efficiency.28,35,38 Continued improvement on shifted color spectrum, not only the shift in peak activation wavelength, but the width of the action spectrum will be necessary in achieving true multicolor control. In addition, as near-infrared light can penetrate much larger brain volume in vivo, opsins sensitive to near-infrared light will enable optogenetic control of larger brain volumes eliminating tissue damage from implanted large fiber optic light sources.
Other Optogenetic Molecular Sensors for Controlling Cellular Signaling
Light-activatable animal (type II) opsins have been used to sensitize neurons to light, such as the three-protein drosophila photoreceptor system,39 rat rhodopsin 4 (RO4),40 and melanopsin.41 In addition, by replacing the intracellular domains of microbial opsins with the intracellular domains of G-protein coupled receptors (GPCRs), GPCRs have been sensitized to light to control beta2 or alpha1 adrenergic receptor signaling pathways.42 The ability to control different ionic flux and signaling pathways expanded the diversity of optogenetic control of different cellular functions.
A number of other strategies involving the combination of chemical and genetic methods have also been developed to sensitize cells to light, including optically uncaging chemical ligands of heterologously expressed receptors,43,44 and photoswitching chemical bonds between modified chemical ligands and mutated ion channels.45,46 However, the use of these approaches in vivo is limited by the need of exogenous chemicals.
Genetic Modification of Specific Cells
A major advantage of optogenetic technology is the ability to control specific cells expressing certain cellular markers, cells experiencing particular activity patterns, or cells projecting to specific brain regions. Such specificity is mainly achieved by the ability to express opsins in the desired cell populations through genetic modification. Genetic modification techniques mainly include whole animal transgenic approaches, viral based gene delivery approaches, and nonviral based gene delivery approaches.
Transgenic Approaches
Transgenic mice represent a versatile and powerful platform to target a variety of cells of interest, in particular in conjunction with the phage derived Cre-LoxP recombination technology.47,48 Cre recombinase catalyzes the recombination between 34bp LoxP recognition sequences flanking genes of interest, which can be used to restrict the expression of genes to cells expressing Cre (Figure 2A). A large number of Cre transgenic mice are available, with targeted Cre expression in specific cells. Upon injection of a virus that mediates Cre-dependent expression of optogenetic molecules, one can target opsins to the same set of cells that also express Cre. Alternatively, Cre transgenic mice can be crossed with transgenic mice expressing opsins in a Cre-dependent fashion. For example, Allen Brain Institute has recently created several such mice, including Ai32 mice with Cre-dependent ChR2(H134R) expression, Ai35 mice with Cre-dependent Arch expression, and Ai39 mice with Cre-dependent Halo3.0/eNpHR3.0 expression. These transgenic mice, when crossed with Cre transgenic mice, can mediate efficient and temporal precise optogenetic activation or silencing in vivo.49
Figure 2.
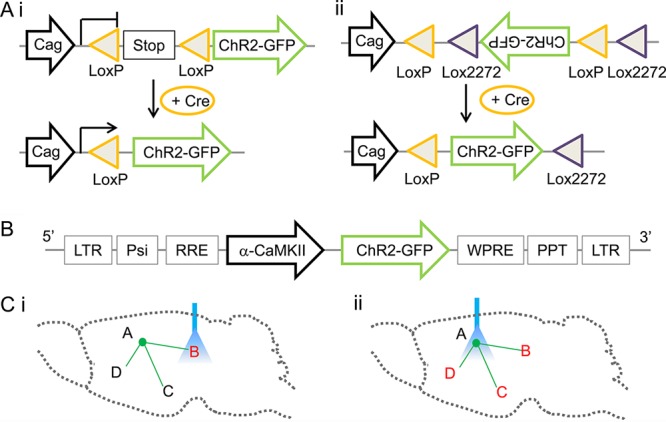
Cell specific targeting. (A) Cell type specific targeting with Cre-LoxP strategies. (Ai) An example demonstrates targeted expression of opsins in Cre expressing cells through removing transcription/translation stop sequences that block the expression of opsins. (Aii) An example demonstrates targeted expression of opsins in Cre expressing cells through flipping the opsin gene that is otherwise in the noncoding orientation (adapted from ref (69)). (B) Cell type specific targeting using short promoters in virus. An example demonstrates selective targeting of cortical excitatory neurons using a lentivirus with a CamKII promoter (adapted from ref (54)). (C) Anatomical pathway specific targeting. (Ci) An example demonstrates light illumination at area B selectively activates A to B projection. (Cii) An example demonstrates light illumination at the cell bodies retrogradely labeled through axon terminals projecting to area B. In this case, all the downstream areas that receive projections from area A are modulated.
Viral Gene Delivery
In genetically intractable species, virus remains the most commonly used method to transduce brain cells. Over the years, viral based gene delivery methods have been well established and widely used in basic research and in human gene therapy trials.8,50 The most commonly used viral vectors, lentivirus and adeno-associated virus (AAV), have been engineered to exhibit little or no toxicity, with excellent transduction efficiency. However, two major limitations remain for viral gene delivery methods. First, the packaging ability of virus is limited, which cannot be easily overcome due to the intrinsic stability of the viral particles. The highly efficient AAV virus can package up to 4.7 kb of DNA, and the slightly less efficient lentivirus can package up to ∼8 kb. Packaging of viral particles is increasingly difficult with larger DNAs, and in reality it is difficult to realize the full packaging capacity of lentivirus or AAV. Herpes simplex virus (HSV) in theory can package 30–50 kb of DNA, but in practice only ∼8 kb of DNA can be efficiently packaged. In addition, there are significant safety concerns for the use of HSV, such as cell death after infection, and HSV fails to mediate persistent expression beyond a couple of weeks.51 Second, different viruses display distinct tropism, because specific membrane receptors are required for viral entry into target cells. As a result, it remains difficult to target specific cells with virus, which has presented a major challenge in realizing the full potential of optogenetics in genetically intractable species.
Nonviral Gene Delivery
A number of nonviral methods have been developed for gene delivery, such as using cationic lipid, cationic polymers, nanoparticles, carbon nanotubes, gene guns, or calcium phosphate.52 However, these methods exhibit low transduction efficiency in vivo in general. A potential advantage of nonviral based gene delivery method is the ability to introduce larger DNAs into cells and thus may enable improved targeting to specific cells. However, continued effort in improving the transduction efficiency of nonviral gene delivery methods is necessary for the use of these methods in vivo.
Cell Type Specific Expression of Opsins
To restrict the expression of opsins to specific cells, one would rely on promoter sequences, which are often huge and include multiple regulatory components spread over a fraction of a chromosome. As a result, transgenic mice, utilizing the entire promoter sequence elements, can achieve high specificity. In contrast, small engineered promoter sequences that viruses can accommodate often fail to restrict the expression to specific cells. In addition, the intrinsic tropism of viruses also influences the expression efficiency in particular cell types. Effort in systematic analysis of various short promoters in several AAV serotypes revealed complex expression patterns in mice, with no clear conclusion on a specific combination of virus and promoter sequences in transducing a particular neuron type.53 Recently, we have demonstrated specific targeting of monkey cortical excitatory neurons, using a short 1.2 kb mouse CamKII promoter in lentivirus54 (Figure 2B). This specificity can be attributed to both the lentivirus tropism as well as the CamKII promoter.
Anatomical Pathway Specific Expression of Opsins
To selectively control synaptic terminals, one could spatially separate the cell bodies and the terminals, so that light will only reach the terminals, but not the cell bodies (Figure 2Ci). Transgenic mice with spatially restricted opsin expression, or virus injected at certain anatomical locations would be able to achieve such specificity. This strategy requires sufficiently strong expression of opsins at the synaptic terminals, but the feasibility has been demonstrated with ChR2 in vitro55,56 and in vivo.57 However, one potential concern is that activation of the targeted synapses and fibers of passage in the illuminated area may lead to antidromic stimulation, which will then activate other downstream targets that the antidromically activated neurons project to.
To label cells that project to a brain region of interest, one could use virus capable of retrograde labeling. So far, lentivirus has shown some promise in retrogradely transducing neurons projecting to the virus injection sites. Most promisingly, one lentivirus EIAV pseudotyped with rabies glycoprotein was found to be transported along axons, labeling large number of cells retrogradely.58 It remains to be determined whether such strategy is capable of expressing sufficient amount of opsin proteins for effective optogenetic modulation. This strategy, however, perturbs the network involving all downstream areas of the optogenetically targeted neurons, in addition to the areas of interest (Figure 2Cii).
Light Illumination and Electrophysiology
The efficiency of optogenetic control of neural activities depends on the absolute amount of light reaching a neuron, the number of opsin molecules presented in the plasma membrane, and the light sensitivity of the opsins. The control precision is also influenced by the intrinsic membrane physiological properties of the neuron and its surrounding neural network environment, which cannot be controlled by experimenters. For example, neurons that receive massive inhibition will be difficult to be activated; neurons with electrically leaky plasma membrane will be difficult to depolarize or hyperpolarize.
Optical Properties of the Brain
The absolute amount of light reaching a neuron depends upon the light power at the tip of light source and the pattern of light propagation in the tissue. Light propagation is determined by the optical properties of the brain tissue, with major considerations being tissue absorption and tissue scattering that reduce the intensity of light as it propagates away from the light source.59 At locations within the close proximity of the light source, where light intensity is at the saturation level for opsin action, the efficiency of optically controlling neurons will not vary by location. But at locations further away from the light source, where light intensity falls below the saturation level, the efficiency of controlling neurons will decrease with distance, and eventually at locations where light power falls below the threshold for opsin activation, light will be unable to modulate neural activities. In addition, the presence of electrodes and optical fibers also influences the pattern of light propagation.
Current opsins operate at the visible light wavelength, ∼450–600 nm, where hemoglobins, oxygenated (HbO2) and deoxygenated (Hb), are the major light absorbers (Figure 3). Light propagation in brain tissue can be estimated with Monte Carlo simulations.59 With Monte Carlo simulation, light of different colors within the visible wavelength range, out of different light sources, such as, LEDs or optical fibers, in general falls off nonlinearly, and falls to ∼1% at locations ∼1 mm away from the tip of the light source.35,60 These simulation results are consistent with our in vivo experimental observations that the magnitude of ChR2 mediated neuron activation correlates with systematic reduction of the light power at the optical fiber tip or stepwise increase of the distance between an optical fiber and an electrode,54 as well as in vitro estimation.61 To increase the effective volume of illumination, it is critical to develop novel opsin molecules that can be sensitized with red light, such as, >650 nm, where both Hb and HbO2 absorption coefficients are drastically reduced (Figure 3).
Figure 3.
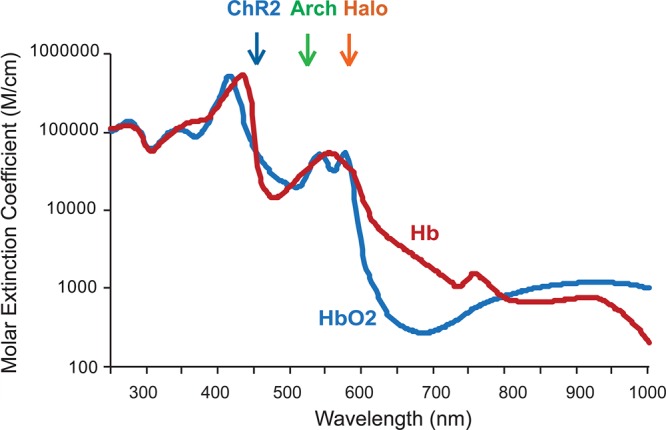
Hemoglobin, oxygenated (HbO2) and deoxygenated (Hb), absorption spectrum. Values are based on those summarized by Scott Prahl, http://omlc.ogi.edu/spectra. The peak excitation wavelengths for ChR2, Arch, and Halo are indicated.
Tissue Damages from Device Insertion and Heat
Various tissue damages occur during optogenetic experiments from device insertion, as well as from heat produced by light. To reduce mechanical damage from device insertion, ideally one would use devices as small as possible, thin optical fibers, thin recording electrodes, and thin viral injection cannulas. For example, a single optical fiber of 200 μm in diameter is equivalent in volume to four optical fibers of 100 μm in diameter. But a fiber array consisting of four optical fibers of 100 μm in diameter illuminates a much larger volume than a single 200 μm fiber. Note, the tissue damage produced by closely positioned fibers within a fiber array will produce greater tissue damage than a single fiber occupying the same volume. Adaption of fiber arrays will be helpful in reducing mechanical tissue damage.62
Heat generated by light could also produce tissue damage, though the exact damage is hard to evaluate. For example, heat alone may influence neurons’ responses, as demonstrated by pulsed infrared light stimulation63 or possibly by pulsed ultrasound stimulation.62,64 Most optogenetic studies use light up to ∼300 mW/mm2, or a few mW of total light power in vivo, without noticing detrimental heat damage.
Optical Artifact on Metal Electrodes
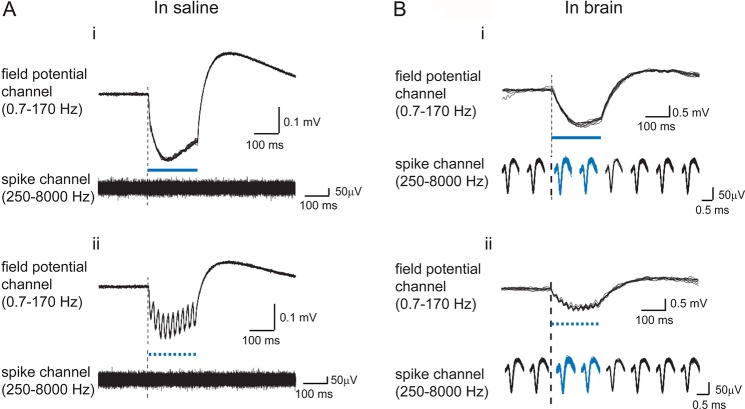
To monitor neural responses on a sub-millisecond time scale, electrophysiological recordings are often performed simultaneously with optical stimulation. However, we and others have observed strong voltage deflection artifact when laser light is directed onto metal electrode tips, in brain or in saline (Figure 4).37,54,65 This effect was clearly observed when the electrode tip was positioned in the blue laser beam in saline, and was also evident in the brain with a radiant flux of 80 mW/mm2, an intensity often needed for in vivo optogenetic experiments, when the tip of the optical fiber is 0.5–1 mm away from the electrode tip. We have observed that the magnitude of the artifact is proportional to the power of light illumination, but varies with the wavelength of the light. For example, we observed stronger voltage deflection artifact with 472 nm blue light than with 532 nm green light or 589 nm yellow light.
Figure 4.
Light mediated artifact on metal electrodes. Optical artifact observed on tungsten electrodes immersed in saline (A) or in brain (B) upon exposure to 200 ms blue light pulses (i) or trains of 10 ms blue light pulses delivered at 50 Hz (ii). Light pulses are indicated by blue dashes. Electrode data was hardware filtered using two data acquisition channels operating in parallel, yielding a low-frequency component (“field potential channel”) and a high-frequency component (“spike channel”). For the “spike channel” traces taken in brain (B), spikes were grouped into 100 ms bins, and then the binned spikes were displayed beneath the corresponding parts of the simultaneously acquired “field potential channel” signal. (Shown are the spikes in 8 such bins: 2 bins before light onset, 2 bins during light delivery period, and 4 bins after light cessation. Adapted from ref (54).)
The light induced artifact is consistent with the Becquerel effect, a classical photoelectrochemical phenomenon first demonstrated by Becquerel in 1839.66,67 Becquerel demonstrated that exposing metal electrodes, such as platinum, gold, and silver, to sunlight produced very small electric current when these metals were positioned in electrolyte. Consistent with the generality of the Becquerel effect, we observed such artifact with metal electrode wires made of stainless steel, platinum–iridium, silver/silver chloride, gold, nichrome, copper, or silicon.54 However, we have never observed such artifact with hollow glass microelectrodes.28,35,54 A few cautions have to be made with glass electrodes in optogenetic experiments. For example, if laser light reaches the Ag/AgCl wire that is in direct contact with the solution inside the glass electrode, light will induce artifact. Similarly, if light reaches the metal ground electrode, this optical artifact will also be picked up by the recording glass electrodes. Even though glass electrodes offer a good way to circumvent the artifact problem at the site of illumination, it is difficult to record from multiple glass electrodes, and the use of glass electrodes for chronic in vivo recording is limited.
Light induced artifact is slow evolving and thus only corrupts local field potential (LFP), but not spike waveforms. However, precautions are needed when laser light is pulsed briefly, that is, less than a couple milliseconds per pulse, which may produce artifact waveforms identical to spike waveforms. If the artifact waveforms produced by brief light pulses are significantly different from the spike waveforms, it is possible to isolate light artifact waveforms through spike sorting. In contrast, LFP that measures slow voltage fluctuations at lower frequencies of a few Hz to tens of Hz cannot be isolated from the slow artifact. Thus, while this artifact typically does not influence the ability to record spikes, it does prevent accurate measurement of LFP at the site of illumination. It is possible that part of the voltage defection recorded in the brain reflects physiological changes in LFP, but it is not yet possible to reliably isolate light evoked physiological responses from the optical artifact.
It might be possible to develop computational methods to remove optical artifact, since the amplitude and the time course of the artifact are stable with repeated light illumination when the electrode and the optical fiber remain at the same location and the light intensity remain constant. However, to isolate the real physiological effect, some computational/experimental methods will have to be used, which are yet to be developed. More promisingly, optimization of the electrode tip surface or electrode material may be proven useful in eliminating optical artifact. Recently, Zorzos et al. drastically attenuated optical artifact by coating the electrode tip with conducting polymer indium tin oxide (ITO).68 Continued advance in improving electrode coating strategy or developing novel electrodes is critical in enabling measurement of LFP at the site of illumination.
Concluding Remarks
Microbial opsin based optogenetic tools have enabled rapid and reversible activation or silencing of neurons in vivo. Such precise control of specific cell populations has enabled a new generation of time-resolved analysis of the causal role of specific cells in neural circuit computation and behavior. Current light-activated opsins, channelrhodopsins, halorhodopsins, and archaerhodopsins, have fulfilled much of the demand to achieve the perturbation needed to examine the sufficiency and necessity of specific cells in neural computation, and optogenetics has been applied to examine a variety of neural circuits responsible for many behaviors. However, it remains important to consider any secondary effect that may have contributed to the experimental observations. For example, changes in intracellular or extracellular ionic concentrations may influence the excitability of the network secondary to the direct perturbation effect produced by optogenetics. In addition, continued development of novel opsins with red-light shifted action spectrum could enable less invasive light illumination, particularly important for translating optogenetics to human therapies. Continued improvement of light activation spectrum would enable true multicolor controls of multiple cell populations or cellular functionalities. Finally, continued improvement of the ability to genetically target specific cells of interest will enable more precise dissection of different neural circuit components.
Acknowledgments
X.H. thanks Dr. Kenneth Rothschild and Dr. John Spudich for helpful comments on the manuscript, and Dr. Richie Kohman for figure illustrations.
X.H. acknowledges funding from NIH (5R00MH085944, 1R01NS081716), Alfred P. Sloan Foundation, Michael J. Fox Foundation, Pew Charitable Trusts Foundation, Boston University Biomedical Engineering Department, and Boston University Photonic Center.
The authors declare no competing financial interest.
References
- Boyden E. S.; Zhang F.; Bamberg E.; Nagel G.; Deisseroth K. (2005) Millisecond-timescale, genetically targeted optical control of neural activity. Nat. Neurosci. 8, 1263–1268. [DOI] [PubMed] [Google Scholar]
- Zhang F.; Vierock J.; Yizhar O.; Fenno L. E.; Tsunoda S.; Kianianmomeni A.; Prigge M.; Berndt A.; Cushman J.; Polle J.; Magnuson J.; Hegemann P.; Deisseroth K. (2011) The microbial opsin family of optogenetic tools. Cell 147, 1446–1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein J. G.; Boyden E. S. (2011) Optogenetic tools for analyzing the neural circuits of behavior. Trends Cognit. Sci. 15, 592–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yizhar O.; Fenno L. E.; Davidson T. J.; Mogri M.; Deisseroth K. (2011) Optogenetics in neural systems. Neuron 71, 9–34. [DOI] [PubMed] [Google Scholar]
- Miesenbock G. (2011) Optogenetic control of cells and circuits. Annu. Rev. Cell Dev. Biol. 27, 731–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knopfel T., and Boyden E. S., Eds. (2012) Optogenetics: Tools for Controlling and Monitoring Neuronal Activity, Vol. 196, Elsevier. [Google Scholar]
- Chow B. Y.; Han X.; Boyden E. S. (2012) Genetically encoded molecular tools for light-driven silencing of targeted neurons. Prog. Brain Res. 196, 49–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X. (2012) Optogenetics in the nonhuman primate. Prog. Brain Res. 196, 215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spudich J. L.; Yang C. S.; Jung K. H.; Spudich E. N. (2000) Retinylidene proteins: structures and functions from archaea to humans. Annu. Rev. Cell Dev. Biol. 16, 365–392. [DOI] [PubMed] [Google Scholar]
- Spudich J. L. (2006) The multitalented microbial sensory rhodopsins. Trends Microbiol. 14, 480–487. [DOI] [PubMed] [Google Scholar]
- Sineshchekov O. A.; Govorunova E. G.; Spudich J. L. (2009) Photosensory functions of channelrhodopsins in native algal cells. Photochem. Photobiol. 85, 556–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihara K.; Umemura T.; Katagiri I.; Kitajima-Ihara T.; Sugiyama Y.; Kimura Y.; Mukohata Y. (1999) Evolution of the archaeal rhodopsins: evolution rate changes by gene duplication and functional differentiation. J. Mol. Biol. 285, 163–174. [DOI] [PubMed] [Google Scholar]
- Boyden E. S. (2011) A history of optogenetics: the development of tools for controlling brain circuits with light. F1000 Biology Reports 3, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagel G.; Szellas T.; Huhn W.; Kateriya S.; Adeishvili N.; Berthold P.; Ollig D.; Hegemann P.; Bamberg E. (2003) Channelrhodopsin-2, a directly light-gated cation-selective membrane channel. Proc. Natl. Acad. Sci. U.S.A. 100, 13940–13945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldbauer K.; Zimmermann D.; Pintschovius V.; Spitz J.; Bamann C.; Bamberg E. (2009) Channelrhodopsin-2 is a leaky proton pump. Proc. Natl. Acad. Sci. U.S.A. 106, 12317–12322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J. Y.; Lin M. Z.; Steinbach P.; Tsien R. Y. (2009) Characterization of engineered channelrhodopsin variants with improved properties and kinetics. Biophys. J. 96, 1803–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berndt A.; Schoenenberger P.; Mattis J.; Tye K. M.; Deisseroth K.; Hegemann P.; Oertner T. G. (2011) High-efficiency channelrhodopsins for fast neuronal stimulation at low light levels. Proc. Natl. Acad. Sci. U.S.A. 108, 7595–7600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagel G.; Brauner M.; Liewald J. F.; Adeishvili N.; Bamberg E.; Gottschalk A. (2005) Light activation of channelrhodopsin-2 in excitable cells of Caenorhabditis elegans triggers rapid behavioral responses. Curr. Biol. 15, 2279–2284. [DOI] [PubMed] [Google Scholar]
- Wang H.; Sugiyama Y.; Hikima T.; Sugano E.; Tomita H.; Takahashi T.; Ishizuka T.; Yawo H. (2009) Molecular determinants differentiating photocurrent properties of two channelrhodopsins from chlamydomonas. J. Biol. Chem. 284, 5685–5696. [DOI] [PubMed] [Google Scholar]
- Wen L.; Wang H.; Tanimoto S.; Egawa R.; Matsuzaka Y.; Mushiake H.; Ishizuka T.; Yawo H. (2010) Opto-current-clamp actuation of cortical neurons using a strategically designed channelrhodopsin. PloS One 5, e12893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunaydin L. A.; Yizhar O.; Berndt A.; Sohal V. S.; Deisseroth K.; Hegemann P. (2010) Ultrafast optogenetic control. Nat. Neurosci. 13, 387–392. [DOI] [PubMed] [Google Scholar]
- Berndt A.; Yizhar O.; Gunaydin L. A.; Hegemann P.; Deisseroth K. (2009) Bi-stable neural state switches. Nat. Neurosci. 12, 229–234. [DOI] [PubMed] [Google Scholar]
- Bamann C.; Gueta R.; Kleinlogel S.; Nagel G.; Bamberg E. (2010) Structural guidance of the photocycle of channelrhodopsin-2 by an interhelical hydrogen bond. Biochemistry 49, 267–278. [DOI] [PubMed] [Google Scholar]
- Zhang F.; Prigge M.; Beyriere F.; Tsunoda S. P.; Mattis J.; Yizhar O.; Hegemann P.; Deisseroth K. (2008) Red-shifted optogenetic excitation: a tool for fast neural control derived from Volvox carteri. Nat. Neurosci. 11, 631–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govorunova E. G.; Spudich E. N.; Lane C. E.; Sineshchekov O. A.; Spudich J. L. (2011) New channelrhodopsin with a red-shifted spectrum and rapid kinetics from Mesostigma viride. mBio 2, e00115–00111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yizhar O.; Fenno L. E.; Prigge M.; Schneider F.; Davidson T. J.; O’Shea D. J.; Sohal V. S.; Goshen I.; Finkelstein J.; Paz J. T.; Stehfest K.; Fudim R.; Ramakrishnan C.; Huguenard J. R.; Hegemann P.; Deisseroth K. (2011) Neocortical excitation/inhibition balance in information processing and social dysfunction. Nature 477, 171–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinlogel S.; Feldbauer K.; Dempski R. E.; Fotis H.; Wood P. G.; Bamann C.; Bamberg E. (2011) Ultra light-sensitive and fast neuronal activation with the Ca(2)+-permeable channelrhodopsin CatCh. Nat. Neurosci 14, 513–518. [DOI] [PubMed] [Google Scholar]
- Han X.; Boyden E. S. (2007) Multiple-color optical activation, silencing, and desynchronization of neural activity, with single-spike temporal resolution. PloS One 2, e299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F.; Wang L. P.; Brauner M.; Liewald J. F.; Kay K.; Watzke N.; Wood P. G.; Bamberg E.; Nagel G.; Gottschalk A.; Deisseroth K. (2007) Multimodal fast optical interrogation of neural circuitry. Nature 446, 633–639. [DOI] [PubMed] [Google Scholar]
- Ma D.; Zerangue N.; Lin Y. F.; Collins A.; Yu M.; Jan Y. N.; Jan L. Y. (2001) Role of ER export signals in controlling surface potassium channel numbers. Science (New York, N.Y.) 291, 316–319. [DOI] [PubMed] [Google Scholar]
- Gradinaru V.; Zhang F.; Ramakrishnan C.; Mattis J.; Prakash R.; Diester I.; Goshen I.; Thompson K. R.; Deisseroth K. (2010) Molecular and cellular approaches for diversifying and extending optogenetics. Cell 141, 154–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gradinaru V.; Thompson K. R.; Deisseroth K. (2008) eNpHR: a Natronomonas halorhodopsin enhanced for optogenetic applications. Brain Cell Biol. 36, 129–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S.; Cunha C.; Zhang F.; Liu Q.; Gloss B.; Deisseroth K.; Augustine G. J.; Feng G. (2008) Improved expression of halorhodopsin for light-induced silencing of neuronal activity. Brain Cell Biol. 36, 141–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamberg E.; Tittor J.; Oesterhelt D. (1993) Light-driven proton or chloride pumping by halorhodopsin. Proc. Natl. Acad. Sci. U.S.A. 90, 639–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow B. Y.; Han X.; Dobry A. S.; Qian X.; Chuong A. S.; Li M.; Henninger M. A.; Belfort G. M.; Lin Y.; Monahan P. E.; Boyden E. S. (2010) High-performance genetically targetable optical neural silencing by light-driven proton pumps. Nature 463, 98–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raimondo J. V.; Kay L.; Ellender T. J.; Akerman C. J. (2012) Optogenetic silencing strategies differ in their effects on inhibitory synaptic transmission. Nat. Neurosci. [Epub ahead of print] 10.1038/nn.3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X.; Chow B. Y.; Zhou H.; Klapoetke N. C.; Chuong A.; Rajimehr R.; Yang A.; Baratta M. V.; Winkle J.; Desimone R.; Boyden E. S. (2011) A high-light sensitivity optical neural silencer: development and application to optogenetic control of non-human primate cortex. Front. Syst. Neurosci. 5, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duschl A.; Lanyi J. K.; Zimanyi L. (1990) Properties and photochemistry of a halorhodopsin from the haloalkalophile, Natronobacterium pharaonis. J. Biol. Chem. 265, 1261–1267. [PubMed] [Google Scholar]
- Zemelman B. V.; Lee G. A.; Ng M.; Miesenbock G. (2002) Selective photostimulation of genetically chARGed neurons. Neuron 33, 15–22. [DOI] [PubMed] [Google Scholar]
- Li X.; Gutierrez D. V.; Hanson M. G.; Han J.; Mark M. D.; Chiel H.; Hegemann P.; Landmesser L. T.; Herlitze S. (2005) Fast noninvasive activation and inhibition of neural and network activity by vertebrate rhodopsin and green algae channelrhodopsin. Proc. Natl. Acad. Sci. U.S.A. 102, 17816–17821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye H.; Daoud-El Baba M.; Peng R. W.; Fussenegger M. (2011) A synthetic optogenetic transcription device enhances blood-glucose homeostasis in mice. Science (New York, N.Y.) 332, 1565–1568. [DOI] [PubMed] [Google Scholar]
- Airan R. D.; Thompson K. R.; Fenno L. E.; Bernstein H.; Deisseroth K. (2009) Temporally precise in vivo control of intracellular signalling. Nature 458, 1025–1029. [DOI] [PubMed] [Google Scholar]
- Lima S. Q.; Miesenbock G. (2005) Remote control of behavior through genetically targeted photostimulation of neurons. Cell 121, 141–152. [DOI] [PubMed] [Google Scholar]
- Zemelman B. V.; Nesnas N.; Lee G. A.; Miesenbock G. (2003) Photochemical gating of heterologous ion channels: remote control over genetically designated populations of neurons. Proc. Natl. Acad. Sci. U.S.A. 100, 1352–1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banghart M.; Borges K.; Isacoff E.; Trauner D.; Kramer R. H. (2004) Light-activated ion channels for remote control of neuronal firing. Nat. Neurosci. 7, 1381–1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janovjak H.; Szobota S.; Wyart C.; Trauner D.; Isacoff E. Y. (2010) A light-gated, potassium-selective glutamate receptor for the optical inhibition of neuronal firing. Nat. Neurosci. 13, 1027–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsien J. Z.; Chen D. F.; Gerber D.; Tom C.; Mercer E. H.; Anderson D. J.; Mayford M.; Kandel E. R.; Tonegawa S. (1996) Subregion- and cell type-restricted gene knockout in mouse brain. Cell 87, 1317–1326. [DOI] [PubMed] [Google Scholar]
- Sauer B.; Henderson N. (1988) Site-specific DNA recombination in mammalian cells by the Cre recombinase of bacteriophage P1. Proc. Natl. Acad. Sci. U.S.A. 85, 5166–5170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen L.; Mao T.; Koch H.; Zhuo J. M.; Berenyi A.; Fujisawa S.; Hsu Y. W.; Garcia A. J. 3rd; Gu X.; Zanella S.; Kidney J.; Gu H.; Mao Y.; Hooks B. M.; Boyden E. S.; Buzsaki G.; Ramirez J. M.; Jones A. R.; Svoboda K.; Han X.; Turner E. E.; Zeng H. (2012) A toolbox of Cre-dependent optogenetic transgenic mice for light-induced activation and silencing. Nat. Neurosci. 15, 793–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waehler R.; Russell S. J.; Curiel D. T. (2007) Engineering targeted viral vectors for gene therapy. Nat. Rev. Genet. 8, 573–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neve R. L., and Carlezon W. A. Jr., Eds. (2002) Gene delivery into the brain using viral vectors, Vol. 20, Nature Publishing Group: New York. [Google Scholar]
- Luo D.; Saltzman W. M. (2000) Synthetic DNA delivery systems. Nat. Biotechnol. 18, 33–37. [DOI] [PubMed] [Google Scholar]
- Nathanson J. L.; Jappelli R.; Scheeff E. D.; Manning G.; Obata K.; Brenner S.; Callaway E. M. (2009) Short Promoters in Viral Vectors Drive Selective Expression in Mammalian Inhibitory Neurons, but do not Restrict Activity to Specific Inhibitory Cell-Types. Front. Neural Circuits 3, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X.; Qian X.; Bernstein J. G.; Zhou H. H.; Franzesi G. T.; Stern P.; Bronson R. T.; Graybiel A. M.; Desimone R.; Boyden E. S. (2009) Millisecond-timescale optical control of neural dynamics in the nonhuman primate brain. Neuron 62, 191–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petreanu L.; Huber D.; Sobczyk A.; Svoboda K. (2007) Channelrhodopsin-2-assisted circuit mapping of long-range callosal projections. Nat. Neurosci. 10, 663–668. [DOI] [PubMed] [Google Scholar]
- Cruikshank S. J.; Urabe H.; Nurmikko A. V.; Connors B. W. (2010) Pathway-Specific Feedforward Circuits between Thalamus and Neocortex Revealed by Selective Optical Stimulation of Axons. Neuron 65, 230–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tye K. M.; Prakash R.; Kim S. Y.; Fenno L. E.; Grosenick L.; Zarabi H.; Thompson K. R.; Gradinaru V.; Ramakrishnan C.; Deisseroth K. (2011) Amygdala circuitry mediating reversible and bidirectional control of anxiety. Nature 471, 358–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazarakis N. D.; Azzouz M.; Rohll J. B.; Ellard F. M.; Wilkes F. J.; Olsen A. L.; Carter E. E.; Barber R. D.; Baban D. F.; Kingsman S. M.; Kingsman A. J.; O’Malley K.; Mitrophanous K. A. (2001) Rabies virus glycoprotein pseudotyping of lentiviral vectors enables retrograde axonal transport and access to the nervous system after peripheral delivery. Hum. Mol. Genet. 10, 2109–2121. [DOI] [PubMed] [Google Scholar]
- Mobley J., and Vo-Dinh T. (2003) Optical properties of Tissue. In Biomedical Photonics Handbook, pp 1–72, CRC Press: Boca Raton, FL [Google Scholar]
- Bernstein J. G.; Han X.; Henninger M. A.; Ko E. Y.; Qian X.; Franzesi G. T.; McConnell J. P.; Stern P.; Desimone R.; Boyden E. S. (2008) Prosthetic systems for therapeutic optical activation and silencing of genetically-targeted neurons. Proc. Soc. Photo-Opt. Instrum. Eng. 6854, 68540H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamantidis A. R.; Zhang F.; Aravanis A. M.; Deisseroth K.; de Lecea L. (2007) Neural substrates of awakening probed with optogenetic control of hypocretin neurons. Nature 450, 420–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein J. G.; Garrity P. A.; Boyden E. S. (2012) Optogenetics and thermogenetics: technologies for controlling the activity of targeted cells within intact neural circuits. Curr. Opin. Neurobiol. 22, 61–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells J.; Kao C.; Mariappan K.; Albea J.; Jansen E. D.; Konrad P.; Mahadevan-Jansen A. (2005) Optical stimulation of neural tissue in vivo. Opt. Lett. 30, 504–506. [DOI] [PubMed] [Google Scholar]
- Tufail Y.; Yoshihiro A.; Pati S.; Li M. M.; Tyler W. J. (2011) Ultrasonic neuromodulation by brain stimulation with transcranial ultrasound. Nat. Protoc. 6, 1453–1470. [DOI] [PubMed] [Google Scholar]
- Ayling O. G.; Harrison T. C.; Boyd J. D.; Goroshkov A.; Murphy T. H. (2009) Automated light-based mapping of motor cortex by photoactivation of channelrhodopsin-2 transgenic mice. Nat. Methods 6, 219–224. [DOI] [PubMed] [Google Scholar]
- Gratzel M. (2001) Photoelectrochemical cells. Nature 414, 338–344. [DOI] [PubMed] [Google Scholar]
- Honda K. (2004) Dawn of the evolution of photoelectrochemistry. J. Photochem. Photobiol. 63–68. [Google Scholar]
- Zorzos A. N.; Dietrich A.; Talei Franzesi G.; Chow B. Y.; Han X.; Fonstad C. G.; Boyden E. S. (2009) Light-proof neural recording electrodes. Soc. Neurosci. 388, GG107. [Google Scholar]
- Atasoy D.; Aponte Y.; Su H. H.; Sternson S. M. (2008) A FLEX switch targets Channelrhodopsin-2 to multiple cell types for imaging and long-range circuit mapping. J. Neurosci. 28, 7025–7030. [DOI] [PMC free article] [PubMed] [Google Scholar]